Significance
The incredible diversity of vivid color patterns in natural organisms (e.g., butterfly wings, tropical fish skins, angiosperm flowers) has fascinated generations of biologists. Yet, neither the molecular mechanisms responsible for the color pattern formation nor the genetic bases underlying natural color pattern variation are well understood. In this study we have identified a causal gene underlying spatial pattern variation of floral pigments between two Mimulus (monkeyflowers) species: the bumblebee-pollinated Mimulus lewisii and the hummingbird-pollinated Mimulus cardinalis. We demonstrate that the competition between anthocyanin and flavonol biosynthesis sets a “prepattern” of pigment distribution in M. lewisii, producing a white region surrounding the throat of the otherwise pink corolla. This is likely to be a common mechanism underlying flower color patterns.
Keywords: flower color pattern, Mimulus, anthocyanins, flavonols, R2R3-MYB
Abstract
Flower color patterns have long served as a model for developmental genetics because pigment phenotypes are visually striking, yet generally not required for plant viability, facilitating the genetic analysis of color and pattern mutants. The evolution of novel flower colors and patterns has played a key role in the adaptive radiation of flowering plants via their specialized interactions with different pollinator guilds (e.g., bees, butterflies, birds), motivating the search for allelic differences affecting flower color pattern in closely related plant species with different pollinators. We have identified LIGHT AREAS1 (LAR1), encoding an R2R3-MYB transcription factor, as the causal gene underlying the spatial pattern variation of floral anthocyanin pigmentation between two sister species of monkeyflower: the bumblebee-pollinated Mimulus lewisii and the hummingbird-pollinated Mimulus cardinalis. We demonstrated that LAR1 positively regulates FLAVONOL SYNTHASE (FLS), essentially eliminating anthocyanin biosynthesis in the white region (i.e., light areas) around the corolla throat of M. lewisii flowers by diverting dihydroflavonol into flavonol biosynthesis from the anthocyanin pigment pathway. FLS is preferentially expressed in the light areas of the M. lewisii flower, thus prepatterning the corolla. LAR1 expression in M. cardinalis flowers is much lower than in M. lewisii, explaining the unpatterned phenotype and recessive inheritance of the M. cardinalis allele. Furthermore, our gene-expression analysis and genetic mapping results suggest that cis-regulatory change at the LAR1 gene played a critical role in the evolution of different pigmentation patterns between the two species.
Many flowers display interesting color patterns (e.g., spots, stripes, picotees, bull’s-eyes) that are precisely programmed during development. Numerous studies have shown that these color patterns are critically important for plant–pollinator interactions (1–8). Among the most captivating examples are deceptive orchids that display floral pigment patterns remarkably similar to female bees or wasps to lure male counterparts for pseudocopulation, thereby achieving pollination (9–11). Despite the obvious aesthetic and ecological significance of these flower color patterns, the molecular mechanisms of pigment pattern formation is not well understood, nor is the genetic basis underlying pattern variation between related species in nature.
From a genetic and developmental viewpoint, the most extensively studied flower color pattern is venation. Studies in snapdragon (Antirrhinum majus) and petunia (Petunia hybrida) have revealed a conserved mechanism for the formation of vein-associated anthocyanin pigmentation pattern in petal epidermis. Pigments are only produced in the overlapping expression domains of the R2R3-MYB and bHLH coregulators of anthocyanin biosynthetic genes; the bHLH expression is confined to the petal epidermis and the R2R3-MYB expression is specific to cells above the vascular bundles (8, 12, 13). However, the molecular basis of variation in this color pattern among natural species is less clear, although in the case of Antirrhinum, the R2R3-MYB, Venosa, was implicated as the causal gene, explaining much of the natural variation (8, 12). Another interesting pigmentation pattern that has been investigated is that of petal spots. Genetic and developmental analyses of petal spot formation in Gorteria diffusa (14), Clarkia gracilis (15), and Lilium spp. (16) have suggested potential genes and mechanisms underlying spot formation, but the lack of functional analyses in these systems has so far prevented a deeper understanding of the precise molecular bases and developmental mechanisms, a prerequisite for understanding the evolution of petal spot variation among species.
One way to simultaneously address how flower color patterns are generated during the development of an individual plant, and how these patterns are diversified among species, is to genetically map the causal locus (or loci) underlying pattern variation between closely related species, and then examine how different alleles generate different patterns during development. Using this approach we have analyzed a spatial pattern variation of anthocyanin pigmentation between two sister species of Mimulus.
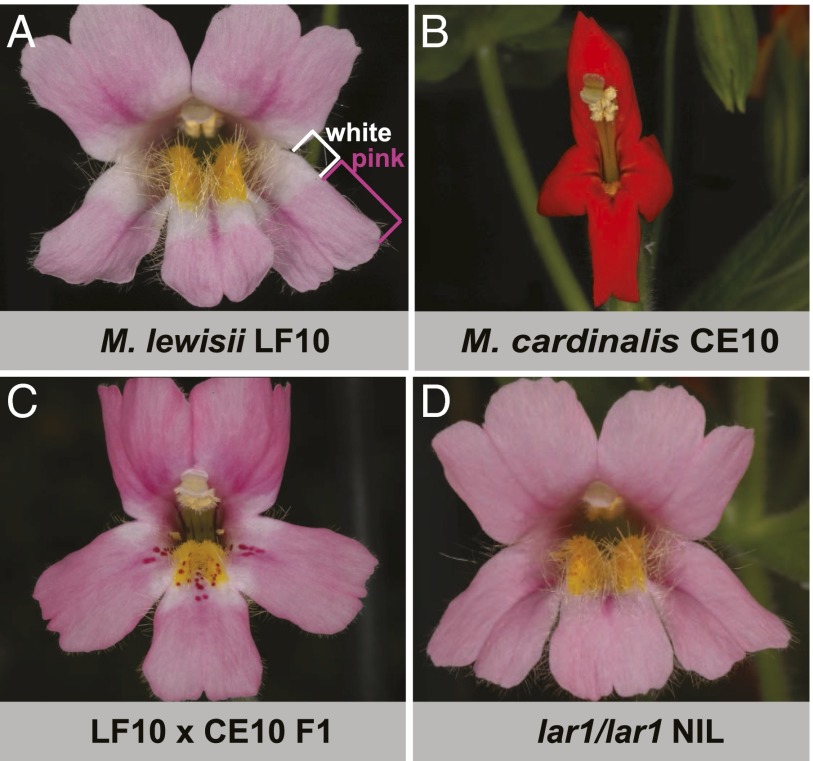
The bumblebee-pollinated Mimulus lewisii bears pink flowers colored by anthocyanins (except the yellow nectar guides as a result of carotenoid pigmentation) (Fig. 1A), whereas the hummingbird-pollinated Mimulus cardinalis produces red flowers as a result of a combination of anthocyanins and carotenoids (Fig. 1B). The petal lobes of M. lewisii display an interesting spatial pattern of anthocyanin pigmentation that is distinct from that seen in M. cardinalis: a white region surrounding the throat of the otherwise pink corolla (Fig. 1A). This is a common floral pigment pattern characterized as marginal picotee, “in which proximal and distal parts of the petals show different colors and the whole flower shows a central spot of one color, encircled by a ring of another” (17). This phenotype has been studied by Hiesey et al. (18) and the white regions surrounding the corolla throat were named “light areas.” The presence vs. absence of the pattern was postulated to be controlled by a single Mendelian locus (18), which we have called LIGHT AREAS1 (LAR1) (18). The M. lewisii allele is dominant, with F1 hybrids between M. lewisii and M. cardinalis showing the light area phenotype (Fig. 1C). Despite being a relatively subtle pattern, the light areas play an important role in bumblebee pollination, as the lack of light areas in a chemically induced M. lewsii floral mutant was shown to significantly decrease bumblebee visitation rate (7).
