Significance
FOF1, or ATP synthase, is often referred to as the “world’s smallest motor.” Similar to automotive engines, it employs a rotating shaft that interacts with mechanical actuators. When operating a combustion engine under load, the bearings exert significant forces on the crankshaft, leading to enhanced mechanical stress. Here, we demonstrate that analogous load-dependent effects occur in molecular motors. When FOF1 pumps protons against a transmembrane gradient, the rotor shaft undergoes structural destabilization attributed to resistive forces in its apical bearing. The effect disappears when the transmembrane gradient opposing proton pumping is short-circuited by an uncoupler, as predicted by fundamental principles of mechanics. Our observations highlight fascinating parallels between engine operation on the macroscale and the nanoscale.
Keywords: molecular motor, molecular bearing, conformational dynamics, destabilized helix, rotational resistance
Abstract
FoF1 is a membrane-bound molecular motor that uses proton-motive force (PMF) to drive the synthesis of ATP from ADP and Pi. Reverse operation generates PMF via ATP hydrolysis. Catalysis in either direction involves rotation of the γε shaft that connects the α3β3 head and the membrane-anchored cn ring. X-ray crystallography and other techniques have provided insights into the structure and function of FoF1 subcomplexes. However, interrogating the conformational dynamics of intact membrane-bound FoF1 during rotational catalysis has proven to be difficult. Here, we use hydrogen/deuterium exchange mass spectrometry to probe the inner workings of FoF1 in its natural membrane-bound state. A pronounced destabilization of the γ C-terminal helix during hydrolysis-driven rotation was observed. This behavior is attributed to torsional stress in γ, arising from γ⋅⋅⋅α3β3 interactions that cause resistance during γ rotation within the apical bearing. Intriguingly, we find that destabilization of γ occurs only when FoF1 operates against a PMF-induced torque; the effect disappears when PMF is eliminated by an uncoupler. This behavior resembles the properties of automotive engines, where bearings inflict greater forces on the crankshaft when operated under load than during idling.
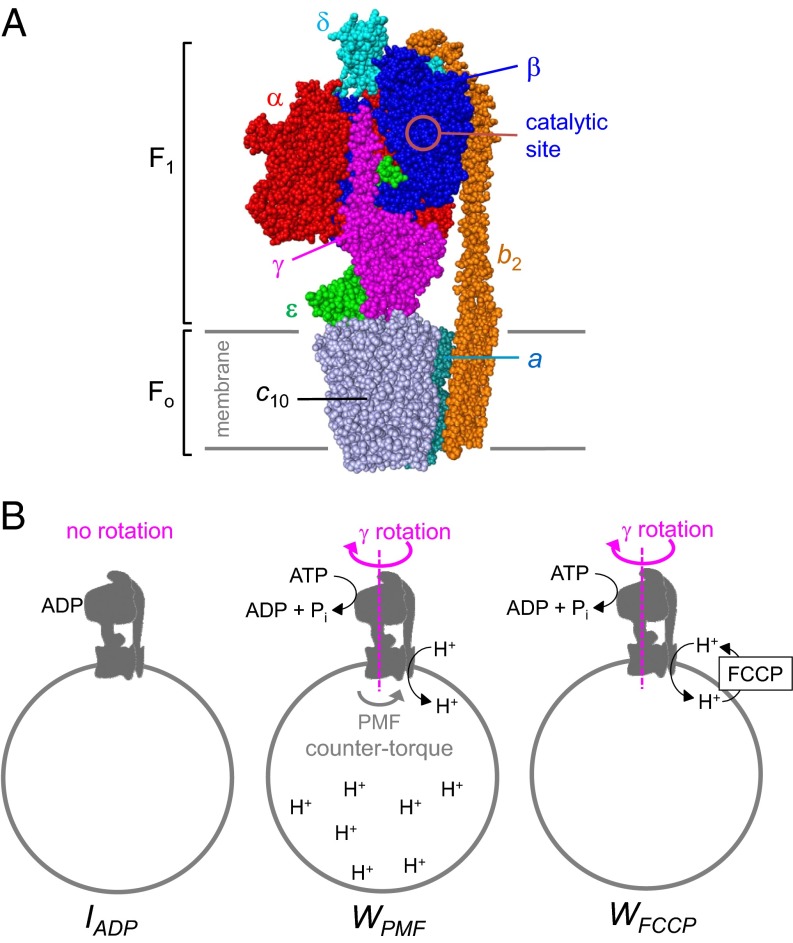
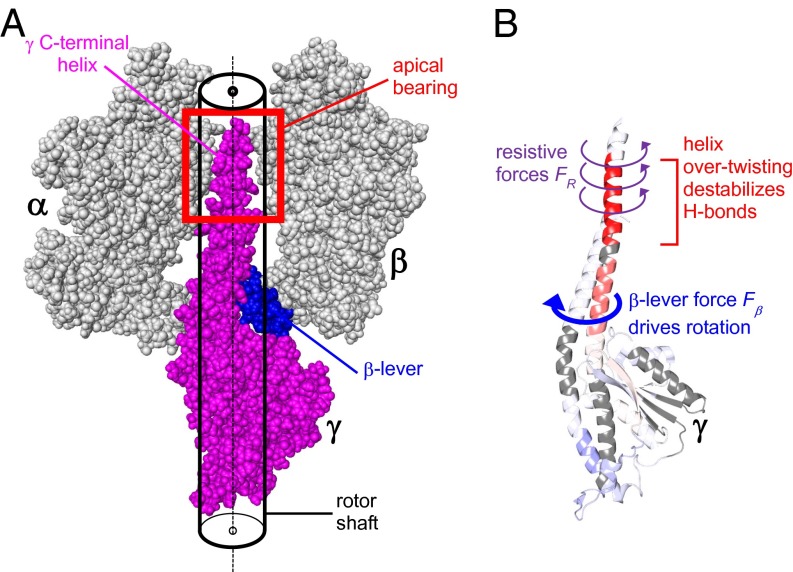
Bacteria, mitochondria, and chloroplasts share a similar FoF1-ATP synthase architecture (1–3) (Fig. 1A). The Escherichia coli enzyme considered in this work has the rotor composition γεc10 (4). The stator consists of the α3β3 catalytic head, the δb2 peripheral stalk, and the a subunit. The γε-rotor shaft connects α3β3 with the ac10 proton translocator (3, 5). The intact molecular motor has a mass of 525 kDa. FoF1 investigations are typically conducted under hydrolysis conditions (1, 6, 7), where γεc10 rotation is driven by the β subunits that cycle through a series of conformations (8). ATP hydrolysis triggers power strokes that involve consecutive interactions of the β-levers with γ (3, 6, 9).
