Abstract
The differential use of protein precursors and their products is a key strategy used during poliovirus replication. To characterize the role of protein precursors during replication, we examined the complementation profiles of mutants that inhibited 3D polymerase or 3C-RNA binding activity. We showed that 3D entered the replication complex in the form of its precursor, P3 (or 3CD), and was cleaved to release active 3D polymerase. Furthermore, our results showed that P3 is the preferred precursor that binds to the 5’CL. Using reciprocal complementation assays, we showed that one molecule of P3 binds the 5’CL and that a second molecule of P3 provides 3D. In addition, we showed that a second molecule of P3 served as the VPg provider. These results support a model in which P3 binds to the 5’CL and recruits additional molecules of P3, which are cleaved to release either 3D or VPg to initiate RNA replication.
Keywords: Poliovirus (PV), reciprocal complementation, poliovirus P3, poliovirus 3CD, 3D polymerase, VPg, 5’Cloverleaf (5’CL), 3C-RNA binding activity, RNA replication complex, (−) strand RNA
Introduction
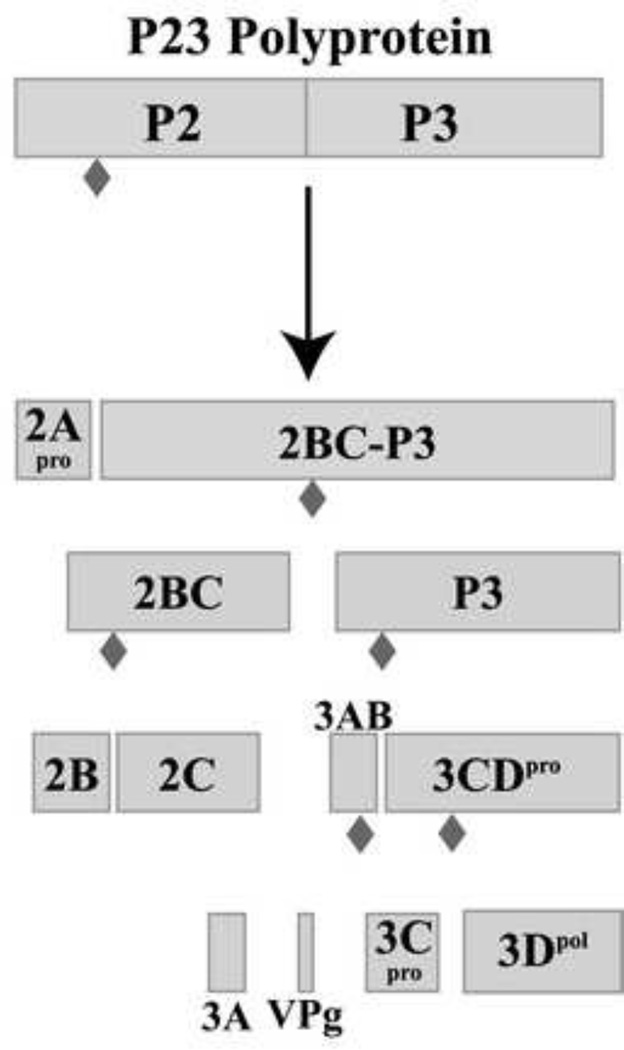
Poliovirus (PV) is a prototypical human enterovirus, which is a member of the small, single-strand (ss), positive sense RNA virus family Picornaviridae. The PV genome, like that of other picornaviruses, is covalently linked to a viral protein, VPg, at the 5’ terminus. The PV genome also contains a conserved RNA structure in its 5’ terminal sequence, the 5’cloverleaf (5’CL), which has been shown to bind both cellular poly(C) binding protein (PCBP) as well as a viral precursor protein, 3CD (Andino et al., 1990b;Andino et al., 1993;Gamarnik and Andino, 1997;Parsley et al., 1997). Translation of the viral genome utilizes an internal ribosomal entry site (IRES) to drive synthesis of a single large open reading frame, which is subsequently processed by the viral proteases 2A, 3C and 3CD (reviewed in Palmenberg, 1990). Initial polyprotein processing occurs co-translationally at the boundary between the structural (P1) and non-structural (P23) proteins by 2A. All subsequent cleavage events are mediated by 3C and 3CD (Wimmer et al., 1993). Previous studies by Lawson and Semler (Lawson and Semler, 1992) showed that the predominant pathway for cleavage of the P23 polyprotein was the membrane-associated pathway starting with the cleavage at the junction of 2A–2B (Fig.1). This generates 2A and the precursor protein, 2BC-P3, which is then rapidly processed to form 2BC and P3. P3 is processed to 3CD and 3AB. Cleavage of 3AB generates 3A and the viral protein primer, VPg (3B). 3CD is processed to yield 3C and the active form of the polymerase, 3D. VPg is uridylylated by the viral polymerase, 3D, to form VPgpUpU which is used as the primer for the initiation of both (−) and (+) strand synthesis.
Fig.1.
Diagram of the poliovirus P23 polyprotein processing cascade showing the precursor and processed viral proteins. The P23 polyprotein is cleaved by the viral protease 3C/3CD at the cleavage sites shown as filled diamonds ( ).
).
Given the minimal coding capacity of these small RNA viruses, the differential use of polyprotein precursors and their products is a key strategy employed by PV to perform the many diverse functions required during viral replication using limited sequence space. An extension of this is the evolution of multiple activities within a single protein or protein precursor. The PV precursor protein, 3CD, exemplifies both of these concepts in that it performs multiple functions as a precursor, and these activities are functionally distinct from its processed products, 3C and 3D. As a precursor, 3CD exhibits no polymerase activity, however its processed product 3D, is the PV RNA-dependent RNA polymerase (RdRp) (Flanegan and Baltimore, 1979;Flanegan and Van Dyke, 1979;Harris et al., 1992). The 3CD precursor also has the ability to bind to stem-loop ‘d’ of the 5’CL. While this ability is partially retained by its processed product 3C, the binding affinity of 3C for the 5’CL is 10-fold lower than that of 3CD (Andino et al., 1993). Although 3CD and 3C are proteases, their cleavage specificities and activity levels are different. This difference is particularly apparent in the processing of the viral capsid precursor (P1) as well as processing at the 3C–3D junction (Parsley et al., 1999). Despite these functional differences, there are very few structural differences between 3C and 3D individually and within the 3CD precursor as determined by x-ray crystallography (Gruez et al., 2008;Marcotte et al., 2007;Thompson and Peersen, 2004). A critical difference, however, is that the N-terminal glycine residue in 3D is buried in a pocket near the base of the fingers domain in the active 3D structure, but it is part of the 3C–3D linker in the inactive 3CD precursor. Studies on the structure of both PV and Coxsackievirus B3 (CVB3) 3D indicate that the buried N-terminal glycine residue is required to activate the picornaviral polymerase (Gruez et al., 2008;Thompson and Peersen, 2004).
Current models of PV replication complex formation invoke genomic circularization mediated by ribonucleoprotein (RNP) complexes formed at the 5’CL and the 3’NTR-poly(A) tail to promote initiation of (−) strand synthesis (Barton et al., 2001;Herold and Andino, 2001;Lyons et al., 2001;Ogram and Flanegan, 2011;Teterina et al., 2001). Viral precursor protein, 3CD, in the presence of PCBP or 3AB, was observed to form RNP complexes with the 5’CL (Andino et al., 1990b;Andino et al., 1993;Harris et al., 1994;Parsley et al., 1997;Xiang et al., 1995). In a previous study, a revertant that contained a second site suppressor mutation in the RNA binding domain of viral protein 3C was isolated from cells transfected with viral RNA with an insertion mutation in stem-loop ‘d’ (Andino et al., 1990b). This finding provided evidence for a direct interaction between 3CD and stem-loop ‘d’ of the 5’CL. Additional studies showed that mutations in stem-loop ‘d’ of the 5’CL, which disrupted 3CD binding, also inhibited PV RNA replication (Andino et al., 1993;Parsley et al., 1997;Xiang et al., 1995;Barton et al., 2001;Vogt and Andino, 2010). Furthermore, mutations in the 3C-RNA binding domain in 3CD disrupted the formation of the 5’CL-RNP complex and inhibited viral RNA replication (Andino et al., 1993;Hammerle et al., 1992;Harris et al., 1994;Blair et al., 1998).
The results of the above studies suggest that the diverse activities associated with 3CD, and presumably other precursor proteins like P3, are required for PV RNA replication. Consistent with this idea, previous studies showed that P3 was required to complement an RNA replication-defective mutation in the 3A region of protein 3AB (Towner et al., 1998). In addition, the results of another study showed that P3 was required for complementation of a VPg-linkage mutant (Liu et al., 2007). Taken together, these findings suggest that viral precursor proteins and their processed products play distinct and essential roles in the assembly of the poliovirus RNA replication complex. However, the precise molecular mechanisms which drive the requirement for the viral precursor proteins have not been delineated. To characterize these mechanisms, we utilized the HeLa S10 translation-replication system to examine the complementation profiles of viral transcript RNAs, which contain either a 3D polymerase mutation or a 3C-RNA binding mutation. These mutant RNAs were assayed for their ability to assemble functional replication complexes and initiate (−) strand synthesis. The results of the experiments with the 3D polymerase mutant showed that 3D is initially assembled into the replication complex in the form of its inactive precursor, P3 or 3CD, and that 3D itself cannot assemble into the complex. The precursor proteins are subsequently cleaved to release active 3D for VPg uridylylation and RNA replication. In the experiments with the 3C-RNA binding mutant, our results showed that P3, and not 3CD, is the preferred precursor that binds to the 5’CL. Furthermore, the results of reciprocal complementation assays demonstrated that the molecule of P3 that binds the 5’CL does not serve as the 3D polymerase provider. Instead, a second molecule of P3, which does not bind RNA, functions as the source of active 3D polymerase. Interestingly, our results also showed that a second molecule of P3 that does not bind RNA served as the source of VPg in the replication complex. These findings support a model in which P3 binds to the 5’CL and then recruits additional molecules of P3, which are then cleaved to release either active 3D or VPg for RNA replication. Taken together, our results illustrate that the viral precursor protein, P3, performs distinct and essential functions in the replication complex.
