Significance
A major goal in genetics is to understand the processes that shape the frequency of new mutations, particularly those causing human disease. Here, we focus on specific mutations in the male germline that, although initially rare, confer a growth or survival advantage to the stem cell, leading to clonal expansion over time: a process similar to early tumor growth and currently described only in humans. Previous studies supporting this “selfish” selection quantified mutations in sperm or testis pieces using methods that destroyed their cellular origins. Here, we pinpoint and identify pathogenic mutations directly within individual seminiferous tubules, the structures that generate spermatozoa. This methodology provides unprecedented precision in documenting the spectrum and prevalence of selfish mutations in men’s testes.
Keywords: mutation, testis, germline, seminiferous tubule, selfish selection
Abstract
De novo point mutations arise predominantly in the male germline and increase in frequency with age, but it has not previously been possible to locate specific, identifiable mutations directly within the seminiferous tubules of human testes. Using microdissection of tubules exhibiting altered expression of the spermatogonial markers MAGEA4, FGFR3, and phospho-AKT, whole genome amplification, and DNA sequencing, we establish an in situ strategy for discovery and analysis of pathogenic de novo mutations. In 14 testes from men aged 39–90 y, we identified 11 distinct gain-of-function mutations in five genes (fibroblast growth factor receptors FGFR2 and FGFR3, tyrosine phosphatase PTPN11, and RAS oncogene homologs HRAS and KRAS) from 16 of 22 tubules analyzed; all mutations have known associations with severe diseases, ranging from congenital or perinatal lethal disorders to somatically acquired cancers. These results support proposed selfish selection of spermatogonial mutations affecting growth factor receptor-RAS signaling, highlight its prevalence in older men, and enable direct visualization of the microscopic anatomy of elongated mutant clones.
Discerning the source of spontaneous germline mutations is fundamental to understanding the causes of many diseases, including monogenic developmental disorders (1) and complex conditions such as autism (2, 3) and schizophrenia (4). Recent whole genome sequencing studies of parent–child trios show that most mutations (such as nucleotide substitutions) originate from the paternal germline and increase in frequency with the father’s age (5, 6), an issue of particular significance given the demographic shift to delayed reproduction in many populations (7). The deduction that the testes of older men harbor a greater burden of mutations, compared with younger men, is consistent with indirect measures of genetic decline, ranging from high indices of arrested germ cell divisions to complete involution of the seminiferous tubules (7–9). Surprisingly, however, it has not previously been possible to trace specific mutations back to their origins within individual germ cells (spermatogonia) of human testes.
One mechanism proposed to contribute to the age-related increase in male mutations is selfish spermatogonial selection, a process equivalent to neoplasia but occurring in the unique context of the germ cell (10). In this process, specific point mutations that confer gain-of-function to components of the growth factor receptor-RAS signaling pathway occur rarely in spermatogonial stem cells of the adult testis but show a steep increase in prevalence with age, attributed to clonal expansion of mutant spermatogonia over time (11–16). Fertilization of the egg by a mutant sperm leads to serious congenital disorders in the next generation, characterized by multiple malformations and, in some cases, a predisposition to malignancy. These disorders include Apert, Crouzon, and Pfeiffer syndromes [caused by FGF receptor 2 (FGFR2) mutations] (17–20), achondroplasia and thanatophoric dysplasia (TD) [FGF receptor 3 (FGFR3)] (21–23), multiple endocrine neoplasia (RET) (24), Noonan syndrome [protein tyrosine phosphatase, non-receptor type 11 (PTPN11)] (25), and Costello syndrome [Harvey rat sarcoma viral oncogene homolog (HRAS)] (25). Consistent with the proposed clonal expansion mechanism, strong gain-of-function mutations of HRAS and FGFR3 have been identified in spermatocytic tumor (seminoma), a testicular tumor characteristically occurring in older men (16, 26). Based on the unexpectedly high birth prevalence of several of the associated congenital disorders, the causative nucleotide substitutions are the most frequently observed spontaneous mutations in the male germline, occurring at levels up to 1,000-fold higher than the background rate (10). Evidence that such “paternal age effect” (PAE) mutations are enriched in either sperm (16, 26–29) or testes (11–15) has required intensive experimental studies targeting specific nucleotide positions; none of these studies preserved the cellular context in which the mutations occurred, meaning that it has not been possible to trace specific mutations back to their origins within individual spermatogonia.
Here, we aimed to isolate pathogenic mutations directly in normal human testes that had been removed for coincidental pathologies (most commonly inguinal hernias) unrelated to either infertility or parenchymal malignancy. We focused this work on the testes of older men, reasoning that PAE mutations would be more readily detected in this age group. In an earlier immunohistochemical survey, we reported that a small fraction of seminiferous tubules (which we termed “immunopositive tubules”) exhibit increased numbers of spermatogonia displaying strong immunoreactivity to antibodies against MAGEA4 (melanoma antigen A4, a spermatogonial marker of unknown function) (30), FGFR3 (a known PAE protein expressed in spermatogonia) (26, 31), and pAKT (phospho–v-akt murine thymoma viral oncogene homolog, a marker of downstream signal activation and a key regulator of spermatogonial stem cell self-renewal) (32, 33). Observing that such increased staining could be traced through multiple serial sections that were physically contiguous (up to 1 mm in length), we speculated that these appearances might correspond to the presence of clonal mutations (34, 35). Here, we have implemented a scalable method targeting >100 genes to identify associated DNA changes in immunopositive tubules. Successful application of this approach, combined with the preservation of tissue architecture, enables the description of the associated mutation spectrum, mapping of the extent of mutant clones, and direct correlation with indices of spermatogenesis.
Results
Proof-of-Principle Study to Identify Mutations in Immunopositive Tubules.
