Significance
Programmed cell death is usually considered a cell-autonomous suicide program, synonymous with apoptosis. Here we demonstrate that a specific example of large-scale nonapoptotic developmental programmed cell death in the Drosophila ovary occurs by an alternative cell death program where surrounding epithelial cells nonautonomously promote the death of the germ line. We find that genes normally required for engulfment of dying cells act to promote the death of the germ line. Developmental programmed cell death in the Drosophila ovary is an intriguing example of nonapoptotic, nonautonomous cell death, providing insight on the diversity of cell death mechanisms.
Keywords: Drosophila, ovary, cell death, phagocytosis, engulfment
Abstract
Programmed cell death (PCD) is usually considered a cell-autonomous suicide program, synonymous with apoptosis. Recent research has revealed that PCD is complex, with at least a dozen cell death modalities. Here, we demonstrate that the large-scale nonapoptotic developmental PCD in the Drosophila ovary occurs by an alternative cell death program where the surrounding follicle cells nonautonomously promote death of the germ line. The phagocytic machinery of the follicle cells, including Draper, cell death abnormality (Ced)-12, and c-Jun N-terminal kinase (JNK), is essential for the death and removal of germ-line–derived nurse cells during late oogenesis. Cell death events including acidification, nuclear envelope permeabilization, and DNA fragmentation of the nurse cells are impaired when phagocytosis is inhibited. Moreover, elimination of a small subset of follicle cells prevents nurse cell death and cytoplasmic dumping. Developmental PCD in the Drosophila ovary is an intriguing example of nonapoptotic, nonautonomous PCD, providing insight on the diversity of cell death mechanisms.
Programmed cell death (PCD) is the genetically controlled elimination of cells that occurs during organismal development and homeostasis. Cells are considered dead when they have undergone irreversible plasma membrane permeabilization or have become completely fragmented (1). Apoptosis is the most well-characterized form of PCD, however there are at least a dozen cell death modalities that are morphologically, biochemically, and genetically distinct (2, 3). Two examples of nonapoptotic cell death are autophagic cell death and necrosis, but there are several alternative cell death mechanisms that are less well understood.
Nonapoptotic PCD occurs on a large scale in the Drosophila ovary. Drosophila females can produce hundreds of eggs during their lifetime, and for every egg that is formed, developmental PCD of supporting nurse cells (NCs) occurs. However, the mechanisms of developmental PCD in the Drosophila ovary are poorly understood. Each egg forms from a 16-cell germ-line cyst, comprised of the single oocyte and 15 NCs that support the oocyte throughout 14 stages of oogenesis (4, 5). Hundreds of somatically derived follicle cells (FCs) surround the germ-line cyst, forming an egg chamber. At stage 11 of oogenesis, NCs rapidly transfer (“dump”) their cytoplasm into the oocyte. Concurrently, the NCs asynchronously undergo developmental PCD, resulting in mature stage 14 egg chambers that no longer contain any NCs (4–6). Interestingly, caspases, proteases associated with apoptosis, play only a minor role in the death of the NCs in late oogenesis (7–9). Furthermore, combined inhibition of caspases and autophagy does not significantly block NC death during late oogenesis (10). To date, defining the major mechanism of developmental PCD in the Drosophila ovary has remained elusive.
An intriguing possibility is that the somatic FCs non–cell-autonomously promote developmental PCD of the NCs during late oogenesis. Non–cell-autonomous regulation of PCD occurs when a cell or group of cells extrinsically initiates or promotes the death of another cell. This concept challenges the idea that PCD is largely a self-regulated, autonomous suicide program in which a cell controls its own demise. One well-characterized example of non–cell-autonomous control of PCD is apoptosis induced by the death ligands Fas or TNF (11, 12).
Another type of non–cell-autonomous PCD is phagoptosis (or primary phagocytosis), in which engulfing cells directly cause the death of other cells via “murder” or “assisted suicide.” Phagoptosis is distinct from the engulfment of cell corpses, as the engulfing cell plays an active role in the death of a cell, rather than simply degrading a cell that died via another mechanism. The defining characteristic of phagoptosis is that inhibition of phagocytosis leads to a failure in cell death (13, 14). Phagoptosis has been demonstrated in activated microglia that phagocytose viable neurons, resulting in their destruction (13–15). Entosis is another example of non–cell-autonomous PCD, often referred to as “cell cannibalism,” in which a viable cell invades another cell, where it is degraded by lysosomes. Entosis is distinct from phagoptosis, as the inhibition of phagocytosis genes does not prevent entosis (16). Phagocytosis has also been shown to promote PCD in Caenorhabditis elegans, although this is an example of assisted suicide, as dying cells also require apoptotic machinery (17, 18).
Genetic studies in C. elegans have identified two partially redundant signaling pathways that control phagocytosis: the cell death abnormality (CED)-1, 6, 7 and CED-2, 5, 12 pathways (19–21). The CED-1, 6, 7 and CED-2, 5, 12 pathways act in parallel to promote the activation of CED-10, a Rac GTPase responsible for cytoskeletal rearrangements that allow for internalization of the cell corpse. In Drosophila, the roles of the Ced-1, 6, 7 and Ced-2, 5, 12 pathways appear to be conserved. The CED-1 ortholog, Draper, is a transmembrane protein that localizes to the surface of the engulfing cell and acts as a receptor to recognize dying cells. Draper was first shown to be required for engulfment of apoptotic neurons in the embryonic central nervous system with mutants displaying lingering cell corpses (22). Additionally, Draper has been shown to be important in several other contexts including the engulfment of severed axons, bacteria, imaginal disc cells, hemocytes, and apoptotic NCs in midoogenesis (23–27). In addition to Draper, other Drosophila engulfment receptors include Croquemort (28) and integrins (29–31). Croquemort is related to CD36, a scavenger receptor involved in engulfment in mammals (32), and integrins also act as engulfment receptors in C. elegans and mammals (33, 34). The upstream activators of the Ced-2, 5, 12 pathway are largely unknown, although integrins may activate the pathway (34). As in C. elegans, it appears that Ced-12 and draper function in separate pathways in Drosophila. Ced-12 and draper have been shown to function in distinct steps in axon clearance (35). In macrophages, Ced-12 has been shown to function in a separate pathway from simu, a bridging molecule that acts upstream of draper (36). A number of other engulfment genes have been identified in Drosophila, and their molecular interactions are under active investigation (36–39).
Given the minor role for apoptosis and autophagic cell death during developmental PCD in the Drosophila ovary, we investigated the possibility that the FCs non–cell-autonomously promote NC death. Previously we showed that FCs of the Drosophila ovary are capable of phagoptosis in midoogenesis when phagocytosis genes are overexpressed (27), and we questioned whether phagocytosis genes might normally function to control cell death in late oogenesis. Indeed, we found that the phagocytosis genes draper and Ced-12/ELMO are required in the FCs for NC removal in late oogenesis and that they function partly in parallel. We also show that the FCs non–cell-autonomously control events associated with the death of the NCs, including nuclear envelope permeabilization, acidification, and DNA fragmentation. Furthermore, the genetic ablation of stretch FCs disrupted all cellular changes associated with developmental PCD of the NCs. Therefore, PCD of the NCs is a unique model of a naturally occurring developmental cell death program that is nonapoptotic and non–cell-autonomously controlled.
Results
Stretch FCs Surround NCs During Late Oogenesis.
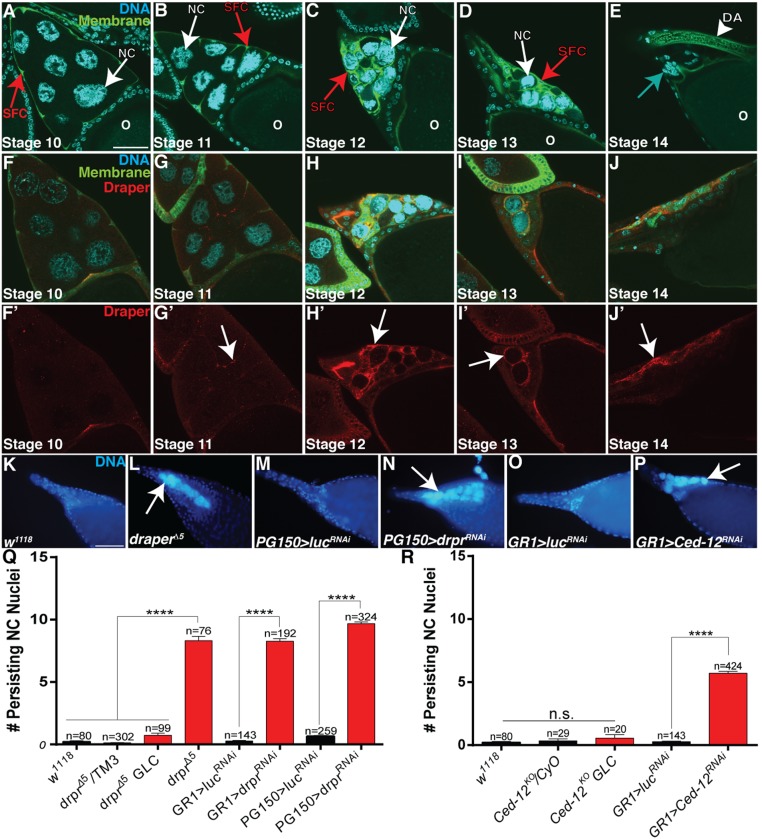
During late oogenesis, a population of ∼50 FCs known as the stretch FCs cover the NCs on the anterior of the egg chamber, and it has been proposed that these FCs phagocytose the NCs following their death (40–42). However, exactly how this is accomplished and whether stretch FCs play a role in the developmental PCD of the NCs remains unclear. To analyze the relationship between stretch FCs and NCs, we expressed a membrane-tethered GFP specifically in stretch FCs (PG150 > mCD8-GFP) (Fig. 1 A–E and Fig. S1A). The stretch FCs were visible in stage 10 (Fig. 1A) and began to project extensions around individual NCs in stage 11 (Fig. 1B). By stage 12, cytoplasmic dumping of the NCs was completed, and the NC nuclei appeared completely enveloped by the stretch FCs (Fig. 1C). The stretch FCs remained around the NC nuclei in stage 13, as they were eliminated (Fig. 1D). By stage 14, characterized by fully formed dorsal appendages (DAs), all NC nuclei were eliminated (Fig. 1E). Therefore, the stretch FCs were intimately associated with the NCs throughout the progression of developmental PCD, raising the intriguing possibility that the stretch FCs play an active role in the death of the NCs.
Fig. 1.
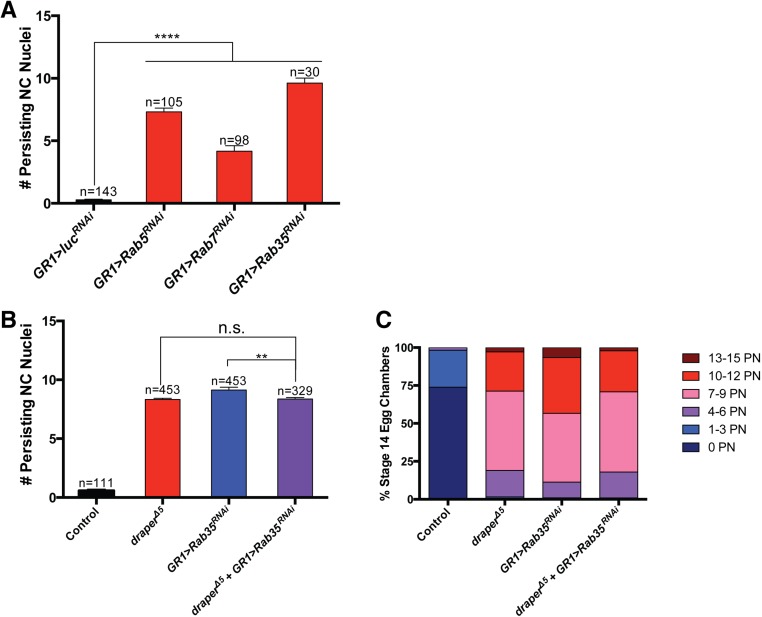
draper is required in the FCs for the removal of NCs during late oogenesis. (A–E) The stretch FCs surround the NCs during late oogenesis. Stage 10–14 egg chambers (PG150-GAL4/+; UAS-mCD8-GFP/+) express GFP specifically in the membranes of the stretch FCs (green, red arrows) and are stained with DAPI to label DNA (cyan). NC nuclei are indicated by white arrows. Oocyte is labeled with “O.” (A) Stretch FCs are apparent on the anterior of a stage 10 egg chamber. (B) Stretch FCs begin to extend around NCs in stage 11. (C) Stretch FCs surround NC nuclei in stage 12. (D) Stretch FCs continue to surround NC nuclei in stage 13. (E) A stage 14 egg chamber no longer contains NC nuclei and has fully formed DAs (arrowhead). Small nuclei are FC nuclei (blue arrow). (F–J’) Draper is enriched on the FC membranes (arrows) in late oogenesis. Stage 10–14 egg chambers (UAS-mCD8-GFP/+; GR1-GAL4/+) are stained with α-Draper antibody (red) and DAPI (cyan) and express GFP specifically in the FC membranes (green). (F and F’) Stage 10 egg chamber has nonspecific staining along the NC/oocyte interface. (G and G’) Draper enrichment becomes apparent in stage 11. (H and H’) Stage 12 egg chamber has enriched Draper on the FC membranes. (I and I’) Stage 13 egg chamber with Draper enrichment on the FC membranes. (J and J’) Stage 14 egg chamber has residual Draper staining. (Scale bar, 50 μm for images A–J’.) (K–P) Persisting NC nuclei (arrows) are present in stage 14 egg chambers with engulfment genes knocked down. Stage 14 egg chambers were stained with DAPI (blue). (Scale bar, 20 μm.) (K) Wild type (w1118). (L) Homozygous draper∆5. (M) Control (PG150-GAL4/+;UAS-luciferaseRNAi/+). (N) draperRNAi expressed specifically in the stretch FCs (PG150-GAL4/+; UAS-draperRNAi/+). (O) Control (GR1-GAL4/UAS-luciferaseRNAi). (P) Ced-12RNAi expressed specifically in the FCs (GR1-GAL4/UAS-Ced-12RNAi). (Q and R) Quantification of persisting NC nuclei in stage 14 egg chambers. GLC, germ-line clone. Data from two distinct RNAi lines were combined for Ced-12. Data presented are mean ± SEM. ****P ≤ 0.0001.
