Abstract
Sea urchins are noted for the absence of neoplastic disease and represent a novel model to investigate cellular and systemic cancer protection mechanisms. Following intracoelomic injection of the DNA alkylating agent methyl methanesulfonate, DNA damage was detected in sea urchin cells and tissues (coelomocytes, muscle, oesophagus, ampullae and gonad) by the alkaline unwinding, fast micromethod. Gene expression analyses of the coelomocytes indicated upregulation of innate immune markers, including genes involved in NF-κB signalling. Results suggest that activation of the innate immune system following DNA damage may contribute to the naturally occurring resistance to neoplastic disease observed in sea urchins.
Keywords: sea urchins, methyl methanesulfonate, DNA damage, innate immune genes
1. Introduction
There is a wide discrepancy in the occurrence of cancer across different animal groups, with high incidence in some animals (e.g. some mammals, fish, bivalves) and others showing low or no incidence (e.g. echinoderms, crustaceans) [1,2]. Sea urchins have been noted for the absence of neoplastic disease [2,3], despite the observation that some species are very long-lived (living more than 100 years) [4], they possess high regenerative capabilities [5] and lack an adaptive immune system [6]. Incidences of tumours in some commercial shellfish are often correlated with exposure to genotoxic chemicals in the wild [7] and some studies have established a causal relationship by exposing bivalves to carcinogens in controlled laboratory experiments [8,9]. Whole animal exposures of sea urchins have not been reported; however, sea urchin coelomocytes (immune cells) treated in vitro are highly resistant to a variety of DNA damaging agents and show efficient DNA repair mechanisms [10,11]. It is unknown whether this resistance to DNA damage is a unique characteristic of the immune cells or whether other cell types and tissues of adult sea urchins are also resistant to genotoxicants.
The immune system plays an important role in detection and ablation of potentially cancerous cells and several studies have uncovered direct links between the DNA damage response and the innate immune system [12,13]. For example, the DNA damage response can activate the transcription factor NF-κB, which initiates transcription of genes involved in innate immunity [14]. The innate immune system of sea urchins is highly complex, with expansion of several immune gene families (i.e. toll-like receptors) and homologues of many key vertebrate immune receptors, regulators and effectors including transcription factors related to the NF-κB family [6,15]. It is clear that innate immunity and the DNA damage response are intimately linked [12,14], and understanding the extent of innate immune response to genotoxic challenge in sea urchins is potentially informative in elucidating mechanisms of natural resistance to carcinogenesis. The aim of this study is to measure DNA damage among different tissues of adult sea urchins and evaluate induction of innate immune genes following in vivo exposure to the genotoxicant methyl methanesulfonate (MMS).
2. Material and methods
All animal collections and experiments complied with the Collecting and Experimental Ethics Policy of the Bermuda Institute of Ocean Sciences. Adult Lytechinus variegatus (50–90 mm) were maintained in flow-through aquaria under ambient temperature and light conditions and were fed macroalgae and lettuce.
Coelomocyte in vitro exposures were carried out according to Reinardy & Bodnar [11]. After exposure, subsamples of cells were removed for analysis of DNA damage in whole coelomocytes, and the remaining cells were pelleted (8000g, 5 min) and stored at −80°C for DNA extraction.
For in vivo exposures, animals were treated by intracoelomic injection (0.5 ml) of MMS diluted in calcium–magnesium-free seawater (CMFSW) [11] to give a final concentration of 0 (CMFSW only), 100 or 300 mg MMS kg−1 body weight. Each animal was held in an individual container with 1 l fresh seawater for 24 h. Three animals were treated simultaneously (0, 100 and 300 mg kg−1), and this experimental set-up was repeated on five consecutive days for a total of n = 5 replicates. Prior to in vivo treatment, coelomic fluid (CF, 200 µl) was collected for initial cell counts and background DNA damage measurement of coelomocytes. CF (200 µl) was collected after 1, 3, 6 and 24 h. After 24 h, animals were dissected and tissues [oesophagus (ES), Aristotle's lantern muscle (ALM), gonad (GON), ampullae (AMP) and coelomocytes (COEL, pelleted from 1.5 ml CF)] were collected. DNA was isolated from tissues (or cell pellets) following the DNAzol ES® Reagent protocol (MRC, USA) and quantified by dsDNA Quant-iT assay (Invitrogen).
