Significance
Combinatorial biosynthesis is enabled by the use of promiscuous enzymes, but also can be applied to the discovery of such biocatalysts. Here, such an approach was used for the investigation of the substrate selectivity of cytochrome P450 enzymes from diterpenoid phytohormone biosynthesis, enabling facile characterization of both multiple substrates and structural analysis of the resulting products. The discovery of one such promiscuous cytochrome P450, reported here, provides insight into the underlying structure–function relationship and also may have biotechnological implications.
Keywords: labdane-related diterpenoids, cytochrome P450 monooxygenases, natural products, enzyme specificity, gibberellins
Abstract
The substrate specificity of enzymes from natural products’ metabolism is a topic of considerable interest, with potential biotechnological use implicit in the discovery of promiscuous enzymes. However, such studies are often limited by the availability of substrates and authentic standards for identification of the resulting products. Here, a modular metabolic engineering system is used in a combinatorial biosynthetic approach toward alleviating this restriction. In particular, for studies of the multiply reactive cytochrome P450, ent-kaurene oxidase (KO), which is involved in production of the diterpenoid plant hormone gibberellin. Many, but not all, plants make a variety of related diterpenes, whose structural similarity to ent-kaurene makes them potential substrates for KO. Use of combinatorial biosynthesis enabled analysis of more than 20 such potential substrates, as well as structural characterization of 12 resulting unknown products, providing some insight into the underlying structure–function relationships. These results highlight the utility of this approach for investigating the substrate specificity of enzymes from complex natural products’ biosynthesis.
We have co-opted natural products, the small molecules produced by living organisms, for a wide variety of purposes (1, 2), many of which are only tangentially related to their original biological function. For this reason, it is desirable to screen related compounds for the activity of interest. One means of accessing such chemical diversity is combinatorial biosynthesis (3). However, this is dependent on the use of promiscuous enzymes that can readily react with a variety of substrates. Unfortunately, investigation of substrate specificity is often hindered by the limited availability of structurally related and metabolically accessible compounds. This is particularly true for enzymes involved in the biosynthesis of more complex molecules, such as hormones and related natural products.
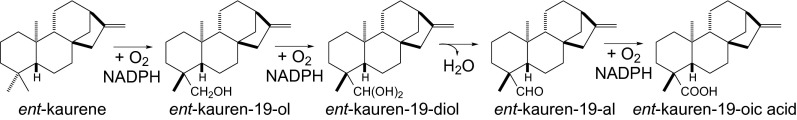
Gibberellins (GAs) are hormones essential for vascular plant growth and development, meeting the criteria for primary metabolites. Accordingly, the genes encoding the enzymes for the GA biosynthetic pathway are present in all tracheophytes (4). Thus, these plants all contain ent-kaurene oxidases (KOs), which are cytochrome P450 monooxygenases (CYPs) from the CYP701 family that catalyze multiple oxidations of the C4α methyl of the tetracyclic olefin intermediate ent-kaurene to form ent-kauren-19-oic acid (Fig. 1) as an early step in GA biosynthesis (5).
Fig. 1.
Conversion of ent-kaurene to ent-kaurenoic acid catalyzed by KOs (13).
Notably, the requirement for GA metabolism provides a reservoir of genes encoding biosynthetic enzymes, which has given rise to the production of many related diterpenoid natural products (6). These are characterized by initial bicyclization of the general diterpenoid precursor (E,E,E)-geranylgeranyl diphosphate (GGPP) to a labdadienyl/copalyl diphosphate (CPP) intermediate by class II diterpene cyclases then termed CPP synthases (CPSs). This has led to designation of the derived natural products as labdane-related diterpenoids (LRDs), and the presence of the corresponding decalin ring structure in almost all of these makes the LRDs easily recognizable (7). In the case of GA biosynthesis, the corresponding intermediate is ent-CPP, with 9R,10R stereochemistry, and this is further cyclized to ent-kaurene by ent-kaurene synthases (KSs) (5). Given the relatively close phylogenetic relationship between KSs and other terpene synthases that act on CPP (of various stereochemistries), these class I diterpene synthases have been termed KS-like (7). Accordingly, the CPS and KS required in all tracheophytes for GA production have served as a genetic reservoir from which more specialized LRD metabolism has repeatedly evolved by gene duplication and neo-functionalization (6).
