Significance
For individuals to develop sexually dimorphic body parts and behavior, their cells must know their sex. In the fruit fly Drosophila melanogaster, this process is carried out by a series of genes ending with fruitless (fru) and doublesex (dsx). We found that both Fru and Dsx regulate the expression of the leucine-rich repeat G protein-coupled receptor 3 (Lgr3) gene in separate sets of neurons, including neurons important for female sexual behavior. Thus, Lgr3 is important for sexual development and is a point of convergence after the fru/dsx split.
Keywords: Lgr3, fruitless, doublesex, Drosophila, enhancer
Abstract
The development of sexually dimorphic morphology and the potential for sexually dimorphic behavior in Drosophila are regulated by the Fruitless (Fru) and Doublesex (Dsx) transcription factors. Several direct targets of Dsx have been identified, but direct Fru targets have not been definitively identified. We show that Drosophila leucine-rich repeat G protein-coupled receptor 3 (Lgr3) is regulated by Fru and Dsx in separate populations of neurons. Lgr3 is a member of the relaxin-receptor family and a receptor for Dilp8, necessary for control of organ growth. Lgr3 expression in the anterior central brain of males is inhibited by the B isoform of Fru, whose DNA binding domain interacts with a short region of an Lgr3 intron. Fru A and C isoform mutants had no observed effect on Lgr3 expression. The female form of Dsx (DsxF) separately up- and down-regulates Lgr3 expression in distinct neurons in the abdominal ganglion through female- and male-specific Lgr3 enhancers. Excitation of neural activity in the DsxF–up-regulated abdominal ganglion neurons inhibits female receptivity, indicating the importance of these neurons for sexual behavior. Coordinated regulation of Lgr3 by Fru and Dsx marks a point of convergence of the two branches of the sex-determination hierarchy.
Most animal species are comprised of female and male individuals, in which sex differences in form and behavior are specified by their genetic makeup. The developmental processes by which genes build sex-specific differences into the nervous system, and hence encode the potential for sex-specific behavior, have long been of interest (1).
In Drosophila melanogaster the assessment of the number of X chromosomes leads to sex-differential splicing of transcripts from genes making up the sex-determination hierarchy, in particular the terminal genes of that hierarchy, fruitless (fru) and doublesex [dsx (2), reviewed in ref. 3]. fru and dsx encode sex-specific Zn-finger transcription factors that alter, either directly or indirectly, the expression of downstream genes to produce the sexually dimorphic elements of flies. The male-specific forms of Fru (FruM) act in a subset of the neurons within the male’s nervous system to establish the potential for social interactions such as courtship behavior and aggression (reviewed in ref. 3). In contrast, Dsx acts in subsets of both neural and nonneural tissues of males and females to regulate behavioral and nonbehavioral aspects of sexual development (reviewed in ref. 3).
Although the mechanisms regulating the production of the sex-specific isoforms of the Fru and Dsx proteins are well-established (4), how these proteins in turn function is only beginning to be elucidated. Several direct Dsx targets and a well-conserved 13-bp Dsx binding site have been identified (5–13). Many Dsx target genes encode well-known transcription factors and cell–cell signaling molecules that function sex-nonspecifically in most tissues in which they are expressed. However, in other tissues, Dsx directs the sex-specific expression of these genes to generate sex-specific aspects of development.
FruM’s regulatory function has thus far proven to be less tractable than that of Dsx. FruM appears to function in a complex with the transcription cofactor Bonus and either histone deacetylase 1 or heterochromatin protein 1a (14). Recent genome-wide screens of RNA expression levels or FruM binding activity have identified potential FruM targets, but have lacked independent confirmation of such regulation (15–17). Thus, our understanding of how FruM specifies the potential for sex-specific behavior remains limited.
Along with the study of the genetic targets of Dsx and FruM, studying the control of neuronal function by genomic enhancer elements has identified behavioral roles for Dsx- and FruM-expressing neurons (3, 18–20). In a screen for additional genomic enhancer elements that drive sexually dimorphic nervous system expression, we identified an enhancer derived from the Lgr3 gene. Lgr3 is a member of the leucine-rich repeat G-protein-coupled receptor (Lgr) family (21). The Drosophila genome contains four members of the Lgr family, including Lgr2, encoded by the rickets gene and necessary for tanning of the adult cuticle in response to the hormone bursicon (22). Lgr-related genes are also present in humans and include those encoding relaxin-family peptide receptors RXFP1 and RXFP2, which among other functions are necessary for normal reproduction in both sexes (reviewed in ref. 23).
We investigated the sex-specific regulation of Lgr3 expression and its functional importance in female sexual behavior. We identified roles for both FruM and Dsx in regulating Lgr3 expression in separate sets of neurons. One isoform of Fru inhibits Lgr3 expression in the male brain, whereas Dsx activates Lgr3 expression in some neurons in the female abdominal ganglion and represses it in others. To better understand the basis of this complex regulation of Lgr3 expression, potential enhancer fragments from the Lgr3 locus were used to narrow down the sites of Fru and Dsx activity. We found that FruM interacts with a specific portion of an Lgr3 intron, suggesting its regulation may be direct. Finally, activation of a subset of Lgr3-expressing abdominal ganglion neurons reduced female receptivity to courtship, indicating the functional importance of sex-specific Lgr3 expression.
Results
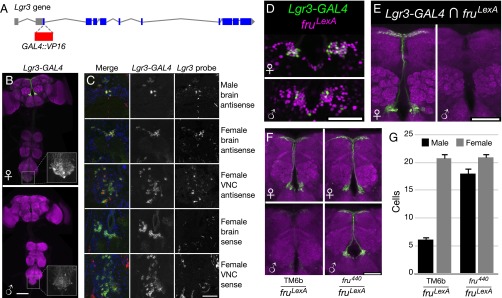
We examined the expression of the Lgr3 gene using a bacterial artificial chromosome (BAC) reporter, Lgr3-GAL4::VP16, encompassing the Lgr3 locus, with the GAL4 DNA-binding domain and VP16 activation domain inserted in place of the first coding exon of Lgr3 (Fig. 1A). Lgr3-GAL4 drives expression of a UAS-GFP reporter in a sexually dimorphic pattern in the brain median bundle and in the abdominal ganglion (Fig. 1B). In the median bundle of both pupae and adults, Lgr3-GAL4 is expressed in significantly more neurons in females than in males (Fig. 1G and Fig. S1A).
Fig. 1.
