Abstract
The Bladder-Exstrophy-Epispadias Complex (BEEC) represents the severe end of the uro-rectal malformation spectrum, and has a profound impact on continence, and on sexual and renal function. While previous reports of familial occurrence, in-creased recurrence among first-degree relatives, high concordance rates among monozygotic twins, and chromosomal aberra-tions were suggestive of causative genetic factors, the recent identification of copy number variations (CNVs), susceptibility regions and genes through the systematic application of array based analysis, candidate gene and genome-wide association studies (GWAS) provide strong evidence. These findings in human BEEC cohorts are underscored by the recent description of BEEC(-like) murine knock-out models. Here, we discuss the current knowledge of the potential molecular mechanisms, mediating abnormal uro-rectal development leading to the BEEC, demonstrating the importance of ISL1-pathway in human and mouse and propose SLC20A1 and CELSR3 as the first BEEC candidate genes, identified through systematic whole-exome sequencing (WES) in BEEC patients.
Keywords: Array, Bladder, Cloacal, Epispadias, Exome, Exstrophy, Pathway
1. INTRODUCTION
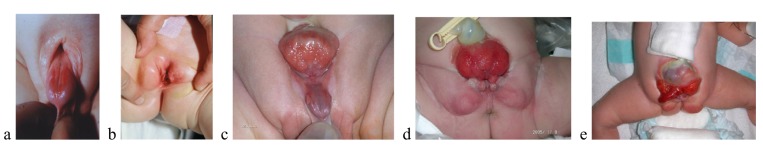
The Bladder-Exstrophy-Epispadias Complex (BEEC) describes a spectrum of malformations, and has a profound impact on continence, and on sexual and renal function. The component findings of BEEC have variable severity, and include Epispadias (E) (Fig. 1; a,b), the mildest phenotype, and Classic bladder exstrophy (CBE) (Fig. 1; c,d), which is the intermediate and most common defect. Persistent cloaca (PC) or Urorectal Septal Malformation Sequence, characterized by a common cloacal cavity (including bladder and intestinal elements), usually imperforate anus, and without exstrophy [1, 2] and Cloacal Exstrophy (CE), which is often referred to as the OEIS complex (omphalocele, exstrophy, imperforate anus, and spinal defects) (Fig. 1; e) [3] may represent the more severe forms of BEEC; however, there are some who consider CE to have a different embryologic origin from CBE [4, 5].
Fig. (1).
a, b) boy and girl with epispadias; c, d) boy and girl with classic exstrophy of the bladder; e) boy with exstrophy of the cloaca(Ebert et al. 2009).
In approximately one third of all BEEC cases, several urological malformations (e.g. ectopic kidney, renal agenesis, hydronephrosis) are present. Vesicoureteral reflux and obstruction of the ureteropelvic junction are observed frequently in both genders, and cryptorchidism is common in males [6]. Other malformations involving the gastrointestinal, skeletal, spinal and genitourinary systems (ambiguous genitalia) are reported frequently in CE [7-9]. Management of the BEEC is primarily surgical, and the main aims are the achievement of secure abdominal wall closure, urinary continence with preservation of renal function, and adequate cosmetic and functional genital reconstruction [10-12]. Following reconstructive surgery of the bladder during the neonatal period, continence rates of around 80% are expected during childhood. Additional surgery might be needed to optimize bladder storage and emptying function. In cases of final reconstruction failure, urinary diversion is necessary. In puberty, genital and reproductive functions are important issues [13, 14]. Psychosocial and psychosexual outcome and adequate health-related quality of life depend on long-term multidisciplinary care [15, 16].
Not a decade ago, the molecular elucidation of such rare and heterogeneous congenital malformation complexes as the BEEC seemed almost impossible. The aim of the present review is to illustrate, how recent systematic applications of array based and next generation sequencing techniques as well as modern functional pathway analyses have started to ultimately decipher the molecular basis even of such rare and heterogeneous congenital diseases as the BEEC.
1.1. Epidemiology
Among children of European descent, the overall birth prevalence for the entire BEEC spectrum has been estimated to be 1 in 10,000 [6]. While BEEC phenotypes have been observed as part of complex malformation syndromes, the majority of cases (approximately 98.5%) are classified as isolated [17-19]. Birth prevalences for the specific subtypes, including terminated pregnancies, have also been estimated. For E, an estimated prevalence of 1 in 117,000 in males and 1 in 484,000 in females has been reported [6]. On the other hand, Lloyd et al. [20] reviewed the American Kids’ Inpatient Database (KID) and found a weighted (1997 – 2009) birth prevalence rate of 9.25 male E births per 100,000 in-hospital live births, with females uncaptured in KID. These authors also noted a 45% increase of E in the birth prevalence during this period.
For CBE, an estimated prevalence of 1 in 37,000 has been reported [21]. Here, Lloyd et al. [20] found a lower value of 1.68 CBE births per 100,000 in KID and a 33% decrease in the birth prevalence from 1997-2009. As for the observed increase of E, the authors suggested a shift in coding practices for the decrease in CBE, rather than a shift in epidemiology [20]. For CE, the estimated prevalence is 1 in 200,000 to 1 in 400, 000 [8]; however, other studies suggest a higher prevalence of 1 in 80,000 to 1 in 159,000 [7, 22-26]. Prevalence ratios did not vary by maternal age [26]. The studies by Martinez-Frias et al. [8] and Caton et al. [25] support, that CE and CBE are different etiologic disorders, based upon their characteristic findings, prevalence and demographic characteristics. Findings of low birth weight, twinning, single umbilical artery, and infant mortality [8, 25], preterm births, and female sex [18] were associated with CE.
Due to the rarity of the phenotype, little is known about possible environmental risk factors. Reports by Wood et al. [27, 28] and Zwink et al. [29] suggest that assisted reproduction is a risk factor. Furthermore, maternal smoking and medical radiation during the first trimester were associated with a more severe phenotype, whereas periconceptional folic acid supplementation appeared to be protective [30]. Reefhuis et al. [31] found a positive association between use of clomid and CE (OR 5.4, 1.0-19.3).
1.2. Inheritance of the BEEC
Although familial occurrence is rare, 30 multiply affected families have been reported, and in some of these, the BEEC appears to follow a Mendelian mode of inheritance [32-35]. Nevertheless, the general consensus in the field is that - in the majority of patients - the genetic basis of the BEEC appears to be multifactorial [18]. For CBE, the reported recurrence risk among siblings ranges between 0.3-2.3%, in cases with non-consanguineous and non-affected parents [36, 37]. In their series, Shapiro et al. [36] reported the recurrence risk for the offspring of affected patients to be 1.4%. Hence, the recurrence risk for the offspring of affected patients and the risk of having a second affected child for parents who are non-consanguineous and non-affected shows an approximate 400-fold increase compared to the general population. Furthermore higher concordance rates among monozygotic compared to dizygotic twin pairs [34] emphasize a probable genetic etiology of the malformation. For CE, there are reports of recurrence in families [38]; increased occurrence among conjoined and monozygotic twins [39-45]; concordant conjoined twins [46]; and discordant dizygotic twins [47], all of which suggest a genetic basis for CE.
2. PATHOGENIC CONCEPT
BEEC has been recognized as one of the more intriguing phenotypes for mouse developmental researchers. The unique features of such a syndrome are its simultaneous onset of abnormalities of the bladder region and the upper (dorsal) part of external (ext.) genitalia.
2.1. Growth Factor Signals and Other Regulators Related with BEEC
Several essential growth factor signals for organogenesis are Hedgehog (Hh) and Wnt signals, which govern many developmental processes. Both signals are detected in the cloacal region, often in the cloacal membrane (CM) [48-50]. Such signals have been recently reported as being composed by the signal cascade, i.e., Hedgehog signal (Sonic Hedgehog (Shh))-Wnt signal-FGF (FGF8) by compound mutant analyses on embryonic ext. genitalia (genital tubercle; GT) and CM [51]. The Hedgehog signal (Shh as the ligand) and Wnt signal have been known to play roles in CM development [49, 52]. Alteration of these signals leads to abnormalities of or absence of CM formation, leading to severe caudal abnormalities [49]. Milder forms of CM abnormality such as partial (e.g., dorsal part) agenesis or abnormal orifice formation related to the CM may lead to BEEC phenotypes often with abnormal URS (urorectal septum) development [10, 52]. The timing of CM disruption was also suggested to influence these phenotypes [10, 52].
In addition to CM disruption, localized alterations in cell death either in the CM and/or in the PCM may have an affect on the phenotypes in mouse models of BEEC [10, 49]. However, the role of cell death, such as apoptosis, is still not well characterized, except for its possible role in the CM [48, 49].