In contrast, KO appears to have been much less frequently diverted to more specialized LRD metabolism, with the only biochemically distinct CYP701 family members characterized to date found in rice (Oryza sativa). In particular, rice contains an expanded family of CYP701A subfamily members (8), at least two of which have distinct biochemical function [i.e., cannot act as KOs (9, 10)] whereas genetic evidence clearly indicates that only one (CYP701A6/OsKO2; O. sativa KO #2) actually functions in GA metabolism (11). In contrast, Arabidopsis thaliana is known to have only a single KO, CYP701A3/AtKO (A. thaliana KO), as required for GA biosynthesis (12). Previous investigation has demonstrated some promiscuity for AtKO, in that it will react with at least two LRD olefins that are structurally distinct yet related to ent-kaurene [i.e., ent-atiserene and ent-beyerene (13)]. However, this promiscuity has not been thoroughly investigated with a wider range of potential substrates, nor compared with any other KO. As with many such enzymes involved in the metabolism of more complex natural products, such studies are hindered by the severely limited access to the relevant compounds, both substrates and products. Here we report the use of combinatorial biosynthesis for such investigations, enabling analysis of more than 20 potential substrates, revealing significant promiscuity for AtKO, whereas OsKO2 was found to be much more specific. Critically, this approach further allowed structural characterization of 12 resulting unknown compounds, providing insight into the underlying structure–function relationship.
Results
Screening Substrate Specificity.
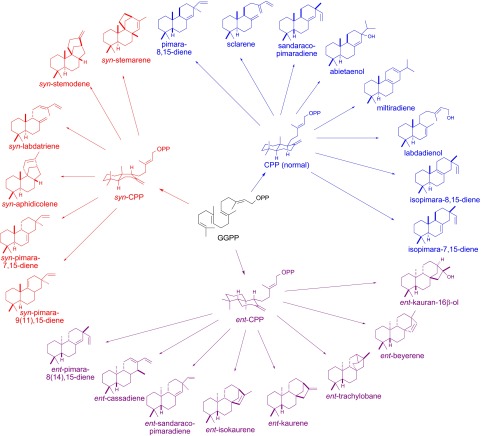
We have developed a modular metabolic engineering system that enables facile transformation of Escherichia coli to engineer the production of any terpene for which the corresponding synthase or synthases are known (14). This has been further coupled to strategies for functional expression of plant CYPs in E. coli, to (re)create terpenoid biosynthetic pathways via a synthetic biology approach (15). Given the difficulty of obtaining an extended range of relevant substrates for in vitro reactions, for example, by (retro)synthesis, here we have adopted this approach to screen AtKO and OsKO2 activity against a range of LRD olefins in E. coli. Specifically, we coexpressed codon-optimized and N-terminally modified constructs for each of these KOs (9, 13), along with a corresponding cytochrome P450 reductase (CPR) from the same species (i.e., AtCPR1 or OsCPR1) (16, 17), with upstream CPS and KS(L) genes from a variety of sources, not only plants but bacteria as well, that together produce an array of more than 20 LRDs (Fig. 2; SI Appendix, Table S1). On the basis of the ability of AtKO to react with a range of LRDs derived from ent-CPP, we further screened both AtKO and OsKO2 against LRD olefins produced via the available alternative stereoisomers [i.e., normal (9S,10S) and syn (9S,10R)].
Fig. 2.
LRDs screened in this study, with origin of the underlying stereochemistry (i.e., CPP stereoisomer) also shown and indicated by color (i.e., CPP and derived diterpenes are depicted in blue, ent-CPP and derived diterpenes in purple, with syn-CPP and derived diterpenes in red).
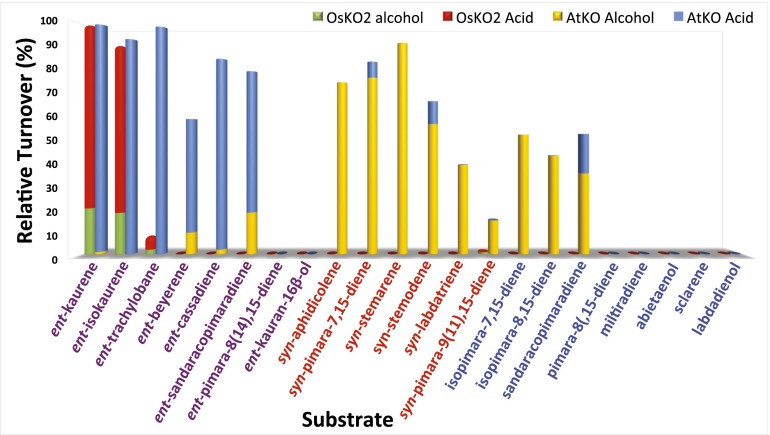
As expected, when either AtKO or OsKO2 was coexpressed in E. coli engineered to produce ent-kaurene, turnover to ent-kauren-19-oic acid was observed (SI Appendix, Fig. S1). Upon wider screening, AtKO was found to react with a much broader range of the tested LRD olefins than OsKO2 (Fig. 3). In particular, OsKO2 only reacted with ent-kaurene, its double-bond isomer ent-isokaurene, and the closely related ent-trachylobane, converting the C4α methyl to a mixture of the alcohol and further oxidized carboxylic acid with each substrate (SI Appendix, Figs. S1–S3). In contrast, AtKO is more promiscuous, reacting with LRDs derived from ent-CPP to transform the C4α methyl to varying ratios of alcohols and derived carboxylic acids (SI Appendix, Figs. S1–S6). In addition, AtKO will even react with LRDs derived from normal or syn-CPP (SI Appendix, Figs. S7–S14), although largely only to the extent of carrying out a single oxygenation reaction to produce hydroxylated derivatives.