Results
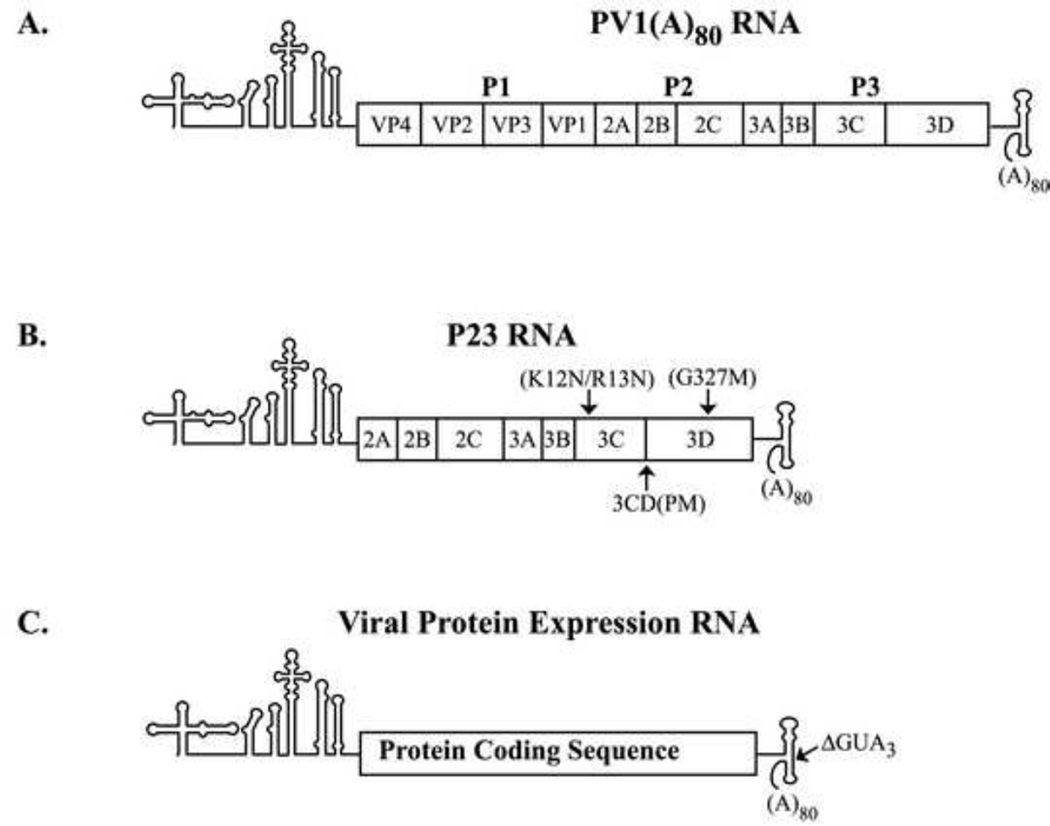
Previous studies have identified several functions associated with the viral protein 3CD that are required for PV RNA replication. The initial goal of this study was to use a genetic complementation analysis to identify specific functions of 3CD that are required to form functional replication complexes to initiate (−) strand synthesis. Specific mutations that disrupted 3D polymerase activity, the processing of 3CD to 3C and 3D and the RNA binding activity of 3C (and 3CD) were constructed in a subgenomic RNA transcript, PV P23 RNA (Fig.2B). P23 RNA encodes all of the viral replication proteins and serves as an efficient template to measure (−) strand synthesis (see Materials and methods). For the complementation analysis, protein expression RNAs that encoded P3, 3CD, 3C or 3D were constructed as described (Fig.2C). (−) Strand synthesis was measured in preinitiation replication complexes (PIRCs) isolated from HeLa S10 translation-RNA replication reactions containing one of the P23 RNA mutants and one of the viral protein expression RNAs. To confirm that equivalent levels of the viral proteins were synthesized in each reaction, translation of the viral transcript RNA was measured in the presence of [35S]-methionine.
Fig.2.
Schematic of the PV1 RNA transcripts used in this study. (A) PV1(A)80 transcript RNA contains the entire poliovirus genomic RNA sequence. This RNA encodes all of the viral proteins and serves as a template for (−) strand synthesis. (B) P23 transcript RNA contains a deletion of the P1 capsid coding region, but encodes the viral replication proteins and serves as a replicon RNA. Mutations constructed in 3C/3D/3CD are indicated by arrows and include the 3C-RNA binding mutant (RBM; K12N/R13N), the 3CD processing mutant (PM; T181K, Q182D, S183G, Q184N), and the 3D polymerase activity mutant (G327M). (C) The viral protein expression RNA contains the specific protein coding sequence flanked by the authentic PV1 5’ NTR, IRES and 3’ NTR and poly(A) tail.
Characterization of 3D polymerase activity mutants
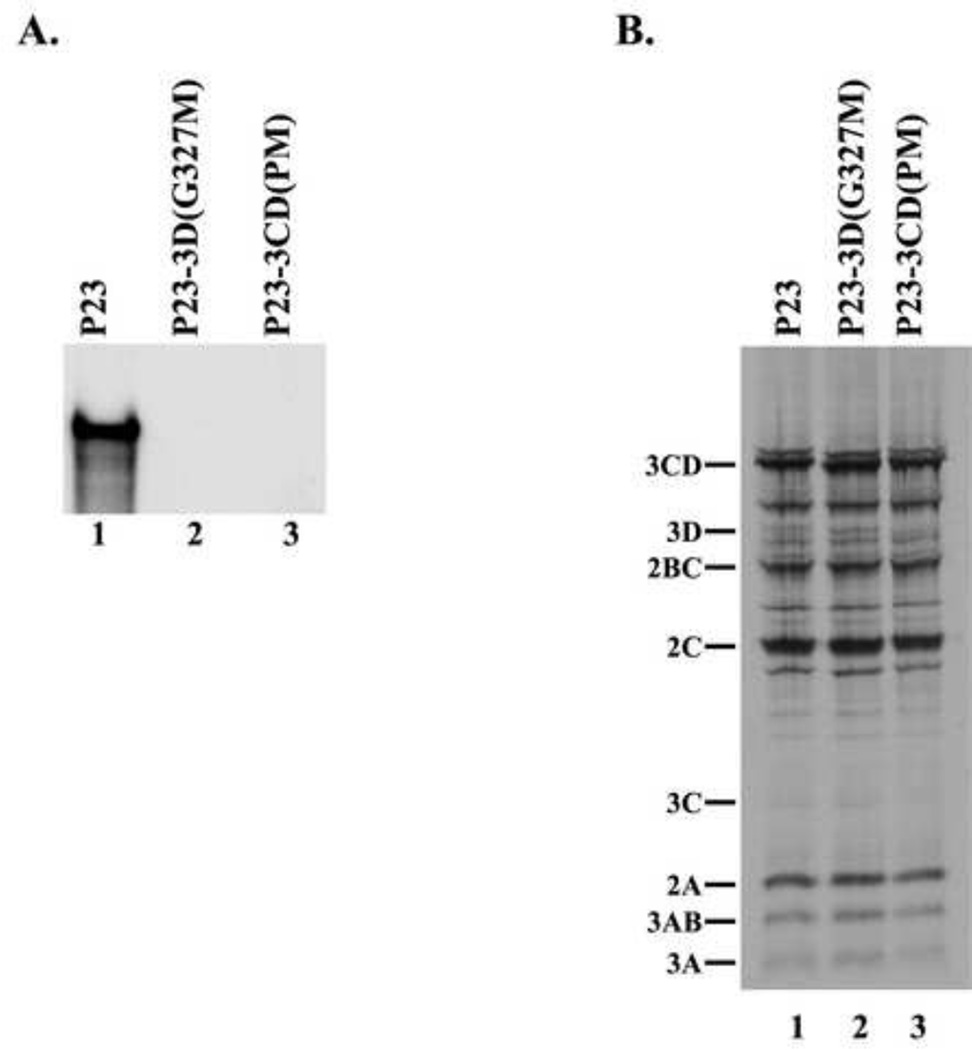
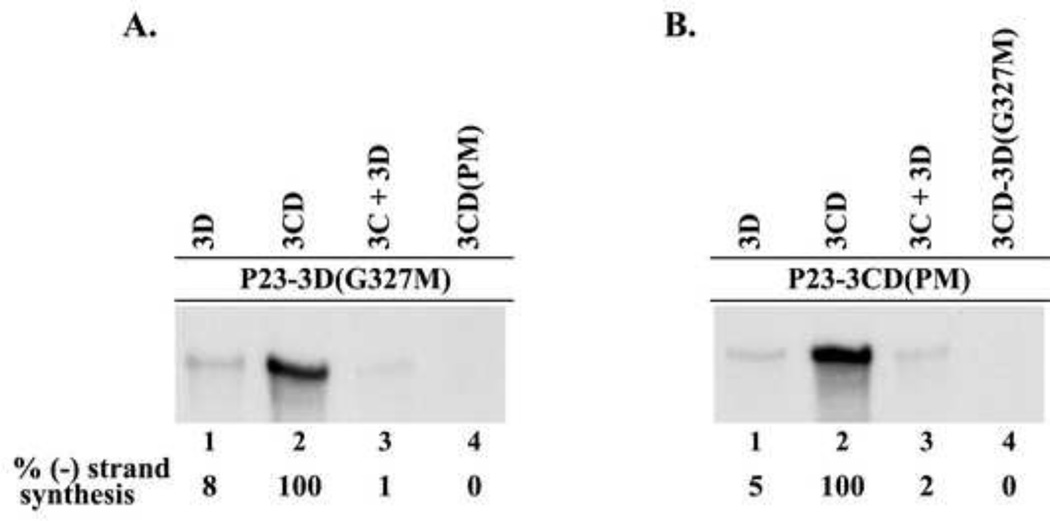
The first mutant, P23-3D(G327M) RNA, contained a mutation in the active site of the 3D, which abolishes all polymerase activity (Jablonski et al., 1991). The second mutant contained changes in the 3C-3D cleavage site, which blocked the proteolytic processing of 3CD to 3C and 3D. Previous studies showed that 3CD does not have any measureable RNA polymerase activity until it undergoes proteolytic processing to release active 3D polymerase (Flanegan and Van Dyke, 1979;Harris et al., 1992). Therefore, blocking the proteolytic processing of 3CD maintains 3D in an inactive state. The 3C–3D cleavage site mutant (P23-3CD(PM)) contained four mutations at positions P1–P4 in 3C, thereby maintaining the integrity of the amino acid sequence in 3D. The P23-3CD(PM) mutant RNA, combined two previously described 3C-3D processing site mutations (T181K, Q182D) (Andino et al., 1990a;Harris et al., 1992) with two additional mutations (S183G, Q184N). This extensive mutagenesis at the 3CD processing site was required to completely inhibit the processing of 3CD (data not shown). Therefore, both the P23-3D(G327M) RNA and the P23-3CD(PM) RNA were not predicted to generate active 3D and thus not support (−) strand synthesis. To confirm that RNA synthesis was inhibited in reactions containing either P23-3D(G327M) RNA or P23-3CD(PM) RNA, we measured (−) strand synthesis in PIRCs isolated from HeLa S10 reactions containing these two mutant RNAs. As expected, 32P-labeled (−) strand RNA was synthesized in the reaction containing wildtype P23 RNA (Fig.3A, lane 1). In contrast, detectable levels of labeled (−) strand RNA were not observed in the reactions containing either P23-3D(G327M) RNA or P23-3CD(PM) RNA (Fig.3A, lane 2–3). These results confirmed that active 3D was not produced in the reactions containing either mutant RNA. In control experiments, we showed that both viral protein synthesis and polyprotein processing were not affected in reactions that contained P23-3D(G327M) RNA (Fig.3B, compare lanes 1 and 2). In reactions containing P23-3CD(PM) RNA, viral protein synthesis was not affected and polyprotein processing was normal except that 3C and 3D were not detected in the reaction, as expected (Fig.3B, compare lanes 1 and 3). These results confirmed that (−) strand synthesis was completely inhibited in reactions that contained either the P23-3D(G327M) or the P23-3CD(PM) RNAs.
Fig.3.
Characterization of mutations inhibiting 3D polymerase activity. (A) 32P-labeled (−) strand RNA synthesis was assayed using PIRCs isolated from HeLa S10 reactions as described in Materials and methods. Reactions contained subgenomic wildtype P23 RNA or P23 RNA with a mutation either in 3D (3D(G327M)) or in 3CD coding region (3CD (PM)). Full length labeled product RNA was analyzed by denaturing CH3HgOH gel electrophoresis and autoradiography. (B) Portions of the HeLa S10 reactions described in (A) were labeled with [35S] methionine to assay for protein synthesis. These reactions were analyzed by SDS-PAGE and autoradiography. Viral proteins are indicated at left.