DNA damage was detected by the fast micromethod, which analyses rates of alkaline unwinding as an indication of the amount of DNA strand breaks [10,11,16], with minor modification for extracted DNA. Samples were assayed in triplicate or quadruplicate by loading 20 µl (50 000 cells or 75–100 ng DNA) into each well. Lysis, alkaline unwinding, fluorescent detection (SpectraMaxM2, Molecular Devices) and strand scission factor (SSF) calculation [SSF = log(%dsDNAsample/%dsDNAcontrol) × (−1)] were as previously described [10,11,16].
RNA extraction, cDNA synthesis and quantitative reverse-transcription PCR (ABI7300) were conducted in coelomocytes as previously described [11]. Primer details are in table 1. PCR efficiency was calculated (E = 10(−1/slope)), control genes were validated by non-significant treatment effect on expression and the geometric mean of control genes was used to calculate relative gene expression [17]:
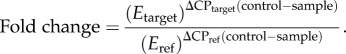 |
Table 1.
Sea urchin (Lytechinus variegatus) gene primer details.
| gene name | gene identifiera | forward primer (5′–3′) | reverse primer (5′–3′) |
|---|---|---|---|
| DNA damage detection | |||
| gadd45 | SPU_026064 | CAAGCAGCAAGAAAACTAGAACCA | AGCCACGTCCACGATTCC |
| innate immune signalling | |||
| traf | SPU_026479 | ATCATCTGGATCTTGGCAAACA | TTTAAGAAACCAACGACGGAGAA |
| ikk2 | SPU_008255 | GGCACCGGAACTATACTCACAAA | GGACGGTTCCAAAACTCCAA |
| ikb | SPU_011197 | TGGCCATTCATCATCAACAAG | GGTAGGCCGAGTCGCAAGT |
| rel | SPU_012203 | GCTCAACCGATGGAGGAAGA | TTTATTCTCTGAAGGCTGGTTGTG |
| nfkb | SPU_008177 | CCCAGCATCGGCTCTACAA | CCATCCAGCGATTCTCTCCTT |
| rae1 | SPU_003645 | TGGCGGTGGTGGGTACA | AAGGCTGGTTTTCAAGCTGGTA |
| tbk1 | SPU_004671 | TGGTCACTTGGAGTAACCTTCTATCA | CATGGGCACGGAATGGA |
| irf4 | SPU_026877 | TCACCACCAACCGCTATCAA | CGCCCTCTCCGGACAAG |
| 185/333 | SPU_019327 | AGGCCGTTCCGGTTCTTC | GCCGGTTCGGTTGTGTCT |
| control genes | |||
| cyclophilin-like-7 | SPU_008305 | CCTCCTTCCACAGGGTTATCC | GTACCGTTGCCCCTGGTAAA |
| rpl8 | SPU_010692 | GCCAACAGGGCCATGGT | TTACGCTTGACCTTGTATTTGAAGTAG |
| profilin | SPU_020197 | TGCAGGCGAGTAAGACAGCTATA | CTCCTTTATTCAAGTTCCCTTGCT |
| actin | SPU_006797 | GGTCAGGTCATCACTATTG | GCTGTTGTATGTGGTCTC |
aGene identifiers from annotated genome of Strongylocentrotus purpuratus (www.echinobase.org).
