Significance
We describe a filamentous virus, Pyrobaculum filamentous virus 1 (PFV1), with a linear double-stranded DNA genome. PFV1 infects hyperthermophilic archaea of the genus Pyrobaculum and displays a virion organization that is unique among filamentous DNA viruses. With its linear genome enclosed in a tripartite shell consisting of two protein layers and an external envelope, PFV1 virion organization bears a superficial resemblance to that of filoviruses, such as Ebola and Marburg viruses, which have negative-sense RNA genomes. This observation suggests that overall similar designs in the viral world have been achieved on multiple independent occasions.
Keywords: hyperthermophilic archaea, virion organization, filamentous viruses
Abstract
Viruses package their genetic material in diverse ways. Most known strategies include encapsulation of nucleic acids into spherical or filamentous virions with icosahedral or helical symmetry, respectively. Filamentous viruses with dsDNA genomes are currently associated exclusively with Archaea. Here, we describe a filamentous hyperthermophilic archaeal virus, Pyrobaculum filamentous virus 1 (PFV1), with a type of virion organization not previously observed in DNA viruses. The PFV1 virion, 400 ± 20 × 32 ± 3 nm, contains an envelope and an inner core consisting of two structural units: a rod-shaped helical nucleocapsid formed of two 14-kDa major virion proteins and a nucleocapsid-encompassing protein sheath composed of a single major virion protein of 18 kDa. The virion organization of PFV1 is superficially similar to that of negative-sense RNA viruses of the family Filoviridae, including Ebola virus and Marburg virus. The linear dsDNA of PFV1 carries 17,714 bp, including 60-bp-long terminal inverted repeats, and contains 39 predicted ORFs, most of which do not show similarities to sequences in public databases. PFV1 is a lytic virus that completely disrupts the host cell membrane at the end of the infection cycle.
Over the past years it has been shown that extreme thermal environments represent a habitat for unique DNA viruses with profound morphotypical diversity, many of which have not been observed in environments with moderate life conditions (1, 2). The peculiarity of these viruses seems to be linked with the specific nature of their hosts, hyperthermophilic members of the phylum Crenarchaeaota of the domain Archaea (1, 3). Nevertheless, in their overall morphology some crenarchaeal viruses, including those with icosahedral and filamentous virions, resemble viruses infecting bacteria and eukaryotes. The exact evolutionary relationship between viruses infecting hosts from different domains of life is often enigmatic. In icosahedral archaeal viruses the major capsid protein has the double β-barrel fold (4, 5), which also is found in the capsid proteins of many bacterial and eukaryotic viruses, pointing toward an evolutionary relationship among these viruses (6–8). In contrast, filamentous viruses infecting hosts from different cellular domains appear to be evolutionarily unrelated. Indeed, bacterial, eukaryotic, and archaeal filamentous viruses contain ssDNA, ssRNA, and dsDNA genomes, respectively (9). Virion organization of bacterial M13-like inoviruses and the plant Tobacco mosaic virus have been studied extensively for decades using various biochemical, biophysical, and structural biology approaches. As a result, a comprehensive picture of how these virions are assembled and how they are organized is available (10–12). In contrast, little is known about the organization of filamentous viruses infecting archaea, with the notable exception of the nonenveloped Sulfolobus islandicus rod-shaped virus 2 (SIRV2), for which a 3D reconstruction at ∼4-Å resolution has been obtained recently (13).
Here we report on the isolation and characterization of a new filamentous dsDNA virus that infects members of the archaeal genus Pyrobaculum. We show that the virus has a unique virion organization, with its linear genome being enclosed in a tripartite shell consisting of two protein layers and an external envelope. Our results provide new insights into the diversity of architectural solutions used by filamentous viruses.
Results
Virus and Host Isolation.
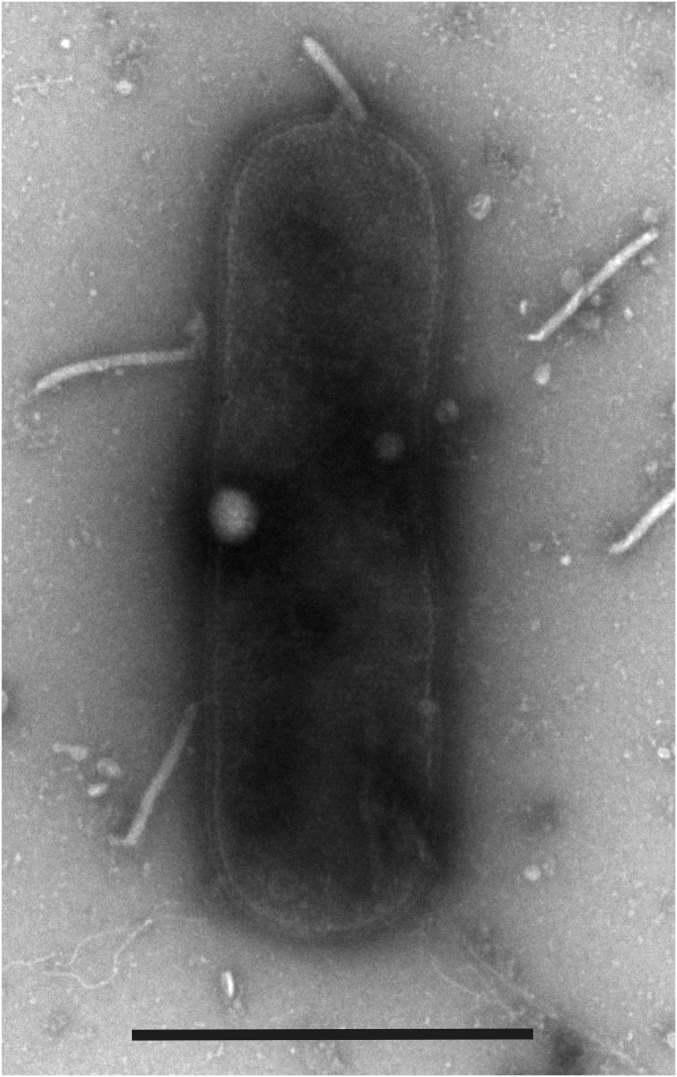
From the environmental sample collected at the Pozzuoli Solfatara, Italy, enrichment cultures were established in conditions known to favor the growth of aerobic members of the archaeal genus Pyrobaculum (14). The virus-like particles (VLPs) were detected in the enrichment culture by transmission electron microscopy (TEM). They were filamentous, uniform in overall appearance, and measured roughly 400 × 30 nm (Fig. S1). Their titer in the enrichment culture did not change after two rounds of 1:100 dilution by culture medium, with further growth of cells, suggesting active replication of the VLPs. For further analysis, the VLPs were collected and concentrated.
Fig. S1.
Transmission electron micrograph of a cell and VLPs present in the enrichment cultures. Negative stain with 2% uranyl acetate. (Scale bar: 1,000 nm.)
Twelve isolates of cells were colony purified from the VLP-producing enrichment culture and were analyzed for the ability to produce VLPs. The presence of VLPs was not detected in cell cultures of these isolates. In all cases, however, the addition of aliquots from the VLP preparation to exponentially growing cell cultures of all 12 isolates, followed by their further growth to the stationary phase, resulted in a dramatic increase of the VLP concentration. The results strongly suggested that the VLPs represent infectious virus particles and that all 12 analyzed isolates could be infected by this virus. One of these isolates was subjected to an additional round of colony purification, and the isolated strain, designated “2GA,” was selected as a standard virus host for all following experiments, unless stated otherwise. Cells of the 2GA isolate are rod-shaped, with an average length of about 4 μm and a width of about 0.7 μm. Analysis of the 16S-rRNA gene sequence revealed that the isolate represents a strain of Pyrobaculum arsenaticum. The sequence, including the intron, was 100% identical to the sequence of P. arsenaticum PZ6 (DSM13514) that previously was isolated from the same hot spring (15).
The filamentous virus that replicated in P. arsenaticum 2GA, after P. arsenaticum 2GA had been infected at an multiplicity of infection (MOI) of ∼0.5, was named “Pyrobaculum filamentous virus 1” (PFV1) (Fig. 1A). The morphology of PFV1 virions was identical to that of the VLPs observed in the enrichment culture (Fig. S1).
Fig. 1.