Fig. S1.
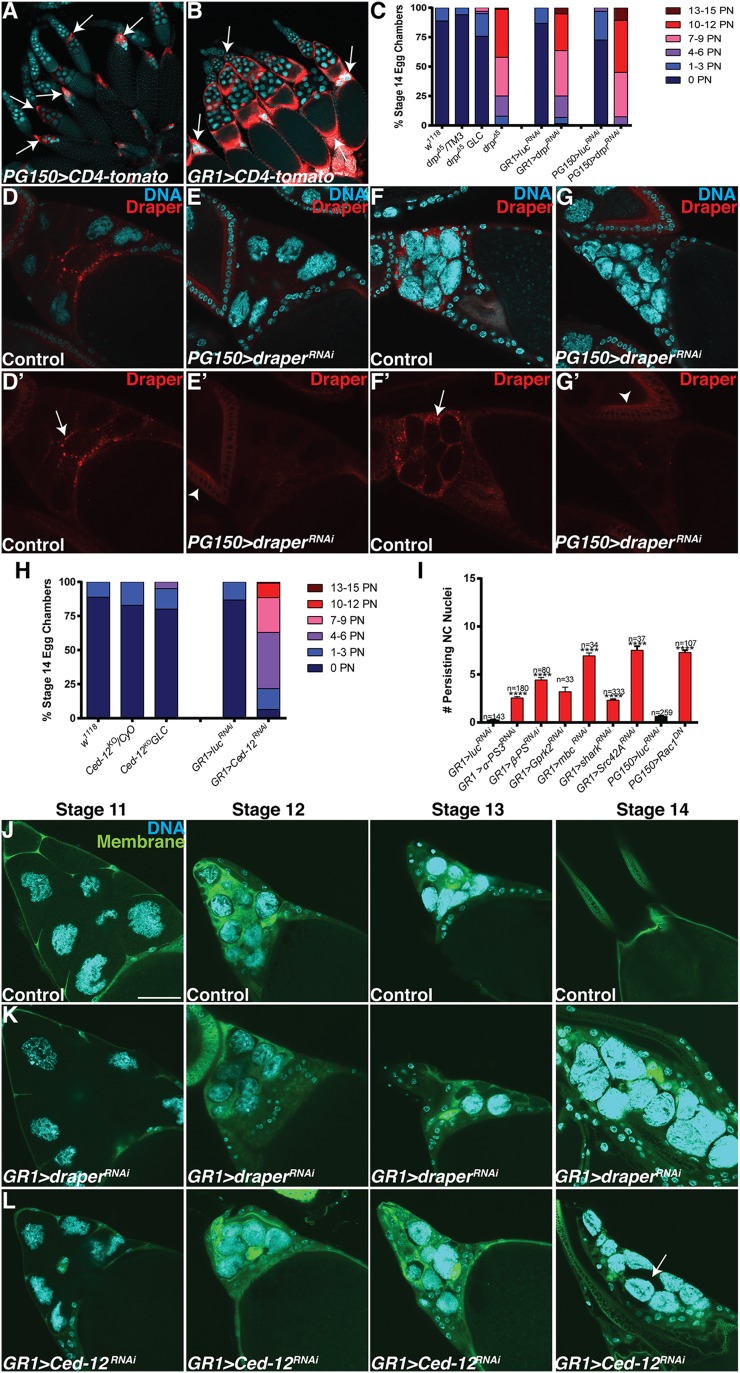
FC GAL4 drivers and analysis of draper, Ced-12, and other engulfment genes. (A and B) FC GAL4 driver expression patterns. Egg chambers express the membrane-tethered protein UAS-CD4-tomato (red) specifically in the FCs and are stained with DAPI (cyan) to label DNA. (A) PG150-GAL4 drives expression of UAS-CD4-tomato (PG150-GAL4/+;UAS-CD4-tomato/+) specifically in the stretch FCs beginning in stage 10 through stage 14 (arrows). (B) GR1-GAL4 drives expression of UAS-CD4-tomato (UAS-CD4-tomato/+; GR1-GAL4/+) in all FCs (arrows) beginning in stage 3. (C) Alternative quantification of data presented in Fig. 1Q. The number of persisting nuclei (PN) per stage 14 egg chamber was categorized into bins of 0 PN, 1–3 PN, 4–6 PN, 7–9 PN, 10–12 PN, and 13–15 PN, and the percentage of stage 14 egg chambers in each bin was calculated. (D–G’) The enrichment of Draper is specific to the FCs. Late stage egg chambers expressing mCD8-GFP (control) or draperRNAi specifically in the stretch FCs were stained together with α-Draper antibody (red, arrows) and DAPI (cyan). (D and D’) Control (PG150-GAL4/+; UAS-mCD8-GFP/+) stage 11 egg chamber shows enriched Draper along membranes that surround the NCs. (E and E’) draper is knocked down specifically in the stretch FCs of a stage 11 egg chamber (PG150-GAL4/+; UAS-draperRNAi/+), and the enrichment of Draper is absent. Basal levels of Draper staining are apparent in an earlier stage egg chamber (arrowhead). (F and F’) Control (PG150-GAL4/+; UAS-mCD8-GFP/+) stage 13 egg chamber shows enriched Draper along membranes that surround the NCs. (G and G’) draper is knocked down specifically in the stretch FCs of a stage 13 egg chamber (PG150-GAL4/+; UAS-draperRNAi/+), and the enrichment of Draper is absent. Basal levels of Draper staining are apparent in an earlier stage egg chamber (arrowhead). (H) Alternative quantification of data presented in Fig. 1R. The number of persisting nuclei (PN) per stage 14 egg chamber was categorized into bins of 0 PN, 1–3 PN, 4–6 PN, 7–9 PN, 10–12 PN, and 13–15 PN, and the percentage of stage 14 egg chambers in each bin was calculated. (I) Quantification of persisting NC nuclei in engulfment gene knockdowns analyzed in the candidate gene screen (Table S1). Data presented are mean ± SEM. ****P ≤ 0.0001. (J–L) Visualization of stretch follicle membranes with mDC8-GFP. (Scale bar, 50 μm.) (J) Stage 11–14 egg chambers from GR1-GAL4 UAS-mCD8-GFP/TM3 (control) flies express membrane–GFP in all FCs (green) and are stained with DAPI (cyan). The stretch FC membranes surround the NCs during late oogenesis. (K) Most FCs from UAS-draperRNAi/+; GR1-GAL4 UAS-mCD8-GFP/+ egg chambers in stages 10–14 of oogenesis appear to surround the NCs. (L) Most FCs from GR1-GAL4 UAS-mCD8-GFP/UAS-Ced-12RNAi egg chambers in stages 11–14 of oogenesis appear to surround the NCs. Occasionally some NCs do not appear to be surrounded by the FCs (arrow).
draper Is Required Specifically in the Stretch FCs for NC Removal.
Given that the stretch FCs completely surrounded the NCs throughout late oogenesis, we investigated whether the phagocytic machinery in the FCs contributed to NC removal. Egg chambers expressing a membrane-tethered GFP specifically in all FCs (GR1 > mCD8-GFP; Fig. S1B) were stained with an antibody against the engulfment receptor Draper (Fig. 1 F–J’). Draper was detected as the stretch FCs surrounded the NCs in stage 11 and appeared to define the path of stretch FC extension (Fig. 1 G and G’). The enrichment of Draper on the FC membranes was most intense in stages 12 and 13 (Fig. 1 H–I’), and some residual Draper staining was observed in stage 14 (Fig. 1 J and J’). The enrichment of Draper was specific to the stretch FCs, because egg chambers with draper knocked down specifically in stretch FCs (PG150 > draperRNAi) lacked Draper staining in late stage egg chambers (Fig. S1 D–G’).
To determine whether draper was required for the removal of NCs during late oogenesis, we analyzed egg chambers from draper∆5 (null) flies. Interestingly, we found a striking number of persisting NC nuclei in stage 14 egg chambers (Fig. 1L), compared with the control (w1118), where NC nuclei were removed normally (Fig. 1K). On average, there were ∼8 persisting NC nuclei in draper∆5 egg chambers compared with 0.23 in the w1118 control (Fig. 1Q). Moreover, 100% of draper∆5 stage 14 egg chambers contained at least one persisting NC nucleus, and >40% had more than 10 persisting nuclei (Fig. S1C). These data show that draper is required for the removal of the NCs.
draper has been shown in several contexts to be required in engulfing cells for corpse clearance (22–27). However, draper was also shown to be required cell-autonomously in the salivary gland for autophagic cell death (43). To determine which cell type required draper during developmental PCD of the NCs, we generated draper∆5 germ-line clones (GLCs) and found that the number of persisting NC nuclei was significantly reduced compared with draper∆5 homozygotes (Fig. 1Q and Fig. S1C), suggesting that draper is required nonautonomously for NC removal. Next, we expressed draperRNAi using FC-specific GAL4 drivers (Fig. S1 A and B). Compared with controls (Fig. 1 M and O), stage 14 egg chambers expressing draperRNAi in all FCs (GR1 > draperRNAi) had a strong persisting NC nuclei phenotype similar to draper∆5 (Fig. 1Q and Fig. S1C). Furthermore, we demonstrated that the requirement for draper was specifically in the stretch FCs (PG150 > draperRNAi) (Fig. 1 M, N, and Q), with ∼55% of egg chambers containing >10 persisting NC nuclei (Fig. S1C). Together, these findings indicate that draper is specifically required in the stretch FCs for NC removal in late oogenesis.
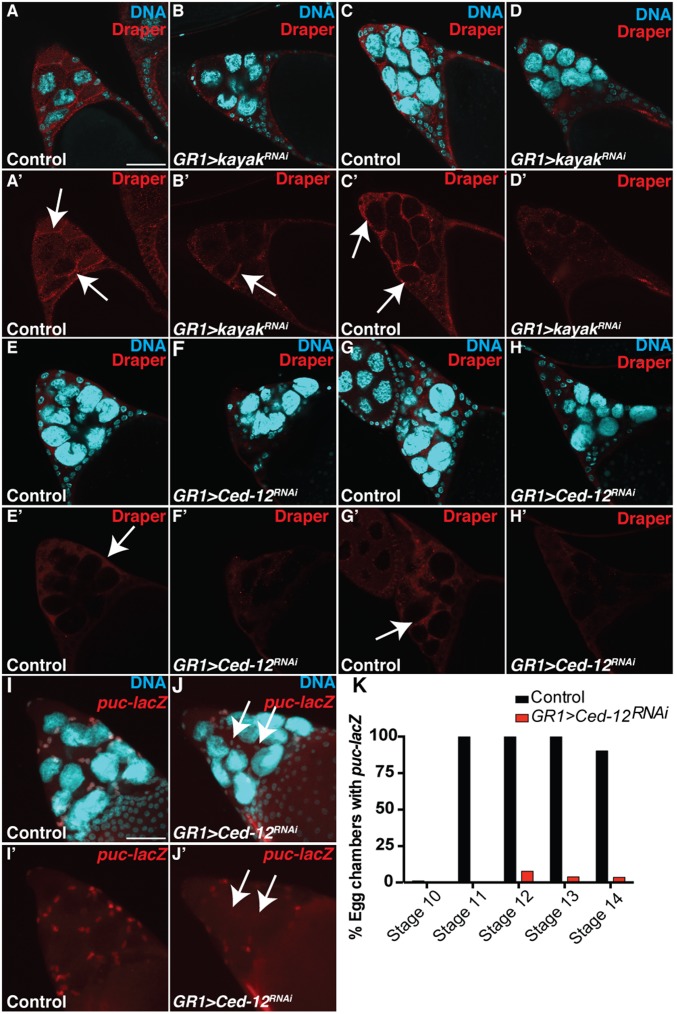
To identify other genes that are required in the FCs for NC removal, we knocked down several candidate genes specifically in the FCs via RNAi or dominant-negative constructs (Table S1). The candidate screen revealed that FC-specific knockdown of several known engulfment genes, including Ced-12, resulted in persisting NC nuclei (Fig. 1 O, P, and R and Fig. S1 H and I), further demonstrating that the engulfment machinery of the FCs is important for the removal of NCs during late oogenesis. To determine if Ced-12 was required in the germ line, we generated GLCs and found that the number of persisting NC nuclei was not significantly different from the w1118 control, demonstrating that Ced-12 is required nonautonomously in the FCs for NC removal (Fig. 1R and Fig. S1H). Although both draper and Ced-12 knockdowns demonstrated a severe disruption to the removal of the NCs, most of the FCs still appeared to surround the NCs, indicating that the FCs were morphologically normal (Fig. S1 J–L). Several other known engulfment genes, including integrins, Gprk2, mbc, shark, Src42A, and Rac1, were found to disrupt NC removal when knocked down in the FCs (Table S1 and Fig. S1I).
Table S1.
