Significance
The systematic discovery of new gene and drug combinations that modulate complex biological phenotypes and human diseases requires scalable and multiplexed screening technologies. We leverage the programmability of the CRISPR-Cas9 system for multiplexed targeting of specific genomic loci and the versatility of the combinatorial genetics en masse (CombiGEM) technology to rapidly assemble barcoded combinatorial genetic perturbation libraries that can be tracked with high-throughput sequencing. CombiGEM-CRISPR enables simple, massively parallel screening of barcoded combinatorial gene perturbations in human cells, and the translation of these hits into effective drug combinations. This approach is broadly applicable for performing pooled combinatorial genetic perturbations to map out how the orchestrated action of genes controls complex phenotypes and to translate these findings into novel drug combinations.
Keywords: CRISPR-Cas, CombiGEM, multifactorial genetics, genetic perturbations, high-throughput screening
Abstract
The orchestrated action of genes controls complex biological phenotypes, yet the systematic discovery of gene and drug combinations that modulate these phenotypes in human cells is labor intensive and challenging to scale. Here, we created a platform for the massively parallel screening of barcoded combinatorial gene perturbations in human cells and translated these hits into effective drug combinations. This technology leverages the simplicity of the CRISPR-Cas9 system for multiplexed targeting of specific genomic loci and the versatility of combinatorial genetics en masse (CombiGEM) to rapidly assemble barcoded combinatorial genetic libraries that can be tracked with high-throughput sequencing. We applied CombiGEM-CRISPR to create a library of 23,409 barcoded dual guide-RNA (gRNA) combinations and then perform a high-throughput pooled screen to identify gene pairs that inhibited ovarian cancer cell growth when they were targeted. We validated the growth-inhibiting effects of specific gene sets, including epigenetic regulators KDM4C/BRD4 and KDM6B/BRD4, via individual assays with CRISPR-Cas–based knockouts and RNA-interference–based knockdowns. We also tested small-molecule drug pairs directed against our pairwise hits and showed that they exerted synergistic antiproliferative effects against ovarian cancer cells. We envision that the CombiGEM-CRISPR platform will be applicable to a broad range of biological settings and will accelerate the systematic identification of genetic combinations and their translation into novel drug combinations that modulate complex human disease phenotypes.
New therapeutic strategies are needed to treat complex human diseases. Because disease phenotypes are often regulated by interwoven genetic networks, exploiting combination therapy to target multiple pathways, as opposed to only single ones, can enhance treatment efficacy (1). However, discovering effective combination therapies for human diseases is challenging with existing methods, due to the cost, effort, and labor required to construct and analyze each combination (2). For example, the National Cancer Institute tested ∼5,000 pairwise combinations of 100 cancer drugs against the NCI-60 panel in a study that took 2 y and cost about USD $4 million (3). Thus, there is a need for technological advances to accelerate the identification of effective combinatorial therapies. Here, we used our combinatorial genetics en masse (CombiGEM)-CRISPR platform to perform rapid pooled screening of pairwise genetic knockouts against genes coding for epigenetic regulators and then translated our screen hits into drug combinations against human ovarian cancer cells.
CRISPR-Cas9 technology has been used for large-scale genetic perturbation screens with single-guide RNA (sgRNA) libraries for gene knockouts (4–7), repression, and activation (8, 9). Despite its simplicity for multiplexed genetic perturbations (10–12), new methods are needed to enable high-throughput CRISPR-Cas9–based screening with combinatorial sets of guide RNAs (gRNAs), which would be broadly useful for studying combinatorial gene functions in multigenic phenotypes and diseases. By using CombiGEM-based DNA assembly (13, 14), we developed a strategy for the simple and efficient assembly of barcoded combinatorial gRNA libraries. These libraries can be delivered into human cells by lentiviruses to create genetically ultradiverse cell populations harboring unique gRNA combinations that can be tracked via barcode sequencing in pooled assays. This strategy, termed CombiGEM-CRISPR, uses one-pot cloning steps to enable the assembly of combinatorial gRNA libraries, thus simplifying and accelerating the workflow toward systematic analysis of combinatorial gene functions.