Sexually dimorphic expression of Lgr3. (A) To create Lgr3-GAL4::VP16, GAL4::VP16 was inserted into a BAC containing the Lgr3 locus, along with ∼1-kb upstream and 6.6-kb downstream sequence, the latter including several neighboring genes. Sequence encoding the GAL4 DNA-binding domain and VP16 activation domain replaced the first coding exon of Lgr3. (B) Lgr3-GAL4 (attP40), UAS-myrGFP (attP2) shows higher expression in the female median bundle compared with the male median bundle. The brain and VNC were imaged in separate 20× tiles and composited. (Inset) Lgr3-GAL4 expression in the abdominal ganglion at a higher gain and approximately 2.4× higher magnification. (Scale bar, 100 µm.) (C) DIG-labeled in situ hybridization of Lgr3 antisense probe (in red) shows correspondence with Lgr3-GAL4 (attP40), UAS-myrGFP (su(Hw)attP5) (in green) in horizontal sections through the male and female brain and abdominal ganglion. The sense probe did not show appreciable overlap with GFP. The membrane-bound GFP generally surrounds the mRNA signal in neurons with both labels. DAPI nuclear label (in blue) is included for reference. (Scale bar, 50 µm.) (D) Lgr3-GAL4, UAS-Stinger-GFP (green) overlaps with fruLexA, LexAop > tdTomato (magenta) in females, but less so in males. Overlap is in white. (Scale bar, 50 µm.) (E) Intersection of Lgr3-GAL4 (attP40) with fruLexA using LexAop2-FlpL (attP40) and UAS > stop > myrGFP (su(Hw)attP1) leads to GFP expression primarily in females (8.6 ± 0.9 cells in females vs. 0.6 ± 0.2 in males, P < 0.001 by Student's t test, n = 12–14 hemispheres). GFP expression occurs only in cells expressing both Lgr3 and fruM. (Scale bar, 50 µm.) (F) Lgr3-GAL4 (attP40), UAS-myrGFP (su(Hw)attP5) shows dependence on FruM. Elimination of FruM function using fru4−40/fruLexA brings male expression close to female levels. (Scale bar, 50 µm.) (G) Cell counts for F. Expression in control females was significantly broader than in control males (20.6 ± 0.7 cells vs. 6.1 ± 0.7, P < 0.001 by ANOVA and Tukey post hoc test, n = 8–10 brain hemispheres). fru4−40/fruLexA male expression (17.9 ± 0.8) was significantly increased from control males (P < 0.001), but still lower than in the female (20.9 ± 0.7), by a small amount (P < 0.01). Error bars indicate SEM.
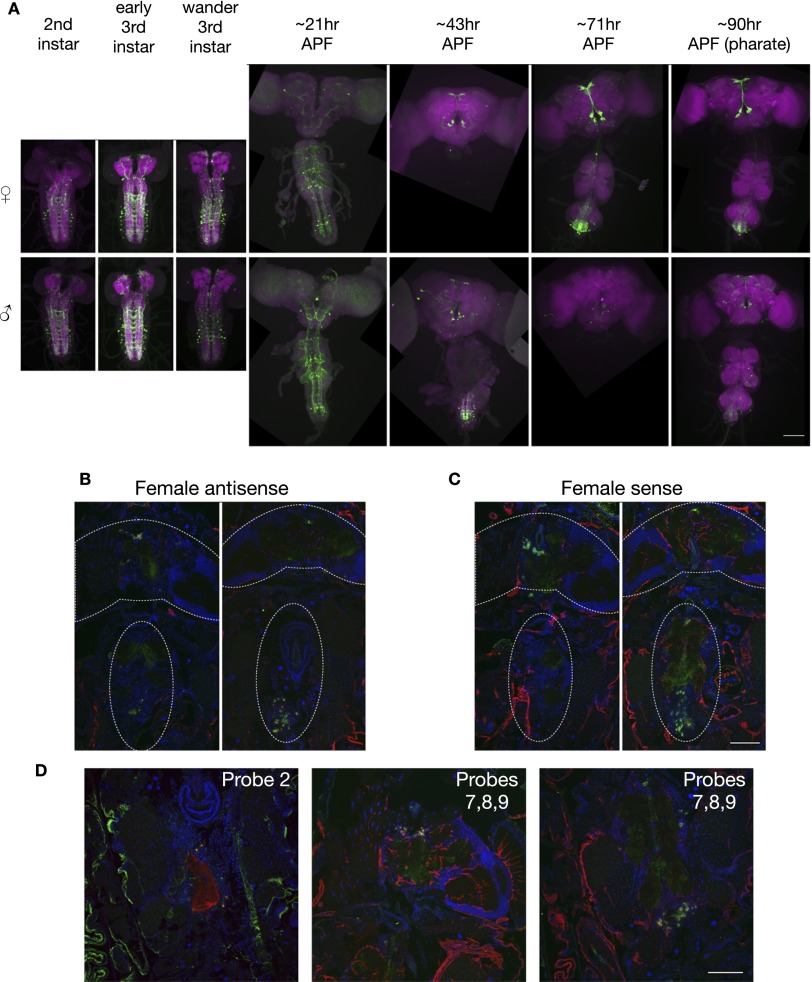
Fig. S1.
(Related to Fig. 1.) Lgr3 in situ and time course. (A) Larval and pupal time course of Lgr3-GAL4 (attP40), UAS-myrGFP (su(Hw)attP5) expression in females and males. Times are approximate. Early pupal stages measured relative to time of white prepupa collection (APF, after puparium formation). VNCs were not obtained for all samples. The brain and VNC were imaged in separate 20× tiles and composited. (Scale bar, 100 µm.) (B) Full in situ hybridization images of antisense probe 3 in female brain and VNC horizontal sections. Approximate locations of brain and VNC are outlined, although some sections contain only part of each. The brain and VNC were imaged in separate 20× tiles and composited. (C) Full in situ hybridization images of sense probes 2 and 3 pooled in females. (Scale bar, 100 µm.) (D) In situ hybridization images of antisense probe 2 and pooled antisense probes 7, 8, and 9 in females. (Scale bar, 100 µm.)