Fig. 1.
Flower phenotypes. (A) Mimulus lewisii inbred line LF10. The petal lobe comprises a proximal white region (i.e., the light area) and a distal pink region, as marked by the white and pink brackets, respectively. (B) M. cardinalis inbred line CE10. (C) F1 between LF10 and CE10. (D) A lar1/lar1 NIL in the LF10 background, lacking the light areas.
Here we show that the LAR1 locus encodes an R2R3-MYB transcription factor that regulates the expression of FLAVONOL SYNTHASE (FLS). The dominant LAR1 allele in M. lewisii causes high expression of FLS in the light areas relative to the pink areas, which presumably redirects metabolite flux from anthocyanin biosynthesis to the production of colorless flavonols, thereby explaining the lack of pink color in the light areas. The recessive lar1 allele in M. cardinalis has very low expression level in the petal lobe because of a cis-regulatory difference, which leads to a low level of FLS expression in the petal lobe and a consequent absence of the spatial patterning.
Results and Discussion
Creation of a High-Resolution Near-Isogenic Line.
In addition to the presence vs. absence of the light areas, F2 hybrids between M. lewisii and M. cardinalis display a wide range of pigment composition and intensity (18). To make phenotype scoring more straightforward and to facilitate downstream gene expression analysis, we created a high-resolution lar1/lar1 near-isogenic line (NIL) in the M. lewisii LF10 genetic background. The recessive M. cardinalis CE10 lar1 allele was introgressed into LF10 through four rounds of backcrossing and selfing, with phenotype-based selection; after each selfing event, a single individual lacking the light areas but otherwise most similar to LF10 was selected for the next round of backcrossing. The resulting BC4S1 lar1/lar1 NIL closely resemble LF10, except in the light area trait (Fig. 1D).
Genetic Mapping of LAR1.
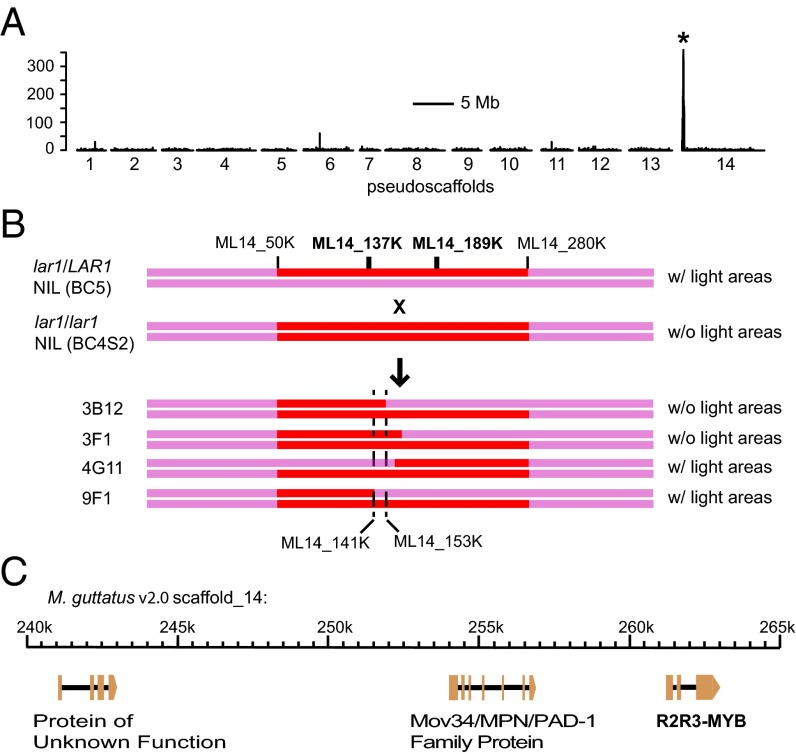
To determine the size and location of the chromosome segment introgressed from CE10 to LF10 and to ultimately identify the LAR1 gene, we generated a fine-mapping population by crossing a BC5 lar1/LAR1 heterozygous plant and a BC4S2 lar1/lar1 homozygous plant (Fig. 2). The offspring segregated 1:1 for the dominant (presence of light areas) and recessive (absence of light areas) flower phenotypes. We pooled DNA samples from 100 lar1/lar1 homozygous offspring and performed a bulk segregant analysis by deep sequencing (Materials and Methods). Based on the homozygous SNP profile after aligning the bulk segregant reads to the LF10 reference genome, the introgressed chromosome segment from CE10 to LF10 was determined to be ∼400 kb on pseudoscaffold 14 (between marker ML14_50K and ML14_280K) (Fig. 2 A and B). Examination of recombination events using the short read data from the 100 lar1/lar1 individuals further narrowed the candidate interval to a smaller region between ML14_137K and ML14_189K (∼100 kb).
Fig. 2.
Genetic mapping of the LAR1 gene. (A) Genome scan for regions that are enriched in homozygous SNPs after aligning the bulk segregant reads to the LF10 reference genome. The y axis indicates the number of homozygous SNPs in each 20-kb window. The peak (indicated by the asterisk) corresponds to an interval between marker ML14_50K and ML14_280K. (B) Cross design to generate the fine-scale mapping population, and the four most informative recombinants that reduced the candidate interval to a small region between markers ML14_141K and ML14_153K. Flower color phenotypes of the parental and recombinant lines are shown on the right. (C) The candidate region corresponds to an ∼25-kb, three-gene interval on M. guttatus v2.0 scaffold 14.
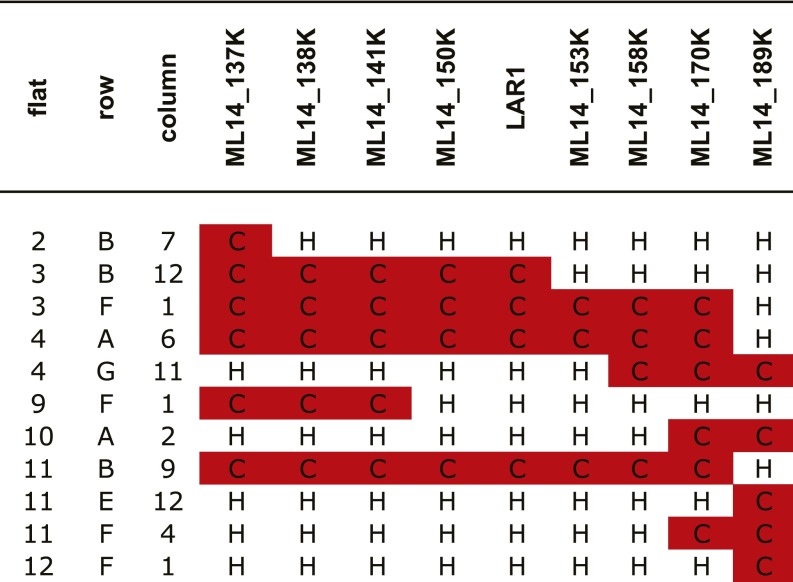
To refine the LAR1 locus, we genotyped 1,664 additional offspring from the “BC5 (lar1/LAR1) × BC4S2 (lar1/lar1)” cross. Eleven individuals had recombination between marker ML14_137K and ML14_189K. Further genotyping of the 11 recombinants using additional markers reduced the LAR1 candidate interval to a 25-kb region containing only three genes (Fig. 2 B and C and Fig. S1). One of the recombinants, 3B12 (Fig. 2B), was selected to generate a lar1/lar1 homozygous NIL for subsequent transgenic experiments and gene expression analysis.