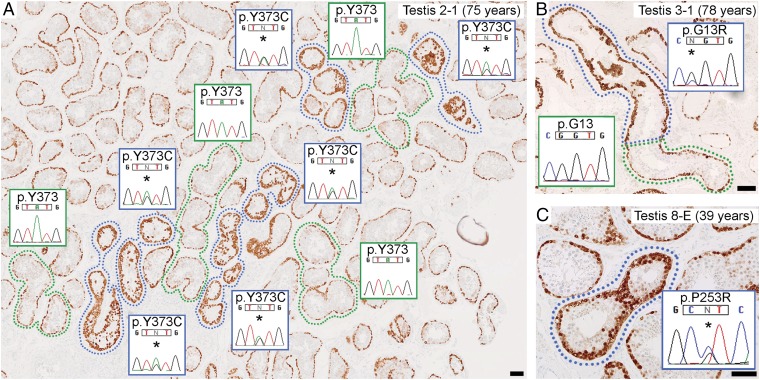
To determine whether immunopositive tubules harbor pathogenic mutations, we performed an initial study on 12 clusters (multiple closely adjacent cross-sections) of seminiferous tubules (11 immunopositive, 1 normal) from four different formalin-fixed, paraffin-embedded (FFPE) testes (testis IDs 1-1, 2-1, 3-1, and 7-1) obtained from donors aged 70–78 y. The indications for removal of the testes and the analytical strategy are detailed in SI Appendix, Table S1 and Fig. S1, respectively. We used laser-capture microdissection (LCM) of thin sections mounted on microscope slides to isolate selected tubules of interest, after which DNA extraction and whole genome amplification (WGA) were performed to increase the amount of target DNA substrate. This material was used to sequence the coding regions of 107 candidate genes (Dataset S1) using HaloPlex target enrichment technology. After excluding known single nucleotide polymorphisms (SNPs) (see SI Appendix, Targeted Sequencing and Variant Calling for full details), many predicted protein-altering variants remained (mean 455, range 231–746); the vast majority of these calls are likely technical artifacts attributable to errors generated during WGA of small amounts of poor quality FFPE DNA (these errors occurred at a frequency of ∼0.2% per nucleotide). To distinguish true mutations from the numerous artifacts, we used an overlap strategy. In testis 1-1, only one variant, FGFR2 c.1024T>A (p.C342S), was shared by the two separate immunopositive tubules but not observed in the single normal tubule. Dideoxy-sequencing of non-WGA DNA extracted from the corresponding tubules of an adjacent section, as well as from six other immunopositive and seven normal neighboring clusters, confirmed that this known pathogenic (Crouzon/Pfeiffer syndrome) (18) mutation (Table 1) was exclusive to the immunopositive tubules (SI Appendix, Fig. S2).
Table 1.
Mutations identified in immunopositive seminiferous tubules
| Testis ID | Age, y | Tubule ID | Mutation identified | Associated germline disorder | Mutation in cancer* |
| 1-1† | 71 | 2 | FGFR2 c.1024T>A (p.C342S)‡§ | Crouzon/Pfeiffer syndrome (18) | No |
| 6 | FGFR2 c.1024T>A (p.C342S)ठ| ||||
| 2-1† | 75 | 7 | FGFR3 c.1118A>G (p.Y373C)§¶ | Thanatophoric dysplasia I# (23) | Yes |
| 11 | FGFR3 c.1118A>G (p.Y373C)§¶ | ||||
| 13 | — | — | — | ||
| 3-1† | 78 | 8 | — | — | — |
| 12 | HRAS c.37G>C (p.G13R)§¶ | None reported | Yes | ||
| 20 | HRAS c.37G>C (p.G13R)§¶ | ||||
| 7-1 | 70 | 1 | — | — | — |
| 4 | — | — | — | ||
| 7 | — | — | — | ||
| 8-E | 39 | A (×3) | FGFR2 c.758C>G (p.P253R) | Apert syndrome (17) | Yes |
| B (×3) | PTPN11 c.215C>T (p.A72V) | None reported | Yes | ||
| 9-L2C | 62 | A (×3) | PTPN11 c.181G>T (p.D61Y)¶ | None reported | Yes |
| 10-F | 63 | B (×3) | FGFR2 c.1019A>G (p.Y340C) | Pfeiffer syndrome# (20) | No |
| 11-H | 70 | A (×3) | FGFR2 c.1019A>G (p.Y340C)¶ | Pfeiffer syndrome# (20) | No |
| 12-E | 71 | A (×3) | KRAS c.182A>G (p.Q61R) | None reported | Yes |
| 12-H | A (×3) | — | — | — | |
| 13-G | 72 | A (×3) | PTPN11 c.215C>T (p.A72V) | None reported | Yes |
| B (×3) | FGFR3 c.742C>T (p.R248C) | Thanatophoric dysplasia I# (22) | Yes | ||
| 14-D | 80 | A (×3) | KRAS c.182A>G (p.Q61R)¶ | None reported | Yes |
| 15-B | 80 | A (×3) | KRAS c.182A>G (p.Q61R) | None reported | Yes |
| 16-D | 87 | A (×3) | FGFR3 c.1948A>G (p.K650E)¶ | Thanatophoric dysplasia II# (22) | Yes |
| B (×3) | FGFR2 c.870G>T (p.W290C) | Pfeiffer syndrome (19) | Yes | ||
| 17-2E | 90 | A (×3) | FGFR3 c.1948A>G (p.K650E)‡¶ | Thanatophoric dysplasia II# (22) | Yes |
Further details are available in SI Appendix, Table S1, and full staining and sequencing data are available in SI Appendix, Figs. S2–S5, S7–S16, and S18.
Present in COSMIC database v67 (36).