3. Results and discussion
MMS is an alkylating agent that adds methyl groups to nucleophilic sites on the DNA bases, creating lesions including 7-methylguanine that are primarily repaired by the base excision repair (BER) pathway [18]. MMS can additionally cause DNA strand breaks by indirect oxidative damage (intracellular production of reactive oxygen species) and targeted nicks in the DNA by BER [19]. MMS-induced DNA damage can be detected by DNA strand break assays such as the fast micromethod, which measures total DNA damage rather than DNA methylation directly and has the advantage of being applicable to both whole cells and isolated DNA [11]. There was a significant concentration-dependent increase in DNA damage in both whole coelomocytes and DNA extracted from frozen samples after in vitro exposure, and in whole coelomocytes after in vivo exposure to MMS (figure 1) (individual data in the electronic supplementary material). There was a trend in the whole coelomocytes to have higher absolute levels of DNA damage compared with levels detected in extracted DNA despite similar estimated DNA input. Results demonstrate the possibility of applying the fast micromethod to extracted DNA as a measure of DNA damage in tissues after in vivo genotoxicant exposure; however, care must be taken when comparing values across different experiments, tissues and methods.
Figure 1.
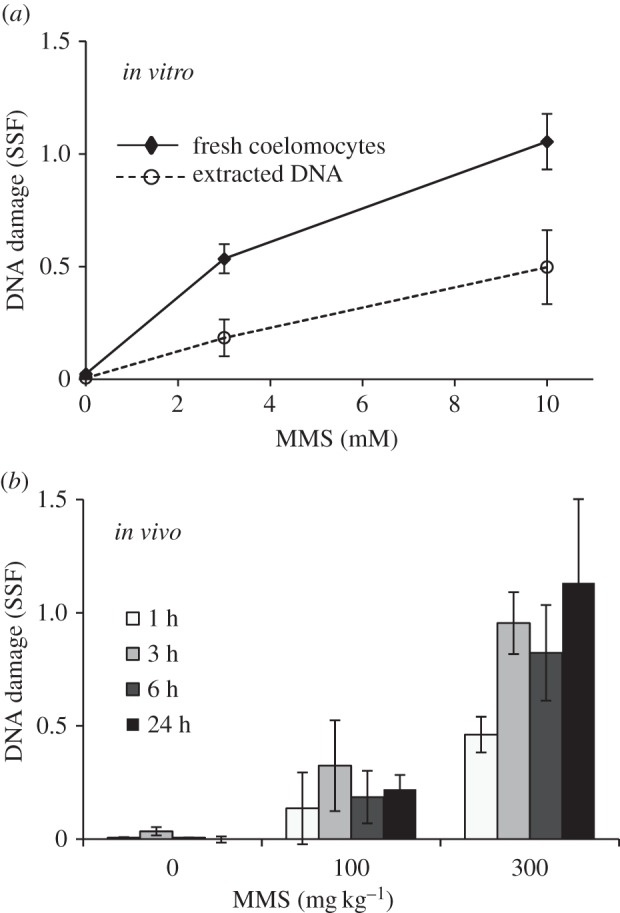
DNA damage in coelomocytes after exposure to MMS. SSF in whole coelomocytes compared with DNA extracted from coelomocytes after 1 h in vitro exposure (a), and in whole coelomocytes sampled over 24 h after in vivo injection with MMS (b). Data are means ± s.e.m., n = 5–8 (in vitro), n = 5 (in vivo), significant concentration response for each timepoint (Kruskal–Wallis, p < 0.05).
The in vitro concentration response in DNA damage for coelomocytes exposed to MMS up to 10 mM was the basis for calculating concentrations of MMS for in vivo exposure. Sea urchin volume was estimated as the volume of a hemisphere (based on animal diameter) and verified by volume of CF collected upon dissection following 24 h exposures. The highest in vivo treatment of 300 mg kg−1 approximated to 5.2–9.6 mM MMS based on estimated hemisphere volume or volume of CF, respectively. Despite uncertain estimation, the exposure range between the in vitro and in vivo experiments was closely matched.
After 24 h exposure, the animals were dissected and DNA extracted from the tissues. There was a concentration-dependent increase in DNA damage in all tissues, significantly higher than controls in gonads and coelomocytes (figure 2a). High variability among individuals may reflect high natural variability in these wild-caught animals, or technical variations in delivery of treatment or sampling of tissues. This is the first study to compare genotoxicant sensitivity in adult sea urchin tissues exposed in vivo, and results suggest similar levels of DNA damage in all tissues. Previous studies have suggested that coelomocytes are highly resistant to DNA damaging agents [10,11], and based on the results of in vivo MMS exposure, it is tempting to speculate that this resistance extends to other sea urchin tissues; however, further studies are required before making this conclusion.