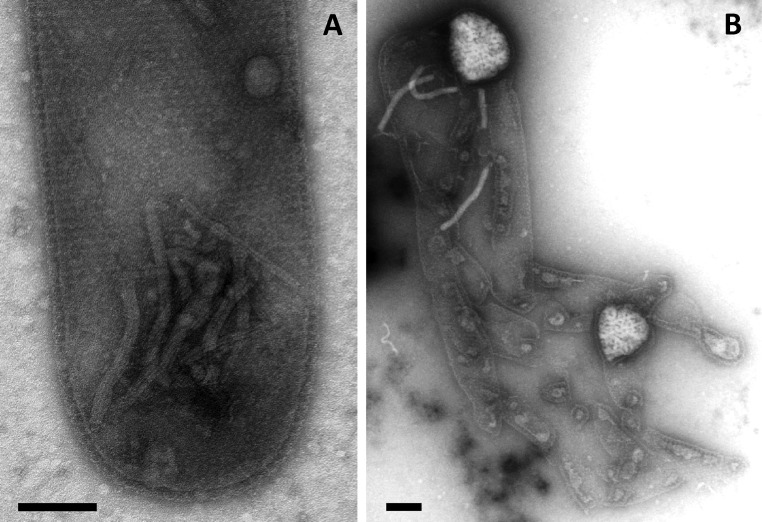
Transmission electron micrographs of PFV1 virions. (A) Purified virions. (B) Virions in a P. arsenaticum 2GA cell. (C) Virions in the process of egress from a lysed P. arsenaticum 2GA cell. Negative stain with 2% uranyl acetate. (Scale bars: 100 nm in A; 1,000 nm in B and C.)
Virus–Host Interactions.
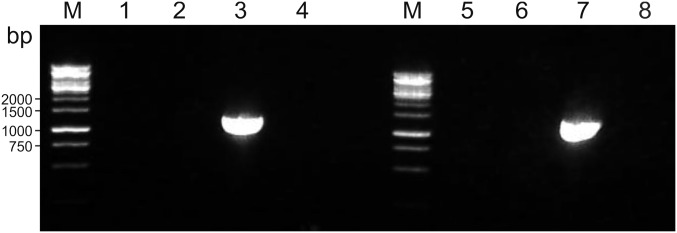
Besides P. arsenaticum 2GA, PFV1 also could infect P. arsenaticum PZ6 and Pyrobaculum oguniense TE7, but Pyrobaculum calidifontis VA1 was resistant to the virus. Notably, P. arsenaticum PZ6, which was originally described as an anaerobic strain (15), could grow in aerobic conditions using DSMZ medium 1090 (14). The result is unexpected, considering the absence of the gene for the cytochrome c biogenesis factor (Pogu_1433)—a part of the cytochrome cluster implicated in the aerobic growth of P. oguniense (16, 17)—in the genomes of P. arsenaticum strains PZ6 and 2GA (Fig. S2). However, it should be noted that the DSMZ medium contains sodium thiosulfate, which is a widely used oxygen scavenger; thus, it is likely that the concentration of oxygen in the medium, if any, was low.
Fig. S2.
Analysis of the presence of the cytochrome c biogenesis factor (Pogu_1433) in the genomes of P. Oguniense strain TE7 and P. arsenaticum strains PZ6 and 2GA by agarose gel electrophoresis of PCR products using cytochrome c biogenesis factor-specific primers. DNA templates: lanes 1 and 5, P. arsenaticum PZ6; lanes 2 and 6, P. arsenaticum 2GA; lanes 3 and 7, P. oguniense TE7; lanes 4 and 8, negative control. PCR primers: lanes 1–4, Set 1; lanes 5–8, Set 2. Lanes M, DNA markers.
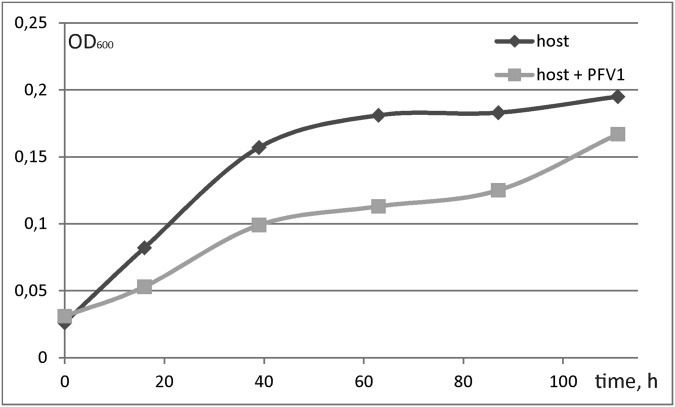
Replication of PFV1 in P. arsenaticum 2GA resulted in cell growth retardation (Fig. S3). Because Pyrobaculum cells could not be propagated as a lawn on solid medium, a plaque assay could not be established for PFV1; therefore we relied on TEM observations for assessment and quantification of virus–host interactions. In the experimental conditions no virions were observed in the cell-free culture of P. arsenaticum 2GA supernatant in the first hours post infection (hpi); the first intracellular virions could be observed around 16 hpi (Fig. S4A). At approximately at 20 hpi the cells were densely packed with virions (Fig. 1B), which were released by 24 hpi, coinciding with the dramatic rupture of the host cell envelope (Fig. 1C and Fig. S4B). Based on the TEM observations, we estimate that the burst size exceeded 100 virions per cell.
Fig. S3.
Growth of uninfected (dark gray) and infected (light gray) cells of P. arsenaticum 2GA.
Fig. S4.
Transmission electron micrographs of cells infected with PFV1. (A) 16 hpi. (B) 24 hpi. Negative stain with 2% uranyl acetate. (Scale bars: 200 nm.)
Virion Organization.
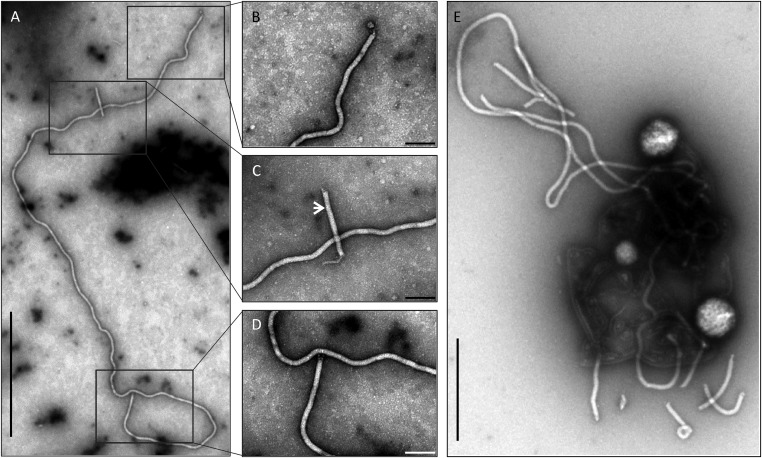
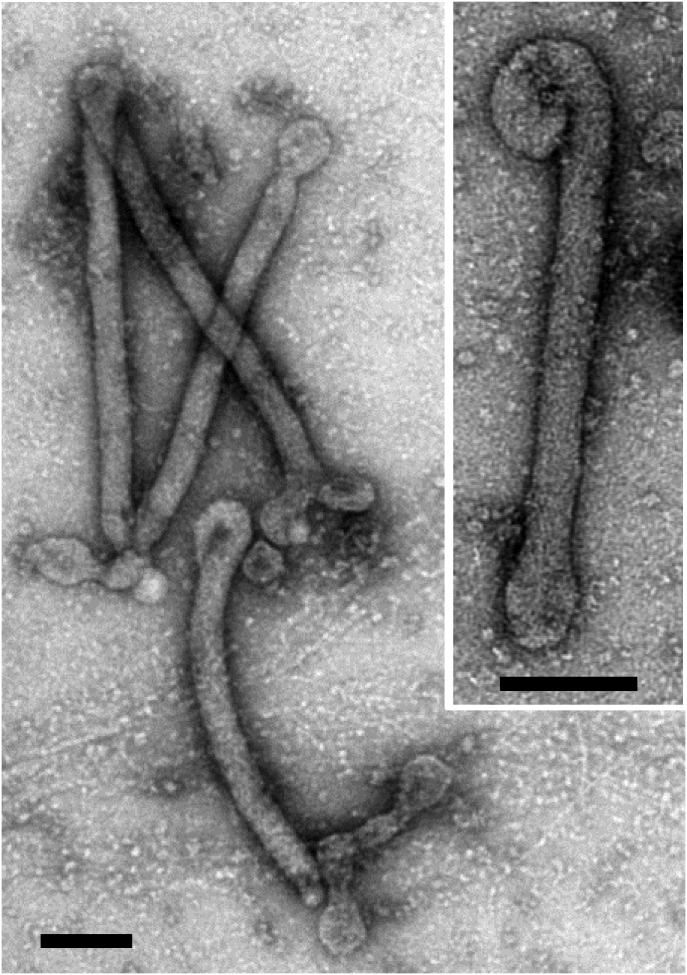
Negatively stained PFV1 virions are flexible, filamentous particles, measuring 400 ± 20 × 32 ± 3 nm (Figs. 1 and 2A). Bundles of thin filaments are attached to both ends of the virions. They measure about 2 nm in width and can reach the length of 80 nm. Very long virions (∼16 times longer than the typical virion) have been observed occasionally (Fig. S5), suggesting that such particles encapsidate more than one unit-length of the genome, as is the case in some other filamentous viruses (18).
Fig. 2.