Fly strain information for genes required in the FCs for NC removal
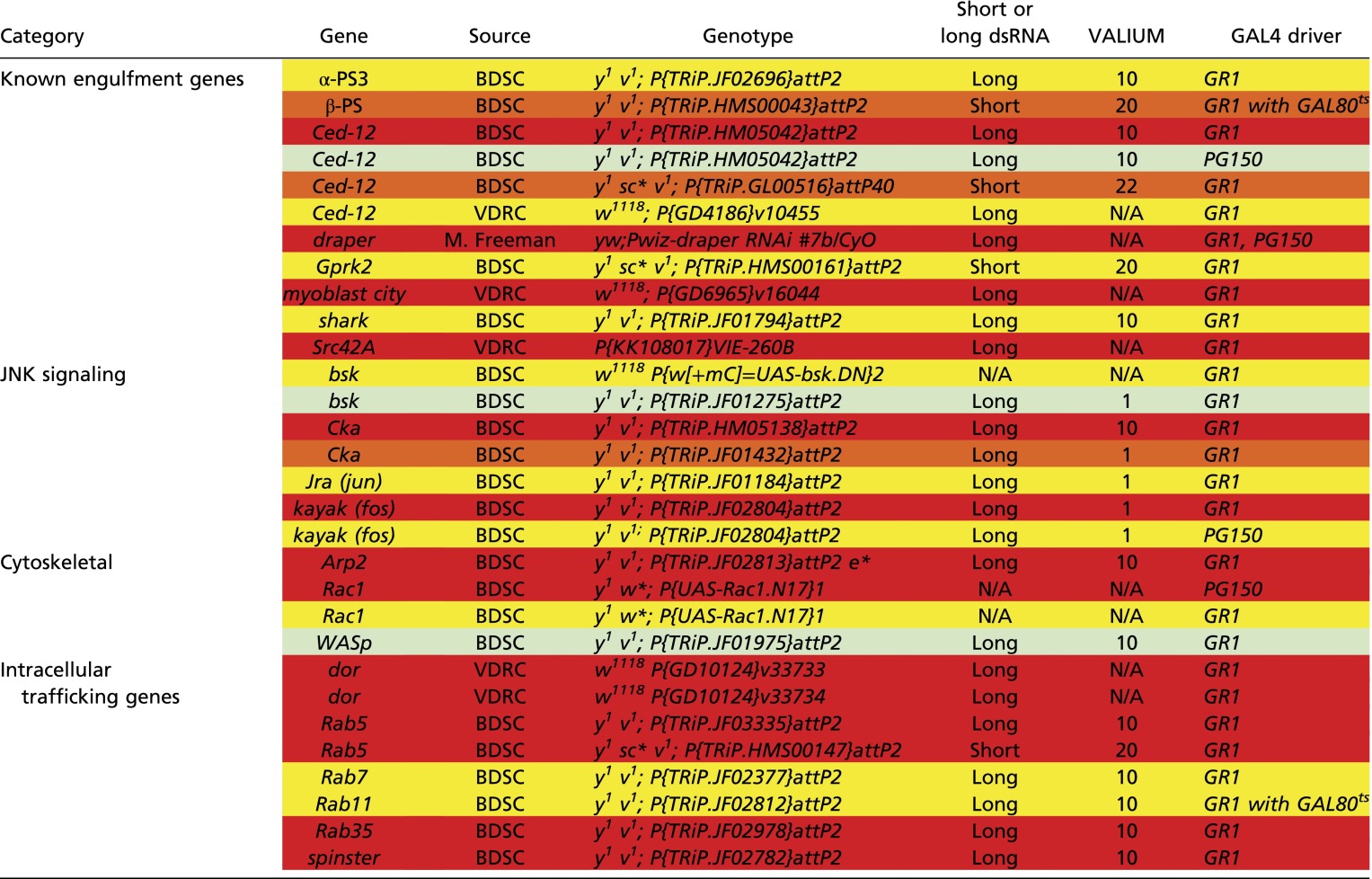 |
Red shading indicates very strong phenotype: 75–100% stage 14 egg chambers have >4 PN. Orange shading indicates strong phenotype: 50–75% stage 14 egg chambers have >4 PN. Yellow shading indicates moderate phenotype: 25–50% stage 14 egg chambers have >4 PN. Light green shading indicates weak phenotype: 10–25% stage 14 egg chambers have >4 PN.
draper and Ced-12 Act in Parallel to Promote Developmental PCD of the NCs.
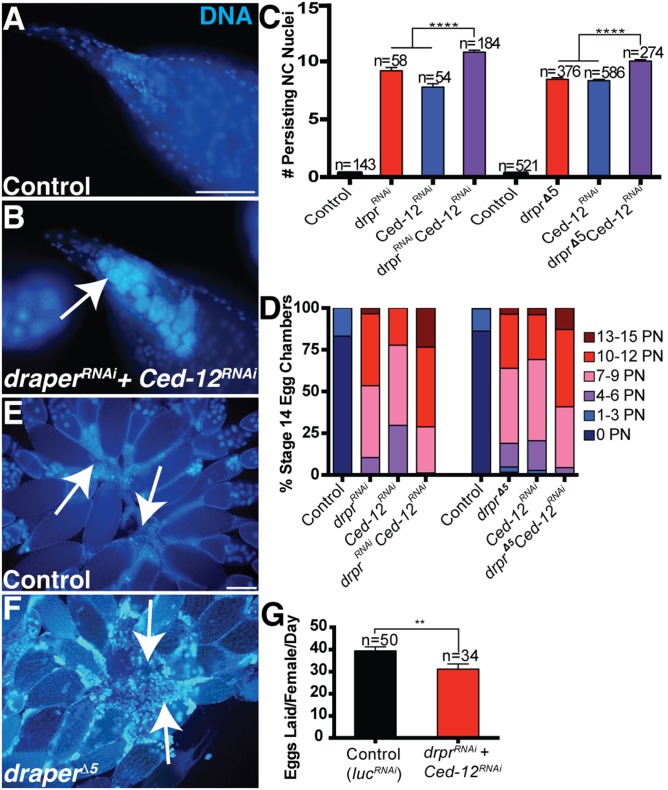
In C. elegans, it has been reported that Ced-12 acts in parallel to Ced-1, the ortholog of draper (20, 44), and evidence in Drosophila suggests that this may be conserved (35, 36). To determine whether draper and Ced-12 functioned in parallel in FCs, we performed double mutant analysis and found that draperRNAi Ced-12RNAi double knockdowns had a more severe persisting nuclei phenotype than either draper or Ced-12 alone (Fig. 2 A–D). Over 20% of stage 14 egg chambers had 13–15 persisting nuclei in draperRNAi Ced-12RNAi double knockdowns (Fig. 2D), the strongest phenotype we encountered. To determine if this interaction could be due to incomplete knockdowns with the RNAi, we repeated this experiment using the null allele, draper∆5, in combination with Ced-12RNAi and observed a similar enhanced phenotype (Fig. 2 C and D). Although the majority of NCs were affected, the knockdown of draper and Ced-12 did not cause a complete disruption to the removal of NCs, raising the possibility that a third pathway (autonomous or non–cell-autonomous) may also contribute. The incomplete disruption to NC removal could also be due to incomplete knockdown of Ced-12 via RNAi. Together, these data suggest that draper and Ced-12 act in parallel pathways to promote NC removal.
Fig. 2.
Ced-12 and draper act in parallel pathways, and mutants show reduced fecundity. (A and B) draperRNAi Ced 12RNAi double knockdowns have a severe persisting nuclei phenotype (arrow). Stage 14 egg chambers are stained with DAPI (blue). (Scale bar, 20 μm.) (A) Control (UAS-GAL4/Sco; MKRS/TM6B) stage 14 egg chamber does not have any persisting NC nuclei. (B) Egg chamber from draperRNAi Ced-12RNAi double knockdown (UAS-GAL4/UAS-draperRNAi; GR1-GAL4 G89/UAS-Ced-12RNAi) exhibits a complete failure in NC removal with 15 persisting NC nuclei. (C) Quantification of persisting NC nuclei in stage 14 egg chambers. draperRNAi Ced-12RNAi knockdown (purple, UAS-GAL4/UAS-draperRNAi; GR1-GAL4 G89/UAS-Ced-12RNAi) has a more severe persisting nuclei phenotype than the draperRNAi knockdown (red, UAS-GAL4/UAS-draperRNAi;GR1-GAL4 G89/MKRS) or Ced-12RNAi knockdown (blue, UAS-GAL4/CyO;GR1-GAL4 G89/UAS-Ced-12RNAi). draper∆5 Ced-12RNAi double knockdown (purple, draper∆5 UAS-Ced-12RNAi/draper∆5 GR1-GAL4) has a more severe persisting NC nuclei phenotype than draper∆5 (red, draper∆5 UAS-Ced-12RNAi/draper∆5) or Ced-12RNAi (blue, draper∆5 UAS-Ced-12RNAi/GR1-GAL4 UAS-mCD8-GFP) single knockdowns. Mixed sibling controls lack either the GAL4 driver or the RNAi constructs. Data presented are mean ± SEM. ****P ≤ 0.0001. (D) Alternative quantification of data presented in Fig. 2C (see Materials and Methods). The draper Ced-12 double knockdowns result in a stronger persisting nuclei phenotype than draper or Ced-12 single knockdowns. (E and F) Intact ovaries stained with DAPI (blue). (Scale bar, 100 μm.) (E) Control [FRT 2A/Df(3L)BSC181] ovary does not contain lingering NC nuclei. The center of the ovary contains FCs that have been shed from mature stage 14 egg chambers as they enter the oviduct (arrows). (F) NC nuclei accumulate in homozygous draper∆5 ovary (arrows). (G) draperRNAi Ced-12RNAi double knockdowns in stretch FCs (PG150-GAL4/+; UAS-GAL4/UAS-draperRNAi; +/UAS- Ced-12RNAi) have reduced egg laying compared with the control (PG150-GAL4/+; UAS-GAL4/+; UAS-luciferaseRNAi/+). Egg laying was quantified in females 21–25 d old. n, number of females. Data presented are average number of eggs laid per female per day ± SEM. **P ≤ 0.01.
The Accumulation of NC Nuclei Inhibits Egg Laying.
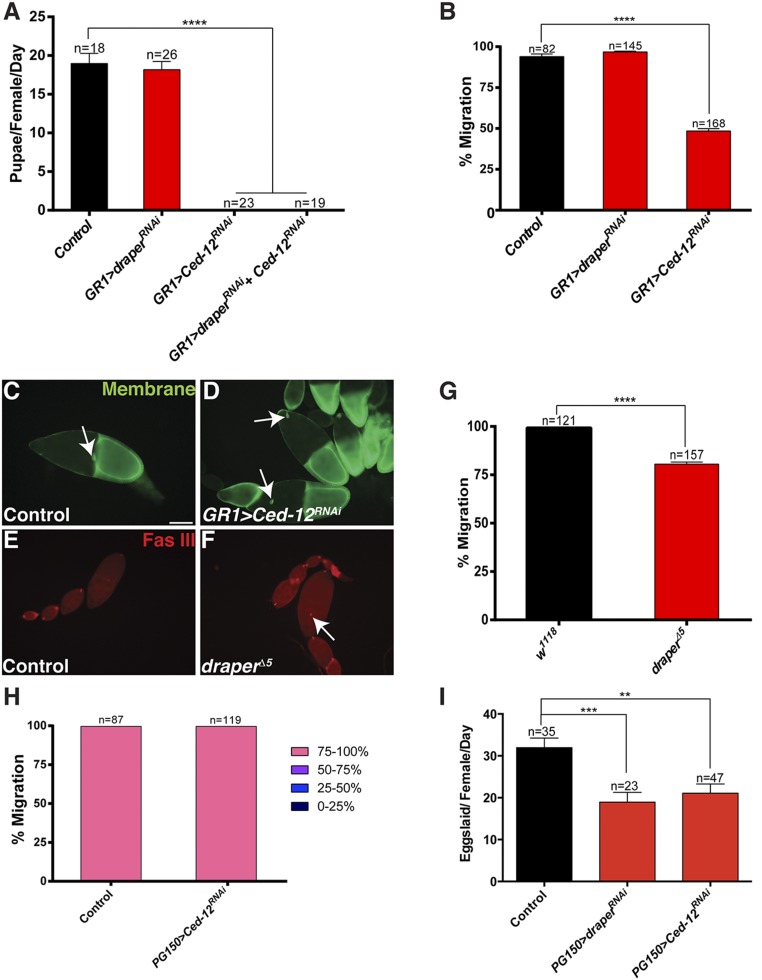
We observed a striking accumulation of NC nuclei in the ovaries (Fig. 2 E and F) of mutants with strong persisting NC nuclei phenotypes. We questioned whether the accumulation of NC nuclei in the ovary could affect the fecundity of the flies. Initially, we tested draper Ced-12 double knockdowns in all FCs because they had the strongest phenotype and found that both the draper Ced-12 double knockdown and the Ced-12 knockdown were completely sterile (Fig. S2A). However, this was likely due to a requirement for Ced-12 during border cell migration (Fig. S2 B–D) (45). Interestingly, we also noticed that draper∆5 mutants displayed defects in border cell migration (Fig. S2 E–G). To avoid the possibility that border cell migration could disrupt egg laying, we used PG150-GAL4 to knock down draper and Ced-12 only in the stretch FCs. We confirmed that Ced-12 did not cause a border cell migration defect when knocked down in the stretch FCs (Fig. S2H). When we measured egg laying in draperRNAi Ced-12RNAi double knockdowns, we found that females laid fewer eggs per day than the control (Fig. 2G). In an independent experiment, we observed defects in egg laying in draperRNAi and Ced-12RNAi single knockdowns (Fig. S2I). Therefore, when phagocytosis genes were inhibited in the FCs, NC nuclei accumulated in the ovary and there was an adverse effect on fecundity.
Fig. S2.