Previously studied in ref. 34.
Apparently homozygous.
Identical mutations found in distinct cross-sections of tubules are likely to be part of the same contiguous tubule.
Identified in candidate screen.
Perinatal lethal.
In the other three testes, no variants were shared by all three immunopositive tubular clusters from an individual testis. Therefore, we prioritized mutations identified in individual tubule clusters based on their association with germline disorders or cancer (36) and validated candidates by dideoxy-sequencing of non-WGA DNA from corresponding tubules of an adjacent section. In testis 2-1, a heterozygous FGFR3 c.1118A>G (p.Y373C) mutation (which causes TD I) (23) was present in two of three immunopositive clusters (one of which was negative by HaloPlex sequencing, suggesting that allelic dropout occurred during WGA) and in multiple other immunopositive tubules in the section; normal-appearing neighboring tubules did not harbor this mutation (Fig. 1A and SI Appendix, Fig. S3). In testis 3-1, a heterozygous HRAS c.37G>C (p.G13R) mutation was present in two of three immunopositive clusters (one of which was HaloPlex screen-negative) (Fig. 1B and SI Appendix, Fig. S4). HRAS p.G13R has not been reported as a germline mutation but occurs as a somatic mutation in numerous tumors (36), including spermatocytic tumor (16). No candidate mutations were confirmed in testis 7-1 (SI Appendix, Fig. S5). Therefore in this initial study, we found that 6 of 11 immunopositive tubules, from three of the four testes, harbored an identifiable pathogenic mutation in a known PAE gene. Tubules shown to have the same mutation (and therefore likely to have a single mutational origin) were physically separated on the FFPE section by up to 4.8 mm, 15.5 mm, and 13.7 mm in testes 1-1, 2-1, and 3-1, respectively (SI Appendix, Figs. S2–S4), placing a lower bound on the length of the mutant clone.
Fig. 1.
Seminiferous tubules strongly immunopositive for MAGEA4 contain pathogenic mutations. (A–C) Thin sections from three FFPE testes showing spermatogonia, marked by MAGEA4 positivity (brown staining), present in a single layer at the periphery of normal tubular cross-sections (green surround or unlabeled). A subset of tubules (immunopositive tubules) display enhanced MAGEA4 staining (blue surround) due to dense clustering of spermatogonia with strong immunoreactivity. Dideoxy-sequencing traces were obtained from non-WGA DNA extracted from microdissected tissue of an adjacent section. (A) Heterozygous FGFR3 c.1118A>G (p.Y373C) mutations (*) are present in immunopositive tubules, but not in neighboring normal tubules. Clusters of mutation-positive tubules likely represent cross-sections of a single convoluted tubule. (B) In a longitudinal section of a tubule showing the transition from normal to strongly immunopositive staining, the heterozygous HRAS c.37C>G (p.G13R) mutation (*) was specific to the immunopositive portion, pinpointing the boundary between nonmutant and mutant cells. (C) Heterozygous FGFR2 c.758C>G (p.P253R) mutation (*) in immunopositive tubule. (Scale bars: 100 µm.)
Mutation Identification Using Triplicate Samples.
To improve success in mutation identification (despite the poor input FFPE DNA quality and quantity), we refined our protocol in a second round of experiments. Key improvements were to select only testes obtained within the previous 6 y, to increase the amount of microdissected tubules in each reaction, to perform independent triplicate analyses of each tubule and intersect the final sequencing data, and to sequence constitutional DNA to exclude inherited variation (SI Appendix, Fig. S6).
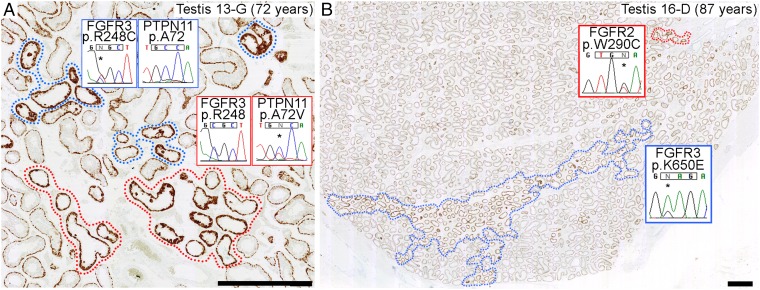
Based on size, appearance, and DNA quality, 10 clusters of immunopositive tubular cross-sections from nine further testes (donor age range 39–87 y) were selected. We sequenced 135 genes, including all oncogenes (37) and gain-of-function cancer predisposition genes (38), using an updated HaloPlex panel (Dataset S1). Candidate variants present in all three tubular replicates, but absent from the matched constitutional DNA, were validated by dideoxy-sequencing of non-WGA DNA microdissected from an adjacent section. In 5 of 10 tubule clusters, mutations that were previously shown to be pathogenic in constitutional disorders and/or cancer were called in each of the triplicates and validated (Table 1): FGFR2 c.758C>G (p.P253R; Apert syndrome) (17) in testis 8-E (Fig. 1C) and c.1019A>G (p.Y340C; Pfeiffer syndrome) (20) in testis 10-F, FGFR3 c.742C>T (p.R248C; TD I) (22) in testis 13-G (Fig. 2A), and KRAS (Kirsten rat sarcoma viral oncogene homolog) c.182A>G (p.Q61R) [oncogenic (36), no germline cases reported] in testes 12-E and 15-B. We observed that four known PAE hotspots in FGFR3 were poorly covered in all samples (median 0–4×) (Dataset S1); targeted Ion PGM resequencing of these regions identified FGFR3 c.1948A>G (p.K650E; TD II) (22) in the triplicate samples from testis 16-D (Fig. 2B). For original data supporting each result, see SI Appendix, Figs. S7–S12.
