Significance
Despite its importance for a host of cellular processes and contribution to neurological, viral, and bacterial disease, the molecular mechanisms underlying the regulation of the heterotetrameric motor kinesin-1 by its light chains and the binding of its cargo are not well understood. Here, we describe how a previously unnoticed intramolecular interaction between the light chain tetratricopeptide repeat domain (KLC2TPR) and a highly conserved peptide motif within an unstructured region of the molecule occludes a key cargo binding site on the light-chain TPR domain. Cargo binding displaces this intramolecular interaction, effecting a global overall conformational change in KLCs that results in a more extended conformation. We propose a model describing how, via this molecular switch, cargo binding regulates the activity of the holoenzyme.
Keywords: kinesin, KLC, TPR domain, microtubule motor, cytoskeleton
Abstract
The light chains (KLCs) of the microtubule motor kinesin-1 bind cargoes and regulate its activity. Through their tetratricopeptide repeat domain (KLCTPR), they can recognize short linear peptide motifs found in many cargo proteins characterized by a central tryptophan flanked by aspartic/glutamic acid residues (W-acidic). Using a fluorescence resonance energy transfer biosensor in combination with X-ray crystallographic, biochemical, and biophysical approaches, we describe how an intramolecular interaction between the KLC2TPR domain and a conserved peptide motif within an unstructured region of the molecule, partly occludes the W-acidic binding site on the TPR domain. Cargo binding displaces this interaction, effecting a global conformational change in KLCs resulting in a more extended conformation. Thus, like the motor-bearing kinesin heavy chains, KLCs exist in a dynamic conformational state that is regulated by self-interaction and cargo binding. We propose a model by which, via this molecular switch, W-acidic cargo binding regulates the activity of the holoenzyme.
The heterotetrameric microtubule (MT) motor kinesin-1 (also known as conventional kinesin) has diverse roles in protein, ribonuclear protein, vesicular, and organelle transport by virtue of its ability to interact with many different cargoes (1, 2). It is also hijacked by pathogens during infection (3). Accumulating evidence suggests a key role for kinesin-1–dependent MT transport in several neurological disorders including Alzheimer’s disease (4). Thus, determining the molecular basis for cargo recognition and regulation of kinesin-1 is important for understanding its role in normal cell function and disease states.
Kinesin-1 is composed of two heavy (KHCs) and two light chains (KLCs) that, in mammalian cells, are encoded by several closely related genes with distinct cell and tissue expression profiles (Kif5A-C and KLC1-4, respectively). The heavy chains have a MT-binding ATPase motor domain at their amino terminus followed by a neck coil and an extended series of coiled coils, separated by a hinge region(s), that results in heavy-chain dimerization (5). The carboxyl-terminal domain of the heavy chains is largely unstructured. The light chains associate with the heavy-chain coiled coils at the carboxyl-terminal portion of the molecule through a series of heptad repeats (6). A highly charged unstructured linker region connects this heavy-chain binding region to a tetratricopeptide repeat domain (KLCTPR) formed of six helix-turn-helix TPR repeats (TPR1-6), followed by a C-terminal region that varies considerably between the different KLCs and splice variants.
In the absence of cargo binding, kinesin-1 exists in a folded, compact state that prevents unnecessary cycles of ATP hydrolysis. This is achieved via an intramolecular interaction in which the C-terminal isoleucine–alanine–lysine (IAK) motif (and flanking amino acids) of a single KHC tail binds at the N-terminal motor dimer interface and participates in a “double-lockdown” mechanism whereby it cross-links the motor domains preventing movement of the neck linker region that is required for ADP release (7–12). In the cargo-bound active state, tail-mediated inhibition is relieved resulting in a more elongated structure that is able to hydrolyze ATP and translocate along MTs (12–16). As well as binding to cargoes, the KLCs are thought to regulate KHC autoinhibition, although the molecular mechanism(s) that couple these two functions are unclear (17). Several studies suggest that KLCs reduce interaction with MTs and help to maintain the autoinhibited state in the absence of cargo (12, 13, 18), whereas in vitro biophysical studies have suggested that the presence of light chains reduces the affinity of the motor domains for the C-terminal autoinhibitory heavy-chain tail through both steric and electrostatic factors (19).
Vesicular cargoes interact via adaptor proteins that can bind to several sites on both KHCs and KLCs, and it is generally thought that these multiple contacts help to stabilize the active state and/or destabilize the inactive state and thus promote cargo-dependent transport (15, 17, 20, 21). It has emerged that diversity of light-chain cargo recognition is accomplished, in part, through TPR domain interaction with short linear peptide motifs (22–25). We have recently described how the TPR domain of KLC2 (KLC2TPR) recognizes one class of these peptides that are characterized by a central tryptophan typically flanked by aspartic or glutamic acid residues (W-acidic). The X-ray structure of KLC2TPR in complex with a W-acidic peptide of the lysosome adaptor SKIP (SKIPWD) shows that these motifs interact with a concave positively charged groove at the KLCTPR N terminus. Both sequence-specific and electrostatic elements contribute to peptide recognition, which is stabilized by residues from TPR2–TPR3 and the internal helix of TPR4 (23). Functional W-acidic motifs have been identified in a growing number of cargo adaptors, including the neuronal protein calsyntenin-1 (CSTN1) that plays a role in the axonal transport of amyloid precursor protein, as well as nesprin-2, gadkin, vaccinia virus A36R, and cayman ataxia protein (BNIP-H), where, in each case, they provide a crucial link between motor and cargo with diverse functions (16, 23, 24, 26–31). It is interesting to note that W-acidic motifs share sequence similarity with the A (acidic) motif of several actin nucleation-promoting factors (NPFs) including WASP, N-WASP, and WAVE1 (32), and the mechanism of binding to KLCTPR is somewhat similar to the interaction of the fission yeast WASP A motif on the Arp2/3 complex (33). Indeed, in the case of gadkin, there also appears to be functional overlap (32).
Here, we describe an intramolecular interaction between KLCTPR and the unstructured region immediately N-terminal to it. This flexible linker features a highly conserved leucine–phenylalanine–proline motif flanked by acidic residues (LFP-acidic) that interacts with KLCTPR partly occluding its W-acidic motif binding site. This autoinhibitory interaction is displaced by cargo binding, resulting in overall conformational changes within the light chains. Thus, paralleling the behavior of KHCs, kinesin-1 KLCs also exist in a dynamic conformational state that is regulated by self-interaction and cargo binding. We propose a model to explain how this previously unnoticed molecular switch may couple KLCTPR–W-acidic peptide recognition to the regulation of kinesin-1 activity.
Results
The KLC Region N-Terminal to Its TPR Domain Features a Conserved LFP-Acidic Motif and Negatively Regulates W-Acidic Cargo Binding.
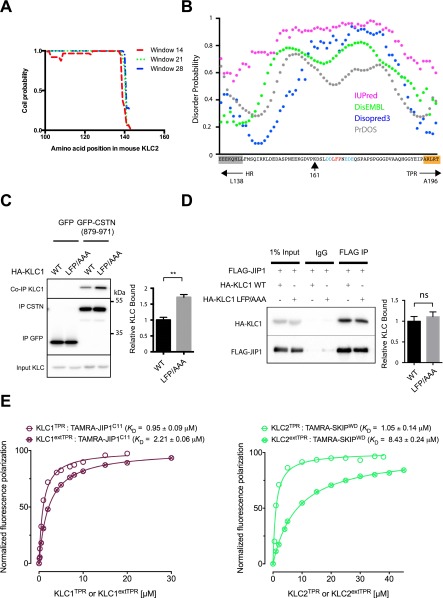
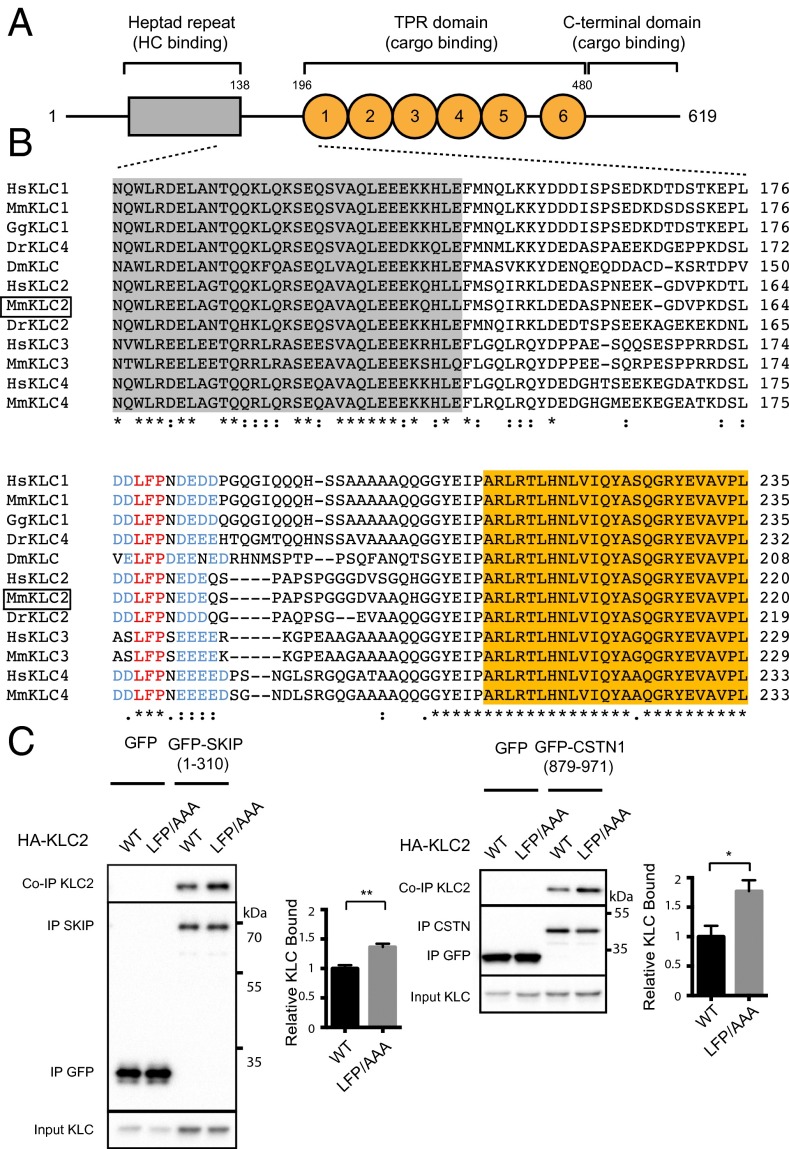
Amino acid sequence alignment of all four KLCs from human and mouse as well as representative kinesin-1 light chains from several diverse species reveals that the heptad repeat region (that interacts with KHC via a predicted coiled coil; Fig. S1A) and the TPR domain (that binds cargoes) are highly conserved, whereas the intervening stretch of highly charged amino acids (F139–P195 in mouse KLC2) is considerably more divergent (Fig. 1 A and B). Within this region, we noticed, however, that a leucine–phenylalanine–proline (LFP) motif (residues 167–169 in mouse KLC2) is totally conserved (in red in Fig. 1B). This short motif that is followed by Asn/Ser and flanked by negatively charged Asp/Glu residues (in blue in Fig. 1B) is present in all KLCs. No function has been ascribed to this conserved LFP-acidic feature. Analyses using a panel of intrinsic-disorder prediction packages (PrDOS, Disopred, IUPred, DisEMBL) indicate that this protein region is likely unstructured (Fig. S1B).
