Abstract
DNA sequences extracted from preserved remains can add considerable resolution to inference of past population dynamics. For example, coalescent-based methods have been used to correlate declines in some arctic megafauna populations with habitat fragmentation during the last ice age. These methods, however, often fail to detect population declines preceding extinction, most likely owing to a combination of sparse sampling, uninformative genetic markers, and models that cannot account for the increasingly structured nature of populations as habitats decline. As ancient DNA research expands to include full-genome analyses, these data will provide greater resolution of the genomic consequences of environmental change and the genetic signatures of extinction.
Keywords: ancient DNA, megafauna, extinction, coalescent, demography
1. Background
DNA extracted from the preserved remains of plants and animals can be used to reconstruct demographic changes over the time during which these changes occurred. Since the first recovery of ancient DNA three decades ago, one focus of ancient DNA research has been to better understand how organisms responded to past periods of environmental change [1,2]. The Late Pleistocene was a period of dramatic changes to climate and habitats worldwide, including the peak of the last ice age around 20 thousand years ago (ka) and subsequent rapid warming, and the expansion of human populations. These events are believed to be responsible for the increased rate of extinction during this period, including the disappearance of most megafauna, mammals weighing greater than 44 kg [3]. Understanding why some species survived while others did not remains an important area of ancient DNA research.
Today, ancient DNA can be isolated from organisms that lived in habitats ranging from the tropics to the Arctic and as long ago as 700 ka [4,5]. Datasets comprising hundreds of individuals have been generated for multiple species, both extinct and extant. Simultaneously, statistical approaches have been developed that incorporate data sampled over time within a coalescent framework to reconstruct demographic history [6,7]. Such approaches can infer, for example, population bottlenecks [8,9] and long-range dispersals [10], and correlate demographic changes with palaeoclimatic events [11]. While in a few instances, ancient DNA data have been able to identify declines [12,13], this requires special circumstances, for example slow declines, small population sizes or dense sampling within the period of extinction. We hypothesize that, for most ancient DNA studies, the failure to detect extinctions is a consequence of both the type of data collected and the limitations of existing statistical models. As the field expands to include palaeogenomics as well as mitochondrial population genetics [14], the capacity of ancient DNA research to detect and therefore better understand extinctions will improve.
2. Early promises of ancient DNA
Many of the earliest ancient DNA studies targeted mitochondrial DNA, which is present in higher copy-number and therefore more likely to be preserved than nuclear DNA. These data were used to test hypotheses about evolutionary relationships between extinct and living species and long-term population stability, providing insights that were not apparent from studying the fossil record. For example, while their fossil records suggested continuity, ancient DNA from brown bears [15], cave bears [16] and caribou [17] revealed local extinctions and replacements, highlighting the dynamic nature of ice age communities. As the size of ancient DNA datasets grew, it became increasingly possible to detect genetic trends that related the changing climate to evolutionary change. Brown bears, for example, were discovered to have lost genetic diversity between 50 and 15 ka, which was interpreted as a consequence of climate changes leading up to the last glacial maximum [2,15].
3. Inferring population history using coalescent-based approaches
As populations change, the signatures of those changes are written into their DNA. Coalescent theory quantifies the demographic history of a population from these genetic signatures [18]. Whereas fossil-based estimates of changes in abundance rely on consistent rates of bone preservation through time, coalescent-based estimates should be less affected by taphonomic biases and therefore provide more robust estimates of changes in population size. Several approaches have been developed that simulate the coalescent process within a Markov chain Monte Carlo framework with time-structured data to infer one or several demographic parameters simultaneously. For example, the software beast [7] can be run with a flexible coalescent model [19] to produce plots of effective population size through time, from which key periods of demographic change can be identified for further analysis. In addition, rejection sampling approaches such as approximate Bayesian computation [20] with serial coalescent simulations [6] can be used to determine specific demographic parameters of interest, such as the timing of population bottlenecks [8] and changes in population structure through time [12].
One of the main findings from coalescent simulations using the skyline family of approaches [21] has been that population sizes are positively correlated with the amount of habitat available [11]. For example, palaeoenvironmental reconstructions showed a rapid decline in bison habitat 30–21 ka, around the same time that coalescent reconstructions of bison demographic history revealed a precipitous decline in mitochondrial diversity [11] (figure 1a). This study found no evidence of population size change for the two extinct species analysed: mammoths and woolly rhinos (figure 1b,c). In fact, only two skyline reconstructions to date have reproduced a decline in population size preceding extinction: cave bears [13], and a more recent analysis of a larger dataset of mammoths [12]. In these analyses, mammoths declined rapidly 25–10 ka, after which populations remained small (figure 1d) and cave bears declined slowly to extinction beginning approximately 50 ka (figure 1e). Interestingly, only the latter of these was able to detect the final decline to extinction.