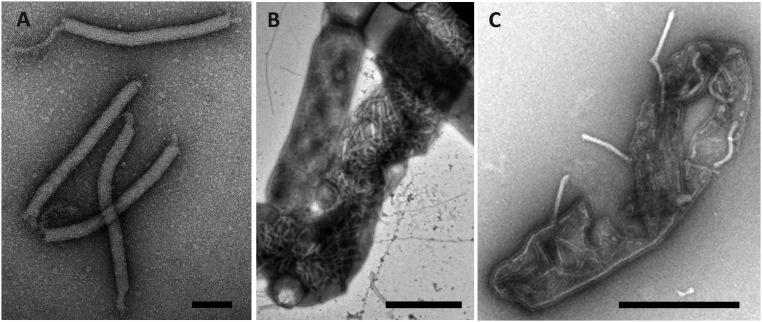
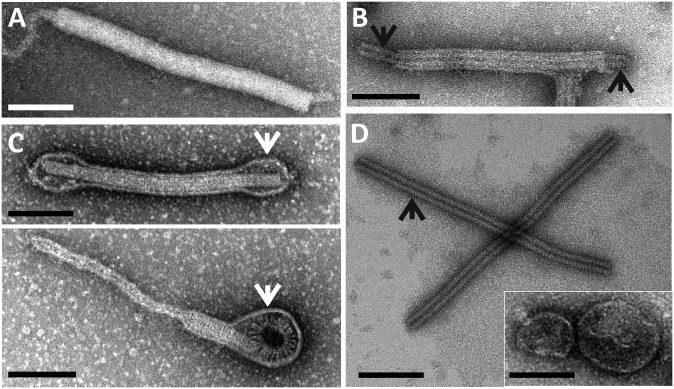
Transmission electron micrographs of native and disrupted PFV1 virions. (A) Native virion. (B) Virion disrupted in the course of purification. (C) Virions treated with Tween-20; white arrows indicate an envelope. (D) Structural entities resulting from virions treated with Triton X-100. Black arrows in B and D indicate a helical nucleocapsid. Negative stain with 2% uranyl acetate. (Scale bars: 100 nm.)
Fig. S5.
Transmission electron micrographs of anomalous PFV1 virions. Boxed areas in A are shown at higher magnification in B–D, as indicted. The white arrow in C indicates the native virion. (E) Egress of an anomalous PFV1 virion from a lysed cell. Negative stain with 2% uranyl acetate. (Scale bars: 2 μm in A; 200 nm in B–D; 1 μm in E.)
Insights into the virion organization were obtained by TEM observations of partially disintegrated particles. PFV1 virions appear to be sensitive to mechanical stress and can be partially disrupted in the course of purification and concentration. Purified PFV1 preparations contained a small proportion of disrupted particles that were devoid of their envelope, revealing a complex structure of the virion core, 30 ± 3 nm in width (Fig. 2B). The core consists of a nucleoprotein filament, a nucleocapsid about 20 nm in width (indicated by black arrows in Fig. 2B), and a surrounding protein sheath. Regular striations in both the nucleoprotein filament and the nucleocapsid most likely result from their helical organization. Mild treatment of PFV1 virions with Tween-20 caused partial detachment of the envelope from the nucleocapsid but not its complete removal. In a few cases, the envelope was detached at both ends of the virion. In most cases, envelope detachment caused the virion ends to curl up, transforming the filamentous virions into “lollipop-like” structures and clearly revealing the helical organization of the core (Fig. 2C). After mild treatment with Nonidet P-40 or Triton X-100 the virions were disassociated into completely unsheathed nucleocapsids and pleomorphic vesicle-like spheres. No other structural entity was produced as a result of such treatment (Fig. 2D). The unsheathed nucleocapsids were thinner than the intact cores (20 ± 5 versus 30 ± 3 nm). Surprisingly, these structures were about 40 nm longer than the intact cores or native virions (440 ± 30 nm versus 400 ± 20 nm) (Fig. 2D), suggesting that the sheath is involved in nucleocapsid compression. However, we do not exclude the possibility that the observed differences in nucleocapsid length in the intact and partially disintegrated states may be artifactual. The PFV1 virions were completely degraded when treated with 0.1% SDS. Treatment with ethanol (50% vol/vol) did not affect the integrity of the virions but did result in their conversion into cane-shaped particles (Fig. S6).
Fig. S6.
Transmission electron micrographs of PFV1 virions treated with ethanol. Negative stain with 2% uranyl acetate. (Scale bar: 100 nm.)
Genome Organization.
Nucleic acid extracted from the purified PFV1 particles was sensitive to DNase I and type II restriction endonucleases (BamHI, EcoRI, and SalI). However, it was not sensitive to RNase. The results indicate that the genome of PFV1 is a dsDNA molecule.
Sequencing of the PFV1 genome yielded a sequence of 17,714 bp, including 60-bp-long terminal inverted repeats (Fig. 3). The genome has a G+C content of 45.4%, which is considerably (55.1%) lower than that of the sequenced P. arsenaticum PZ6 genome (GenBank accession no. CP000660). The PFV1 genome contains 39 predicted ORFs that are larger than 40 codons (Fig. 3). These are tightly arranged and occupy 93.1% of the genome. All except one of the ORFs are encoded on the same strand. Thirty-six ORFs use AUG as a start codon. Ten of the PFV1 ORFs were predicted to encode proteins with one or more transmembrane domains (Fig. 3 and Table S1).
Fig. 3.
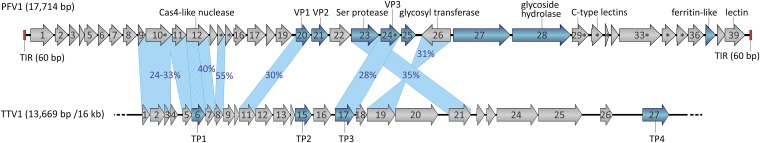
A genome map of PFV1 compared with the partial genome map of the virus TTV1. ORFs are indicated by arrows, which indicate the direction of transcription. Genes encoding the structural proteins are shown in blue; those encoding predicted membrane proteins are indicated by asterisks. Terminal inverted repeats (TIR) are depicted by red rectangles. The incompleteness of the TTV1 genome is illustrated by the broken line. The genes shared by PFV1 and TTV1 are connected by blue shading, and the identity between the corresponding proteins is indicated. The relationship between the cas4 gene of PFV1 and the TP1 gene of TTV1 is indicated by hatched shading.
Table S1.
Summary of predicted ORFs in the PFV1 genome
| ORF | Coordinates | Length, aa | TMDs | Predicted function | HHpred/CD hit | Blast hit | Identity (%), E-value |
| ORF1-196 | 317.0.754 | 145 | |||||
| ORF2-113 | 762.0.1103 | 113 | |||||
| ORF3-88 | 1106.0.1372 | 88 | |||||
| ORF4-52 | 1348.0.1506 | 52 | |||||
| ORF5-112 | 1484.0.1822 | 112 | |||||
| ORF6-93 | 1816.0.2097 | 93 | |||||
| ORF7-110 | 2094.0.2426 | 110 | |||||
| ORF8-120 | 2423.0.2785 | 120 | |||||
| ORF9-65 | 2786.0.2983 | 65 | TTV1 (CAA32969) | 21/64 (33), 3e-02 | |||
| ORF10-214 | 2976.0.3620 | 214 | 1 | N terminal similar to TTV1 (CAA32970); C terminal similar to TTV1 (CAA32971) | 25/84 (30), 2e-06; 20/53 (38), 9e-05 | ||
| ORF11-122 | 3617.0.3985 | 122 | TTV1 (CAA32973) | 19/79 (24), 2e-05 | |||
| ORF12-189 | 3966.0.4535 | 189 | Cas4-like nuclease | HHpred hit to Cas4 from Pyrobaculum calidifontis JCM 11548 (4r5q), P = 98.1; CD hit to Cas4 (TIGR00372), E = 2.4e-05 | TTV1_orf7-74 | 29/72 (40), 1e-08 | |
| ORF13-65 | 4537.0.4734 | 65 | |||||
| ORF14-63 | 4727.0.4918 | 63 | 2 | TTV1 (CAA32979) | 12/22 (55), 4e-04 | ||
| ORF15-67 | 4915.0.5118 | 67 | 3 | HHpred hit to DUF4536 (PF15055), P = 92 | |||
| ORF16-109 | 5122.0.5451 | 109 | |||||
| ORF17-155 | 5453.0.5920 | 155 | |||||
| ORF18-78 | 5917.0.6153 | 78 | |||||
| ORF19-139 | 6150.0.6569 | 139 | |||||
| ORF20-129 | 6640.0.7029 | 129 | Nucleoprotein VP1 | TTV1 (CAA32982) | 39/131 (30), 6e-04 | ||
| ORF21-145 | 7026.0.7463 | 145 | Nucleoprotein VP2 | A stretch of 14 Gly residues at the C terminus, also present in TTV1_orf12 (CAA32983) | |||
| ORF22-174 | 7465.0.7989 | 174 | |||||
| ORF23-230 | 7986.0.8678 | 230 | Trypsin-like Ser proteases | HHpred hits to various trypsin-like Ser proteases (PF13365), e.g., from Human astrovirus1 (2w5e), P = 96.7 | TTV1 (CAA32996); hyperthermophilic Archaeal virus 1 (YP_003773430) | 64/184(35); 9e-22; 57/184 (31); 6e-11 | |
| ORF24-166 | 8709.0.9209 | 166 | 2 | Sheath protein VP3 | TTV1_TP3 (CAA32989) | 42/152 (28), 2e-07 | |