Fig. 1.
Subunit architecture of FoF1 ATP synthase from E. coli. (A) Composite model assembled from subcomplex structures [Protein Data Bank (PDB) ID codes 3OAA, 3J0J, 1C17, 2XOK, and 2WSS]. One pair of αβ subunits facing the observer was removed to illustrate how γ extends into the apical bearing formed by the α3β3 head. During rotational catalysis, ε (partially obstructed by γ) is likely folded down toward c10, rather than being extended as shown here (10). (B) Cartoon of membrane vesicle-bound FoF1 under different experimental conditions. IADP represents ADP-inhibited FoF1; WPMF represents catalytically active FoF1 that pumps protons against a PMF-mediated countertorque; and WFCCP refers to catalytically active FoF1 that pumps protons in the presence of the uncoupler FCCP, which prevents PMF buildup.
X-ray crystallography (5, 8, 10, 11) and cryoelectron microscopy (12, 13) have provided valuable snapshots of subcomplexes and solubilized FoF1. Molecular dynamics (MD) simulations (14–16) can help interpret such data in a dynamic context. However, there is scant information on how the conformation of FoF1 is affected by internal and external forces while working under physiological conditions, where ATP/ADP interconversion is coupled with transmembrane proton transport in the presence of proton-motive force (PMF). With few exceptions (1, 3, 17), previous experiments on the rotary mechanism have focused on isolated F1.
Of particular interest is the question of how moving protein surfaces within molecular motors interact with each other. The irregular topologies and specific binding interactions of biomolecular interfaces complicate a rigorous discussion of dissipative effects in terms of classical friction models (18). It seems likely that sliding motions at such interfaces will be associated with resistance as individual residues clash with one another, and as transient binding interactions are formed and disrupted (19). Rotation of γ within the α3β3 apical bearing of FoF1 represents a prime example of such a scenario. Along these lines, recent MD simulations provided evidence for friction during torque-driven rotation of γ (15). More generally, both solvent friction and internal friction have been discussed in the context of conformational changes for monomeric proteins (19–21). For addressing these issues in the context of FoF1, it is necessary to interrogate the structure and dynamics of the molecular motor in situ while maintaining the system under different catalytically active and inhibited conditions.
Hydrogen/deuterium exchange (HDX) mass spectrometry (MS) is a sensitive approach for probing changes in protein structure and dynamics in response to external stimuli (22, 23). This technique monitors backbone deuteration in the presence of D2O. Rigid protein segments exchange more slowly than regions that are not as structurally stable (24). Under continuous-labeling conditions, the deuteration kinetics can be monitored in a spatially resolved fashion by subjecting aliquots to peptic digestion, followed by liquid chromatography and MS analysis of the resulting peptides. The use of HDX/MS is well established for protein binding experiments and several other types of investigations (22, 23); however, this technique is underused for mechanistic studies on molecular machines (25). Ryrie and Jagendorf (26) monitored hydrogen/tritium exchange in chloroplast FoF1 four decades ago, but those experiments did not yield spatially resolved information.
Here, we use HDX/MS to compare the conformational dynamics of FoF1 under various catalytically active and inhibited conditions. The experiments were conducted using vesicles that represent the natural E. coli membrane environment. Our data reveal that the γ C-terminal helix experiences load-dependent destabilization. This effect reveals that rotation of the helix within the α3β3 apical bearing is hindered by interchain contacts. We believe this investigation to be the first in situ HDX/MS investigation of a catalytically active rotational molecular motor.
Results and Discussion
HDX/MS of FoF1 in Membrane Vesicles.
Deuteration measurements were conducted on FoF1 in inside-out E. coli membrane vesicles, with focus on three conditions (Fig. 1B): (i) The presence of ADP (without ATP) produces the inhibited state IADP, where Mg⋅ADP and azide remain permanently bound in at least one catalytic site (27); (ii) the WPMF state is catalytically active (“working”; i.e., protons are pumped into the vesicle against PMF); and (iii) WFCCP also represents catalytically active FoF1, but PMF buildup is prevented by the uncoupler carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP). An ATP regeneration system was used to ensure that WPMF and WFCCP underwent ATP hydrolysis with kcat = 11 ± 1 s−1, resulting in ∼30,000 catalytic events per enzyme during the 45-min HDX time window. This value corresponds to ∼10,000 complete γ rotations for both WPMF and WFCCP, whereas rotation is blocked under IADP conditions.
Control measurements were extended to two additional types of samples: adenosine 5′-(β,γ-imido)triphosphate (AMP-PNP)–inhibited FoF1, as well as Mg2+-depleted preparations (SI Appendix, Fig. S1). These control measurements confirmed that HDX/MS is capable of pinpointing changes in structure and dynamics of vesicle-bound FoF1 with high fidelity (SI Appendix, Fig. S1A).