Viral proteins 3CD or P3 were required to rescue replication of 3D(G327M) or 3CD(PM) mutant RNAs
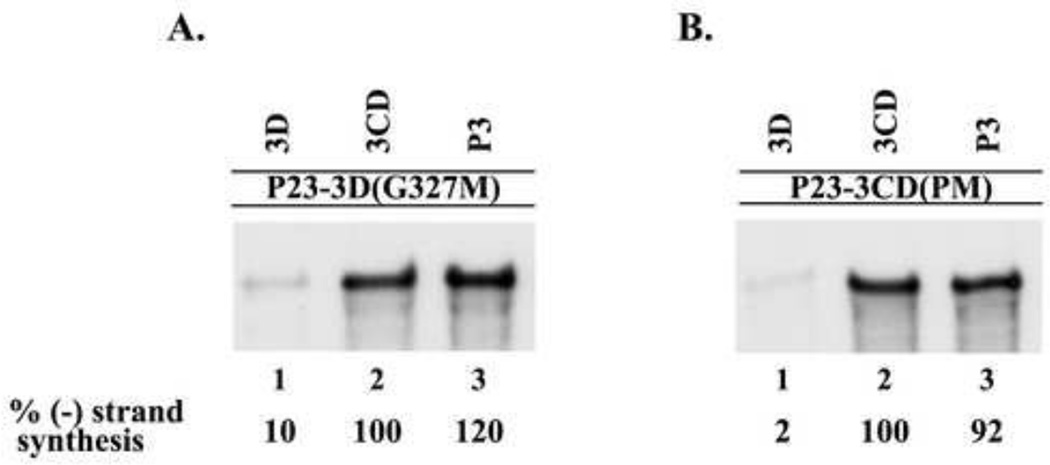
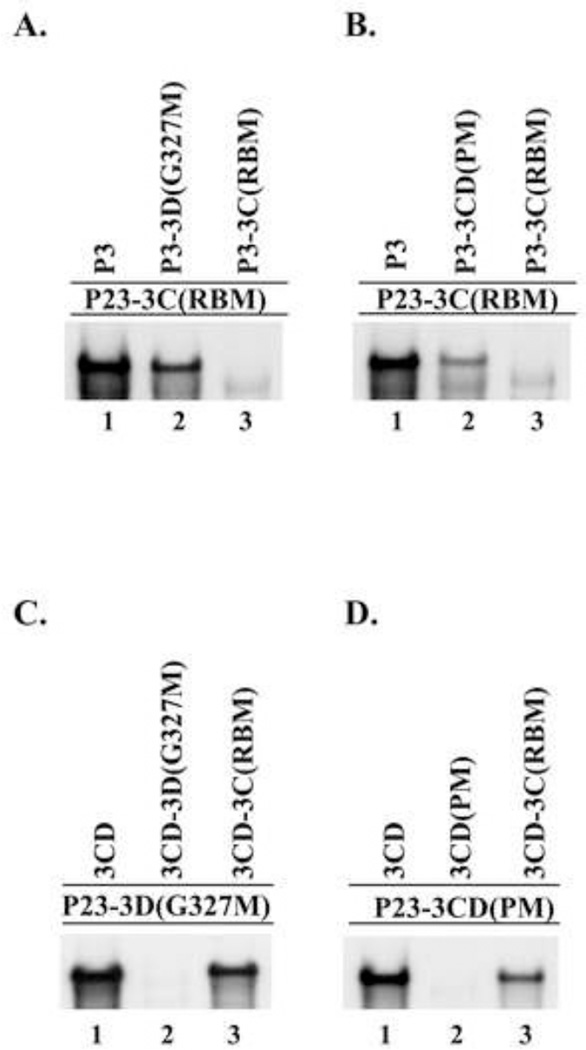
A complementation analysis was performed to identify the specific viral protein, or precursor proteins, that were required to efficiently rescue the replication of P23-3D(G327M) RNA. (−) Strand synthesis was measured in reactions that contained P23-3D(G327M) RNA and a protein expression RNA that encoded either active 3D or a 3D containing precursor proteins, 3CD or P3 (see Materials and methods). In reactions that contained P23-3D(G327M) RNA and wildtype 3D, only a very low level of (−) strand synthesis was observed (Fig.4A, lane 1). In contrast, a high level of (−) strand synthesis was observed in the reactions that contained either wildtype 3CD or P3 (Fig.4A, lanes 2–3). These results demonstrated that (−) strand synthesis was efficiently rescued on P23-3D(G327M) RNA templates in reactions that contained either 3CD or P3.
Fig.4.
Viral proteins 3CD or P3 were required to rescue replication of 3D(G327M) or 3CD(PM) mutant RNAs. (−) Strand RNA synthesis was assayed using PIRCs isolated from HeLa S10 reactions as described in Materials and methods. (A) Reactions contained P23-3D(G327M) RNA as a template and a second complementing RNA expressing the indicated protein. (B) Reactions contained P23-3CD(PM) RNA as template and a second RNA expressing the indicated protein. All complementing RNAs contain the ΔGUA3 mutation which inhibits (−) strand synthesis. Labeled product RNA was analyzed as described in Fig.3. The amount of the labeled product RNAs synthesized in each reaction was quantitated using ImageJ as described in Materials and methods.
We predicted that similar complementation results would be observed in reactions that contained P23-3CD(PM) RNA. (−) Strand synthesis was assayed in reactions that contained P23-3CD(PM) RNA and an expression RNA that encoded either wildtype 3D, 3CD or P3. In the reaction that contained the P23-3CD(PM) RNA and 3D, only very low levels of (−) strand synthesis were observed (Fig.4B, lane 1). In contrast, high levels of (−) strand syntheses were observed in reactions that contained either 3CD or P3 (Fig.4B, lanes 2–3). These results showed that (−) strand synthesis on P23-3CD(PM) RNA was efficiently restored in reactions that contained either wildtype 3CD or P3. As predicted, these findings were consistent with the results observed in the assays performed with the P23-3D(G327M) RNA. These results indicated that 3CD or P3, but not 3D itself, was able to efficiently complement the replication of mutant RNAs that do not encode active 3D polymerase.
Finally, we determined if the presence of additional 3C would enhance the ability of 3D to restore (−) strand synthesis in reactions containing either P23-3D(G327M) RNA or P23-3CD(PM) RNA. To do this, (−) strand synthesis was assayed in reactions that contained one of the mutant RNA templates and both the 3C and 3D expression RNAs. Low levels of (−) strand synthesis were again observed in the presence of 3D (5–8%) compared to the levels observed with 3CD (Figs. 5A and 5B, lanes 1–2). The presence of additional 3C in the assay did not enhance the low levels of (−) strand synthesis observed with 3D alone (Figs. 5A and 5B, lanes 1 and 3). As expected, 3CD(PM) did not complement the replication of P23-3D(G327M) RNA (Fig.5A, lane 4) and 3CD(G327M) did not complement the replication of P23-3CD(PM) RNA (Fig.5B, lane 4). Taken together, the results of these complementation assays showed that low levels of (−) strand synthesis were observed with 3D (2–10%) compared to the levels observed with 3CD. In addition, similar levels of (−) strand synthesis were observed in the presence of P3 (92–120%) compared to the levels observed with 3CD. In summary, these results indicated that active 3D polymerase is provided by the processing of precursor proteins P3 or 3CD in viral RNA replication complexes.
Fig.5.
The presence of additional 3C did not enhance the ability of 3D to restore (−) strand synthesis. (−) Strand synthesis was assayed using PIRCs isolated from HeLa S10 reactions as described in Materials and methods. Reactions contained P23-3D(G327M) RNA (A) or P23-3CD(PM) RNA (B) and complementing RNAs expressing the indicated protein(s). Labeled product RNA was analyzed as described in Fig.3 and quantitated as described in Fig.4.
Characterization of a 3C-RNA binding mutant
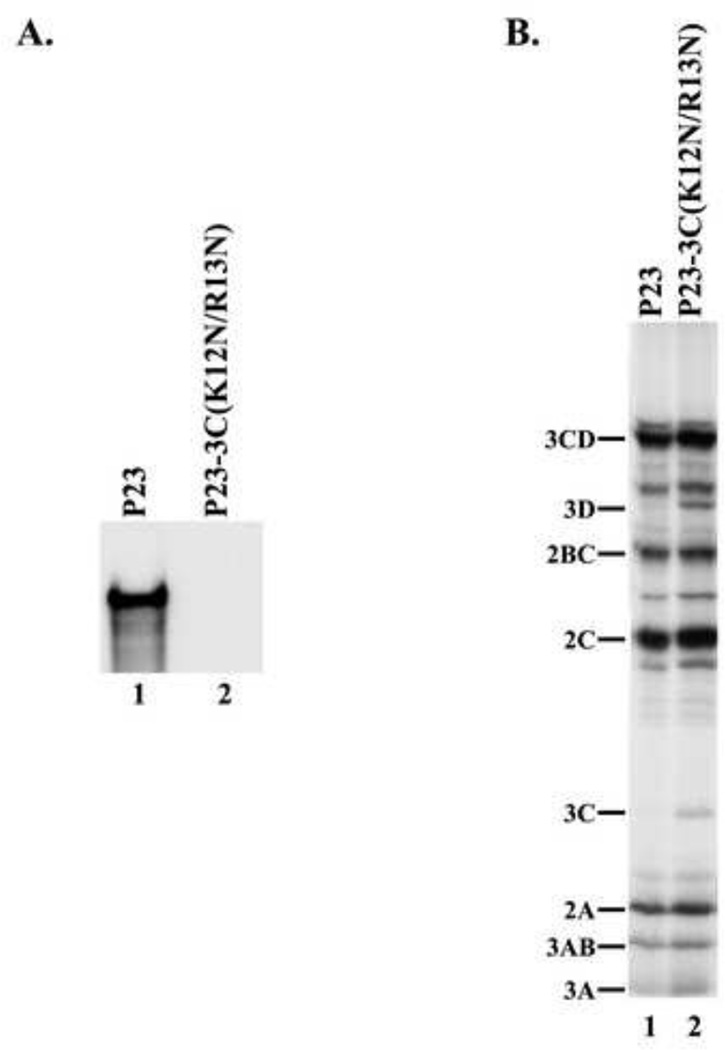
We next determined the effect of a 3C-RNA binding mutant on (−) strand RNA synthesis. A mutant that combined two previously described RNA binding mutations in the N-terminal RNA binding region of 3C was constructed in P23 RNA (i.e., P23-3C(K12N/R13N)) and used in this study (Blair et al., 1998). We determined the effect of this 3C-RNA binding mutant on (−) strand synthesis. As expected, labeled (−) strand RNA was synthesized in the reaction that contained wildtype P23 RNA (Fig.6A, lane 1). In contrast, (−) strand synthesis was completely inhibited in the reaction that contained P23-3C(K12N/R13N) RNA (Fig.6A, lane 2). Viral RNA translation was also measured in these reactions and similar levels of labeled viral proteins were observed in reactions containing either wildtype or mutant RNAs (Fig.6B, lanes 1–2). In the reaction that contained the P23-3C(K12N/R13N) RNA, viral protein processing was normal except for a small increase in 3C and 3D (Fig.6B, lane 2). These results demonstrated that (−) strand synthesis was completely inhibited by this 3C-RNA binding mutant.