Fig. 3.
Relative activity of AtKO versus OsKO2 with screened LRDs. Shown is turnover observed with recombinant AtKO (and AtCPR1) or OsKO2 (and OsCPR1) in E. coli also metabolically engineered to produce the noted LRD.
Determination of Hydroxylation Positions.
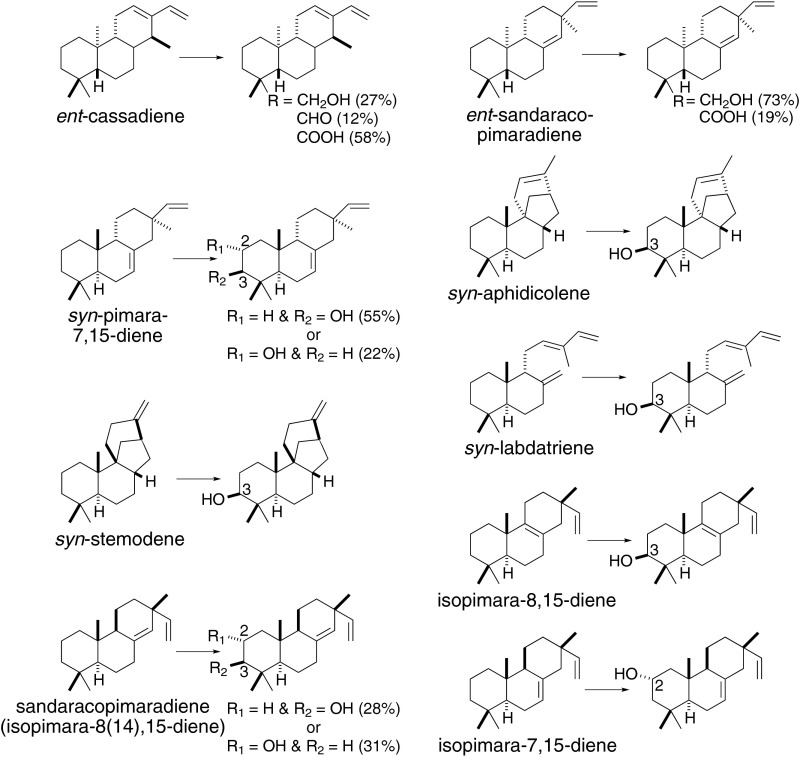
Although we have authentic standards for the C19 oxygenated products mentioned here (e.g., ent-kauren-19-oic, ent-isokauren-19-oic, ent-trachyloban-19-oic acids), as well as ent-beyeren-19-ol and ent-beyeren-19-oic acid, for many of the observed AtKO products, no such standards are available. Accordingly, to determine the regio- and stereochemistry of the catalyzed reaction, we carried out structural characterization of these compounds by NMR. To obtain sufficient amounts for this purpose, we increased flux to terpenoid metabolism in the engineered cultures by further including a compatible plasmid that enables conversion of mevalonate to isoprenoid precursors (18). The resulting major product or products were then isolated and subjected to NMR structural analysis, elucidating their composition (Fig. 4). Through these studies, it was found that AtKO targets the C4α methyl group in ent-CPP-derived LRDs, including ent-cassadiene and ent-sandaracopimaradiene, which are converted to a mixture of the alcohol and derived carboxylic acid (SI Appendix, Figs. S15 and S16 and Tables S2 and S3). In contrast, with LRDs derived from normal or syn-CPP, AtKO targets the neighboring C3 and C2 positions. Specifically, we found that AtKO largely hydroxylates syn-aphidicolene, syn-labda-8(17),12E,14-triene, syn-pimara-7,15-diene, and syn-stemod-13(17)-ene at the C3β position. In addition, however, with syn-pimara-7,15-diene, AtKO also yields small amounts of the C2α-hydroxyl, and with syn-stemodene, converts small amounts of the C4β methyl to a carboxylate (SI Appendix, Figs. S17–S20 and Tables S4–S8). Similarly, AtKO hydroxylates isopimara-8,15-diene at the C3β position and hydroxylates isopimara-7,15-diene at the C2α position, with a minor amount of further oxidization to the ketone also observed and sandaracopimaradiene [isopimara-8(14),15-diene] at both the C2α and C3β positions (SI Appendix, Figs. S21–S23 and Tables S9–S14).