The use of natural E. coli membranes resulted in peptic digests containing a large number of peptides from proteins other than FoF1. Ribosomal proteins, in particular, were present in high abundance, despite the use of sucrose gradient ultracentrifugation during membrane isolation (details are provided in SI Appendix, Methods). A multidimensional analysis work flow using peptide separation based on m/z, retention time, and ion mobility drift time was required to cope with these highly complex samples (SI Appendix, Fig. S2). These digestion and analysis conditions consistently yielded a total of 203 FoF1 peptides with signal-to-noise ratios that were adequate for providing highly reproducible HDX/MS data. Sequence coverage for the extramembrane subunits of FoF1 was high (α, 83%; β, 81%; γ, 74%; δ, 77%; ε, 48%; b, 58%). Unfortunately, only a few peptides were detected for membrane-embedded subunits, such that a meaningful characterization of a and c was not possible. Low digestion yields for transmembrane segments are common in HDX/MS (28). The use of natural membranes in this work is particularly challenging (29). Notably, most earlier membrane protein HDX/MS investigations used detergent-solubilized samples (30) or purified membrane surrogates, such as nanodiscs (31).
Deuteration Behavior of Selected Peptides.
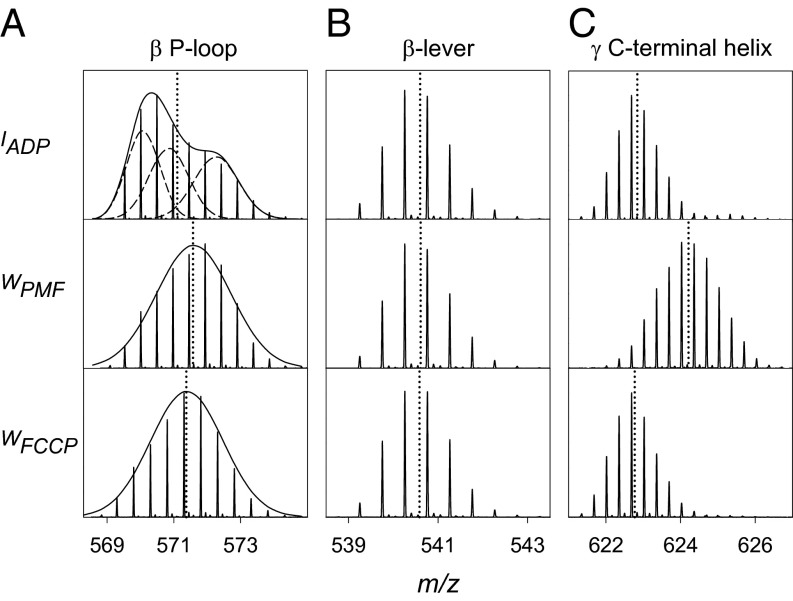
It is instructive to look initially at selected unprocessed HDX/MS data. The P-loop peptide β148FGGAGVGKTVNM159 is involved in nucleotide binding (8). In the IADP state, this region displays asymmetrical HDX distributions (Fig. 2A) that are consistent with the three stable catalytic site conformations βtight, βloose, and βopen (32). We attribute the most extensively deuterated P-loop component to βloose, which is known to adopt a distorted conformation with several disrupted H-bonds (8). Rotational averaging under WPMF and WFCCP conditions causes coalescence into a unimodal HDX envelope (Fig. 2A). Similar effects were observed for two other peptides close to the catalytic sites: β177AGVGERTREGNDF189 and α360FNAGIRPAVNPGIS373. Regions that are insensitive to changes in conditions include the β-levers (Fig. 2B). The γ C-terminal helix shows greatly enhanced HDX for WPMF, but not for WFCCP or IADP (Fig. 2C). Results for ε are illustrated in SI Appendix, Fig. S3.
Fig. 2.
HDX mass spectra of selected peptides in the IADP, WPMF, and WFCCP states. (A) Data for a peptide comprising the active site P-loop (β148FGGAGVGKTVNM159) after 1.5 min of deuteration. Gaussian deconvolution of the IADP spectrum reveals the presence of three equally populated noninterconverting conformers, attributed to βtight, βloose, and βopen. (B) β-Lever region (β380DELSEEDKL388) at t = 45 min. (C) γ C-terminal region (γ260LQLVYNKARQASITQE275) at t = 45 min. Vertical dotted lines represent centroid m/z values.
Comprehensive Overview of HDX Patterns.
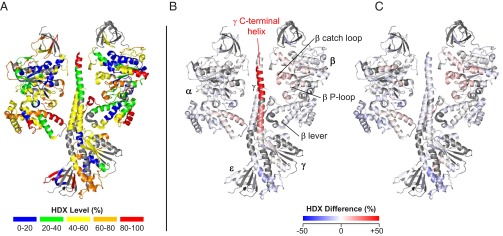
For discussing the HDX properties of FoF1, it is convenient to adopt IADP as a reference state (Fig. 3A), yielding the difference maps of Fig. 3 B and C. WPMF conditions are known to induce elastic deformation of the γ globular bottom domain (33), because the β-lever action is opposed by a c10 countertorque (3). For WFCCP, this mechanical stress is greatly reduced because the c10 countertorque is eliminated. These considerations suggest that FoF1 power transmission elements might exhibit PMF-dependent HDX characteristics. Unexpectedly, Fig. 3 B and C reveals that the HDX behavior of key power transmission elements [γ bottom domain and β-levers (3)] is largely identical with and without PMF (Fig. 3 B and C). We conclude that much of the FoF1 H-bonding network resists perturbation by intramolecular forces encountered during rotational catalysis.
Fig. 3.