Results
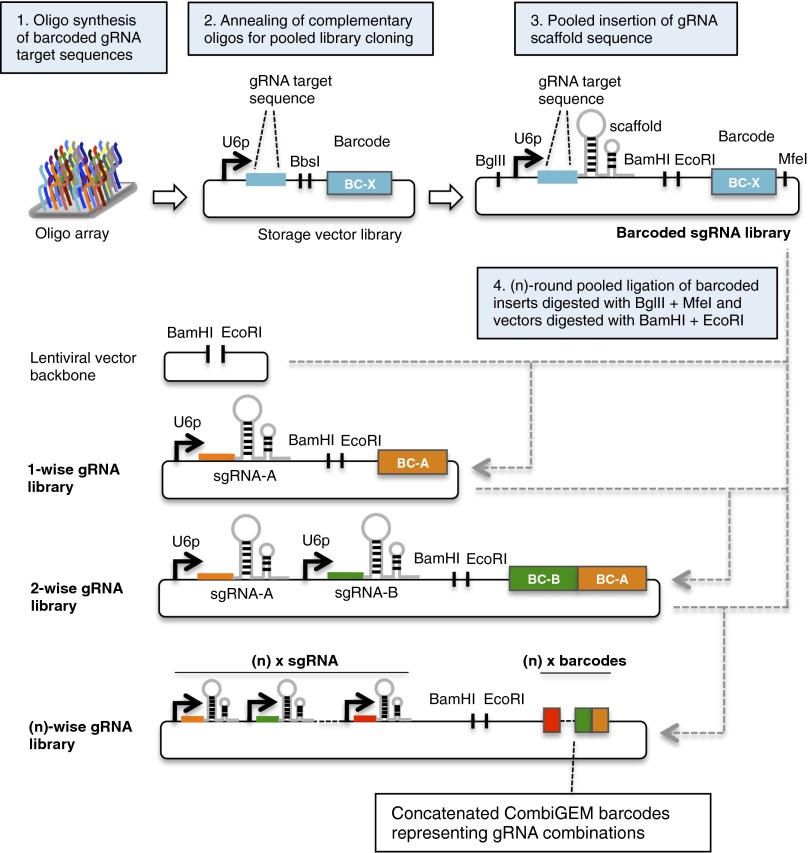
To create the initial barcoded sgRNA library, an array of oligo pairs encoding a library of barcoded gRNA target sequences was first synthesized, annealed, and pooled in equal ratios for cloning downstream of a U6 promoter in the storage vector (Fig. 1 and SI Appendix, Fig. S1). Subsequently, the scaffold sequence for the gRNAs was inserted into the storage vector library in a single-pot ligation reaction. We then applied the CombiGEM method for assembly of combinatorial gRNA libraries (Fig. 1). Within the barcoded sgRNA construct, BamHI and EcoRI sites were positioned in between the gRNA sequence and its barcode, whereas BglII and MfeI sites were located at the ends. Strategic positioning of these restriction enzyme sites resulted in the segregation of the barcode from its gRNA sequence upon enzymatic digestion and the concatenation of barcodes representing their respective gRNAs upon ligation of inserts. To construct the one-wise library, pooled inserts of the barcoded sgRNA expression units were prepared by restriction digestion of the storage vectors with BglII and MfeI and joined to their compatible DNA ends in the lentiviral destination vector, which was digested with BamHI and EcoRI. The one-wise library then served as the destination vector for the next round of pooled insertion of the barcoded sgRNA expression units to generate the two-wise library, in which barcodes representing each sgRNA were localized to one end of each lentiviral construct. This process can be iteratively repeated to generate higher-order barcoded combinatorial gRNA libraries. The identity of the combinatorial gRNAs can be tracked by high-throughput sequencing of the concatenated barcodes, which are unique for each combination.
Fig. 1.
Strategy for assembling barcoded combinatorial gRNA libraries. Barcoded gRNA oligo pairs were synthesized, annealed, and cloned in storage vectors in pooled format. Oligos with the gRNA scaffold sequence were inserted into the pooled storage vector library to create the barcoded sgRNA library. Detailed assembly steps are described in SI Appendix, Fig. S1. The CombiGEM strategy was used to build the combinatorial gRNA library. Pooled barcoded sgRNA inserts prepared from the sgRNA library with BglII and MfeI digestion were ligated via compatible overhangs generated in the destination vectors with BamHI and EcoRI digestion. Iterative one-pot ligation created (n)-wise gRNA libraries with unique barcodes corresponding to the gRNAs concatenated at one end, thus enabling tracking of individual combinatorial members within pooled populations via next-generation sequencing.
To evaluate the functionality of our lentiviral combinatorial gRNA expression system, we built gRNA combinations targeting green fluorescent protein (GFP) and red fluorescent protein (RFP) sequences (SI Appendix, Dataset S1) and determined the combinatorial gene perturbation phenotypes using flow cytometry (SI Appendix, Fig. S2 A–D) and fluorescence microscopy (SI Appendix, Fig. S2E). Lentiviruses carrying dual RFP and GFP reporters together with the barcoded combinatorial gRNA expression units were used to infect human ovarian cancer cells (OVCAR8-ADR) (15) stably expressing human codon-optimized Cas9 nuclease (OVCAR8-ADR-Cas9) (SI Appendix, Fig. S2A). We anticipated that active gRNAs would target the GFP and RFP sequences, and generate indels to knockout the expression of GFP and RFP. Efficient repression of GFP and RFP fluorescence levels was observed, as the GFP and RFP double-negative population was the major one observed in cells carrying both Cas9 nuclease and double gRNA expression units at both days 4 and 8 postinfection (∼83–97% of the total population), compared with <0.7% in the vector control (SI Appendix, Fig. S2 C and D). This repression was not observed in control cell lines expressing the gRNAs targeting GFP and/or RFP but without Cas9 nuclease (SI Appendix, Fig. S2B). The specificity of gene perturbation was confirmed, as cells harboring GFP-targeting sgRNA exhibited loss of the GFP signal but not the RFP signal, and vice versa for cells with the RFP-targeting sgRNAs (SI Appendix, Fig. S2E). These results demonstrate the ability of lentiviral vectors to encode combinatorial gRNA constructs that can repress the expression of multiple genes simultaneously within a single human cell.