Fig. S1.
(A) Coil probability plot using PCOILS (toolkit.tuebingen.mpg.de/pcoils) showing predicted end of the KHC interacting heptad repeat region in KLC2. (B) Disorder probability plot using four different disorder prediction servers (IUPred, magenta; DisEMBL, green; Disopred3, blue; PrDOS, gray) showing predicted disorder in the HR–TPR linker region. Predicted end of heptad repeat (HR L138) and start of TPR1 (A196) are highlighted in blue and orange, respectively. LFP motif is in red. Residue K161 that is the start KLC2extTPR construct is also indicated. (C) Western blot analysis of GFP-TRAP immunoprecipitation experiments from transfected HeLa cells showing enhanced binding between GFP-CSTN1 (879–971) and HA-KLC1 when the LFP motif is mutated to AAA. Graphs show quantification of relative binding from three independent experiments. Error bars show SEM. **P < 0.01. (D) Western blot analysis of FLAG-immunoprecipitation experiment from transfected HeLa cells showing comparable binding between FLAG-JIP1 and HA-KLC1, with or without the LFP > AAA mutation. Graph shows quantification of relative binding from three independent experiments. Error bars show SEM. ns, not statistically significant. (E, Left) FP measurements showing effect of amino-terminal extension of the KLC1extTPR to residue 173 (KLC1extTPR) on interaction with a TAMRA-conjugated peptide derived from the C terminus of JIP1 (JIPC11). (Right) Panel from Fig. 3B is reproduced for comparison.
Fig. 1.
Mutational disruption of a conserved leucine–phenylalanine–proline (LFP) motif in KLC2 enhances binding to W-acidic cargoes. (A) Schematic showing domain organization of KLC2 (numbering refers to mouse protein). The heptad repeat region (HR) that interacts with KHC is shown in gray, and the six TPR repeats comprising the TPR domain are represented by orange circles. (B) Multiple sequence alignment using Clustal W showing the region linking the HR and the first TPR repeat in KLCs 1–4 from human and mouse as well as representative fly (Dm), zebrafish (Dr), or chicken (Gg) homologs as annotated in the Homologene database. The highly conserved HR and TPR regions are highlighted in gray and orange, respectively. An asterisk (*) indicates a completely conserved residue, whereas a colon (:) indicates strong conservation of residue properties. A universally conserved LFP motif (red), located within the otherwise-divergent linker region is highlighted in red as well as flanking conserved acidic residues in (blue). (C) Western blot analysis of GFP-TRAP immunoprecipitation experiments from transfected HeLa cells showing enhanced binding between GFP-SKIP (1–310) (Left) or GFP-CSTN1 (879–971) (Right) to HA-KLC2 when the LFP triplet is replaced to AAA. Graphs show quantification of relative binding from three independent experiments. Error bars show SEM. **P < 0.01, *P < 0.05.
To examine the role of the LFP-acidic motif in light-chain function, we performed GFP-TRAP immunoprecipitation experiments from HeLa cells with two independent W-acidic cargo proteins using full-length wild-type KLC2 or KLC2 where the LFP triplet was replaced by AAA. This revealed that disruption of this sequence enhances binding to both the N-terminal domain of SKIP (amino acids 1–310) and the cytoplasmic domain of CSTN1 (amino acids 879–971) by 36% and 78%, respectively (Fig. 1C). Consistently, the LFP/AAA replacement in KLC1 enhanced binding to CSTN1 by 72% (Fig. S1B). However, disruption of this motif did not affect binding to the non–W-acidic cargo JIP1, which mutagenesis data suggest binds to a distinct site on KLC1TPR (Fig. S1D) (22, 34). Given that the LFP motif is located N-terminal to the TPR domain, we reflected that this could imply a mechanism of cross talk between this linker region and the W-acidic binding site involving residues from TPR2-3 and the internal helix of TPR4 (23). We considered the possibility that this cross talk could be mediated by an intramolecular interaction.
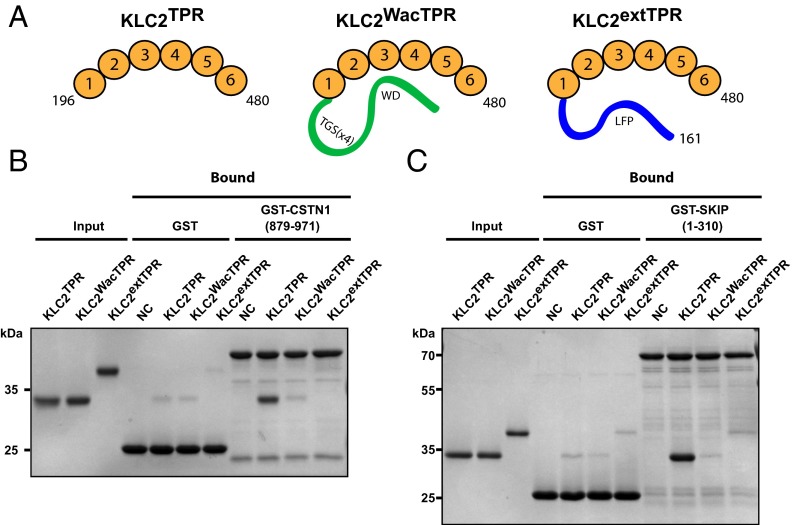
To directly examine the contribution of this region to TPR domain cargo binding in vitro, we performed pull-down assays using purified GST-SKIP (1–310) or GST-CSTN1 (879–971). We compared binding to KLC2TPR, KLC2TPR with a N-terminal extension to include the LFP-acidic motif (KLC2 161–480, KLC2extTPR) or KLC2TPR fused N-terminally to a competitor W-acidic peptide from SKIP (KLC2WacTPR) (Fig. 2A). For both SKIP and CSTN1, amino-terminal extension of the TPR domain up to residue 161, to include the LFP-acidic motif, inhibits binding to the W-acidic motif-containing cargoes in a manner comparable to inclusion of a direct competitor peptide attached by a flexible linker (Fig. 2 B and C). This strongly supports the proposition that this linker region can compete with W-acidic cargo for a binding site on the TPR domain through an intramolecular interaction.
Fig. 2.
The LFP-acidic motif-containing HR–TPR linker region inhibits W-acidic cargo binding in vitro. (A) Schematic showing KLC2TPR, KLC2WacTPR, and KLC2extTPR proteins used in this work. KLCTPR consists of residues (196–480) of KLC2 comprising its six TPR repeats; KLCextTPR consists of residues (161–480) encompassing the six TPR repeats and an N-terminal extension (ext) to include the conserved LFP-acidic motif. KLCWacTPR comprises residues (196–480) of KLC2 with the first W-acidic motif of SKIP (STNLEWDDSAI, amino acids 202–212) coupled by a flexible (TGS)4 linker. (B and C) GST–pull-down experiment showing interaction between the above TPR proteins with (B) GST-CSTN1 (879–971) or (C) GST-SKIP (1–310). The TPR domain alone shows robust binding to both W-acidic cargo proteins, but binding is reduced to background levels by inclusion of the N-terminal region carrying the LFP-acidic motif. Similarly, inclusion of a competitor peptide sequence also inhibits the interaction.
The KLC Region N-Terminal to Its TPR Domain Directly Interacts in an LFP Motif-Dependent Manner with the TPR Domain.
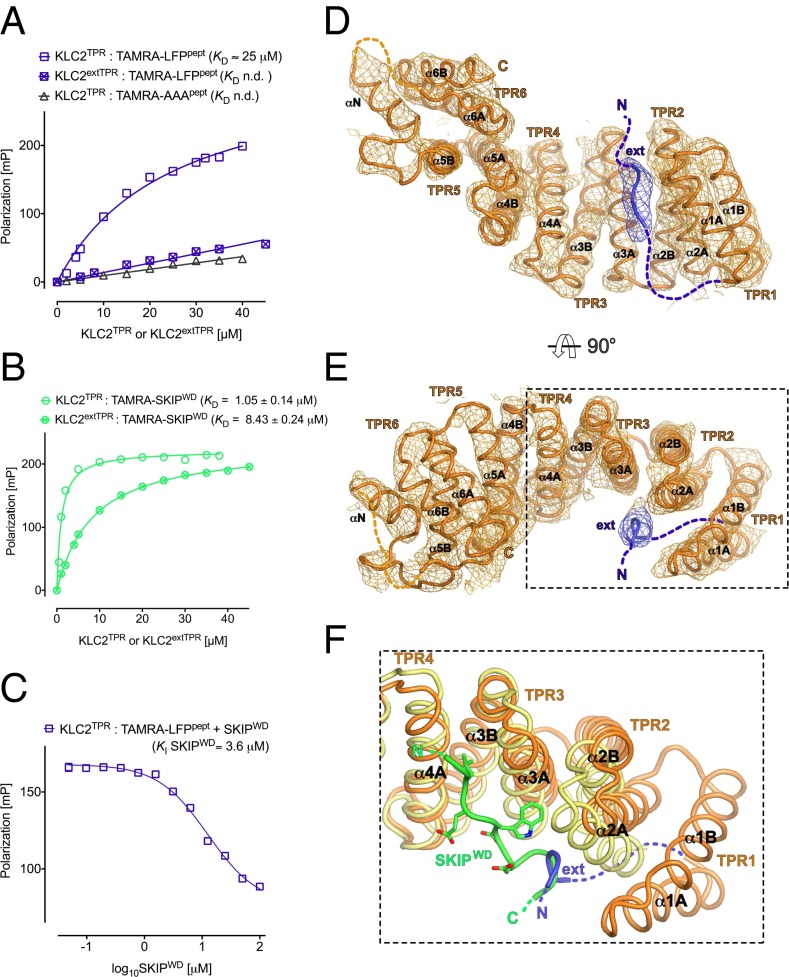
Next, we used a biophysical assay to test whether the LFP-acidic motif can interact with purified KLC2 proteins. Fluorescence polarization (FP) measurements using a 14-aa N-terminally carboxytetramethylrhodamine (TAMRA)-conjugated peptide comprising the LFP motif and its flanking residues (LFPpept, DSLDDLFPNEDEQS) showed that this sequence binds to KLC2TPR with a dissociation constant (KD) estimated at ∼25 μM (squares in Fig. 3A). This interaction requires the LFP sequence because an equivalent peptide, in which this triplet was replaced by AAA (AAApept, DSLDDAAANEDEQS), did not bind to KLC2TPR (triangles in Fig. 3A). Further supporting a self-interaction model, binding between LFPpept and KLC2TPR was also strongly inhibited by the inclusion of the extended endogenous N-terminal sequence (crossed squares in Fig. 3A). Similarly, and consistent with our GST–pull-down analysis, the presence of the N-terminal extension also reduced affinity for a TAMRA-conjugated W-acidic cargo peptide from SKIP (SKIPWD, STNLEWDDSAI; KD increased from 1.05 ± 0.14 to 8.43 ± 0.24 μM) (Fig. 3B). Moreover, a competition assay in which increasing amounts of nonlabeled SKIPWD were titrated into a KLC2TPR:TAMRA-LFPpept complex revealed a concentration-dependent decrease in FP (Fig. 3C). From this experiment, we estimated a KI value (35) for the unlabeled SKIPWD peptide of 3.6 μM. This is in good agreement with the KD value for TAMRA-SKIPWD supporting a model in which W-acidic binding displaces the bound LFP peptide. In a separate experiment, we also validated our immunoprecipitation analysis that indicated that KLC1 LFP/AAA replacement has little effect on JIP1 binding (Fig. S1D). Consistent with this, we found that inclusion of an N-terminal extension of KLC1TPR (KLC1extTPR) only marginally reduced its affinity for a TAMRA-conjugated peptide comprising the TPR binding C-terminal 11 aa of JIP1 (YTCPTEDIYLE) (KD increased from 0.95 ± 0.09 to 2.21 ± 0.06 μM; Fig. S1E).