Fig. 4.
AtKO catalyzes hydroxylations of multiple LRDs. Shown are the reactions characterized here, as elucidated by structural analysis of the resulting major product or products.
Relative Kinetic Analysis.
To enable comparison of the reaction rates of the different substrates by AtKO, in vitro assays were carried out with a subset of alternative substrates, also obtained via the metabolic engineering system. Given the variability in the relationship between catalytic activity and the amount of “correctly folded” CYP measured by carbon monoxide binding difference spectra observed between enzymatic preparations, we first attempted to carry out competition assays as previously described (19). However, AtKO reacts very efficiently with ent-kaurene and invariably preferentially converts this to ent-kaurenoic acid before reacting with the alternative (LRD) substrate. Accordingly, to compare catalytic efficiency, we normalized our substrate turnover to ent-kaurene; that is, each enzyme preparation was assayed with ent-kaurene and a standard amount of such measured activity used in assays with the alternative substrates. AtKO exhibited much lower affinity for its nonnative substrates, with KM ranging from 30 to 50 µM, representing >10-fold increases over the 2 µM KM for ent-kaurene (13). Moreover, Vmax was generally ∼10-fold less than that observed with ent-kaurene as well, indicating that AtKO exhibits at least two order of magnitude decreases in catalytic efficiency with these alternative substrates (Table 1).
Table 1.
AtKO comparative kinetic data
| Substrate | KM, µM | Vmax, relative to ent-kaurene |
| ent-cassadiene | 50 ± 7 | 0.14 |
| ent-sandaracopimaradiene | 46 ± 24 | 0.11 |
| syn-aphidicolene | 50 ± 14 | 0.12 |
| Isopimara-7,15-diene | 30 ± 8 | 0.11 |
Molecular Phylogenetic Analysis.
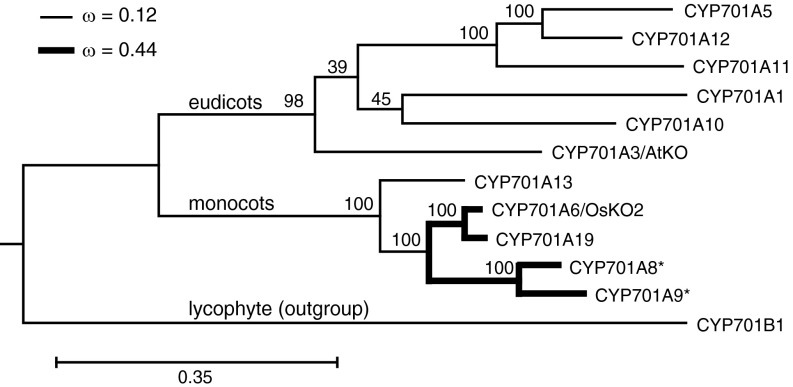
The difference between the activity exhibited by AtKO and OsKO2 may reflect their distinct evolutionary contexts. In particular, although Arabidopsis seems to be representative of the Brassicaceae plant family, which does not produce any more specialized LRDs, only GA, and contains just one CYP701 family member, rice is representative of the Poaceae plant family, where it has been indicated that more specialized LRDs arose early, with widespread retention (20), and rice contains a number of CYP701A subfamily members with distinct function (9–11). This presumably exerted differing selective pressures on these genes, which was examined by molecular phylogenetic analysis of all characterized members of the CYP701 family (Fig. 5; SI Appendix, Table S14). As might be expected, AtKO groups with other dicot KOs, whereas OsKO2 groups with its paralogs (i.e., the other rice CYP701A subfamily members) and the only other characterized cereal KO; specifically, that from barley, CYP701A13/HvKO. To detect adaptive evolution, the ratios of the nonsynonymous and synonymous substitution rates (dN/dS or ω) were estimated for this gene family, not only for overall and pairwise comparisons, but also for lineage and/or site-specific models (21). Across the CYP701 family, purifying selection is dominant (ω < 0.2), nor is this appreciably different for AtKO relative to the rest of the family, as indicated by the essentially identical values for ω, as well as log-likelihood score, for this distinct branch model. In contrast, the paralogs from rice (CYP701A6, 8, 9, 19) are clearly under different selective pressure than the rest of the family (P < 0.001), with ω = 0.44 for these, versus 0.12 for the remainder of the family. Additional analysis further strongly supported variation in selective pressure along the coding sequence, again with P < 0.001 from a likelihood ratio test. However, these sites do not generally appear to be under positive selection, as the relevant null test exhibits an identical log-likelihood score. Instead, this branch-site model indicates that a proportion of the sites are released from the usual purifying selection pressure, with ω changing from <0.1 to 1.0 (i.e., neutral) for ∼25% of the residues in the rice CYP701A paralogs. Nevertheless, this analysis does suggest that a few residues may be under positive selection pressure, which may still be possible, given the limited number of available sequences. Indeed, the distinct function of two of the rice CYP701A subfamily members, CYP701A8 and CYP701A9 (9, 10), almost certainly required such differentiating positive selection pressure (although this is not evident from the relevant branch model analysis, and comparison of log likelihood scores indicate that selective pressure differs for all of the rice paralogs, rather than only these two).