Fig. 2.
Independent mutations can populate adjacent MAGEA4 immunopositive tubules, and extensive regional spread can occur. (A) Mutually exclusive FGFR3 c.742C>T (p.R248C) (blue surround) and PTPN11 c.215C>T (p.A72V) (red surround) mutations (*) in neighboring tubules with similar immunopositive appearance. (B) Low magnification view showing extensive region of tubules with FGFR3 c.1948A>G (p.K650E) mutation (*) (blue surround). Immunopositive tubules outside this region harbor a different nucleotide substitution, FGFR2 c.870G>T (p.W290C) (*) (red surround). (Scale bars: 1 mm.)
Next, for the six testes in which a mutation was identified, we determined by dideoxy-sequencing whether other immunopositive tubules within the same FFPE section harbored the identical mutation. Although some mutations were found in extensive regions, indicating substantial clonal expansion (Fig. 2B), in the five testes for which additional immunopositive tubules were present in the same FFPE section, at least one of these tubules did not carry the original mutation. We selected three such tubular clusters for a further round of HaloPlex sequencing using the same triplicate strategy and identified a mutation different from that present in the original immunopositive tubule in every case; the mutations (Table 1) were FGFR2 c.870G>T (p.W290C; Pfeiffer syndrome) (19) in testis 16-D (Fig. 2B), and PTPN11 c.215C>T [p.A72V; oncogenic (36), no germline cases reported] in testes 8-E and 13-G (Fig. 2A). For original data supporting these results, see SI Appendix, Figs. S12, S7, and S9, respectively.
For the four samples in which the triplicate intersection approach had not directly identified a mutation, we hypothesized that allelic dropout may have occurred during WGA (as observed in the proof-of-principle study). Based on the previous results, which had identified mutations only in the known PAE genes and KRAS, we examined the HaloPlex sequencing data for variants at known hotspots within these six genes that were present in one or two of the triplicates, and subsequently dideoxy-sequenced these candidates using non-WGA DNA. Mutations were confirmed in three of the four samples (Table 1): PTPN11 c.181G>T [p.D61Y; oncogenic (36), no germline cases reported] in testis 9-L2C, FGFR2 p.Y340C in testis 11-H, and KRAS p.Q61R in testis 14-D. No candidates were confirmed in testis 12-H. For original data supporting these results, see SI Appendix, Figs. S13–S16.
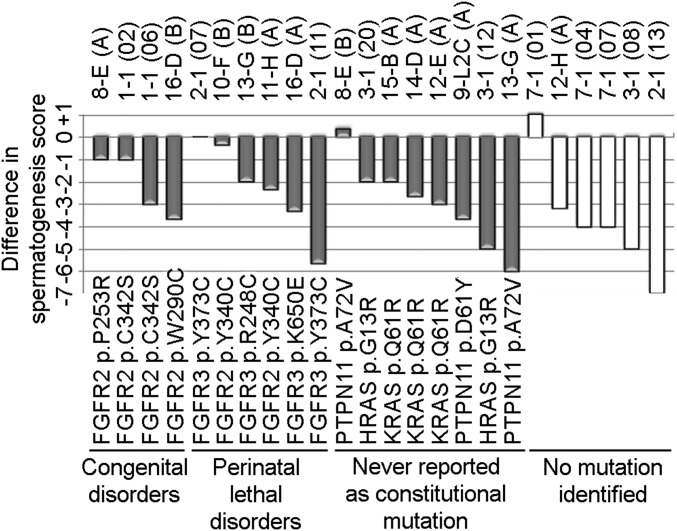
Combining the two studies, we identified 15 distinct mutational events (involving 11 different pathogenic substitutions), all of which were specific to tubules with an immunopositive appearance. To assess whether the immunopositive tubules analyzed exhibited altered cellular dynamics of spermatogenesis, they were compared with adjacent normal-staining tubules using Johnsen’s scoring criteria (39). In most cases, immunopositive tubules showed significantly impaired spermatogenesis compared with the neighboring normal tubules, irrespective of mutation status (Fig. 3 and SI Appendix, Fig. S17).
Fig. 3.
Analysis of spermatogenesis in immunopositive tubules. The difference in Johnsen scores between paired immunopositive and adjacent normal tubules is plotted for each pair; Johnsen scores range between 1 (no seminiferous epithelium) and 10 (full spermatogenesis). In pairwise analysis, the Johnsen score is significantly lower in immunopositive than in normal-staining tubules (P = 5.2 × 10−5, Wilcoxon signed rank test).
Apparent Survival Advantage of Mutant Germ Cells in an Atrophic Testis.
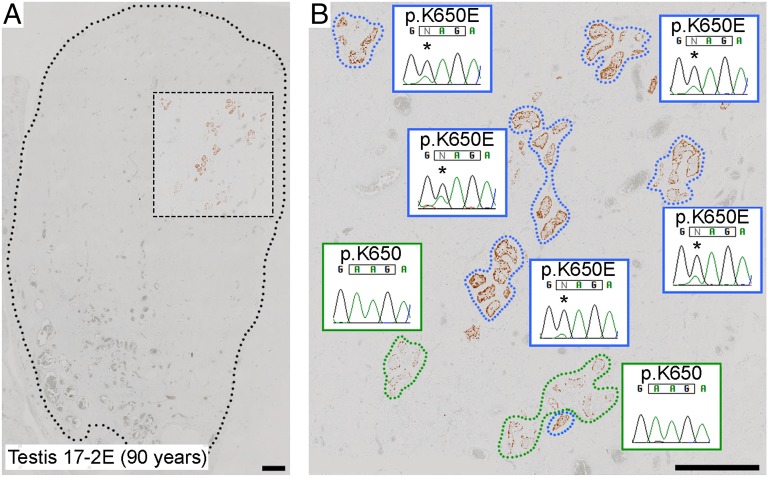
In addition to the macroscopically normal testes, we analyzed a testis from a 90-y-old man (17-2E) that showed evidence of severe atrophy owing to strangulation in an inguinal hernia. Consistent with the clinical presentation, few identifiable tubules containing germ cells remained; surprisingly, however, the majority of the cross-sections had an immunopositive appearance (Fig. 4A and SI Appendix, Fig. S18). Ion PGM sequencing of WGA triplicates of one immunopositive cluster, targeted to 37 mutational hotspots across the five PAE genes (SI Appendix, Table S2), identified an FGFR3 p.K650E mutation (22) that was validated by dideoxy-sequencing in all other immunopositive cross-sections (Fig. 4B).