(A) HDX levels of IADP for an HDX period of 45 min. Colors represent deuteration percentages as indicated in the legend; these data were derived from the centroids of the corresponding isotope distributions. To simplify the graphic representation, this figure only displays HDX/MS data for one α subunit and one β subunit, as well as γ and ε (PDB ID code 3OAA). (B) Deuteration difference map of WPMF vs. IADP. (C) Deuteration difference map of WFCCP vs. IADP. Dark red coloring of the γ C-terminal helix in B highlights dramatically enhanced deuteration of γ260LQLVYNKARQASITQEL276 under WPMF compared with IADP conditions. Regions not covered by peptide mapping are shown in dark gray.
Interestingly, major PMF-dependent HDX changes are seen for parts of the γ C-terminal helix, with deuteration levels that are ∼40% higher under WPMF conditions than in the WFCCP and IADP states (Figs. 3 B and C and 4). This effect is most pronounced for the range γ260LQLVYNKARQASITQEL276, which appears in dark red in Fig. 3B. The corresponding region is covered by five partially overlapping peptides (SI Appendix, Fig. S5). The greatly enhanced deuteration of this γ region reveals the occurrence of structural perturbations when rotational catalysis proceeds in the presence of PMF (Fig. 3B). No such destabilization is observed when the rotor shaft is stationary (IADP) or when rotation takes place in the absence of PMF (WFCCP; Fig. 3C).
Fig. 4.
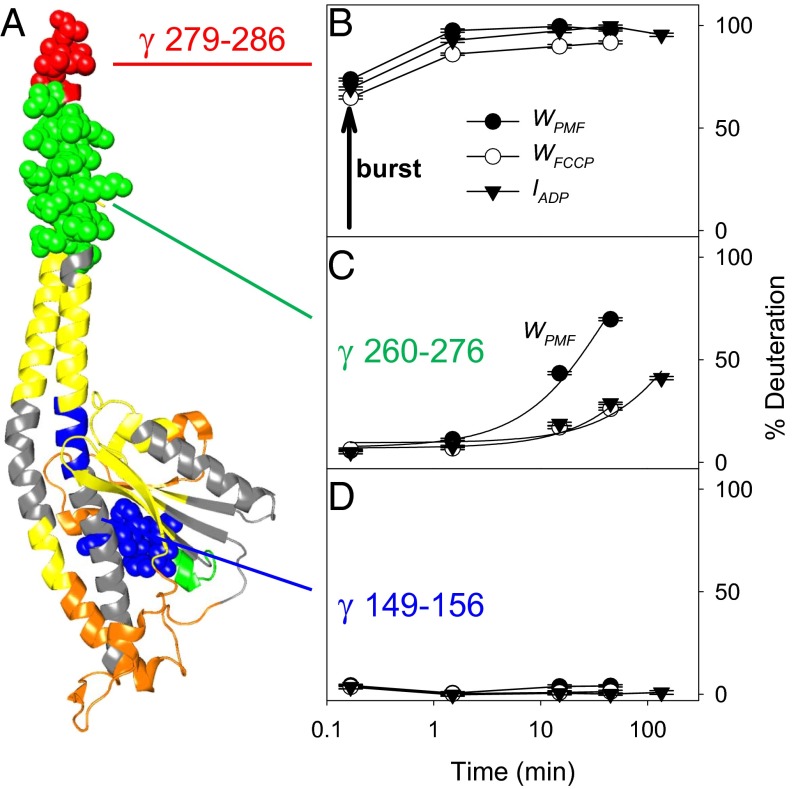
Deuteration behavior of selected γ segments. (A) X-ray structure of γ, colored as in Fig. 3A. Three peptides are highlighted in space-fill representation: γ279IVSGAAAV286, γ260LQLVYNKARQASITQEL276, and γ149IGPVKVML156. (B–D) HDX kinetics of these peptides are shown for IADP, WPMF, and WFCCP conditions. Lines in C are single-exponential fits, subject to the constraint that the deuteration level for t = ∞ is 100%. The corresponding rate constants are 6 × 10−5 s−1, 46 × 10−5 s−1, and 9 × 10−5 s−1, respectively. The vertical arrow in B represents burst phase labeling with an amplitude of ∼70%.
Characterizing the Structure of PMF-Destabilized γ.
For exploring the nature of PMF-induced structural perturbations in the γ C-terminal helix, it is helpful to consider the deuteration behavior of γ in the context of classical HDX theory (24, 34, 35). Backbone NH groups in regions that are permanently disordered are known to undergo rapid HDX with an overall rate constant kHDX that approaches the “chemical” rate constant of kch ≈ 30 s−1 at pD 8 (35). Such rapid deuteration causes a burst phase in continuous labeling experiments. This behavior is displayed by the extreme C terminus of γ (279IVSGAAAV286; Fig. 4B), which shows a burst amplitude of ∼70%. In other words, γ279IVSGAAAV286 is largely disordered in solution, regardless of experimental conditions.
Deuteration of amide NH groups in folded regions is mediated by transient H-bond opening/closing fluctuations, as envisioned by the native state mechanism (24, 35):
where kcl >> kop. In the EX2 regime encountered here, the resulting overall rate constant is kHDX = (kop/kcl) × kch and the free energy associated with H-bond opening is . Amides that follow this native state mechanism do not show a burst phase (i.e., deuteration for short HDX times is close to zero). For extremely stable regions, kop/kcl is so small that deuteration remains virtually undetectable throughout the entire experimental time window. Such a case is encountered for γ149IGPVKVML156, which is part of the globular γ bottom domain (Fig. 4D).