Hedgehog and Wnt signals have been also known to affect the nearby Peri cloacal mesenchyme (PCM) through epithelial-mesenchymal interaction. By such interaction, the PCM is influenced by the CM to develop its characteristic mesenchymal gene expression, such as Bmp (bone morphogenetic protein) 4 [49]. In addition to these changes in gene expression, regulation of mesenchymal differentiation has been suggested. Thus, alteration of hedgehog signals by several genetic combinations has been shown to display BEEC like phenotypes. In fact, several mouse hedgehog-signal mutants display BEEC-related abnormalities. Developmental genes such as Alx4 (paired type homeobox gene) have also been shown to genetically interact with hedgehog signals [53]. Alx4 has been shown to interact with the hedgehog pathway, particularly in limb formation and digit development by the ZPA (zone of polarizing activity). Alx4 mutant mice have been shown to display several defects affecting the coordinated developmental process in the formation of the ventral body wall and external genitalia, like bladder abnormalities with epispadias-like phenotypes together with upper GT defects.
Similar phenotypes are described in Gli3 mutant mice, which is a negative transcriptional regulator of hedgehog signaling [53]. Various genetic mutants of Alx4 and/or Gli3 have been shown to display defective body wall closure. In such mutants, abnormal bladder and ext. genitalia phenotypes with reduced Alx4 or together with reduced Gli3 are partly rescued in some genetic backgrounds by “reducing” hedgehog (Shh) gene dosage [53]. Thus, the orchestration of the hedgehog signaling pathway with its modifier-genes needs to be further investigated.
In contrast to BEEC in mouse models, there have been several genes reported to be responsible for the development of body wall abnormalities including body wall closure. Hedgehog signaling has also been shown to be essential for body wall closure with abnormalities related to omphalocele [54]. However, the down-stream target genes remain to be elucidated.
In addition to growth factor signals, there have been several genes found to be responsible for body wall abnormalities including body wall hernia or gastroschisis [55]. Currently, only a few genes have been identified as being responsible for BEEC like phenotypes. One such example is the Ap2 gene, which has been suggested as one of the essential genes for body wall closure. Ap2alpha-deficient mice have been reported as showing phenotypes related to body wall closure and upper GT formation [56]. In addition to Ap2, TP63 has been also suggested as showing similar phenotypes. TP63 is an essential regulator of skin development and epithelial formation. In addition, the TP63 KO mouse displayed defective body wall and upper GT phenotypes [57]. However, it remains still unclear whether the reported mutants display highly penetrant BEEC-like phenotypes.
Most recently, Chari et al. [58] demonstrated a potential cross regulation of the Shh and TP63 pathways in keratinocytes that maintains epidermal homeostasis by controlling both, proliferation and differentiation.
Although both genes have been relatively well characterized with respect to their interacting genes during epithelial development, little is known regarding their interacting genes and, as outlined in chapter 5, their contribution to BEEC-like phenotypes.
2.2. Developmental Cell Lineages Related with BEEC
Orchestrated and complex in vivo phenotypes can be also analyzed in mouse models using cell lineage analysis. Hedgehog and Wnt signals have been known to affect the nearby mesenchyme, the Peri-cloacal mesenchyme (PCM) through epithelial-mesenchymal interaction. An important breakthrough for mouse BEEC models was the use of tissue lineage analysis using hedgehog (HH) signal-responding cell analysis [59]. The lineage for hedgehog-responding cells was monitored by Gli1 gene-marked cell imaging. Mesenchyme close to the developing cloaca (PCM) was shown to contribute to the bladder region and upper part of the external genitalia [59]. This study opened the door for lineage analysis in the mouse, which was essential to understand syndromic abnormalities such as BEEC.
Another line of work should also be noted for its progress. Several cell lineages including the cloacal region have been suggested to contribute to bladder (in addition to other pelvic organs) and ext. genitalia. One lineage includes the Isl1 type developmental gene-positive cell lineage. The Isl1 gene, also discovered in a genome-wide association study and meta-analysis as a top CBE candidate gene (see chapter 5), has been suggested to regulate limb development as it was determined to be upstream of Hand - Shh in hindlimb formation. Recently Isl1 was shown to be expressed in hindlimb and GT (genital tubercle) [60]. In fact, alteration of the Bmp 4 (Bone morphogenesis protein) gene in this cell lineage resulted in abnormalities in ext. genitalia and bladder.
As suggested previously, Shh is expressed in the CM and Bmp4 is expressed in the nearby mesenchyme (PCM). The Isl1 gene can mark a rather broad area of PCM, GT and caudal embryonic regions together with hindlimb [60]. How the alteration of Bmp signals in this Isl1 cell lineage can lead to defective caudal mesenchyme remains to be investigated. Generally, epithelial (endoderm)-derived hedgehog signals “relayed” toward mesenchymal genes such as Bmp4 have been suggested [61]. Regarding Bmp signals, Msx genes have been long suggested as interacting with Bmp signals and its mutants show body wall abnormalities [62].
Regarding cell lineage contributions, the possible connection between bladder organogenesis and umbilical cord for body wall formation remains to be elucidated.
The umbilical cord (UC) is located adjacent to the cloacal membrane/cloaca, and an unstable or defective cloacal membrane/cloaca could be one of the causes of such syndromes. Proper umbilical cord development is essential for development of the umbilical region, connecting the maternal environment and the embryo. Some developmental genes are suggested to function within such processes.
In the lower body region, cells derived from the anterior (upper) part of body wall, the aPCM region, have been reported to contribute to the upper part of genitalia [53]. Such cell lineages can also explain simultaneous onset of BEEC phenotypes. Dye induced cell marking experiments revealed the presence of cell migration from the umbilical cord region (aPCM) or infra-umbilical mesenchyme region towards upper part of ext. genitalia. Genetically marked experiments or genetic modulation of such cell populations would be necessary to verify such significance. Part of these candidate genes were reported in the case of Alx4 but some other genes showed similar expression patterns, uncluding Tbx4, Tbx3, Pitx1, and Mab21l2 genes [53]. Redundancy among such genes may exist for their developmental functions.
2.3. Merits and Perspectives on Mouse Models for BEEC
Complex in vivo phenotypes can be also analyzed in mutant mouse models. Given that such phenotypes often “phenocopy” those of human patients, there are often no questions regarding the usefulness of analyzing mutant mouse models. However, essential genes and growth factor signaling pathways simultaneously governing multiple tissues are often expressed in the very early embryo (for example gastrula). Thus, introduction of a mutation in these important regulatory genes is often expected to lead to the early embryonic lethality prior to organogenesis. In general, conditional gene inactivation systems would be effective and necessary to analyze such processes. However, cell lineages are only currently being identified for both genital and bladder regions. Therefore, conditional mutant approaches based on cell lineages such as Bmp signals in Isl1 cells, or other effector genes in hedgehog responding cells, will be necessary in the future.
Another factor to consider is the potential species-specific differences related to BEEC phenotypes between mice and humans. General remarks on BEEC phenotypes between human and mouse are species dependent morphogenic events. It should be noted that reproductive organ formation, particularly for GT (genital tubercle; embryonic ext. genitalia) formation, displays species variations. Although there are no questions regarding the importance of mouse model analysis, one should also pay attention to potential differences between mouse and human phenotypes. Such differences are partly derived from the anatomical/developmental differences of the two species. The upper (dorsal) part of the ext. genitalia is suggested as formed by species-specific developmental processes, such as the formation of penile bones, mesenchymal and prepucial differentiation. Hence, the extent of phenotypic similarity one can identify with respect to GT formation including its abnormalities, such as epispadias, requires attention. Fundamental developmental pathways for upper GT formation could be conserved between human and mice. Some pelvic organs, pelvic urethral curvature and pelvic muscles also show species dependent changes.
Regarding such “fundamental” developmental processes, BEEC phenotypes should be regarded not as just simple simultaneous bladder and genital region abnormalities but they may offer good models to understand organogenesis. Organs should develop in coordination. Thus, it is expected that multiple phenotypes may arise during such processes. The broader spectrum of BEEC and research on these embryologic mechanisms may include the caudal end of phenotypes including the perineum plus anterior portion of body wall, and midline abnormalities including bladder regions. Continuous efforts to analyze mouse mutant series in addition to cell lineage analysis will gradually provide insights into understanding these intriguing phenotypes.
3. COPY NUMBER VARIATIONS (CNVs)
Since the BEEC represents a rare and severe birth defect, with life-long impairments including reduced reproduction rate, one can assume a significant fraction of de novo events among these patients. The experience in medical genetics suggests that the mutational events will comprise genomic alterations of different size ranging from small changes affecting single nucleotides to large alterations resulting in losses or gains of several thousand to millions of base pairs.