Fig. S1.
Fine-scale recombination mapping that located LAR1 to the interval between ML14_141K and ML14_153K, which contains only three genes. H: heterozygous for LF10 and CE10; C: homozygous for CE10.
The LAR1 Locus Encodes an R2R3-MYB Transcription Factor.
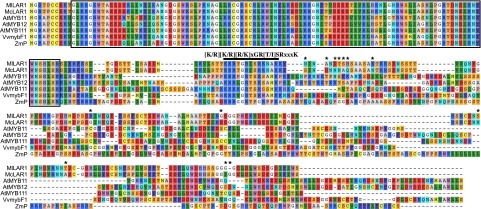
One of the three genes in the 25-kb interval encodes a subgroup-7 R2R3-MYB, defined by a signature motif (“[K/R][K/R][R/K]xGR[T/I]SRxxxK”) downstream of the R2R3 MYB domain (Figs. 2C and 3). This subgroup of MYB transcription factors is known to regulate flavonol biosynthesis in Arabidopsis, grapevine, and tomato (19–22), and regulate the biosynthesis of 3-deoxyflavonoids and phlobaphenes in maize (23). Both flavonol and anthocyanin biosynthesis require the same intermediate substrate, dihydroflavonol. The enzyme flavonol synthase (FLS) converts this substrate to colorless flavonols, whereas the enzyme dihydroflavonol 4-reductase (DFR) directs it to the anthocyanin biosynthetic pathway. Interplay between flavonol and anthocyanin biosynthesis could potentially generate the observed spatial pattern of anthocyanin pigmentation, and therefore, this R2R3-MYB was considered the best candidate gene for LAR1.
Fig. 3.
Alignment of the candidate R2R3-MYB amino acid sequences of M. lewisii, M. cardinalis, and their homologs in other species. The boxed region is the conserved R2R3 MYB DNA binding domain. The bar above the alignment indicates the signature motif defining the subgroup-7 MYBs. Note that this motif is slightly different from the one defined by Stracke et al. (33), which was based on Arabidopsis sequences only. Arabidopsis sequences were retrieved from the TAIR site (www.arabidopsis.org/); the other two sequences were retrieved from GenBank (VvmybF1: FJ948477; ZmP: P27898). The 13-aa differences (marked by asterisks) between M. lewisii and M. cardinalis are all located in nonconserved regions.
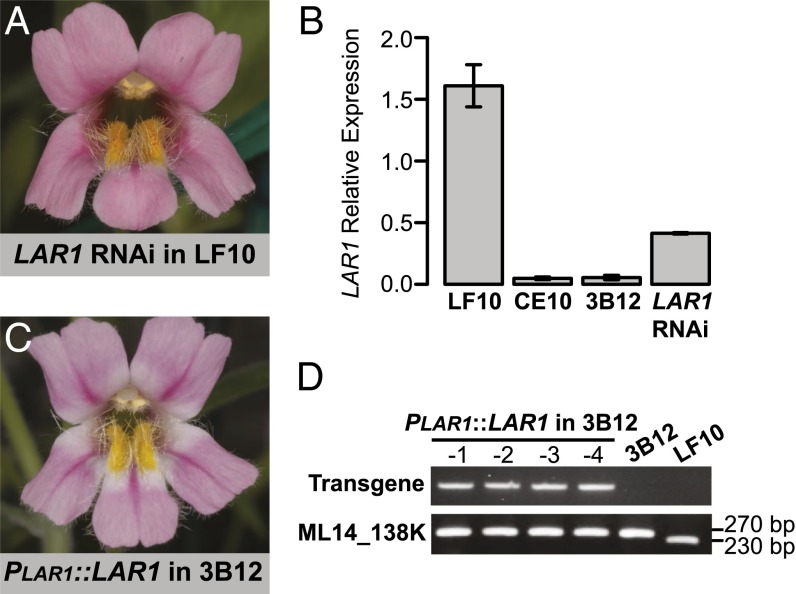
To verify the LAR1 gene identity, we performed two transgenic experiments: (i) Knocking down the expression of this R2R3-MYB in LF10 by RNA interference (RNAi) is expected to recapitulate the lar1/lar1 phenotype (i.e., lack of light areas). We obtained seven independent RNAi lines, five of which are indistinguishable from the lar1/lar1 NIL (Fig. 4A); the other two showed an intermediate phenotype. Quantitative RT-PCR (qRT-PCR) of two strong RNAi lines confirmed that the expression level of this R2R3-MYB was knocked down to ∼25% of the wild-type level in the petal lobes at the 10-mm corolla stage (Fig. 4B) (unless otherwise noted, “petal lobe” herein means the entire lobe, including both the pink and the white areas as marked in Fig. 1A, but does not include the corolla tube). We chose this developmental stage for qRT-PCR because the LAR1 expression level starts decreasing after 10-mm (Fig. S2A) and petal lobes are relatively easy to cut from the corolla at this stage. (ii) Introducing the dominant LF10 allele into the lar1/lar1 NIL should restore the light areas. We built a rescue construct that contains ∼1.5-kb upstream sequence from the ATG initiation codon and the full-length gene (including all three exons and the two introns but excluding 3′UTR). Four independent 3B12 plants were transformed with the rescue construct and the light area phenotype was fully rescued in these transgenic lines (Fig. 4C). Genotyping these lines showed that they all contain the transgene and are in the lar1/lar1 genetic background (Fig. 4D). Taken together, these results strongly suggest that this subgroup-7 R2R3-MYB is indeed the causal gene underlying the LAR1 locus.
Fig. 4.
Transgenic characterization of the R2R3-MYB gene. (A) A strong LAR1 RNAi line in the LF10 background phenocopies the lar1/lar1 NIL. (B) qRT-PCR of LAR1 in the petal lobes of LF10, CE10, 3B12 NIL, and two RNAi lines at the 10-mm corolla stage. MlUBC was used as the reference gene. Error bars represent 1 SD from three biological replicates (except LAR1 RNAi, which had only two biological replicates). (C) The 3B12 plants transformed with a genomic copy of the dominant LF10 LAR1 allele show the light area phenotype. (D) Molecular validation of the rescue lines, which have the lar1/lar1 genotype and contain the transgene.