Fig. 3.
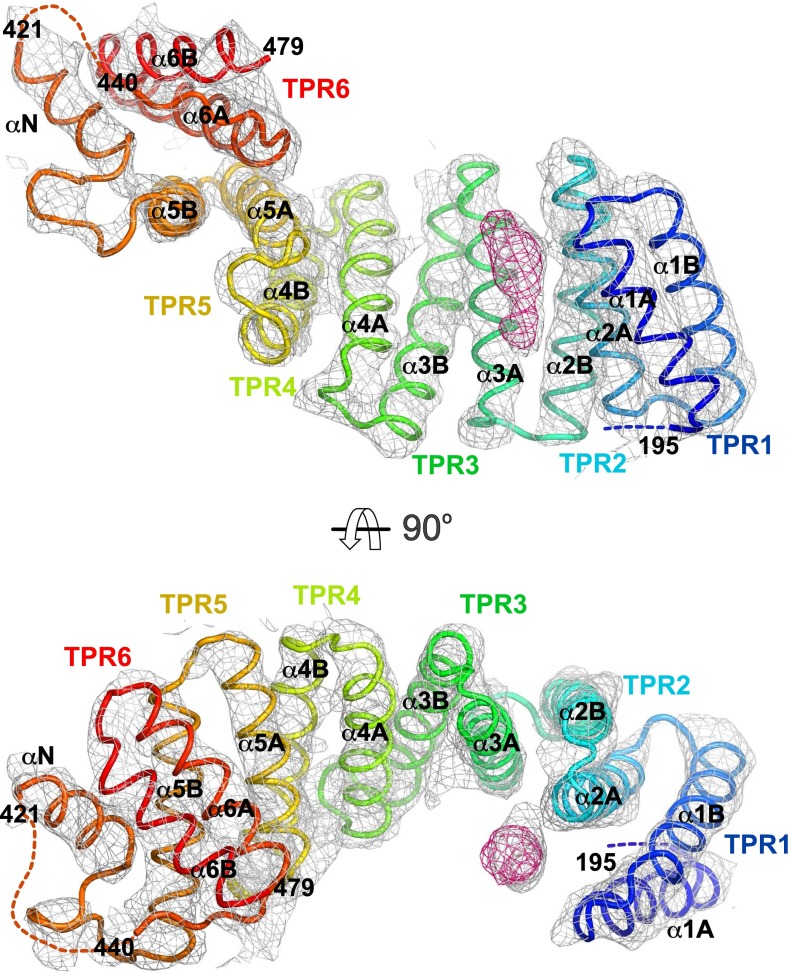
The HR–TPR linker region interacts in an LFP-dependent manner with KLC2TPR at a binding site partly overlapping with that of cargo W-acidic motifs. (A) Fluorescence polarization (FP) measurements showing concentration and LFP-dependent interaction between TAMRA-conjugated LFPpept (DSLDDLFPNEDEQS) and KLC2TPR. Mutation of the LFP motif to AAA (DSLDDAAANEDEQS) essentially eliminates binding. Amino-terminal extension of the TPR domain to residue 161 (KLCextTPR) also inhibits interaction with LFPpept. (B) FP measurements showing amino-terminal extension of the TPR domain to residue 161 (KLCextTPR) inhibits interaction with SKIPWD compared with KLC2TPR alone. (C) FP experiment titrating increasing amounts of nonlabeled SKIPWD in a KLC2TPR:LFPpept complex showing that binding of LFPpept and SKIPWD to KLC2TPR are mutually exclusive. (D and E) Illustrated cartoon representations of the KLC2extTPR X-ray structure in two orthogonal orientations. The peptide originating from the extended N-terminal region (ext in blue) is bound to the KLC2TPR domain (orange). 2mFo-DFc electron density map is shown at the 1.2σ level. Individual TPR repeats composed by helix1-turn-helix2 elements are highlighted. The non-TPR helix between TPR5 and TPR6 is labeled αN. (F) Superposition between KLC2extTPR and cargo-bound SKIPWD:KLC2TPR (3ZFW) structures in the same orientation as E. The SKIPWD W-acidic cargo peptide is shown in green, and the cargo-bound KLC2TPR is shown in yellow. The KLC2extTPR structure is color-coded as in D and E. The SKIPWD and ext peptides binding sites on KLC2TPR partly overlap.
To further define the interaction site on KLC2TPR, we crystallized the extended KLC2extTPR protein. After substantial crystal screening, we were able to obtain a dataset at the 4-Å resolution (data collection and refinement statistics in Table S1). This allowed the modeling of the complete KLC2TPR domain, extending the available structures for this isoform, which lack either the complete TPR1 repeat (3ZFW) or its first α-helix (3CEQ). In the course of crystallographic refinement, difference Fourier maps also revealed the presence of elongated electron density in close proximity of the C-terminal end of the first α-helix of the TPR2 repeat (α2A) (Fig. S2). In all three independent molecules present in the crystallographic asymmetric unit, this residual density was satisfactorily modeled as a short peptide stretch (5 aa in molecules A and B and 2 aa in molecule C) in an extended conformation (Fig. 3 D and E). Although the limited resolution of the data does not allow for exact amino acid identification, given that our biochemical and biophysical results support an LFP-dependent self-interaction model, the bound peptide (ext in Fig. 3 D and E) almost certainly originates from the extended flexible region N-terminal to the TPR and involves the LFP-acidic motif. A structural comparison between autoinhibited and cargo-bound KLC2TPR highlights that stabilization of the ext peptide involving the C-terminal portion of α2A does not trigger the conformational change at the N-terminal TPR region observed upon W-acidic cargo binding (23) (Fig. 3F). In the latter case, upon W-acidic recognition, an “induced-fit” rigid jaw movement closes TPR2-3 engendering the binding surface and pockets for the SKIPWD peptide. Such movement is not seen in KLC2extTPR. Thus, the interactions involved in ext:KLC2TPR stabilization, appear different from those critical for SKIPWD binding. However, the X-ray structures do reveal a partial overlap between the W-acidic SKIPWD and the ext binding sites on the KLC2TPR domain. In particular, residues immediately C-terminal to the W-acidic motif (SAI in SKIPWD) essentially occupy the same topological location as ext on the KLC2TPR receptor (Fig. 3F). Thus, crystallographic analysis fully supports the notion that the autoinhibited (ext-bound) and the W-acidic cargo-bound states are mutually exclusive.
Table S1.
Data collection and refinement statistics
| Dataset | KLC2extTPR |
| Data collection | |
| Beam line | I24 (DLS) |
| Wavelength, Å | 0.96861 |
| Resolution range, Å | 42.88–4.00 |
| Highest res. bin, Å | (4.10–4.00) |
| Space group | C2 |
| Cell dimensions | |
| a, b, c, Å | 148.70, 86.28, 111.74 |
| β, ° | 98.4 |
| Unique reflections | 11,813 |
| (893) | |
| Overall redundancy | 3.0 |
| (3.1) | |
| Completeness, % | 98.4 |
| (98.8) | |
| Rmerge, % | 12.4 |
| (121.8) | |
| Rpim (I), % | 10.0 |
| (100.52) | |
| 〈I/σ(I)〉 | 3.9 |
| (0.6) | |
| Wilson B factor, Å2 | 216 |
| Refinement | |
| PDB ID code | 5FJY |
| Rfactor, %/Rfree, % | 23.4/26.1 |
| rms bond lengths, Å | 0.010 |
| rms bond angles, ° | 1.17 |
Fig. S2.
Illustrated cartoon representations in two orthogonal orientations of the KLC2extTPR (KLC2 residues 161–480) X-ray structure during crystallographic refinement. 2mFo-DFc and mFo-DFc electron density maps are shown in gray (1.2σ level) and in magenta (+3σ level), respectively. The KLC2TPR model (residues 195–421 and 440–479) used at this stage of refinement is shown as a tube in rainbow color (blue to red) going from the amino terminus to the carboxyl terminus. Broken lines indicate amino acid stretches absent in the model. Individual TPR repeats composed by helix1-turn-helix2 elements are highlighted. The non-TPR helix between TPR5 and TPR6 is labeled αN. Elongated and unaccounted for electron density that is believed to arise from a portion of the (161–195) extension N-terminal to KLC2TPR is seen in proximity of α2A.
In Vivo Conformational Dynamics of KLC Are Governed by Self-Interaction and Cargo Binding.