Figure 1.
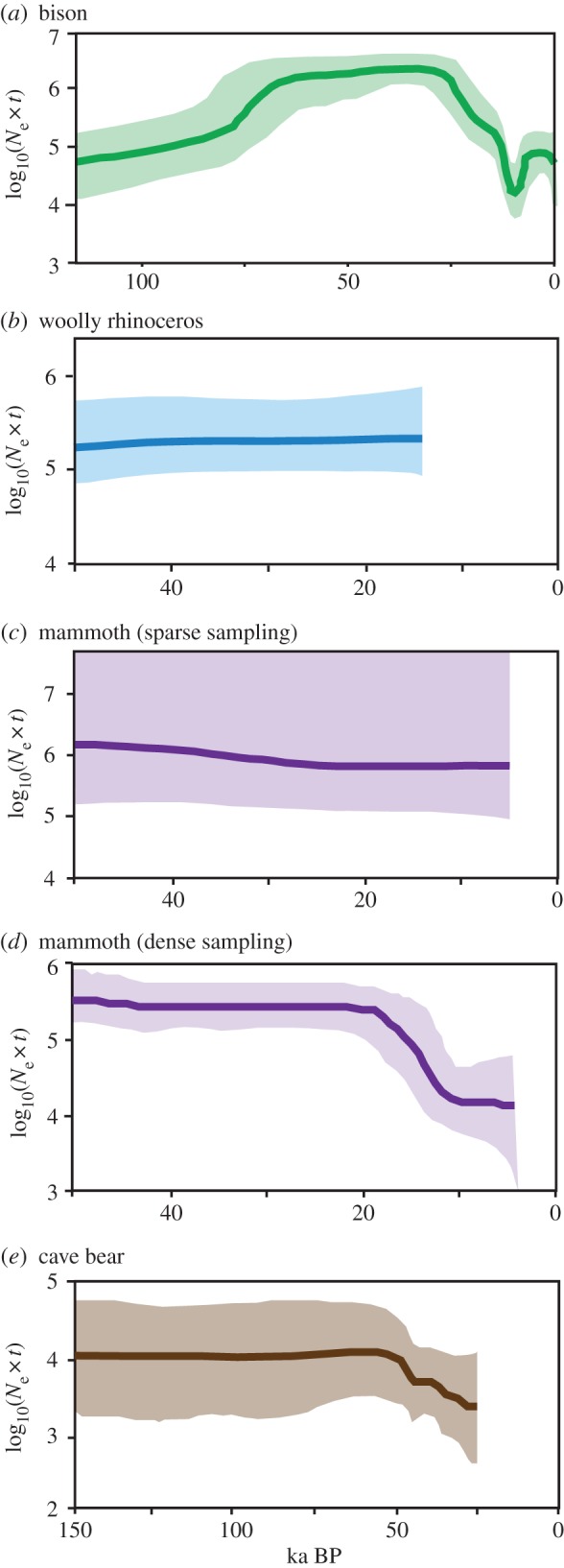
The variable power of skyline plots to reveal past population size changes and extinctions. Five datasets are used, modified from the original publications. In each plot, the x axis is thousands of years before present, and the y axis reflects the mean (solid line) and 95% highest posterior density estimate of the product of the effective population size (Ne) and the generation length in years (t). (a) A large dataset of bison mitochondrial DNA sequences reveals population decline followed by recovery [11,19], highlighting the power of skyline approaches to reveal past demography. Smaller datasets, such as (b) woolly rhinos [11] and (c) mammoths [11], have less power to detect population size change. (d) A more extensive dataset of mammoth mitochondrial DNA sequences later recovered population decline, but the final extinction event is not observed [12]. (e) A dataset of cave bears reveals a slow and steady decline preceding extinction; Bayes factor tests support the flexible model over a constant population size [13]. (Online version in colour.)
Several hypotheses may explain why extinction is difficult to detect using a coalescent approach. First, datasets may be too small or too sparsely sampled. The mammoth dataset used for figure 1c was sparsely sampled across a broad geographical range, and uninformative with respect to demography. However, increased sampling provided sufficient information to infer periods of demographic change (figure 1d). The mode of extinction is also important with respect to sampling. In bison, the genetic signature of the bottleneck approximately 13 ka is preserved in individuals postdating the bottleneck. To detect extinction, and in particular rapid extinction, the decline must be captured as it happens, and the last remaining individuals must be sampled, which may be impossible. In fact, detecting extinction using coalescent-based approaches may require special circumstances, for example slow declines, thorough sampling, and high levels of gene flow throughout decline. This combination of effects may explain why the final decline is detected in cave bears (figure 1e).