To verify that the expression of the Lgr3-GAL4 reporter accurately reflects expression of the Lgr3 gene, we compared Lgr3-GAL4’s expression pattern to the pattern of in situ hybridization of Lgr3 probes to transcripts in sections of males and females (Fig. 1C and Fig. S1 B and C). Lgr3 antisense probe signal colocalized with Lgr3-GAL4 expression in both males and females, whereas the sense probe control lacked any observable coexpression (Fig. 1C). Expression of both GFP and the in situ probe appeared greater in females than males. Additional antisense probes targeting other regions of Lgr3 showed a similar pattern (Fig. S1D). Although otherwise coexpressed, hybridization signal in the abdominal ganglion was present in a few cells beyond the Lgr3-GAL4 pattern in both sexes.
FruM Inhibits Expression of Lgr3 in the Male Median Bundle.
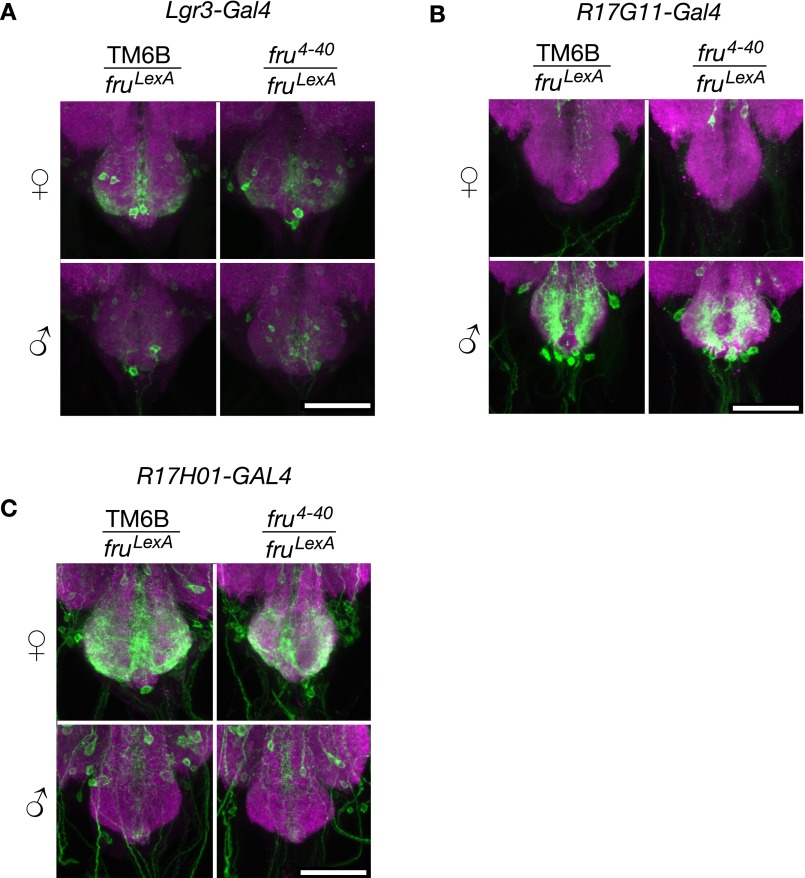
To understand the basis of sexually dimorphic Lgr3 expression in the median bundle, we examined the dependence of Lgr3-GAL4 expression on FruM and Dsx. Dsx expression has not been detected in the median bundle (24, 25), and dsx loss-of-function mutants had no effect on Lgr3-GAL4 expression in the median bundle (Fig. S2). In contrast, FruM is expressed in the median bundle (26), and we observed coexpression of a LexA reporter for fruM (fruLexA) (27) and Lgr3-GAL4 when examined with fluorescent nuclear reporters in the median bundle (Fig. 1D). Similar coexpression was seen when LexAop2-FlpL and UAS > stop > myrGFP were used to perform Flippase-mediated genetic intersection, in which fru-LexA and Lgr3-GAL4 are both needed to cause Flp to remove the stop codon in UAS > stop > myrGFP and the production of GFP (Fig. 1E). In a fruM loss-of-function mutant (fru4-40/fruLexA), Lgr3-GAL4, UAS-GFP expression in males was restored to nearly female levels, indicating that wild-type FruM likely inhibits Lgr3-GAL4 expression in males (Fig. 1 F and G).
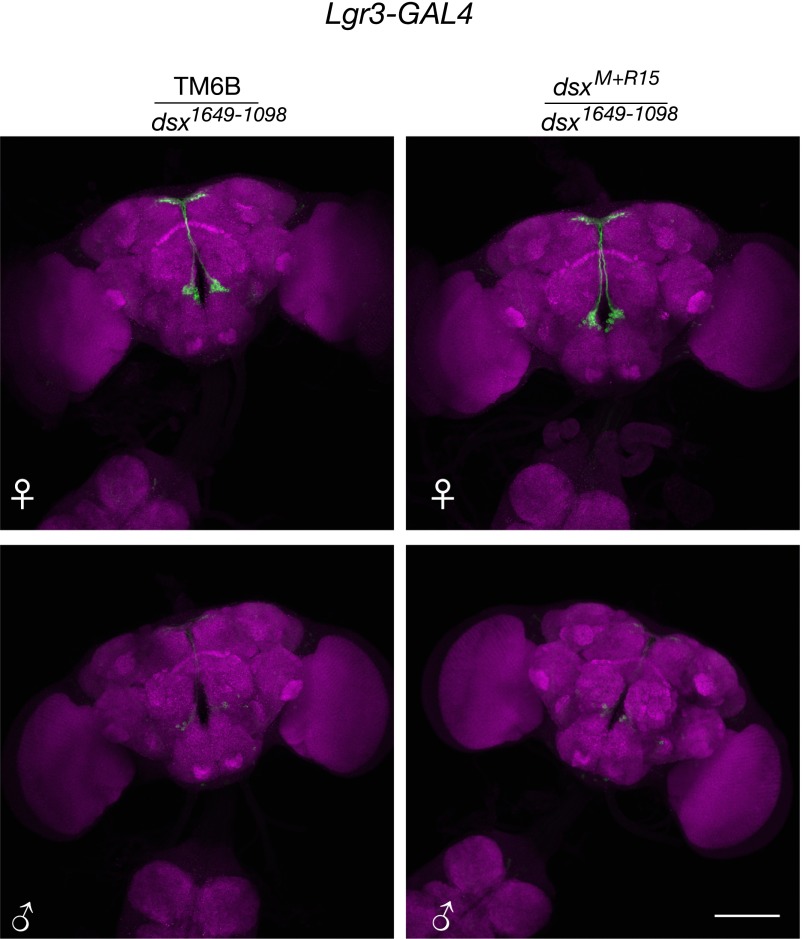
Fig. S2.