Ced-12 knockdowns and draper∆5 mutants have reduced fecundity and defects in border cell migration. (A) The knockdown of Ced-12 in all FCs results in sterility. Ced-12 single knockdown (UAS-GAL4/CyO; GR1-GAL4 G89/UAS-Ced-12RNAi) and draperRNAi Ced-12RNAi double knockdowns (UAS-GAL4/UAS-draperRNAi; GR1-GAL4 G89/UAS-Ced-12RNAi) do not produce any progeny. The control (UAS-GAL4/UAS-draperRNAi; TM6B/UAS-Ced-12RNAi) and draper single knockdown (UAS-GAL4/UAS-draperRNAi; GR1-GAL4 G89/MKRS) produce similar numbers of progeny. These data represent progeny produced over the entire lifetime of the females tested. n, number of females. Data presented are average pupae per female per day ± SEM. ****P < 0.0001. (B–D) Border cell migration is defective when Ced-12 is knocked down in all FCs. (B) Ced-12 knockdowns (GR1-GAL4 UAS-mCD8-GFP/UAS-Ced-12RNAi) have impaired border cell migration compared with control (GR1-GAL4 UAS-mCD8-GFP/TM3). Border cell migration in draper knockdown (UAS-draperRNAi/+; GR1-GAL4/+) is normal. Data presented are mean % migration ± SEM. ****P ≤ 0.0001. (C and D) Egg chambers express membrane GFP in the FCs (green), including the border cells (arrows). (Scale bar, 50 µm.) (C) Border cells in GR1-GAL4 UAS-mCD8-GFP/TM3 (control) stage 10 egg chambers successfully migrate to the NC/oocyte interface. (D) GR1-GAL4 UAS-mCD8-GFP/UAS-Ced-12RNAi egg chambers have impaired border cell migration. (E–G) Border cell migration is impaired in draper∆5 mutants. (E and F) Egg chambers stained with anti-FasIII (red) to mark polar cells within the border cell cluster (arrows). (E) w1118 control stage 10 egg chamber has normal border cell migration. (F) Border cell migration is delayed in draper∆5 mutant. (G) Quantification of border cell migration. Data presented are mean % migration ± SEM. ****P ≤ 0.0001. (H) Border cell migration is not delayed when Ced-12 is knocked down only in the stretch FCs. (I) draperRNAi (PG150-GAL4/+; UAS-draperRNAi/+) and Ced-12RNAi (PG150-GAL4/+; UAS-Ced-12RNAi/+) single knockdowns have reduced egg laying compared with the control (PG150-GAL4/+; UAS-luciferaseRNAi/+). Egg laying was quantified in females 21–25 d old. n, number of females. Data presented are average number of eggs laid per female per day ± SEM. ***P ≤ 0.001 or **P ≤ 0.01.
Lysosomal and Intracellular Trafficking Genes Are Required in the FCs for NC Removal.
Previous work has demonstrated a role for lysosomal trafficking genes such as deep orange (dor) in developmental NC death (46). dor is a member of the HOPS complex that localizes to endosomes and is important for delivery of material to lysosomes (47). As we previously reported (46), dor hypomorphs have defects in NC removal (Fig. 3 A, B, and E). Interestingly, we found that the requirement for dor is in the FCs. dor GLCs largely showed normal NC removal (Fig. 3 C and E), and the expression of dorRNAi specifically in the FCs (GR1 > dorRNAi) resulted in persisting NC nuclei (Fig. 3 D and E). Intracellular trafficking genes—Rab5, Rab7, and Rab35—were also found to be required in the FCs for the elimination of the NCs (Fig. S3A and Table S1). To determine whether draper acts in the same pathway or in a parallel pathway to intracellular trafficking genes, we analyzed draper∆5 Rab35RNAi double mutants. We found that the double mutants were not significantly different from draper∆5 (Fig. S3 B and C). This suggests that draper acts in the same pathway as intracellular trafficking proteins, like its ortholog ced-1 (48).
Fig. 3.
The FC lysosomes play a role in NC removal. (A–D) dor is required in the FCs for NC removal. Stage 14 egg chambers were stained with DAPI (blue). (A) Control (w1118) stage 14 egg chamber does not have persisting NC nuclei. (B) dor hypomorph (dor4/dor4) stage 14 egg chamber has persisting NC nuclei (arrow). (C) dor4 GLC stage 14 egg chamber is normal. (D) Knockdown of dor specifically in the FCs (GR1-GAL4/UAS-dorRNAi) causes persisting nuclei (arrow). (Scale bar, 20 μm.) (E) Quantification of persisting nuclei. dorRNAi quantification includes two distinct RNAi lines. Data presented are mean ± SEM. ****P < 0.0001. (F and F’) LysoTracker staining (red) on egg chambers that express membrane-GFP (green) specifically in stretch FCs (PG150-GAL4/+; UAS-mCD8-GFP/+). (Scale bar, 50 μm.) (F) Two NC nuclei are acidified in a stage 12 egg chamber (arrows), and LysoTracker puncta are present within the stretch FC membranes (arrowhead). (F’) Zoom of image pictured in F shows LysoTracker puncta in the stretch FC membranes. (G–H’) Stage 13 egg chambers stained with DAPI (cyan) and LysoTracker (red). (G and G’) Control (UAS-GAL4/Sco; MKRS/TM6B) stage 13 egg chamber has acidified NC nuclei (arrow) and LysoTracker puncta (arrowhead). (H and H’) draper Ced-12 double knockdown (UAS-GAL4/UAS-draperRNAi; GR1-GAL4 G89/UAS-Ced-12RNAi) stage 13 egg chamber does not contain acidified NC nuclei or LysoTracker puncta. (I) The percentage of late stage egg chambers (stages 11–13) with punctate or nuclear LysoTracker staining was quantified. Control is mixed siblings lacking either the GAL4 driver or the RNAi construct. Other genotypes are draper∆5, draperRNAi (UAS-GAL4/UAS-draperRNAi; GR1-GAL4 G89/MKRS), Ced-12RNAi (UAS-GAL4/Sco; GR1-GAL4 UAS-Ced-12RNAi/GR1-GAL4 G89 or TM6B), and draper Ced-12 double knockdown (UAS-GAL4/UAS-draperRNAi; GR1-GAL4 UAS-Ced-12RNAi/GR1-GAL4 G89 or TM6B).
Fig. S3.
Intracellular trafficking genes are required in the FCs for NC removal and act in the same pathway as draper. (A) Quantification of persisting nuclei in Rab5, Rab7, and Rab35 FC knockdowns identified in the candidate screen. The quantification of GR1-GAL4/UAS-luciferaseRNAi was used as the control for multiple experiments and was first presented in Fig. 1Q. Data presented are mean ± SEM. ****P ≤ 0.0001. (B and C) draper and Rab35 act in the same pathway to promote NC removal. (B) Quantification of persisting NC nuclei in draper∆5 Rab35RNAi double knockdowns. draper∆5 Rab35RNAi double knockdown (purple, draper∆5 UAS-Rab35RNAi/draper∆5 GR1-GAL4) has a similar persisting NC nuclei phenotype to draper∆5 (red, draper∆5 UAS-Rab35RNAi/draper∆5) but is significantly weaker than Rab35RNAi (blue, draper∆5 UAS-Rab35RNAi/GR1-GAL4 UAS-mCD8-GFP). Mixed sibling controls lack either the GAL4 driver or the RNAi constructs. Data presented are mean ± SEM. ****P ≤ 0.0001. (C) Alternative quantification of data presented in Fig. S3B. The number of persisting nuclei (PN) per stage 14 egg chamber was categorized into bins of 0 PN, 1–3 PN, 4–6 PN, 7–9 PN, 10–12 PN, and 13–15 PN, and the percentage of stage 14 egg chambers in each bin was calculated.
draper and Ced-12 Are Required in the FCs for the Acidification of the NCs.
We previously showed that punctate LysoTracker staining, likely labeling lysosomes, surrounds NC nuclei in late oogenesis and progresses to complete acidification of NC nuclei (46, 49). Using double labeling, we found that most LysoTracker puncta overlapped with the membranes of stretch FCs (Fig. 3 F and F’), raising the possibility that lysosomes from the stretch FCs play an active role in NC death. Electron microscopy has also demonstrated that lysosomes are present in the FCs that border the NCs, but not in FCs surrounding the oocyte, indicating that they may be important for NC elimination (40). By stages 12 and 13, the majority of wild-type egg chambers contained LysoTracker-positive nuclei and puncta (Fig. 3 G, G’, and I). However, LysoTracker labeling in draperRNAi and Ced-12RNAi single knockdowns was severely reduced in stage 12–13 egg chambers compared with the control (Fig. 3I). Strikingly, draperRNAi Ced-12RNAi double knockdowns showed a complete disruption to NC acidification (Fig. 3 H and I). Therefore, the engulfment machinery in the FCs is required for acidification of NCs during developmental PCD. However, whether the FC lysosomes actively contribute to the death process or are involved in the phagocytic processing of the NCs remains unknown.
draper Is Required in the Stretch FCs for Fragmentation of the NC Nuclei.
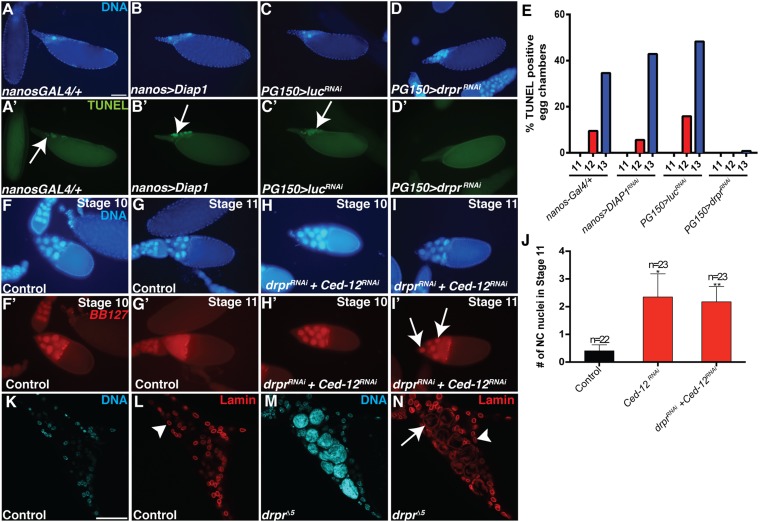
DNA fragmentation is an important step in the destruction of a cell and is considered to be a hallmark of apoptotic cell death (50). Typically, DNA fragmentation is executed by the autonomous activation of endonucleases, such as caspase-activated DNase (CAD), and is considered to be the “point of no return” in the death process (46, 51). TUNEL (terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling) staining is used to label 3′-OH ends of fragmented DNA in apoptotic cells (52, 53). During developmental PCD of NCs in late oogenesis, NC nuclei become TUNEL-positive, indicating that DNA fragmentation occurs (41, 53–56). Consistent with previous findings, we observed that NC nuclei became TUNEL-positive, especially in stage 13 (Fig. 4 A, A’, C, C’, and E). Interestingly, TUNEL-labeled NC nuclei were often not detectable by DAPI. Quantification revealed that 35–50% of control stage 13 egg chambers (nanos-GAL4/+ and PG150 > luciferaseRNAi) contained TUNEL-positive NC nuclei (Fig. 4E). Consistent with findings that developmental PCD of the NCs is largely caspase-independent (7–10), we found that overexpression of the caspase inhibitor Diap1 in the NCs (nanos > UASp-Diap1) did not disrupt DNA fragmentation in late oogenesis (Fig. 4 B, B’, and E). In contrast, we found that TUNEL staining was completely absent in stages 12 and 13 when draper was knocked down in the stretch FCs (Fig. 4 D and E). These data demonstrate that DNA fragmentation is inhibited when draper is knocked down in the FCs, implicating a role for the FCs in the death of the NCs.
Fig. 4.
The FCs nonautonomously contribute to NC death events. (A–D’) Stage 13 egg chambers stained with DAPI (blue) and TUNEL (green) as a marker for DNA fragmentation. (Scale bar, 50 μm.) (A and A’) Control (nanos-GAL4/+) egg chamber contains several TUNEL-positive NC nuclei (arrow), although only one nucleus is visible by DAPI staining. (B and B’) Overexpression of Diap1 (nanos-GAL4/UAS-Diap1) shows several TUNEL-positive nuclei (arrow). (C and C’) Control (PG150-GAL4/+; UAS-luciferaseRNAi/+) egg chamber has many TUNEL-positive NC nuclei (arrow). (D and D’) The knockdown of draper specifically in the stretch FCs (PG150-GAL4/+; UAS-draperRNAi/+) shows no TUNEL staining. (E) The percentage of egg chambers containing TUNEL-positive nuclei in the late stages of oogenesis (stages 11–13) was quantified for each genotype. (F–I’) Egg chambers expressing the BB127 lacZ enhancer trap are stained with DAPI (blue) and α–β-Gal antibody (red) to detect nuclear leakage. BB127 labels both NCs (large nuclei) and centripetal FCs (small nuclei). (F–G’) Control egg chambers (mixed siblings, BB127/+; Sco or UAS-draperRNAi/CyO or UAS-GAL4; +/MKRS) have NC nuclear β-Gal in stage 10B (F and F’) and cytoplasmic β-gal in stage 11 (G and G’). (H–I’) draper Ced-12 double knockdowns (BB127/+; UAS-draperRNAi/CyO or UAS-GAL4; +/GR1-GAL4 UAS-Ced-12RNAi) have nuclear β-Gal in stage 10B (H and H’). In this stage 11 egg chamber, β-Gal staining is still largely nuclear (arrows, I and I’). (J) The average number of intact NC nuclei in stage 11 egg chambers was quantified. Data presented are mean ± SEM. *P < 0.05, **P ≤ 0.01. (K–N) Stage 14 egg chambers stained with DAPI (cyan) and Lamin Dm0 ADL67.10 antibody (red). (K and L) Control egg chambers are devoid of NC nuclei, and Lamin staining appears at the periphery of remaining FC nuclei (L, arrowhead). (M and N) draper∆5 mutant egg chambers have Lamin associated with persisting NC nuclei (N, arrow) in addition to labeling FC nuclei (N, arrowhead).
draper and Ced-12 Act in the FCs to Promote NC Nuclear Permeability During Developmental PCD.