Fig.6.
Characterization of a 3C-RNA binding mutant. (A) (−) Strand synthesis was assayed using PIRCs isolated from HeLa S10 reactions as described in Materials and methods. Reactions contained either wildtype P23 RNA or a 3C-RNA binding mutant (P23-3C(K12N/R13N)). Labeled product RNA was analyzed as described in Fig.3. (B) Translation of the transcript RNA was measured as described in Fig.3B.
Viral protein P3 is the preferred 3C precursor that rescues replication of a 3C-RNA binding mutant
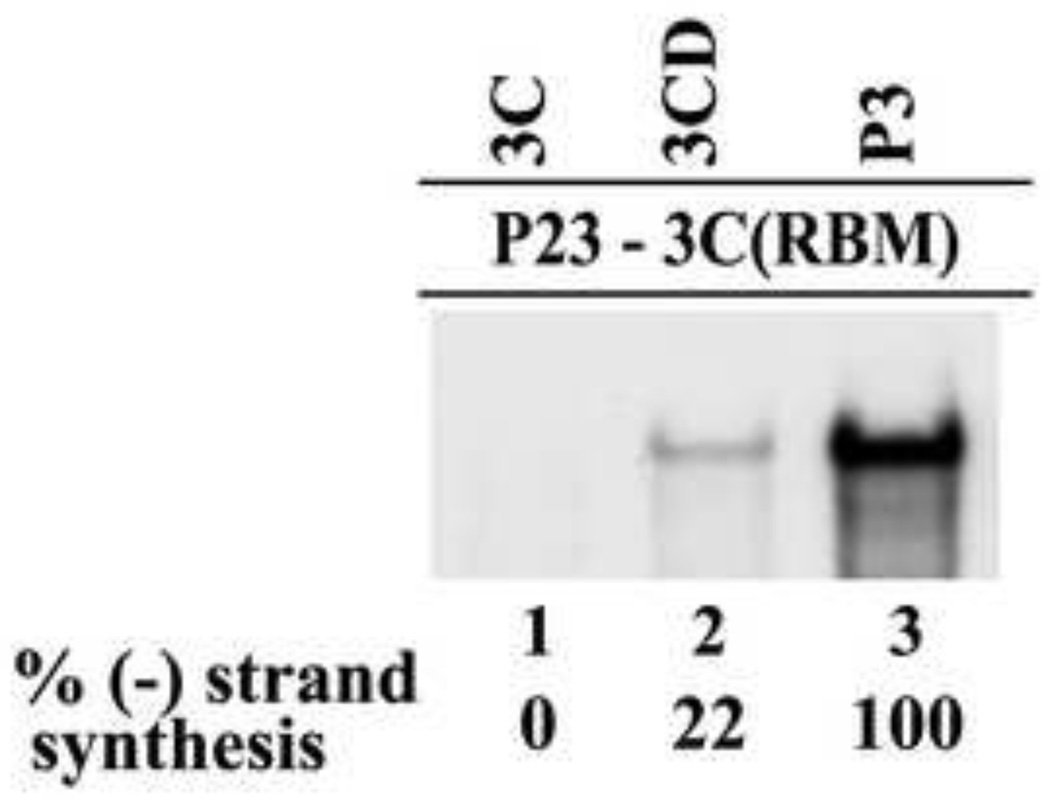
Complementation assays provide an experimental approach to determine the relative ability of 3C versus 3C containing precursors, 3CD and P3 to rescue the replication of a 3C-RNA binding mutant. To test this, complementation assays were performed in reactions that contained the RNA binding mutant P23-3C(K12N/R13N) RNA (designated as P23-3C(RBM)) in combination with expression RNAs that encoded 3C, 3CD or P3. (−) Strand synthesis was not detected in the reaction containing 3C (Fig.7, lane 1). In addition, (−) strand synthesis was not observed even when 3C was expressed together with 3D (data not shown). In contrast, measureable levels of (−) strand synthesis were observed in the presence of 3CD and P3 (Fig.7, lanes 2–3). Interestingly, higher levels of (−) strand synthesis were observed in the presence of P3 compared to 3CD (Fig.7, lanes 2–3). The level of (−) strand synthesis observed with 3CD was about 22% of the level observed with P3. Taken together, these results demonstrated that P3 is the preferred 3C precursor that is required to efficiently rescue replication of P23-3C(RBM) RNA.
Fig.7.
Viral protein P3 was required to rescue replication of P23-3C(RBM). (−) Strand synthesis was assayed using PIRCs isolated from HeLa S10 reactions as described in Materials and methods. Reactions contained P23 RNA containing a 3C-RNA binding mutation (3C(K12N/R13N or 3C(RBM)) and a complementing RNA expressing the indicated protein. Labeled product RNA was analyzed as described in Fig.3 and quantitated as described in Fig.4.
P3 binds to the 5’CL more efficiently than 3CD
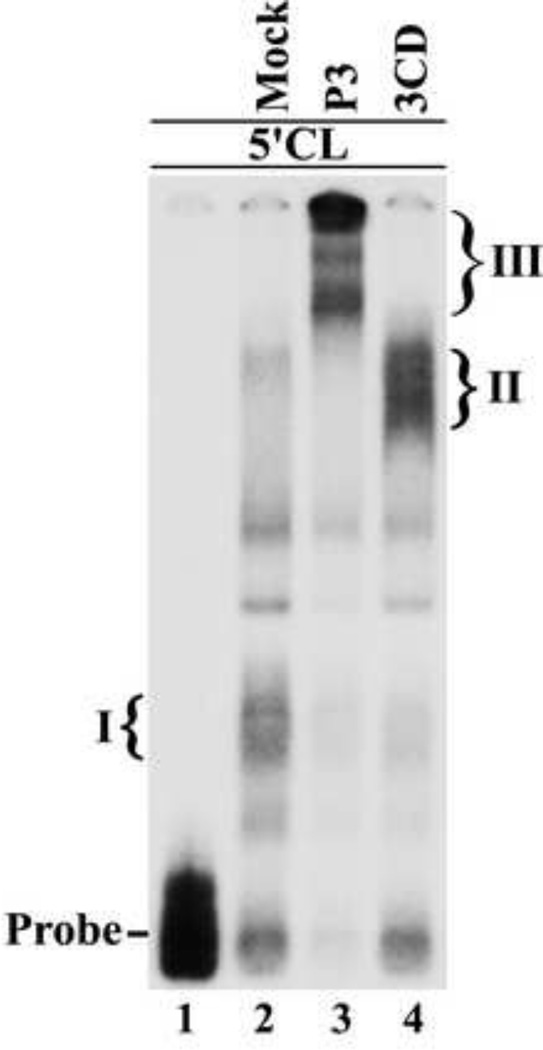
The finding that P3 was the preferred 3C containing precursor that rescued the replication of P23-3C(RBM) RNA suggested that P3 binds the 5’CL more efficiently than 3CD. To compare the binding of 3CD and P3 to the 5’CL, we performed electrophoretic mobility shift assays (EMSAs) using the 5’CL RNA as probe in the presence of P3 or 3CD. Since P3 and 3CD are proteolytically cleaved to their respective cleavage products, we engineered two previously described mutations in the active site of 3C protease (E71Q and C147S) to prevent cleavage of P3 and 3CD (Kean et al., 1993). EMSAs were performed with equimolar amounts of P3 and 3CD as described in Materials and methods. In mock reactions, the 5’CL formed the previously characterized complex with PCBP (Complex I) (Fig.8, compare lanes 1–2) (Andino et al., 1990a). In the reaction containing 3CD, a slower migrating complex (Complex II) was observed, which was previously shown to contain 3CD and PCBP (Andino et al., 1990a) (Fig.8, lane 4). Interestingly, a larger and slower migrating complex (Complex III) was observed in reactions containing P3 (Fig.8, lane 3). Finally, we observed that more of the 5’CL probe was shifted in the presence of P3 compared to 3CD (Fig.8, lanes 3–4). This result suggested that P3 binds to the 5’CL with higher affinity than 3CD, which was consistent with the results of the complementation assays with the 3C-RNA binding mutant.
Fig.8.
Viral protein P3 protein binds the 5’CL RNA. Electrophoretic mobility shift assay (EMSA) using labeled 5’CL RNA probe as described in Materials and methods. The position of the RNA probe is shown in lane 1. The probe was incubated with an aliquot of a mock reaction (lane 2) or with P3 (lane 3) or 3CD (lane 4). The P3 and 3CD used in these reaction contained mutations in the active site of 3C protease to inhibit processing of P3 and 3CD. The previously described RNP complex formed with the 5’CL RNA and PCBP is labeled as complex I (lane 2). The RNP complex formed with the 5’CL RNA and 3CD is labeled as complex II (lane 4).The complex formed with the 5’CL RNA and P3 is labeled as complex III (lane 3).