Fig. 5.
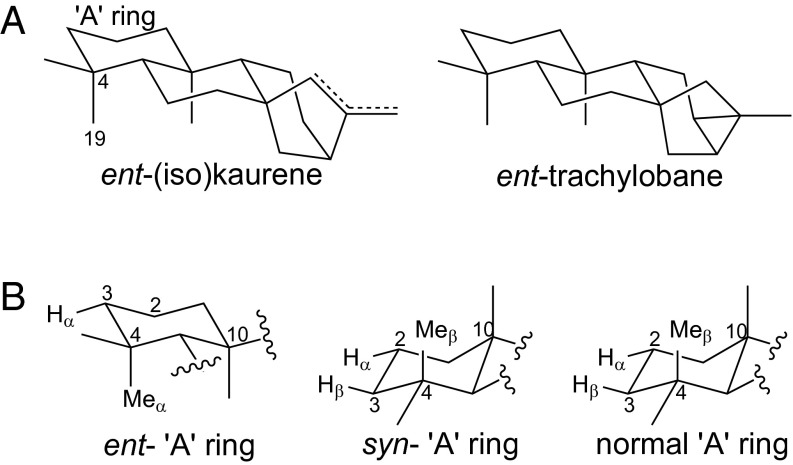
Structure–function relationship for the activity exhibited by (A) OsKO2 and (B) AtKO. (A) 3D rendering illustrating the close structural relationship between ent-(iso)kaurene and ent-trachylobane, the only substrates accepted by OsKO2. (B) Configuration of the “A” ring from the three underlying stereoisomers (i.e., of CPP) illustrating targeting of the same two roughly apposed sites with normal and syn-CPP derived LRDs that may be related to their similar configuration in this region. Previous work has demonstrated that other non-KO CYP701A subfamily members (i.e., from rice) target the C3α position with ent- and syn-CPP-derived LRDs (9, 10).
Discussion
Although enzymatic promiscuity enables combinatorial biosynthesis, investigation of this has been limited, particularly for more complex natural products’ metabolism, by the availability of substrates and authentic standards for identification of the resulting products. As demonstrated here, combinatorial biosynthesis also provides a robust approach for discovery and characterization of promiscuous enzymes. Reflecting the underlying (bio)chemical logic of plant natural product biosynthesis (22), here various diterpene synthases were combined to produce a range of scaffolds that can be acted upon by tailoring enzymes such as plant CYPs like the KOs from GA biosynthesis investigated here. Indeed, this approach enabled investigation of more than 20 potential substrates (Fig. 2), along with structural resolution of 12 resulting unknown products (Fig. 4).
Notably, this approach revealed that although OsKO2 is quite specific for ent-(iso)kaurene, and only also reacts with the structurally closely related ent-trachylobane (Fig. 5A), AtKO exhibits much broader promiscuity, albeit with restrictions that provide some insight into the limits of its active site to accommodate alternative substrates. For example, with the eight LRD substrates derived from ent-CPP, AtKO reacts with all but ent-pimara-8(14),15-diene, which differs from ent-sandaracopimaradiene only in configuration of the methyl and vinyl groups at C13, and 16α-hydroxy-ent-kaurane, which differs from ent-(iso)kaurene only in the presence of the hydroxyl group. From this, it appears that differences in LRDs distal from the targeted C4α methyl affect the ability of AtKO to accommodate these as substrates. Nevertheless, with all six reactive LRDs derived from ent-CPP, AtKO specifically targets this methyl group.