Fig. 4.
Retention of mutant seminiferous tubules in severely atrophic testis. (A) Low magnification view of MAGEA4-stained atrophic testis with the dotted line demarcating the testicular parenchyma. Tubules with spermatogonia (identified by brown stain) are rare and present only in the outlined box. (B) Higher magnification of boxed region, demonstrating that most of the tubular cross-sections containing spermatogonia have an immunopositive appearance (blue surround) and carry the apparently homozygous FGFR3 c.1948A>G (p.K650E) mutation (*). The normally stained tubules (green surround) do not carry the mutation at a detectable level. (Scale bars: 1 mm.)
Discussion
Using scalable methods, we have identified mutations at their source in testes from 14 men aged 39–90 y. Although there were variable ischemic changes in some of the testes, most were histologically normal (SI Appendix, Table S1). The spermatogonial origin of these mutations is indicated by the strong immunoreactivity of cells within putative clones to MAGEA4, FGFR3, and pAKT, antigens characteristic of premeiotic germ cells (30–33). Microdissection of putative clones, with the aim of maximizing the relative mutant DNA content of the sample (up to ±50%), enabled the simultaneous screening of ∼300,000 nucleotides in over 100 candidate genes, in comparison with only 19 nucleotides (across 6 genes) that were investigated in all previous sperm (16, 26–29) and testis piece (11–15) studies combined. This strategy, which successfully identified pathogenic mutations in 76% of the abnormal tubules sequenced, advances understanding of the pathophysiology of male-driven mutation in two key respects. First, we can document the range of mutations that are likely to be subject to selfish selection over a broader spectrum than has hitherto been possible in a single experiment; second, we can visualize the geographical extent of mutant clones and make deductions about the functional consequences of the mutations.
In addition to detecting well-established PAE mutations in FGFR2 (c.758C>G; p.P253R) and FGFR3 (c.1948A>G; p.K650E) that were previously validated experimentally in sperm and/or testes (12, 26, 29), we widened the spectrum of mutations found in testes for four of the five recognized PAE genes (FGFR2, FGFR3, HRAS, and PTPN11; we found no mutations in RET). Although we surveyed >100 additional candidate genes, mutations were found in only one of these genes, KRAS [immunopositive tubules from three different testes contained the identical oncogenic c.182A>G (p.Q61R) mutation]. Like its paralog HRAS, KRAS encodes a canonical component of RAS signal transduction; although KRAS does not formally qualify as a PAE gene because the parental origin of KRAS-associated Noonan syndrome mutations (25) has not been investigated, the observation of KRAS mutations in immunopositive tubules implicates dysregulation of growth factor receptor-RAS signaling, the same mechanism invoked for classical PAE mutations (10).
Overall, we conclude that, although the majority of mutations identified have not previously been studied in either testes or sperm, there is close correspondence between the spectrum of mutant clones identified in situ in testes and the documented properties of PAE mutations. Here, we can visualize these expanded clones for the first time, to our knowledge. Immediate conclusions from this work are that mutant clones can readily be detected in testes from older men, that distinct clones sometimes occur in close proximity to each other (Fig. 2 and SI Appendix, Figs. S7, S9, and S12), and that their boundaries with normal regions of tubules can be identified (Fig. 1B). Importantly, clonal growth occurs one-dimensionally, along the highly convoluted tubules (40), reminiscent of normal spermatogonial clonal dynamics in the murine testis (41, 42) and consistent with a previous experimental study (43). The lower bounds of the length of mutant clones found in the proof-of-principle study (4.4–15.3 mm), correspond to 1–4 × 10−5 of the estimated total length (350-400 m) of seminiferous tubules in a human testis (44), similar to figures for mutation prevalence previously attributed to selfish spermatogonial selection (10), but much higher than the background germline mutation rate (5, 6). The majority of the mutations were present in the heterozygous state (taking into account dilution by nonmutant cells including Sertoli and extratubular cells); we propose that these mutations are necessary and sufficient to drive the clonal growth observed. In two cases, loss-of-heterozygosity of the mutation (in testes 1-1 and 17-2E) was present: these additional events may represent early stages of clonal evolution toward spermatocytic tumor (26, 35).
Activation of signaling through RAS may confer either proliferative or survival advantage to cells (33, 43); in many contexts, including selfish spermatogonial selection, it can be difficult to disentangle the contributions of these two processes. Our finding that the few remaining seminiferous tubules in the atrophic, presumed chronically ischemic, testis mostly harbored a pathogenic FGFR3 mutation (Fig. 4) points to the contribution of a survival advantage in this case. Analogously, the presence of the FGFR3 p.G380R mutation was proposed to account for the unusual preservation of active spermatogenesis and fertility in a patient with Klinefelter syndrome, who also had achondroplasia (45).