Using CombiGEM-CRISPR, we sought to discover combinatorial epigenetic perturbations with anticancer phenotypes, because diverse epigenetic modifications tend to act cooperatively to regulate gene expression patterns (16) and combinatorial epigenetic modulation is emerging as a promising strategy for cancer therapeutics (17, 18). We constructed a library of 153 barcoded sgRNAs targeting a set of 50 genes that encode epigenetic regulators (three sgRNAs per gene) and three control sgRNAs based on the GeCKOv2 library (4) (SI Appendix, Dataset S1). Using the Drug–Gene Interaction database (DGIdb) (19) and recent literature (20, 21), we confirmed that at least 26 of the 50 genes in our library are known drug targets. Epigenetic protein families belong to druggable classes of enzymes or cofactors against which an increasing list of drugs is undergoing preclinical or clinical development (20).
We confirmed the expression of these 50 genes in OVCAR8-ADR cells using qRT-PCR (SI Appendix, Dataset S2). We then created a two-wise (153 × 153 sgRNAs = 23,409 total combinations) pooled barcoded gRNA library using CombiGEM. Lentiviral pools were produced to deliver the library into OVCAR8-ADR-Cas9 cells, and genomic DNA from the pooled cell populations was isolated for unbiased barcode amplification by PCR. Illumina HiSeq was used to quantify the representation of individual barcoded combinations in the plasmid pools stored in Escherichia coli and also in the infected human cell pools (SI Appendix, Fig. S3 A–D). We achieved high coverage for the two-wise library within both the plasmid and infected cell pools from ∼23–34 million reads per sample (SI Appendix, Fig. S3B), and a relatively even distribution of barcoded gRNA combinations was observed (SI Appendix, Fig. S3 A and B). Furthermore, we observed highly correlated barcode representation between the plasmid and infected cell pools (SI Appendix, Fig. S3C), as well as high reproducibility in barcodes represented in biological replicates for infected cell pools (SI Appendix, Fig. S3D). Thus, CombiGEM-CRISPR can be used to efficiently assemble and deliver barcoded combinatorial gRNA libraries into human cells.
Previous CRISPR-Cas9–based gene knockout screens have demonstrated high genomic modification efficiencies after about 6–12 d postexpression of Cas9 and sgRNA in human cells. However, it is important to evaluate genomic modification efficiencies in the specific cell types to be studied due to variations among gRNAs and cell types (4–6). To confirm the ability of the CRISPR-Cas system to edit endogenous genes in OVCAR8-ADR-Cas9 cells, we performed surveyor assays to detect mutations at genomic loci targeted by eight randomly chosen gRNAs from our library. We observed cleavage of DNA mismatches for all of the gRNA-targeted loci at day 12 postinfection (SI Appendix, Fig. S4 A–C). We further determined the simultaneous cleavage efficiency at multiple loci in our dual gRNA system, and observed comparable levels of cleavage in cells expressing individual gRNAs or double gRNAs (SI Appendix, Fig. S4 A and D). Depletion of targeted protein levels in individual gRNA- and double gRNA-expressing cells was also detected (SI Appendix, Fig. S4E). These results suggest that our multiplexed system does not hamper the activity of gRNAs.
We next estimated indel generation efficiency by performing deep sequencing at targeted genomic loci. Consistent with previous reports (4–6), we observed large variations in the rates of generating indels (i.e., 14–93%; SI Appendix, Fig. S5A) and frameshift mutations (i.e., 52–95% of all indels; SI Appendix, Fig. S5B) among different gRNAs. In addition, gRNAs that were validated in a previous study with A375 melanoma cells (4) displayed reduced activity (e.g., for NF1-sg4 and MED12-sg1 sgRNAs) and differential indel generation preferences (e.g., for the NF1-sg1 sgRNA) in OVCAR8-ADR-Cas9 cells (SI Appendix, Fig. S5C). Such discrepancies could be partially due to variations in chromatin accessibility at target loci (22) and DNA break repair mechanisms (23) that can vary among cell types. Continual efforts in gRNA design optimization, including improving on-target cleavage rates (24) and minimizing off-target cleavage, should enable the creation of more efficient gRNA sets that will improve their applicability for large-scale genetic perturbation screening in a broad range of cell types. We further assessed indel generation by gRNAs in our multiplexed system. Our deep sequencing analysis detected largely comparable indel generation frequencies and preferences for the same gRNA expressed under the sgRNA or double gRNA systems (SI Appendix, Fig. S5 D and E). To distinguish dual-cleavage events directed by double gRNAs within a single cell from cleavage events distributed across the population, we isolated clones derived from single cells infected with double gRNA constructs and were able to detect cells with insertions, deletions, or mutations in both targeted genomic loci (SI Appendix, Fig. S6 A–C and Dataset S3). Our results indicate that our combinatorial gRNA library can be used to generate double genetic mutants in OVCAR8-ADR-Cas9 cells. However, we believe that improvements in the efficiency of CRISPR-Cas reagents for gene knockouts would yield higher-quality CombiGEM-CRISPR libraries.