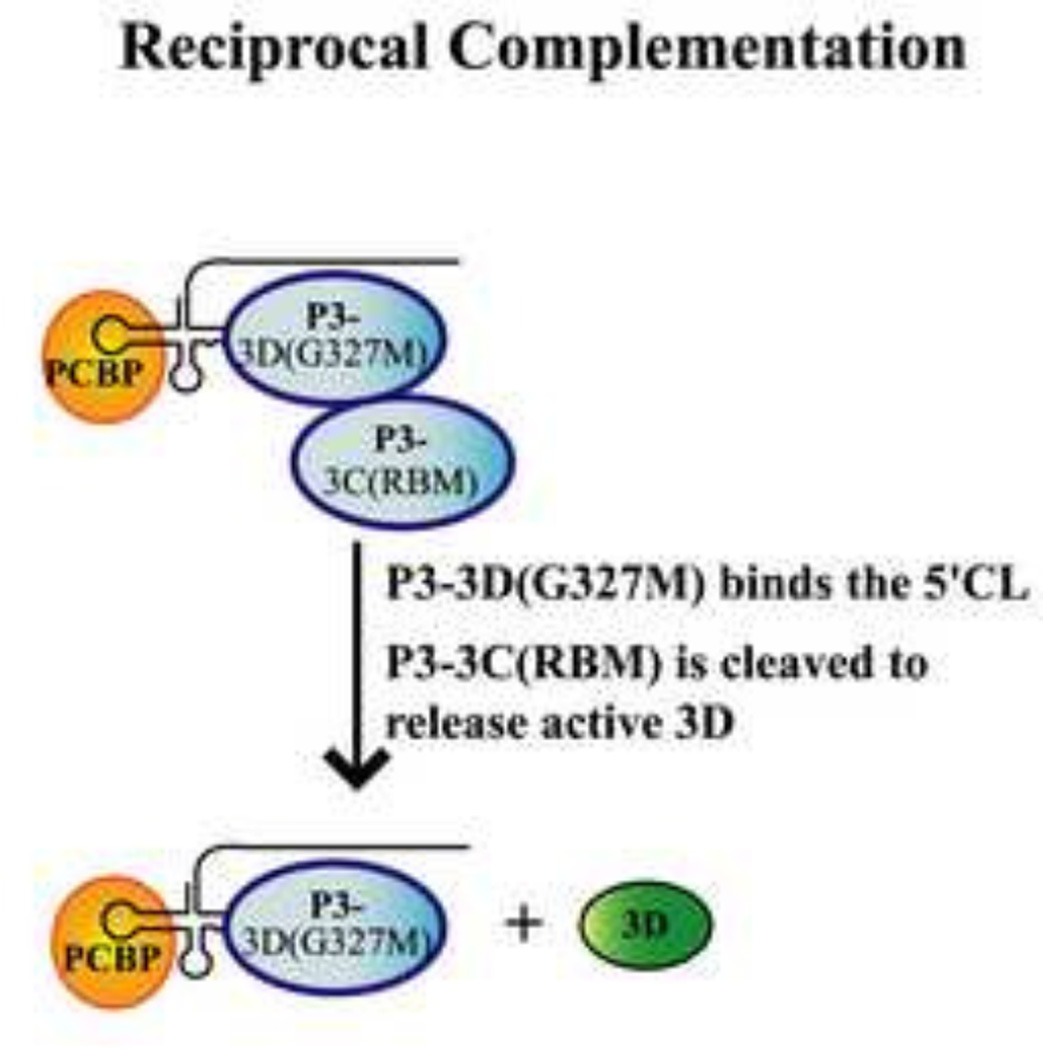
Reciprocal complementation is observed between a 3D polymerase mutant and a 3C-RNA binding mutant
The above complementation analysis showed that P3 was required to efficiently complement mutations that inhibited 3C-RNA binding activity. In addition, P3 (or 3CD) was required to efficiently complement mutations that inhibited 3D polymerase activity. To determine if the RNA binding and the polymerase activities are provided by the same or different molecules of P3, we performed reciprocal complementation assays. The rationale for this approach is shown in Fig.9 for a reaction containing P3-3D(G327M) and P3-3C(RBM). The P3-3D(G327M) can bind to the 5’CL but cannot provide active 3D polymerase. On the other hand, P3-3C(RBM) cannot bind to the 5’CL but can provide active 3D polymerase after proteolytic processing. Therefore, if (−) strand synthesis is observed, this suggests that the RNA binding and the polymerase activities are provided by different molecules of P3. In contrast, if (−) strand synthesis is not observed, this suggests that both activities are provided by the same molecule of P3. To distinguish between these two possibilities, we measured (−) strand synthesis in reactions containing P23-3C(RBM) RNA as the template in the presence of P3-3D(G327M) or P3-3CD(PM) expression RNAs. As expected, (−) strand synthesis was observed in the presence of wildtype P3 but not with P3-3C(RBM) (Figs.10A–B, lanes 1 and 3) . Importantly, (−) strand synthesis was also observed in the reactions containing P3-3D(G327M) (Fig.10A, lane 2) or P3-3CD(PM) (Fig.10B, lane 2). These results demonstrated that both polymerase mutants were able to efficiently complement the replication of the 3C-RNA binding mutant RNA.
Fig.9.
Model showing reciprocal complementation between a 3C-RNA binding mutant and a 3D polymerase mutant in the same reaction. In this model, the following multi-step process is predicted to occur during reciprocal complementation: (1) the P3-3D(G327M) provides the RNA binding activity and binds the 5’CL along with PCBP. (2) The P3-3C(RBM) is recruited to this complex and provides 3D polymerase activity after undergoing proteolytic processing.
Fig.10.
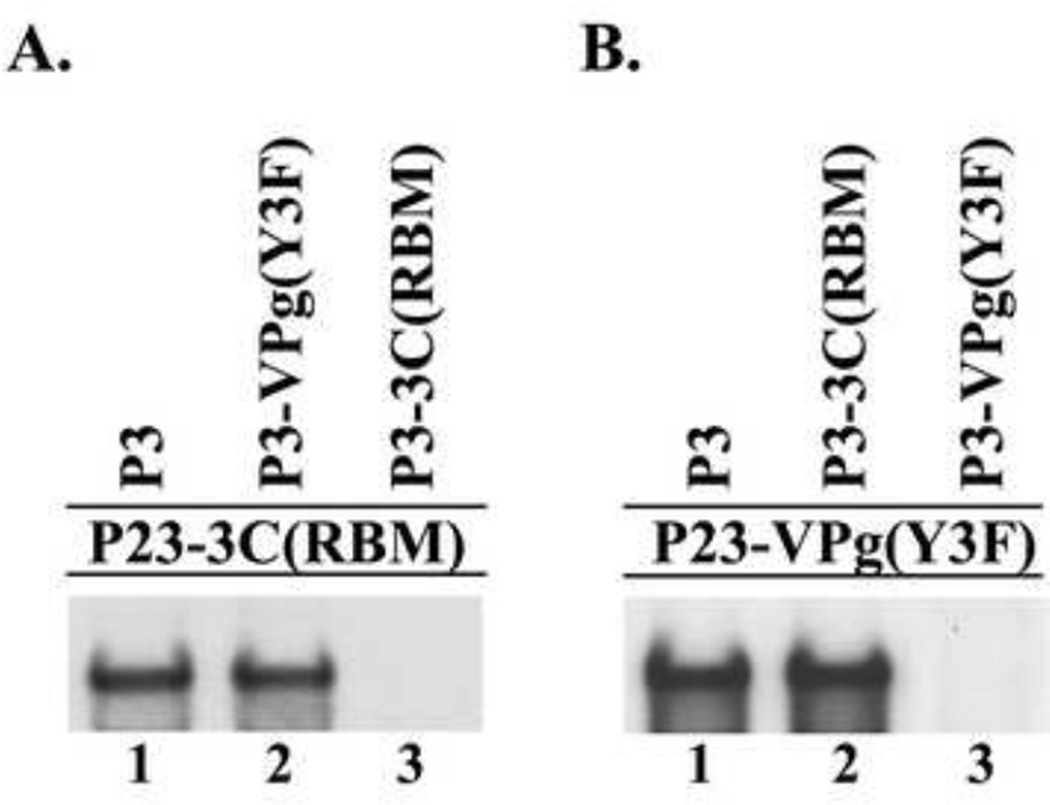
Reciprocal complementation between a 3C-RNA binding mutant and a 3D polymerase mutant. (−) Strand synthesis was assayed using PIRCs isolated from HeLa S10 reactions as described in Materials and methods. (A) and (B) Reactions contain P23-3C(RBM) template RNA and a second complementing RNA expressing the indicated protein. (C) Reactions contained P23-3D(G327M)) template RNA and a second RNA expressing the indicated protein. (D) Reactions contained P23-3CD(PM) template RNA and a second RNA expressing the indicated protein. Labeled product RNA was analyzed as described in Fig.3.
Similar reciprocal complementation experiments were performed in reactions containing either P23-3D(G327M) or P23-3CD(PM) template RNAs in the presence of expression RNAs that encoded wildtype 3CD, 3CD-3D(G327M) or 3CD-3C(RBM). In these experiments, 3CD expression RNAs were used since either P3 or 3CD complemented the replication of mutant polymerase template RNAs at about the same levels (Fig.4). In reactions containing P23-3D(G327M), efficient complementation was observed in the presence of 3CD or 3CD-3C(RBM) but not in the presence of the 3CD-3D(G327M) (Fig.10C, lanes 1–3). Likewise, in reactions containing P23-3CD(PM), efficient complementation was observed with 3CD or 3CD-3C(RBM), but not in the reaction containing 3CD(PM) (Fig.10D, lanes 1–3). These results demonstrated that the RNA binding mutant was able to complement both 3D polymerase mutants efficiently. In summary, our reciprocal complementation analysis showed that one molecule of P3 binds RNA and recruits a second molecule of P3 (or 3CD) which provides active 3D polymerase, but does not bind RNA.
Reciprocal complementation is observed between a 3C-RNA binding mutant and a VPg-linkage site mutant
The results presented above demonstrated that P3 was required to efficiently complement a 3C-RNA binding mutant. In addition, the results of a previous study showed that P3 is the preferred VPg-precursor that is required to efficiently complement a VPg-linkage site mutant (Liu et al., 2007). Therefore, to determine if RNA binding activity and VPg are provided by the same or different molecules of P3, we performed reciprocal complementation assays containing P3-3C(RBM) and P3-VPg(Y3F) (Murray and Barton, 2003;Reuer et al., 1990). In this assay, the P3-VPg(Y3F) can bind to the 5’CL but cannot provide active VPg. In contrast, the P3-3C(RBM) cannot bind to the 5’CL but can provide active VPg after proteolytic processing. Therefore, if (−) strand synthesis is observed in this assay, this indicates that the RNA binding activity and active VPg are provided by different molecules of P3. Alternatively, if (−) strand synthesis is not observed, this suggests that both activities are provided by the same molecule of P3. To distinguish between these two possibilities, we measured (−) strand synthesis in reactions containing P23-3C(RBM) RNA as the template in the presence of P3-VPg(Y3F) expression RNA. As expected, (−) strand synthesis was observed in the presence of wildtype P3 but not with P3-3C(RBM) (Fig.11A, lanes 1 and 3). Interestingly, (−) strand synthesis was also observed in the reaction containing P3-VPg(Y3F) (Fig.11A, lane 2). These results demonstrated that the P3-VPg(Y3F) mutant was able to efficiently complement the replication of the P23-3C(RBM) RNA template. This result indicated that the P3-3C(RBM), which cannot bind RNA, served as a VPg provider in this experiment. These findings indicated that the first molecule of P3, which binds the 5’CL, does not serve as the VPg provider and that the second molecule of P3 serves as the VPg provider in the replication complex.
Fig.11.
Reciprocal complementation between a 3C-RNA binding mutant and a VPg-linkage site mutant. (−) Strand synthesis was assayed using PIRCs isolated from HeLa S10 reactions as described in Materials and methods. (A) Reactions contain P23-3C(RBM) template RNA and a second complementing RNA expressing the indicated protein. (B) Reactions contain P23-VPg(Y3F) template RNA and a second RNA expressing the indicated protein. Labeled product RNA was analyzed as described in Fig.3.
A similar reciprocal complementation experiments was performed using P23-VPg(Y3F) as the template RNA in the presence of P3-3C(RBM) RNA expression RNAs. As expected, efficient (−) strand synthesis was observed in the presence of P3 but not in reactions containing P3-VPg(Y3F) expression RNA (Fig.11B, lanes 1 and 3). Significantly, efficient (−) strand synthesis was observed in the reaction containing P3-3C(RBM) (Fig.11B, lane 2). This result showed that the RNA binding mutant was able to rescue the replication of the VPg(Y3F) mutant. Taken together, the results of these reciprocal complementation experiments showed that the P3, which binds the 5’CL, does not serve as the VPg provider. Instead, a second molecule of P3, which does not bind RNA, served as the VPg provider in the RNA replication complex.
Discussion
The work presented here illustrates the multifunctional nature of the viral precursor protein, P3, particularly as it pertains to the initiation of viral RNA synthesis. By performing trans-complementation assays using the HeLa S10 translation-replication system, we have further defined the role of P3, and its processed products in the formation of a functional PV replication complex. Using this approach, we showed that 3D polymerase is recruited to the replication complex in the form of its precursors, P3 or 3CD, and that RNA binding activity is not a prerequisite for this function. Furthermore, our findings showed that P3 is the preferred precursor protein that is required to form the 5’CL-RNP complex. Results of our reciprocal complementation assays showed that the P3 that binds to the 5’CL does not function as the polymerase provider. A second molecule of P3 (or 3CD) is recruited to the 5’CL-RNP complex and serves as the precursor of active 3D polymerase. In addition, our results also showed that a second molecule of P3 functions as the VPg provider in the replication complex. Based on these findings, we propose a model in which P3 binds to the 5’CL and recruits additional P3 molecules via protein-protein interactions, which are then cleaved to provide active 3D and VPg for VPg uridylylation and the initiation of RNA replication.