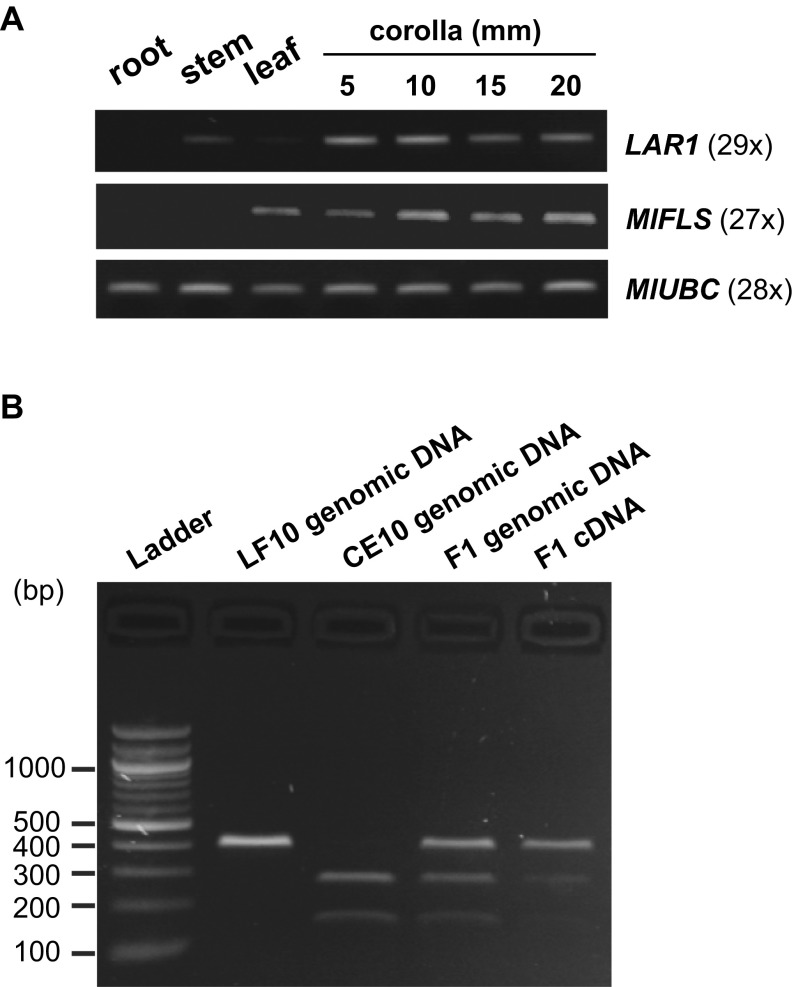
Fig. S2.
Gene expression analyses of LAR1 and MlFLS. (A) RT-PCR of LAR1 and MlFLS in various tissue types and four stages of corolla development. In this case, whole corollas (including the corolla tube and the petal lobes) were used. LAR1 expression is relatively high in early developmental stages and starts decreasing after 10-mm. MlFLS expression is relatively low in early corolla and becomes strong at 10-mm and later stages. MlUBC was used as a reference gene to control for cDNA concentration and quality. MlFLS expression is detectable in the leaf but LAR1 is not, indicating that MlFLS may be regulated by other transcription factors in the leaf. (B) Assay of relative expression level of the M. lewisii (LF10) and M. cardinalis (CE10) LAR1 alleles in the F1 using allele-specific restriction enzyme digestion. A 400-bp fragment in the third exon of LAR1 was amplified (29 PCR cycles) from the F1 genomic DNA and petal lobe cDNA (10-mm stage) using the primer pair 5′-TCAGGTCCATTACAGGTGACTC-3′ and 5′-TGTTTTCRCATCGAGACCTCAC-3′. The restriction enzyme AluI cuts the CE10 allele into two smaller fragments (270 bp and 130 bp), but does not cut the LF10 allele. The F1 genomic DNA has equal representation of the LF10 allele and the CE10 allele. Note that the smaller sizes of the digested CE10 bands accounts for their lower brightness compared with the undigested LF10 band (second to the last lane). For the F1 cDNA, the CE10 allele is barely detectable (last lane) compared with the LF10 allele, indicating that the latter is preferentially expressed in the F1. The LF10 and CE10 genomic DNA amplicons were used as control for complete restriction digestion.
Cis-Regulatory Change Causes LAR1 Allelic Difference.
Having identified the causal gene, we next asked: What is the molecular nature of the allelic difference underlying the phenotypic difference between M. lewisii LF10 and M. cardinalis CE10? We first compared the amino acid sequences of the LF10 and CE10 alleles. There are 13 amino acid substitutions between the two alleles (Fig. 3). However, all these substitutions are located in hypervariable regions (marked by asterisks in Fig. 3), and are therefore unlikely to have functional significance.
Alternatively, if the recessive M. cardinalis allele is the result of cis-regulatory change, the LAR1 mRNA level should be lower in M. cardinalis than in M. lewisii flowers. qRT-PCR experiments on the LAR1 gene in LF10, CE10, and 3B12 petal lobes at the 10-mm corolla stage strongly support this hypothesis. The LAR1 transcript level is ∼40-fold lower in CE10 than in LF10 (Fig. 4B). A similarly low level of LAR1 expression in the NIL background (Fig. 4B) suggests that the expression difference is unlikely to be caused by trans-acting factors located somewhere else in the genome. More importantly, the fine-mapping results suggest that whatever the causal mutations are, they must locate in the 25-kb candidate interval, thus ruling out trans-acting factors outside the 25-kb interval as potential causes of the gene expression change. Furthermore, allele-specific restriction enzyme digestion of a LAR1 gene fragment amplified from the F1 hybrid cDNA also showed preferential expression of the LF10 allele over the CE10 allele (Fig. S2B), indicating a promoter activity (i.e., cis-element) difference between LF10 and CE10.
Although we cannot completely rule out the possibility that the coding DNA changes may also play a role, our results from the RNAi experiment and gene-expression analyses allow us to conclude that cis-regulatory changes in the LAR1 gene are sufficient to explain the allelic difference between the two species. These results on the functional divergence of the LAR1 alleles between M. lewisii and M. cardinalis contribute to a rapidly growing body of evidence that support the critical role of cis-regulatory changes in phenotypic diversification (reviewed in refs. 24 and 25).
Interplay Between Flavonol and Anthocyanin Biosynthesis Underlies the Light Area Pattern Formation.
The allelic difference between M. lewisii and M. cardinalis suggests that expression of LAR1 is required to form the light areas around the corolla throat. However, what is the underlying mechanism for the pattern formation?
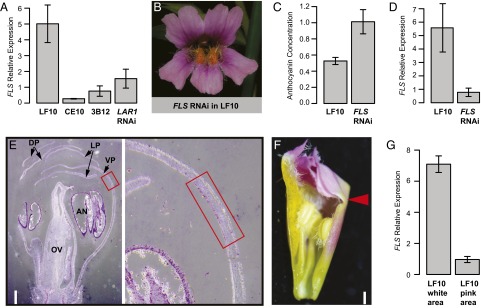
The lack of anthocyanins in the light areas indicates that LAR1 represses anthocyanin biosynthesis, but previous studies have repeatedly demonstrated that LAR1 homologs in other species function as transcriptional activators instead of repressors (19–23). However, if LAR1 positively regulates FLS expression—as its orthologs in Arabidopsis, tomato, and grapevine do (19–23)—and FLS competes with DFR for the limited supply of dihydroflavonol to produce colorless flavonols (26), the net effect would be repression of anthocyanin production by LAR1. To test this idea, we examined the relative expression level of FLS in the petal lobes (10-mm corolla stage) of LF10, CE10, the 3B12 lar1/lar1 NIL, and the LAR1 RNAi lines. qRT-PCR experiments revealed that FLS is expressed at significantly lower levels in CE10, 3B12, and LAR RNAi lines relative to LF10 (Fig. 5A), indicating that LAR1 is a positive regulator of FLS in Mimulus.
Fig. 5.