In contrast, with the reactive LRDs derived from normal and syn-CPP, AtKO largely targets C2α and/or C3β, which are roughly apposed but on the opposite “side” of the “A” ring from the C4α methyl targeted by AtKO in ent-CPP-derived LRDs. Indeed, at least in one case (i.e., syn-stemodene), the only other identified target is the C4β methyl. This highlights the effect of the configuration of the “A” ring on the orientation of the LRDs in the active site, and in particular, the configuration of the methyl substituent on C10, as the β-configuration of this is the most prominent feature in common between the LRDs derived from normal and syn-CPP (Fig. 5B). In addition, although AtKO reacts with all of the tested syn-CPP derived LRDs, it exhibits some selectivity with those derived from normal CPP. Specifically, it will only react with isopimaradienes, which have a β-methyl (and α-vinyl) at C13. This further emphasizes the effect that such distal differences in structure have on substrate binding in the AtKO active site. Finally, the preferential yield of alcohols, rather than more oxidized products with these alternative substrates, may stem from the previously noted reduced affinity of AtKO for even the natural ent-kauren-19-ol intermediate relative to the olefin precursor ent-kaurene (or more oxidized ent-kauren-19-al intermediate) (13).
The contrast between the quite strict specificity of OsKO2 and the promiscuity exhibited by AtKO is intriguing (Fig. 3). Even beyond the question of the basis for their contrasting substrate specificity, given the presence of alternative substrates over substantial evolutionary timescales in rice, but not Arabidopsis (i.e., in the Poaceae vs. Brassicaceae), and the physiological function of both OsKO2 and AtKO in primary (GA) biosynthesis, this observed difference, although obviously limited in scope, is consistent with the previously proposed general hypothesis that such evolutionary context (i.e., metabolic background) plays a role in shaping enzymatic specificity (23). Further evidence may be obtained by investigating the substrate specificity of a wider array of KOs from other plant species with distinct ranges of LRD metabolism in the future. In addition, it might then be of interest to examine the effect that substitution of OsKO2 with a more promiscuous KO (such as AtKO) has on rice plants. In any case, to the extent that such promiscuity reflects the activity of KO from plants with limited LRD metabolism, such activity provides a means for immediate production of more elaborated metabolites from novel diterpenes that might arise (e.g., by gene duplication and neo-functionalization of upstream diterpene synthases from GA biosynthesis). These natural products would be more likely to exhibit biological activity (6), and such latent biosynthetic capacity then may (partially) underlie the broad diversity of LRD natural products observed in plants, consistent with a recently suggested mechanism for the evolution of more specialized metabolism in plants more generally (24).
Regardless of evolutionary basis, although the promiscuity of AtKO revealed here is coupled to somewhat reduced catalytic efficiency in vitro, its ability to convert alternative substrates largely to hydroxylated products suggests the observed activity might be of use in industrial biotechnology approaches. Indeed, the demonstrated incorporation of membrane-associated CYPs such as the KOs into a modular bacterial metabolic engineering system via a synthetic biology approach emphasizes the utility of such microbial host production systems. Moreover, the results reported here further highlight the usefulness of applying combinatorial biosynthesis for investigating the substrate specificity of enzymes from complex natural products’ metabolism.
Materials and Methods
Combinatorial Biosynthesis.
The substrate specificity of AtKO and OsKO2 was assessed by use of our previously described modular metabolic engineering system for production of LRDs in E. coli (14). The relevant combinations of diterpene synthases are described in the SI Appendix (SI Appendix, Table S1). This system is further amendable to incorporation of CYPs, in particular, using synthetic genes, codon-optimized for expression in E. coli along with replacement of the N-terminal transmembrane helix sequence with a leader peptide (15), as has been previously reported for AtKO and OsKO2 (9, 13). Accordingly, it was possible to coexpress either KO, along with the requisite CPR from the same species, in E. coli also engineered to make any one (or two) of more than 20 different LRDs (see SI Appendix for more details). Organic (hexane) extracts of these recombinant cultures were then analyzed by GC-MS to screen for KO activity. Moreover, in those cases where the resulting products could not be identified from the available authentic standards, it was then possible to produce sufficient quantities for structural analysis via NMR by increasing flux toward isoprenoid metabolism, as well as simply increasing culture volumes, much as previously described (25).
Chemical Structure Identification.
Unknown products were purified via flash chromatography and HPLC, much as has been previously described (9, 10, 17, 19, 26–28), with further details available in the SI Appendix. The resulting compounds were then resuspended in deuterated chloroform (CDCl3) for structural analysis by NMR, as described in more detail in the SI Appendix. Note that the acquired 1D 1H and 13C spectra can be found in the SI Appendix, Figs. S24–S35. In certain cases, the identity of the observed carboxylic acid products was verified by feeding the structurally characterized hydroxylated precursor (i.e., for ent-cassadien-19-oic and ent-sandaracopimaradien-19-oic acids, as described in more detail in the SI Appendix).
Analysis of Catalytic Efficiency.