In accordance with this hypothesis, several earlier studies using conventional karyotyping report larger structural chromosomal aberrations in BEEC [35]. In 2004, Thauvin-Robinet and colleagues reported an infant with CE carrying a relatively large 9q34.1-qter deletion [63]. With the introduction of array based techniques over the past decade additional microaberrations have been found in BEEC patients including patients with severe CE phenotypes as well as the milder CBE phenotype. In 2005, Kosaki et al. reported the first smaller deletion in a CE patient comprising chromosomal region 3q12.2-q13.2 spanning 13Mb [64]. While this aberration was still visible on conventional karyotyping, in 2010 El-Hattab et al. reported another, much smaller deletion (2.4Mb) in a CE patient comprising chromosomal region 1p36.33 [65]. This aberration was not visible on conventional karyotyping and the deleted region harbored approximately 70 RefSeq genes. Among these genes, the authors discussed NOC2L, DVL1, the human homolog of the Drosophila dishevelled gene (dsh), encoding a cytoplasmic phosphoprotein that functions as a mediator of the WNT signaling pathway [66, 67], and MMP23B as possible candidate genes. Despite these reports, no single chromosome abnormality has been shown to be associated with CE.
Independently, in 2010, Draaken et al. [68] and Lundin et al. [69] identified two unrelated CBE patients carrying a 22q11.2 microduplication using array-based analysis. Subsequently, Draaken et al. [70], using multiplex ligation-dependent probe amplification and microarray-based analyses identified four additional patients with 22q11.21 duplications in 244 unrelated BEEC patients, including 217 CBE patients. The identified duplications were of variable size. Pooling data of all three studies (eight duplications in 313 CBE patients) yielded a combined odds ratio of 31.86 (95% confidence interval, 4.24-1407.97). Array-based sequence capture and high-throughput targeted re-sequencing established, that all breakpoints resided within the low-copy repeats 22A to 22D. Comparison of the eight duplications revealed a 414 kb phenocritical region, harboring 12 validated RefSeq genes. Characterization of these 12 candidate genes through whole-mount in situ hybridization (WISH) of mouse embryos at embryonic day 9.5 and 10.5 suggested CRKL, THAP7, and LZTR1 as CBE candidate genes. A ninth CBE patient carrying a 22q11.21 duplication was reported by Pierquin and Uwineza [71].
In 2011, Vlangos et al. reported their copy number variation (CNV) study in a cohort of 13 CE patients using two different platforms, a 250K SNP array and a 244K Oligonucleotide array [72]. The oligonucleotide array showed increased sensitivity over the SNP array; however, the majority of CNVs that were detected were observed multiple times in the Database of Genome Variants (http://dgv.tcag.ca/ dgv/app/home). Other CNVs did not contain any known genes or were determined to be inherited from an unaffected parent. The authors concluded from this study that CE is unlikely to be caused by a single recurrent genomic CNV. In the same study, 14 candidate genes were analyzed based on their known or predicted role in caudal development in humans or mice [72]. Ten novel variants were found in seven genes, six of which were within introns and did not uncover a cryptic splice site. The remaining variants included a missense variant in the IHH gene and an 18bp duplication in the FGF8 gene, both of which were carried by an unaffected parent. Two variants were located in the 5’ untranslated region of the WNT3 gene, and the only available parent for analysis did not carry the variant. Based on these data, the authors concluded that none of the variants that were identified provided a likely genetic explanation for the CE phenotype in these patients. However, it could not be ruled out that some of the rare variants could be associated with the CE phenotype and have reduced penetrance in the parents.
Further array-based molecular karyotyping by Draaken et al. [73] in an additional 110 individuals with BEEC identified one additional de novo 0.9 Mb 19p13.12 microduplication, involving 20 validated RefSeq genes. WISH of the corresponding mouse orthologous genes on embryos at gestational days 9.5 and 10.5, with particular emphasis on the ventrolateral trunk, showed that only transcripts for orthologs of human CASP14, AKAP8, SYDE1, BRD4, CYP4F22, and WIZ were found to be specifically expressed in the hindgut and/or caudal region. Of these, WIZ was prioritized over the other genes, showing expression in the developing genital region. However, subsequent Sanger sequencing of WIZ in the complete BEEC cohort did not reveal any pathogenic alterations affecting the coding region of WIZ.
In 2014, Söderhäll et al. [74] reported on a case with CBE and an unbalanced X chromosome rearrangement that was further characterized with array-based CGH analysis. The X chromosome rearrangement consisted of a gain of chromosomal material in region Xq26.3->qter and loss in region Xp22.12->pter. This aberration was also carried by the patients’ mother and sister, neither with CBE. All three have disproportionate short stature, as expected due to the deletion of one of the copies of the SHOX gene. X-inactivation studies revealed a complete skewed inactivation pattern in carriers. Crossover events in the maternal germline furthermore resulted in different genetic material on the rearranged X chromosome between the index patient and her sister suggesting an X-linked genetic risk factor for CBE. Most recently, Jorgez et al. [75] reported on a 10-year-old boy with CBE carrying a 66 kb deletion, which deleted only one copy of OTX1. The boy was identified in a cohort of 30 BEEC patients using NimbleGen 36720 aCGH. None of the remaining 29 BEEC subjects or the 85 controls without GU defects presented with CNVs in OTX1.
4. IDENTIFICATION OF SINGLE GENE MUTATIONS USING WHOLE-EXOME SEQUENCING (WES)
The previous findings from CNV analyses provide a proof of concept that rare de novo mutations contribute to the formation of the BEEC in humans. To identify smaller de novo dominant mutations/small CNVs, the use of next-generation sequencing, specifically whole-exome sequencing (WES) seems plausible. Yet, to the best of our knowledge, no study has been published to date on WES in the elucidation of the BEEC. Here, we present our first approach to the discovery of BEEC candidate genes using WES. Besides the aim to identify smaller de novo dominant mutations, we also sought to identify inherited autosomal-recessive mutations.
WES was performed in eight patients with CE, representing the most severe form of the BEEC spectrum. The CE phenotype was chosen, since to our knowledge, no report of adult patients with CE and biological children has been published [35]. This is probably due to the severe malformation of the external and internal reproductive system as well as the co-occurring defects (imperforate anus, spinal defects, renal anomalies). The lack of reproduction in CE patients is suggestive of an underlying dominant de novo event or rare autosomal-recessive disease causing genes. Prior to exome sequencing, all patients underwent CNV analysis to exclude the presence of larger causative CNVs [68, 73]. All eight case-parent trios were analyzed by WES as outlined elsewhere [76]. All variants detected with a frequency >1% in our in-house data were not further followed. We first searched for heterozygous de novo variants and identified five, yet unknown, non-synonymous amino acid substitutions in three patients (Table 1). These variants were confirmed by Sanger sequencing and were all absent in the respective parents. Several publicly available prediction programs consistently characterized three of these variants in PRPF38A, PRPF8 and SLC20A1 as disease causing (Table 1). While Slc20a1 is expressed in the mouse urinary tract system (UTS) at embryonic Theiler Stage 22 implicating a developmental role in mouse UTS, expression was regionally restricted. Further, extensive data mining in human, mouse and zebrafish databases provided no evidence for the involvement of PRPF38A or PRPF8 in BEEC etiology. Moreover, the p.Arg1681Trp variant identified in PRPF8, encoding the pre-mRNA-processing-splicing factor 8, is likely to be a neutral variant, since mutations in this gene have been associated with autosomal dominant retinitis pigmentosa type 13, a feature not observed in our CE patient. Hence, here only the p.Gly237Arg substitution in SLC20A1 remains the most promising pathogenic variant found.
Table 1.