Figure 2.
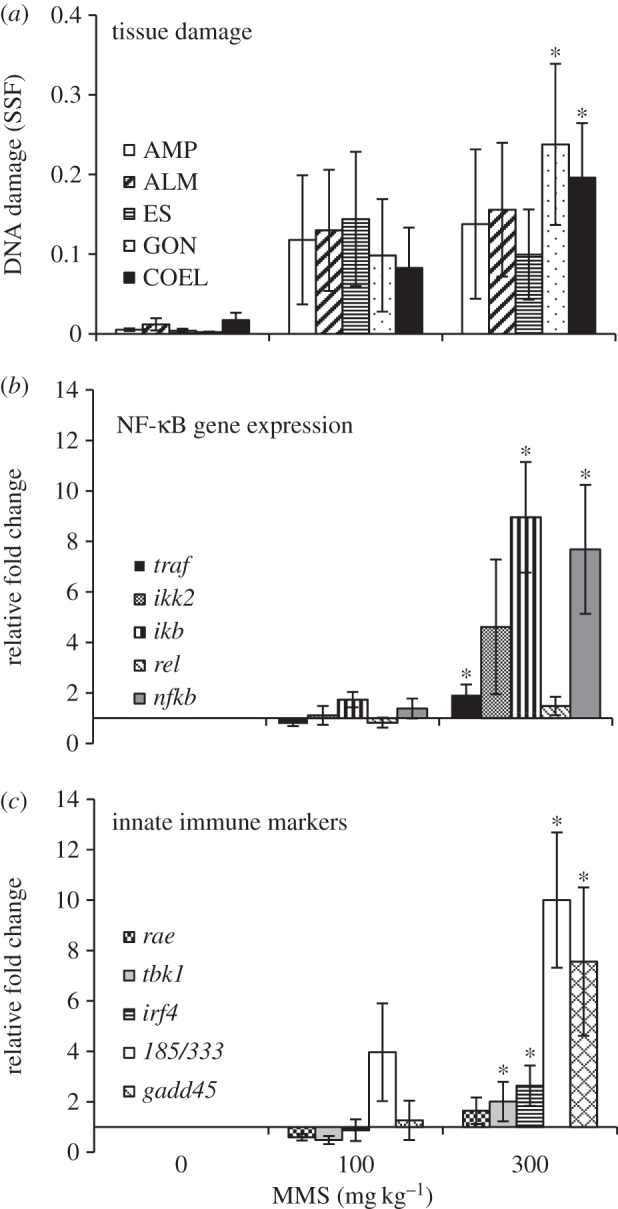
DNA damage in tissues and gene expression in coelomocytes after in vivo exposure to MMS. SSF in DNA extracted from tissues (a), and expression of genes of the NF-κB pathway (b) and other selected innate immune genes (c) in coelomocytes, after 24 h exposure. Data are means ± s.e.m., n = 5 (n = 4, 300 mg kg−1, b and c). Asterisks denote significantly higher than control animals (Kruskal–Wallis, post hoc multiple range test).