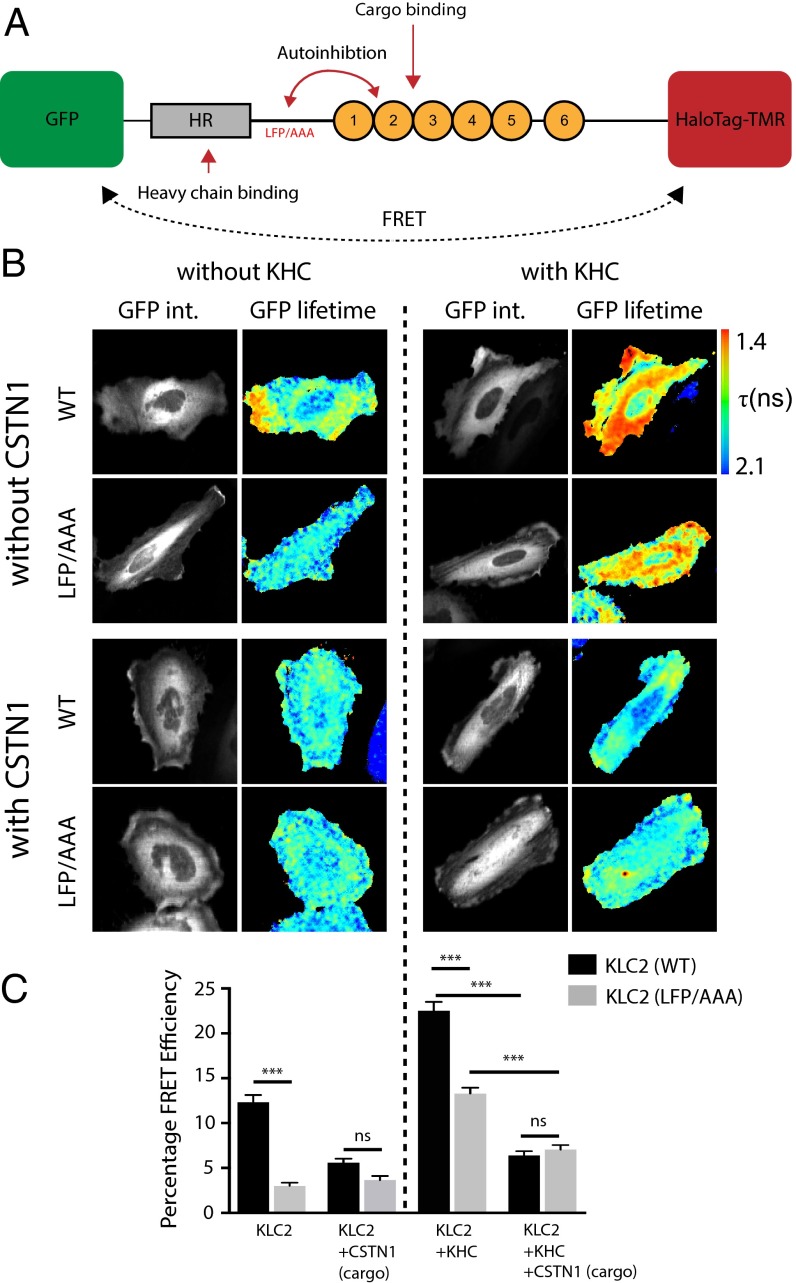
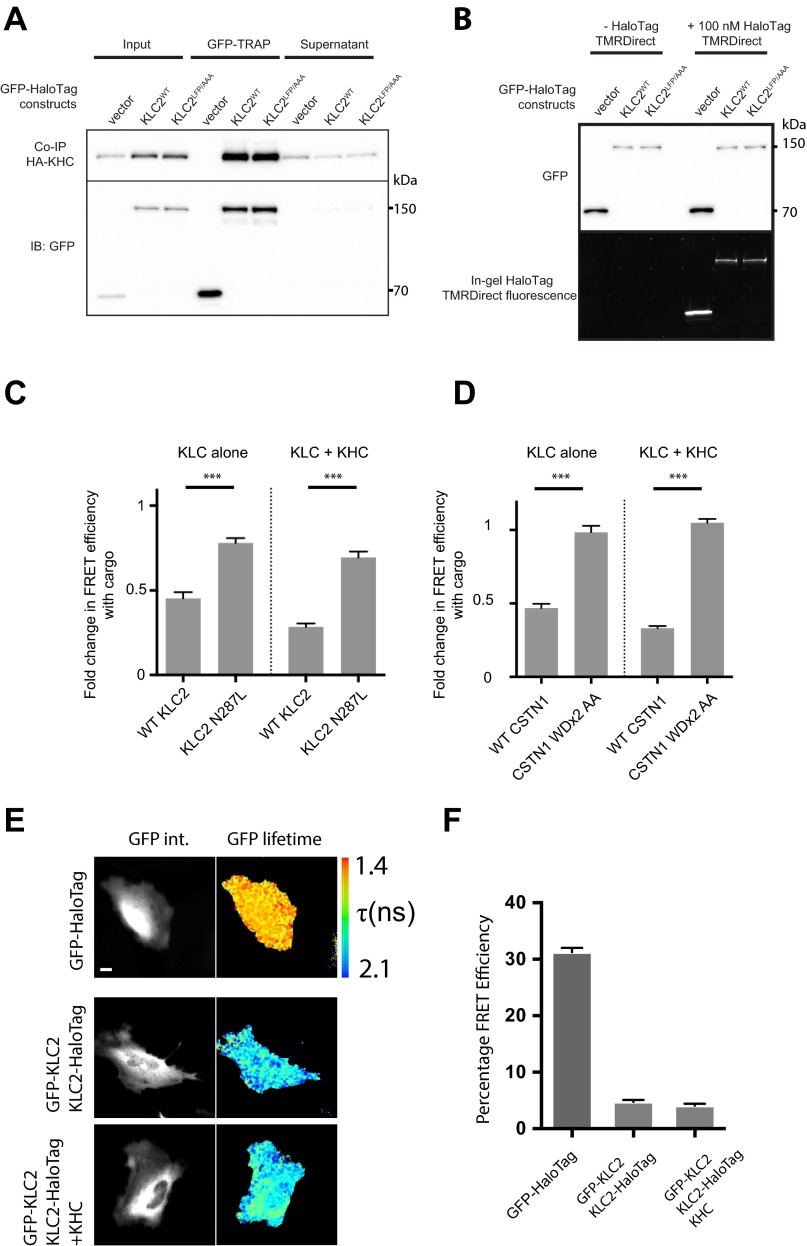
The above data imply that an intramolecular interaction within KLC2 mediated, at least in part, by amino acids in LFPpept, must be displaced for W-acidic cargo binding to occur. This could have an effect on the overall conformation of the light chain. To test this hypothesis in vivo, we designed a KLC2 fluorescence resonance energy transfer (FRET) conformation biosensor (Fig. 4A). A similar approach has been used previously to study conformational change within kinesin-1, but not to assess conformational changes within the light chains themselves (13). To achieve this, we coupled the amino terminus of full-length KLC2 to a donor eGFP and the carboxyl terminus to a HaloTag that allows in-cell conjugation to a tetramethylrhodamine (TMR) FRET acceptor. We then used fluorescence lifetime imaging microscopy (FLIM) to quantify changes in the efficiency of energy transfer from GFP to HaloTag-TMR. These measures subsequently inform on quantitative conformational changes in KLC caused by the modulation of the distance between the N- and C-terminal fluorophores. Western blot analysis of TMR-labeled cell extracts confirmed specific targeting of TMR to the GFP-KLC2-HaloTag biosensor and comparable labeling of wild-type and mutant proteins. Immunoprecipitation showed that the biosensor retained capacity to interact with KHC (Kif5C) (Fig. S3 A and B). Expression and labeling of the wild-type biosensor in HeLa cells (without addition of exogenous KHC or cargo) gave a robust baseline FRET efficiency of 12.3 ± 0.8% (Fig. 4 B and C). This was significantly reduced by the replacement of the LFP motif with AAA (3.0 ± 0.3%), demonstrating that the LFP triplet contributes to a relatively compact KLC2 conformation (compare first two columns on graph in Fig. 4C). Coexpression of a W-acidic cargo [myc-tagged cytoplasmic domain of CSTN1 (869–971)] reduced wild-type KLC2 FRET efficiency by a similar extent (5.6 ± 0.5%) but did not significantly affect FRET in the LFP/AAA background (3.6 ± 0.5%, compare columns 3 and 4 on graph in Fig. 4C). Importantly, introduction of the N287L substitution in KLC2 that disrupts W-acidic cargo binding (23) or mutation of the W-acidic residues in CSTN1 (16, 27, 28), suppressed the cargo-dependent response (Fig. S3 C and D). In comparison, FRET efficiency for GFP directly coupled to HaloTag-TMR was 31.3 ± 0.76%, whereas expression of the two fluorophores on separate polypeptide chains [N-terminal labeled (GFP-KLC2) and C-terminal labeled (KLC2-Halo)] resulted in low levels of FRET (4.76 ± 1.26%), indicating that the biosensor predominantly reports on intramolecular interaction within KLC2 (Fig. S3 E and F). To determine whether these cargo/LFP-dependent changes also occur in the context of the kinesin-1 tetramer, equivalent experiments were carried out in the presence of KHC. Inclusion of the heavy chain resulted in an increase in baseline FRET efficiency to 22.5 ± 1.0%, indicating that binding to KHC helps to support a more compact KLC conformation. FRET efficiency was significantly lower in the LFP/AAA replacement biosensor (13.3 ± 0.6%) (columns 5 and 6 on graph in Fig. 4C). Cotransfection of cargo reduced FRET efficiency to comparable levels in both backgrounds (6.4 ± 0.5% vs. 7.1 ± 0.5%, columns 7 and 8 on graph in Fig. 4C) showing that KLC undergoes significant cargo-dependent conformational change in the context of the kinesin-1 tetramer. This cargo-dependent change in FRET was again suppressed by the KLC2 N287L variant and W-acidic mutations in CSTN (Fig. S3 C and D), and separation of the fluorophores on different polypeptides resulted in only minimal levels of FRET (4.09 ± 1.23%), indicating that the measured FRET efficiency remains predominantly intramolecular in the context of the holoenzyme (Fig. S3 E and F).
Fig. 4.