Although our protocol enables a wide survey of genes to be undertaken, it is striking that a majority of the 16 mutations separately identified are associated with severe cellular phenotypes, with only three of the FGFR2 mutations expected to be compatible with long-term survival of the offspring; the remaining mutations cause either perinatal lethal disorders (6/16) or have not previously been reported as constitutional mutations (7/16) (Table 1). The last group (including all mutations in PTPN11, HRAS, and KRAS) are allelic to neurocardiofaciocutaneous syndromes (25) but either encode distinct amino acid substitutions or involve mutations at different positions that are oncogenic when acquired in somatic tissue (36). Therefore these mutations may lead to early embryonic lethality; alternatively, they might be incompatible with formation of mature sperm through focal germ cell arrest (8), as supported by the significantly impaired spermatogenesis associated with immunopositive tubules (Fig. 3). The tendency to identify severe mutations likely reflects preferential isolation of tubules with strong immunoreactivity. Consistent with this interpretation, we did not detect any tubules containing the FGFR3 c.1138G>A (p.G380R; achondroplasia) mutation, thought to represent the most frequent point mutation in the human germline (21), which is associated with mild pathway dysregulation (46) and probably a more subtle immunoreactive profile. These findings will stimulate efforts to increase the sensitivity for isolating mutations with milder functional consequences: for example, using different antibodies combined with analysis of frozen tissue to improve DNA quality. Further increases in sensitivity will be required to determine the extent to which selfish selection has a more pervasive effect on male-biased mutation in disease (10, 47).
In summary, for the first time, to our knowledge, this experimental approach traces the origin of de novo pathogenic mutations to specific germ cells of the human male, illustrating a fundamental principle in Mendelian genetics not previously described in any model organism. Recent studies have highlighted the widespread occurrence of somatically acquired mutant clones in a variety of tissues [for example blood (48, 49) and skin (50, 51)]; here, we have illustrated analogous phenomena, but in the unique context of the germline, with its implications for altering the genetic constitution of offspring.
Materials and Methods
Testis Samples.
Ethical approval was provided by the Oxfordshire Research Ethics Committee A (C03.076), and all patients had given informed written consent for research use of pathological samples. Archival blocks of formalin-fixed paraffin-embedded (FFPE) testes were sectioned and screened by immunohistochemistry using spermatogonial cell markers as described (34). Overall histology was recorded, and spermatogenesis in immunopositive and neighboring normal tubular cross-sections was scored, blinded to immunopositive and mutational status, using Johnsen’s criteria (39).
Immunohistochemical Screening and Microdissection.
Initially, one to two 5-µm sections from each FFPE block were stained using a MAGEA4 antibody (which, empirically, we have found to be a robust screening marker for initial identification of candidate clones) and scanned for atypical expression (i.e., “immunopositive status”) (34). Further sections were obtained from FFPE blocks in which immunopositive tubules were detected (SI Appendix, Table S3), and alternate sections were immunostained with MAGEA4, FGFR3, and phospho-AKT (pAKT) antibodies or mounted on laser-capture microdissection (LCM) slides. In the proof-of-principle study (four testes), one to four neighboring cross-sections of tubules from three geographically distinct regions of the slide were isolated using a laser capture microscope (Zeiss). For the triplicate study (10 FFPE blocks obtained from nine testes), three biological replicates of tubular cross-sections from the same region were obtained from serial LCM slides. DNA extraction and WGA were performed for each sample independently. Corresponding tubules from adjacent slides were microdissected and processed without WGA for dideoxy-sequence validation. Constitutional DNA control was obtained by pooling material from three whole sections.
Targeted Sequencing and Variant Calling.
Coding regions of 107 (proof-of-principle study) or 135 (main study) genes were captured using a HaloPlex custom design panel (Agilent Technologies) and sequenced on the HiSeq 2000 platform (Illumina). After processing reads, somatic variants were called using VarScan (52) and annotated with ANNOVAR (53). Two poorly covered regions of FGFR3 were amplified by PCR from WGA material and sequenced with Ion PGM (Life Technologies); a similar method was used to sequence nine mutation hotspots in DNA from an atrophic testis (17-2E). In the proof-of-principle study, common SNPs [frequency of ≥0.001 in ESP6500 (54) or 1000 Genomes Project (55)] were removed, and candidate variants were prioritized based on known disease or cancer association (36). For the triplicate study, variants called in all three replicates, but not present in matched constitutional DNA, were validated by dideoxy-sequencing. In samples without triplicate consensus calls, variants in PAE genes and KRAS were screened by dideoxy-sequencing. For a detailed description of the methods, see SI Appendix, SI Text.
Supplementary Material
Acknowledgments
We thank Linda Godfrey, Alethea Hutchison, Divija Jatavallabhula, Jane Niederer, Gareth Turner, and the Oxford Centre for Histopathology Research (OCHRe) for sourcing and processing samples; Giulio Spagnoli for the MAGEA4 antibody; Hayley Davis, Simon Leedham, and Ian Tomlinson for assistance with laser capture microdissection; John Frankland and Tim Rostron for dideoxy-sequencing; Kerry Miller and Indira Taylor for Ion PGM sequencing; and Lorna Witty and the High-Throughput Genomics core at the Wellcome Trust Centre for Human Genetics (funded by Wellcome Trust Grant 090532) for generation of Illumina sequencing data. OCHRe is supported by the NIHR Oxford Biomedical Research Centre. This work was funded by Wellcome Trust Grants 091182 (to A.G. and A.O.M.W.) and 102731 (to A.O.M.W.) and Simons Foundation Grant 332759 (to A.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1521325113/-/DCSupplemental.