To explore potential links between the DNA damage response and the innate immune system, expression of a number of genes was investigated in coelomocytes 24 h following in vivo MMS exposure. These genes were selected to cover a range of innate immune pathways to indicate links between DNA damage induction and innate immune response. Out of the five animals in the highest treatment group, one individual consistently did not respond by upregulation of any genes of interest, in contrast with the consistent pattern of gene expression presented in the other four animals. Significant upregulation of gadd45 (figure 2c) indicates stress signalling and DNA damage detection, and GADD45 also plays an important anti-tumour surveillance role in immunity [20,21]. Genes involved in NF-κB signalling were selected because of the critical importance of NF-κB in immune regulation, and development and progression of cancer [13]. Immune challenge and genotoxic stress have been shown to activate NF-κB signalling via transmembrane cell receptors or stress-activated protein kinases (e.g. Jun N-terminal kinases) that recruit factors such as TNF-receptor associated factor to activate and phosphorylate IκB kinase complex to initiate downstream NF-κB nuclear translocation and gene transcription response [22]. While NF-κB-mediated response is well characterized for DNA double strand breaks, the response to genotoxic alkylating agents is not well characterized [14]. The key genes selected from the NF-κB pathway (traf, ikb, nfkb) were all significantly upregulated in coelomocytes from the highest treatment group (300 mg kg−1 MMS), but the trend in upregulation was not significant for ikk2 and rel (figure 2b). Results indicate a response in the NF-κB pathway; however, induction of other innate immune genes suggests a general response beyond the NF-κB pathway. Other genes that responded to the genotoxicant treatment were TANK-binding kinase 1 (tbk1) and irf4, which are associated with DNA-PK in surveying DNA and signalling IRF3-dependent nuclear transcription [23], non-significant upregulation of rae1, an NKG2D ligand involved in protein binding [13], and a significant upregulation of 185/333 (figure 2c). 185/333 is a diverse gene family in sea urchins, with around 100 different alleles, extensive post-translational protein diversity and responsive to bacterial lipopolysaccharide challenge, but the cellular function of the proteins, and the evolutionary significance of the high diversity, are still unknown [24]. These results indicate a far-reaching innate immune response following MMS-induced genotoxic stress in sea urchins; however, further studies are required to rule out possible indirect effects of methylation of other nucleophilic cellular components. Investigations of the genome-wide response to MMS and other DNA damaging agents are needed to build on this foundation and explore the extent of interactions between the DNA damage response and the innate immune system in resistance to neoplasm.
Supplementary Material
Ethics
All animal collections and experiments complied with the Collecting and Experimental Ethics Policy of the Bermuda Institute of Ocean Sciences.
Data accessibility
All data can be found as electronic supplementary material accompanying this manuscript.
Authors' contributions
H.C.R. and A.G.B. conceived and designed the study; H.C.R. and J.C. performed experiments; H.C.R. analysed the data; H.C.R., A.G.B. and J.C. wrote the manuscript. All authors agree to be held accountable for the work and approve the final version to be published.
Competing interests
We declare we have no competing interests.
Funding
This work received funding from the Bermuda Program (J.C.), the Christian Humann Foundation and a Bermuda Charitable Trust.
References
- 1.McAloose D, Newton AL. 2009. Wildlife cancer: a conservation perspective. Nat. Rev. Cancer 9, 517–526. ( 10.1038/nrc2665) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robert J. 2010. Comparative study of tumorigenesis and tumor immunity in invertebrates and nonmammalian vertebrates. Dev. Compar. Immunol. 34, 915–925. ( 10.1016/j.dci.2010.05.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jangoux M. 1987. Diseases of Echinodermata. 4. Structural abnormalities and general considerations on biotic diseases. Dis. Aquat. Organ. 3, 221–229. ( 10.3354/dao003221) [DOI] [Google Scholar]
- 4.Ebert T, Southon J. 2003. Red sea urchins (Strongylocentrotus franciscanus) can live over 100 years: confirmation with A-bomb 14carbon. Fish. Bull. 101, 915–922. [Google Scholar]