We initiated a pooled combinatorial genetic screen with OVCAR8-ADR-Cas9 cells to identify gRNA combinations that regulate cancer cell proliferation. We constructed a mathematical model to map out how relative changes in abundances of each library member within a population depend on various parameters (SI Appendix, SI Materials and Methods and Fig. S7 A and B). We simulated populations containing heterogeneous subpopulations that harbor different gRNA combinations. Specifically, we defined specific percentages of the overall population at the start of the simulation as harboring subpopulations with antiproliferative (fs) and proproliferative (ff) gRNA combinations. Within each subpopulation, a fraction of cells was mutated by the CRISPR-Cas9 system (p) at the start of the simulation, resulting in a modified doubling time (Tdoubling,m). Our model indicated that the representation of barcoded cells with an antiproliferative gRNA set in the entire cell population can be depleted by about 23–97% under simulated conditions (i.e., (fs), and (ff) = 2%, 5%, or 10%; p = 0.2, 0.4, 0.6, 0.8, or 1.0; (Tdoubling,m) = 36, 48, or 60 h) (SI Appendix, Fig. S7B). In general, increasing mutation efficiencies, increasing doubling times for antiproliferative cells, decreasing doubling times for proproliferative cells, as well as increasing the percentage of proproliferative combinations in the population (SI Appendix, Fig. S7C), are expected to result in greater barcode depletion of antiproliferative barcodes in the overall population.
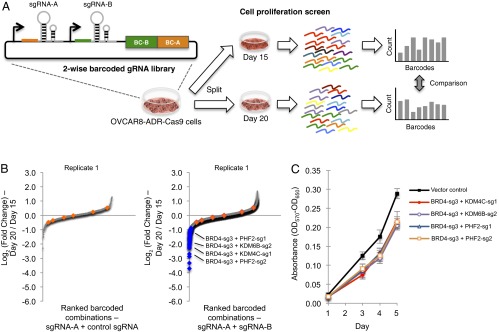
In the experimental screen, we cultured OVCAR8-ADR-Cas9 cell populations infected with the two-wise combinatorial gRNA library for 15 and 20 d and isolated their genomic DNA for unbiased amplification and quantification of the integrated barcodes (Fig. 2A and SI Appendix, Fig. S3 E and F). Because phenotypes resulting from progressive epigenetic alterations following targeted gene inactivation are expected to take time to manifest (25), barcode abundances (normalized per million reads) between day 15 and day 20 groups were compared with yield log2 (barcode count ratios) values (Fig. 2A and SI Appendix, Fig. S3 E and F), based on similar time windows used in previous studies on the antiproliferative effects resulting from epigenetic perturbations (25, 26). To reduce variability, combinations with less than ∼100 absolute reads in the day 15 group were filtered out, and the log2 ratios of the two possible arrangements for each gRNA pair (i.e., sgRNA-A + sgRNA-B and sgRNA-B + sgRNA-A) were averaged (SI Appendix, Fig. S8). Less than 4.4% of all combinations were detected at <100 absolute reads in the day 15 group in both sets of experiments. The correlation between the log2 ratios of the two possible arrangements for each gRNA pair could be improved by increasing the fold representation of cells per combination in the pooled screen to reduce experimental noise, as previously noted in pairwise genetic perturbation screens (27). Log2 ratios for each gRNA combination were determined for two biological replicates and ranked (Fig. 2B and SI Appendix, Fig. S9A). The majority of the gRNA combinations did not exhibit significant changes in barcode representations between day 15 and day 20 groups, including three control gRNAs from the GeCKOv2 library (4) that do not have on-target loci in the human genome as internal controls. We defined 61 gRNA combinations as top hits that exerted considerable antiproliferative effects (log2 ratio < −0.90) in both biological replicates (Q-value <0.01; SI Appendix, Fig. S9 B and C and Dataset S4), yielding potential sets of genes to investigate further for their ability to suppress the growth of cancer cells. A potential caveat to consider when comparing barcode abundances between day 15 and day 20 groups is that cells with strongly synthetic-lethal gene combinations inactivated could be eliminated before day 15, leading to false negatives. To account for this, we compared log2 ratios between day 15 and day 5 groups, and did not identify any hits (SI Appendix, Fig. S9D). We speculate that this could be due to latency in proliferation changes resulting from the time required to alter epigenetic marks and gene expression after knockouts occur (25, 26). This analysis highlights the need to identify optimal time windows for performing barcode comparisons to facilitate hit identification in high-throughput pooled screens.
Fig. 2.