Active 3D polymerase is recruited into the replication complex in the form of its inactive precursors, P3 or 3CD
Although 3CD contains the entire 3D protein, it does not have polymerase activity (Flanegan and Baltimore, 1979;Flanegan and Van Dyke, 1979;Harris et al., 1992). The activation of 3D is most likely due to changes in positioning of the N-terminus of 3D that occur subsequent to processing of the cleavage site between 3C and 3D (Hobson et al., 2001;Marcotte et al., 2007;Rothstein et al., 1988;Thompson and Peersen, 2004). Therefore, 3CD appears to function as a proenzyme, which can be specifically activated by proteolytic processing to release 3D. Our results showed that replication of viral RNAs with mutations that inhibit polymerase activity could not be efficiently rescued when 3D polymerase was provided in trans. Efficient replication of the mutant RNAs was only observed in the presence of P3 or 3CD. Interestingly, equivalent levels of replication were observed in the presence of either P3 or 3CD (Fig.4). This indicated that 3CD was the minimal precursor required to rescue the replication of a polymerase mutant. These results indicated that 3D gains the ability to enter the replication complex when it is covalently linked to 3C. This suggests that the 3C in 3CD (or P3) provides a function that is required to recruit the 3D precursors into the replication complex. These inactive precursors are then processed in the replication complex to release active 3D polymerase and initiate viral RNA replication.
P3 is the precursor protein that is required to form the 5’CL-RNP complex
Previous studies have demonstrated the importance of 3CD binding to stem-loop ‘d’ of the 5’CL during PV RNA replication (Andino et al., 1993;Parsley et al., 1997;Xiang et al., 1995). Three distinct regions in the 3C protein are required for the binding of 3C and 3CD to the 5’CL (Andino et al., 1990a;Bergmann et al., 1997;Blair et al., 1998;Matthews et al., 1994;Mosimann et al., 1997). This includes an N-terminal region (Y6, K12, R13), a central region (K82, F83, R84, D85, I86, R87), and a C-terminal region (T154, G155, K156). In this study, we showed that mutations in the N-terminal RNA binding domain (3C(K12N/R13N)) completely inhibited RNA replication. In complementation assays, we observed that 3C was unable to rescue the replication of this mutant RNA when provided in trans. In contrast, both 3CD and P3 complemented the replication of this RNA binding mutant. Surprisingly, the level of replication was about five times higher in the reactions containing P3 compared to 3CD. This result suggested that P3 binds to the 5’CL more efficiently than 3CD, which was confirmed using EMSAs. Taken together, these findings suggest that binding of 3CD to the 5’CL is enhanced when it is covalently linked to 3AB in P3. In addition, 3AB as part of P3 may facilitate the formation of a functional membrane-associated 5’CL RNP complex. In summary, the results suggest that P3 is the preferred 3C precursor protein, which binds to the 5’CL to form the 5’CL-RNP complex that is required for viral RNA replication.
Model showing the role of the P3 precursor protein in the assembly of a functional 5’CL RNP complex
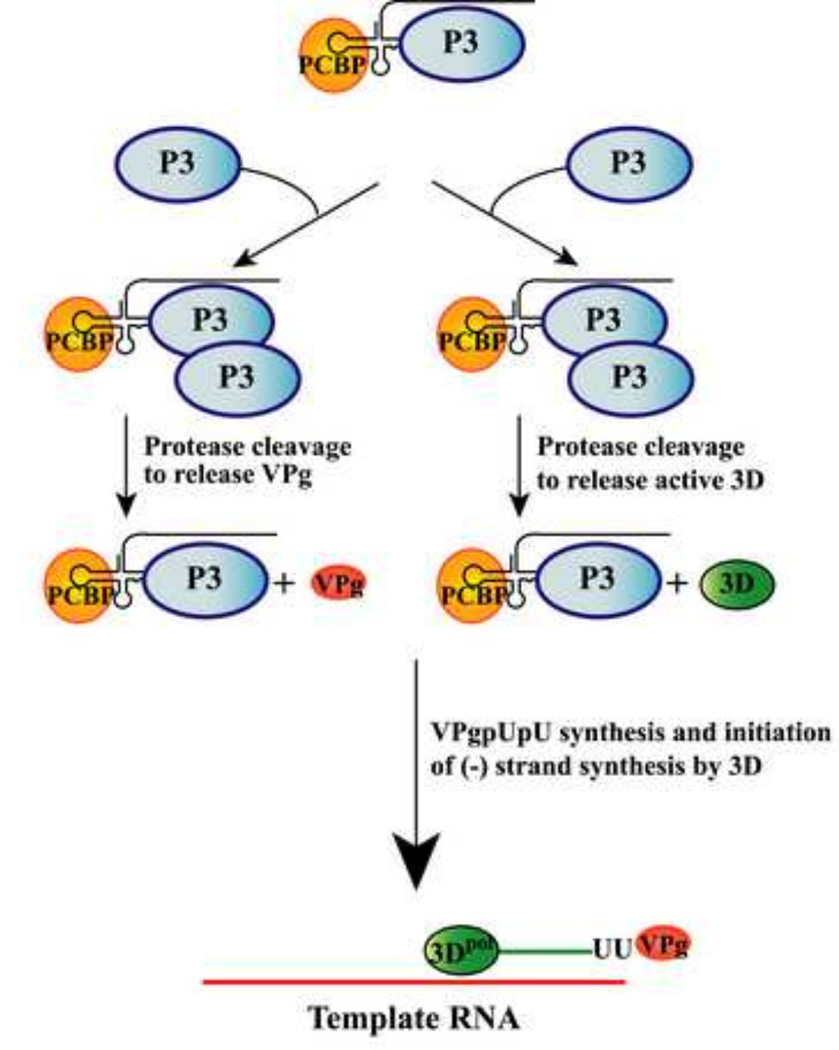
Using reciprocal complementation assays, we demonstrated that a polymerase mutant complemented the replication of an RNA binding mutant and vice versa. These findings support a model in which the RNA binding activity is provided by one molecule of P3 and the polymerase activity is provided by the cleavage of a second molecule of P3 (or 3CD) (Fig.12). In addition, we showed that replication of an RNA binding mutant was rescued by a VPg(Y3F) mutant in reciprocal complementation assays. These results are also consistent with a model in which one molecule of P3 binds the 5’CL and a second molecule of P3 is recruited to the complex and cleaved to provide VPg (Fig.12).
Fig.12.
Model showing the role of P3 precursor protein in the multi-step process of replication complex assembly. The cellular protein, PCBP, and the viral precursor protein, P3, bind to the 5’CL and form the 5’CL-RNP complex. Pathway on the right: A second molecule of P3 is recruited to the 5’CL-RNP complex via protein-protein interactions. This P3 serves as the 3D provider after being cleaved to release active 3D polymerase in the replication complex. Pathway on the left: A second molecule of P3, which serves as the VPg provider, is recruited to the 5’CL-RNP complex. This P3 is cleaved to release VPg. The 3D polymerase is then used to uridylylate VPg and to initiate and elongate VPgpUpU-primed (−) strand synthesis.
In this model, P3 along with PCBP binds to the 5’CL to form a functional 5’CL-RNP complex, which recruits a second molecule of P3 presumably via protein-protein interactions. Based on the results of our reciprocal complementation assays, the P3 bound to the 5’CL requires RNA binding activity, but does not serve as the precursor for either 3D polymerase or VPg. A second molecule of P3, which is recruited to the 5’CL-RNP complex, serves as the 3D provider in the replication complex (Fig.12, pathway on the right). This idea is based on our results which showed that P3 or 3CD, but not 3D, efficiently complemented a polymerase mutant. This suggests that 3D is unable to form a stable interaction with the P3 bound to the 5’CL unless 3D is covalently linked to 3C. The gain in function provided by 3C in this case may facilitate protein-protein interactions between the P3 molecules shown in the model. Likewise, a second molecule of P3 is recruited to the 5’CL-RNP complex to serve as the VPg provider in the replication complex (Fig.12, pathway on the left). In this case, P3 may also facilitate the delivery of VPg to the replication complex by protein-protein interactions with the P3 bound to the 5’CL. Finally, the model suggests that proteolytic cleavage of the second molecule of P3 is required to release active 3D polymerase or VPg to the complex. In the model, it is proposed that the bound P3 provides the protease activity required for the cleavage of the second molecule of P3. This allows for the activation of the polymerase and the release of VPg at the optimal time and location in the replication complex. The active 3D is then used to uridylylate VPg and to initiate VPgpUpU-primed (−) strand synthesis (Fig.12). In summary, this model illustrates the multi-step process in which viral proteins are initially assembled as precursor proteins to form a functional 5’CL-RNP complex. The precursor proteins along with their cleaved products are then used to perform discrete and essential functions during viral RNA replication.
Materials and methods
Poliovirus cDNA clones
A previously described cDNA clone of the Mahoney strain of type 1 poliovirus, designated pT7-PV1(A)80, was used as the parent clone for all poliovirus based constructs used in all studies herein (Fig.2A) (Barton et al., 2001). (i) pP23 is a previously described construct with a deletion of the P1 capsid coding region (Jurgens and Flanegan, 2003). RNA transcripts of this construct, P23 RNA, express all essential replication proteins from the P2 and P3 regions of the viral genome. (ii) pP3ΔGUA3 was derived from a previously described construct (pP3) which contains the coding region for the entire P3 polyprotein precursor (Jurgens and Flanegan, 2003). The ΔGUA3 deletion was engineered into the pP3 by inserting a Avr II-Mlu 1 fragment derived from pT7-PV1(A)80ΔGUA3 that has been previously described (Jurgens and Flanegan, 2003). This 5-nt deletion in the 3’ NTR (ΔGUAAA)inhibits (−) strand synthesis without affecting translation (Barton et al., 2001;Jurgens and Flanegan, 2003). Transcripts of this construct, P3ΔGUA3 RNA, function as a helper RNA expressing the P3 polyprotein precursor. (iii) p3CDΔGUA3 was constructed by inserting the 3CD (nt 5438–7375) protein coding sequence in-frame into the Msc I site of pDJB2 using the approach previously described (Jurgens and Flanegan, 2003). The ΔGUA3 mutation was transferred into this clone as described above. Transcripts of this construct, 3CDΔGUA3 RNA, serve as a helper RNA expressing 3CD, which has both RNA-binding and protease activities. (iv) p3DΔGUA3 was constructed by inserting the 3D (nt 5987–7375) coding sequence inframe into the Msc I site of pDJB2, and the ΔGUA3 mutation was transferred into this plasmid as described above. Transcripts of this construct, 3DΔGUA3 RNA, express 3D, which was shown in previous studies to be fully active in polymerase elongation assays in vitro using a poliovirion RNA template and oligo(U) primer (Eisner Smerage, 1998). (v) p3CΔGUA3 was constructed by inserting two stops codons at the end of the 3C coding sequence (nt 5438 – 5986) in p3CDΔGUA3 Transcripts of this construct, 3CΔGUA3 RNA, express proteolytically active 3C.