(Related to Fig. 1.) Dsx regulation of Lgr3-GAL4 brain expression. Lgr3-GAL4 (attP40), UAS-myrGFP (su(Hw)attP5) expression in the female and male brain. Left column is heterozygous for the TM6B balancer and either dsxM+R15 or dsx1649-1098. Right column is heterozygous for dsxM+R15 and dsx1649-1098. (Scale bar, 100 µm.)
Differences in the numbers of cells expressing Lgr3-GAL4 could be a result of Lgr3-GAL4 expression being altered in existing cells, or to the death of some of those cells. Prior studies of FruM expression reported cell number dimorphism in other subsets of FruM neurons, but not in the median bundle (28, 29). Cachero et al. (28) in particular counted the cells produced by neuroblast clones in both sexes and did not detect a difference. We verified that the number of FruM-expressing median bundle neurons did not significantly differ between females and males (66.6 ± 2.3 cells vs. 68.1 ± 1.8, P = 0.6 by Student's t test, n = 22–26 hemispheres). Thus, differences in Lgr3-GAL4 median bundle expression do not appear to result from cell death.
FruM Acts Through a Small Region of an Lgr3 Intron.
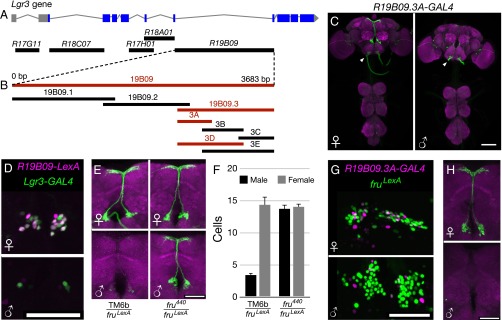
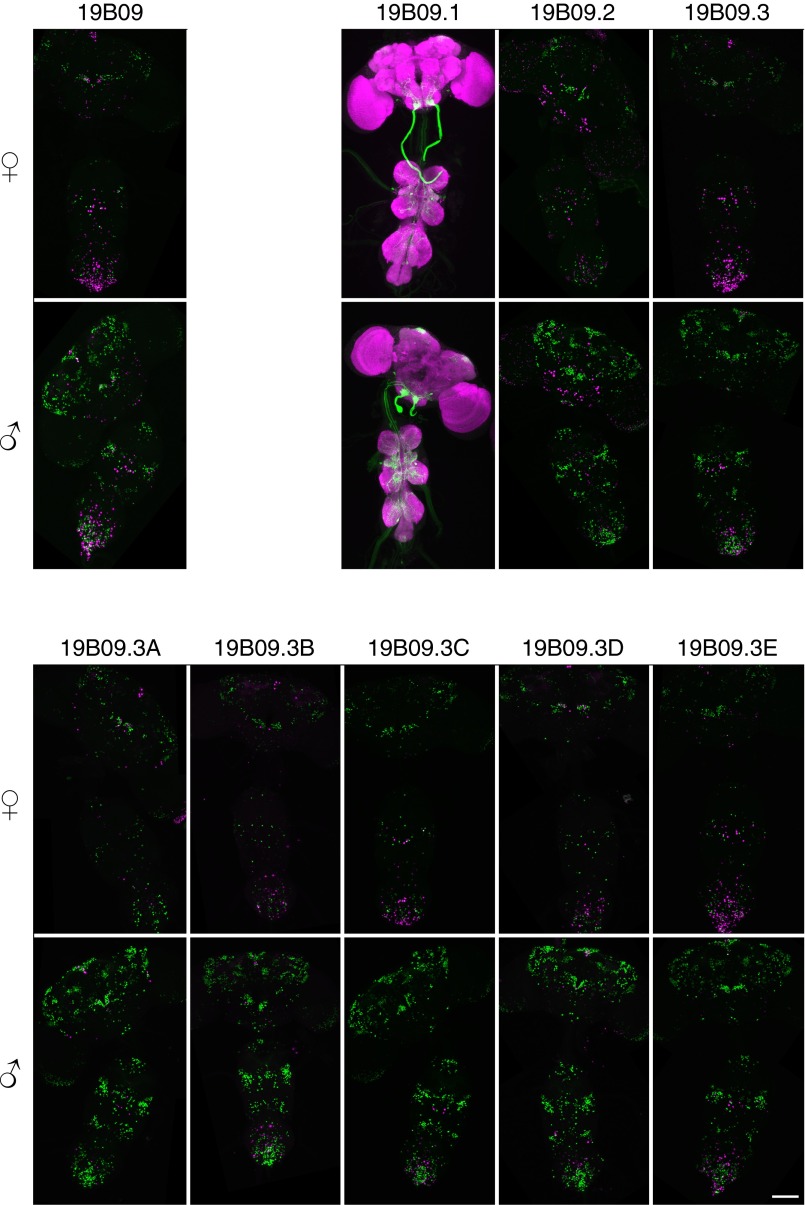
To determine the region of the Lgr3 locus through which FruM regulates Lgr3 median bundle expression, we asked whether various fragments of the Lgr3 gene contained enhancer elements sufficient to confer FruM-dependent enhancer-GAL4 expression (Fig. 2A) (30). Of the five Lgr3 fragments tested in this manner, only R19B09-GAL4, a fragment from the largest Lgr3 intron, conferred sexually dimorphic median bundle expression similar to that exhibited by Lgr3-GAL4 (Fig. S3). R19B09 thus likely contains the enhancer sequences necessary for the median bundle expression observed with Lgr3. To further localize these enhancer sequences, we subdivided the 3,683-bp R19B09-GAL4 reporter into smaller fragments, each of which was assayed for sexually dimorphic GAL4 expression (Fig. 2B and Fig. S3). Two rounds of such subdivisions yielded a 484-bp reporter termed R19B09.3A-GAL4 that maintained dimorphic median bundle expression (Fig. 2 B–D). As with Lgr3-GAL4, the expression of R19B09.3A-GAL4 both colocalizes with fru-LexA in females and is inhibited by FruM in males (Fig. 2 E–H).
Fig. 2.