| ORF25-125 | 9212.0.9589 | 125 | |||||
| ORF26-242 | Complement (9700.0.10428) | 242 | Glycosyltransferase (GT-A fold; cd02510) | HHpred hit to various Gtases (cd02510), P = 97.4 | TTV1 (CAA32992); Los Azufres archaeal virus 1 (AHA92064) | 67/215 (31); 7e-09; 33/92(36); 1e-04 | |
| ORF27-471 | 10402.0.11889 | 495 | |||||
| ORF28-481 | 11918.0.13363 | 481 | O-glycoside hydrolase (5-bladed β-propeller fold; family 43) | N-terminal region (amino acids 56–150): HHpred hit to a noncatalytic jelly-roll Lyase_N domain (PF09092) required for binding of the protein to long glycosaminoglycan chains, P = 94. C-terminal region (amino acids 111–400): HHpred hits to various family 43 O-glycoside hydrolases (PF04616), e.g., from Bacteroides thetaiotaomicron (3qz4), P = 95.5. | Microvirga lupini (WP_036352061) | 75/268(28); 2e-07 | |
| ORF29-143 | 13374.0.13805 | 143 | 1 | Carbohydrate-binding protein (C-type lectin-like fold; PF02369) | HHpred hit to integrin-binding protein (1cwv), P = 95.4 | ||
| ORF30-100 | 13847.0.14149 | 100 | 1 | Carbohydrate-binding protein (C-type lectin-like fold) | HHpred hit to integrin-binding protein (1cwv), P = 99.2 | Candidatus Methanoperedens nitroreducens (KCZ71275) | 39/81(48); 2e-07 |
| ORF31-40 | 14176.0.14298 | 40 | 1 | ||||
| ORF32-68 | 14323.0.14529 | 68 | |||||
| ORF33-343 | 14526.0.15557 | 343 | 2 | ||||
| ORF34-119 | 15587.0.15925 | 112 | 3 | ||||
| ORF35-96 | 15925.0.16215 | 96 | 3 | ||||
| ORF36-135 | 16203.0.16538 | 111 | |||||
| ORF37-87 | 16568.0.16831 | 87 | Ferritin (cd01045) | Numerous hits to ferritins from various archaea and bacteria, e.g., 3k6c (P = 99.8), 2e0z (P = 99.8), 1vjx (P = 99.3) | Pyrococcus furiosus DSM 3638 (NP_578921); Thermoproteus tenax Kra 1 (YP_004892222) | 30/83 (36), 9e-10; 32/79 (41), 2e-06 | |
| ORF38-71 | 16848.0.17063 | 71 | |||||
| ORF39-171 | 17047.0.17550 | 167 | Carbohydrate-binding protein (lectin) | HHpred hits to eukaryotic carbohydrate-binding lectins from Polyporus squamosus (3phz, P = 99.9) and Marasmius oreades (3ef2, P = 99.5) | Candidatus Nanosalina sp. J07AB43 (EGQ44217); Environmental halophage eHP-11 (AFH21856) | 33/107 (31), 1e-09; 33/99 (33), 6e-08 |
PFV1 gene products that share significant similarity (BLASTP cutoff of E = 1e-05) with sequences in the nonredundant protein database are indicated by boldface type. TMD, transmembrane domain.
Homology searches using the BLASTP program (19) revealed that only seven (18%) PFV1 gene products are significantly similar (cutoff of E = 1e-05) to sequences in the nonredundant protein database (Table S1). The closest homologs for three of these proteins (ORFs 23, 24, and 26) were encoded in the partially sequenced genome of Thermoproteus tenax virus 1 (TTV1), a virus infecting hyperthermophilic archaeon Thermoproteus tenax (20) (see below).
Notably, the enveloped filamentous virions of TTV1 are superficially similar to those of PFV1, and one of the three ORFs (ORF24) shared between the two viruses encodes a structural protein, TP3, previously detected in TTV1 virions (20). To investigate the relationship between PFV1 and TTV1 further, all PFV1 putative proteins were compared directly with those of TTV1 using BLASTP. This approach allowed the identification of six additional putative protein pairs (with pairwise identities of 24–55%) shared between these viruses (Fig. 3 and Table S1). Two of the genes were split in TTV1 as compared with PFV1. Most notably, the Cas4 nuclease-encoding gene, split in TTV1, is intact in PFV1 (ORF12). Strikingly, in TTV1 the fragment encoding the N-terminal domain of Cas4 has been exapted to function as one of the nucleocapsid proteins (TP1) (21). The intact Cas4 gene in PFV1 suggests that this virus has the ancestral genome arrangement with respect to that of TTV1. Similarly, ORF10 of PFV1 corresponds to ORFs 2 and 3 in the TTV1 genome (Fig. 3). Although we cannot exclude the possibility that some of the apparently split genes in the TTV1 genome result from sequencing errors, the experimentally determined molecular weight of the nucleocapsid protein TP1 is consistent with the one calculated from the sequence (20).
A combination of BLASTP and a more sensitive hidden Markov model (HMM)-based HHpred (homology detection and structure prediction by HMM) (22) analyses allowed the assignment of putative functions to less than a quarter of all PFV1 ORFs (nine ORFs, 23%) (Table S1). Five of the ORFs (26, 28, 29, 30, and 39) were predicted to be involved in carbohydrate metabolism. ORF26 codes for a putative glycosyltransferase (P = 97.4), and the gene product of ORF28 is a putative O-glycoside hydrolase containing a five-bladed β-propeller fold (P = 95.5). The N-terminal region (amino acids 15–146) of the latter putative protein also contains a noncatalytic carbohydrate-binding domain. ORFs 29, 30, and 39, encode small, lectin-like carbohydrate-binding proteins (Table S1). As mentioned above, ORF12 encodes a Cas4-like nuclease, which may participate in certain stages of genome replication and repair (23) or in the degradation of the host chromosome (24), as has been suggested for another crenarchaeal virus, SIRV2 (25). The product of ORF37 was confidently predicted by both BLASTP (E = 9e-10) and HHpred (P = 99.8) to be a ferritin protein, possibly involved in iron homeostasis and oxidative stress response. Finally, ORF23 encodes a putative trypsin-like serine protease (P = 96.7). Notably, the latter gene is located immediately upstream of ORF24, which encodes a homolog of the TTV1 structural protein TP3 (Fig. 3).
Virion Composition.
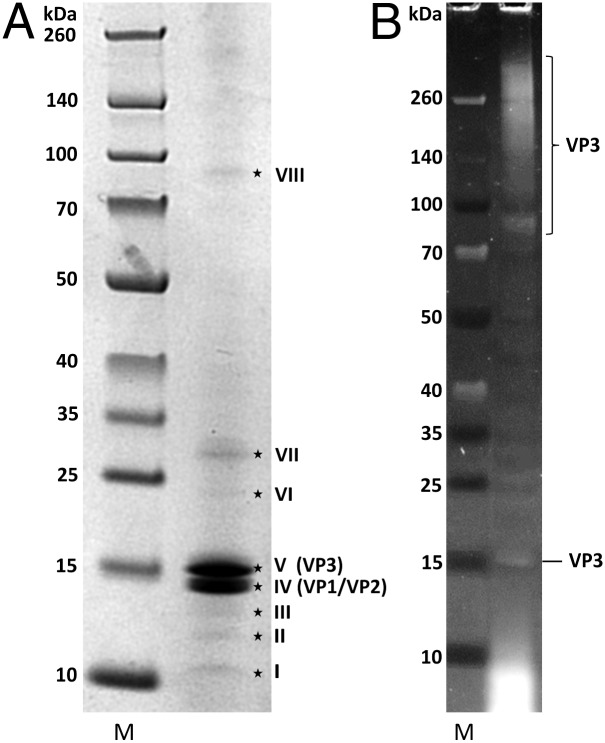
To determine the structural proteins of PFV1, highly purified virions were analyzed by SDS/PAGE. Two prominent bands of equal intensity (bands IV and V), and six weaker bands (I, II, III, VI, VII, and VIII), were observed on the gel stained with Coomassie Brilliant Blue (Fig. 4A). The analysis by LC-MS/MS revealed that band IV contained two proteins with molecular masses of about 14 kDa, the viral proteins (VP) 1 and VP2, encoded by ORF20 and ORF21, respectively (Fig. 3). Both proteins also were present in band III. Bands V, VII, and VIII contained protein VP3 with molecular mass of about 18 kDa, encoded by ORF24 (Fig. 3). VP3 also was present in the area between band VIII and the stacking gel (Fig. 4A). The incidence of VP3 at multiple positions on the gel could be caused by the occurrence of multimers of VP3 or its random aggregation, as well as by different extents of posttranslational modification of the protein. The weak bands (Fig. 4A) contained minor virion proteins, encoded by ORF23 (band VI), ORF25 (band II), ORF27 (band VII), ORF28 (band VII), and ORF37 (band I).