- 5.Reinardy HC, Emerson CE, Manley JM, Bodnar AG. 2015. Tissue regeneration and biomineralization in sea urchins: role of Notch signaling and presence of stem cell markers. PLoS ONE 10, e133860 ( 10.1371/journal.pone.0133860) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hibino T, et al. 2006. The immune gene repertoire encoded in the purple sea urchin genome. Dev. Biol. 300, 349–365. ( 10.1016/j.ydbio.2006.08.065) [DOI] [PubMed] [Google Scholar]
- 7.Van Beneden RJ. 1994. Molecular analysis of bivalve tumors: models for environmental/genetic interactions. Environ. Health Perspect. 102, 81–83. ( 10.1289/ehp.94102s1281) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gardner GR, Pruell RJ, Malcolm AR. 1992. Chemical induction of tumors in oysters by a mixture of aromatic and chlorinated hydrocarbons, amines and metals. Mar. Environ. Res. 34, 59–63. ( 10.1016/0141-1136(92)90083-X) [DOI] [Google Scholar]
- 9.Khudoley VV, Syrenko OA. 1978. Tumor induction by N-nitroso compounds in bivalve mollusks Unio pictorum. Cancer Lett. 4, 349–354. ( 10.1016/S0304-3835(78)95722-1) [DOI] [PubMed] [Google Scholar]
- 10.El-Bibany AH, Bodnar AG, Reinardy HC. 2014. Comparative DNA damage and repair in echinoderm coelomocytes exposed to genotoxicants. PLoS ONE 9, e107815 ( 10.1371/journal.pone.0107815) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reinardy HC, Bodnar AG. 2015. Profiling DNA damage and repair capacity in sea urchin larvae and coelomocytes exposed to genotoxicants. Mutagenesis 30, 829–839. ( 10.1093/mutage/gev052) [DOI] [PubMed] [Google Scholar]
- 12.Ermolaeva MA, Segref A, Dakhovnik A, Ou H-L, Schneider JI, Utermohlen O, Hoppe T, Schumacher B. 2013. DNA damage in germ cells induces an innate immune response that triggers systemic stress resistance. Nature 501, 416–420. ( 10.1038/nature12452) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gasser S, Orsulic S, Brown EJ, Raulet DH. 2005. The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature 436, 1186–1190. ( 10.1038/nature03884) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu ZH, Miyamoto S. 2007. Many faces of NF-κB signaling induced by genotoxic stress. J. Mol. Med. 85, 1187–1202. ( 10.1007/s00109-007-0227-9) [DOI] [PubMed] [Google Scholar]
- 15.Buckley KM, Rast JP. 2012. Dynamic evolution of toll-like receptor multigene families in echinoderms. Front. Immunol. 3, 136 ( 10.3389/fimmu.2012.00136) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schröder HC, Batel R, Schwertner H, Boreiko O, Muller WE. 2006. Fast micromethod DNA single-strand-break assay. Methods Mol. Biol. 314, 287–305. ( 10.1385/1-59259-973-7:287) [DOI] [PubMed] [Google Scholar]
- 17.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, p45e. ( 10.1093/nar/29.9.e45) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wyatt MD, Pittman DL. 2006. Methylating agents and DNA repair responses: methylated bases and sources of strand breaks. Chem. Res. Toxicol. 19, 1580–1594. ( 10.1021/tx060164e) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoeijmakers JHJ. 2001. DNA repair mechanisms. Maturitas 38, 17–22. ( 10.1016/S0378-5122(00)00188-2) [DOI] [PubMed] [Google Scholar]
- 20.Moskalev AA, Smit-McBride Z, Shaposhnikov MV, Plyusnina EN, Zhavoronkov A, Budovsky A, Tacutu R, Fraifeld VE. 2012. Gadd45 proteins: relevance to aging, longevity and age-related pathologies. Ageing Res. Rev. 11, 51–66. ( 10.1016/j.arr.2011.09.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ju S, Zhu Y, Liu L, Dai S, Li C, Chen E, He Y, Zhang X, Lu B. 2009. Gadd45b and Gadd45g are important for anti-tumor immune responses. Eur. J. Immunol. 39, 3010–3018. ( 10.1002/eji.200839154) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baldwin AS. 1996. The NF-κB and IκB proteins: new discoveries and insights. Annu. Rev. Immunol. 14, 649–683. ( 10.1146/annurev.immunol.14.1.649) [DOI] [PubMed] [Google Scholar]
- 23.Ferguson BJ, Mansur DS, Peters NE, Ren H, Smith GL. 2012. DNA-PK is a DNA sensor for IRF-3-dependent innate immunity. eLife 1, e00047 ( 10.7554/eLife.00047) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghosh J et al. 2010. Sp185/333: a novel family of genes and proteins involved in the purple sea urchin immune response. Dev. Comp. Immunol. 34, 235–245. ( 10.1016/j.dci.2009.10.008) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data can be found as electronic supplementary material accompanying this manuscript.