Heterozygous de novo and compound heterozygote variants detected by WES in a total of eight CE patients
| Patient ID | Complete Phenotype of the CE Patients | Chromosomal Location | Gene | Gene Function (Associated Disease) | cDNA Position, Substitution and Consequence | Known SNP (dbSNP142), Minor Allele Frequency (MAF) | Predicted Consequence1 | Transmission | |
|---|---|---|---|---|---|---|---|---|---|
| Heterozygous de novo variants | |||||||||
| 19 | bladder exstrophy, anal atresia, inguinal hernia, abnormal penis, enlargement of the pubic symphysis | 1p32.2 | PRPF38A | pre-mRNA-processing factor 38A | c.514G>A, p.Glu72Lys | - | D,D,D,D | - | |
| 341 | omphalocele, tethered cord, Arnold-Chiari malformation, ventricular septal defect, permanent foramen ovale, os sacrum dysplasia, bipartite scrotum, inguinal hernia, epilepsy | 14q21.2 | MIA2 | melanoma inhibitory activity protein 2 | c.896A>C, p.Glu299Ala | - | B,B,D,- | - | |
| 17p13.3 | PRPF8 | pre-mRNA-processing-splicing factor 8 (AD2 retinitis pigmentosa type 13) | c.5041C>T, p.Arg1681Trp | - | D,D,D,- | - | |||
| 19q13.33 | NUCB1 | nucleobindin 1 | c.281A>G, p.His94Arg | - | D,B,D,B | - | |||
| 407 | omphalocele, anal atresia, tethered cord, lipoma, atrial septal defect, duplex kidney (right), sacrum dysgenesis | 2q13 | SLC20A1 | sodium-dependent phosphate transporter 1 | c.709G>A, p.Gly237Arg | - | D,D,D,- | - | |
| Compound heterozygous variants | |||||||||
| 19 | see above | 14q23.3 | SPTB | erythrocytic spectrin beta-1 (AD2 spherocytosis type 2; AR3 forms of elliptocytosis 3 and anemia, neonatal hemolytic, fatal and near-fatal) | c.2480G>A, p.Arg827Glnc.3282G>A, p.Glu1095Lys | rs150013838 (MAF: 0.051)rs201653621 (MAF: 0.046) | B,B,B,-D,B,D,- | paternalmaternal | |
| 131 | cloacal exstrophy, lipomeningocele (lumbosacral) with tethered cord, uterus duplex, os sacrum dysplasia, hip dysplasia, malformed feet | 4p16.1 | WFS1 | wolframin-1 (AD2 forms of Wolfram syndrome type 1, Wolfram-like syndrome and deafness) | c.41A>G, p.Gln14Argc.2452C>T, p.Arg818Cys | rs142651446 (MAF: 0.00184)rs35932623 (MAF: 0.0042) | B,B,B,BD,D,D,D4 | maternalpaternal | |
| 406 | bladder exstrophy, omphalocele, anal atresia without fistula, tethered cord, bilateral hydronephrosis, megaureter, uterus duplex, vaginal septum | 3p21.31 | CELSR3 | cadherin, EGF.like, laminin G-like, seven pass G-type receptor 3 | c.5470G>A, p.Val1824Metc.6950T>C, p.Met2317Tyr | rs141484230 (MAF: 0.00079)rs200387155 (MAF ?) | B,D,B,DD,D,B,D | maternalpaternal | |
| 407 | see above | 9q34.13 | SETX | probable helicase senataxin (AR3 spino-cerebellar ataxia-1; AD2 amyotrophic lateral sclerosis 4, juvenile) | c.472T>G, p.Leu158Valc.3287A>G, p.His1096Arg | rs145438764 (MAF: 0.0016)- | B,B,D,DB,B,B,B | maternalpaternal | |
1prediction from publically available programs in the order Mutation Taster, MutPred, PolyPhen-2 and SIFT: B, benign or polymorphism; D, disease causing or probably damaging;
2AD, autosomal dominant
3AR, autosomal recessive
4benign according to the NCBI ClinVar database
We then focused on rare recessive, homozygous or compound heterozygous variants in the patients. Whereas none of the patients were found to be homozygous for any of the variants, we detected compound heterozygosity in four genes in different patients (Table 1). Sanger sequencing confirmed all these variants and confirmed maternal and paternal transmittance for each of the two variants. For all four genes, at least one variant was predicted to be benign. In three patients, compound heterozygosity was found in the genes encoding wolframin-1, senataxin and erythrocytic spectrin beta-I, respectively. Mutations in these genes are found to be associated either with different forms of autosomal dominant Wolfram syndrome, autosomal recessive spinocerebellar ataxia-1/autosomal dominant juvenile amyotrophic lateral sclerosis type 4, or manifesting clinically as types of hemolytic anemia, features not observed in these isolated CE patients. Hence, here only the variants in the cadherin receptor gene CELSR3 seem to remain as candidates for disease association. This gene is involved in planar cell polarity signaling, organizing multicilia in individual cells (single-cell polarity) [77].
Early somatic mutations in mesenchymal bladder precursors, as well as epigenetic events in relevant tissues might also contribute to the disease. Moreover, our previous twin study provided evidence for environmental effects on disease formation [34]. Altogether, one may expect a model of gene-gene and/or gene-environment (or -epigenetic) interaction, underlying BEEC etiology.
Further investigation of both, SLC20A1 and CELSR3 is warranted, including screening of larger CE patient cohorts in order to identify additional mutation carrying patients as well as functional murine and zebrafish studies.
5. ANALYSIS OF CANDIDATE GENES AND GENOME WIDE ASSOCIATION STUDIES (GWAS) IN THE CONTEXT OF MULTIFACTORIAL PHENOTYPES
As outlined above, the general consensus in the field is that, in the majority of BEEC patients, the genetic basis appears to be multifactorial [18]. To account for this multifactorial genetic basis, candidate gene association studies and more recently GWAS provided the basis to investigate the contribution of candidate genes or regions to the formation of multifactorial phenotypes. For the BEEC, only a few candidate gene association studies have been performed to date [78-80] with the most important being the one conducted by Wilkins et al. [80]. Based on their previous observations, an inactivation of the N-terminal truncated isoform of p63, deltaNp63, in mouse embryos, caused the formation of a CBE phenotype in a subset of litters [57]. Wilkins et al. investigated the contribution of TP63 to the formation of human BEEC [80]. The authors hypothesized that TP63 is involved in human BEEC pathogenesis. Re-sequencing of the deltaNp63 promoter in 163 BEEC patients and 285 ethnicity-matched controls showed seven single nucleotide polymorphisms and four insertion/deletion (indel) polymorphisms. These indel polymorphisms were associated with an increased risk of BEEC. Significantly the sites of indel polymorphisms differed between Caucasian and non-Caucasian populations. A 12-base-pair deletion was associated with an increased risk only in Caucasian patients whereas a 4-base-pair insertion was only associated with BEEC in non-Caucasian patients. This association however, was inconsistent over the different exstrophy populations (Asian, Australian, Canadian) tested. In an attempt to replicate this finding, Qi et al. [79] performed a candidate gene association study of TP63 in 154 Caucasian BEEC patients and their unaffected parents. Using 109 selected tagging SNPs localized within TP63 with a minor allele frequency >0.01, they found nominally significant associations between BEEC and six SNPs and four haplotype blocks including or near these significant SNPs; however, none of these results remained significant after correction for multiple testing. Moreover, Darling et al. [81] did not observe any TP63 promoter mutation in 112 BEEC patients. On the other hand, we detected that PERP, a tetraspan protein and a direct TP63 downstream target, was 22.2-fold overexpressed in human bladder sets [82]. PERP specifically localizes to the desmosomes, where it extends through the plasma membrane to link neighboring cells and whole-mount in situ hybridization studies have demonstrated prominent PERP expression in bladder [83].
Our GWAS (see below) however, detected no evidence for an involvement of TP63, PERP or any other desmosomal gene at a genome-wide level [84]. Hence, the desmosomal and cytoskeletal dysregulation observed in postnatal tissue [82] may not reflect the early embryonic situation. Otherwise, the dysregulation of various TP63 isoforms in newborn exstrophy bladder tissue might be due to defects in factors regulating alternative TP63 expression and splicing. Hence, supporting evidence for a contribution of TP63 and desmosomal genes in the etiology of BEEC needs further investigation.
The first GWAS, comprising 218 cases and 865 controls and 78 trios in total, all of European descent identified suggestive evidence for association with CBE across the authors` discovery and follow-up samples. Interestingly, WNT3 and Wnt9b have been both associated with urogenital anomalies in humans and mice respectively [85, 86]. The most associated SNP within this region resides 4 kb next to the WNT3 promoter (http://promoter.cdb.riken.jp/), a region highly conserved among amniotes. Of note, variants in the WNT3 5’UTR were also identified in the CE cohort reported by Vlangos et al. [72]. Nakamura et al. identified several potential transcription factor-binding motifs to exist within the WNT3 promoter region [87], several of which were previously found to be differentially expressed in human newborn bladder exstrophy tissue and known to be important for promotion of the embryonic urorectal septation process [82, 88]. Finally, this region has been shown to contain regulatory elements which regulate Wnt signaling via p63 [89]. Thus, it is tempting to speculate that there are regulatory domains within this intergenic region that are able to modulate Wnt signaling via a conserved WNT3-WNT9B-p63 regulatory module in the context of urorectal and urogenital development.
In our second GWAS we analyzed 110 CBE patients and 1,177 controls of European origin. Here, an association was found with a region of approximately 220kb on chromosome 5q11.1. This region harbors the human ISL1 (ISL LIM homeobox 1) gene. Multiple markers in this region showed evidence for association with CBE, including 84 markers with genome-wide significance [84]. We then performed a meta-analysis using data from our first GWAS (see above). This meta-analysis also implicated the 5q11.1 locus in CBE risk. A total of 138 markers at this locus reached genome-wide significance in the meta-analysis, and the most significant marker (rs9291768) achieved a P value of 2.13 x 10-12. Murine expression analyses detected Isl1 expression in the genital region within the critical time frame for human CBE development. Genital regions with Isl1 expression included the peri-cloacal mesenchyme and the urorectal septum.