In vivo conformational dynamics of kinesin light chain are controlled by self-interaction and cargo binding. (A) Schematic showing KLC2 FRET biosensor with an N-terminal eGFP and a C-terminal HaloTag that allows covalent coupling of TMR. Red arrows indicate mechanistic questions addressed using the biosensor. (B) Multiphoton fluorescent lifetime images of FRET between GFP and TMR-HaloTag. “GFP int.” are multiphoton GFP intensity images, whereas lifetime image refers to the fluorescence lifetime of GFP (τ) and is represented by a pseudocolor scale. In these images, a reduction in lifetime (change in color from blue to red) indicates FRET and therefore close association of GFP and TMR-HaloTag. (C) Graphs show data from 15 cells expressed as FRET efficiency (SI Materials and Methods). (Scale bar: 10 μm.) Error bars are SEM. ***P < 0.001. (Left) Light-chain biosensor alone [with and without cotransfection of myc-CSTN (879–971) cargo]; (Right) equivalent experiments where exogenous KHC is included.
Fig. S3.
(A) GFP-TRAP immunoprecipitation experiment showing that GFP-KLC2-HaloTag (wild type or LFP > AAA) efficiently coimmunoprecipitates with HA-KHC. (B) GFP Western blot and in-gel HaloTag-TMR fluorescence of whole-cell TMR-labeled extracts expressing GFP-HaloTag vector and GFP-KLC2-HaloTag constructs. (C and D) Graphs showing fold change in FRET efficiency following cargo addition (myc-CSTN1 879–971) in the indicated conditions. C shows effect of the N287L mutation in KLC2 that inhibits W-acidic cargo binding; D shows effect of mutating both W-acidic (WD) motifs in CSTN to AA. Data are from 15 cells per condition. Error bars show SEM. ***P < 0.001. (E) Multiphoton fluorescent lifetime images of FRET between GFP and TMR-HaloTag for GFP directly coupled to TMR (Top) or GFP-KLC2 and KLC2-Halo with and without exogenous KHC (Bottom). “GFP int.” are multiphoton GFP intensity images, whereas lifetime image refers to the fluorescence lifetime of GFP (τ) and is represented by a pseudocolor scale. In these images, a reduction in lifetime (change in color from blue to red) indicates FRET and therefore close association of GFP and TMR-HaloTag. (F) Graphs show data from 16 cells expressed as FRET efficiency. (Scale bar: 10 μm.) Error bars are SEM.
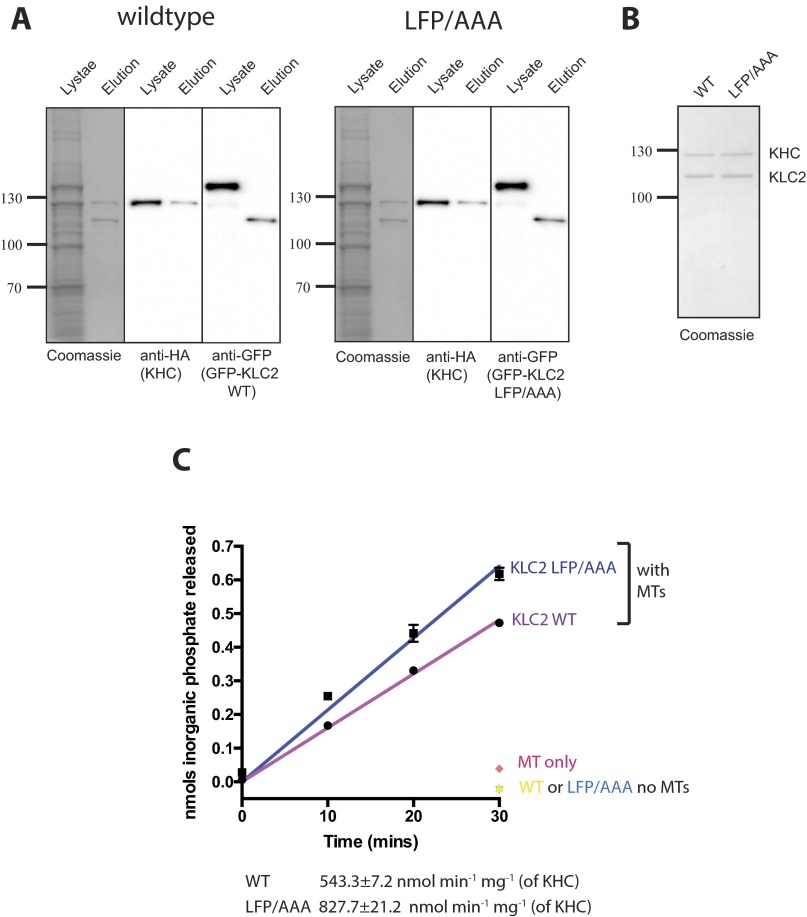
To examine the effect of LFP motif-mediated intramolecular interaction in KLC on the MT-dependent ATPase activity associated with KHC, kinesin-1 containing either wild-type or LFP/AAA KLC2 was expressed in, and isolated from, mammalian cells by covalent coupling of GFP-KLC2-Halo to HaloLink resin and subsequent release by TEV protease cleavage of the HaloTag (Fig. S4 A and B). As assessed by measurement of inorganic phosphate resulting from hydrolysis of ATP, these kinesin-1 preparations had no detectable ATPase activity in the absence of MTs (Fig. S4C). However, in the presence of MTs, the ATPase rate of wild-type kinesin-1 was 543.3 ± 7.2 nmol⋅min−1⋅mg−1 (of KHC). The LFP/AAA substitution resulted in an increase in the ATPase rate to 827.7 ± 21.2 nmol⋅min−1⋅mg−1, demonstrating that this intramolecular interaction within the light chain can modulate KHC-associated ATPase activity.
Fig. S4.
(A) Coomassie-stained SDS/PAGE gels and Western blot analysis showing kinesin-1 (KLC wild type) and KLC2 LFP/AAA purifications. Whole-cell extracts and TEV-cleaved samples are shown. Note increase in mobility of KLC2 following TEV protease cleavage. (B) Coomassie-stained gel showing direct comparison between wild-type and LFP/AAA preparations. (C) Graph showing microtubule (MT)-stimulated ATPase activity of wild-type and LFP/AAA kinesin-1 preparations at 10-, 20-, and 30-min time points following ATP addition. The y axis shows nanomoles of inorganic phosphate released. Control reactions without MTs or MTs alone are shown as a single point at the 30-min time point. Values are mean of three replicates, and error bars show SEM.
Taken together, these data show that the LFP motif-mediated intramolecular interaction within KLC regulates KLC conformation in the context of the holoenzyme in vivo. This autoinhibitory interaction and conformation are themselves directly governed by W-acidic cargo binding.
Discussion
Despite its importance for a host of cellular processes and contribution to neurological, viral and bacterial disease, the molecular mechanisms underlying the regulation of kinesin-1 by its light chains and the binding of its cargo are not well understood. The data presented here provide conceptual insight into the molecular events that occur both in its regulated state and following W-acidic cargo recognition. We show that, like the kinesin heavy chain, the light chains of kinesin-1 exist in a dynamic conformational state that is regulated by self-interaction and cargo binding. We highlight how recognition of short linear peptide motifs by the TPR domain can be transduced and amplified to result in larger scale modification of the organizational state of the light chain. Thus, we uncover unanticipated mechanistic parallels between the heavy- and light-chain components of the kinesin-1 tetramer.
Such an intramolecular interaction mediated by a TPR domain is reminiscent of the interaction of the N-arm helix of the mitochondrial outer membrane protein Fis1 with its TPR domain (36), although in this case the self-interacting helix forms an important component of the binding interface for other proteins. A closer mechanistic analogy perhaps lies in the regulation of protein phosphatase 5 (Ppp5) activity by its TPR domain where, in its autoinhibited state, the TPR domain engages with the catalytic channel of Ppp5 (37). This conformation is stabilized by the C-terminal αJ helix that contacts a region of the Hsp90 binding groove on the TPR domain. Binding of the C-terminal MEEVD peptide of Hsp90 disrupts this interaction, relieving autoinhibition and activating the phosphatase. The notion that TPR domain function is not restricted to that of a protein–protein interaction module and that peptide binding can be transduced through conformational change to control function is supported by studies of the dimeric bacterial transcription factor and virulence regulator PlcR where TPR binding of the PapR signal peptide is propagated to control the DNA binding properties of helix-turn-helix DNA binding domains (38). We note some functional parallels in another transport/trafficking system with a recent report describing how the interaction of the AP2 β2 hinge (containing a clathrin-binding motif) with the core of the molecule that is disrupted (in an allosteric manner in this case) by phospholipid and cargo binding, promoting clathrin binding activity (39).
In the context of the present study, it is interesting to note that several reports have demonstrated that fusion of W-acidic motifs to otherwise non–kinesin-1 binding proteins is sufficient to promote kinesin-1–dependent transport/dispersion of specific cellular compartments, such as lysosomes (16, 24, 40). The strong implication of these studies is that W-acidic motifs have an intrinsic capacity to promote (at least partial) relief of kinesin-1 autoinhibition. However, given that kinesin-1 ATPase activity is predominantly controlled by an intramolecular interaction between the motor domains and the IAK C-terminal region of a single heavy chain, it was not obvious how a TPR domain–peptide interaction could contribute to this change. Our present data suggest a possible model. Here, we show that mutational disruption of the LFP motif in KLC enhances the inherent MT-stimulated ATPase activity of purified kinesin-1. Rice and colleague (19) demonstrated that the heptad repeat–TPR linker region contributes to light-chain–mediated destabilization of the heavy-chain carboxyl-tail/MT interaction in vitro, implying a mechanism of communication between the linker and heavy-chain tail. Moreover, the same study reported that the TPR domain itself contributes to a light-chain–mediated reduction in the affinity of the KHC head for its C-terminal tail, through a mechanism requiring steric and electrostatic factors. Incorporating that analysis, we propose a model whereby the W-acidic motif-mediated displacement of the highly charged linker from the TPR will result in interactions (predominantly electrostatic) with the heavy-chain tail that combine with steric changes resulting from a large-scale change in conformation of the light chains to promote the cargo-dependent transition of holoenzyme to its active state (Fig. S5).
Fig. S5.