The γ C-terminal helix region that undergoes PMF-dependent destabilization (γ260LQLVYNKARQASITQEL276) shows deuteration kinetics in-between these two extremes (Fig. 4C). None of the profiles in Fig. 4C exhibits a burst phase, implying that residues γ 260–276 remain predominantly folded, with occasional H-bond opening/closing transitions. These transient opening events are much more extensive under WPMF conditions than for WFCCP and IADP. The heavy extent of deuteration and greatly increased HDX rate of WPMF are readily apparent in Fig. 4C. The kHDX values estimated from the exponential fits of Fig. 4C imply that hydrogen bonds in this region get destabilized by ∼4 kJ⋅mol−1 (1–2 kBT) on average, from ΔG° ≈ 32 kJ⋅mol−1 in the WFCCP state to 28 kJ⋅mol−1 per hydrogen bond under WPMF conditions.
Evidence for Hindered Rotation of γ.
What causes the destabilization of the γ C-terminal helix? Any mechanistic explanation has to address the observation that rotation, per se, is insufficient for causing this destabilization (WFCCP; Fig. 3C). Instead, destabilization is observed only when FoF1 operates against a PMF-mediated countertorque (WPMF; Fig. 3B). Different reasons may be considered when trying to account for this behavior:
-
i)
Direct power transmission: The most strongly destabilized region (Fig. 3B, dark red) is not directly involved in β/γ power transmission (3, 36, 37). Even FoF1 constructs with severely truncated γ C termini can still generate significant torque (36, 38, 39). Those previous findings imply that the PMF-induced destabilization of the γ C-terminal helix seen in our experiments is not a direct power transmission effect.
-
ii)
Reverse rotation during dwell periods: Individual power strokes cause γ to turn 120° in a clockwise direction, when viewed from the top of the α3β3 crown (40). Each power stroke is followed by a dwell, during which none of the β-levers actively drives γ rotation (6). One may contemplate whether PMF can cause reverse (counterclockwise) rotation of c10 by a small angle (<<120°) during these dwells, as suggested by some early experiments (41). Could such reverse rotation events destabilize the γ C-terminal helix? The γεc10 rotor is remarkably compliant (“soft”) in the globular bottom domain of γ, as required for elastic power transmission (3). Small reverse rotation events would likely be absorbed in this compliant region, such that their effects would not be felt at the opposite end of γ. Thus, it appears implausible that reverse rotation events could be responsible for the PMF-dependent destabilization of the γ C-terminal helix.
-
iii)
Permanent stalling of the γ C terminus: Hilbers et al. (37) demonstrated that rotational catalysis proceeds unimpeded in F1 constructs that had the γ C terminus cross-linked with α via an engineered disulfide bridge. In those constructs, the torque provided by α3β3 permanently unfolds parts of the γ C-terminal helix, causing γ to undergo ϕ/ψ swivel motions instead of rotating as a whole (37). One has to consider the possibility that comparable effects are encountered for wild-type FoF1 in our WPMF experiments, (i.e., complete stalling of the γ C terminus and unraveling of the γ C-terminal helix). Catalysis in the cross-linked constructs of Hilbers et al. (37) proceeds with permanent opening of approximately eight H-bonds in the γ C-terminal region. Such a situation would cause an HDX burst phase. The absence of a burst in Fig. 4C argues against permanent unfolding, making it unlikely that PMF induces complete stalling of the γ C terminus in our experiments. The work of Hilbers et al. (37) nonetheless provides important clues for the current discussion. Specifically, one can consider a scenario where γ is not permanently stalled, but experiences hindrance as the C-terminal helix rotates within the α3β3 apical bearing. In the following section, we make the case that such a model is consistent with our data.
-
iv)
Rotational resistance in the apical bearing: The movement of closely spaced protein surfaces relative to each other is accompanied by side-chain clashes and transient binding/dissociation events (19), likely giving rise to intermittent motion in a stick-slip regime (18). MD simulations indicate that such friction-like effects are also encountered during rotation of the γ C-terminal helix within the α3β3 apical bearing (15). The simulated γ rotation in the study by Okazaki and Hummer (15) is orders of magnitude faster than in our experiments, thereby complicating comparisons of the data obtained. Nonetheless, our findings support the basic conclusion of Okazaki and Hummer (15) that rotation of γ is associated with mechanical resistance. The FoF1 apical bearing region is highlighted in Fig. 5A; it includes the sleeve surrounding the extreme γ C terminus (8), as well as adjacent elements, such as the β-catch loops (42, 43). Rotation will be accompanied by steric clashes as γ side chains are forced past the side chains of α3β3 within the tight annular gap of the bearing (15). Intermolecular H-bonds and other noncovalent γ⋅⋅⋅α3β3 linkages (43, 44) may amplify these resistive effects. Fig. 5B illustrates how the resulting resistive forces (FR, purple) will hinder rotation of γ. These FR oppose the β-lever force (Fβ, blue), thereby causing overtwisting of the γ C-terminal helix. The torsional stress generated under these conditions will destabilize backbone H-bonds such that HDX rates are enhanced (Fig. 5B). Protein-protein contacts will also hinder rotation elsewhere along the shaft, but the apical region is most vulnerable because the apical region is where γ tapers from a coiled coil into a single helix.
Fig. 5.
(A) Close-up view of γ/α3β3 contacts (PDB ID code 3OAA after hydrogen addition; only one αβ pair is shown). Surface roughness in the apical bearing causes FR during γ rotation. (B) Interplay between FR exerted by the apical bearing and Fβ induces torsional stress in the γ C-terminal helix during each power stroke. We propose that this torsional stress destabilizes H-bonds, and thereby accelerates HDX in this region. The coloring of γ in B is consistent with the coloring of γ in Fig. 3B.