High-throughput screen identifies gRNA combinations that inhibit cancer cell proliferation. (A) OVCAR8-ADR-Cas9 cells infected with the barcoded two-wise gRNA library were cultured for 15 and 20 d. Barcode representations within the cell pools were quantified using Illumina HiSeq. (B) Two-wise gRNA combinations that modulated proliferation were ranked by log2 ratios between their normalized barcode counts in 20-d versus 15-d cultured cells (Right). To enable comparisons between the two-wise gRNA combinations with their sgRNA counterparts, the same data for sgRNAs paired with control sgRNAs (Left) are also plotted. Combinations with control gRNA pairs are highlighted in orange. The antiproliferative effects of gRNA combinations that were confirmed in another biological replicate are highlighted in blue (SI Appendix, Fig. S9). The labeled gRNA combinations were further validated in this study. (C) Individual validation of two-wise combinations that modulated cancer cell growth. OVCAR8-ADR-Cas9 cell populations individually infected with lentiviruses expressing the indicated two-wise gRNA combinations were cultured for 15 d, and an equal number of cells was then replated and cultured for additional time periods as indicated. Cell viability was measured by the MTT assay, and characterized by absorbance measurements (OD570–OD650) (n = 3). Data represent mean ± SD.
CRISPR-Cas9–based screens, like other genetic screens, can lead to false discovery due to off-target effects and false-positive hits (28, 29). Thus, we performed individual validation experiments to verify the phenotype-modifying effects of genetic perturbations identified as hits from our screen. We validated screen hits by demonstrating their ability to inhibit the proliferation of OVCAR8-ADR-Cas9 cells in individual (nonpooled) cell growth assays using the corresponding gRNA pairs delivered via lentiviruses (Fig. 2C and SI Appendix, Fig. S10). In addition, false-negative hits could be anticipated in the screen (28, 30) due to the presence of inefficient gRNAs (SI Appendix, Fig. S5), which could be addressed with an experimentally validated set of gRNAs with optimized on-target efficiencies.
Global alterations of epigenetic landscapes observed in cancer progression (31) and the reversible nature of epigenetic states (32) suggest that targeting multiple epigenetic regulators could help to suppress cancer growth. Interestingly, we observed that many gRNAs targeting genes encoding epigenetic regulators exhibited stronger antiproliferative effects when used in combination with other gene-targeting gRNAs than when they were used in combination with control gRNAs (Fig. 2B and SI Appendix, Fig. S9A). With individual validation assays, we confirmed that specific gRNA pairs (Fig. 3A and SI Appendix, Fig. S4) targeting KDM4C and BRD4 simultaneously led to synergistic reductions in cancer cell growth. We also assessed the off-target activity of these gRNAs with deep sequencing, which revealed a low indel generation rate (i.e., 0.15–0.38%) at the exonic off-target genomic loci computationally predicted by the CRISPR design (33) and CCTop (34) tools for the two gRNAs (SI Appendix, Fig. S11). In addition, we observed reduced growth in a single-cell–derived clone harboring both KDM4C and BRD4 frameshift mutations (SI Appendix, Fig. S12). Collectively, we established and validated an experimental pipeline for the systematic screening of barcoded combinatorial gRNAs that are capable of exerting antiproliferative effects on ovarian cancer cells.
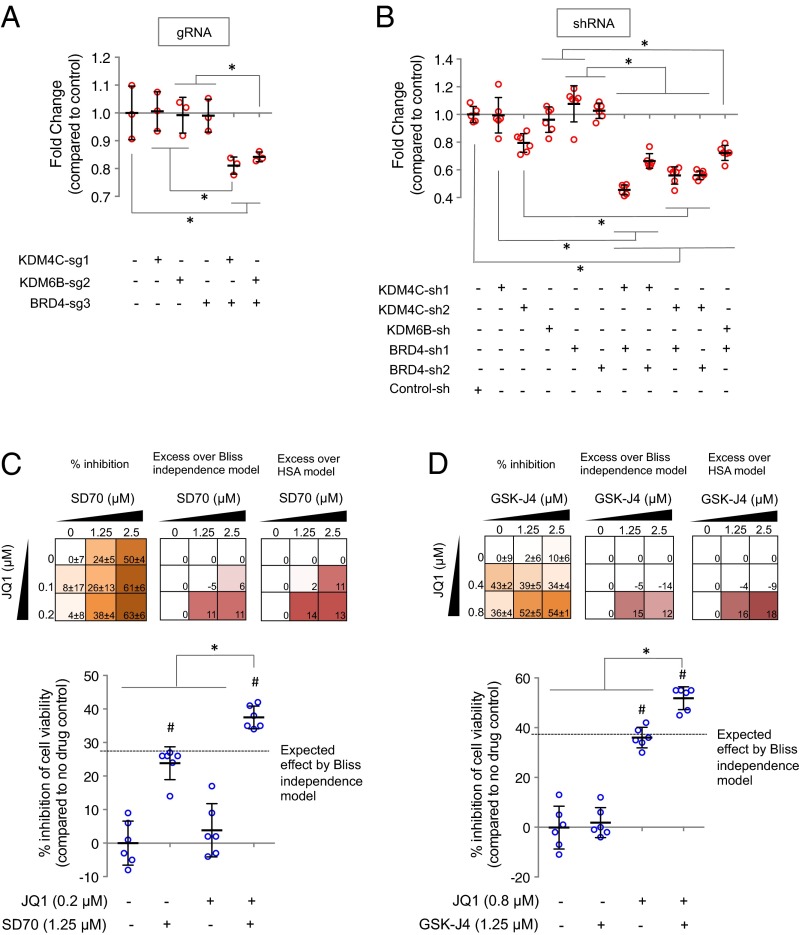
Fig. 3.