Hence, our second GWAS identified the first genome-wide significant locus for CBE at chromosomal region 5q11.1, and provided strong evidence for the hypothesis that ISL1 is the responsible candidate gene in this region.
6. CONCLUSION
The systematic and comprehensive application of modern array based and next generation sequencing techniques in large BEEC cohorts has started to identify putative disease causing genes and regions in the human genome for both Mendelian and multifactorial BEEC phenotypes. These studies will ultimately provide new diagnostic possibilities, and allow precise estimation of recurrence risk in affected families. Parallel functional analysis of the respective embryonic pathways provides a more and more profound understanding of the molecular biological mechanisms underlying the normal and disturbed embryology of the human urogenital system. Furthermore, understanding the respective embryonic pathways will subsequently help to explain related urorectal and genitourinary malformations, such as anorectal malformations, vesicoureteral reflux, ectopic pelvic kidney, renal agenesis, and cryptorchidism.
CONFLICT OF INTEREST
The author(s) confirm that this article content has no conflict of interest.
ACKNOWLEDGEMENTS
This work was supported by the grant RE 1723/1-1, TH 1327/1-1 from the German Research Foundation (Deutsche Forschungsgemeinschaft, DFG). K.K.-N. is supported by the Intramural Research Program of the National Human Genome Research Institute. All authors (H.R., K.K.-N., C.K., H.T., G.Y., and M.L.) wrote this manuscript. The ideas and opinions expressed in this paper are those of the authors only and do not necessarily represent any position or policy of the National Institutes of Health or any other institution organization to which any of the authors are affiliated.
REFERENCES
- 1.Escobar L.F., Weaver D.D., Bixler D., Hodes M.E., Mitchell M. Urorectal septum malformation sequence. Report of six cases and embryological analysis. Am. J. Dis. Child. 1987;141(9):1021–1024. doi: 10.1001/archpedi.1987.04460090098038. [DOI] [PubMed] [Google Scholar]
- 2.Wheeler P.G., Weaver D.D., Obeime M.O., Vance G.H., Bull M.J., Escobar L.F. Urorectal septum malformation sequence: report of thirteen additional cases and review of the literature. Am. J. Med. Genet. 1997;73(4):456–462. doi: 10.1002/(SICI)1096-8628(19971231)73:4<456::AID-AJMG15>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 3.Carey J.C. Exstrophy of the cloaca and the OEIS complex: one and the same. Am. J. Med. Genet. 2001;99(4):270. doi: 10.1002/ajmg.1211. [DOI] [PubMed] [Google Scholar]
- 4.Carey J.C., Greenbaum B., Hall B.D. The OEIS complex (omphalocele, exstrophy, imperforate anus, spinal defects). Birth Defects Orig. Artic. Ser. 1978;14(6B):253–263. [PubMed] [Google Scholar]
- 5.Mildenberger H., Kluth D., Dziuba M. Embryology of bladder exstrophy. J. Pediatr. Surg. 1988;23(2):166–170. doi: 10.1016/S0022-3468(88)80150-7. [DOI] [PubMed] [Google Scholar]
- 6.Gearhart J.P. Exstrophy, epispadias, and other bladder anomalies. In: Walsh P.C., Retik A.B., Vaughan E.D., Wein A.J., editors. Campbell's Urology. 8th ed. Philadelphia: WB Saunders Co; 2002. pp. 2136–2196. [Google Scholar]
- 7.Keppler-Noreuil K.M. OEIS complex (omphalocele-exstrophy-imperforate anus-spinal defects): a review of 14 cases. Am. J. Med. Genet. 2001;99(4):271–279. doi: 10.1002/1096-8628(2001)9999:9999<00::AID-AJMG1094>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 8.Martínez-Frías M.L., Bermejo E., Rodríguez-Pinilla E., Frías J.L. Exstrophy of the cloaca and exstrophy of the bladder: two different expressions of a primary developmental field defect. Am. J. Med. Genet. 2001;99(4):261–269. doi: 10.1002/ajmg.1210. [DOI] [PubMed] [Google Scholar]
- 9.van der Putte S.C., Spliet W.G., Nikkels P.G. Common (“classical”) and covered cloacal exstrophy: a histopathological study and a reconstruction of the pathogenesis. Pediatr. Dev. Pathol. 2008;11(6):430–442. doi: 10.2350/07-06-0292.1. [DOI] [PubMed] [Google Scholar]
- 10.Ebert A.K., Reutter H., Ludwig M., Rösch W.H. The exstrophy-epispadias complex. Orphanet J. Rare Dis. 2009;4(23):23. doi: 10.1186/1750-1172-4-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inouye B.M., Tourchi A., Di Carlo H.N., Young E.E., Mhlanga J., Ko J.S., Sponseller P.D., Gearhart J.P. Safety and efficacy of staged pelvic osteotomies in the modern treatment of cloacal exstrophy. J. Pediatr. Urol. 2014;10(6):1244–1248. doi: 10.1016/j.jpurol.2014.06.018. [DOI] [PubMed] [Google Scholar]
- 12.Shah B.B., Di Carlo H., Goldstein S.D., Pierorazio P.M., Inouye B.M., Massanyi E.Z., Kern A., Koshy J., Sponseller P., Gearhart J.P. Initial bladder closure of the cloacal exstrophy complex: outcome related risk factors and keys to success. J. Pediatr. Surg. 2014;49(6):1036–1039. doi: 10.1016/j.jpedsurg.2014.01.047. [DOI] [PubMed] [Google Scholar]
- 13.Ebert A.K., Bals-Pratsch M., Seifert B., Reutter H., Rösch W.H. Genital and reproductive function in males after functional reconstruction of the exstrophy-epispadias complex--long-term results. Urology. 2008;72(3):566–569. doi: 10.1016/j.urology.2007.11.166. [DOI] [PubMed] [Google Scholar]
- 14.Ebert A.K., Falkert A., Hofstädter A., Reutter H., Rösch W.H. Pregnancy management in women within the bladder-exstrophy-epispadias complex (BEEC) after continent urinary diversion. Arch. Gynecol. Obstet. 2011;284(4):1043–1046. doi: 10.1007/s00404-011-1945-3. [DOI] [PubMed] [Google Scholar]
- 15.Reutter H., Lee C., Grässer M.F., Noeker M. [Subjective developmental outcome in bladder exstrophy and epispadias. A pilot study]. Urologe A. 2005;44(1):57–63. doi: 10.1007/s00120-004-0739-7. [Subjective developmental outcome in bladder exstrophy and epispadias. A pilot study]. [DOI] [PubMed] [Google Scholar]
- 16.Lee C., Reutter H.M., Grässer M.F., Fisch M., Noeker M. Gender-associated differences in the psychosocial and developmental outcome in patients affected with the bladder exstrophy-epispadias complex. BJU Int. 2006;97(2):349–353. doi: 10.1111/j.1464-410X.2005.05910.x. [DOI] [PubMed] [Google Scholar]
- 17.Anonymous Epidemiology of bladder exstrophy and epispadias: a communication from the International Clearinghouse for Birth Defects Monitoring Systems. Teratology. 1987;36(2):221–227. doi: 10.1002/tera.1420360210. [DOI] [PubMed] [Google Scholar]
- 18.Boyadjiev S.A., Dodson J.L., Radford C.L., Ashrafi G.H., Beaty T.H., Mathews R.I., Broman K.W., Gearhart J.P. Clinical and molecular characterization of the bladder exstrophy-epispadias complex: analysis of 232 families. BJU Int. 2004;94(9):1337–1343. doi: 10.1111/j.1464-410X.2004.05170.x. [DOI] [PubMed] [Google Scholar]
- 19.Gambhir L., Höller T., Müller M., Schott G., Vogt H., Detlefsen B., Ebert A.K., Fisch M., Beaudoin S., Stein R., Boyadjiev S.A., Gearhart J.P., Rösch W., Utsch B., Boemers T.M., Reutter H., Ludwig M. Epidemiological survey of 214 families with bladder exstrophy-epispadias complex. J. Urol. 2008;179(4):1539–1543. doi: 10.1016/j.juro.2007.11.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lloyd J.C., Wiener J.S., Gargollo P.C., Inman B.A., Ross S.S., Routh J.C. Contemporary epidemiological trends in complex congenital genitourinary anomalies. J. Urol. 2013;190(4) Suppl.:1590–1595. doi: 10.1016/j.juro.2013.04.034. [DOI] [PubMed] [Google Scholar]