Fig. 4.
SDS/PAGE of PFV1 proteins. (A) Proteins stained with Coomassie Brilliant Blue. Roman numerals indicate bands of proteins with identified genes. (B) Proteins stained with ProQ-Emerald. M, molecular mass standards.
Potential glycosylation of virion proteins was analyzed using ProQ-Emerald Glycoprotein stain (Fig. 4B). Only VP3 could be identified in the stained bands. The smear in the lower part of the gel in Fig. 4B contained no PFV1 proteins and might correspond to glycolipids.
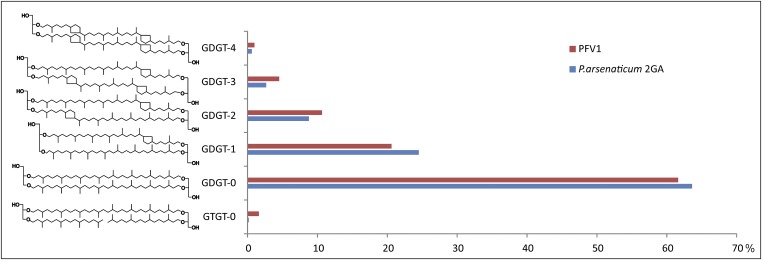
Analysis of virion organization by TEM suggested the presence of an envelope. To determine if PFV1 virions contain lipids and to compare their content with that of the host, we performed lipid analysis by LC-MS/MS (SI Materials and Methods). The analysis revealed similar lipid composition in both CsCl gradient-purified PFV1 virions and membranes extracted from P. arsenaticum 2GA (Fig. S7), suggesting a nonselective acquisition of the host lipids by the virus. In both cases, the membranes contained archaea-specific sn-2,3-dibiphytanyl diglycerol tetraethers (also known as “glycerol dibiphytanyl glycerol tetraether,” GDGT) and small amount of the glycerol trialkyl glycerol tetraether (GTGT) lipids. However, we cannot exclude the possibility that the lipid composition of the PFV1 preparation is affected by contamination by cellular components. Unfortunately, because of the low yield of PFV1, it was not feasible to perform further purification steps to exclude the latter possibility.
Fig. S7.
Analysis of lipids of P. arsenaticum strain 2GA and PFV1. (Left) Structures of identified lipids. (Right) Relative distribution (in %) of core lipid species identified by HPLC-atmosphere pressure chemical ionization (APCI)-MS in P. arsenaticum cells (blue) and purified PFV1 virions (red).
Location of the Major Virion Proteins.
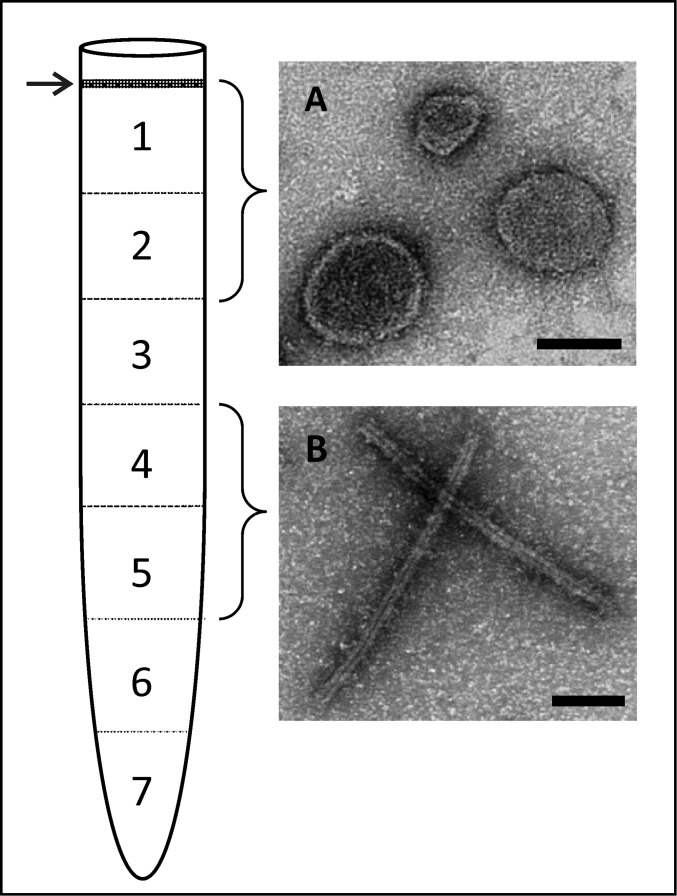
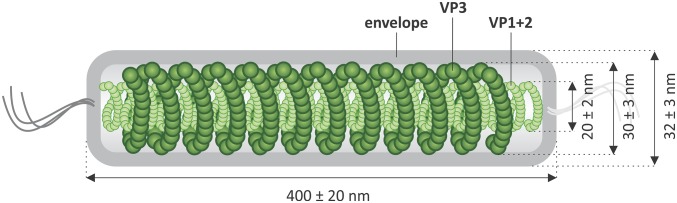
To understand the architecture of PFV1 virions further and to reveal the location of the major structural proteins, virions were dissociated by Triton X-100 treatment into two types of entities shown in Fig. 2D: irregular spheres and filaments. These two types of structures could be separated by isopycnic centrifugation in CsCl gradient in the presence of the detergent (Fig. S8). The spherical particles, likely corresponding to the viral envelope, were found in fractions collected close to the top of the centrifuge tube, in which mass spectrometry analysis revealed the presence of solely VP3 (Fig. S8). Consistently, VP3 is the only structural protein with predicted transmembrane domains (Table S1). This major virion protein apparently forms the sheath, which, along with the outer envelope, is a constituent of the spherical particles. The unsheathed helical filaments were present in the fractions collected from the middle of the centrifuge tube (Fig. S8). Mass spectrometry analysis revealed that these fractions contain proteins VP1 and VP2, which apparently are the principal protein constituents of the helical nucleocapsid. The results of the analysis of virion organization and the location of the major virion proteins are summarized as a schematic model in Fig. 5.
Fig. S8.
Separation by CsCl gradient centrifugation of PFV1 virion constituents obtained by Triton X-100 treatment. In the scheme on the left, the arrow indicates the visible band, and numbers correspond to collected fractions. (A) Electron micrographs of particles observed in the visible band and fractions 1 and 2. (B) Electron micrographs of particles observed in fractions 4 and 5. Negative stain with 2% uranyl acetate. (Scale bars: 100 nm.)
Fig. 5.
A scheme showing the organization of the PFV1 virion.
SI Materials and Methods
Purification and Analysis of Cellular DNA.
DNA was extracted from cells of the P. arsenaticum strains 2GA and PZ and P. oguniense TE7 using the Wizard Genomic DNA Purification Kit (Promega). The 16S rRNA genes were amplified by PCR using A8F forward (52) and U1525R reverse (53) primers. Additional primers for gap closing were created using CLC Genomics Workbench.
To verify the presence of the cytochrome c biogenesis factor (Pogu_1433) in the genomes of Pyrobaculum strains, two sets of PCR primers were used, one (Set 1) for the amplification of the first part of the genome and the other (Set 2) amplifying the rest of the gene. Primers for Set 1 were CytC10–27 forward: ACGGGCTACTTGTTATTG and CytC1213–1231 reverse: TGGTCAATATGTGCGACTTA. Primers for Set 2 were CytC1194–1213 forward: CGTTTTGACTTT-TTCGCCTA and CytC2609–2627 reverse: AGTATAAACAACAGCGCCA.
Purification and Analysis of PFV1 DNA.
Nucleic acid was extracted from purified virions as described by Häring et al. (54). Before DNA extraction, the virion preparation was treated with DNase I (Roche) to remove cellular DNA. DNase I was inactivated by adding EDTA to a final concentration of 20 mM and heating at 90 °C for 10 min. To identify the nature of the extracted nucleic acid, it was treated with the DNase I and type II restriction endonucleases BamHI, EcoRI, and SalI (Fermentas) to digest dsDNA and with RNase I (Roche) to digest ssRNA.
PFV1 DNA was sequenced at the sequencing facility of the University of Cambridge, Cambridge, UK, and the sequence was assembled using CLC Genomics Workbench software package (CLC Bio, Inc.).
ORFs were predicted using GLIMMER version v3.02 (55) and GeneMark.hmm (version 3.25) (56). The validity of each predicted ORF was confirmed manually by searching for a putative ribosome-binding site about 5 nt upstream of the start codon. The in silico-translated protein sequences were used as queries to search for sequence homologs in the nonredundant protein database at the National Center for Biotechnology Information using BLASTP with an upper threshold E-value of 1e-5. Conserved protein domains were identified using CD-search (57). Searches for distant homologs were performed using HHpred (22). Transmembrane domains were predicted using TMHMM (www.cbs.dtu.dk/services/TMHMM/).