References
- 1.Deciphering Developmental Disorders Study Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519:223–228. doi: 10.1038/nature14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iossifov I, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515(7526):216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Roak BJ, et al. Recurrent de novo mutations implicate novel genes underlying simplex autism risk. Nat Commun. 2014;5:5595. doi: 10.1038/ncomms6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoischen A, Krumm N, Eichler EE. Prioritization of neurodevelopmental disease genes by discovery of new mutations. Nat Neurosci. 2014;17(6):764–772. doi: 10.1038/nn.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kong A, et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488(7412):471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Besenbacher S, et al. Novel variation and de novo mutation rates in population-wide de novo assembled Danish trios. Nat Commun. 2015;6:5969. doi: 10.1038/ncomms6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paul C, Robaire B. Ageing of the male germ line. Nat Rev Urol. 2013;10(4):227–234. doi: 10.1038/nrurol.2013.18. [DOI] [PubMed] [Google Scholar]
- 8.Paniagua R, Nistal M, Amat P, Rodriguez MC, Martin A. Seminiferous tubule involution in elderly men. Biol Reprod. 1987;36(4):939–947. doi: 10.1095/biolreprod36.4.939. [DOI] [PubMed] [Google Scholar]
- 9.Miething A. Arrested germ cell divisions in the ageing human testis. Andrologia. 2005;37(1):10–16. doi: 10.1111/j.1439-0272.2004.00645.x. [DOI] [PubMed] [Google Scholar]
- 10.Goriely A, Wilkie AOM. Paternal age effect mutations and selfish spermatogonial selection: Causes and consequences for human disease. Am J Hum Genet. 2012;90(2):175–200. doi: 10.1016/j.ajhg.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qin J, et al. The molecular anatomy of spontaneous germline mutations in human testes. PLoS Biol. 2007;5(9):e224. doi: 10.1371/journal.pbio.0050224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi SK, Yoon SR, Calabrese P, Arnheim N. A germ-line-selective advantage rather than an increased mutation rate can explain some unexpectedly common human disease mutations. Proc Natl Acad Sci USA. 2008;105(29):10143–10148. doi: 10.1073/pnas.0801267105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi SK, Yoon SR, Calabrese P, Arnheim N. Positive selection for new disease mutations in the human germline: Evidence from the heritable cancer syndrome multiple endocrine neoplasia type 2B. PLoS Genet. 2012;8(2):e1002420. doi: 10.1371/journal.pgen.1002420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shinde DN, et al. New evidence for positive selection helps explain the paternal age effect observed in achondroplasia. Hum Mol Genet. 2013;22(20):4117–4126. doi: 10.1093/hmg/ddt260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoon SR, et al. Age-dependent germline mosaicism of the most common Noonan syndrome mutation shows the signature of germline selection. Am J Hum Genet. 2013;92(6):917–926. doi: 10.1016/j.ajhg.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giannoulatou E, et al. Contributions of intrinsic mutation rate and selfish selection to levels of de novo HRAS mutations in the paternal germline. Proc Natl Acad Sci USA. 2013;110(50):20152–20157. doi: 10.1073/pnas.1311381110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilkie AOM, et al. Apert syndrome results from localized mutations of FGFR2 and is allelic with Crouzon syndrome. Nat Genet. 1995;9(2):165–172. doi: 10.1038/ng0295-165. [DOI] [PubMed] [Google Scholar]
- 18.Tartaglia M, et al. Jackson-Weiss syndrome: Identification of two novel FGFR2 missense mutations shared with Crouzon and Pfeiffer craniosynostotic disorders. Hum Genet. 1997;101(1):47–50. doi: 10.1007/s004390050584. [DOI] [PubMed] [Google Scholar]
- 19.Schaefer F, Anderson C, Can B, Say B. Novel mutation in the FGFR2 gene at the same codon as the Crouzon syndrome mutations in a severe Pfeiffer syndrome type 2 case. Am J Med Genet. 1998;75(3):252–255. doi: 10.1002/(sici)1096-8628(19980123)75:3<252::aid-ajmg4>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 20.Khonsari RH, et al. Central nervous system malformations and deformations in FGFR2-related craniosynostosis. Am J Med Genet A. 2012;158A(11):2797–2806. doi: 10.1002/ajmg.a.35598. [DOI] [PubMed] [Google Scholar]
- 21.Bellus GA, et al. Achondroplasia is defined by recurrent G380R mutations of FGFR3. Am J Hum Genet. 1995;56(2):368–373. [PMC free article] [PubMed] [Google Scholar]
- 22.Tavormina PL, et al. Thanatophoric dysplasia (types I and II) caused by distinct mutations in fibroblast growth factor receptor 3. Nat Genet. 1995;9(3):321–328. doi: 10.1038/ng0395-321. [DOI] [PubMed] [Google Scholar]
- 23.Rousseau F, et al. Missense FGFR3 mutations create cysteine residues in thanatophoric dwarfism type I (TD1) Hum Mol Genet. 1996;5(4):509–512. doi: 10.1093/hmg/5.4.509. [DOI] [PubMed] [Google Scholar]
- 24.Carlson KM, et al. Parent-of-origin effects in multiple endocrine neoplasia type 2B. Am J Hum Genet. 1994;55(6):1076–1082. [PMC free article] [PubMed] [Google Scholar]
- 25.Tartaglia M, Gelb BD, Zenker M. Noonan syndrome and clinically related disorders. Best Pract Res Clin Endocrinol Metab. 2011;25(1):161–179. doi: 10.1016/j.beem.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goriely A, et al. Activating mutations in FGFR3 and HRAS reveal a shared genetic origin for congenital disorders and testicular tumors. Nat Genet. 2009;41(11):1247–1252. doi: 10.1038/ng.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goriely A, McVean GAT, Röjmyr M, Ingemarsson B, Wilkie AOM. Evidence for selective advantage of pathogenic FGFR2 mutations in the male germ line. Science. 2003;301(5633):643–646. doi: 10.1126/science.1085710. [DOI] [PubMed] [Google Scholar]