Combinatorial inhibition of KDM4C and BRD4, as well as KDM6B and BRD4, inhibits human ovarian cancer cell growth. (A and B) OVCAR8-ADR-Cas9 cells infected with lentiviruses expressing the indicated single or combinatorial gRNAs (A) or OVCAR8-ADR cells coinfected with lentiviruses expressing the indicated shRNAs (B) were cultured for 15 d and 9 d, respectively. Equal numbers of infected cells were then replated and cultured for an additional 5 d (A) and 4 d (B). Cell viabilities relative to control sgRNA (A) or shRNA (B) were determined by the MTT assay. (C and D) OVCAR8-ADR cells were treated with the indicated drugs for 5 d (C) and 7 d (D). JQ1 (36), SD70 (35), and GSK-J4 (37) are small-molecule inhibitors of BRD4, KDM4C, and KDM6B/6A, respectively. Percentage inhibition of cell growth relative to no drug control was determined by the MTT assay. The calculated excess inhibition over the predicted Bliss independence and HSA models was shown for each drug combination pair. Data represent mean ± SD (n = 3 for A; n = 6 for B–D) from biological replicates. *P < 0.05 and #P < 0.05 represent significant differences between the indicated samples and between drug-treated versus no drug control samples, respectively.
We further used RNA interference and small-molecule drugs to modulate genes encoding epigenetic regulators and confirm the screening-based phenotypes. Expression of multiple shRNA pairs targeting KDM4C and BRD4 (Fig. 3B and SI Appendix, Fig. S13) and cotreatment with the small-molecule KDM4C inhibitor SD70 (35) and small-molecule BRD4 inhibitor JQ1 (36) (Fig. 3C) inhibited the proliferation of OVCAR8-ADR cells synergistically. Similarly, gRNA pairs (Fig. 3A and SI Appendix, Figs. S4 and S11) and shRNA pairs (Fig. 3B and SI Appendix, Fig. S13) that simultaneously targeted KDM6B and BRD4 exhibited synergy, as did cotreatment with the small-molecule KDM6B/6A inhibitor GSK-J4 (37) and JQ1 (Fig. 3D). Synergy between both of these pairwise combinations of small-molecule drugs was confirmed by both the Bliss independence (38) and the highest single agent (HSA) (39) models (Fig. 3 C and D). These multiple confirmatory strategies suggest that the observed antiproliferation effects were likely caused by the dual inhibition of multiple genes rather than off-target effects. Our approach thus facilitates the identification of previously unidentified interacting gene pairs that inhibit cancer cell proliferation, and the potential development of synergistic drug therapies.
Discussion
In summary, we established CombiGEM-CRISPR as a technology platform for assembling barcoded combinatorial gRNA libraries and facilitating pooled genetic perturbation screening that can be translated into novel drug combinations. This platform expands the utility of CRISPR-Cas9–based systems for performing systematic multiplexed genetic perturbation screens in high throughput. Here, we applied CombiGEM-CRISPR to perform a massively parallel combinatorial CRISPR-Cas screen and successfully isolated gene pairs for which simultaneous inhibition via CRISPR-Cas knockouts, RNA interference, and small molecules led to reduced ovarian cancer cell growth. High-throughput screening of combinatorial genetic perturbations by CombiGEM-CRISPR can expedite the identification of novel drug combinations with desired therapeutic effects by targeting libraries of gRNAs against druggable genes (19, 40). In this study, we further investigated two of our genetic hits (i.e., KDM4C + BRD4 and KDM6B + BRD4) with readily available small-molecule drugs and confirmed their synergistic efficacy against ovarian cancer cells. These drugs have been used previously for treating other cancer cell types in mouse models with limited toxicity (35, 36, 41, 42), suggesting that, in combination, these drugs could be viable therapeutic candidates. Our approach advances upon previously described combinatorial drug screening platforms (2, 39, 43, 44) by using multiplexed and pooled screens to reduce the cost, time, and effort required.
This strategy can also help identify new areas for biological inquiry, such as studies into the mechanisms that underlie observed phenotypes. For example, we analyzed gene expression patterns in cell populations infected with lentiviruses encoding gRNAs targeting both KDM4C and BRD4, or KDM6B and BRD4 (SI Appendix, Fig. S14A). Significantly perturbed genes were associated with gene sets involved in cancer-related pathways, including TNFα/NFκB signaling, p53 pathways, and apoptosis (SI Appendix, Fig. S14B). In addition, the combinatorial effects of epigenetic perturbations are complex and can vary across different cell types (SI Appendix, Fig. S15). In future work, assembled and barcoded combinatorial libraries could be directly delivered into a variety of cell types to rapidly dissect how combinatorial genetic effects vary based on genetic background via the same screening pipeline.