To further investigate whether the FC engulfment machinery affected the death of the NCs, we examined permeability of the NC nuclear envelope, one of the earliest indications of developmental PCD (57, 58). To visualize NC nuclear permeability, we used the BB127 lacZ enhancer trap that specifically labels NC nuclei (and centripetal FCs). Consistent with previous findings (54, 57, 58), we observed that control NC nuclei became permeable between stages 10B and 11, with β-Gal transitioning from a nuclear (Fig. 4 F and F’) to cytoplasmic localization (Fig. 4 G and G’). However, Ced-12RNAi and draperRNAi Ced-12RNAi double knockdowns displayed some stage 11 egg chambers with intact NC nuclei, suggesting that nuclear envelope permeability was delayed (Fig. 4 H–J). Eventually, all NC nuclei became permeable, indicating that that the genes were incompletely knocked down or that another pathway can promote NC nuclear permeabilization. These data suggest that the engulfment machinery in the FCs is important for the permeabilization of the NC nuclear envelope, an initial event during PCD. We also found that the NC nuclear lamina remained largely intact, surrounding the persisting nuclei in draper mutants (Fig. 4 K–N).
The Stretch FCs Are Required for NC Death and Dumping.
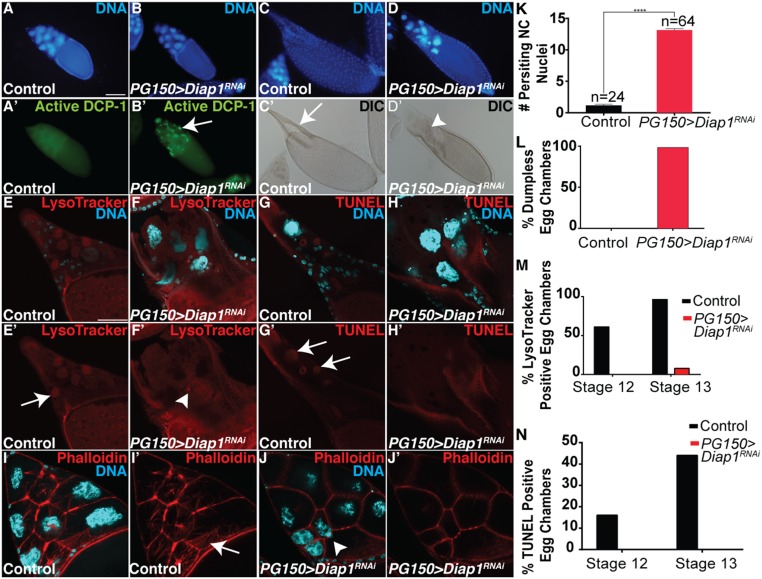
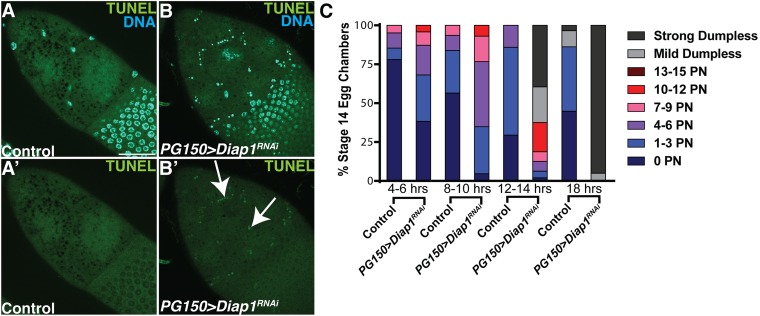
To directly address whether the stretch FCs were required for the developmental PCD of the NCs, we eliminated the stretch FCs via the expression of RNAi against the caspase inhibitor Diap1 (PG150 > Diap1RNAi). The presence of pyknotic FC nuclei, cleaved caspase Dcp-1 staining, and TUNEL confirmed that the stretch FCs were indeed dying (Fig. 5 A–B’ and Fig. S4 A–B’). The genetic ablation of the stretch FCs resulted in nonautonomous effects on the NCs: They failed to dump their cytoplasm into the oocyte or undergo developmental PCD (Fig. 5 C–D’). This disruption to NC dumping and PCD was nearly complete, with 98% of egg chambers exhibiting a failure in NC dumping and an average of 13 persisting nuclei per egg chamber (Fig. 5 K and L). Interestingly, we observed a correlation between increasing the time of Diap1RNAi expression and the severity of egg chamber defects. Egg chambers expressing Diap1RNAi for 4–10 h had increasing numbers of persisting NC nuclei, however egg chambers expressing Diap1RNAi for 12–14 h had a mild dumpless phenotype that shifted to strong dumpless at 18 h (Fig. S4C). To further investigate which NC death events required stretch FCs, we stained PG150 > Diap1RNAi egg chambers with LysoTracker and TUNEL. We observed that acidification (Fig. 5 E–F’ and M) and DNA fragmentation (Fig. 5 G, H, and N) of the NCs were strongly inhibited. A dramatic event that occurs in wild-type egg chambers just before dumping is the formation of actin bundles in the cytoplasm (57) (Fig. 5I). Phalloidin staining revealed that these actin bundle networks failed to form when Diap1RNAi was expressed in stretch FCs (Fig. 5J). These data demonstrate that the FCs nonautonomously control multiple events associated with the dumping and death of the NCs.
Fig. 5.
The stretch FCs are required for NC dumping and developmental PCD. (A–B’) Stage 10 egg chambers stained with DAPI (blue) and α-cleaved Dcp-1 (green). (A and A’) Control (FM7/+;tub-GAL80ts/+;UAS-Diap1RNAi/+) has no detectable Dcp-1. (Scale bar, 50 μm.) (B and B’) Egg chamber expressing Diap1RNAi in the stretch FCs (PG150-GAL4/+;GAL80ts/+; UAS-Diap1RNAi/+) exhibits α-cleaved Dcp-1 in stretch FCs (arrow). (C–D’) Stage 14 egg chambers stained with DAPI (blue). (C and C’) Control egg chamber has DAs and no persisting nuclei or NC cytoplasm (arrow). (D and D’) Stage 14 PG150 > Diap1RNAi egg chamber failed to undergo NC dumping (arrowhead). (E–F’) Stage 13 egg chambers stained with DAPI (cyan) and LysoTracker (red). (E and E’) Control egg chamber with nuclear LysoTracker staining (arrow). (Scale bar, 50 μm.) (F and F’) PG150 > Diap1RNAi egg chamber with LysoTracker staining is absent in NC nuclei, although it is present in some FCs (arrowhead). (G–H’) Stage 13 egg chambers stained with DAPI (cyan) and TUNEL (red). (G and G’) Control egg chamber with TUNEL-positive NC nuclei (arrow). (H and H’) PG150 > Diap1RNAi egg chamber with no TUNEL-positive NC nuclei. (I–J’) Stage 11 egg chambers stained with DAPI (cyan) and phalloidin (red). (I and I’) Phalloidin labels cytoplasmic actin network in control egg chamber (arrow). (J and J’) PG150 > Diap1RNAi egg chamber lacks cytoplasmic actin network, and an NC nucleus is stuck in a ring canal (arrowhead). (K) Quantification of persisting NC nuclei. Data presented are mean ± SEM. ****P ≤ 0.0001. (L) Quantification of cytoplasmic dumping. (M) Quantification of LysoTracker staining. (N) Quantification of TUNEL.
Fig. S4.
Characterization of egg chambers expressing Diap1RNAi in stretch FCs. (A–B’) Stage 10 egg chambers stained with DAPI (blue) and TUNEL (green). (A and A’) Control (FM7/+; tub-GAL80ts/+; UAS-Diap1RNAi/+) has no detectable TUNEL. (B and B’) Egg chamber that expresses Diap1RNAi specifically in the stretch FCs (PG150-GAL4/+; GAL80ts; UAS-Diap1RNAi/+) exhibits TUNEL localized to pyknotic stretch FC nuclei (arrows). (Scale bar, 50 μm.) (C) Time course of persisting NC nuclei and dumpless phenotypes. Flies were kept at 29 °C for increasing time points to degrade Gal80ts and induce Diap1RNAi expression. Phenotypes of control (FM7/+; tub-GAL80ts/+; UAS-Diap1RNAi/+) and PG150 > Diap1RNAi stage 14 egg chambers were scored.
The JNK Signaling Pathway Is Required Specifically in the FCs for NC Removal.
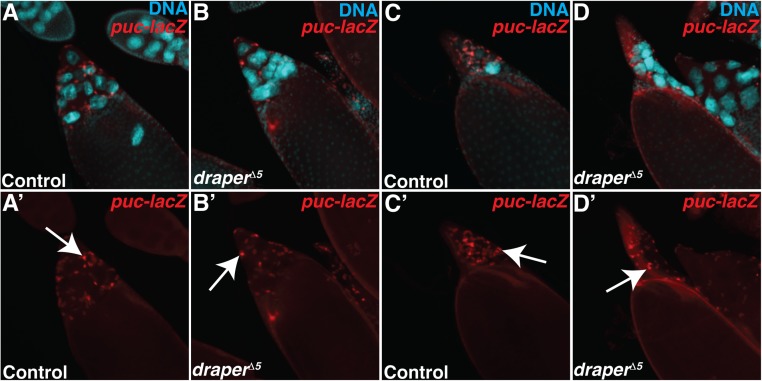
Previous work has demonstrated a role for the JNK pathway in the engulfment of apoptotic cells. For example, JNK is required in the FCs during engulfment of NCs in response to starvation during midoogenesis (27), in imaginal disc cells undergoing cell competition (59), and in glia during engulfment of degenerating axons (60). The JNK signaling pathway is activated in the anterior FCs during late oogenesis (61–63), but whether it plays a role in developmental PCD of the NCs was unknown. Therefore, we investigated whether the JNK pathway was an important component of signaling in the stretch FCs for developmental PCD of the NCs. Using the JNK reporter puc-lacZ, we confirmed that the JNK pathway was activated in the stretch FCs during late oogenesis (Fig. 6 A–D’). Up-regulation of the JNK pathway first became apparent in the FCs in stage 11 (Fig. 6 A and A’ and Fig. S5A) and was most intense in stages 12 and 13 (Fig. 6 B–C’), with FCs showing puc-lacZ directly surrounding individual NC nuclei (Fig. 6 C and C’). NC death is not synchronous, and it may be that several stretch FCs work together to eliminate NCs one at a time. The activation of the JNK pathway paralleled the timing of the enrichment of Draper on the FC membranes (Fig. 1 F–J’), suggesting that JNK may also play an important role in NC removal.
Fig. 6.
JNK signaling is activated and required in the FCs during developmental PCD of the NCs. (A–D’) puc-lacZ/TM3 was used as a reporter for JNK activity (arrows). Stage 11–14 egg chambers are stained with α–β-Gal (red) and DAPI (cyan). puc-lacZ is initially detected in FC nuclei during stage 11 (A and A’) and becomes most highly expressed in stages 12 and 13 (B and B’ and C and C’). puc-lacZ is also apparent in some FCs along the DA in stage 14 (D and D’). (Scale bar, 50 μm.) These data are quantified in Fig. S5A. (E–H) Persisting nuclei (arrows) are observed in stage 14 egg chambers of JNK pathway knockdowns stained with DAPI (blue). (E) GR1-GAL4/UAS-luciferaseRNAi control does not have persisting NC nuclei. (F–H) Persisting NC nuclei are found in (F) GR1-GAL4/UAS-kayakRNAi, (G) GR1-GAL4/UAS-jraRNAi, and (H) GR1-GAL4/UAS-CkaRNAi. (Scale bar, 20 μm.) (I) Quantification of persisting nuclei in JNK pathway knockdowns, including bskDN, kayakRNAi, jraRNAi, and CkaRNAi (data from two distinct Cka RNAi lines were combined), compared with the control GR1-GAL4/UAS-luciferaseRNAi. Data presented are mean ± SEM. ****P ≤ 0.0001.
Fig. S5.
JNK is activated and required in the FCs for NC removal. (A) Quantification of puc-lacZ in stage 10–14 control egg chambers. The number of egg chambers with activated puc-lacZ was taken as a percentage of the total number of egg chambers in each stage. Egg chambers were considered to have activated puc-lacZ when >10 FCs were stained with α–β-gal. (B) Alternative quantification of data presented in Fig. 6I. The number of persisting nuclei (PN) per stage 14 egg chamber was categorized into bins of 0 PN, 1–3 PN, 4–6 PN, 7–9 PN, 10–12 PN, and 13–15 PN, and the percentage of stage 14 egg chambers in each bin was calculated.
To test whether the JNK pathway was required in the FCs for elimination of the NCs, we inactivated several members of the JNK pathway. Expression of RNAi against kayak or jra, which encode components of the dimeric transcription factor AP-1, caused a strong persisting nuclei phenotype in stage 14 egg chambers (Fig. 6 E–G and I and Fig. S5B). Consistent with our findings, Dequier et al. previously reported that kayak hypomorphs contain persisting NC nuclei (63). Additionally, knockdown of the AP-1 scaffold Cka or overexpression of a dominant-negative form of Drosophila JNK (basket, bsk) led to persisting NC nuclei (Fig. 6 H and I). Together, these data suggest that the JNK signaling pathway is required in the FCs for NC removal.
Ced-12 Acts Upstream of JNK to Promote Draper Enrichment.