Analysis of PFV1 Proteins.
The purified PFV1 preparation was treated with trichloroacetic acid (20% final volume), and the precipitated proteins were washed three times with ice-cold acetone, were dissolved in TA buffer, and were analyzed by SDS/PAGE. Proteins were stained with either Coomassie Blue using InstantBlue (Expedeon) or SYPRO Ruby Protein Gel Stain (Life Technologies). Glycoproteins were stained using Pro-Q Emerald 300 Gel and Blot stain (Life Technology).
Stained proteins were excised from the gel, and in-gel tryptic digestion was performed at the Pasteur Proteomics Platform (Institut Pasteur) as described previously (58). Trypsin-digested peptides were analyzed by nano LC-MS/MS using an Ultimate 3000 system (Dionex) coupled to an LTQ-Orbitrap Velos system (Thermo Fisher Scientific) operated in data-dependent acquisition mode with XCalibur software (Thermo Fisher Scientific). Peptide masses were searched against annotated PFV1 proteins using Andromeda (59) with MaxQuant software, version 1.3.0.5 (60).
Analysis of Lipids of P. arsenaticum 2GA Cells and PFV1 Virions.
The freeze-dried P. arsenaticum 2GA cells preparation and the PFV1 virion preparation were directly acid hydrolyzed by refluxing with 5% HCl in methanol for 3 h following Pitcher et al. (61) to release GDGT lipids, and GDGT lipids were analyzed by HPLCy/atmospheric pressure chemical ionization-mass spectrometry according to Hopmans et al. (62). The mass spectrometer was operated in single-ion mode (SIM) to monitor GDGTs with 0–8 cyclopentane moieties and the C46 GDGT standard. Relative abundances of GDGTs were determined by integrating peak areas of the SIM signal. The signal of the C46 GDGT standard was corrected for the difference in ionization efficiency using a 1:1 mixture of the standard and purified GDGT crenarchaeol.
Separation of Virion Fragments.
Virion fragments obtained by Triton X-100 treatment in a 5-mL assay were separated by isopycnic gradient centrifugation in CsCl in the presence 0.5% Triton X-100. After 50 µL was collected from the top of the tube, which contained the visible opalescent band, the gradient was fractionated into seven portions, 650 µL each. Before TEM analysis, Triton X-100 was removed from the fractions by incubation for 2 h at room temperature with Bio-Beads (Bio-Rad). All fractions were dialyzed against 20 mM TA buffer, pH 6.5, and their protein content was analyzed by mass spectrometry.
Discussion
All known filamentous viruses with dsDNA genomes infect hyperthermophilic archaea. They are classified into four different families: Lipothrixviridae (26), Rudiviridae (25), Clavaviridae (27), and Spiraviridae (28). The two former families include evolutionarily related viruses and are grouped into the order Ligamenvirales (29). In this paper we have described the isolation and characterization of a novel hyperthermophilic archaeal virus, PFV1. The only previously known virus that appears to be related to PFV1 is TTV1 (20): (i) TTV1 and PFV1 virions are enveloped rod-shaped particles, measuring about 400–500 nm in length and 32–38 nm in width; (ii) the genomes of the two viruses share nine homologous genes (23% of the PFV1 gene content), including a gene for one structural protein; (iii) both viruses are lytic and leave the host cell after its complete disruption; (iv) both viruses infect neutrophilic hyperthermophilic archaea of the order Thermoproteales. Thus, in all likelihood, TTV1 and PFV1 have evolved from a common ancestor.
Although historically TTV1 has been classified as belonging to the family Lipothrixviridae (genus Alphalipothrixvirus) based on its overall morphology and the presence of the external envelope, it does not share any homologous gene with the rest of the members of this virus family (genera Beta-, Gamma-, and Deltalipothrixvirus) (26). Even the major virion proteins of TTV1 are unrelated to those of other lipothrixviruses. Unfortunately, the TTV1 genome has not been sequenced completely (∼85% has been reported) (30–32), and the virus is not available in culture collections, so the validity of its taxonomic placement cannot be investigated. Comparison of TTV1 with PFV1 and, in particular, the dissociation experiments carried out with PFV1 now clearly show that the two viruses form a group that is unrelated to other known viruses.
TEM observations on structural entities generated by the dissociation of PFV1 virions and the examination of their protein constituents (Fig. 2 and Fig. S8) depict the virion organization and location of major virion proteins VP1, VP2, and VP3 (Fig. 5). The basic structural element of the virion, the inner helical nucleocapsid (20 ± 5 nm in width), consists of the linear dsDNA genome and multiple copies of DNA-binding proteins VP1 and VP2. It is surrounded by a helical sheath apparently composed of protein VP3. The nucleocapsid and the sheath together constitute the helical core of the virion (30 ± 5 nm in width), which is further encased in the envelope (Fig. 5). The structural organization of the PFV1 virion is in many ways similar to that of the TTV1 virion as described by Janekovic et al. (20). In the latter case, however, the sheath that surrounds the nucleocapsid has seemingly been overlooked, although the indirect evidence discussed below suggests its existence.
In the studies on the TTV1 virion, the protein TP3 was solubilized by detergent treatment, and in the liberated form it self-assembled into long filaments ∼30 nm wide (20). It was hypothesized that such filaments could be constituents of the TTV1 virion, intertwining with the helical nucleocapsid (20). In the PFV1 virion, we indeed have observed the presence of the 30-nm-wide nucleocapsid-surrounding sheath, supporting the hypothesis that a similar structural constituent exists in the TTV1 virion. Moreover, the sheath is formed from multiple copies of the protein VP3, which is homologous to the TP3 protein of TTV1.
One of the most unexpected findings of our study is that, despite pronounced structural and genomic similarities, viruses TTV1 and PFV1 differ significantly in the protein composition of their virions. Unexpectedly, VP1 and VP2, the principal constituents of the PFV1 nucleocapsid, are not homologous to nucleoproteins TP1 and TP2 of TTV1. Nevertheless, a homolog of VP1 (30% identical) is encoded by ORF11 of TTV1. The latter protein was not identified in the TTV1 virions, suggesting that VP1 and the product of TTV1 ORF11 play distinct roles in the infection cycles of the two viruses. Even more strikingly, recent sequence analysis of the TTV1 structural proteins has shown that one of the two nucleocapsid proteins, TP1, has evolved from the inactivated Cas4 nuclease, a conserved component of the CRISPR-Cas immunity systems (21). To our knowledge, this shift in function is the first example of exaptation of an enzyme to serve as a virus capsid protein. Unlike TTV1, PFV1 contains an intact Cas4-encoding gene, suggesting that the TP1 nucleoprotein of TTV1 has evolved relatively recently and functionally has replaced one of the ancestral nucleoproteins found in PFV1. This observation suggests that in genetically and structurally related viruses, such as PFV1 and TTV1, nucleoprotein filaments might be constructed using unrelated DNA-binding proteins. Importantly, this finding might be more general in virus evolution than currently appreciated. Different members of the family Bunyaviridae also form helical nucleocapsids using unrelated nucleocapsid proteins. Specifically, nucleoprotein of Crimean-Congo hemorrhagic fever virus (genus Nairovirus in the family Bunyaviridae) is homologous to the corresponding protein of Lassa fever virus (Arenaviridae) (33), whereas nucleocapsid proteins of bunyaviruses from the genus Phlebovirus display a distinct protein fold (34, 35). It also has been reported that phleboviral-like nucleocapsid protein has been transferred between viruses with negative-strand RNA and dsRNA genomes (36). These observations suggest that nucleocapsid protein genes are subject to horizontal transfer between widely different viruses and are not suitable phylogenetic markers for delineating viral families. Accordingly, we propose that VP3 forming the nucleocapsid surface shell (Fig. 5) represents the sole signature protein of PFV1-like viruses.
The fourth protein of the TTV1 virion, TP4, is a minor constituent of the virion and is thought to be involved in formation of the virion termini (20). In the PFV1 virion, along with the three major proteins, VP1, VP2, and VP3, we detected five minor proteins, none of which is homologous to TP4 of TTV1. However, some of these minor proteins might form terminal filaments of PFV1. Examples of interchangeable gene cassettes encoding receptor-binding proteins are known from several different families of archaeal viruses, including Lipothrixviridae, Sphaerolipoviridae, and Fuselloviridae (37–40). Most of the minor structural proteins of PFV1 are encoded in the vicinity of the three major virion proteins (Fig. 3), suggesting that they may be coexpressed. Functions could be predicted for three of these proteins. ORF25 encodes a trypsin-like serine protease, which could be involved in proteolytic virion maturation, as is the case in various bacterial and eukaryotic viruses (41), whereas a glycoside hydrolase encoded by ORF28 could facilitate virus entry by processing the host surface proteins, which typically are heavily glycosylated in archaea (42). In contrast, the role of the ferritin-like protein (ORF37) in the virion remains enigmatic.