Scenario iv where γ encounters rotational resistance can explain the observed PMF dependence. An eccentric force acting on a shaft results in a stable rotation axis only if the shaft is supported by suitable bearings (45). This general principle applies equally to piston-driven crankshaft rotation and to β-lever–driven γ rotation in FoF1. Combustion engine measurements revealed that forces exerted by bearings on the crankshaft are much greater under load than during idling (45). Surface protrusions on either side of the bearing/shaft interface are more likely to interact with each other in the presence of a load, where the pressure inside the bearing is significantly enhanced (45, 46). In extreme cases, these interactions can induce material degradation (46). Our HDX data suggest that analogous effects are experienced by the γ-rotor shaft. The apical α3β3 bearing will exert greater mechanical pressure on the γ C-terminal helix in the presence of a PMF load. This bearing pressure will promote interlocking of side chains in the annular gap, thereby enhancing FR (purple arrows in Fig. 5B). As indicated in Fig. 5B, the resulting interplay of FR and Fβ causes overtwisting of the γ C-terminal helix during each power stroke, thereby destabilizing H-bonds and increasing HDX rates. Bearing pressure is lower in the absence of PMF, such that contacts at the γ/α3β3 interface become less tight. The resulting drop in FR diminishes the extent of helix overtwisting during power strokes, thereby lowering HDX levels under WFCCP conditions. A semiquantitative framework that further dissects the interplay between PMF and γ overtwisting is outlined in SI Appendix, Fig. S6.
In summary, the PMF-induced destabilization of the γ C-terminal helix is a nontrivial phenomenon. Of all the explanations considered above, scenario iv is the most plausible one. It attributes the observed destabilization to rotational resistance, consistent with crystallographically detected contacts between γ and α3β3 (43, 44), and with nonspecific friction seen in MD simulations (15).
Conclusions
FoF1-ATP synthase in a cellular environment normally operates in the presence of PMF. Thus, the findings of this work imply that a certain degree of rotational resistance is intrinsic to FoF1 operation in vivo. Our experiments could only explore ATP hydrolysis-driven rotation, but resistance will likely also be encountered during ATP synthesis.
The HDX kinetics indicate that ∼16 hydrogen bonds in the γ C-terminal region of FoF1 become destabilized by ∼4 kJ⋅mol−1 under WPMF conditions, corresponding to an overall free energy “penalty” of ∼64 kJ⋅mol−1, which is roughly equivalent to the hydrolysis of two ATP molecules (3). Evidently, it is impossible that this amount of free energy is invested during each single ATP hydrolysis event, or during each 360° rotation of γ. Instead, we propose that this 64 kJ⋅mol−1 destabilization reflects accumulated torsional stress in γ that builds up gradually and that persists over extended time periods while FoF1 is catalytically active. In other words, the rotational resistance encountered under WPMF conditions prevents γ from returning to a torsionally relaxed conformation between power strokes. This scenario is consistent with the well-established view that torsional elasticity allows γ to serve as an energy reservoir (3, 33). The functional significance of strong interactions in the apical bearing is revealed by the findings that disruption of contacts in this region by either truncation of γ or specific amino acid changes to β or γ impairs or prevents ATP-dependent H+-pumping in membrane vesicles and oxidative phosphorylation in vivo (42, 43, 47).
Experiments on isolated F1 suggest a catalytic efficiency close to 100% (3, 48). Other data indicate that the efficiency is somewhat lower (6), leaving room for dissipative phenomena. Our proposal of persisting torsional stress in the γ C-terminal helix is consistent with highly efficient operation of FoF1. The total free energy converted by each FoF1 enzyme in our experiments (with 30,000 turnover events) is ∼106 kJ⋅mol−1. Thus, the percentage of free energy associated with torsional stress accumulation is negligibly small (on the order of 64 kJ⋅mol−1/106 kJ⋅mol−1).
Wild-type FoF1 under WPMF conditions shares interesting parallels with the disulfide constructs used by Hilbers et al. (37), although the extent of the effects encountered in either case is different: (i) WPMF experiences mechanical resistance during rotation of the γ C-terminal helix, whereas the disulfide constructs operate with a permanently stalled γ C terminus; (ii) rotational obstacles in WPMF give rise to structural destabilization of the γ C-terminal helix, whereas the disulfide constructs experience complete unfolding of a large helix segment; and (iii) WPMF conditions carry a moderate free energy penalty of ∼64 kJ⋅mol−1, but the persisting nature of the structural destabilization allows wild-type FoF1 to perform highly efficient catalysis. Unraveling of the helix in the disulfide constructs carries a very large free energy penalty that does not interfere with catalysis due to the persisting nature of the structural change (the helix does not refold after the initial unraveling event). Overall, the behavior displayed by the permanently stalled constructs of Hilbers et al. (37) bolsters the validity of our conclusions; their study demonstrates that efficient catalysis is possible even under conditions that are much more severe than the gentle rotational resistance deduced from our HDX/MS data for wild-type WPMF.
The load-dependent rotor destabilization seen here for γ represents a prototypical power train feature (45, 46). Our findings highlight the fact that rotor bearings in macroscopic engines and molecular motors share common operational features. HDX/MS is well suited for interrogating these and other aspects of protein-based nanomachines. In the future, it will be interesting to apply this approach to other types of molecular motors, including flagellar systems.
Methods
The deuteration kinetics of FoF1 were monitored under various inhibited (I) and working (W) conditions [IADP, WPMF, WFCCP, IAMP-PNP, and the Mg2+-depleted inhibited state (IMg-dep)]. Inside-out E. coli membrane vesicles were exposed to 90% D2O-based labeling buffer. Aliquots were removed at various time points. Peptic digestion was carried out offline under acid-quench conditions, and peptides were separated using reverse-phase chromatography on a nanoAcquity UPLC system (Waters). Ion mobility separation and mass analysis were conducted on a Synapt G2 electrospray quadrupole time-of-flight mass spectrometer (Waters). Details regarding sample preparation and experimental methods are provided in SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by the Canadian Institutes of Health Research (Grant FRN 10237 to S.D.D.) and the Natural Sciences and Engineering Council of Canada (Grant DG 217080-2013 to L.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1520464113/-/DCSupplemental.