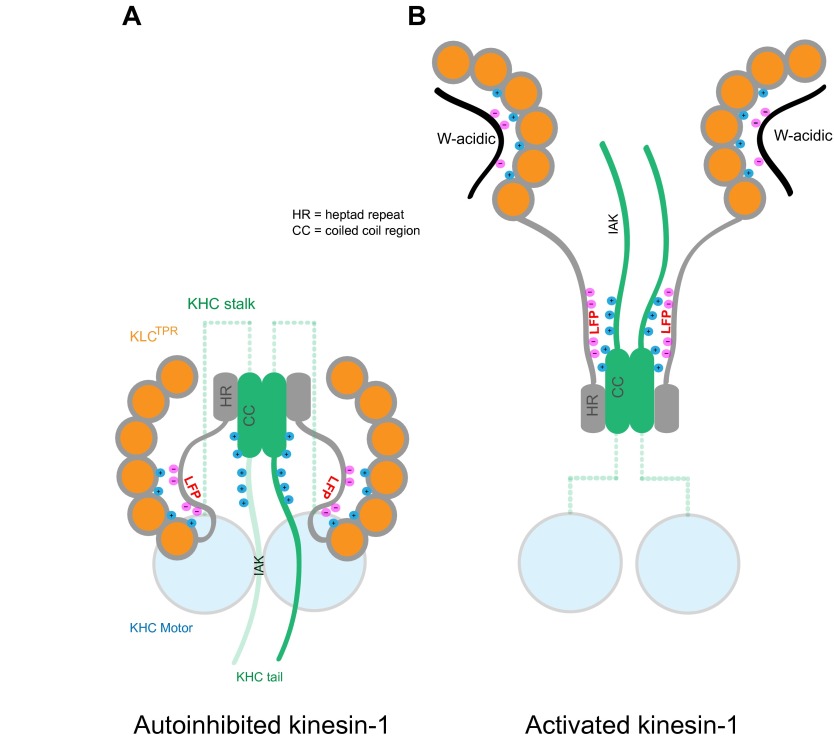
Model showing representation of KHCs (green) and KLCs (gray/orange) in the autoinhibited (A) and W-acidic cargo-bound states (B). In A, both KHCs and KLCs are engaged in a double autoinhibited conformation mediated by IAK motif in KHC and LFP motifs in KLC, with the amino and carboxyl termini of KLCs in close proximity. In B, W-acidic cargo binding displaces the LFP-acidic/ext sequence. This triggers a large-scale conformational change in KLC that results in separation of the amino and carboxyl termini of the light chains. We propose that resulting electrostatic interactions between the displaced ext/linker region and KHC combined with steric changes from TPR domain reorientation may destabilize the tail-mediated KHC inhibition and contribute to activation of the holoenzyme.
If so, it would seem likely that this mechanism can be generalized to other W-acidic motif-containing cargo given their shared binding determinants on the KLCTPR (23, 30). The strong sequence conservation of the LFP motif and our similar observation for CSTN1 and KLC1 (Fig. S1C) suggests that this will also apply to other light-chain isoforms. However, only a subset of kinesin-1 cargoes contain W-acidic motifs, and it is clear that KLCTPR has the capacity to interact with other peptides, including the C-terminal 11 aa of JIP1 (JIP1C11, sequence YTCPTEDIYLE) (22). Despite this, the LFP/AAA replacement does not affect binding to JIP1 in immunoprecipitation experiments (Fig. S1D) and N-terminal extension of the KLC1TPR only has a marginal impact on its affinity for JIP1C11. The JIP1C11 peptide is also insufficient to promote activation of transport (16). Indeed, JIP1 requires interactions with the coiled-coil heavy chain and the heavy-chain tail binding protein Fez1 for full activity (15, 17). Although the precise JIP1C11 binding site on the KLCTPR still awaits structural definition, mutagenesis experiments highlight a particularly crucial role for residues on TPR4/5 that are distinct from the primary determinants of W-acidic binding on TPR2/3 (22, 23). Thus, it may be that the site of peptide binding on the KLCTPR has different functional outcomes, and that this is in part due to its capacity to displace the LFP motif-containing linker region, resulting in differential requirements for supporting interactions to promote kinesin-1 activity. It is therefore likely that there are multiple pathways to activation of kinesin-1 depending upon site(s) of cargo binding.
The dissociation constant (KD) of ∼25 μM (or the equivalent association constant KA = 1/KD ∼ 0.04 μM−1) measured here by FP between TAMRA-LFPpept and KLC2TPR reflects a bimolecular binding process. However, the proposed mechanism of KLC autoinhibition is unimolecular, as the LFPpept is covalently linked to the TPR domain. The balance between the latched (autoinhibited) and unlatched (available to W-acidic cargoes) states is therefore regulated by the intramolecular association constant KAi. This has been shown to be related to KA by KAi = p(d)KA (Eq. S1, SI Materials and Methods) where p(d) is known as the effective concentration, the ligand concentration that would be required to achieve the same fraction of bound state in a bimolecular interaction (41, 42). From the structure of KLC2extTPR, we can estimate the value of p(d) at ∼18.2 mM (Eq. S2, SI Materials and Methods). Thus, using Eq. S1, we obtain KAi ∼ 728. The implication of this is that the equilibrium fraction of cargo-available KLC2TPR is only ∼0.14%. This suggests that autoinhibition of the light chains tightly regulates cargo binding. A similarly stringent intramolecular regulation has been observed for the myristoylated N-terminal latching to the C-terminal lobe of c-Abl that maintains the kinase in an inactive state (43, 44). The low fraction of cargo-available KLC2TPR raises the obvious question of how significant W-acidic cargo binding is achieved. The X-ray structures of cargo-bound KLC2TPR and autoinhibited KLC2extTPR highlight that only a partial overlap exists between the W-acidic motif and the ext autoinhibitory peptide. In particular, the binding region for the most N-terminal part of the W-acidic motif (N-terminal to the central tryptophan residue) appears accessible even in the ext-bound state (Fig. 3F). Thus, one possible mechanism for the relief of KLC autoinhibition is the initial recognition of a portion of the W-acidic cargo motif at this topological location. This could be sufficient to initiate the induced-fit adaptation of the TPR domain, which, in turn, may drive destabilization of ext binding.
Therefore, as well as acting as a component of a pathway to kinesin-1 activation, our findings also suggest that the intramolecular interaction between the linker region and KLCTPR may serve to buffer cargo-binding sensitivity and so provide a point of regulatory access to control the proper loading and unloading of cargo. It is conceivable that binding of other proteins or posttranslational modifications of the light chains could serve to stabilize or destabilize this self-interacting state and thus regulate cargo-binding properties of the TPR domain in a spatial and temporal manner.
SI Materials and Methods
Plasmids.
Full-length, wild-type mouse kinesin light chain-2 (KLC2) and rat kinesin-1 heavy chain (KHC, Kif5C) were cloned into the mammalian expression vector CB6-HA. The W-acidic kinesin-1 cargo proteins, SKIP (amino acids 1–310) and calsyntenin-1 (CSTN1) (amino acids 879–971), were subcloned into GST-(pMW-GST) or myc-(CB6-myc) expression vectors from CB6-GFP expression constructs described previously (24). WD/AA mutations in CSTN1 were introduced by site-directed mutagenesis. CB6-HA-KLC1 (isoform A) was described previously (23). The TPR domain of KLC2 alone (KLC2TPR, amino acids A196–S480), a longer version containing the conserved LFP motif (KLC2extTPR, K161–S480), and equivalent constructs for KLC1 (KLC1TPR, A211–S495, and KLC1extTPR, K173–S495) as well as a chimera of the first W-acidic motif of SKIP (amino acids 202–212) fused to the N terminus of KLC2TPR via a TGSx4 linker (SKIPWacTPR) were cloned into the bacterial expression vector pET28His-Thrombin. A GFP-KLC2-HaloTag· FRET biosensor was generated by inserting GFP-KLC2 into a C-terminal HaloTag vector carrying a TEV protease cleavage site between KLC2 and HaloTag (Promega). Control GFP-KLC2 or KLC2-Halo expression constructs were derived from this plasmid. Plasmids with mutations that disrupt the LFP motif in KLC2 (L167, F168, P169 to AAA), or equivalent mutations in KLC1 (179–181), as well as binding between KLC2 and its cargoes (N287L) were generated by site-directed mutagenesis. All plasmids were verified by DNA sequencing.
Cell Culture, Transfection, and Immunoprecipitation.
HeLa cells were maintained in DMEM supplemented with 10% (vol/vol) FBS, l-glutamine, and penicillin/streptomycin, and cultured in a humidified 5% CO2 environment. To determine the impact of the LFP motif on KLC cargo binding, 1 × 106 HeLa cells were plated on 10-cm dishes and transfected with HA-KLC2WT or HA-KLC2LFP/AAA and GFP, GFP-SKIP (1–310), or GFP-CSTN1 (879–971). After 16 h, transfected cells were lysed in 1 mL of 25 mM Hepes, pH 7.5, 150 mM NaCl, 0.5% Nonidet P-40, 0.5× Triton X-100 containing a protease inhibitor mixture (Roche) for 10 min before centrifugation at 13,000 × g for 10 min at 4 °C. The resulting supernatant was incubated with 15 μL of prewashed GFP-Trap (Chromotek) beads for 90 min. Beads were washed four times in assay buffer (25 mM Hepes, pH 7.5, 150 mM NaCl), resuspended in 100 μL of buffer and 25 μL of SDS-loading buffer before boiling. Twenty microliters of samples were subjected to SDS/PAGE and analyzed by Western blot using antibodies against GFP and HA. Twenty microliters of total cell lysate was loaded for analysis of input levels. To immunoprecipitate FLAG-JIP/KLC1 complexes, lysates from cells expressing FLAG-JIP1 (15) and HA-KLC1WT, HA-KLC1LFP/AAA, were incubated with 100 µL of protein A-agarose bound to 5 μg of anti-FLAG (F1804; Sigma) or negative IgG for 4 h at 4 °C. Beads were washed four times in assay buffer, resuspended in 100 μL of buffer and 25 μL of SDS-loading buffer before boiling. Twenty microliters of samples were subjected to SDS/PAGE and analyzed by Western blot using antibodies against FLAG and HA. Ten microliters of total cell lysate was loaded for analysis of input levels.
FRET/FLIM Sample Preparation and Data Acquisition.
For FRET studies, 1 × 104 HeLa cells were seeded onto 13-mm fibronectin-coated coverslips in 24-well plates. Cells were transfected with combinations of plasmids expressing GFP-KLC2-HaloTag (WT, LFP/AAA, or N287L), HA-KHC and myc-CSTN1 (869–971) (WT or binding-deficient mutants). After 6 h posttransfection, medium was replaced with fresh medium containing HaloTag TMRDirect ligand (Promega) at a 1:1,000 dilution according to manufacturer’s instructions. After an overnight incubation, cells were fixed in 4% paraformaldehyde and permeabilized in 0.2% (wt/vol) Triton X-100 in PBS. After quenching with 1 mg/mL sodium borohydride in PBS for 10 min at room temperature (RT), cells were washed in PBS and mounted in Mowiol containing 2.5% (wt/vol) Dabco (Sigma-Aldrich). Time domain FLIM was performed with a multiphoton microscope system (with TE2000 microscope; Nikon) described in detail previously (45). Fluorescence lifetime imaging capability was provided by time-correlated single-photon counting electronics (SPC-700; Becker and Hickl). A 40× objective was used throughout (Plan Fluor N.A. 1.3; CFI 60; Nikon), and data were collected at 500 ± 20 nm through a bandpass filter (35-5040; Coherent). Acquisition times of the order of 300 s at a low 890-nm excitation power were used to achieve sufficient photon statistics for fitting, while avoiding either pulse pile up or significant photobleaching. Histogram data are plotted as mean FRET efficiency from specified numbers of cells per sample over two/three independent experiments. Lifetime images of example cells are presented using a pseudocolor scale, whereby blue depicts normal GFP lifetime (no FRET) and red depicts lower GFP lifetime (areas of high FRET).