During the engulfment of dying NCs by the FCs in midoogenesis in response to starvation, JNK was found to act downstream of Draper in a feed-forward loop to maintain Draper enrichment on the FC membranes (27). Similarly, during phagocytosis of axonal debris by glia, JNK was required to increase Draper in glia to levels sufficient for clearance (60). Thus, we investigated whether the enrichment of Draper on FC membranes in late oogenesis required JNK activity. We found that kayak was not required for the initial localization of Draper on FC membranes in stage 11 (Fig. 7 A–B’), but it was necessary for enrichment of Draper in stages 12 and 13 (Fig. 7 C–D’). Thus, JNK signaling may act to maintain Draper on the FC membranes. These data are similar to previous findings showing that JNK is required for sustaining Draper levels in engulfing cells (27, 60). The inverse experiment examining JNK activity in draper mutants revealed that draper was not required for activation of JNK (Fig. S6 A–D’), unlike engulfment in midoogenesis (27). Interestingly, we found that Ced-12 was required for the enrichment of Draper on FC membranes throughout late oogenesis (Fig. 7 E–H’). FC membranes in Ced-12 knockdowns surrounded NCs normally (Fig. S1L); thus, defects in Draper enrichment were not due to morphological abnormalities in FCs.
Fig. 7.
Ced-12 acts upstream of JNK in late oogenesis. (A–D’) Control (PG150-GAL4/+; UAS-mCD8-GFP/+) and kayak knockdown (GR1-GAL4/UAS-kayakRNAi) egg chambers were stained with α-Draper antibody (red) and DAPI (cyan). (Scale bar, 50 μm.) (A–B’) The initial expression of Draper during stage 11 is unaffected. (C–D’) The enrichment of Draper on the FC membranes in kayak knockdown in stage 12 (D and D’) is reduced compared with the control (C and C’). (E–H’) Control (GR1-GAL4/UAS-mCD8-GFP) and Ced-12 knockdown (GR1-GAL4/UAS-Ced-12RNAi) egg chambers were stained with α-Draper antibody (red) and DAPI (cyan). (E–F’) The initial enrichment of Draper on the FC membranes during stage 11 is not apparent in the Ced-12 knockdown. (G–H’) The enrichment of Draper on the FC membranes in the Ced-12 knockdown in stage 12 (H and H’) is absent compared with the control (G and G’). (I–J’) Projection images (34 slices) of stage 11 egg chambers stained with DAPI (cyan) and α–β-Gal (red) as a readout of JNK activity. (I and I’) Most stretch FCs in control puc-lacZ/TM3 egg chambers express β-Gal. (J and J’) puc-lacZ/GR1-GAL4 UAS-Ced-12RNAi egg chamber FCs have reduced β-Gal expression (arrows). (K) Quantification of puc-lacZ in Ced-12 knockdowns (GR1-GAL4 Ced12RNAi/puc-lacZ) compared with the control (puc-lacZ/TM3). Egg chambers were considered to have activated puc-lacZ when >10 FCs were stained with α–β-gal.
Fig. S6.
Draper does not regulate JNK signaling in late oogenesis. (A–D’) Projection images of control (puc-lacZ/TM3) and draper∆5 (+/CyO; puc-lacZ draper∆5/draper∆5) egg chambers stained with DAPI (cyan) and anti–β-Gal (red) to detect JNK activity (arrows). puc-lacZ is detected in the FCs of both control and draper∆5 egg chambers. (A and A’) Control stage 11. (B and B’) draper∆5 stage 11. (C and C’) Control stage 13. (D and D’) draper∆5 stage 13.
Our data indicate that unlike midoogenesis (27), JNK is activated by a pathway distinct from Draper during late oogenesis. Indeed, we found that Ced-12 is required to activate JNK: Egg chambers with Ced-12 knocked down in the FCs showed reduced expression of puc-lacZ (Fig. 7 I–K). Taken together, we propose that Ced-12 acts upstream of JNK, which leads to increased Draper in FCs to promote the removal of NCs.
Discussion
The nonautonomous control of PCD has wide-ranging implications. Before this study, the role of phagocytic machinery in promoting cell death has been documented in systems where phagocytosis is artificially activated (15, 27, 64) or acts in cooperation with apoptosis (17, 18, 65). In this work, we have demonstrated that a naturally occurring example of nonapoptotic PCD fails to occur properly when the phagocytic machinery is disrupted in surrounding cells.
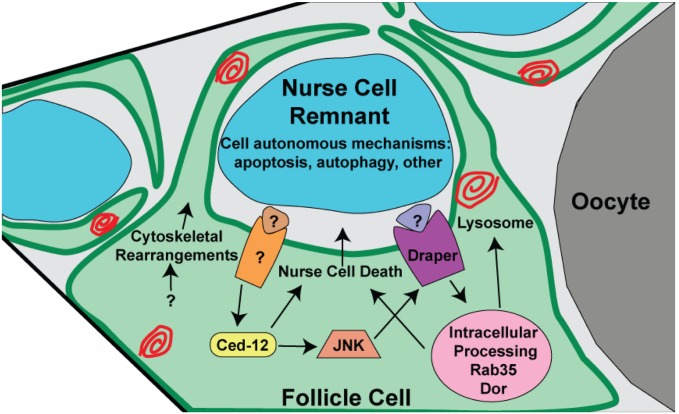
In the adult female fly, hundreds of NCs die every day via developmental PCD, but previous work ruled out major roles for autonomous apoptosis and autophagic cell death mechanisms during late oogenesis (7, 9, 10, 55). We and others have previously shown that FCs can be genetically induced to perform phagoptosis during midoogenesis (27, 64). Therefore, we investigated whether the phagocytic machinery of the FCs might promote PCD and removal of NCs that occurs naturally during late oogenesis. Indeed, we found that there is a failure in NC removal when phagocytosis is disrupted in the somatic FCs that directly surround the NCs. We showed that the phagocytic genes draper and Ced-12 act in at least two pathways to complete the process of NC removal in late oogenesis (Fig. S7), and we identified several other genes that are important in the FCs for NC removal. We demonstrated that the events associated with the death of the NCs, including permeabilization of the nuclear envelope, acidification, lamin degradation, and DNA fragmentation, are impaired when phagocytosis genes are inhibited in the FCs. Furthermore, genetic ablation of the stretch FCs caused a near complete failure in NC death, emphasizing their central role in NC death. This work suggests that the FCs nonautonomously promote the death and removal of the NCs, likely via phagoptosis.
Fig. S7.
Model for NC death in late oogenesis. Stretch FCs surround NC remnants during late oogenesis and nonautonomously promote their death. Draper becomes activated by an unidentified signal to promote intracellular trafficking and NC death. Ced-12 is activated by an unknown engulfment receptor and activates JNK, which promotes enrichment of Draper, causing NC death. Ced-12 additionally has a function independent of Draper. Nonautonomous mechanisms play a major role in promoting NC death, but NCs also use autonomous cell death mechanisms.
Although disruption of the phagocytic machinery largely prevented removal of the NCs, analysis of cellular events was necessary to determine whether the FCs affected the death of NCs. One indication that death of the NCs is nonautonomously controlled is that acidification of the NC nuclei is completely disrupted when draper and Ced-12 were knocked down in the FCs. These data need to be interpreted carefully, as acidification is indicative not necessarily of cell death but perhaps degradation of a cell corpse. The presence of lysosomes within the stretch FCs and the block to acidification of NCs when phagocytosis is impaired in the FCs raises many questions about the role of the lysosomes in NC death. Lysosomes may be exocytosed from the FCs, releasing their contents to promote NC death. For example, T cells have been shown to exocytose lysosomes to destroy pathogens (66, 67). Alternatively, lysosomes could be delivered to the plasma membrane to allow for rapid membrane growth as the FCs stretch around the NCs (68). An open question is how FCs are capable of destroying the much larger NCs; perhaps stretch FCs fuse together to surround individual NCs. Our findings indicate that several stretch FCs surround the NCs and may cooperate to remove individual NCs in a stepwise manner. Previously, we found a requirement for spinster in the germ line during dumping and NC death, indicating that lysosomes may have both autonomous and nonautonomous roles (46). Further investigation is needed to clarify the role(s) of the lysosomes during NC death.
Further support for our conclusions that death of the NCs is nonautonomously controlled is that the NCs fail to become TUNEL-positive when the phagocytic machinery is disrupted in the FCs. In general, the initial steps of DNA fragmentation during PCD are thought to occur cell-autonomously; however, previous studies have indicated that DNA fragmentation can be carried out by engulfing cells. In mammals, apoptotic thymocytes from mice expressing caspase-resistant ICAD (inhibitor of CAD) were resistant to DNA fragmentation but became TUNEL-positive once engulfed by macrophages (69). One explanation is that lysosomal enzymes in engulfing cells may contribute to DNA fragmentation. Interestingly, it was demonstrated in C. elegans that ced-1 engulfment mutants had a reduced number of corpses with TUNEL-positive nuclei (70). In the ovary, we found that DNA fragmentation was strongly disrupted in wild-type NCs when phagocytosis genes were inhibited in the FCs. The Nomenclature Committee on Cell Death has suggested that cells should only be considered dead when they “either exhibit irreversible plasma membrane permeabilization or have undergone complete fragmentation” (1). We found that many NC nuclei remained intact in engulfment mutants, retaining their nuclear lamina and failing to undergo DNA fragmentation, indicating that death was disrupted.
The first indication that the NCs are dying occurs between stages 10 and 11 when the NC nuclei become permeable (57). This event is clearly separable from engulfment, as it occurs before the onset of dumping and envelopment by FCs. We found that permeability of the NC nuclear envelope was delayed when draper and Ced-12 were knocked down specifically in the FCs. Ultimately, the NC nuclei did become permeable, suggesting that other pathways can also promote NC nuclear permeabilization. These other pathways are likely to initiate from the stretch FCs, as ablation of the stretch FCs blocked all NC death.
Ablation of the stretch FCs caused a failure in NC dumping, which occurs concurrently with NC death. Many genes have been identified as important for NC dumping, particularly genes affecting the ring canals and actin cytoskeleton (71). For example, spaghetti squash encodes the regulatory light chain of myosin II and must be phosphorylated to promote NC dumping. It has been proposed that the FCs release a signal that activates the kinase that phosphorylates regulatory light chain of myosin II (72). Although both NC death and dumping were disrupted when the FCs were ablated, it is unlikely that dumping is the signal for death because NC nuclear permeability, a cell death event, is apparent before the onset of dumping. Furthermore, NCs in other dumpless mutants (e.g., chickadee) have TUNEL-positive NC nuclei (55, 56). It remains to be determined how the stretch FCs promote the precise developmental timing of NC death. Moreover, how the FCs become activated to promote dumping and remove NCs is not clear; an upstream signal could originate from the oocyte, in the NCs, or from a source extrinsic to the ovary.
Although our experiments aimed at disrupting the phagocytic machinery in the FCs caused a severe block to NC death and removal, it did not cause a total block. When draper and Ced-12 were knocked down in the FCs, there was an average of ∼11 persisting NC nuclei, meaning that on average four NCs per egg chamber were successfully eliminated. Knockdown of integrins specifically in the stretch FCs also led to persisting nuclei, raising the possibility that integrin signaling and/or other pathways act in parallel to draper and Ced-12. Alternatively, some cells that fail to be removed by the normal phagocytic mechanism could die by an another pathway such as necrosis. Although previous work has ruled out some autonomous mechanisms including apoptosis and autophagic cell death as major contributors to NC death, mutants do show weak phenotypes, suggesting that these processes play a minor role. Furthermore, there are likely unknown cell-autonomous effectors of developmental NC death.
Previously we determined that Draper acted upstream of JNK in FCs for the phagocytosis of NCs that die in midoogenesis in response to starvation (27). Our findings here differ in several ways. First, we have found that phagocytosis genes are required for NC death in addition to clearance in late oogenesis, whereas the role is limited to clearance in midoogenesis. Second, we have found that Draper does not regulate JNK activity in late oogenesis, in contrast to midoogenesis. Moreover, we have found that Ced-12 acts upstream of JNK, leading to increased Draper protein, in late oogenesis (Fig. S7). However, our double mutant analysis (Fig. 2) indicates that Ced-12 also has a role in NC removal that is independent of Draper. These findings suggest that the roles of JNK, Ced-12, and Draper may differ depending on whether the cell is promoting phagoptosis (cell death) or phagocytosis (clearance).
Germ-line cysts are found in the ovary and testis of many organisms. Interestingly, non–cell-autonomous control over cell death of the germ line has been suggested to occur in several species. For example, the somatic sheath cells of the C. elegans gonad promote apoptotic death of the germ cells in conjunction with autonomous cell death mechanisms (65). In the developing mammalian ovary, germ-cell cysts break apart and oocytes become surrounded by somatic cells. At the same time, ∼2/3 of the oocytes undergo cell death. Deletion of Notch2 in the somatic cells causes reduced oocyte death and defects in the breakdown of germ-cell cysts, resulting in reduced fertility (73). Similarly, we found that failure of the NCs to appropriately undergo PCD reduced egg laying of Drosophila females. The exact mechanism of how a failure in developmental PCD of the NCs leads to reduced egg laying is unknown. It is possible that accumulation of NC nuclei blocks the entrance to the oviducts, or there could be feedback signaling to the stem cells that affects the rate of egg production. Therefore, non–cell-autonomous PCD in the germ line may be evolutionarily conserved and play a critical role in the reproductive success of organisms. Overall, we demonstrated that the phagocytosis machinery in FCs promotes the death and removal of NCs during the late stages of Drosophila oogenesis. To our knowledge, this is the first example of developmental PCD that is both nonapoptotic and non–cell-autonomously controlled.
Materials and Methods
Fly Strains and Manipulations.