R19B09.3A-GAL4 recapitulates dimorphic Lgr3 median bundle expression. (A) Rubin GAL4 enhancer fragment lines contain sequences from the indicated regions of the Lgr3 locus. (B) R19B09-GAL4 was subdivided in two rounds to identify a smaller region driving dimorphic median bundle expression. Fragments in red showed dimorphic median bundle expression, whereas those in black had other patterns of expression (Fig. S3). (C) Expression of R19B09.3A-GAL4, UAS-myrGFP (attP40) is sexually dimorphic in the female and male median bundle. UAS-GFP in (attP40) has basal expression in the labial nerve (arrowheads) (44). The brain and VNC were imaged in separate 20× tiles and composited. (Scale bar, 100 µm.) (D) R19B09-LexA, LexAop2-IVS-nlstdTomato (VK22) colocalizes with Lgr3-GAL4, UAS-Stinger-GFP in females, but both have low male expression. (Scale bar, 50 µm.) (E) R19B09.3A-GAL4 (attP2), UAS-myrGFP (attP40) shows dependence on FruM. Elimination of FruM function using fru4-40/fruLexA brings male expression to female levels. (Scale bar, 50 µm.) (F) Cell counts for E. Expression in control females was significantly broader than in males (14.2 ± 0.8 cells vs. 3.4 ± 0.5, P < 0.001 by ANOVA and Tukey post hoc test, n = 4–10 brain hemispheres). fru4-40/fruLexA male expression was indistinguishable from females (13.6 ± 0.5 vs. 13.9 ± 0.5, P > 0.5). Error bars indicate SEM. (G) R19B09.3A-GAL4, UAS-RedStinger colocalizes with fruLexA, LexAop-Stinger-GFP in females but not males. (Scale bar, 50 µm.) (H) Intersection of R19B09.3A-GAL4 (attP2) with fruLexA using LexAop2-FlpL (attP40) and UAS > stop > myrGFP (su(Hw)attP1) shows expression in females but not in males. (Scale bar, 50 µm.)
Fig. S3.
(Related to Fig. 2.) R19B09 subdivision expression. Expression of subdivisions of R19B09 was measured in females and males. R19B09.1-GAL4 is shown driving UAS-myrGFP attP40 in green, with neuropil in magenta. All other GAL4 lines are shown driving UAS-RedStinger, shown in magenta, with fruLexA driving LexAop-Stinger-GFP shown in green. The brain and VNC were imaged in separate 20× tiles and composited.
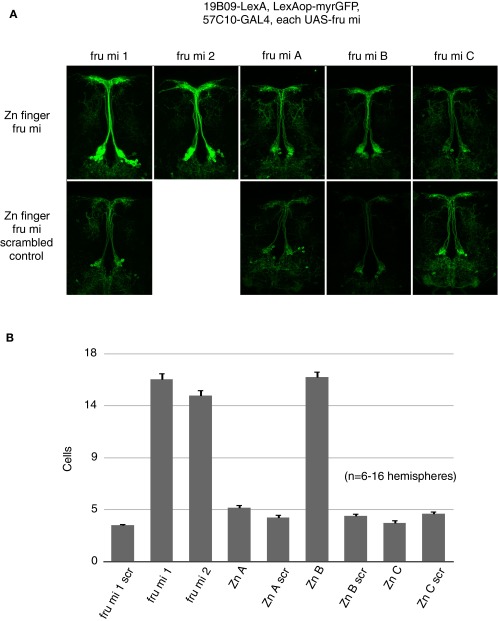
The fru gene encodes multiple transcripts through the use of alternative promoters in combination with sex-specific and sex-nonspecific alternative pre-mRNA splicing. The fru mRNAs thus generated are translated into proteins with different C-terminal Zn-finger DNA-binding domains. Of these, isoforms A, B, and C have detectable nervous system expression and are candidates for regulators of Lgr3 expression in the male nervous system (31). We examined the expression of R19B09.3A in flies individually mutant for the A, B, or C isoforms (13, 28), and found that R19B09.3A expression was only affected by the absence of FruB (Fig. 3 A and B). FruA and FruC mutants had no effect on R19B09.3A expression, whereas in FruB mutant males R19B09.3A expression is increased to female levels, suggesting that FruB negatively affects R19B09.3A-dependent expression. Using pan-neuronally expressed microRNAs targeting transcripts of the A–C fru isoforms, we similarly observed that knockdown only of FruB brought male expression to female levels (Fig. S4). Thus, FruB appears likely to inhibit Lgr3 median bundle expression via this fragment of an Lgr3 intron.
Fig. 3.
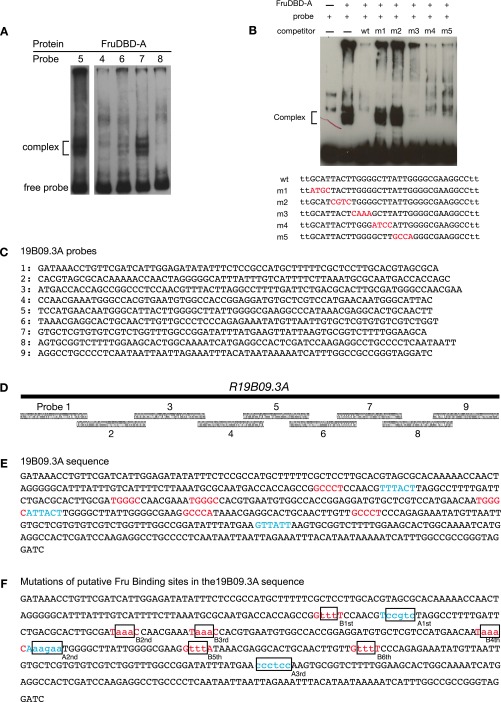
FruB regulates R19B09.3A-GAL4 expression. (A) Expression of R19B09.3A-GAL4, UAS-myrGFP (attP40) in females and males carrying heteroallelic combinations of fruLexA with either a wild-type, fru∆A, fru∆B, or fru∆C fru isoform mutant as indicated. (Scale bar, 50 µm.) (B) Cell counts for A. Expression in males was significantly lower than in females for flies carrying heteroallelic combinations of fruLexA with either wild-type, fru∆A, or fru∆C (P < 0.001 by Mann–Whitney u test with Bonferroni correction on each male/female pair, n = 18–30 hemispheres). However, in fruLexA/fru∆B individuals the expression of R19B09.3A-GAL4, UAS-myrGFP (attP40) did not differ between males and females (P > 0.1). Error bars indicate SEM. (C) EMSA showing binding of purified Fru-DBD-B protein to probe #3 from R19B09.3A (Fig. S5 C and D). A decreasing gradient in the amount of protein was used to show the binding kinetics. The bands for free probe and protein-DNA complex are indicated. (D) Tiled substitutions in a shorter probe derived from probe #3 were used as competitors in the EMSA to identify the specific position of Fru-DBD-B binding. The sequence of the labeled probe and the mutant sequences used as competitors are listed below the gel with the substituted nucleotides indicated in red lowercase. (E) Single base substitutions in the probe were used as competitors in the EMSA to examine the contribution of each position to the Fru-DBD-B binding site.