Mutant cDNA clones
p3CD-3D(G327M)ΔGUA3, pP3-3D(G327M)ΔGUA3 and pP23-3D(G327M) were engineered by transferring the BstB I-Avr II fragment (containing the G327M mutation) from pT7-PV1(A)80-3D(G327M) to p3CDΔGUA3, pP3ΔGUA3 or pP23. Transcripts of these constructs 3CD-3D(G327M)ΔGUA3 RNA, P3-3D(G327M)ΔGUA3 RNA or P23-3D(G327M) RNA express viral proteins, 3CD, P3 or P23, respectively, which contain the G327M mutation in 3D. The G327M mutation in the essential YGDD motif in the catalytic site of 3D abolishes all polymerase activity (Jablonski et al., 1991) (ii) p3CD(PM)ΔGUA3 was created by mutagenic PCR, using p3CDΔGUA3 as a template. This mutant combines two previously described processing site mutations (T181K, Q182D) with two additional mutations (S183G, Q184N) and was designed to completely abrogate 3C-3D processing (Andino et al., 1993;Blom et al., 1996;Harris et al., 1992). Transcripts of this construct 3CD(PM)ΔGUA3 RNA express 3CD, which retains RNA binding and protease activity, but is not processed into 3C and active 3D. (iii) pP23-3CD(PM) and pP3-3CD(PM)ΔGUA3 were created by transferring the Bgl II-BstB I restriction fragment from p3CD(PM)ΔGUA3 into the corresponding sites of pP23 and pP3ΔGUA3. (iv) p3CD-3C(K12N,R13N)ΔGUA3, pP3-3C(K12,R13N)ΔGUA3, and pP23-3C(K12N,R13N) were created by mutagenic PCR, using p3CDΔGUA3, pP3ΔGUA3 or pP23 as templates, respectively. These mutations were previously shown to inhibit the RNA binding ability of 3C (Blair et al., 1998). (vi) pP3-3C(E71Q,C147S)ΔGUA3, p3CD-3C(E71Q,C147S) were constructed by mutagenic PCR using pP3ΔGUA3 and p3CDΔGUA3 as templates, respectively. These individual mutations were previously shown to inhibit 3C protease activity (Kean et al., 1993). We combined these mutations and confirmed that 3C protease activity was completely inhibited in reactions containing these mutant RNAs. (vii) pP23-3B(Y3F) and pP3-3B(Y3F) were constructed by site directed mutagenesis using pP23ΔGUA3 and pP3ΔGUA3 as templates. The tyrosine (Y) at position 3 in 3B(VPg), which is the linkage site for VPg uridylylation, was changed to phenylalanine (F). This mutation was previously shown to inhibit VPgpUpU synthesis and viral replication (Murray and Barton, 2003;Reuer et al., 1990). Transcripts of these constructs are designated as P23-3B(Y3F) RNA and P3-3B(Y3F) RNA.
Preparation of RNA transcripts
Prior to in vitro transcription, the run-off transcription template was prepared by digesting the desired plasmid DNA with Mlu I. Digestion with this enzyme resulted in linearization of the circular plasmid DNA via a single cut immediately following the poliovirus 3’NTR-poly(A) tail. The DNA in the restriction digest reactions was extracted three times with phenol:chloroform, three times with chloroform and then ethanol precipitated. The Mlu I cut template DNA was then resuspended in Tris-EDTA buffer (10 mM Tris HCl (pH 8), 1 mM EDTA) at 0.5 µg/µl and stored at −20°C.
Transcription reactions were performed as previously described (Barton et al., 1996). Briefly, these reactions contained 1× transcription buffer [40 mM Tris HCl (pH 8), 6 mM MgCl2, 2 mM spermidine], 10 mM DTT, 0.4 U/µl RNasin (Promega), 1 mM of each NTP (ATP, CTP, GTP and UTP), and 15 ng/µl linearized DNA. Approximately 1µl of purified bacterially expressed T7 polymerase was used per 100 µl transcription reaction. Reactions were incubated at 37°C for 2 h and were stopped by the addition of 2.5 volumes of 0.5% SDS buffer [10 mM Tris HCl (pH 7.5), 100 mM NaCl, 1 mM EDTA, 0.5% sodium dodecyl sulfate]. The RNA transcripts were phenol: chloroform extracted three times, chloroform extracted three times, and subsequently precipitated by the addition of three volumes of 100% ethanol and incubated overnight at −20°C. The RNA transcripts were further purified by desalting over Sephadex G-50 (GE Healthcare) gel filtration resin (0.5 × 13 cm column). The transcript RNAs were then stored in ethanol at −20°C and were precipitated immediately prior to their use in the HeLa S10 translation-replication reaction.
HeLa S10 translation-RNA replication reactions
HeLa S10 extracts and HeLa cell translation initiation factors were prepared as described by Barton et al (Barton et al., 1996). HeLa S10 translation-replication reactions were performed as previously described with some modifications (Barton et al., 1996). Briefly, P23 RNAs (wildtype or mutant RNA as indicated) (4 pmol total) were added to 100 µl reaction mixtures in the presence of 2 mM guanidine HCl, and were incubated for 4 h at 34°C. For the complementation experiments, equimolar amounts of the template and expression RNAs (4 pmol total) were added to 100 µl reaction mixtures at the start of the incubation. Preinitiation-replication complexes (PIRCs) were isolated from these reactions by centrifugation and were resuspended in a replication assay buffer containing [α-32P] CTP and 50 µg/ml puromycin as previously described (Barton et al., 1995;Barton et al., 1996;Barton et al., 2002). The resuspended PIRCs were incubated at 37°C for 1 h, and the resulting 32P-labeled (−) strand product RNA was purified by phenol: chloroform extraction and ethanol precipitation. The P23 RNA transcripts contain two 5’ terminal non-viral G’s, which have been shown in previous studies to inhibit positive-strand but not (−) strand initiation (Barton et al., 1996;Herold and Andino, 2000;Morasco et al., 2003). Therefore, 32P-labled (−) strand RNA synthesis was specifically measured in these assays.
Analysis of 32P-labeled (−) strand RNA
Purified labeled (−) strand product RNA was analyzed by electrophoresis in CH3HgOH -1% agarose gels as previously described (Spear et al., 2008). In these denaturing gels, RNA structure is completely disrupted and full-length (−) strand RNA runs as a single band of labeled RNA. To ensure that equivalent amounts of RNA were loaded in each lane, the gels were stained with ethidium bromide to verify that equivalent amounts of the 18S and 28S ribosomal RNAs were recovered in each reaction. The labeled product RNAs were detected by autoradiography of the dried gel. The scans of the autoradiograms were quantitated using the image processing and analysis program, ImageJ, developed by the National Institutes of Health.
Translation of viral RNA transcripts
To confirm that equivalent levels of proteins were synthesized in each reaction, translation reactions were performed in parallel with RNA replication reactions as previously described (Barton et al., 1996). Viral protein synthesis was measured by labeling of the viral proteins using [35S]-methionine. Briefly, a 10 µl aliquot of the above described 100 µl HeLa S10 translation-replication reaction was removed and incubated at 34°C for 4 h in the presence of 11 µCi of [35S]-methionine. Following this incubation, a 5 µl aliquot was removed and added to 45 µl of 1× Laemmli sample buffer (20% v/v glycerol, 2% w/v SDS, 62.5 mM Tris HCl [pH 6.8], 72 mM β-mercaptoethanol, 0.1% w/v bromophenol blue). Samples were heated to 95°C for 5 min, and were analyzed by SDS-PAGE on a 9–18% gradient gel. Labeled viral protein products were visualized by autoradiography of the dried gel. It is important to note, that the processing of poliovirus 3CD to 3C and 3D is known to be slow compared to other viral precursor proteins. Therefore, relatively low levels of released 3C and 3D are observed in these translation reactions at 4 h. However, as shown in this manuscript these levels of 3C and 3D were sufficient to support efficient (−) strand synthesis in these cell-free reactions.
RNA Electrophoretic Mobility Shift Assays
To generate the 5’CL RNA probe, plasmid pP23 was linearized with Hga1, which cleaves at nucleotide position 118 at the 5’ end of the viral genome. Labeled 5’CL RNA was made by T7 transcription using the linearized DNA template in the presence of [α-32P]CTP (400 Ci/mmol). Probes were purified using NucAway Spin Columns (Ambion) and ethanol precipitated. The Electrophoretic mobility shift assays (EMSAs) contained 32P-labeled 5’CL RNA (20 fmol) and were performed as previously described (Andino et al., 1990a;Silvestri et al., 2006;Spear et al., 2008). We used P3 and 3CD, which contained mutations (E71Q,C147S) in the active site of 3C to inhibit 3C protease activity. HeLa S10 translation reactions containing either P3 or 3CD expression RNAs were incubated for 3 h at 34°C. The mock reaction did not contain expression RNA. The relative amounts of P3 and 3CD proteins synthesized in the reactions was measured by labeling of the proteins using [35S]-methionine in a parallel reaction. The labeled proteins were analyzed by SDS-PAGE on a 12.5% gel and quantitated using a PhosphorImager. The amount of labeled protein synthesized in each reaction was determined and the relative molar amount of each protein was calculated based on the number of methionines. The amount of the translation reaction that was added to the EMSAs was adjusted to ensure that equimolar amounts of P3 and 3CD were present in each assay. The 5’CL-RNP complexes were analyzed on a 5% native polyacrylamide gel and visualized by autoradiography.
Highlights.
Poliovirus 3D polymerase enters the replication complex in the form of its precursor, P3 (or 3CD).
P3, and not 3CD, is the preferred precursor that binds to the 5’CL to form the 5’CL-RNP complex.
Different molecules of P3 provide the polymerase, VPg and RNA binding activities in the replication complex.
Model is proposed in which P3 binds to the 5’CL and recruits additional molecules of P3, which are processed to 3D polymerase and VPg to initiate RNA replication.