Molecular mechanism underlying the light area pattern formation. (A) LAR1 positively regulates FLS expression in the petal lobe, as shown by qRT-PCR of FLS in LF10, CE10, 3B12 NIL, and LAR1 RNAi lines at the 10-mm corolla stage. Error bars in all qRT-PCR panels (A, D, G) represent 1 SD from three biological replicates (except LAR1 RNAi lines, for which only two replicates were assayed). (B) RNAi knock-down of the FLS alone in LF10 is sufficient to restore anthocyanin pigments in the light areas. (C) Anthocyanin concentration in the petal lobe. One disk was punched from each dorsal petal with a 1.5-mL Eppendorf tube, anthocyanins were extracted with 0.3 mL of methanol/0.1% HCl, and absorbance was recorded at 525 nm. Error bars represent 1 SD from eight samples. (D) FLS mRNA abundance was knocked-down ∼sevenfold in the petal lobes of three FLS RNAi lines at the 10-mm corolla stage. (E) In situ hybridization shows that in the wild-type LF10, FLS is preferentially expressed in the light areas relative to the pink region. Red boxes mark the light area region (the transition zone between the corolla tube and limb, see F). (Left) Full view of a flower bud. (Scale bar, 0.5 mm; right panel is a 5× zoom-in of the light area region.) Significance of the FLS signal in the anthers is unclear. AN, anther; DP, dorsal petal; LP, lateral petal; OV, ovary; VP, ventral petal;. (F) A corolla bud with one of the lateral petals removed. The arrowhead marks the light area as the transition zone between the corolla tube and limb. (Scale bar, 1 mm.) (G) qRT-PCR of FLS in the white vs. pink portion of the LF10 petal lobes at the 15-mm corolla stage.
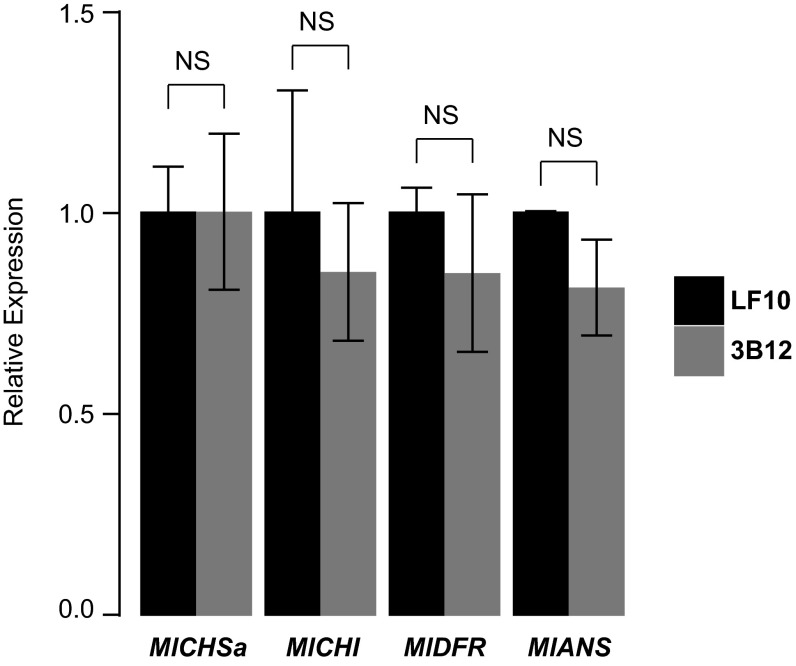
On the other hand, the low expression level of LAR1 in 3B12 does not seem to affect the expression of anthocyanin biosynthetic genes, such as Chalcone synthase (CHS), Chalcone isomerase (CHI), DFR, or Anthocyanidin synthase (ANS) (Fig. S3), which suggests that proper expression of FLS alone might be sufficient to generate the light area pattern. To test this hypothesis, we decided to knock down FLS expression in LF10. We obtained 16 independent FLS RNAi lines, all of which display a striking phenotype: the white region is completely filled with anthocyanins, and the overall petal lobe anthocyanin content is twice that of the wild-type (Fig. 5 B and C). qRT-PCR assay of three RNAi lines showed an average of ∼sevenfold down-regulation of FLS in the petal lobes of these RNAi lines compared with the wild-type at the 10-mm corolla stage (Fig. 5D).
Fig. S3.
qRT-PCR of four anthocyanin biosynthetic genes in the petal lobe of 3B12 (the lar1/lar1 NIL) compared with wild-type LF10 at the 10-mm corolla stage. Expression level was standardized to LF10. None of these genes shows significant difference between wild-type and 3B12 NIL (two-tailed t test: P > 0.1). Error bars represent 1 SD from three biological replicates.
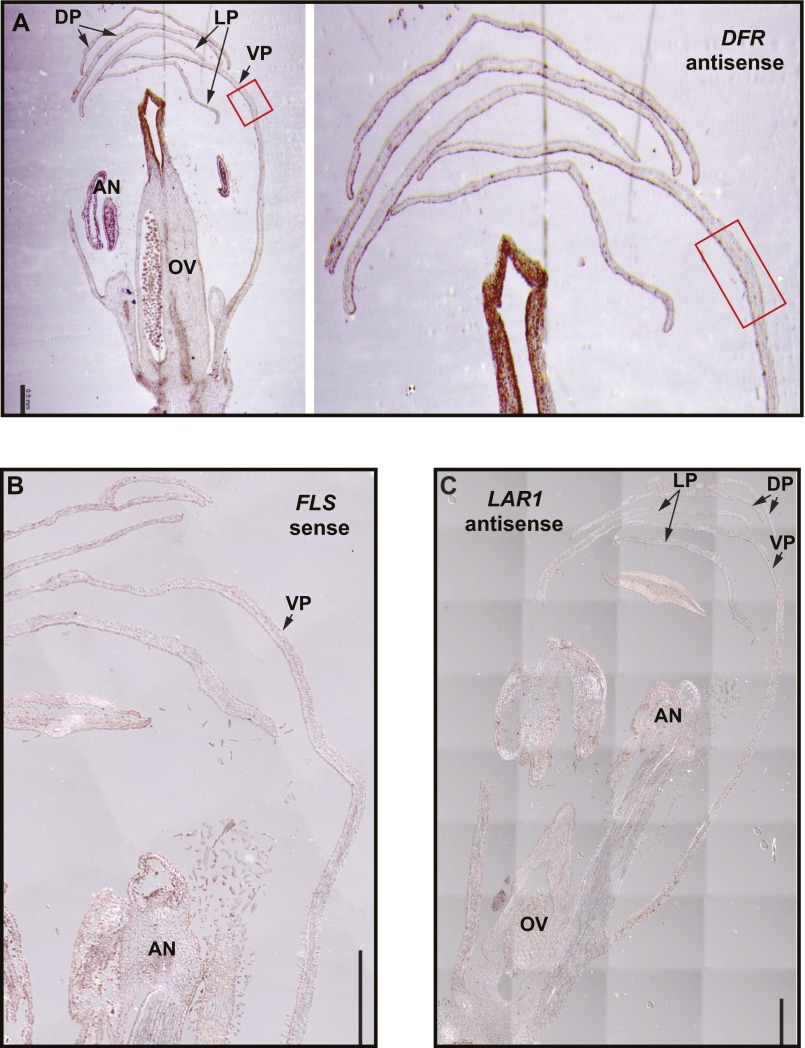
Having established that FLS is the key determinant of the pigment pattern, we reasoned that in the wild-type LF10, FLS should be expressed more strongly in the white region compared with the pink region of the petal lobe. To test this prediction, we examined spatial expression pattern of FLS in flower buds by in situ hybridization (7-mm corolla stage was used for the convenience of tissue fixation and embedding). FLS is indeed preferentially expressed in the light areas (Fig. 5 E and F) (the sense probe control does not display this pattern, as shown in Fig. S4). This expression pattern is corroborated by qRT-PCR using tissue dissected from the white vs. pink portion of the petal lobe at the 15-mm corolla stage (Fig. 5G) (for this experiment we chose the 15-mm stage because the light areas are too small to dissect at the 10-mm stage). In contrast, DFR expression level is not conspicuously different between the white and the pink region (Fig. S4). We also tried to examine the spatial pattern of LAR1 expression, but failed to detect any signal (Fig. S4), likely because of the relatively low expression level of LAR1 (it took at least two more cycles for LAR1 to reach a similar band intensity as FLS during RT-PCR) (Fig. S2).