FRET Data Analysis.
Data were analyzed as previously described (45). Bulk measurements of FRET efficiency (i.e., intensity-based methods) cannot distinguish between an increase in FRET efficiency (i.e., coupling efficiency) and an increase in FRET population (concentration of FRET species) because the two parameters are not resolved. Measurements of FRET based on analysis of the fluorescence lifetime of the donor can resolve this issue. The assumption that noninteracting and interacting fractions are present allows the determination of the efficiency of interaction. The FRET efficiency is related to the molecular donor–acceptor separation and the fluorescence lifetime of the interacting fraction by the following:
where is the Förster radius, is the molecular separation, is the lifetime of the interacting fraction, and is the lifetime of the donor in the absence of acceptor. and can also be taken to be the lifetime of the interacting fraction and noninteracting fraction, respectively. All data were analyzed using TRI2 software (developed by P. Barber, Gray Cancer Institute, London, UK). Histogram data presented here are derived from biexponential analysis and plotted as mean FRET efficiency from specified numbers of cells per sample from at least two independent experiments.
Protein Expression and Purification.
Proteins were expressed in Escherichia coli BL21(DE3) cells. Briefly, single colonies were picked and grown at 37 °C overnight. Small-scale overnight bacterial cultures were used to inoculate 2 × 1-L cultures that were incubated at 37 °C until they reached an OD600 of 0.5. The temperature was then lowered to 16 °C, and protein synthesis was induced by the addition of 300 μM isopropyl β-d-1-thiogalactopyranoside for 16 h. Cells were harvested by centrifugation at 5,000 × g for 15 min at 4 °C. For GST-tagged proteins, cells were resuspended in lysis buffer (25 mM Hepes, pH 7.5, 500 mM NaCl, 5 mM β-mercaptoethanol) supplemented with protease inhibitor mixture (Roche). Lysis buffer containing 20 mM imidazole was used to purify His-tagged proteins. Cell lysis was accomplished by sonication. Insoluble material was sedimented by centrifugation at 16,500 × g for 1 h at 4 °C. GST-tagged proteins were obtained via batch purification using Glutathione Sepharose beads (GE Life Sciences), and His-tagged proteins were purified using His-trap FF columns (GE Life Sciences). Purified proteins from both methods were dialyzed overnight against glutathione or imidazole-free lysis buffer, respectively. His-tagged protein samples were further purified by size-exclusion chromatography (SEC) on a 16/60 HiLoad Superdex 75 column (GE Healthcare). For crystallization experiments, the His-tag was cleaved from KLC constructs and removed from solution using a thrombin cleavage/capture kit (Merck Millipore) according to the manufacturer’s instructions and was followed by a second round of SEC.
X-Ray Crystallography.
Untagged KLC2extTPR (KLC2 161–480) was concentrated at 4.8 mg/mL in 20 mM Hepes, pH 7, 500 mM NaCl, 5 mM 2-mercaptoethanol, and crystals were obtained in the PGA screen (Molecular Dimensions) in the presence of 0.1 M Na-cacodylate, pH 6.5, 0.3 M Na-malonate, and 8% (wt/vol) PGA-LM. For data collection, crystals were cryoprotected in reservoir enriched with 20% (vol/vol) ethylene glycol. A complete dataset at the 4-Å resolution was measured at the I24 beam line of Diamond Light Source (United Kingdom), and data were processed using the xia2 package in space group C2 (a = 148.70 Å, b = 86.28 Å, c = 111.74 Å, α = 90°, β = 98.4°, γ = 90°). The structure was solved by the molecular replacement (MR) technique using the software packages Phaser and Molrep of the CCP4 suite. An initial estimation of the Matthews’ coefficient suggested the presence of three to five KLC2 molecules in the a.u. with a solvent content ranging from 38% (five molecules) to 63% (three molecules). Using an ensemble defined by KLC2TPR monomers from 3CEQ and 3ZFW Phaser positioned two molecules. The TFZ score (8.7) together with the presence of additional electron density consistent with the additional α1A helix not present in the search probe suggested this to be a genuine solution. Inspection of residual electron density suggested the presence of an additional KLC2 molecule. This was positioned using a second MR search with Molrep using the previous solution after rigid-body refinement as fixed model. Model building and crystallographic refinement was performed using COOT and Buster 2.0 (Global Phasing), respectively. A summary of data collection and refinement statistics are shown in Table S1. Structural images were prepared with PyMol (Schrödinger).
GST Pull Down.
GST-SKIP 1–310 or GST-CSTN 869–971 (0.5 nmol of protein per reaction) bound to Glutathione Sepharose beads was incubated with 1 mL of 1.0 μM His-KLC2TPR, His-KLC2extTPR, or His-SKIPWacTPR proteins for 2 h at 4 °C. Beads were washed four times with assay buffer (25 mM Hepes, pH 7.5, 150 mM NaCl, 5 mM β-mercaptoethanol, 25 mM imidazole), resuspended in 100 μL of buffer and 25 μL of SDS-loading buffer before boiling. Twenty microliters of each sample were loaded, and the SDS/PAGE gel was stained with InstantBlue Protein Stain (Expedeon).
Purification of HaloTag Protein from Mammalian Cells and Kinesin ATPase Activity Assay.
A total of 106 293T cells were seeded onto 10-cm dishes. Cells were transfected with plasmids expressing HA-KHC and GFP-KLC2-HaloTag (WT or LFP/AAA). A total of four plates of cells were transfected per sample. Purification of HaloTag proteins was performed using the HaloTag Mammalian Protein Detection and Purification System (Promega). Briefly, 16 h posttransfection, cells were washed with PBS and collected with gentle scraping and centrifugation at 200 × g for 5 min. Cell pellets were resuspended in 1 mL of lysis buffer (50 mM Tris⋅HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100, 0.1% sodium deoxycholate) supplemented with protease inhibitor followed by incubation with rotating at RT for 15 min. The lysates were diluted 1:3 with HaloTag protein purification buffer (50 mM Hepes, pH 7.5, 150 mM NaCl, 1 mM DTT, 0.005% IGEPAL CA-630). After centrifugation at 10,000 × g for 30 min at 4 °C, the supernatants were added to 200 µL of equilibrated HaloLink resin and incubated at RT for 1.5 h with end-over-end rotation. After protein binding, the beads were washed three times in HaloTag protein purification buffer and bound proteins were released by HaloTEV protease cleavage. The ATPase activity of purified HA-KHC and GFP-KLC2 (WT or LFP/AAA) complexes was measured with the Kinesin ATPase End-point Biochem Kit (Cytoskeleton) according to the manufacturer’s instructions. Thirty nanograms of purified proteins (KHC) were incubated with 2 µg of MTs and reaction buffer (15 mM Pipes, pH 7.0, 5 mM MgCl2) in a 30-µL volume. A final concentration of 0.3 mM ATP was added per reaction and left to proceed for the indicated times before reactions were terminated with the addition of 70 µL of CytoPhos. Absorbance at 650 nm was measured after 10-min incubation using a BMG Labtech PolarStar Omega plate reader. The amount of ATP hydrolysis was calculated based on inorganic phosphate standards. ATPase rates and associated errors are derived from the slope of a straight line fitted using Prism. Data points are mean of three replicates, and error bars show SEM.
Intramolecular Binding Analysis.
Under the assumption that binding of the ext peptide to the TPR domain does not interfere with the statistical distribution of the flexible linker consisting of L peptide bonds that connects them at an end-to-end distance d, the intramolecular association constant is related to the bimolecular association constant by the following equation:
| [S1] |
where is the effective concentration (41, 42, 44). Using the worm-like chain model, a good approximation for is given by the following:
| [S2] |
In the equation above, and are the contour length and the persistence length, respectively. The contour length can be calculated as , where = 3.8 Å is the nearest Cα–Cα distance. The persistence length is ∼3 Å for peptide linkers (9–11). In the case of KLC2extTPR, application of Eq. S2 using = 25 and = 17.5 Å gives an effective concentration of 18.2 mM. The value of has been determined from the crystallographic structure using the Cα positions of the most C-terminal residue of the ext peptide and P195 positioned at the beginning of α1A (TPR1) as end-to-end extremes of the flexible linker. The value for has been derived from the MmKLC2 sequence assuming that ext is LFPpept. This appears a reasonable assumption on the basis of our biophysical (fluorescence polarization), biochemical, and cellular data.
Analysis of Intrinsic Disorder in KLC2.
A panel of software was used to analyze the heptad repeat–TPR linker regions in KLC2: PrDOS (prdos.hgc.jp/cgi-bin/top.cgi), Disopred3 (bioinf.cs.ucl.ac.uk/psipred/), IUPred (iupred.enzim.hu/), and DisEMBL (dis.embl.de/).
Materials and Methods
N-terminal TAMRA-conjugated peptides and nonconjugated peptides used for FP and competition measurements were supplied by Bio-Synthesis. Sequences were as follows: SKIPWD, STNLEWDDSAI; LFPpept, DSLDDLFPNEDEQS; AAApept, DSLDDAAANEDEQS; and JIPC11, YTCPTEDIYLE. Measurements were performed on a BMG Labtech PolarStar Omega plate reader at 20 °C by incubating 300 nM TAMRA-labeled peptides with the indicated protein at increasing concentrations in 25 mM Hepes, pH 7.5, 150 mM NaCl, and 5 mM β-mercaptoethanol. Estimation of the equilibrium dissociation constant (KD) for the different peptides was performed assuming a one-site specific-binding model. For competition experiments, a mixture of TAMRA-LFPpept and KLC2TPR at 300 nM and 12 μM, respectively, were incubated with increasing concentrations of unlabeled SKIPWD peptide in buffer supplemented by 5% (vol/vol) DMSO. The concentration-dependent decrease in FP signal was fitted to a sigmoidal equation to derive IC50. Analyses and Ki estimation were performed using the Prism package (GraphPad Software). All data points are the mean of three replicates.