- 28.Goriely A, et al. Gain-of-function amino acid substitutions drive positive selection of FGFR2 mutations in human spermatogonia. Proc Natl Acad Sci USA. 2005;102(17):6051–6056. doi: 10.1073/pnas.0500267102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoon SR, et al. The ups and downs of mutation frequencies during aging can account for the Apert syndrome paternal age effect. PLoS Genet. 2009;5(7):e1000558. doi: 10.1371/journal.pgen.1000558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aubry F, et al. MAGE-A4, a germ cell specific marker, is expressed differentially in testicular tumors. Cancer. 2001;92(11):2778–2785. doi: 10.1002/1097-0142(20011201)92:11<2778::aid-cncr10125>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 31.von Kopylow K, Staege H, Schulze W, Will H, Kirchhoff C. Fibroblast growth factor receptor 3 is highly expressed in rarely dividing human type A spermatogonia. Histochem Cell Biol. 2012;138(5):759–772. doi: 10.1007/s00418-012-0991-7. [DOI] [PubMed] [Google Scholar]
- 32.Lee J, et al. Akt mediates self-renewal division of mouse spermatogonial stem cells. Development. 2007;134(10):1853–1859. doi: 10.1242/dev.003004. [DOI] [PubMed] [Google Scholar]
- 33.Lee J, et al. Genetic reconstruction of mouse spermatogonial stem cell self-renewal in vitro by Ras-cyclin D2 activation. Cell Stem Cell. 2009;5(1):76–86. doi: 10.1016/j.stem.2009.04.020. [DOI] [PubMed] [Google Scholar]
- 34.Lim J, et al. Selfish spermatogonial selection: Evidence from an immunohistochemical screen in testes of elderly men. PLoS One. 2012;7(8):e42382. doi: 10.1371/journal.pone.0042382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maher GJ, Goriely A, Wilkie AOM. Cellular evidence for selfish spermatogonial selection in aged human testes. Andrology. 2014;2(3):304–314. doi: 10.1111/j.2047-2927.2013.00175.x. [DOI] [PubMed] [Google Scholar]
- 36.Forbes SA, et al. COSMIC: Exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43(Database issue) D1:D805–D811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rahman N. Realizing the promise of cancer predisposition genes. Nature. 2014;505(7483):302–308. doi: 10.1038/nature12981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnsen SG. Testicular biopsy score count--a method for registration of spermatogenesis in human testes: Normal values and results in 335 hypogonadal males. Hormones. 1970;1(1):2–25. doi: 10.1159/000178170. [DOI] [PubMed] [Google Scholar]
- 40.Nakata H, et al. Three-dimensional structure of seminiferous tubules in the adult mouse. J Anat. 2015;227(5):686–694. doi: 10.1111/joa.12375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klein AM, Nakagawa T, Ichikawa R, Yoshida S, Simons BD. Mouse germ line stem cells undergo rapid and stochastic turnover. Cell Stem Cell. 2010;7(2):214–224. doi: 10.1016/j.stem.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 42.Hara K, et al. Mouse spermatogenic stem cells continually interconvert between equipotent singly isolated and syncytial states. Cell Stem Cell. 2014;14(5):658–672. doi: 10.1016/j.stem.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin LA, Assif N, Gilbert M, Wijewarnasuriya D, Seandel M. Enhanced fitness of adult spermatogonial stem cells bearing a paternal age-associated FGFR2 mutation. Stem Cell Rep. 2014;3(2):219–226. doi: 10.1016/j.stemcr.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Glass J. Testes and epididymes. In: Standring S, editor. Gray’s Anatomy: The Anatomical Basis of Clinical Practice. 39th Ed. Churchill Livingston; Edinburgh: 2005. pp. 1304–1310. [Google Scholar]
- 45.Juul A, et al. Preserved fertility in a non-mosaic Klinefelter patient with a mutation in the fibroblast growth factor receptor 3 gene: Case report. Hum Reprod. 2007;22(7):1907–1911. doi: 10.1093/humrep/dem126. [DOI] [PubMed] [Google Scholar]
- 46.Naski MC, Wang Q, Xu J, Ornitz DM. Graded activation of fibroblast growth factor receptor 3 by mutations causing achondroplasia and thanatophoric dysplasia. Nat Genet. 1996;13(2):233–237. doi: 10.1038/ng0696-233. [DOI] [PubMed] [Google Scholar]
- 47.Goriely A, McGrath JJ, Hultman CM, Wilkie AOM, Malaspina D. “Selfish spermatogonial selection”: A novel mechanism for the association between advanced paternal age and neurodevelopmental disorders. Am J Psychiatry. 2013;170(6):599–608. doi: 10.1176/appi.ajp.2013.12101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jaiswal S, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371(26):2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McKerrell T, et al. Understanding Society Scientific Group Leukemia-associated somatic mutations drive distinct patterns of age-related clonal hemopoiesis. Cell Reports. 2015;10(8):1239–1245. doi: 10.1016/j.celrep.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hafner C, et al. Spectrum of FGFR3 mutations in multiple intraindividual seborrheic keratoses. J Invest Dermatol. 2007;127(8):1883–1885. doi: 10.1038/sj.jid.5700804. [DOI] [PubMed] [Google Scholar]
- 51.Martincorena I, et al. High burden and pervasive positive selection of somatic mutations in normal human skin. Science. 2015;348(6237):880–886. doi: 10.1126/science.aaa6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koboldt DC, et al. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22(3):568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP), Seattle, WA. Available at evs.gs.washington.edu/EVS/. Accessed January 31, 2015.
- 55.Abecasis GR, et al. 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.