Population structure will also affect the capacity of coalescent-based approaches to detect extinction. For several arctic species, the average genetic distance between individuals increased as populations declined [11], probably reflecting increasing population fragmentation. If all subpopulations decline simultaneously in the absence of gene flow, coalescent inference that assumes no structure will show little or no evidence of decline.
Finally, choice of genetic locus will affect the detection of extinction. Although it has been the most common locus used in ancient DNA studies, mitochondrial DNA may be a poor candidate for demographic inference in some species. The decline in mammoths (figure 1d) corresponds to the loss of two of three mitochondrial lineages and not to their eventual extinction [12]. As mitochondria are a single genetic locus and mitochondrial diversity is not necessarily indicative of population structure [22], the dynamics of mitochondria may not reflect the species' population history.
4. The future
Our capacity to infer demographic history accurately has improved considerably in recent years. Lower sequencing costs and technical improvements in ancient DNA extraction are increasing both the size and geographical range of datasets, and techniques such as hybridization capture [23] are facilitating the use of multiple loci. As these datasets grow, it will become increasingly important to account for geographical structure, and several approaches have been developed to this end. Within the beast framework, for example, it is possible to explore the phylogeographic history of the samples explicitly as part of demographic reconstruction [24]. This approach has been used to show that a group of maternally related brown bears dispersed out of Alaska approximately 30 ka and contributed mitochondrial DNA to populations across Asia and Europe by approximately 14 ka [10]. While these approaches are potentially powerful, they, like other methods, are limited by available data. In the specific case of bears, the strong mitochondrial population structure is not observed in nuclear data [25], probably owing to male-mediated gene flow. Nonetheless, the relatively fast rate of mitochondrial compared with nuclear genome evolution makes mitochondrial DNA useful for exploring recent demographic change.
Nuclear genomes contain millions to billions of independently inherited loci and, as such, have increased power to infer demographic history compared with mitochondrial DNA. As complete genomes are sequenced from extinct species, approaches such as pairwise sequentially Markovian coalescent (PSMC) modelling [26] have been used to reconstruct long-term demographic histories of ancient horses [27] and mammoths [22], and to speculate about palaeoclimatic events that may have instigated population growth or decline. Unfortunately, PSMC has little power to detect extinction, because the number of recent coalescent events retained in a single genome is small [26]. Other approaches, however, can be used to recover signatures of extinction from full-genome data. For example, a comparison of heterozygosity between one of the last surviving Wrangel Island mammoths and a 45 ka mammoth from mainland Siberia found evidence of increased inbreeding in the Wrangel population as it approached extinction [22]. As palaeogenomic data become more widely available, it will become increasingly feasible to apply sophisticated population genetics tests to these data. When assessed in the context of the palaeoenvironmental record, these data will provide new insights into the genomic consequences of environmental change and the genetic signatures of extinction.
5. Conclusion
Ancient DNA can reveal how past periods of environmental change affected the genetic diversity of species. Many insights have come from the use of coalescent simulations to infer demographic history. Denser sampling around the time of extinction and the development of more sophisticated coalescent models may improve our ability to infer the onset of declines to extinction. However, the principal advances are likely to come from the increasing availability of palaeogenomes, which can be analysed for other genetic signatures of extinction, such as long runs of homozygosity, excesses of non-synonymous substitutions and higher genetic loads.
Authors' contributions
Both authors participated in the design and writing of this manuscript.
Competing interests
We have no competing interests.
Funding
B.S. and D.C. received support from the Packard Foundation and NSF ARC-1417036.