- 21.Wiesel A., Queisser-Luft A., Clementi M., Bianca S., Stoll C., EUROSCAN Study Group Prenatal detection of congenital renal malformations by fetal ultrasonographic examination: an analysis of 709,030 births in 12 European countries. Eur. J. Med. Genet. 2005;48(2):131–144. doi: 10.1016/j.ejmg.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 22.Hayden P.W., Chapman W.H., Stevenson J.K. Exstrophy of the cloaca. Am. J. Dis. Child. 1973;125(6):879–883. doi: 10.1001/archpedi.1973.04160060077018. [DOI] [PubMed] [Google Scholar]
- 23.Petersen E.M., Nelson M.M. Three cases of exstrophy of the cloaca. Am. J. Med. Genet. 1982;11(4):483. doi: 10.1002/ajmg.1320110415. [DOI] [PubMed] [Google Scholar]
- 24.Evans J.A., Darvill K.D., Trevenen C., Rockman-Greenberg C. Cloacal exstrophy and related abdominal wall defects in Manitoba: incidence and demographic factors. Clin. Genet. 1985;27(3):241–251. doi: 10.1111/j.1399-0004.1985.tb00215.x. [DOI] [PubMed] [Google Scholar]
- 25.Caton A.R., Bloom A., Druschel C.M., Kirby R.S. Epidemiology of bladder and cloacal exstrophies in New York State, 1983-1999. Birth Defects Res. A Clin. Mol. Teratol. 2007;79(11):781–787. doi: 10.1002/bdra.20402. [DOI] [PubMed] [Google Scholar]
- 26.Feldkamp M.L., Botto L.D., Amar E., Bakker M.K., Bermejo-Sánchez E., Bianca S., Canfield M.A., Castilla E.E., Clementi M., Csaky-Szunyogh M., Leoncini E., Li Z., Lowry R.B., Mastroiacovo P., Merlob P., Morgan M., Mutchinick O.M., Rissmann A., Ritvanen A., Siffel C., Carey J.C. Cloacal exstrophy: an epidemiologic study from the International Clearinghouse for Birth Defects Surveillance and Research. Am. J. Med. Genet. C. Semin. Med. Genet. 2011;157C(4):333–343. doi: 10.1002/ajmg.c.30317. [DOI] [PubMed] [Google Scholar]
- 27.Wood H.M., Trock B.J., Gearhart J.P. In vitro fertilization and the cloacal-bladder exstrophy-epispadias complex: is there an association? J. Urol. 2003;169(4):1512–1515. doi: 10.1097/01.ju.0000054984.76384.66. [DOI] [PubMed] [Google Scholar]
- 28.Wood H.M., Babineau D., Gearhart J.P. In vitro fertilization and the cloacal/bladder exstrophy-epispadias complex: a continuing association. J. Pediatr. Urol. 2007;3(4):305–310. doi: 10.1016/j.jpurol.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 29.Zwink N., Jenetzky E., Hirsch K., Reifferscheid P., Schmiedeke E., Schmidt D., Reckin S., Obermayr F., Boemers T.M., Stein R., Reutter H., Rösch W.H., Brenner H., Ebert A.K. Assisted reproductive techniques and risk of exstrophy-epispadias complex: a German case-control study. J. Urol. 2013;189(4):1524–1529. doi: 10.1016/j.juro.2012.11.108. [DOI] [PubMed] [Google Scholar]
- 30.Reutter H., Boyadjiev S.A., Gambhir L., Ebert A.K., Rösch W.H., Stein R., Schröder A., Boemers T.M., Bartels E., Vogt H., Utsch B., Müller M., Detlefsen B., Zwink N., Rogenhofer S., Gobet R., Beckers G.M., Bökenkamp A., Kajbafzadeh A.M., Jaureguizar E., Draaken M., Lakshmanan Y., Gearhart J.P., Ludwig M., Nöthen M.M., Jenetzky E. Phenotype severity in the bladder exstrophy-epispadias complex: analysis of genetic and nongenetic contributing factors in 441 families from North America and Europe. J. Pediatr. 2011;159(5):825–831.e1. doi: 10.1016/j.jpeds.2011.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reefhuis J., Honein M.A., Schieve L.A., Rasmussen S.A., National Birth Defects Prevention Study Use of clomiphene citrate and birth defects, National Birth Defects Prevention Study, 1997-2005. Hum. Reprod. 2011;26(2):451–457. doi: 10.1093/humrep/deq313. [DOI] [PubMed] [Google Scholar]
- 32.Reutter H., Shapiro E., Gruen J.R. Seven new cases of familial isolated bladder exstrophy and epispadias complex (BEEC) and review of the literature. Am. J. Med. Genet. A. 2003;120A(2):215–221. doi: 10.1002/ajmg.a.20057. [DOI] [PubMed] [Google Scholar]
- 33.Ludwig M., Utsch B., Reutter H. [Genetic and molecular biological aspects of the bladder exstrophy-epispadias complex (BEEC)]. Urologe A. 2005;44(9):1037–1038, 1040-1044. doi: 10.1007/s00120-005-0863-z. [Genetic and molecular biological aspects of the bladder exstrophy-epispadias complex (BEEC)]. [DOI] [PubMed] [Google Scholar]
- 34.Reutter H., Qi L., Gearhart J.P., Boemers T., Ebert A.K., Rösch W., Ludwig M., Boyadjiev S.A. Concordance analyses of twins with bladder exstrophy-epispadias complex suggest genetic etiology. Am. J. Med. Genet. A. 2007;143A(22):2751–2756. doi: 10.1002/ajmg.a.31975. [DOI] [PubMed] [Google Scholar]
- 35.Ludwig M., Ching B., Reutter H., Boyadjiev S.A. Bladder exstrophy-epispadias complex. Birth Defects Res. A Clin. Mol. Teratol. 2009;85(6):509–522. doi: 10.1002/bdra.20557. [DOI] [PubMed] [Google Scholar]
- 36.Shapiro E., Lepor H., Jeffs R.D. The inheritance of the exstrophy-epispadias complex. J. Urol. 1984;132(2):308–310. doi: 10.1016/s0022-5347(17)49605-4. [DOI] [PubMed] [Google Scholar]
- 37.Messelink E.J., Aronson D.C., Knuist M., Heij H.A., Vos A. Four cases of bladder exstrophy in two families. J. Med. Genet. 1994;31(6):490–492. doi: 10.1136/jmg.31.6.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith N.M., Chambers H.M., Furness M.E., Haan E.A. The OEIS complex (omphalocele-exstrophy-imperforate anus-spinal defects): recurrence in sibs. J. Med. Genet. 1992;29(10):730–732. doi: 10.1136/jmg.29.10.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Redman J.F., Seibert J.J., Page B.C. Cloacal exstrophy in identical twins. Urology. 1981;17(1):73–74. doi: 10.1016/0090-4295(81)90017-0. [DOI] [PubMed] [Google Scholar]
- 40.McLaughlin J.F., Marks W.M., Jones G. Prospective management of exstrophy of the cloaca and myelocystocele following prenatal ultrasound recognition of neural tube defects in identical twins. Am. J. Med. Genet. 1984;19(4):721–727. doi: 10.1002/ajmg.1320190412. [DOI] [PubMed] [Google Scholar]
- 41.Lee D.H., Cottrell J.R., Sanders R.C., Meyers C.M., Wulfsberg E.A., Sun C.C. OEIS complex (omphalocele-exstrophy-imperforate anus-spinal defects) in monozygotic twins. Am. J. Med. Genet. 1999;84(1):29–33. doi: 10.1002/(SICI)1096-8628(19990507)84:1<29::AID-AJMG7>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 42.Goldfischer E.R., Almond P.S., Statter M.B., Miller G., Arensman R.M., Cromie W.J. Omphalopagus twins with covered cloacal exstrophy. J. Urol. 1997;157(3):1004–1005. doi: 10.1016/S0022-5347(01)65132-2. [DOI] [PubMed] [Google Scholar]
- 43.Casale P., Grady R.W., Waldhausen J.H., Joyner B.D., Wright J., Mitchell M.E. Cloacal exstrophy variants. Can blighted conjoined twinning play a role? J. Urol. 2004;172(3):1103–1106. doi: 10.1097/01.ju.0000142108.62457.81. [DOI] [PubMed] [Google Scholar]
- 44.Siebert J.R., Rutledge J.C., Kapur R.P. Association of cloacal anomalies, caudal duplication, and twinning. Pediatr. Dev. Pathol. 2005;8(3):339–354. doi: 10.1007/s10024-005-1157-6. [DOI] [PubMed] [Google Scholar]
- 45.Tihtonen K., Lagerstedt A., Kähkönen M., Kirkinen P. Diamniotic omphalopagus conjoined twins in a diamniotic pregnancy. Fetal Diagn. Ther. 2009;25(3):343–345. doi: 10.1159/000235882. [DOI] [PubMed] [Google Scholar]