PFV1, like TTV1, is a lytic virus. The other two examples of lytic viruses of hyperthermophilic archaea are the rod-shaped SIRV2 and the icosahedral Sulfolobus turreted icosahedral virus 1 (STIV). The two viruses exploit a unique mechanism of virion egress through virus-encoded pyramidal structures that are formed on the host cell surface (43–46). The egress mechanism of PFV1 appears to be completely different. No specific structures are formed on the surface of infected cells. Moreover, the cell membrane at the later stages of the infection cycle appears to be slashed by long, straight cuts not observed previously for any archaeal virus–host system (Fig. 1B). It is likely that some of the PFV1 proteins with a predicted transmembrane domain are involved in this process (Table S1).
It is interesting that, although unique among DNA viruses, the virion organization of PFV1 is superficially similar to that of members of the family Filoviridae, including Ebola virus and Marburg virus, which possess negative-sense RNA genomes (47, 48). Similar to PFV1, filamentous virions of filoviruses consist of a helical nucleocapsid, a matrix layer, and a lipid membrane. The matrix protein, similar to VP3 of PFV1, mediates the interaction between the nucleocapsid and the envelope (47, 48). This observation again suggests that overall similar designs in the evolutionarily unrelated viruses have been achieved convergently on multiple independent occasions.
Because of the unique genomic and architectural features of PFV1 and TTV1, we propose that the two viruses be considered representatives of a new viral family, which we tentatively name “Tristromaviridae” (from the Greek tria, three, and stroma, a layer).
Materials and Methods
Enrichment Culture.
The environmental sample was collected from a hot spring with neutral pH (88 °C, pH 6) in Pozzuoli, Italy (40°49′45′′ N and 14°8′50′′ E). We used 5 mL of the sample to inoculate 100 mL of the DSMZ medium 1090 (14) containing 10 g Tryptone, 1 g yeast extract, and 3 g Na2S2O3 × 5H2O without pH adjustment. The enrichment culture was incubated for 4 d at 90 °C in aerobic conditions without shaking in flasks topped with Steristoppers (Heinz Herenz).
Isolation of PFV1 and Host Strains.
VLPs were collected from cell-free culture supernatant by filtration through a nitrocellulose filter membrane with a pore size of 0.22 µm (Merck Millipore). Single Pyrobaculum strains were colony-purified from the VLP-producing enrichment culture by plating on Exedia cellulose plates (Kyokuto Pharmaceutical Industrial), infiltrated with medium 1090, and incubated for 5 d at 90 °C.
P. arsenaticum PZ6 (DSM 13514), P. calidifontis VA1 (DSM 21063), and P. oguniense TE7 (DSM 13380) were purchased from the DSMZ. The two former strains were grown in DSMZ medium 1090, as described above. Cells of P. oguniense were grown in DSMZ medium 390 (49). In all cases, the pH value was adjusted to about 7.
The analyzed strain of PFV1 was obtained by infecting cells of P. arsenaticum 2GA with freshly prepared filamentous VLPs at an MOI of ∼0.5. The virions were collected from cell-free culture supernatant by precipitation with polyethylene glycol 6000 and were purified by isopycnic gradient centrifugation in CsCl, as described previously (50).
TEM.
The samples were analyzed by TEM as described previously (27). PFV1 titer was estimated by counting the number of virions in meshes on copper grids. As a standard, we used a preparation of the SIRV2 with the titer determined by plaque assay (51).
Dissociation of PFV1 Virions.
Virus particles were dissociated by incubation in Tris-acetate (TA) buffer with Tween-20 (0.5% final concentration) or Triton X-100 (0.5% final concentration) for 4 h at room temperature. Virion fragments obtained by Triton X-100 treatment were separated by gradient centrifugation and analyzed as described in SI Materials and Methods.
DNA and protein purification and analyses were performed as described in SI Materials and Methods. The complete genome sequence of PFV1 has been deposited in the GenBank database under the accession no KU307456.
Acknowledgments
We thank Mery Pina for help in collecting environmental samples; Soizick Lucas-Staat and Jérémie Chaligné for assistance in experiments; Patrick Forterre for helpful discussions; Magalie Duchateau (Pasteur Proteomics Platform) for help with proteomics analyses; and Denise Dorhout and Irene Rijpstra (NIOZ) for lipid analysis. This work was supported by the Agence National de la Recherche program BLANC, projects “REPVIR” and “EXOVIR.” T.M. was supported also by Bourse du Gouvernement Français (Dossier 2008661) and Allocations Pasteur-Weizmann.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. KU307456).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1518929113/-/DCSupplemental.
References
- 1.Prangishvili D. The wonderful world of archaeal viruses. Annu Rev Microbiol. 2013;67:565–585. doi: 10.1146/annurev-micro-092412-155633. [DOI] [PubMed] [Google Scholar]
- 2.Krupovic M, Prangishvili D, Hendrix RW, Bamford DH. Genomics of bacterial and archaeal viruses: Dynamics within the prokaryotic virosphere. Microbiol Mol Biol Rev. 2011;75(4):610–635. doi: 10.1128/MMBR.00011-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prangishvili D. Archaeal viruses: Living fossils of the ancient virosphere? Ann N Y Acad Sci. 2015;1341:35–40. doi: 10.1111/nyas.12710. [DOI] [PubMed] [Google Scholar]
- 4.Veesler D, et al. Atomic structure of the 75 MDa extremophile Sulfolobus turreted icosahedral virus determined by CryoEM and X-ray crystallography. Proc Natl Acad Sci USA. 2013;110(14):5504–5509. doi: 10.1073/pnas.1300601110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Happonen LJ, et al. Familial relationships in hyperthermo- and acidophilic archaeal viruses. J Virol. 2010;84(9):4747–4754. doi: 10.1128/JVI.02156-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abrescia NG, Bamford DH, Grimes JM, Stuart DI. Structure unifies the viral universe. Annu Rev Biochem. 2012;81:795–822. doi: 10.1146/annurev-biochem-060910-095130. [DOI] [PubMed] [Google Scholar]
- 7.Krupovic M, Bamford DH. Virus evolution: How far does the double beta-barrel viral lineage extend? Nat Rev Microbiol. 2008;6(12):941–948. doi: 10.1038/nrmicro2033. [DOI] [PubMed] [Google Scholar]
- 8.Krupovic M, Bamford DH. Double-stranded DNA viruses: 20 families and only five different architectural principles for virion assembly. Curr Opin Virol. 2011;1(2):118–124. doi: 10.1016/j.coviro.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 9.King AM, Adams MJ, Lefkowitz EJ. Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier, London; 2011. [Google Scholar]
- 10.Klug A. The tobacco mosaic virus particle: Structure and assembly. Philos Trans R Soc Lond B Biol Sci. 1999;354(1383):531–535. doi: 10.1098/rstb.1999.0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marvin DA, Symmons MF, Straus SK. Structure and assembly of filamentous bacteriophages. Prog Biophys Mol Biol. 2014;114(2):80–122. doi: 10.1016/j.pbiomolbio.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 12.Stubbs G, Kendall A. Helical viruses. Adv Exp Med Biol. 2012;726:631–658. doi: 10.1007/978-1-4614-0980-9_28. [DOI] [PubMed] [Google Scholar]
- 13.DiMaio F, et al. Virology. A virus that infects a hyperthermophile encapsidates A-form DNA. Science. 2015;348(6237):914–917. doi: 10.1126/science.aaa4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amo T, et al. Pyrobaculum calidifontis sp. nov., a novel hyperthermophilic archaeon that grows in atmospheric air. Archaea. 2002;1(2):113–121. doi: 10.1155/2002/616075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huber R, Sacher M, Vollmann A, Huber H, Rose D. Respiration of arsenate and selenate by hyperthermophilic archaea. Syst Appl Microbiol. 2000;23(3):305–314. doi: 10.1016/S0723-2020(00)80058-2. [DOI] [PubMed] [Google Scholar]
- 16.Bernick DL, et al. Complete genome sequence of Pyrobaculum oguniense. Stand Genomic Sci. 2012;6(3):336–345. doi: 10.4056/sigs.2645906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nunoura T, Sako Y, Wakagi T, Uchida A. Regulation of the aerobic respiratory chain in the facultatively aerobic and hyperthermophilic archaeon Pyrobaculum oguniense. Microbiology. 2003;149(Pt 3):673–688. doi: 10.1099/mic.0.26000-0. [DOI] [PubMed] [Google Scholar]
- 18.Rasched I, Oberer E. Ff coliphages: Structural and functional relationships. Microbiol Rev. 1986;50(4):401–427. doi: 10.1128/mr.50.4.401-427.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janekovic D, et al. TTV1, TTV2 and TTV3, a family of viruses of the extremely thermophilic, anaerobic, sulfur reducing archaebacterium Thermoproteus tenax. Mol Gen Genet. 1983;192(1-2):39–45. [Google Scholar]