The utility of CombiGEM-CRISPR will be further enhanced as CRISPR-Cas technology continues to be improved in terms of enhanced cleavage efficiency (24, 40, 45) and reduced off-target effects (46–48). Variations in gRNA efficiency and indel generation preference can result in quantitative differences in creating disruptive mutations between gRNAs targeting the same gene. In addition, mixed genotypes generated by the CRISPR-Cas genome editing can generate variability in pooled screens. As new rules for designing highly efficient frameshift-mutation-creating gRNAs are established in the future (28), CombiGEM-CRISPR could be potentially applied to perform large-scale studies of epistasis for interrogating gene–gene interactions. Previous efforts have laid powerful experimental and computational foundations for performing systematic genetic interaction analysis in yeast and human cells (49, 50). In addition to generating gene knockouts, the CombiGEM-CRISPR platform could be used for high-order combinatorial gene activation and repression studies by incorporating gRNAs and deactivated Cas9 variants repurposed as transcriptional and epigenetic regulators (8, 9, 51, 52). This technology could also be used to interrogate the function of large genomic deletions (53) and rearrangements (54, 55) with barcoded gRNA pairs. Thus, CombiGEM-CRISPR provides a facile approach to uncover gene and drug combinations that exert desired biological responses, especially for phenotypes that require more than a single perturbation to be manifested due to underlying complex biological networks.
Materials and Methods
Methods for vector construction, assembly of the barcoded combinatorial sgRNA library pool, cell culture, lentivirus production and transduction, sample preparation for barcode sequencing, barcode sequencing data analysis, cell viability assay, drug synergy quantification, flow cytometry, fluorescence microscopy, quantitative PCR, immunoblot analysis, surveyor assay, sequencing analysis for indel detection, RNA-Seq and data analysis, and the mathematical modeling of cell proliferation in a mixed population, are provided in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank members of the T.K.L. laboratory for helpful discussions, T. Ochiya for OVCAR8-ADR cells, the MIT BioMicro Center for technical support on Illumina Sequencing, and C. Liu for technical assistance. This work was supported by the NIH (DP2OD008435, P50GM098792, and R01 NS089076), the Office of Naval Research (N00014-13-1-0424), the Defense Threat Reduction Agency (HDTRA1-15-1-0050), and the Ellison Foundation New Scholar in Aging Award. A.S.L.W. and C.H.C. were supported by the Croucher Foundation and a Natural Sciences and Engineering Research Council of Canada postgraduate fellowship, respectively.
Footnotes
Conflict of interest statement: T.K.L., A.S.L.W., and G.C.G.C. have filed a patent application based on this work with the US Patent and Trademark Office.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE71074).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1517883113/-/DCSupplemental.
References
- 1.Al-Lazikani B, Banerji U, Workman P. Combinatorial drug therapy for cancer in the post-genomic era. Nat Biotechnol. 2012;30(7):679–692. doi: 10.1038/nbt.2284. [DOI] [PubMed] [Google Scholar]
- 2.Kummar S, et al. Utilizing targeted cancer therapeutic agents in combination: Novel approaches and urgent requirements. Nat Rev Drug Discov. 2010;9(11):843–856. doi: 10.1038/nrd3216. [DOI] [PubMed] [Google Scholar]
- 3.Holbeck S. 2014 Cell-based screens of drug combinations at NCI. National Institutes of Health. Available at deainfo.nci.nih.gov/advisory/ctac/1114/5%20-%20Holbeck.pdf. Accessed January 14, 2016.
- 4.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343(6166):84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343(6166):80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou Y, et al. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014;509(7501):487–491. doi: 10.1038/nature13166. [DOI] [PubMed] [Google Scholar]
- 7.Koike-Yusa H, Li Y, Tan E-P, Velasco-Herrera MdelC, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol. 2014;32(3):267–273. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- 8.Gilbert LA, et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 2014;159(3):647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Konermann S, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517(7536):583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilbert LA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154(2):442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng AA, Ding H, Lu TK. Enhanced killing of antibiotic-resistant bacteria enabled by massively parallel combinatorial genetics. Proc Natl Acad Sci USA. 2014;111(34):12462–12467. doi: 10.1073/pnas.1400093111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong ASL, Choi GCG, Cheng AA, Purcell O, Lu TK. Massively parallel high-order combinatorial genetics in human cells. Nat Biotechnol. 2015;33(9):952–961. doi: 10.1038/nbt.3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Honma K, et al. RPN2 gene confers docetaxel resistance in breast cancer. Nat Med. 2008;14(9):939–948. doi: 10.1038/nm.1858. [DOI] [PubMed] [Google Scholar]
- 16.Wang Z, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40(7):897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dawson MA, Kouzarides T. Cancer epigenetics: From mechanism to therapy. Cell. 2012;150(1):12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 18.Juergens RA, et al. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer Discov. 2011;1(7):598–607. doi: 10.1158/2159-8290.CD-11-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffith M, et al. DGIdb: Mining the druggable genome. Nat Methods. 2013;10(12):1209–1210. doi: 10.1038/nmeth.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: A new frontier for drug discovery. Nat Rev Drug Discov. 2012;11(5):384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- 21.Rodriguez R, Miller KM. Unravelling the genomic targets of small molecules using high-throughput sequencing. Nat Rev Genet. 2014;15(12):783–796. doi: 10.1038/nrg3796. [DOI] [PubMed] [Google Scholar]