Fig. S4.
(Related to Fig. 3.) fru microRNA effects on R19B09-LexA expression. (A) Expression of different isoforms of fru transcript were inhibited by pan-neuronal microRNA expression and expression of R19B09-LexA in the male central brain was observed, with the genotype R19B09-LexA, LexAop-myrGFP, R57C10-GAL4, fru mi X. fru mi 1 and 2 target male-specific fru transcripts, whereas fru mi A–C target fru Zn finger isoforms. Scrambled controls for most fru microRNAs were included. Images were captured at 20× magnification. (B) Quantification of genotypes shown in A. n = 6–16 brain hemispheres per genotype. Error bars indicate SEM.
Having delimited a relatively short Lgr3 fragment that confers regulation by FruM, we asked whether this regulation is direct. To do so, we performed EMSAs examining the binding of the Fru A, B, and C DNA-binding domains (DBDs) to probes tiled across the R19B09.3A fragment (Fig. 3 C–E and Fig. S5). We observed several regions that are bound by Fru-DBD-A (Fig. S5A) or Fru-DBD-B domains (Fig. 3C), suggesting Fru DBDs can directly bind sequences from the Lgr3 gene in vitro.
Fig. S5.
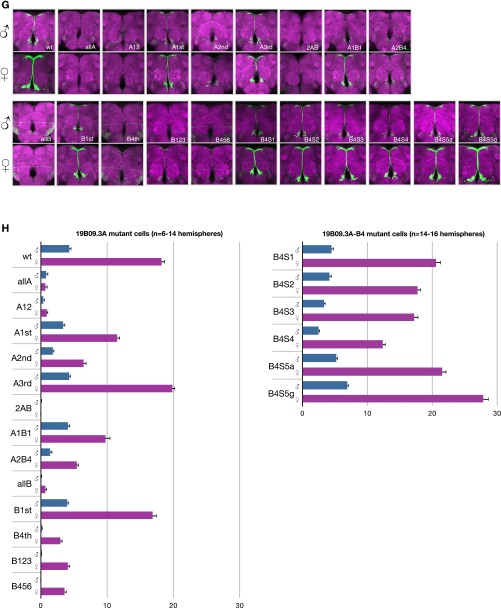
(Related to Fig. 3.) R19B09.3A Fru binding and mutations. (A) Fru-DBD-A shows binding to R19B09.3A probes #5 and #7 but weak or no binding to other probes by EMSA. There appear to be multiple bands for the DNA–protein complex (as indicated), most likely a result of the partial degradation of the fusion protein. Data are shown for five of the nine tested probes. (B) Fru-DBD-A binding to a shorter probe derived from probe #5 was assayed in competition with probes containing tiled substitutions. The labeled probe consisted of the wild-type (wt) sequence in the lower part of the figure. The mutant sequences used as competitors are listed below with the substituted nucleotides indicated in red. The wild-type competitor effectively competed with probe binding, but m1 and m2 mutations prevented competition, suggesting these regions are necessary for Fru-DBD-A binding. (C) Sequences of probes tiling R19B09.3A used for EMSA. (D) Diagram of probes from C tiling across R19B09.3A. (E) Summary of the potential Fru-DBD-A and Fru-DBD-B binding sites identified by EMSA in R19B09.3A. The binding sites for Fru-DBD-A are indicated in red and the binding sites for Fru-DBD-B are indicated in blue. (F) Mutations made for the Fru binding sites in the R19B09.3A sequence. The positions of binding sites for Fru-DBD-A are indicated in red and the positions of binding sites for Fru-DBD-B are indicated in blue. The mutated nucleotides at each binding site are indicated in lowercase. (G) Mutations were made in various combinations to assay effects in vivo based on EMSA results, as labeled. For example, “allA” included mutations at all Fru-DBD-A sites, whereas “A12” mutated Fru-DBD-A sites 1 and 2. “B4S1” through “B4S5g” were single base pair changes across the Fru-DBD-B binding site 4, with “B4S5a” being a mutation of C to A and “B4S5g” being a mutation of C to G. Images were captured at 20× magnification. (H) Cell counts from G. Error bars indicate SEM.
We subdivided each bound probe roughly into thirds and used additional EMSAs to further refine the regions important for Fru binding. In addition, we designed competitor oligos with mutations tiled across the smaller probes (Fig. 3D and Fig. S5B). Competitors with mutations at residues unimportant for binding outcompete labeled probes, reducing or eliminating the labeled probe/protein complex. Competitors with mutations at residues important for binding leave the complex unaffected, allowing identification of necessary bases (e.g., competitor m4 in Fig. 3D). We made single base mutations in the competitor to provide maximal resolution of binding specificity (Fig. 3E). Potential Fru binding sites have been explored previously (15, 16). The sites we identified generally agree with the consensus sites previously reported. Notably, although Dalton et al. (15) identified Fru binding motifs in Lgr3, it did not rise above their thresholds for responding to Fru expression, perhaps because of the small number of neurons affected.
Anticipating that binding at multiple sites may be necessary for Fru function, we mutated our identified sites individually and in multiple combinations in the R19B09.3A reporter (Fig. S5 E and F). Examining these combinations in females and males identified several sites necessary for expression in both sexes, but no tested single mutation or combination thereof increases expression in males to female-like levels (Fig. S5 G and H). As the fourth FruB binding site has particularly strong effects on expression, we explored further single base changes across it, but observed minimal effects on expression (Fig. S5 G and H). We hypothesize that a transcriptional activator may bind the same sites as Fru, such that mutations of the shared binding site reduce or eliminate expression in both sexes. Alternatively, the full-length FruM protein in vivo could require a longer binding site or additional cofactors for binding.
DsxF Activates and Inhibits Lgr3 Expression in Different Abdominal Ganglion Neurons.