- 46.Métneki J., Czeizel A. Conjoined twins in Hungary, 1970-1986. Acta Genet. Med. Gemellol. (Roma) 1989;38(3-4):285–299. doi: 10.1017/s0001566000002695. [DOI] [PubMed] [Google Scholar]
- 47.Bruch S.W., Adzick N.S., Goldstein R.B., Harrison M.R. Challenging the embryogenesis of cloacal exstrophy. J. Pediatr. Surg. 1996;31(6):768–770. doi: 10.1016/S0022-3468(96)90128-1. [DOI] [PubMed] [Google Scholar]
- 48.Haraguchi R., Suzuki K., Murakami R., Sakai M., Kamikawa M., Kengaku M., Sekine K., Kawano H., Kato S., Ueno N., Yamada G. Molecular analysis of external genitalia formation: the role of fibroblast growth factor (Fgf) genes during genital tubercle formation. Development. 2000;127(11):2471–2479. doi: 10.1242/dev.127.11.2471. [DOI] [PubMed] [Google Scholar]
- 49.Haraguchi R., Mo R., Hui C., Motoyama J., Makino S., Shiroishi T., Gaffield W., Yamada G. Unique functions of Sonic hedgehog signaling during external genitalia development. Development. 2001;128(21):4241–4250. doi: 10.1242/dev.128.21.4241. [DOI] [PubMed] [Google Scholar]
- 50.Perriton C.L., Powles N., Chiang C., Maconochie M.K., Cohn M.J. Sonic hedgehog signaling from the urethral epithelium controls external genital development. Dev. Biol. 2002;247(1):26–46. doi: 10.1006/dbio.2002.0668. [DOI] [PubMed] [Google Scholar]
- 51.Miyagawa S., Moon A., Haraguchi R., Inoue C., Harada M., Nakahara C., Suzuki K., Matsumaru D., Kaneko T., Matsuo I., Yang L., Taketo M.M., Iguchi T., Evans S.M., Yamada G. Dosage dependent hedgehog signals integrated with 1 Wnt/β-catenin regulate embryonic external genitalia formation as an appendicular program. Development. 2009;136(23):3969–3978. doi: 10.1242/dev.039438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miyagawa S., Harada M., Matsumaru D., Tanaka K., Inoue C., Nakahara C., Haraguchi R., Matsushita S., Suzuki K., Nakagata N., Ng R.C., Akita K., Lui V.C., Yamada G. Disruption of the temporally regulated cloaca endodermal β-catenin signaling causes anorectal malformations. Cell Death Differ. 2014;21(6):990–997. doi: 10.1038/cdd.2014.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matsumaru D., Haraguchi R., Moon A.M., Satoh Y., Nakagata N., Yamamura K., Takahashi N., Kitazawa S., Yamada G. Genetic analysis of the role of Alx4 in the coordination of lower body and external genitalia formation. Eur. J. Hum. Genet. 2014;22(3):350–357. doi: 10.1038/ejhg.2013.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Matsumaru D., Haraguchi R., Miyagawa S., Motoyama J., Nakagata N., Meijlink F., Yamada G. Genetic analysis of Hedgehog signaling in ventral body wall development and the onset of omphalocele formation. PLoS One. 2011;6(1):e16260. doi: 10.1371/journal.pone.0016260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Williams T. Animal models of ventral body wall closure defects: a personal perspective on gastroschisis. Am. J. Med. Genet. C. Semin. Med. Genet. 2008;148C(3):186–191. doi: 10.1002/ajmg.c.30179. [DOI] [PubMed] [Google Scholar]
- 56.Brewer S., Williams T. Loss of AP-2α impacts multiple aspects of ventral body wall development and closure. Dev. Biol. 2004;267(2):399–417. doi: 10.1016/j.ydbio.2003.11.021. [DOI] [PubMed] [Google Scholar]
- 57.Cheng W., Jacobs W.B., Zhang J.J., Moro A., Park J.H., Kushida M., Qiu W., Mills A.A., Kim P.C. DeltaNp63 plays an anti-apoptotic role in ventral bladder development. Development. 2006;133(23):4783–4792. doi: 10.1242/dev.02621. [DOI] [PubMed] [Google Scholar]
- 58.Chari N.S., Romano R.A., Koster M.I., Jaks V., Roop D., Flores E.R., Teglund S., Sinha S., Gruber W., Aberger F., Medeiros L.J., Toftgard R., McDonnell T.J. Interaction between the TP63 and SHH pathways is an important determinant of epidermal homeostasis. Cell Death Differ. 2013;20(8):1080–1088. doi: 10.1038/cdd.2013.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haraguchi R., Motoyama J., Sasaki H., Satoh Y., Miyagawa S., Nakagata N., Moon A., Yamada G. Molecular analysis of coordinated bladder and urogenital organ formation by Hedgehog signaling. Development. 2007;134(3):525–533. doi: 10.1242/dev.02736. [DOI] [PubMed] [Google Scholar]
- 60.Suzuki K., Adachi Y., Numata T., Nakada S., Yanagita M., Nakagata N., Evans S.M., Graf D., Economides A., Haraguchi R., Moon A.M., Yamada G. Reduced BMP signaling results in hindlimb fusion with lethal pelvic/urogenital organ aplasia: a new mouse model of sirenomelia. PLoS One. 2012;7(9):e43453. doi: 10.1371/journal.pone.0043453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haraguchi R., Matsumaru D., Nakagata N., Miyagawa S., Suzuki K., Kitazawa S., Yamada G. The hedgehog signal induced modulation of bone morphogenetic protein signaling: an essential signaling relay for urinary tract morphogenesis. PLoS One. 2012;7(7):e42245. doi: 10.1371/journal.pone.0042245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ogi H., Suzuki K., Ogino Y., Kamimura M., Miyado M., Ying X., Zhang Z., Shinohara M., Chen Y., Yamada G. Ventral abdominal wall dysmorphogenesis of Msx1/Msx2 double-mutant mice. Anat. Rec. A Discov. Mol. Cell. Evol. Biol. 2005;284(1):424–430. doi: 10.1002/ar.a.20180. [DOI] [PubMed] [Google Scholar]
- 63.Thauvin-Robinet C., Faivre L., Cusin V., Khau Van Kien P., Callier P., Parker K.L., Fellous M., Borgnon J., Gounot E., Huet F., Sapin E., Mugneret F. Cloacal exstrophy in an infant with 9q34.1-qter deletion resulting from a de novo unbalanced translocation between chromosome 9q and Yq. Am. J. Med. Genet. A. 2004;126A(3):303–307. doi: 10.1002/ajmg.a.20596. [DOI] [PubMed] [Google Scholar]
- 64.Kosaki R., Fukuhara Y., Kosuga M., Okuyama T., Kawashima N., Honna T., Ueoka K., Kosaki K. OEIS complex with del(3)(q12.2q13.2). Am. J. Med. Genet. A. 2005;135(2):224–226. doi: 10.1002/ajmg.a.30733. [DOI] [PubMed] [Google Scholar]
- 65.El-Hattab A.W., Skorupski J.C., Hsieh M.H., Breman A.M., Patel A., Cheung S.W., Craigen W.J. OEIS complex associated with chromosome 1p36 deletion: a case report and review. Am. J. Med. Genet. A. 2010;152A(2):504–511. doi: 10.1002/ajmg.a.33226. [DOI] [PubMed] [Google Scholar]
- 66.Rosso S.B., Sussman D., Wynshaw-Boris A., Salinas P.C. Wnt signaling through Dishevelled, Rac and JNK regulates dendritic development. Nat. Neurosci. 2005;8(1):34–42. doi: 10.1038/nn1374. [DOI] [PubMed] [Google Scholar]
- 67.Bilic J., Huang Y.L., Davidson G., Zimmermann T., Cruciat C.M., Bienz M., Niehrs C. Wnt induces LRP6 signalosomes and promotes dishevelled-dependent LRP6 phosphorylation. Science. 2007;316(5831):1619–1622. doi: 10.1126/science.1137065. [DOI] [PubMed] [Google Scholar]
- 68.Draaken M., Reutter H., Schramm C., Bartels E., Boemers T.M., Ebert A.K., Rösch W., Schröder A., Stein R., Moebus S., Stienen D., Hoffmann P., Nöthen M.M., Ludwig M. Microduplications at 22q11.21 are associated with non-syndromic classic bladder exstrophy. Eur. J. Med. Genet. 2010;53(2):55–60. doi: 10.1016/j.ejmg.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 69.Lundin J., Söderhäll C., Lundén L., Hammarsjö A., White I., Schoumans J., Läckgren G., Kockum C.C., Nordenskjöld A. 22q11.2 microduplication in two patients with bladder exstrophy and hearing impairment. Eur. J. Med. Genet. 2010;53(2):61–65. doi: 10.1016/j.ejmg.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 70.Draaken M., Baudisch F., Timmermann B., Kuhl H., Kerick M., Proske J., Wittler L., Pennimpede T., Ebert A.K., Rösch W., Stein R., Bartels E., von Lowtzow C., Boemers T.M., Herms S., Gearhart J.P., Lakshmanan Y., Kockum C.C., Holmdahl G., Läckgren G., Nordenskjöld A., Boyadjiev S.A., Herrmann B.G., Nöthen M.M., Ludwig M., Reutter H. Classic bladder exstrophy: Frequent 22q11.21 duplications and definition of a 414 kb phenocritical region. Birth Defects Res. A Clin. Mol. Teratol. 2014;100(6):512–517. doi: 10.1002/bdra.23249. [DOI] [PubMed] [Google Scholar]