References
- 1.Hewitt G. 2000. The genetic legacy of the Quaternary ice ages. Nature 405, 907–913. ( 10.1038/35016000) [DOI] [PubMed] [Google Scholar]
- 2.Leonard JA, Wayne RK, Cooper A. 2000. Population genetics of ice age brown bears. Proc. Natl Acad. Sci. USA 97, 1651–1654. ( 10.1073/pnas.040453097) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koch PL, Barnosky AD. 2006. Late quaternary extinctions: state of the debate. Annu. Rev. Ecol. Evol. S 37, 215–250. ( 10.1146/annurev.ecolsys.34.011802.132415) [DOI] [Google Scholar]
- 4.Karanth KP, Delefosse T, Rakotosamimanana B, Parsons TJ, Yoder AD. 2005. Ancient DNA from giant extinct lemurs confirms single origin of Malagasy primates. Proc. Natl Acad. Sci. USA 102, 5090–5095. ( 10.1073/pnas.0408354102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orlando L, et al. 2013. Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature 499, 74–78. ( 10.1038/nature12323) [DOI] [PubMed] [Google Scholar]
- 6.Anderson CN, Ramakrishnan U, Chan YL, Hadly EA. 2005. Serial SimCoal: a population genetics model for data from multiple populations and points in time. Bioinformatics 21, 1733–1734. ( 10.1093/bioinformatics/bti154) [DOI] [PubMed] [Google Scholar]
- 7.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973. ( 10.1093/molbev/mss075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan YL, Anderson CN, Hadly EA. 2006. Bayesian estimation of the timing and severity of a population bottleneck from ancient DNA. PLoS Genet. 2, e59 ( 10.1371/journal.pgen.0020059) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shapiro B, et al. 2004. Rise and fall of the Beringian steppe bison. Science 306, 1561–1565. ( 10.1126/science.1101074) [DOI] [PubMed] [Google Scholar]
- 10.Edwards CJ, et al. 2011. Ancient hybridization and an Irish origin for the modern polar bear matriline. Curr. Biol. 21, 1251–1258. ( 10.1016/j.cub.2011.05.058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lorenzen ED, et al. 2011. Species-specific responses of Late Quaternary megafauna to climate and humans. Nature 479, 359–364. ( 10.1038/nature10574) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palkopoulou E, et al. 2013. Holarctic genetic structure and range dynamics in the woolly mammoth. Proc. R. Soc. B 280, 20131910 ( 10.1098/rspb.2013.1910) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stiller M, et al. 2010. Withering away—25,000 years of genetic decline preceded cave bear extinction. Mol. Biol. Evol. 27, 975–978. ( 10.1093/molbev/msq083) [DOI] [PubMed] [Google Scholar]
- 14.Shapiro B, Hofreiter M. 2014. A paleogenomic perspective on evolution and gene function: new insights from ancient DNA. Science 343, 1236573 ( 10.1126/science.1236573) [DOI] [PubMed] [Google Scholar]
- 15.Barnes I, Matheus P, Shapiro B, Jensen D, Cooper A. 2002. Dynamics of Pleistocene population extinctions in Beringian brown bears. Science 295, 2267–2270. ( 10.1126/science.1067814) [DOI] [PubMed] [Google Scholar]
- 16.Hofreiter M, Munzel S, Conard NJ, Pollack J, Slatkin M, Weiss G, Paabo S. 2007. Sudden replacement of cave bear mitochondrial DNA in the late Pleistocene. Curr. Biol. 17, R122–R123. ( 10.1016/j.cub.2007.01.026) [DOI] [PubMed] [Google Scholar]
- 17.Kuhn TS, McFarlane KA, Groves P, Mooers AØ, Shapiro B. 2010. Modern and ancient DNA reveal recent partial replacement of caribou in the southwest Yukon. Mol. Ecol. 19, 1312–1323. ( 10.1111/j.1365-294X.2010.04565.x) [DOI] [PubMed] [Google Scholar]
- 18.Kingman JFC. 1982. The coalescent. Stoch. Proc. Appl. 13, 235–248. ( 10.1016/0304-4149(82)90011-4) [DOI] [Google Scholar]
- 19.Drummond AJ, Rambaut A, Shapiro B, Pybus OG. 2005. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 22, 1185–1192. ( 10.1093/molbev/msi103) [DOI] [PubMed] [Google Scholar]
- 20.Beaumont MA, Zhang W, Balding DJ. 2002. Approximate Bayesian computation in population genetics. Genetics 162, 2025–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho SY, Shapiro B. 2011. Skyline-plot methods for estimating demographic history from nucleotide sequences. Mol. Ecol. Resour. 11, 423–434. ( 10.1111/j.1755-0998.2011.02988.x) [DOI] [PubMed] [Google Scholar]
- 22.Palkopoulou E, et al. 2015. Complete genomes reveal signatures of demographic and genetic declines in the woolly mammoth. Curr. Biol. 25, 1395–1400. ( 10.1016/j.cub.2015.04.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carpenter ML, et al. 2013. Pulling out the 1%: whole-genome capture for the targeted enrichment of ancient DNA sequencing libraries. Am. J. Hum. Genet. 93, 852–864. ( 10.1016/j.ajhg.2013.10.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemey P, Rambaut A, Drummond AJ, Suchard MA. 2009. Bayesian phylogeography finds its roots. PLoS Comput. Biol. 5, e1000520 ( 10.1371/journal.pcbi.1000520) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cahill JA, et al. 2013. Genomic evidence for island population conversion resolves conflicting theories of polar bear evolution. PLoS Genet. 9, e1003345 ( 10.1371/journal.pgen.1003345) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, Durbin R. 2011. Inference of human population history from individual whole-genome sequences. Nature 475, 493–496. ( 10.1038/nature10231) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schubert M, et al. 2014. Prehistoric genomes reveal the genetic foundation and cost of horse domestication. Proc. Natl Acad. Sci. USA 111, E5661–E5669. ( 10.1073/pnas.1416991111) [DOI] [PMC free article] [PubMed] [Google Scholar]