Although the sexually dimorphic pattern of Lgr3 expression in the median bundle is relatively straightforward, with females having broader expression than males, we observed a more complex Lgr3-GAL4 pattern in the abdominal ganglion, where both females and males express Lgr3-GAL4, but in different patterns (Fig. 4A). Mutants in fruM did not appear to change these Lgr3-GAL4 abdominal ganglion expression patterns (Fig. S6A), but mutations in dsx did alter these patterns (Fig. 4A). As with Lgr3 expression in the brain median bundle, we examined the expression of Lgr3 enhancer fragment lines (Fig. 2A) to determine if any confer sex-specific expression in the abdominal ganglion. Of the five lines examined, R17G11-GAL4 and R17H01-GAL4 replicate subsets of the Lgr3 pattern in the abdominal ganglion. As before, we examined how these two enhancer fragments relate to Lgr3, fru, and dsx to determine the basis of their sexually dimorphic expression and how it relates to the sexual dimorphisms seen in Lgr3.
Fig. 4.
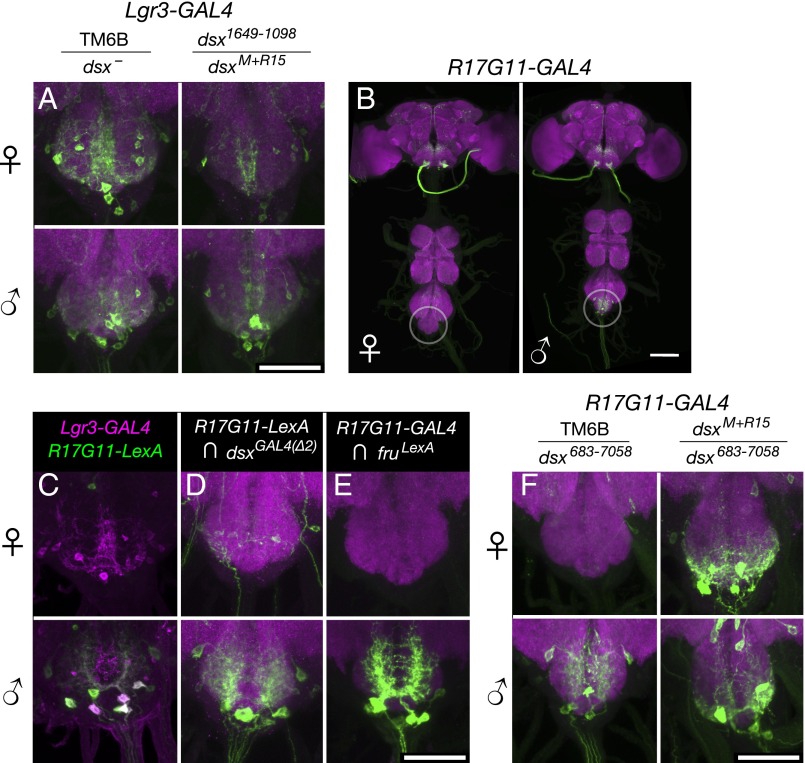
Lgr3-GAL4 and R17G11-GAL4 abdominal ganglion expression depend on DsxF. (A) Lgr3-GAL4 (attP40), UAS-myrGFP (su(Hw)attP5) expression in the female and male abdominal ganglion. Left column is heterozygous for the TM6B balancer and either dsxM+R15 or dsx1649−1098. Right column is heterozygous for dsxM+R15 and dsx1649−1098. (Scale bar, 50 µm.) (B) R17G11-GAL4 (attP2), UAS-myrGFP (attP40) expression in females and males. Sexually dimorphic abdominal ganglion expression is circled. The brain and VNC were imaged in separate 20× tiles and composited. (Scale bar, 100 µm.) (C) Expression of R17G11-LexA::p65 (attP40) and Lgr3-GAL4 (VK33) driving UAS-IVS-mCD8::RFP (attP18) and LexAop2-mCD8::GFP (su(Hw)attP8) in females and males. Coexpression is seen in males but not females. (D) Intersection of R17G11-LexA::p65 (attP40) with dsxGAL4(∆2) using LexAop2-FlpL (attP40) and UAS > stop > myrGFP (su(Hw)attP1). (E) Intersection of R17G11-GAL4 (attP2) with fruLexA using LexAop2-FlpL (attP40) and UAS > stop > myrGFP (su(Hw)attP1). (Scale bar, 50 µm.) (F) R17G11-GAL4 (attP2), UAS-myrGFP (attP40) expression in the female and male abdominal ganglion with dsx683−7058 and either the TM6B balancer or dsxM+R15. (Scale bar, 50 µm.)
Fig. S6.
Fru regulation of Lgr3 abdominal ganglion expression. (A) Lgr3-GAL4 (attP40), UAS-myrGFP (su(Hw)attP5) expression in the female and male abdominal ganglion. Left column is heterozygous for the TM6B balancer and fruLexA. Right column is heterozygous for fru4−40 and fruLexA. (Scale bar, 50 µm.) (B) R17G11-GAL4 (attP2), UAS-myrGFP (attP40) expression in the female and male abdominal ganglion. Left column is heterozygous for the TM6B balancer and fruLexA. Right column is heterozygous for fru4-40 and fruLexA. (Scale bar, 50 µm.) (C) R17H01-GAL4 (attP2), UAS-myrGFP (attP40) expression in the female and male abdominal ganglion. Left column is heterozygous for the TM6B balancer and fruLexA. Right column is heterozygous for fru4-40 and fruLexA. (Scale bar, 50 µm.)
The first of the two fragments mentioned above, R17G11-GAL4, is expressed in several abdominal ganglion cells in males, but not in females (Fig. 4B). R17G11-GAL4 also directs nondimorphic expression in the brain. To compare the abdominal ganglion expression driven by the R17G11 enhancer fragment to that driven by Lgr3-GAL4 and dsxGAL4(∆2), we replaced the GAL4 sequence in R17G11-GAL4 with LexA to make R17G11-LexA. R17G11-LexA showed coexpression with Lgr3-GAL4 in the male abdominal ganglion, but not in the female (Fig. 4C). We examined the intersection between R17G11-LexA and dsxGAL4(∆2) as well as between R17G11-GAL4 and fru-LexA, and in both cases we observed expression in the male abdominal ganglion, but little or none in females (Fig. 4 D and E). Examination of R17G11-GAL4 expression in fruM and dsx loss-of-function mutants indicated that Dsx (Fig. 4F), but not FruM (Fig. S6B), regulates R17G11. Unlike Fru, Dsx has functional forms in both sexes. As loss-of-function mutations of dsx led to expression of R17G11-GAL4 in females, it appears that the female form of Dsx, DsxF, either inhibits R17G11 expression or the survival of these neurons in females.