Unless otherwise indicated, flies were obtained from the Bloomington Stock Center and raised at 25 °C on standard cornmeal/molasses food. RNAi and dominant-negative lines are listed in Table S1. draper∆5, a null allele (22), was provided by Estee Kurant, Technion Israel Institute of Technology, Haifa, Israel. All recombinants were confirmed by PCR. PWIZ-draperRNAi #7b (23) was provided by Mary Logan and Marc Freeman, University of Massachusetts, Worcester, MA. Ced-12KO, a null allele (74), was provided by Erika Geisbrecht, Kansas State University, Manhattan, KS. dor4 is a partial loss-of-function allele (47). GR1-GAL4 (75) was used to drive expression specifically in all FCs after stage 3 (27) and was provided by Trudi Schüpbach, Princeton University, Princeton. PG150-GAL4 (76) was used to drive expression specifically in the stretch FCs and was provided by Ellen LeMosy, Augusta University, Augusta, GA (77). Typically, GR1-GAL4 was used for experiments so that RNAi was expressed in FCs for a longer time during egg chamber development. PG150-GAL4 was used when we wished to limit expression to stretch FCs. GLCs were generated using the ovoD method as described (78). nanos-GAL4 was used to drive the overexpression of UASp-Diap1 specifically in the germ line (7). The reporter for JNK activity was puc-lacZA251.1F3 ry/TM3 (79). To obtain well-developed ovaries, females <20 d old (unless otherwise noted) were conditioned on fresh yeast paste for at least 48 h, changing yeast paste every 24 h. See SI Materials and Methods for more details.
Staining Methods.
See SI Materials and Methods for more details. Standard antibody staining techniques were used as described in ref. 53. LysoTracker Red DND-99 (1:50, Invitrogen) was used to detect acidified compartments (49). We used the DeadEnd Fluorometric TUNEL system (Promega) (53). Samples were mounted in VectaShield with DAPI (Vector Labs) and imaged on an Olympus FV10i confocal microscope or Olympus BX60 upright fluorescence microscope. Images were processed and compiled in Image J, Adobe Photoshop, and Adobe Illustrator.
Quantifications and Statistics.
We used GraphPad Prism to graph and analyze all of our data. The unpaired t test was used for all statistical analyses. Persisting NC nuclei (PN) in stage 14 egg chambers were quantified in “bins” of 0 PN, 1–3 PN, 4–6 PN, 7–9 PN, 10–12 PN, or 13–15 PN and presented as a percentage of total stage 14 egg chambers. To calculate the average number of PN, the median number in each bin (e.g., two in the 1–3 bin) was used. With the exception of fecundity analyses, “n” always refers to the number of egg chambers quantified. To quantify NC nuclear permeability, the number of NC nuclei that were still intact in stage 11 were counted.
SI Materials and Methods
Fly Strains and Manipulations.
Age-matched siblings lacking either the driver or RNAi construct were used as controls. When two UAS-RNAi constructs were expressed during our double mutant analyses, we included UAS-GAL4 so that GAL4 was not limiting with two UAS targets. For the expression of Diap1RNAi in the stretch FCs (Fig. 5), flies were raised at 18 °C and moved to 29 °C for 4–18 h to inactivate GAL80ts. FRT2A was recombined onto the draper∆5 chromosome, and a recombinant was isolated and confirmed by PCR. The draper FRT homozygous mutant showed the same ovary phenotype in trans to a deficiency [Df(3L)BSC181], and this line was used for all further studies.
Fecundity Analysis.
Newly eclosed females were mated to CantonS males (1:1 ratio, <10 flies per vial), aged for 20 d on a normal diet, and transferred to fresh food every 5 d. We chose to analyze 20-d-old flies to allow time for NCs to accumulate in the ovary. The 20-d-old flies were transferred into vials containing grape juice agar with three yeast granules per female every 24 h until the flies were 25 d old. The number of eggs laid were counted at the same time each day. If a male escaped or died, it was replaced with another CantonS male. If a female died, it was scored as half a fly for that day. After day 25, flies were conditioned on yeast paste for 1–3 d, dissected, and stained with DAPI. To score fecundity using pupae, 1–4 females and an equal number of males were transferred to new vials daily, and the number of pupae were counted.
Antibody Staining.
Dissected ovaries were fixed for 20 min in Grace’s fix: 375 μL Grace’s media, 125 μL 16% (wt/vol) paraformaldehyde (opened <1 wk), and 250 μL heptane. Fixed ovary tissue was washed 3× over 30 min in PBT (1× PBS + 0.1% TritonX). If desired, samples were incubated in rhodamine phalloidin (1:500 in PBT; Molecular Probes) for 1 h, protected from light. Tissue was blocked for 1 h at room temperature (RT) in PBANG [PBT + 0.5% BSA + 5% (vol/vol) Normal Goat Serum] and incubated in 1° antibody diluted in PBANG overnight at 4 °C. After rinsing 2× with PBT, samples were washed 4× over 2 h in PBT + 0.5% BSA, changing the wash every 30 min. The 2° antibody was diluted 1:200 in PBANG, added to samples for 1 h, and protected from light. Again, samples were washed 4× over 2 h in PBT + 0.5% BSA, changing the wash every 30 min. After desired staining, samples were incubated in 1–2 drops of VectaShield with DAPI (Vector Labs) overnight at 4 °C before mounting on slides.
Primary antibodies were α-Draper (used at 1:500; Marc Freeman, University of Massachusetts, Worcester, MA), α-Draper 5D14 [1:50; Developmental Studies Hybridoma Bank (DSHB)], α–β-Gal (1:400; Promega), α-FasIII (1:20; DSHB), α–Dcp-1 (1:100; Cell Signaling Technology), and α-Lamin Dm0 ADL67.10 (1:2; DSHB). Secondary antibodies included goat–α-mouse Cy3 (1:200; Jackson ImmunoResearch) and goat–α-rabbit AlexaFluor488 (1:200; Invitrogen).
LysoTracker Staining.
After dissection, ovaries were incubated with LysoTracker Red DND-99 (Invitrogen) 1:50 in 1× PBS for 3 min while periodically flicking the tube. After 3 min, LysoTracker solution was removed and ovaries were washed for 10 min in 1× PBS three times. Next, tissue was fixed for 20 min with Grace’s Fix as described above. Fix was removed, and ovary tissue was washed 3× in PBT over 15 min. After the removing wash, samples were stained with DAPI, mounted, and imaged as described above. For more details, see ref. 49. For TUNEL and LysoTracker quantifications, stage 12–13 egg chambers were staged by DA thickness and scored for staining. Stage 12 egg chambers had no DA formation, stage 13 had minimal to moderately thick DAs, and stage 14 had DAs of normal thickness.
TUNEL Staining.
Fixed ovary tissue was washed 3× over 30 min (10 min per wash) in PBT and permeabilized with 1× PBS + 0.2% TritonX for 15 min. Ovary tissue was washed twice in PBT, 5 min per wash. Using reagents from the DeadEnd Fluorometric TUNEL system (Promega), ovary tissue was washed in ∼40 μL of equilibration buffer for 10 min. Next, 51 μL of Terminal deoxynucleotidyl transferase (Tdt) reaction mix was added to each sample (45 μL of equilibration buffer, 5 μL of nucleotide mix, and 1 μL of Tdt enzyme). Samples were incubated for 3 h at 37 °C and protected from light. Then, >300 μL of 2× SSC was added for 1 min to stop the reaction, and a second wash with 2× SSC was performed for 15 min. After the stop reaction, samples were washed 3× for 10 min each in PBT. Samples were stained with DAPI, mounted, and imaged as described above. For more details, see ref. 53.
Quantification of Border Cell Migration.
To perform the quantification of border cell migration (Fig. S2), the distance between the anterior tip of the egg chamber and NC/oocyte interface was measured as the total. The distance between the anterior tip of the egg chamber and the center of the border cell cluster was calculated as a percentage of the total distance for each egg chamber. The data were presented as average percent migration (Fig. S2 B and G) or categorized into bins (Fig. S2H).
Acknowledgments
We thank Horacio Frydman, Angela Ho, Chip Celenza, and laboratory members for helpful discussions and comments and Todd Blute for help with microscopy. We thank the Bloomington Stock Center, the Transgenic RNAi Project at Harvard Medical School (NIH R01-GM084947), the VDRC (Vienna Drosophila RNAi Center), Estee Kurant, Marc Freeman, Mary Logan, Trudi Schüpbach, Erika Geisbrecht, and Ellen LeMosy for fly strains, and we thank the Developmental Studies Hybridoma Bank and Marc Freeman for antibodies. We thank our funding sources: NIH Grants R01 GM060574 (+American Recovery and Reinvestment Act supplement) and R01 GM094452 (to K.M.), NIH Fellowship F31 GM099425 (to J.I.E.), National Science Foundation Northeast Alliance for Graduate Education and the Professoriate (to A.A.M.), and a Beckman Foundation award (to C.E.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1522830113/-/DCSupplemental.
References
- 1.Galluzzi L, et al. Essential versus accessory aspects of cell death: Recommendations of the NCCD 2015. Cell Death Differ. 2015;22(1):58–73. doi: 10.1038/cdd.2014.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kroemer G, et al. Nomenclature Committee on Cell Death 2009 Classification of cell death: Recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 2009;16(1):3–11. doi: 10.1038/cdd.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galluzzi L, et al. Molecular definitions of cell death subroutines: Recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ. 2012;19(1):107–120. doi: 10.1038/cdd.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spradling AC. Developmental genetics of oogenesis. In: Bate M, Martinez Arias A, editors. The Development of Drosophila melanogaster. Vol I. Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 1993. pp. 1–70. [Google Scholar]
- 5.King RC. Ovarian Development in Drosophila melanogaster. Academic Press; New York: 1970. [Google Scholar]
- 6.Jenkins VK, Timmons AK, McCall K. Diversity of cell death pathways: Insight from the fly ovary. Trends Cell Biol. 2013;23(11):567–574. doi: 10.1016/j.tcb.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peterson JS, Barkett M, McCall K. Stage-specific regulation of caspase activity in drosophila oogenesis. Dev Biol. 2003;260(1):113–123. doi: 10.1016/s0012-1606(03)00240-9. [DOI] [PubMed] [Google Scholar]
- 8.Baum JS, Arama E, Steller H, McCall K. The Drosophila caspases Strica and Dronc function redundantly in programmed cell death during oogenesis. Cell Death Differ. 2007;14(8):1508–1517. doi: 10.1038/sj.cdd.4402155. [DOI] [PubMed] [Google Scholar]
- 9.Mazzalupo S, Cooley L. Illuminating the role of caspases during Drosophila oogenesis. Cell Death Differ. 2006;13(11):1950–1959. doi: 10.1038/sj.cdd.4401892. [DOI] [PubMed] [Google Scholar]
- 10.Peterson JS, McCall K. Combined inhibition of autophagy and caspases fails to prevent developmental nurse cell death in the Drosophila melanogaster ovary. PLoS One. 2013;8(9):e76046. doi: 10.1371/journal.pone.0076046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pérez-Garijo A, Fuchs Y, Steller H. Apoptotic cells can induce non-autonomous apoptosis through the TNF pathway. eLife. 2013;2:e01004. doi: 10.7554/eLife.01004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson NS, Dixit V, Ashkenazi A. Death receptor signal transducers: Nodes of coordination in immune signaling networks. Nat Immunol. 2009;10(4):348–355. doi: 10.1038/ni.1714. [DOI] [PubMed] [Google Scholar]
- 13.Brown GC, Neher JJ. Eaten alive! Cell death by primary phagocytosis: ‘Phagoptosis’. Trends Biochem Sci. 2012;37(8):325–332. doi: 10.1016/j.tibs.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 14.Brown GC, Neher JJ. Microglial phagocytosis of live neurons. Nat Rev Neurosci. 2014;15(4):209–216. doi: 10.1038/nrn3710. [DOI] [PubMed] [Google Scholar]
- 15.Neher JJ, et al. Inhibition of microglial phagocytosis is sufficient to prevent inflammatory neuronal death. J Immunol. 2011;186(8):4973–4983. doi: 10.4049/jimmunol.1003600. [DOI] [PubMed] [Google Scholar]
- 16.Overholtzer M, et al. A nonapoptotic cell death process, entosis, that occurs by cell-in-cell invasion. Cell. 2007;131(5):966–979. doi: 10.1016/j.cell.2007.10.040. [DOI] [PubMed] [Google Scholar]
- 17.Reddien PW, Cameron S, Horvitz HR. Phagocytosis promotes programmed cell death in C. elegans. Nature. 2001;412(6843):198–202. doi: 10.1038/35084096. [DOI] [PubMed] [Google Scholar]
- 18.Hoeppner DJ, Hengartner MO, Schnabel R. Engulfment genes cooperate with ced-3 to promote cell death in Caenorhabditis elegans. Nature. 2001;412(6843):202–206. doi: 10.1038/35084103. [DOI] [PubMed] [Google Scholar]
- 19.Mangahas PM, Zhou Z. Clearance of apoptotic cells in Caenorhabditis elegans. Semin Cell Dev Biol. 2005;16(2):295–306. doi: 10.1016/j.semcdb.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 20.Reddien PW, Horvitz HR. The engulfment process of programmed cell death in Caenorhabditis elegans. Annu Rev Cell Dev Biol. 2004;20:193–221. doi: 10.1146/annurev.cellbio.20.022003.114619. [DOI] [PubMed] [Google Scholar]
- 21.Fullard JF, Kale A, Baker NE. Clearance of apoptotic corpses. Apoptosis. 2009;14(8):1029–1037. doi: 10.1007/s10495-009-0335-9. [DOI] [PubMed] [Google Scholar]