- 71.Pierquin G., Uwineza A. 22q11.2 microduplication in a patient with bladder exstrophy and delayed psychomotor development. Eur. J. Hum. Genet. 2012;20(Suppl. 1):89. [abstr]. [Google Scholar]
- 72.Vlangos C.N., Siuniak A., Ackley T., van Bokhoven H., Veltman J., Iyer R., Park J.M., Keppler-Noreuil K., Keegan C.E. Comprehensive genetic analysis of OEIS complex reveals no evidence for a recurrent microdeletion or duplication. Am. J. Med. Genet. A. 2011;155A(1):38–49. doi: 10.1002/ajmg.a.33757. [DOI] [PubMed] [Google Scholar]
- 73.Draaken M., Mughal S.S., Pennimpede T., Wolter S., Wittler L., Ebert A.K., Rösch W., Stein R., Bartels E., Schmidt D., Boemers T.M., Schmiedeke E., Hoffmann P., Moebus S., Herrmann B.G., Nöthen M.M., Reutter H., Ludwig M. Isolated bladder exstrophy associated with a de novo 0.9 Mb microduplication on chromosome 19p13.12. Birth Defects Res. A Clin. Mol. Teratol. 2013;97(3):133–139. doi: 10.1002/bdra.23112. [DOI] [PubMed] [Google Scholar]
- 74.Söderhäll C., Lundin J., Lagerstedt-Robinson K., Grigelioniene G., Lackgren G., Kockum C.C., Nordenskjöld A. A case with bladder exstrophy and unbalanced X chromosome rearrangement. Eur. J. Pediatr. Surg. 2014;24(4):353–359. doi: 10.1055/s-0033-1349056. [DOI] [PubMed] [Google Scholar]
- 75.Jorgez C.J., Rosenfeld J.A., Wilken N.R., Vangapandu H.V., Sahin A., Pham D., Carvalho C.M., Bandholz A., Miller A., Weaver D.D., Burton B., Babu D., Bamforth J.S., Wilks T., Flynn D.P., Roeder E., Patel A., Cheung S.W., Lupski J.R., Lamb D.J. Genitourinary defects associated with genomic deletions in 2p15 encompassing OTX1. PLoS One. 2014;9(9):e107028. doi: 10.1371/journal.pone.0107028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Basmanav F.B., Oprisoreanu A.M., Pasternack S.M., Thiele H., Fritz G., Wenzel J., Größer L., Wehner M., Wolf S., Fagerberg C., Bygum A., Altmüller J., Rütten A., Parmentier L., El Shabrawi-Caelen L., Hafner C., Nürnberg P., Kruse R., Schoch S., Hanneken S., Betz R.C. Mutations in POGLUT1, encoding protein O-glucosyltransferase 1, cause autosomal-dominant Dowling-Degos disease. Am. J. Hum. Genet. 2014;94(1):135–143. doi: 10.1016/j.ajhg.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Boutin C., Labedan P., Dimidschstein J., Richard F., Cremer H., André P., Yang Y., Montcouquiol M., Goffinet A.M., Tissir F. A dual role for planar cell polarity genes in ciliated cells. Proc. Natl. Acad. Sci. USA. 2014;111(30):E3129–E3138. doi: 10.1073/pnas.1404988111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Reutter H., Becker T., Ludwig M., Schäfer N., Detlefsen B., Beaudoin S., Fisch M., Ebert A.K., Rösch W., Nöthen M.M., Boemers T.M., Betz R.C. Family-based association study of the MTHFR polymorphism C677T in the bladder-exstrophy-epispadias-complex. Am. J. Med. Genet. A. 2006;140(22):2506–2509. doi: 10.1002/ajmg.a.31484. [DOI] [PubMed] [Google Scholar]
- 79.Qi L., Wang M., Yagnik G., Mattheisen M., Gearhart J.P., Lakshmanan Y., Ebert A.K., Rösch W., Ludwig M., Draaken M., Reutter H., Boyadjiev S.A. Candidate gene association study implicates p63 in the etiology of nonsyndromic bladder-exstrophy-epispadias complex. Birth Defects Res. A Clin. Mol. Teratol. 2013;97(12):759–763. doi: 10.1002/bdra.23161. [DOI] [PubMed] [Google Scholar]
- 80.Wilkins S., Zhang K.W., Mahfuz I., Quantin R., D’Cruz N., Hutson J., Ee M., Bagli D., Aitken K., Fong F.N., Ng P.K., Tsui S.K., Fung W.Y., Banu T., Thakre A., Johar K., Jaureguizar E., Li L., Cheng W. Insertion/deletion polymorphisms in the ΔNp63 promoter are a risk factor for bladder exstrophy epispadias complex. PLoS Genet. 2012;8(12):e1003070. doi: 10.1371/journal.pgen.1003070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Darling T., Mahfuz I., White S.J., Cheng W. No TAP63 promoter mutation is detected in bladder exstrophy-epispadias complex patients. J. Pediatr. Surg. 2013;48(12):2393–2400. doi: 10.1016/j.jpedsurg.2013.08.012. [DOI] [PubMed] [Google Scholar]
- 82.Qi L., Chen K., Hur D.J., Yagnik G., Lakshmanan Y., Kotch L.E., Ashrafi G.H., Martinez-Murillo F., Kowalski J., Naydenov C., Wittler L., Gearhart J.P., Draaken M., Reutter H., Ludwig M., Boyadjiev S.A. Genome-wide expression profiling of urinary bladder implicates desmosomal and cytoskeletal dysregulation in the bladder exstrophy-epispadias complex. Int. J. Mol. Med. 2011;27(6):755–765. doi: 10.3892/ijmm.2011.654. [DOI] [PubMed] [Google Scholar]
- 83.Ihrie R.A., Marques M.R., Nguyen B.T., Horner J.S., Papazoglu C., Bronson R.T., Mills A.A., Attardi L.D. Perp is a p63-regulated gene essential for epithelial integrity. Cell. 2005;120(6):843–856. doi: 10.1016/j.cell.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 84.Draaken M., Knapp M., Pennimpede T., Schmidt J.M., Ebert A.K., Rösch W., Stein R., Utsch B., Hirsch K., Boemers T.M., Mangold E., Heilmann S., Ludwig K.U., Jenetzky E., Zwink N., Moebus S., Herrmann B.G., Mattheisen M., Nöthen M.M., Ludwig M., Reutter H. Genome-wide association study and meta-analysis identify ISL1 as genome-wide significant susceptibility gene for bladder exstrophy. PLoS Genet. 2015;11(3):e1005024. doi: 10.1371/journal.pgen.1005024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Niemann S., Zhao C., Pascu F., Stahl U., Aulepp U., Niswander L., Weber J.L., Müller U. Homozygous WNT3 mutation causes tetra-amelia in a large consanguineous family. Am. J. Hum. Genet. 2004;74(3):558–563. doi: 10.1086/382196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Carroll T.J., Park J.S., Hayashi S., Majumdar A., McMahon A.P. Wnt9b plays a central role in the regulation of mesenchymal to epithelial transitions underlying organogenesis of the mammalian urogenital system. Dev. Cell. 2005;9(2):283–292. doi: 10.1016/j.devcel.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 87.Nakamura Y., Tsiairis C.D., Özbek S., Holstein T.W. Autoregulatory and repressive inputs localize Hydra Wnt3 to the head organizer. Proc. Natl. Acad. Sci. USA. 2011;108(22):9137–9142. doi: 10.1073/pnas.1018109108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kang S., Graham J.M., Jr, Olney A.H., Biesecker L.G. GLI3 frameshift mutations cause autosomal dominant Pallister-Hall syndrome. Nat. Genet. 1997;15(3):266–268. doi: 10.1038/ng0397-266. [DOI] [PubMed] [Google Scholar]
- 89.Ferretti E., Li B., Zewdu R., Wells V., Hebert J.M., Karner C., Anderson M.J., Williams T., Dixon J., Dixon M.J., Depew M.J., Selleri L. A conserved Pbx-Wnt-p63-Irf6 regulatory module controls face morphogenesis by promoting epithelial apoptosis. Dev. Cell. 2011;21(4):627–641. doi: 10.1016/j.devcel.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]