Given the variability between the characteristic CO binding difference spectra usually used to quantify CYP concentrations and the actually observed enzymatic activity in vitro, catalytic efficiency was determined by relative kinetic analyses. This was limited to assays with AtKO, as OsKO2 was not sufficiently stable enough for reliable use in vitro. These assays were carried out with cell-free extracts, fed the relevant substrate (i.e., those shown in Table 1), with comparison of the observed activity to that observed with parallel assays fed the native ent-kaurene substrate, enabling normalization of Vmax (i.e., to the rate observed with ent-kaurene, as presented in Table 1). A more detailed description of these studies can be found in the SI Appendix.
Molecular Phylogenetic Analysis.
The ORFs coding for the characterized CYP701 family members were obtained from GenBank (SI Appendix, Table S14). Alignment, phylogenetic tree construction, and overall (as well as pairwise) analyses of evolutionary pressure were carried out using the MEGA 6.06 software package for Mac (29), all of which supported only purifying selection. Lineage-specific adaptive evolution was investigated using the PAML 4.8 software package for Mac (30), particularly the CODEML program, used largely as outlined (31). The fit of various models to the evolutionary history of the CYP701 gene family was assessed by comparison of the calculated log likelihood scores, with statistical significance estimated by likelihood ratio tests where appropriate (i.e., between models with different degrees of freedom). Such comparisons supported a distinct branch model in which the clade of rice paralogs are under less stringent purifying selection than the rest of the CYP701 family (as depicted in Fig. 6), with further branch site analysis indicating this is a result of release of such purifying selection on a subsection of sites/residues, albeit while also suggesting that a few residues may be under positive selection (see the SI Appendix for more details).
Fig. 6.
Molecular phylogenetic analysis of the CYP701 family, based on full-length ORFs for the functionally characterized members (SI Appendix, Table S14). Shown is a rooted phylogenetic tree, with the corresponding bootstrap values (from 100 replicates). Also indicated are the branches that delineate the relevant classes of plants from which these CYP701 family members originate. The thicker lines delineate the clade of rice paralogs that lineage-specific adaptive evolutionary analysis strongly indicates is under distinct selective pressure relative to the rest of the family. More detailed branch-site analysis further indicates that this is a result of release of purifying selection on a subset (∼25%) of the sites/residues in the rice CYP701 family members, two of which are functionally distinct (these are indicated by *).
Supplementary Material
Acknowledgments
This work was supported by grants from the National Science Foundation (MCB0919735) and National Institutes of Health (GM109773) (to R.J.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1512096113/-/DCSupplemental.
References
- 1.Newman DJ, Cragg GM. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod. 2012;75(3):311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cantrell CL, Dayan FE, Duke SO. Natural products as sources for new pesticides. J Nat Prod. 2012;75(6):1231–1242. doi: 10.1021/np300024u. [DOI] [PubMed] [Google Scholar]
- 3.Zhou H, Xie X, Tang Y. Engineering natural products using combinatorial biosynthesis and biocatalysis. Curr Opin Biotechnol. 2008;19(6):590–596. doi: 10.1016/j.copbio.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 4.Peters RJ. Gibberellin phytohormone metabolism. In: Bach T, Rohmer M, editors. Isoprenoid Synthesis in Plants and Microorganisms: New Concepts and Experimental Approaches. Springer; New York: 2013. pp. 233–249. [Google Scholar]
- 5.Hedden P, Thomas SG. Gibberellin biosynthesis and its regulation. Biochem J. 2012;444(1):11–25. doi: 10.1042/BJ20120245. [DOI] [PubMed] [Google Scholar]
- 6.Zi J, Mafu S, Peters RJ. To gibberellins and beyond! Surveying the evolution of (di)terpenoid metabolism. Annu Rev Plant Biol. 2014;65:259–286. doi: 10.1146/annurev-arplant-050213-035705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peters RJ. Two rings in them all: The labdane-related diterpenoids. Nat Prod Rep. 2010;27(11):1521–1530. doi: 10.1039/c0np00019a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Itoh H, et al. A rice semi-dwarf gene, Tan-Ginbozu (D35), encodes the gibberellin biosynthesis enzyme, ent-kaurene oxidase. Plant Mol Biol. 2004;54(4):533–547. doi: 10.1023/B:PLAN.0000038261.21060.47. [DOI] [PubMed] [Google Scholar]