- 22.Wu X, et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol. 2014;32(7):670–676. doi: 10.1038/nbt.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghezraoui H, et al. Chromosomal translocations in human cells are generated by canonical nonhomologous end-joining. Mol Cell. 2014;55(6):829–842. doi: 10.1016/j.molcel.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doench JG, et al. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol. 2014;32(12):1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knutson SK, et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat Chem Biol. 2012;8(11):890–896. doi: 10.1038/nchembio.1084. [DOI] [PubMed] [Google Scholar]
- 26.Daigle SR, et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell. 2011;20(1):53–65. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kampmann M, Bassik MC, Weissman JS. Functional genomics platform for pooled screening and generation of mammalian genetic interaction maps. Nat Protoc. 2014;9(8):1825–1847. doi: 10.1038/nprot.2014.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet. 2015;16(5):299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohr SE, Smith JA, Shamu CE, Neumüller RA, Perrimon N. RNAi screening comes of age: Improved techniques and complementary approaches. Nat Rev Mol Cell Biol. 2014;15(9):591–600. doi: 10.1038/nrm3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Booker M, et al. False negative rates in Drosophila cell-based RNAi screens: A case study. BMC Genomics. 2011;12(1):50. doi: 10.1186/1471-2164-12-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128(4):683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoo CB, Jones PA. Epigenetic therapy of cancer: Past, present and future. Nat Rev Drug Discov. 2006;5(1):37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- 33.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8(11):2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stemmer M, Thumberger T, Del Sol Keyer M, Wittbrodt J, Mateo JL. CCTop: An intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS One. 2015;10(4):e0124633. doi: 10.1371/journal.pone.0124633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin C, et al. Chem-seq permits identification of genomic targets of drugs against androgen receptor regulation selected by functional phenotypic screens. Proc Natl Acad Sci USA. 2014;111(25):9235–9240. doi: 10.1073/pnas.1404303111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Asangani IA, et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510(7504):278–282. doi: 10.1038/nature13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kruidenier L, et al. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature. 2012;488(7411):404–408. doi: 10.1038/nature11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bliss CI. The toxicity of poisons applied jointly. Ann Appl Biol. 1939;26:585–615. [Google Scholar]
- 39.Borisy AA, et al. Systematic discovery of multicomponent therapeutics. Proc Natl Acad Sci USA. 2003;100(13):7977–7982. doi: 10.1073/pnas.1337088100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi J, et al. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat Biotechnol. 2015;33(6):661–667. doi: 10.1038/nbt.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hashizume R, et al. Pharmacologic inhibition of histone demethylation as a therapy for pediatric brainstem glioma. Nat Med. 2014;20(12):1394–1396. doi: 10.1038/nm.3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bolden JE, et al. Inducible in vivo silencing of Brd4 identifies potential toxicities of sustained BET protein inhibition. Cell Reports. 2014;8(6):1919–1929. doi: 10.1016/j.celrep.2014.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mathews Griner LA, et al. High-throughput combinatorial screening identifies drugs that cooperate with ibrutinib to kill activated B-cell-like diffuse large B-cell lymphoma cells. Proc Natl Acad Sci USA. 2014;111(6):2349–2354. doi: 10.1073/pnas.1311846111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim J, et al. A programmable microfluidic cell array for combinatorial drug screening. Lab Chip. 2012;12(10):1813–1822. doi: 10.1039/c2lc21202a. [DOI] [PubMed] [Google Scholar]
- 45.Chari R, Mali P, Moosburner M, Church GM. Unraveling CRISPR-Cas9 genome engineering parameters via a library-on-library approach. Nat Methods. 2015;12(9):823–826. doi: 10.1038/nmeth.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pattanayak V, et al. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol. 2013;31(9):839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuscu C, Arslan S, Singh R, Thorpe J, Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotechnol. 2014;32(7):677–683. doi: 10.1038/nbt.2916. [DOI] [PubMed] [Google Scholar]
- 48.Mali P, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31(9):833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bassik MC, et al. A systematic mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell. 2013;152(4):909–922. doi: 10.1016/j.cell.2013.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Costanzo M, et al. The genetic landscape of a cell. Science. 2010;327(5964):425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hilton IB, et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol. 2015;33(5):510–517. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kearns NA, et al. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat Methods. 2015;12(5):401–403. doi: 10.1038/nmeth.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Essletzbichler P, et al. Megabase-scale deletion using CRISPR/Cas9 to generate a fully haploid human cell line. Genome Res. 2014;24(12):2059–2065. doi: 10.1101/gr.177220.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blasco RB, et al. Simple and rapid in vivo generation of chromosomal rearrangements using CRISPR/Cas9 technology. Cell Reports. 2014;9(4):1219–1227. doi: 10.1016/j.celrep.2014.10.051. [DOI] [PubMed] [Google Scholar]
- 55.Choi PS, Meyerson M. Targeted genomic rearrangements using CRISPR/Cas technology. Nat Commun. 2014;5:3728. doi: 10.1038/ncomms4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.