See SI Materials and Methods for additional materials and methods.
Acknowledgments
This work was supported by Biotechnology and Biological Sciences Research Council Project Grant BB/L006774/1 (to M.P.D. and R.A.S.), as well as a Wellcome Trust Research Career Development Fellowship and London Law Trust Medal Fellowship (to M.P.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 5FJY).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1520817113/-/DCSupplemental.
References
- 1.Vale RD. The molecular motor toolbox for intracellular transport. Cell. 2003;112(4):467–480. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- 2.Vale RD, Reese TS, Sheetz MP. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell. 1985;42(1):39–50. doi: 10.1016/s0092-8674(85)80099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dodding MP, Way M. Coupling viruses to dynein and kinesin-1. EMBO J. 2011;30(17):3527–3539. doi: 10.1038/emboj.2011.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morihara T, et al. Transcriptome analysis of distinct mouse strains reveals kinesin light chain-1 splicing as an amyloid-β accumulation modifier. Proc Natl Acad Sci USA. 2014;111(7):2638–2643. doi: 10.1073/pnas.1307345111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hackney DD. Jump-starting kinesin. J Cell Biol. 2007;176(1):7–9. doi: 10.1083/jcb.200611082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diefenbach RJ, Mackay JP, Armati PJ, Cunningham AL. The C-terminal region of the stalk domain of ubiquitous human kinesin heavy chain contains the binding site for kinesin light chain. Biochemistry. 1998;37(47):16663–16670. doi: 10.1021/bi981163r. [DOI] [PubMed] [Google Scholar]
- 7.Kaan HYK, Hackney DD, Kozielski F. The structure of the kinesin-1 motor-tail complex reveals the mechanism of autoinhibition. Science. 2011;333(6044):883–885. doi: 10.1126/science.1204824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dietrich KA, et al. The kinesin-1 motor protein is regulated by a direct interaction of its head and tail. Proc Natl Acad Sci USA. 2008;105(26):8938–8943. doi: 10.1073/pnas.0803575105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hackney DD, Baek N, Snyder AC. Half-site inhibition of dimeric kinesin head domains by monomeric tail domains. Biochemistry. 2009;48(15):3448–3456. doi: 10.1021/bi8022575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wong YL, Dietrich KA, Naber N, Cooke R, Rice SE. The Kinesin-1 tail conformationally restricts the nucleotide pocket. Biophys J. 2009;96(7):2799–2807. doi: 10.1016/j.bpj.2008.11.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hackney DD, Stock MF. Kinesin’s IAK tail domain inhibits initial microtubule-stimulated ADP release. Nat Cell Biol. 2000;2(5):257–260. doi: 10.1038/35010525. [DOI] [PubMed] [Google Scholar]
- 12.Friedman DS, Vale RD. Single-molecule analysis of kinesin motility reveals regulation by the cargo-binding tail domain. Nat Cell Biol. 1999;1(5):293–297. doi: 10.1038/13008. [DOI] [PubMed] [Google Scholar]
- 13.Cai D, Hoppe AD, Swanson JA, Verhey KJ. Kinesin-1 structural organization and conformational changes revealed by FRET stoichiometry in live cells. J Cell Biol. 2007;176(1):51–63. doi: 10.1083/jcb.200605097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hackney DD, Levitt JD, Suhan J. Kinesin undergoes a 9 S to 6 S conformational transition. J Biol Chem. 1992;267(12):8696–8701. [PubMed] [Google Scholar]
- 15.Blasius TL, Cai D, Jih GT, Toret CP, Verhey KJ. Two binding partners cooperate to activate the molecular motor Kinesin-1. J Cell Biol. 2007;176(1):11–17. doi: 10.1083/jcb.200605099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawano T, et al. A small peptide sequence is sufficient for initiating kinesin-1 activation through part of TPR region of KLC1. Traffic. 2012;13(6):834–848. doi: 10.1111/j.1600-0854.2012.01350.x. [DOI] [PubMed] [Google Scholar]
- 17.Fu M-M, Holzbaur ELF. Integrated regulation of motor-driven organelle transport by scaffolding proteins. Trends Cell Biol. 2014;24(10):564–574. doi: 10.1016/j.tcb.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verhey KJ, et al. Light chain-dependent regulation of Kinesin’s interaction with microtubules. J Cell Biol. 1998;143(4):1053–1066. doi: 10.1083/jcb.143.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong YL, Rice SE. Kinesin’s light chains inhibit the head- and microtubule-binding activity of its tail. Proc Natl Acad Sci USA. 2010;107(26):11781–11786. doi: 10.1073/pnas.1005854107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu M-M, Holzbaur ELF. JIP1 regulates the directionality of APP axonal transport by coordinating kinesin and dynein motors. J Cell Biol. 2013;202(3):495–508. doi: 10.1083/jcb.201302078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verhey KJ, Hammond JW. Traffic control: Regulation of kinesin motors. Nat Rev Mol Cell Biol. 2009;10(11):765–777. doi: 10.1038/nrm2782. [DOI] [PubMed] [Google Scholar]
- 22.Zhu H, et al. Crystal structures of the tetratricopeptide repeat domains of kinesin light chains: Insight into cargo recognition mechanisms. PLoS One. 2012;7(3):e33943. doi: 10.1371/journal.pone.0033943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pernigo S, Lamprecht A, Steiner RA, Dodding MP. Structural basis for kinesin-1:cargo recognition. Science. 2013;340(6130):356–359. doi: 10.1126/science.1234264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dodding MP, Mitter R, Humphries AC, Way M. A kinesin-1 binding motif in vaccinia virus that is widespread throughout the human genome. EMBO J. 2011;30(22):4523–4538. doi: 10.1038/emboj.2011.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verhey KJ, et al. Cargo of kinesin identified as JIP scaffolding proteins and associated signaling molecules. J Cell Biol. 2001;152(5):959–970. doi: 10.1083/jcb.152.5.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aoyama T, et al. Cayman ataxia protein caytaxin is transported by kinesin along neurites through binding to kinesin light chains. J Cell Sci. 2009;122(Pt 22):4177–4185. doi: 10.1242/jcs.048579. [DOI] [PubMed] [Google Scholar]
- 27.Konecna A, et al. Calsyntenin-1 docks vesicular cargo to kinesin-1. Mol Biol Cell. 2006;17(8):3651–3663. doi: 10.1091/mbc.E06-02-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Araki Y, et al. The novel cargo Alcadein induces vesicle association of kinesin-1 motor components and activates axonal transport. EMBO J. 2007;26(6):1475–1486. doi: 10.1038/sj.emboj.7601609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosa-Ferreira C, Munro S. Arl8 and SKIP act together to link lysosomes to kinesin-1. Dev Cell. 2011;21(6):1171–1178. doi: 10.1016/j.devcel.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson MH, Holzbaur ELF. Nesprins anchor kinesin-1 motors to the nucleus to drive nuclear distribution in muscle cells. Development. 2015;142(1):218–228. doi: 10.1242/dev.114769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidt MR, et al. Regulation of endosomal membrane traffic by a Gadkin/AP-1/kinesin KIF5 complex. Proc Natl Acad Sci USA. 2009;106(36):15344–15349. doi: 10.1073/pnas.0904268106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maritzen T, et al. Gadkin negatively regulates cell spreading and motility via sequestration of the actin-nucleating ARP2/3 complex. Proc Natl Acad Sci USA. 2012;109(26):10382–10387. doi: 10.1073/pnas.1206468109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ti S-C, Jurgenson CT, Nolen BJ, Pollard TD. Structural and biochemical characterization of two binding sites for nucleation-promoting factor WASp-VCA on Arp2/3 complex. Proc Natl Acad Sci USA. 2011;108(33):E463–E471. doi: 10.1073/pnas.1100125108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hammond JW, Griffin K, Jih GT, Stuckey J, Verhey KJ. Co-operative versus independent transport of different cargoes by Kinesin-1. Traffic. 2008;9(5):725–741. doi: 10.1111/j.1600-0854.2008.00722.x. [DOI] [PubMed] [Google Scholar]
- 35.Nikolovska-Coleska Z, et al. Development and optimization of a binding assay for the XIAP BIR3 domain using fluorescence polarization. Anal Biochem. 2004;332(2):261–273. doi: 10.1016/j.ab.2004.05.055. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y, Chan DC. Structural basis for recruitment of mitochondrial fission complexes by Fis1. Proc Natl Acad Sci USA. 2007;104(47):18526–18530. doi: 10.1073/pnas.0706441104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang J, et al. Molecular basis for TPR domain-mediated regulation of protein phosphatase 5. EMBO J. 2005;24(1):1–10. doi: 10.1038/sj.emboj.7600496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grenha R, et al. Structural basis for the activation mechanism of the PlcR virulence regulator by the quorum-sensing signal peptide PapR. Proc Natl Acad Sci USA. 2013;110(3):1047–1052. doi: 10.1073/pnas.1213770110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kelly BT, et al. Clathrin adaptors. AP2 controls clathrin polymerization with a membrane-activated switch. Science. 2014;345(6195):459–463. doi: 10.1126/science.1254836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pu J, et al. BORC, a multisubunit complex that regulates lysosome positioning. Dev Cell. 2015;33(2):176–188. doi: 10.1016/j.devcel.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou HX. The affinity-enhancing roles of flexible linkers in two-domain DNA-binding proteins. Biochemistry. 2001;40(50):15069–15073. doi: 10.1021/bi015795g. [DOI] [PubMed] [Google Scholar]
- 42.Zhou H-X. Quantitative relation between intermolecular and intramolecular binding of pro-rich peptides to SH3 domains. Biophys J. 2006;91(9):3170–3181. doi: 10.1529/biophysj.106.090258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hantschel O, et al. A myristoyl/phosphotyrosine switch regulates c-Abl. Cell. 2003;112(6):845–857. doi: 10.1016/s0092-8674(03)00191-0. [DOI] [PubMed] [Google Scholar]
- 44.Zhou H-X. How often does the myristoylated N-terminal latch of c-Abl come off? FEBS Lett. 2003;552(2-3):160–162. doi: 10.1016/s0014-5793(03)00911-6. [DOI] [PubMed] [Google Scholar]
- 45.Parsons M, Messent AJ, Humphries JD, Deakin NO, Humphries MJ. Quantification of integrin receptor agonism by fluorescence lifetime imaging. J Cell Sci. 2008;121(Pt 3):265–271. doi: 10.1242/jcs.018440. [DOI] [PMC free article] [PubMed] [Google Scholar]