References
- 1.Sielaff H, Borsch M. Twisting and subunit rotation in single F(O)(F1)-ATP synthase. Philos Trans R Soc B Biol Sci. 2013;368(1611):20120024. doi: 10.1098/rstb.2012.0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wächter A, et al. Two rotary motors in F-ATP synthase are elastically coupled by a flexible rotor and a stiff stator stalk. Proc Natl Acad Sci USA. 2011;108(10):3924–3929. doi: 10.1073/pnas.1011581108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Junge W, Nelson N. ATP synthase. Annu Rev Biochem. 2015;84:631–657. doi: 10.1146/annurev-biochem-060614-034124. [DOI] [PubMed] [Google Scholar]
- 4.Ballhausen B, Altendorf K, Deckers-Hebestreit G. Constant c10 ring stoichiometry in the Escherichia coli ATP synthase analyzed by cross-linking. J Bacteriol. 2009;191(7):2400–2404. doi: 10.1128/JB.01390-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Symersky J, et al. Structure of the c(10) ring of the yeast mitochondrial ATP synthase in the open conformation. Nat Struct Mol Biol. 2012;19(5):485–491. doi: 10.1038/nsmb.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin JL, Ishmukhametov R, Hornung T, Ahmad Z, Frasch WD. Anatomy of F1-ATPase powered rotation. Proc Natl Acad Sci USA. 2014;111(10):3715–3720. doi: 10.1073/pnas.1317784111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uchihashi T, Iino R, Ando T, Noji H. High-speed atomic force microscopy reveals rotary catalysis of rotorless F₁-ATPase. Science. 2011;333(6043):755–758. doi: 10.1126/science.1205510. [DOI] [PubMed] [Google Scholar]
- 8.Abrahams JP, Leslie AGW, Lutter R, Walker JE. Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370(6491):621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 9.Mnatsakanyan N, Krishnakumar AM, Suzuki T, Weber J. The role of the betaDELSEED-loop of ATP synthase. J Biol Chem. 2009;284(17):11336–11345. doi: 10.1074/jbc.M900374200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cingolani G, Duncan TM. Structure of the ATP synthase catalytic complex (F(1)) from Escherichia coli in an autoinhibited conformation. Nat Struct Mol Biol. 2011;18(6):701–707. doi: 10.1038/nsmb.2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morales-Rios E, Montgomery MG, Leslie AGW, Walker JE. Structure of ATP synthase from Paracoccus denitrificans determined by X-ray crystallography at 4.0 Å resolution. Proc Natl Acad Sci USA. 2015;112(43):13231–13236. doi: 10.1073/pnas.1517542112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baker LA, Watt IN, Runswick MJ, Walker JE, Rubinstein JL. Arrangement of subunits in intact mammalian mitochondrial ATP synthase determined by cryo-EM. Proc Natl Acad Sci USA. 2012;109(29):11675–11680. doi: 10.1073/pnas.1204935109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allegretti M, et al. Horizontal membrane-intrinsic α-helices in the stator a-subunit of an F-type ATP synthase. Nature. 2015;521(7551):237–240. doi: 10.1038/nature14185. [DOI] [PubMed] [Google Scholar]
- 14.Nam K, Pu J, Karplus M. Trapping the ATP binding state leads to a detailed understanding of the F1-ATPase mechanism. Proc Natl Acad Sci USA. 2014;111(50):17851–17856. doi: 10.1073/pnas.1419486111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okazaki K, Hummer G. Elasticity, friction, and pathway of γ-subunit rotation in FoF1-ATP synthase. Proc Natl Acad Sci USA. 2015;112(34):10720–10725. doi: 10.1073/pnas.1500691112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mukherjee S, Warshel A. Dissecting the role of the γ-subunit in the rotary-chemical coupling and torque generation of F1-ATPase. Proc Natl Acad Sci USA. 2015;112(9):2746–2751. doi: 10.1073/pnas.1500979112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sambongi Y, et al. Mechanical rotation of the c subunit oligomer in ATP synthase (F0F1): Direct observation. Science. 1999;286(5445):1722–1724. doi: 10.1126/science.286.5445.1722. [DOI] [PubMed] [Google Scholar]
- 18.Gangloff D, Bylinskii A, Counts I, Jhe W, Vuletic V. Velocity tuning of friction with two trapped atoms. Nat Phys. 2015;11(11):915–919. [Google Scholar]
- 19.Soranno A, et al. Quantifying internal friction in unfolded and intrinsically disordered proteins with single-molecule spectroscopy. Proc Natl Acad Sci USA. 2012;109(44):17800–17806. doi: 10.1073/pnas.1117368109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chung HS, Eaton WA. Single-molecule fluorescence probes dynamics of barrier crossing. Nature. 2013;502(7473):685–688. doi: 10.1038/nature12649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bieri O, et al. The speed limit for protein folding measured by triplet-triplet energy transfer. Proc Natl Acad Sci USA. 1999;96(17):9597–9601. doi: 10.1073/pnas.96.17.9597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pirrone GF, Iacob RE, Engen JR. Applications of hydrogen/deuterium exchange MS from 2012 to 2014. Anal Chem. 2015;87(1):99–118. doi: 10.1021/ac5040242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaltashov IA, Bobst CE, Abzalimov RR. Mass spectrometry-based methods to study protein architecture and dynamics. Protein Sci. 2013;22(5):530–544. doi: 10.1002/pro.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Englander SW, Mayne L, Krishna MMG. Protein folding and misfolding: mechanism and principles. Q Rev Biophys. 2007;40(4):287–326. doi: 10.1017/S0033583508004654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan Y, Brown L, Konermann L. Hydrogen exchange mass spectrometry of bacteriorhodopsin reveals light-induced changes in the structural dynamics of a biomolecular machine. J Am Chem Soc. 2011;133(50):20237–20244. doi: 10.1021/ja206197h. [DOI] [PubMed] [Google Scholar]