- 22.Freeman MR, Delrow J, Kim J, Johnson E, Doe CQ. Unwrapping glial biology: Gcm target genes regulating glial development, diversification, and function. Neuron. 2003;38(4):567–580. doi: 10.1016/s0896-6273(03)00289-7. [DOI] [PubMed] [Google Scholar]
- 23.MacDonald JM, et al. The Drosophila cell corpse engulfment receptor Draper mediates glial clearance of severed axons. Neuron. 2006;50(6):869–881. doi: 10.1016/j.neuron.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 24.Manaka J, et al. Draper-mediated and phosphatidylserine-independent phagocytosis of apoptotic cells by Drosophila hemocytes/macrophages. J Biol Chem. 2004;279(46):48466–48476. doi: 10.1074/jbc.M408597200. [DOI] [PubMed] [Google Scholar]
- 25.Cuttell L, et al. Undertaker, a Drosophila Junctophilin, links Draper-mediated phagocytosis and calcium homeostasis. Cell. 2008;135(3):524–534. doi: 10.1016/j.cell.2008.08.033. [DOI] [PubMed] [Google Scholar]
- 26.Li W, Baker NE. Engulfment is required for cell competition. Cell. 2007;129(6):1215–1225. doi: 10.1016/j.cell.2007.03.054. [DOI] [PubMed] [Google Scholar]
- 27.Etchegaray JI, et al. Draper acts through the JNK pathway to control synchronous engulfment of dying germline cells by follicular epithelial cells. Development. 2012;139(21):4029–4039. doi: 10.1242/dev.082776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Franc NC, Heitzler P, Ezekowitz RA, White K. Requirement for croquemort in phagocytosis of apoptotic cells in Drosophila. Science. 1999;284(5422):1991–1994. doi: 10.1126/science.284.5422.1991. [DOI] [PubMed] [Google Scholar]
- 29.Meehan TL, Kleinsorge SE, Timmons AK, Taylor JD, McCall K. Polarization of the epithelial layer and apical localization of integrins are required for engulfment of apoptotic cells in the Drosophila ovary. Dis Model Mech. 2015;8(12):1603–1614. doi: 10.1242/dmm.021998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagaosa K, et al. Integrin βν-mediated phagocytosis of apoptotic cells in Drosophila embryos. J Biol Chem. 2011;286(29):25770–25777. doi: 10.1074/jbc.M110.204503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nonaka S, Nagaosa K, Mori T, Shiratsuchi A, Nakanishi Y. Integrin αPS3/βν-mediated phagocytosis of apoptotic cells and bacteria in Drosophila. J Biol Chem. 2013;288(15):10374–10380. doi: 10.1074/jbc.M113.451427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Savill J, Hogg N, Ren Y, Haslett C. Thrombospondin cooperates with CD36 and the vitronectin receptor in macrophage recognition of neutrophils undergoing apoptosis. J Clin Invest. 1992;90(4):1513–1522. doi: 10.1172/JCI116019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Savill J, Dransfield I, Hogg N, Haslett C. Vitronectin receptor-mediated phagocytosis of cells undergoing apoptosis. Nature. 1990;343(6254):170–173. doi: 10.1038/343170a0. [DOI] [PubMed] [Google Scholar]
- 34.Hsu TY, Wu YC. Engulfment of apoptotic cells in C. elegans is mediated by integrin alpha/SRC signaling. Curr Biol. 2010;20(6):477–486. doi: 10.1016/j.cub.2010.01.062. [DOI] [PubMed] [Google Scholar]
- 35.Ziegenfuss JS, et al. Draper-dependent glial phagocytic activity is mediated by Src and Syk family kinase signalling. Nature. 2008;453(7197):935–939. doi: 10.1038/nature06901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Goethem E, Silva EA, Xiao H, Franc NC. The Drosophila TRPP cation channel, PKD2 and Dmel/Ced-12 act in genetically distinct pathways during apoptotic cell clearance. PLoS One. 2012;7(2):e31488. doi: 10.1371/journal.pone.0031488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doherty J, et al. PI3K signaling and Stat92E converge to modulate glial responsiveness to axonal injury. PLoS Biol. 2014;12(11):e1001985. doi: 10.1371/journal.pbio.1001985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiao H, et al. The Pallbearer E3 ligase promotes actin remodeling via RAC in efferocytosis by degrading the ribosomal protein S6. Dev Cell. 2015;32(1):19–30. doi: 10.1016/j.devcel.2014.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han C, et al. Epidermal cells are the primary phagocytes in the fragmentation and clearance of degenerating dendrites in Drosophila. Neuron. 2014;81(3):544–560. doi: 10.1016/j.neuron.2013.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cummings MR, King RC. Ultrastructural changes in nurse and follicle cells during late stages of oogenesis in Drosophila melanogaster. Z Zellforsch Mikrosk Anat. 1970;110(1):1–8. doi: 10.1007/BF00343981. [DOI] [PubMed] [Google Scholar]
- 41.Nezis IP, Stravopodis DJ, Papassideri I, Robert-Nicoud M, Margaritis LH. Stage-specific apoptotic patterns during Drosophila oogenesis. Eur J Cell Biol. 2000;79(9):610–620. doi: 10.1078/0171-9335-00088. [DOI] [PubMed] [Google Scholar]
- 42.Tran DH, Berg CA. bullwinkle and shark regulate dorsal-appendage morphogenesis in Drosophila oogenesis. Development. 2003;130(25):6273–6282. doi: 10.1242/dev.00854. [DOI] [PubMed] [Google Scholar]
- 43.McPhee CK, Logan MA, Freeman MR, Baehrecke EH. Activation of autophagy during cell death requires the engulfment receptor Draper. Nature. 2010;465(7301):1093–1096. doi: 10.1038/nature09127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gumienny TL, et al. CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell. 2001;107(1):27–41. doi: 10.1016/s0092-8674(01)00520-7. [DOI] [PubMed] [Google Scholar]
- 45.Geisbrecht ER, et al. Drosophila ELMO/CED-12 interacts with Myoblast city to direct myoblast fusion and ommatidial organization. Dev Biol. 2008;314(1):137–149. doi: 10.1016/j.ydbio.2007.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bass BP, et al. Cell-autonomous requirement for DNaseII in nonapoptotic cell death. Cell Death Differ. 2009;16(10):1362–1371. doi: 10.1038/cdd.2009.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sevrioukov EA, He JP, Moghrabi N, Sunio A, Krämer H. A role for the deep orange and carnation eye color genes in lysosomal delivery in Drosophila. Mol Cell. 1999;4(4):479–486. doi: 10.1016/s1097-2765(00)80199-9. [DOI] [PubMed] [Google Scholar]
- 48.Yu X, Lu N, Zhou Z. Phagocytic receptor CED-1 initiates a signaling pathway for degrading engulfed apoptotic cells. PLoS Biol. 2008;6(3):e61. doi: 10.1371/journal.pbio.0060061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Timmons AK, Meehan TL, Gartmond TD, McCall K. Use of necrotic markers in the Drosophila ovary. Methods Mol Biol. 2013;1004:215–228. doi: 10.1007/978-1-62703-383-1_16. [DOI] [PubMed] [Google Scholar]
- 50.Wyllie AH, Kerr JF, Currie AR. Cell death: The significance of apoptosis. Int Rev Cytol. 1980;68:251–306. doi: 10.1016/s0074-7696(08)62312-8. [DOI] [PubMed] [Google Scholar]
- 51.Nagata S. DNA degradation in development and programmed cell death. Annu Rev Immunol. 2005;23:853–875. doi: 10.1146/annurev.immunol.23.021704.115811. [DOI] [PubMed] [Google Scholar]
- 52.Gavrieli Y, Sherman Y, Ben-Sasson SA. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J Cell Biol. 1992;119(3):493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sarkissian T, Timmons A, Arya R, Abdelwahid E, White K. Detecting apoptosis in Drosophila tissues and cells. Methods. 2014;68(1):89–96. doi: 10.1016/j.ymeth.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCall K, Steller H. Requirement for DCP-1 caspase during Drosophila oogenesis. Science. 1998;279(5348):230–234. doi: 10.1126/science.279.5348.230. [DOI] [PubMed] [Google Scholar]
- 55.Foley K, Cooley L. Apoptosis in late stage Drosophila nurse cells does not require genes within the H99 deficiency. Development. 1998;125(6):1075–1082. doi: 10.1242/dev.125.6.1075. [DOI] [PubMed] [Google Scholar]
- 56.Cavaliere V, Taddei C, Gargiulo G. Apoptosis of nurse cells at the late stages of oogenesis of Drosophila melanogaster. Dev Genes Evol. 1998;208(2):106–112. doi: 10.1007/s004270050160. [DOI] [PubMed] [Google Scholar]
- 57.Cooley L, Verheyen E, Ayers K. chickadee encodes a profilin required for intercellular cytoplasm transport during Drosophila oogenesis. Cell. 1992;69(1):173–184. doi: 10.1016/0092-8674(92)90128-y. [DOI] [PubMed] [Google Scholar]
- 58.Buszczak M, Cooley L. Eggs to die for: Cell death during Drosophila oogenesis. Cell Death Differ. 2000;7(11):1071–1074. doi: 10.1038/sj.cdd.4400755. [DOI] [PubMed] [Google Scholar]
- 59.Ohsawa S, et al. Elimination of oncogenic neighbors by JNK-mediated engulfment in Drosophila. Dev Cell. 2011;20(3):315–328. doi: 10.1016/j.devcel.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 60.Macdonald JM, Doherty J, Hackett R, Freeman MR. The c-Jun kinase signaling cascade promotes glial engulfment activity through activation of draper and phagocytic function. Cell Death Differ. 2013;20(9):1140–1148. doi: 10.1038/cdd.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suzanne M, Perrimon N, Noselli S. The Drosophila JNK pathway controls the morphogenesis of the egg dorsal appendages and micropyle. Dev Biol. 2001;237(2):282–294. doi: 10.1006/dbio.2001.0384. [DOI] [PubMed] [Google Scholar]
- 62.Dobens LL, Martín-Blanco E, Martínez-Arias A, Kafatos FC, Raftery LA. Drosophila puckered regulates Fos/Jun levels during follicle cell morphogenesis. Development. 2001;128(10):1845–1856. doi: 10.1242/dev.128.10.1845. [DOI] [PubMed] [Google Scholar]
- 63.Dequier E, et al. Top-DER- and Dpp-dependent requirements for the Drosophila fos/kayak gene in follicular epithelium morphogenesis. Mech Dev. 2001;106(1-2):47–60. doi: 10.1016/s0925-4773(01)00418-x. [DOI] [PubMed] [Google Scholar]
- 64.Thomson TC, Johnson J. Inducible somatic oocyte destruction in response to rapamycin requires wild-type regulation of follicle cell epithelial polarity. Cell Death Differ. 2010;17(11):1717–1727. doi: 10.1038/cdd.2010.49. [DOI] [PubMed] [Google Scholar]
- 65.Li X, Johnson RW, Park D, Chin-Sang I, Chamberlin HM. Somatic gonad sheath cells and Eph receptor signaling promote germ-cell death in C. elegans. Cell Death Differ. 2012;19(6):1080–1089. doi: 10.1038/cdd.2011.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peters PJ, et al. Cytotoxic T lymphocyte granules are secretory lysosomes, containing both perforin and granzymes. J Exp Med. 1991;173(5):1099–1109. doi: 10.1084/jem.173.5.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Samie MA, Xu H. Lysosomal exocytosis and lipid storage disorders. J Lipid Res. 2014;55(6):995–1009. doi: 10.1194/jlr.R046896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Samie M, et al. A TRP channel in the lysosome regulates large particle phagocytosis via focal exocytosis. Dev Cell. 2013;26(5):511–524. doi: 10.1016/j.devcel.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McIlroy D, et al. An auxiliary mode of apoptotic DNA fragmentation provided by phagocytes. Genes Dev. 2000;14(5):549–558. [PMC free article] [PubMed] [Google Scholar]
- 70.Wu YC, Stanfield GM, Horvitz HR. NUC-1, a Caenorhabditis elegans DNase II homolog, functions in an intermediate step of DNA degradation during apoptosis. Genes Dev. 2000;14(5):536–548. [PMC free article] [PubMed] [Google Scholar]
- 71.Hudson AM, Cooley L. Understanding the function of actin-binding proteins through genetic analysis of Drosophila oogenesis. Annu Rev Genet. 2002;36:455–488. doi: 10.1146/annurev.genet.36.052802.114101. [DOI] [PubMed] [Google Scholar]
- 72.Wheatley S, Kulkarni S, Karess R. Drosophila nonmuscle myosin II is required for rapid cytoplasmic transport during oogenesis and for axial nuclear migration in early embryos. Development. 1995;121(6):1937–1946. doi: 10.1242/dev.121.6.1937. [DOI] [PubMed] [Google Scholar]
- 73.Xu J, Gridley T. Notch2 is required in somatic cells for breakdown of ovarian germ-cell nests and formation of primordial follicles. BMC Biol. 2013;11:13. doi: 10.1186/1741-7007-11-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bianco A, et al. Two distinct modes of guidance signalling during collective migration of border cells. Nature. 2007;448(7151):362–365. doi: 10.1038/nature05965. [DOI] [PubMed] [Google Scholar]
- 75.Goentoro LA, Yakoby N, Goodhouse J, Schüpbach T, Shvartsman SY. Quantitative analysis of the GAL4/UAS system in Drosophila oogenesis. Genesis. 2006;44(2):66–74. doi: 10.1002/gene.20184. [DOI] [PubMed] [Google Scholar]
- 76.Bourbon HM, et al. A P-insertion screen identifying novel X-linked essential genes in Drosophila. Mech Dev. 2002;110(1-2):71–83. doi: 10.1016/s0925-4773(01)00566-4. [DOI] [PubMed] [Google Scholar]
- 77.Dinkins MB, Fratto VM, Lemosy EK. Integrin alpha chains exhibit distinct temporal and spatial localization patterns in epithelial cells of the Drosophila ovary. Dev Dyn. 2008;237(12):3927–3939. doi: 10.1002/dvdy.21802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chou TB, Perrimon N. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics. 1996;144(4):1673–1679. doi: 10.1093/genetics/144.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Martín-Blanco E, et al. puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila. Genes Dev. 1998;12(4):557–570. doi: 10.1101/gad.12.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]