Fig. S4.
In situ hybridizations using DFR antisense probe (A), FLS sense probe (B), and LAR1 antisense probe (C) at 7-mm corolla stage. Red boxes in A mark the light area region (the transition zone between the corolla tube and corolla limb) of the ventral petal. The right panel in A is a 3× zoom-in of the light area region. In contrast to the FLS expression pattern (Fig. 5E), DFR expression shows no conspicuous difference between the light areas and the pink areas. AN, anther; DP, dorsal petal; LP, lateral petal; OV, ovary; VP, ventral petal. (Scale bars, 0.5 mm.)
Our results highlight the importance of substrate competition in generating spatial patterns with color contrast (e.g., pink areas vs. white areas). The fact that knocking down FLS alone in M. lewisii can fully restore anthocyanin pigments in the light areas (Fig. 5B), shows that the expression of anthocyanin biosynthetic genes in the light areas must be sufficient to produce anthocyanins: it is the competition of flavonol biosynthesis that redirects the metabolic flux from anthocyanin biosynthesis into colorless flavonols, rendering an acyanic area. A similar mechanism was first proposed by Saito et al. (17, 27) to explain the floral pigment pattern in the Petunia cultivar “Baccara Rose Morn,” although no causal relationship between genotype and phenotype was established. In addition to Mimulus and Petunia, this particular type of marginal picotee (i.e., a central acyanic region in an otherwise cyanic corolla limb) is found in many other horticultural plants (e.g., Pacifica Burgundy Halo Vinca, Cherry Meidiland Rose, Drummond’s Phlox “Tapestry Mix,” Japanese Morning Glory “Cameo Elegance”) and natural species (e.g., Abronia umbellate, Geranium phaeum, Oenothera speciosa, Primula allionii). As such, the competition between flavonol and anthocyanin biosynthesis is likely to be a common mechanism underlying floral pigment patterning.
Finally, it should be noted that the FLS RNAi lines display more anthocyanins across the entire petal lobe compared with the wild-type (which is expected because there is low FLS expression in the pink region of the petal lobe as well) (Fig. 5G), whereas the lar1/lar1 NIL or LAR1 RNAi lines do not (Figs. 1D and 4A). These different outcomes suggest that FLS is unlikely to be the only target of LAR1 in the M. lewisii flower; LAR1 probably also regulates some other aspects of anthocyanin production, modification, or transport to fine-tune the final anthocyanin distribution.
The strategy of genetically mapping the causal gene and then molecularly characterizing the functions of different alleles potentially can be used to study pigment pattern variation (or any phenotypic variation for that matter) in many other systems. The results from such efforts will not only contribute to our understanding of pattern formation mechanisms, but also help to elucidate how these mechanisms are “tinkered” with during evolution to generate variation among species.
Materials and Methods
Plant Materials and Growth Condition.
The M. lewisii inbred line LF10, M. cardinalis inbred line CE10, and greenhouse growth conditions were described previously (28).
Bulk Segregant Analysis by Deep Sequencing.
A BC4S1 individual homozygous for the M. cardinalis allele (lar1/lar1) and, therefore, lacking the light areas (but otherwise closely resembling LF10) was backcrossed to LF10 to generate a BC5 (lar1/LAR1) plant. The fine-mapping population was produced from the cross between the BC5 (lar1/LAR1) and BC4S2 (lar1/lar1), segregating 1:1 for the dominant (with light areas) and recessive (without light areas) flower phenotype.
For the bulk segregant analysis, we grew 250 plants to flowering and sampled 100 segregants without light areas (lar1/lar1). Total genomic DNA was isolated from each of the 100 samples using the BIO 101 System FastDNA kit (Qbiogene), and then pooled together with equal representation from each sample. A small-insert library (∼420 bp) was prepared for the pooled sample, and 100-bp paired-end reads were generated by an Illumina HiSEq. 2000.
We reasoned that if we align the Illumina short reads back to the LF10 reference genome, the vast majority of the genome should be devoid of authentic SNPs (some artifactual “noisy” SNPs may exist as a result of assembly error in the reference genome or nonspecific mapping of the short reads), because these regions are already homozygous for the LF10 allele in both parental lines (i.e., BC5 and BC4S2). In contrast, in the chromosome segment introgressed from CE10 to LF10, there should be many SNPs because of the difference between LF10 and CE10 alleles. In addition, most of these SNPs should have a near 100% frequency (i.e., homozygous for the CE10 allele), because the 100 segregants selected for sequencing all have the lar1/lar1 phenotype. To produce this SNP profile, we first assembled the LF10 genome contigs (28) into 14 “pseudoscaffolds” by aligning the contigs to the 14 chromosomal-level superscaffolds of Mimulus guttatus (29) (phytozome.jgi.doe.gov/pz/#!info?alias=Org_Mguttatus), as described in Yuan et al. (30). The ∼215 million Illumina short reads (∼40-fold average coverage; NCBI SRA274748) were mapped to the 14 LF10 “pseudoscaffolds” with CLC Genomics Workbench 5.0, with the minimum read-length fraction set to 0.8 and minimum similarity set to 0.9. A total of 6,863 homozygous SNPs (100% SNP frequency) were detected. To search for regions that are highly enriched in homozygous SNPs, the LF10 “pseudoscaffolds” were binned into 20-kb intervals, and the numbers of homozygous SNPs in each 20-kb interval were plotted in a bar graph (Fig. 2A).
Fine-Scale Genetic Mapping.
Very young seedlings from the fine-mapping population “BC5 (lar1/LAR1) x BC4S2 (lar1/lar1)” were genotyped at the flanking markers ML14_137K and ML14_189K (∼100 kb) by direct PCR of 0.5-mm leaf punches using Finnzyme’s Phire Plant Direct PCR Kit (Thermo Scientific). Recombinants (n = 11) were transplanted and grown to flowering for phenotyping and further genotyping using six additional markers (Table S1).
Table S1.
Primers used for fine mapping
| Primer | Sequence (5′-3′) | LF10 amplicon (bp) | CE10 amplicon (bp) |
| ML14_137K_F1 | GCTTATGCAAGTGGCTGTGTTATC | 190 | 155 |
| ML14_137K_R1 | CTTGGCTTCCTCCATCATATTGAC | ||
| ML14_138K_F1 | TGGAAGCACCGTGAGTCATGTCATATC | 230 | 270 |
| ML14_138K_R1 | GTGTTCTTCAATGTATATGGAGCAGAG | ||
| ML14_141K_F1 | CCCAAGATTCCGGCTGATTACGT | 190 | 155 |
| ML14_141K_R1 | GTCCCTTTGATTTGCAGGCCGTT | ||
| ML14_150K_F1 | CTCATTCGATTGGAACGAGCTC | 230 | 160 |
| ML14_150K_R1 | ATGGCTGCTGGAAACCAAGACCAA | ||
| ML14_153K_F1 | AGATGTCGGAGCGATATTCAAC | 190 | 220 |
| ML14_153K_R1 | ATGGGATACTGGTCGCCGGCGAAGA | ||
| ML14_158K_F1 | GGCTTGGACTCTGCAGTCTGT | 93 | 120 |
| ML14_158K_R1 | TTCATCGTTGTTTTCCGACGAC | ||
| ML14_170K_F1 | AGCGATGAAGTAATAGTAACGAC | 340 | 280 |
| ML14_170K_R1 | GCAGTACTGCCACCTGCTACAA | ||
| ML14_189K_F1 | ATGAGTGACAGTACTCCGGTCA | 440 | 270 |
| ML14_189K_R1 | GACGGAAGATGGGATATTGGTC |
Plasmid Construction and Plant Transformation.