- 26.Ryrie IJ, Jagendorf AT. An energy-linked conformational change in the coupling factor in chloroplasts. Studies with hydrogen exchange. J Biol Chem. 1971;246(11):3771–3774. [PubMed] [Google Scholar]
- 27.Bowler MW, Montgomery MG, Leslie AGW, Walker JE. How azide inhibits ATP hydrolysis by the F-ATPases. Proc Natl Acad Sci USA. 2006;103(23):8646–8649. doi: 10.1073/pnas.0602915103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joh NH, et al. Modest stabilization by most hydrogen-bonded side-chain interactions in membrane proteins. Nature. 2008;453(7199):1266–1270. doi: 10.1038/nature06977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rey M, Forest E, Pelosi L. Exploring the conformational dynamics of the bovine ADP/ATP carrier in mitochondria. Biochemistry. 2012;51(48):9727–9735. doi: 10.1021/bi300759x. [DOI] [PubMed] [Google Scholar]
- 30.West GM, et al. Ligand-dependent perturbation of the conformational ensemble for the GPCR β2 adrenergic receptor revealed by HDX. Structure. 2011;19(10):1424–1432. doi: 10.1016/j.str.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hebling CM, et al. Conformational analysis of membrane proteins in phospholipid bilayer nanodiscs by hydrogen exchange mass spectrometry. Anal Chem. 2010;82(13):5415–5419. doi: 10.1021/ac100962c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boyer PD. The ATP synthase--a splendid molecular machine. Annu Rev Biochem. 1997;66:717–749. doi: 10.1146/annurev.biochem.66.1.717. [DOI] [PubMed] [Google Scholar]
- 33.Sielaff H, et al. Domain compliance and elastic power transmission in rotary F(O)F(1)-ATPase. Proc Natl Acad Sci USA. 2008;105(46):17760–17765. doi: 10.1073/pnas.0807683105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hvidt A, Nielsen SO. Hydrogen exchange in proteins. Adv Protein Chem. 1966;21:287–386. doi: 10.1016/s0065-3233(08)60129-1. [DOI] [PubMed] [Google Scholar]
- 35.Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17(1):75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hossain MD, et al. The rotor tip inside a bearing of a thermophilic F1-ATPase is dispensable for torque generation. Biophys J. 2006;90(11):4195–4203. doi: 10.1529/biophysj.105.079087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hilbers F, Junge W, Sielaff H. The torque of rotary F-ATPase can unfold subunit gamma if rotor and stator are cross-linked. PLoS One. 2013;8(1):e53754. doi: 10.1371/journal.pone.0053754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Junge W, Sielaff H, Engelbrecht S. Torque generation and elastic power transmission in the rotary F(O)F(1)-ATPase. Nature. 2009;459(7245):364–370. doi: 10.1038/nature08145. [DOI] [PubMed] [Google Scholar]
- 39.Furuike S, et al. Axle-less F1-ATPase rotates in the correct direction. Science. 2008;319(5865):955–958. doi: 10.1126/science.1151343. [DOI] [PubMed] [Google Scholar]
- 40.Noji H, Yasuda R, Yoshida M, Kinosita K., Jr Direct observation of the rotation of F1-ATPase. Nature. 1997;386(6622):299–302. doi: 10.1038/386299a0. [DOI] [PubMed] [Google Scholar]
- 41.Chang TM, Penefsky HS. Energy-dependent enhancement of aurovertin fluorescence. An indicator of conformational changes in beef heart mitochondrial adenosine triphosphatase. J Biol Chem. 1974;249(4):1090–1098. [PubMed] [Google Scholar]
- 42.Greene MD, Frasch WD. Interactions among gamma R268, gamma Q269, and the beta subunit catch loop of Escherichia coli F1-ATPase are important for catalytic activity. J Biol Chem. 2003;278(51):51594–51598. doi: 10.1074/jbc.M309948200. [DOI] [PubMed] [Google Scholar]
- 43.Boltz KW, Frasch WD. Interactions of gamma T273 and gamma E275 with the beta subunit PSAV segment that links the gamma subunit to the catalytic site Walker homology B aspartate are important to the function of Escherichia coli F1F0 ATP synthase. Biochemistry. 2005;44(27):9497–9506. doi: 10.1021/bi050070o. [DOI] [PubMed] [Google Scholar]
- 44.Bowler MW, Montgomery MG, Leslie AGW, Walker JE. Ground state structure of F1-ATPase from bovine heart mitochondria at 1.9 A resolution. J Biol Chem. 2007;282(19):14238–14242. doi: 10.1074/jbc.M700203200. [DOI] [PubMed] [Google Scholar]
- 45.Kapulainen M, et al. Fibre optic sensors for long-term monitoring of oil film pressure in diesel engine main bearing. Tribol Lett. 2014;56(1):47–54. [Google Scholar]
- 46.Wang QA. Seizure failure of journal-bearing conformal contacts. Wear. 1997;210(1-2):8–16. [Google Scholar]
- 47.Iwamoto A, Miki J, Maeda M, Futai M. H(+)-ATPase gamma subunit of Escherichia coli. Role of the conserved carboxyl-terminal region. J Biol Chem. 1990;265(9):5043–5048. [PubMed] [Google Scholar]
- 48.Yasuda R, Noji H, Kinosita K, Jr, Yoshida M. F1-ATPase is a highly efficient molecular motor that rotates with discrete 120 degrees steps. Cell. 1998;93(7):1117–1124. doi: 10.1016/s0092-8674(00)81456-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.