- 21.Krupovic M, Cvirkaite-Krupovic V, Prangishvili D, Koonin EV. Evolution of an archaeal virus nucleocapsid protein from the CRISPR-associated Cas4 nuclease. Biol Direct. 2015;10:65. doi: 10.1186/s13062-015-0093-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33(Web Server issue):W244–248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo Y, Kragelund BB, White MF, Peng X. Functional characterization of a conserved archaeal viral operon revealing single-stranded DNA binding, annealing and nuclease activities. J Mol Biol. 2015;427(12):2179–2191. doi: 10.1016/j.jmb.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 24.Gardner AF, Prangishvili D, Jack WE. Characterization of Sulfolobus islandicus rod-shaped virus 2 gp19, a single-strand specific endonuclease. Extremophiles. 2011;15(5):619–624. doi: 10.1007/s00792-011-0385-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prangishvili D, Koonin EV, Krupovic M. Genomics and biology of Rudiviruses, a model for the study of virus-host interactions in Archaea. Biochem Soc Trans. 2013;41(1):443–450. doi: 10.1042/BST20120313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prangishvili D. In: Lipothrixviridae. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Elsevier Academic; San Diego: 2011. pp. 211–221. [Google Scholar]
- 27.Mochizuki T, et al. Diversity of viruses of the hyperthermophilic archaeal genus Aeropyrum, and isolation of the Aeropyrum pernix bacilliform virus 1, APBV1, the first representative of the family Clavaviridae. Virology. 2010;402(2):347–354. doi: 10.1016/j.virol.2010.03.046. [DOI] [PubMed] [Google Scholar]
- 28.Mochizuki T, et al. Archaeal virus with exceptional virion architecture and the largest single-stranded DNA genome. Proc Natl Acad Sci USA. 2012;109(33):13386–13391. doi: 10.1073/pnas.1203668109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prangishvili D, Krupovic M. A new proposed taxon for double-stranded DNA viruses, the order “Ligamenvirales”. Arch Virol. 2012;157(4):791–795. doi: 10.1007/s00705-012-1229-7. [DOI] [PubMed] [Google Scholar]
- 30.Neumann H, Schwass V, Eckerskorn C, Zillig W. Identification and characterization of the genes encoding three structural proteins of the Thermoproteus tenax virus TTV1. Mol Gen Genet. 1989;217(1):105–110. doi: 10.1007/BF00330948. [DOI] [PubMed] [Google Scholar]
- 31.Neumann H, Zillig W. Coat protein TP4 of the virus TTV1: Primary structure of the gene and the protein. Nucleic Acids Res. 1989;17(22):9475. doi: 10.1093/nar/17.22.9475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neumann H, Zillig W. The TTV1-encoded viral protein TPX: Primary structure of the gene and the protein. Nucleic Acids Res. 1990;18(1):195. doi: 10.1093/nar/18.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo Y, et al. Crimean-Congo hemorrhagic fever virus nucleoprotein reveals endonuclease activity in bunyaviruses. Proc Natl Acad Sci USA. 2012;109(13):5046–5051. doi: 10.1073/pnas.1200808109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raymond DD, Piper ME, Gerrard SR, Skiniotis G, Smith JL. Phleboviruses encapsidate their genomes by sequestering RNA bases. Proc Natl Acad Sci USA. 2012;109(47):19208–19213. doi: 10.1073/pnas.1213553109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiao L, et al. Structure of severe fever with thrombocytopenia syndrome virus nucleocapsid protein in complex with suramin reveals therapeutic potential. J Virol. 2013;87(12):6829–6839. doi: 10.1128/JVI.00672-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krupovic M, Dolja VV, Koonin EV. Plant viruses of the Amalgaviridae family evolved via recombination between viruses with double-stranded and negative-strand RNA genomes. Biol Direct. 2015;10:12. doi: 10.1186/s13062-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pawlowski A, Rissanen I, Bamford JK, Krupovic M, Jalasvuori M. Gammasphaerolipovirus, a newly proposed bacteriophage genus, unifies viruses of halophilic archaea and thermophilic bacteria within the novel family Sphaerolipoviridae. Arch Virol. 2014;159(6):1541–1554. doi: 10.1007/s00705-013-1970-6. [DOI] [PubMed] [Google Scholar]
- 38.Redder P, et al. Four newly isolated fuselloviruses from extreme geothermal environments reveal unusual morphologies and a possible interviral recombination mechanism. Environ Microbiol. 2009;11(11):2849–2862. doi: 10.1111/j.1462-2920.2009.02009.x. [DOI] [PubMed] [Google Scholar]
- 39.Pina M, Bize A, Forterre P, Prangishvili D. The archeoviruses. FEMS Microbiol Rev. 2011;35(6):1035–1054. doi: 10.1111/j.1574-6976.2011.00280.x. [DOI] [PubMed] [Google Scholar]
- 40.Jaakkola ST, et al. Closely related archaeal Haloarcula hispanica icosahedral viruses HHIV-2 and SH1 have nonhomologous genes encoding host recognition functions. J Virol. 2012;86(9):4734–4742. doi: 10.1128/JVI.06666-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mateu MG. Assembly, stability and dynamics of virus capsids. Arch Biochem Biophys. 2013;531(1-2):65–79. doi: 10.1016/j.abb.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 42.Jarrell KF, et al. N-linked glycosylation in Archaea: A structural, functional, and genetic analysis. Microbiol Mol Biol Rev. 2014;78(2):304–341. doi: 10.1128/MMBR.00052-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bize A, et al. A unique virus release mechanism in the Archaea. Proc Natl Acad Sci USA. 2009;106(27):11306–11311. doi: 10.1073/pnas.0901238106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brumfield SK, et al. Particle assembly and ultrastructural features associated with replication of the lytic archaeal virus sulfolobus turreted icosahedral virus. J Virol. 2009;83(12):5964–5970. doi: 10.1128/JVI.02668-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quax TE, et al. Simple and elegant design of a virion egress structure in Archaea. Proc Natl Acad Sci USA. 2011;108(8):3354–3359. doi: 10.1073/pnas.1018052108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Daum B, et al. Self-assembly of the general membrane-remodeling protein PVAP into sevenfold virus-associated pyramids. Proc Natl Acad Sci USA. 2014;111(10):3829–3834. doi: 10.1073/pnas.1319245111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bharat TA, et al. Cryo-electron tomography of Marburg virus particles and their morphogenesis within infected cells. PLoS Biol. 2011;9(11):e1001196. doi: 10.1371/journal.pbio.1001196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bharat TA, et al. Structural dissection of Ebola virus and its assembly determinants using cryo-electron tomography. Proc Natl Acad Sci USA. 2012;109(11):4275–4280. doi: 10.1073/pnas.1120453109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sako Y, Nunoura T, Uchida A. Pyrobaculum oguniense sp. nov., a novel facultatively aerobic and hyperthermophilic archaeon growing at up to 97 degrees C. Int J Syst Evol Microbiol. 2001;51(Pt 2):303–309. doi: 10.1099/00207713-51-2-303. [DOI] [PubMed] [Google Scholar]
- 50.Bettstetter M, Peng X, Garrett RA, Prangishvili D. AFV1, a novel virus infecting hyperthermophilic archaea of the genus acidianus. Virology. 2003;315(1):68–79. doi: 10.1016/s0042-6822(03)00481-1. [DOI] [PubMed] [Google Scholar]
- 51.Prangishvili D, et al. A novel virus family, the Rudiviridae: Structure, virus-host interactions and genome variability of the sulfolobus viruses SIRV1 and SIRV2. Genetics. 1999;152(4):1387–1396. doi: 10.1093/genetics/152.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kolganova TV, Kuznetsov BB, Turova TP. [Designing and testing oligonucleotide primers for amplification and sequencing of archaeal 16S rRNA genes] Mikrobiologiia. 2002;71(2):283–286. [PubMed] [Google Scholar]
- 53.Lane DJ. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic Acids Techniques in Bacterial Systematics. John Wiley & Sons; Chichester: 1991. pp. 115–147. [Google Scholar]
- 54.Häring M, et al. Morphology and genome organization of the virus PSV of the hyperthermophilic archaeal genera Pyrobaculum and Thermoproteus: A novel virus family, the Globuloviridae. Virology. 2004;323(2):233–242. doi: 10.1016/j.virol.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 55.Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27(23):4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Besemer J, Lomsadze A, Borodovsky M. GeneMarkS: A self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001;29(12):2607–2618. doi: 10.1093/nar/29.12.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marchler-Bauer A, et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015;43(Database issue):D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilm M, et al. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature. 1996;379(6564):466–469. doi: 10.1038/379466a0. [DOI] [PubMed] [Google Scholar]
- 59.Cox J, et al. Andromeda: A peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10(4):1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 60.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 61.Pitcher A, et al. Core and intact polar glycerol dibiphytanyl glycerol tetraether lipids of ammonia-oxidizing archaea enriched from marine and estuarine sediments. Appl Environ Microbiol. 2011;77(10):3468–3477. doi: 10.1128/AEM.02758-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hopmans EC, Schouten S, Sinninghe Damsté JS. The effect of improved chromatography on GDGT based paleoproxies. Org Geochem. 2016;93:1–6. [Google Scholar]