RNAi plasmids were constructed with a 193-bp and 343-bp fragment amplified from the coding regions of LAR1 and MlFLS, respectively, following the protocol described in Yuan et al. (28). To ensure target specificity, the fragment included in each RNAi plasmid was BLASTed against the LF10 genome assembly with an E-value cut-off of 0.1 so that no other genomic regions perfectly match this fragment for a contiguous block longer than 16 bp. The PLAR1::LAR1 rescue plasmid was constructed by cloning the 1,674-bp full-length LF10 LAR1 genomic DNA (without the stop codon) and 1,483-bp upstream regulatory sequence into the pEarleyGate 302 vector, following Earley et al. (31). This vector does not contain any built-in promoter sequences. Expression of the LAR1 transgene was entirely driven by the 1,483-bp upstream regulatory sequence. Primers used for amplifying the corresponding DNA fragments are listed in Table S2.
Table S2.
Primers used for plasmid constructions
| Primer | Sequence (5′-3′) | Fragment size (bp) |
| LAR1_RNAi_F | GTTCTAGACCATGGAGGTGACTCCACCACACTCTCTG | 193 |
| LAR1_RNAi_R | GTGGATCCGGCGCGCCACCCTCGTTTTGCACCACAGTTGT | |
| FLS_RNAi_F | GTTCTAGACCATGGGTATCCCAAATGAAGTCATTTCA | 343 |
| FLS_RNAi_R | GTGGATCCGGCGCGCCGCCACCTTCAATTCTTGTTCTT | |
| LAR1_ProF1 | CACCTTTGTGAGGACAACAACACTGC | 3,157 (promoter: 1,483; gene: 1,674) |
| LAR1_cdsR | CGACCAAATGAAGATATCACCTTC |
The sequences in bold contain the restriction sites. The sequence underlined (“CACC”) is the 4-bp sequence necessary for pENTR/D-TOPO cloning.
The final plasmid constructs were verified by sequencing and then transformed into Agrobacterium tumefaciens strain GV3101 for subsequent plant transformation, as described in Yuan et al. (28).
Expression Analyses by RT-PCR.
Total RNA was isolated using the Spectrum Plant Total RNA Kit (Sigma-Aldrich) and then treated with amplification grade DNaseI (Invitrogen). cDNA was synthesized from 1 μg of the DNase-treated RNA using the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen), then diluted 10-fold before PCR. The M. lewisii ortholog of Arabidopsis thaliana ubiquitin-conjugating enzyme gene (At5g25760), MlUBC, was used as a reference gene as described in Yuan et al. (28). qRT-PCR was performed using iQ SYBR Green Supermix (Bio-Rad) in a CFX96 Touch Real-Time PCR Detection System (Bio-Rad). Samples were amplified for 40 cycles of 95 °C for 15 s and 60 °C for 30 s. Reactions were run with three biological replicates except the LAR1 RNAi lines, for which only two replicates (two independent lines) were analyzed. Amplification efficiencies for each primer pair were determined using critical threshold values obtained from a dilution series (1:4, 1:20, 1:100, 1:500). Gene-specific primers used for RT-PCR are listed in Table S3.
Table S3.
RT-PCR primers used in this study
| Primer | Forward (5′-3′) | Reverse (5′-3′) |
| LAR1 | TCAACGAGCAAGAATTTAGCAG | CGACCAAATGAAGATATCACCTTC |
| MlFLS | CTTATCATACACATTGGCGACCA | CAAGACATCCTCGTACTCTCCT |
| MlCHS | GCACCGTCCTCCGCATGGCCAAG | TGCCCGACGAGACTGTCCAAATG |
| MlCHI | AAACTTCCCGCCTGGGAGCTCCA | TACACCATGCCTCCCAATGATCGA |
| MlDFR | TCGAGGATCCCACAGCACAAGGA | TGGCTTCTCTAAACATGTCCTCCA |
| MlANS | CCCAAGATTCCGGCTGATTACGT | TCGTGTTCTAATCCTAGTCCGGT |
| MlUBC | GGCTTGGACTCTGCAGTCTGT | TCTTCGGCATGGCAGCAAGTC |
In Situ Hybridization.
Flower buds (with calyx removed) were fixed in PBS pH 7.5 containing 4% (wt/vol) paraformaldehyde overnight at 4 °C. After dehydration, samples were embedded in paraffin (VWR) using a Tissue-TEK VIP processor (Sakura). Next, 8-µm sections were mounted on polysine slides, dewaxed in histoclear, and hydrated through a decreasing ethanol series. RNA in situ hybridization of slides was carried out as described in Coen et al. (32). Riboprobes against FLS, DFR, and LAR1 were synthesized by firstly amplifying each fragment using primers in Table S4 and cloning each fragment into pCR4-TOPO TA vector (Life Technologies) following the manufacturer’s instructions. The cloned fragments were amplified using a forward specific primer and the M13F primer (pCR4-TOPO kit). Antisense probes were obtained by RNA transcription using the T7 promoter (Roche) and DIG-UTP (Roche), according to the manufacturer’s instructions. The probes were hydrolyzed for 60 min at 60 °C using a 200 mM carbonate buffer pH 10.2 solution.
Table S4.
Primers used for amplifying in situ hybridization probes
| Primer | Sequence (5′-3′) | Fragment size (bp) |
| MlFLS_insitu_F | AGGATCGATAAACGAAGCTAG | 527 |
| MlFLS_insitu_R | CGTTAGGTACGAGGATTGTGA | |
| MlDFR_insitu_F | ATGGATTTCGAATCCGAAGATC | 557 |
| MlDFR_insitu_R | TCAGGCCATTTGTCTCTGATC | |
| LAR1_insitu_F | AGGTGACTCCACCACACTCTCTG | 441 |
| LAR1_insitu_R | GTGGTCCCCACAATCCGTATTGTT |
Acknowledgments
We thank Brian Watson, Doug Ewing, Jeanette Milne, Paul Beeman, Clinton Morse, and Matt Opel for extraordinary plant care in the University of Washington and University of Connecticut greenhouses; Prof. Enrico Coen for providing the experimental environment and support for A.B.R to carry out the in situ hybridizations; and two anonymous reviewers for valuable comments on the manuscript. This work was supported by National Science Foundation Frontiers in Integrative Biological Research Grant 0328636 (to H.D.B.); National Institutes of Health Grant 5R01GM088805 (to H.D.B.); and University of Connecticut Start-up funds (Y.-W.Y.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper has been deposited in the NCBI Short Read Archive (accession no. SRA274748) and the GenBank database (accession nos. KT224453–KT224456).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1515294113/-/DCSupplemental.