Acknowledgments
This work was supported by Public Health Service grants AI15539 and AI32123 from the National Institute of Allergy and Infectious Diseases. A. Spear received support from Public Health Service training grant AI007110. We thank Brian O'Donnell for excellent technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Andino R, Rieckhof GE, Achacoso PL, Baltimore D. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5'-end of viral RNA. EMBO J. 1993;12:3587–3598. doi: 10.1002/j.1460-2075.1993.tb06032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andino R, Rieckhof GE, Baltimore D. A functional ribonucleoprotein complex forms around the 5' end of poliovirus RNA. Cell. 1990a;63:369–380. doi: 10.1016/0092-8674(90)90170-j. [DOI] [PubMed] [Google Scholar]
- Andino R, Rieckhof GE, Trono D, Baltimore D. Substitutions in the protease (3Cpro) gene of poliovirus can suppress a mutation in the 5' noncoding region. J. Virol. 1990;64:607–612. doi: 10.1128/jvi.64.2.607-612.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton DJ, Black EP, Flanegan JB. Complete replication of poliovirus in vitro: preinitiation RNA replication complexes require soluble cellular factors for the synthesis of VPg-linked RNA. J. Virol. 1995;69:5516–5527. doi: 10.1128/jvi.69.9.5516-5527.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton DJ, Morasco BJ, Flanegan JB. Assays for poliovirus polymerase, 3Dpol, and authentic RNA replication in HeLa S10 extracts. Methods Enzymol. 1996;275:35–57. doi: 10.1016/s0076-6879(96)75005-x. [DOI] [PubMed] [Google Scholar]
- Barton DJ, Morasco BJ, Smerage LE, Flanegan JB. Poliovirus RNA Replication and Genetic Complementation in Cell-Free Reactions. In: Semler BL, Wimmer E, editors. Molecular Biology of Picornaviruses. Washington, D.C: ASM Press; 2002. pp. 461–469. [Google Scholar]
- Barton DJ, O'Donnell BJ, Flanegan JB. 5' cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 2001;20:1439–1448. doi: 10.1093/emboj/20.6.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann EM, Mosimann SC, Chernaia MM, Malcolm BA, James MN. The refined crystal structure of the 3C gene product from hepatitis A virus: specific proteinase activity and RNA recognition. J. of Virol. 1997;71:2436–2448. doi: 10.1128/jvi.71.3.2436-2448.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair WS, Parsley TB, Bogerd HP, Towner JS, Semler BL, Cullen BR. Utilization of a mammalian cell-based RNA binding assay to characterize the RNA binding properties of picornavirus 3C proteinases. RNA. 1998;4:215–225. [PMC free article] [PubMed] [Google Scholar]
- Blom N, Hansen J, Blaas D, Brunak S. Cleavage site analysis in picornaviral polyproteins: discovering cellular targets by neural networks. Protein Sci. 1996;5:2203–2216. doi: 10.1002/pro.5560051107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisner Smerage L. Ph.D. dissertation. Gainesville, FL: University of Florida; 1998. Poliovirus RNA replication: Separation of initiation and elongation functions of the viral polymerase using a cell-free system. ufdc.ufl.edu/AA00022315/00001. [Google Scholar]
- Flanegan JB, Baltimore D. Poliovirus polyuridylic acid polymerase and RNA replicase have the same viral polypeptide. J. Virol. 1979;29:352–360. doi: 10.1128/jvi.29.1.352-360.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanegan JB, Van Dyke TA. Isolation of a soluble and template-dependent poliovirus RNA polymerase that copies virion RNA in vitro. J. Virol. 1979;32:155–161. doi: 10.1128/jvi.32.1.155-161.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamarnik AV, Andino R. Two functional complexes formed by KH domain containing proteins with the 5' noncoding region of poliovirus RNA. RNA. 1997;3:882–892. [PMC free article] [PubMed] [Google Scholar]
- Gruez A, Selisko B, Roberts M, Bricogne G, Bussetta C, Jabafi I, Coutard B, De Palma AM, Neyts J, Canard B. The crystal structure of Coxsackievirus B3 RNA-dependent RNA polymerase in complex with its protein primer VPg confirms the existence of a second VPg binding site on Picornaviridae polymerases. J. Virol. 2008;82:9577–9590. doi: 10.1128/JVI.00631-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerle T, Molla A, Wimmer E. Mutational analysis of the proposed FG loop of poliovirus proteinase 3C identifies amino acids that are necessary for 3CD cleavage and might be determinants of a function distinct from proteolytic activity. J. Virol. 1992;66:6028–6034. doi: 10.1128/jvi.66.10.6028-6034.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KS, Reddigari SR, Nicklin MJH, Hammerle T, Wimmer E. Purification and characterization of poliovirus polypeptide 3CD, a proteinase and a precursor for RNA polymerase. J. Virol. 1992;66:7481–7489. doi: 10.1128/jvi.66.12.7481-7489.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KS, Xiang W, Alexander L, Lane WS, Paul AV, Wimmer E. Interaction of poliovirus polypeptide 3CDpro with the 5' and 3' termini of the poliovirus genome: Identification of viral and cellular cofactors needed for efficient binding. J. Biol. Chem. 1994;269:27004–27014. [PubMed] [Google Scholar]
- Herold J, Andino R. Poliovirus requires a precise 5' end for efficient positive-strand RNA synthesis. J. Virol. 2000;74:6394–6400. doi: 10.1128/jvi.74.14.6394-6400.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold J, Andino R. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Molecular Cell. 2001;7:581–591. doi: 10.1016/S1097-2765(01)00205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobson SD, Rosenblum ES, Richards OC, Richmond K, Kirkegaard K, Schultz SC. Oligomeric structures of poliovirus polymerase are important for function. EMBO J. 2001;20:1153–1163. doi: 10.1093/emboj/20.5.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonski SA, Luo M, Morrow CD. Enzymatic activity of poliovirus RNA polymerase mutants with single amino acid changes in the conserved YGDD amino acid motif. J. Virol. 1991;65:4565–4572. doi: 10.1128/jvi.65.9.4565-4572.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurgens C, Flanegan JB. Initiation of poliovirus negative-strand RNA synthesis requires precursor forms of p2 proteins. J. Virol. 2003;77:1075–1083. doi: 10.1128/JVI.77.2.1075-1083.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kean KM, Howell MT, Grunert S, Girard M, Jackson RJ. Substitution mutations at the putative catalytic triad of the poliovirus 3C protease have differential effects on cleavage at different sites. Virology. 1993;194:360–364. doi: 10.1006/viro.1993.1268. [DOI] [PubMed] [Google Scholar]
- Lawson MA, Semler BL. Alternate poliovirus nonstructural protein processing cascades generated by primary sites of 3C proteinase cleavage. Virology. 1992;191:309–320. doi: 10.1016/0042-6822(92)90193-s. [DOI] [PubMed] [Google Scholar]
- Liu Y, Franco D, Paul AV, Wimmer E. Tyrosine 3 of poliovirus terminal peptide VPg(3B) has an essential function in RNA replication in the context of its precursor protein 3AB. J. Virol. 2007;81:5669–5684. doi: 10.1128/JVI.02350-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons T, Murray KE, Roberts AW, Barton DJ. Poliovirus 5'-Terminal Cloverleaf RNA is required in cis for VPg uridylylation and the initiation of negative-strand RNA synthesis. J. Virol. 2001;75:10696–10708. doi: 10.1128/JVI.75.22.10696-10708.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotte LL, Wass AB, Gohara DW, Pathak HB, Arnold JJ, Filman DJ, Cameron CE, Hogle JM. Crystal structure of poliovirus 3CD protein: virally encoded protease and precursor to the RNA-dependent RNA polymerase. J. Virol. 2007;81:3583–3596. doi: 10.1128/JVI.02306-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews DA, Smith WW, Ferre RA, Condon B, Budahazi G, Sisson W, Villafranca JE, Janson CA, McElroy HE, Gribskov CL. Structure of human rhinovirus 3C protease reveals a trypsin-like polypeptide fold, RNA-binding site, and means for cleaving precursor polyprotein. Cell. 1994;77:761–771. doi: 10.1016/0092-8674(94)90059-0. [DOI] [PubMed] [Google Scholar]
- Morasco BJ, Sharma N, Parilla J, Flanegan JB. Poliovirus cre(2C)-dependent synthesis of VPgpUpU is required for positive- but not negative-strand RNA synthesis. J. Virol. 2003;77:5136–5144. doi: 10.1128/JVI.77.9.5136-5144.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosimann SC, Cherney MM, Sia S, Plotch S, James MN. Refined X-ray crystallographic structure of the poliovirus 3C gene product. J. Mol. Biol. 1997;273:1032–1047. doi: 10.1006/jmbi.1997.1306. [DOI] [PubMed] [Google Scholar]
- Murray KE, Barton DJ. Poliovirus CRE-dependent VPg uridylylation is required for positive-strand RNA synthesis but not for negative-strand RNA synthesis. J. Virol. 2003;77:4739–4750. doi: 10.1128/JVI.77.8.4739-4750.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogram SA, Flanegan JB. Non-template functions of viral RNA in picornavirus replication. Curr. Opin. Virol. 2011;1:339–346. doi: 10.1016/j.coviro.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmenberg AC. Proteolytic processing of picornaviral polyprotein. Annu. Rev. Microbiol. 1990;44:603–623. doi: 10.1146/annurev.mi.44.100190.003131. [DOI] [PubMed] [Google Scholar]
- Parsley TB, Cornell CT, Semler BL. Modulation of the RNA binding and protein processing activities of poliovirus polypeptide 3CD by the viral RNA polymerase domain. J. Biol. Chem. 1999;274:12867–13876. doi: 10.1074/jbc.274.18.12867. [DOI] [PubMed] [Google Scholar]
- Parsley TB, Towner JS, Blyn LB, Ehrenfeld E, Semler BL. Poly (rC) binding protein 2 forms a ternary complex with the 5'-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA. 1997;3:1124–1134. [PMC free article] [PubMed] [Google Scholar]
- Reuer Q, Kuhn RJ, Wimmer E. Characterization of poliovirus clones containing lethal and nonlethal mutations in the genome-linked protein VPg. J. Virol. 1990;64:2967–2975. doi: 10.1128/jvi.64.6.2967-2975.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein MA, Richards OC, Amin C, Ehrenfeld E. Enzymatic activity of poliovirus RNA polymerase synthesized in Escherichia coli from viral cDNA. Virology. 1988;164:301–308. doi: 10.1016/0042-6822(88)90542-9. [DOI] [PubMed] [Google Scholar]
- Silvestri LS, Parilla JM, Morasco BJ, Ogram SA, Flanegan JB. Relationship between poliovirus negative-strand RNA synthesis and the length of the 3' poly(A) tail. Virology. 2006;345:509–519. doi: 10.1016/j.virol.2005.10.019. [DOI] [PubMed] [Google Scholar]
- Spear A, Sharma N, Flanegan JB. Protein-RNA tethering: the role of poly(C) binding protein 2 in poliovirus RNA replication. Virology. 2008;374:280–291. doi: 10.1016/j.virol.2007.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teterina NL, Egger D, Bienz K, Brown DM, Semler BL, Ehrenfeld E. Requirements for assembly of poliovirus replication complexes and negative-strand RNA synthesis. J Virol. 2001;75:3841–3850. doi: 10.1128/JVI.75.8.3841-3850.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AA, Peersen OB. Structural basis for proteolysis-dependent activation of the poliovirus RNA-dependent RNA polymerase. EMBO J. 2004;23:3462–3471. doi: 10.1038/sj.emboj.7600357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towner JS, Mazanet MM, Semler BL. Rescue of defective poliovirus RNA replication by 3AB-containing precursor polyproteins. J. Virol. 1998;72:7191–7200. doi: 10.1128/jvi.72.9.7191-7200.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt DA, Andino R. An RNA element at the 5'-end of the poliovirus genome functions as a general promoter for RNA synthesis. PLoS. Pathog. 2010;6:e1000936. doi: 10.1371/journal.ppat.1000936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer E, Hellen CUT, Cao X. Genetics of poliovirus. Annu. Rev. Genetics. 1993;27:353–436. doi: 10.1146/annurev.ge.27.120193.002033. [DOI] [PubMed] [Google Scholar]
- Xiang W, Harris KS, Alexander L, Wimmer E. Interaction between the 5'-terminal cloverleaf and 3AB/3CDpro of poliovirus is essential for RNA replication. J. Virol. 1995;69:3658–3667. doi: 10.1128/jvi.69.6.3658-3667.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]