- 9.Wang Q, Hillwig ML, Wu Y, Peters RJ. CYP701A8: A rice ent-kaurene oxidase paralog diverted to more specialized diterpenoid metabolism. Plant Physiol. 2012;158(3):1418–1425. doi: 10.1104/pp.111.187518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kitaoka N, Wu Y, Xu M, Peters RJ. Optimization of recombinant expression enables discovery of novel cytochrome P450 activity in rice diterpenoid biosynthesis. Appl Microbiol Biotechnol. 2015;99(18):7549–7558. doi: 10.1007/s00253-015-6496-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sakamoto T, et al. An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol. 2004;134(4):1642–1653. doi: 10.1104/pp.103.033696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Helliwell CA, et al. Cloning of the Arabidopsis ent-kaurene oxidase gene GA3. Proc Natl Acad Sci USA. 1998;95(15):9019–9024. doi: 10.1073/pnas.95.15.9019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morrone D, Chen X, Coates RM, Peters RJ. Characterization of the kaurene oxidase CYP701A3, a multifunctional cytochrome P450 from gibberellin biosynthesis. Biochem J. 2010;431(3):337–344. doi: 10.1042/BJ20100597. [DOI] [PubMed] [Google Scholar]
- 14.Cyr A, Wilderman PR, Determan M, Peters RJ. A modular approach for facile biosynthesis of labdane-related diterpenes. J Am Chem Soc. 2007;129(21):6684–6685. doi: 10.1021/ja071158n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kitaoka N, Lu X, Yang B, Peters RJ. The application of synthetic biology to elucidation of plant mono-, sesqui-, and diterpenoid metabolism. Mol Plant. 2015;8(1):6–16. doi: 10.1016/j.molp.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Urban P, Mignotte C, Kazmaier M, Delorme F, Pompon D. Cloning, yeast expression, and characterization of the coupling of two distantly related Arabidopsis thaliana NADPH-cytochrome P450 reductases with P450 CYP73A5. J Biol Chem. 1997;272(31):19176–19186. doi: 10.1074/jbc.272.31.19176. [DOI] [PubMed] [Google Scholar]
- 17.Swaminathan S, Morrone D, Wang Q, Fulton DB, Peters RJ. CYP76M7 is an ent-cassadiene C11α-hydroxylase defining a second multifunctional diterpenoid biosynthetic gene cluster in rice. Plant Cell. 2009;21(10):3315–3325. doi: 10.1105/tpc.108.063677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin VJJ, Pitera DJ, Withers ST, Newman JD, Keasling JD. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotechnol. 2003;21(7):796–802. doi: 10.1038/nbt833. [DOI] [PubMed] [Google Scholar]
- 19.Wu Y, Wang Q, Hillwig ML, Peters RJ. Picking sides: Distinct roles for CYP76M6 and CYP76M8 in rice oryzalexin biosynthesis. Biochem J. 2013;454(2):209–216. doi: 10.1042/BJ20130574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmelz EA, et al. Biosynthesis, elicitation and roles of monocot terpenoid phytoalexins. Plant J. 2014;79(4):659–678. doi: 10.1111/tpj.12436. [DOI] [PubMed] [Google Scholar]
- 21.Anisimova M, Liberles D. Dectecting and understanding natural selection. In: Cannarozzi G, Schneider A, editors. Codon Evolution: Mechanisms and Models. Oxford University Press; New York: 2012. [Google Scholar]
- 22.Anarat-Cappillino G, Sattely ES. The chemical logic of plant natural product biosynthesis. Curr Opin Plant Biol. 2014;19:51–58. doi: 10.1016/j.pbi.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tawfik DS. Accuracy-rate tradeoffs: How do enzymes meet demands of selectivity and catalytic efficiency? Curr Opin Chem Biol. 2014;21:73–80. doi: 10.1016/j.cbpa.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 24.Weng JK, Philippe RN, Noel JP. The rise of chemodiversity in plants. Science. 2012;336(6089):1667–1670. doi: 10.1126/science.1217411. [DOI] [PubMed] [Google Scholar]
- 25.Morrone D, et al. Increasing diterpene yield with a modular metabolic engineering system in E. coli: Comparison of MEV and MEP isoprenoid precursor pathway engineering. Appl Microbiol Biotechnol. 2010;85(6):1893–1906. doi: 10.1007/s00253-009-2219-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Q, et al. Characterization of CYP76M5-8 indicates metabolic plasticity within a plant biosynthetic gene cluster. J Biol Chem. 2012;287(9):6159–6168. doi: 10.1074/jbc.M111.305599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Q, Hillwig ML, Peters RJ. CYP99A3: Functional identification of a diterpene oxidase from the momilactone biosynthetic gene cluster in rice. Plant J. 2011;65(1):87–95. doi: 10.1111/j.1365-313X.2010.04408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu Y, Hillwig ML, Wang Q, Peters RJ. Parsing a multifunctional biosynthetic gene cluster from rice: Biochemical characterization of CYP71Z6 & 7. FEBS Lett. 2011;585(21):3446–3451. doi: 10.1016/j.febslet.2011.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 31.Bielawski JP. 2013. Detecting the signatures of adaptive evolution in protein-coding genes. Curr Protoc Mol Biol Chapter 19:Unit 19.1.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.