References
- 1.Johnson S, Midgley J. Fly pollination of Gorteria diffusa (Asteraceae), and a possible mimetic function for dark spots on the capitulum. Am J Bot. 1997;84(4):429. [PubMed] [Google Scholar]
- 2.Medel R, Botto-Mahan C, Kalin-Arroyo M. Pollinator-mediated selection on the nectar guide phenotype in the Andean monkey flower, Mimulus luteus. Ecology. 2003;84(7):1721–1732. [Google Scholar]
- 3.Lunau K, Fieselmann G, Heuschen B, van de Loo A. Visual targeting of components of floral colour patterns in flower-naïve bumblebees (Bombus terrestris; Apidae) Naturwissenschaften. 2006;93(7):325–328. doi: 10.1007/s00114-006-0105-2. [DOI] [PubMed] [Google Scholar]
- 4.Shi J, et al. Pollination by deceit in Paphiopedilum barbigerum (Orchidaceae): A staminode exploits the innate colour preferences of hoverflies (Syrphidae) Plant Biol (Stuttg) 2009;11(1):17–28. doi: 10.1111/j.1438-8677.2008.00120.x. [DOI] [PubMed] [Google Scholar]
- 5.Gaskett AC. Orchid pollination by sexual deception: Pollinator perspectives. Biol Rev Camb Philos Soc. 2011;86(1):33–75. doi: 10.1111/j.1469-185X.2010.00134.x. [DOI] [PubMed] [Google Scholar]
- 6.Leonard AS, Papaj DR. ‘X’ marks the spot: The possible benefits of nectar guides to bees and plants. Funct Ecol. 2011;25(6):1293–1301. [Google Scholar]
- 7.Owen CR, Bradshaw HD. Induced mutations affecting pollinator choice in Mimulus lewisii (Phrymaceae) Arthropod-Plant Interact. 2011;5(3):235–244. [Google Scholar]
- 8.Shang Y, et al. The molecular basis for venation patterning of pigmentation and its effect on pollinator attraction in flowers of Antirrhinum. New Phytol. 2011;189(2):602–615. doi: 10.1111/j.1469-8137.2010.03498.x. [DOI] [PubMed] [Google Scholar]
- 9.Schiestl FP. On the success of a swindle: Pollination by deception in orchids. Naturwissenschaften. 2005;92(6):255–264. doi: 10.1007/s00114-005-0636-y. [DOI] [PubMed] [Google Scholar]
- 10.Streinzer M, Paulus HF, Spaethe J. Floral colour signal increases short-range detectability of a sexually deceptive orchid to its bee pollinator. J Exp Biol. 2009;212(Pt 9):1365–1370. doi: 10.1242/jeb.027482. [DOI] [PubMed] [Google Scholar]
- 11.Gaskett AC, Herberstein ME. Colour mimicry and sexual deception by Tongue orchids (Cryptostylis) Naturwissenschaften. 2010;97(1):97–102. doi: 10.1007/s00114-009-0611-0. [DOI] [PubMed] [Google Scholar]
- 12.Schwinn K, et al. A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. Plant Cell. 2006;18(4):831–851. doi: 10.1105/tpc.105.039255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Albert NW, et al. Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J. 2011;65(5):771–784. doi: 10.1111/j.1365-313X.2010.04465.x. [DOI] [PubMed] [Google Scholar]
- 14.Thomas MM, Rudall PJ, Ellis AG, Savolainen V, Glover BJ. Development of a complex floral trait: The pollinator-attracting petal spots of the beetle daisy, Gorteria diffusa (Asteraceae) Am J Bot. 2009;96(12):2184–2196. doi: 10.3732/ajb.0900079. [DOI] [PubMed] [Google Scholar]
- 15.Martins TR, Berg JJ, Blinka S, Rausher MD, Baum DA. Precise spatio-temporal regulation of the anthocyanin biosynthetic pathway leads to petal spot formation in Clarkia gracilis (Onagraceae) New Phytol. 2013;197(3):958–969. doi: 10.1111/nph.12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamagishi M, Toda S, Tasaki K. The novel allele of the LhMYB12 gene is involved in splatter-type spot formation on the flower tepals of Asiatic hybrid lilies (Lilium spp.) New Phytol. 2014;201(3):1009–1020. doi: 10.1111/nph.12572. [DOI] [PubMed] [Google Scholar]
- 17.Saito R, et al. Regulation of anthocyanin biosynthesis involved in the formation of marginal picotee petals in Petunia. Plant Sci. 2006;170(4):828–834. [Google Scholar]
- 18.Hiesey W, Nobs MA, Björkman O. 1971. Experimental Studies on the Nature of Species. V. Biosystematics, Genetics, and Physiological Ecology of the Erythranthe section of Mimulus. Carnegie Institute of Washington publication no. 628 (Carnegie Institute, Washington, DC)
- 19.Mehrtens F, Kranz H, Bednarek P, Weisshaar B. The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol. 2005;138(2):1083–1096. doi: 10.1104/pp.104.058032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stracke R, et al. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 2007;50(4):660–677. doi: 10.1111/j.1365-313X.2007.03078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo J, et al. AtMYB12 regulates caffeoyl quinic acid and flavonol synthesis in tomato: Expression in fruit results in very high levels of both types of polyphenol. Plant J. 2008;56(2):316–326. doi: 10.1111/j.1365-313X.2008.03597.x. [DOI] [PubMed] [Google Scholar]
- 22.Czemmel S, et al. The grapevine R2R3-MYB transcription factor VvMYBF1 regulates flavonol synthesis in developing grape berries. Plant Physiol. 2009;151(3):1513–1530. doi: 10.1104/pp.109.142059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grotewold E, Drummond BJ, Bowen B, Peterson T. The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell. 1994;76(3):543–553. doi: 10.1016/0092-8674(94)90117-1. [DOI] [PubMed] [Google Scholar]
- 24.Stern DL. Evolution, Development, and the Predictable Genome. Roberts and Co.; Greenwood Village, CO: 2011. [Google Scholar]
- 25.Martin A, Orgogozo V. The Loci of repeated evolution: A catalog of genetic hotspots of phenotypic variation. Evolution. 2013;67(5):1235–1250. doi: 10.1111/evo.12081. [DOI] [PubMed] [Google Scholar]
- 26.Davies KM, et al. Enhancing anthocyanin production by altering competition for substrate between flavonol synthase and dihydroflavonol 4-reductase. Euphytica. 2003;131(3):259–268. [Google Scholar]
- 27.Saito R, Kuchitsu K, Ozeki Y, Nakayama M. Spatiotemporal metabolic regulation of anthocyanin and related compounds during the development of marginal picotee petals in Petunia hybrida (Solanaceae) J Plant Res. 2007;120(4):563–568. doi: 10.1007/s10265-007-0086-z. [DOI] [PubMed] [Google Scholar]
- 28.Yuan Y-W, Sagawa JM, Young RC, Christensen BJ, Bradshaw HD., Jr Genetic dissection of a major anthocyanin QTL contributing to pollinator-mediated reproductive isolation between sister species of Mimulus. Genetics. 2013;194(1):255–263. doi: 10.1534/genetics.112.146852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hellsten U, et al. Fine-scale variation in meiotic recombination in Mimulus inferred from population shotgun sequencing. Proc Natl Acad Sci USA. 2013;110(48):19478–19482. doi: 10.1073/pnas.1319032110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan YW, Sagawa JM, Di Stilio VS, Bradshaw HD., Jr Bulk segregant analysis of an induced floral mutant identifies a MIXTA-like R2R3 MYB controlling nectar guide formation in Mimulus lewisii. Genetics. 2013;194(2):523–528. doi: 10.1534/genetics.113.151225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Earley KW, et al. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45(4):616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- 32.Coen ES, et al. floricaula: a homeotic gene required for flower development in Antirrhinum majus. Cell. 1990;63(6):1311–1322. doi: 10.1016/0092-8674(90)90426-f. [DOI] [PubMed] [Google Scholar]
- 33.Stracke R, Werber M, Weisshaar B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol. 2001;4(5):447–456. doi: 10.1016/s1369-5266(00)00199-0. [DOI] [PubMed] [Google Scholar]