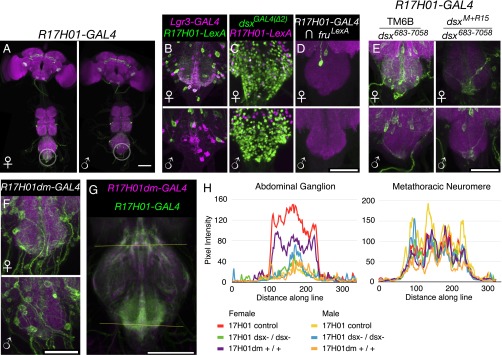
The second of the two fragments mentioned above, R17H01-GAL4, showed a pattern of regulation in the abdominal ganglion opposite to that of R17G11-GAL4. R17H01-GAL4 has expression in several abdominal ganglion cells in females, but fewer cells in males (Fig. 5A). As with R17G11-GAL4, we replaced the GAL4 in R17H01-GAL4 with LexA to make R17H01-LexA to compare it with Lgr3-GAL4 and dsxGAL4(∆2). Coexpression of R17H01-LexA with Lgr3-GAL4 was seen in females, but not in males (Fig. 5B). Comparisons of the expression patterns of R17H01-LexA with dsxGAL4(∆2) and R17H01-GAL4 with fru-LexA indicated minimal coexpression with fruM, but coexpression with dsx especially in females (Fig. 5 C and D). Examination of R17H01-GAL4 expression in fruM and dsx loss-of-function mutants indicated that Dsx (Fig. 5E), but not FruM (Fig. S6C), regulates R17H01-GAL4. Loss-of-function mutations of dsx largely eliminate R17H01-GAL4 expression in the female abdominal ganglion but have no clear effect in males. Thus, it appears that DsxF again plays the controlling role, but in this case it activates R17H01 expression in females, as opposed to the inhibition of R17G11. The effects of DsxF on Lgr3-GAL4 expression in the abdominal ganglion may be the simple additive sum of its effects on R17H01 and R17G11, with each enhancer driving expression in the appropriate subsets of neurons.
Fig. 5.
R17H01-GAL4 expression depends on DsxF. (A) R17H01-GAL4 (attP2), UAS-myrGFP (attP40) expression in females and males. Sexually dimorphic abdominal ganglion expression is circled. The brain and VNC were imaged in separate 20× tiles and composited. (Scale bar, 100 µm.) (B) Expression of R17H01-LexA::p65 (attP2) and Lgr3-GAL4 (VK33) driving UAS-IVS-mCD8::RFP (attP18) and LexAop2-mCD8::GFP (su(Hw)attP8) in females and males. Coexpression is seen in females but not males. (C) R17H01-LexA::p65 (attP2), LexAop2-IVS-nlstdTomato (VK22) has some colocalization with dsxGAL4(∆2), UAS-Stinger-GFP in females, but less so in males. (D) Intersection of R17H01-GAL4 (attP2) with fruLexA using LexAop2-FlpL (attP40) and UAS > stop > myrGFP (su(Hw)attP1). (Scale bar, 50 µm.) (E) R17H01-GAL4 (attP2), UAS-myrGFP (attP40) expression in the female and male abdominal ganglion with dsx683−7058 and either the TM6B balancer or dsxM+R15. (Scale bar, 50 µm.) (F) R17H01dm-GAL4 (attP2), UAS-myrGFP (attP40) expression in the female and male abdominal ganglion. (Scale bar, 50 µm.) (G) Line scans of R17H01-GAL4 and R17H01dm-GAL4 expression in the metathoracic neuromere (upper line) and abdominal ganglion (lower line). R17H01-GAL4 is shown in green, and R17H01dm-GAL4 is in magenta. For each group (see H, below), five to seven VNCs were registered based on nc82, averaged together, and partially z-projected in preparation for line scans. (Scale bar, 100 µm.) (H) Line scan profiles of R17H01-GAL4 and R17H01dm-GAL4 expression in the abdominal ganglion and metathoracic neuromere. R17H01dm-GAL4, UAS-myrGFP (attP2) females and males were compared with a mixture of R17H01-GAL4, UAS-myrGFP (attP40) heterozygous controls (dsx1649−1098/+, dsx683−7058/+, and fruLexA/+) and R17H01-GAL4, UAS-myrGFP (attP40) dsx mutants (dsx683−7058/dsxM+R15, dsx1649−1098/dsxM+R15, and dsx1649−1098/dsx1).
The above results indicate that Dsx regulates Lgr3 expression in the abdominal ganglion, but do not reveal whether this regulation is direct, or a result of indirect effects such as Dsx-dependent cell death. We attempted to address this question, as described below. Previous work identified regions of likely Dsx DNA-binding activity and a consensus 13-bp Dsx binding motif (GCAACAATGTTGC) (5, 13, 32). The Lgr3 locus contains three potential Dsx binding sites: a 9/13-bp partial binding site match in R17G11 (GagACAATGTgaC, with mismatches in lowercase), and two 10- and 11-bp partial matching sites in R17H01 (GCAACAtTGaaGt and GttACAtTGTTGC). We focused on R17H01 because of its closer matches to the 13-bp motif identified previously (5). We mutated these putative binding sites to disrupt Dsx binding, and hence suggest whether Dsx directly regulates R17H01 expression. Mutation of the R17H01-GAL4 Dsx binding sites (changed to GCAgtgccaggGT and GTTACgccaccGC, respectively, with mutated bases in lowercase) created R17H01dm-GAL4, where “dm” denotes the presence of mutated DSX binding sites.
R17H01dm-GAL4 driving UAS-GFP yielded a moderate decrease in female abdominal ganglion expression compared with R17H01-GAL4 and no change in males (Fig. 5 F–H). Because the change in expression in Fig. 5F is subtle, several ventral nerve cords (VNCs) were registered together, and expression of R17H01dm-GAL4 in the abdominal ganglion was compared with R17H01-GAL4 in wild-type and dsx loss-of-function mutants (Fig. 5G). To quantitate the changes in expression, a line scan analysis was performed along the two lines shown in Fig. 5G, the upper control line and the lower line across the abdominal ganglion (Fig. 5H). R17H01dm-GAL4 shows a reduction in expression intensity across the female abdominal ganglion, but not across the mesothoracic neuromere. This reduction was intermediate between control and dsx loss-of-function mutant R17H01-GAL4 females. Thus, it appears that DsxF may directly regulate a fraction of R17H01-GAL4, and hence Lgr3, expression. However, other aspects of Lgr3 abdominal ganglion expression may be regulated indirectly or by enhancers in R17H01 outside the Dsx binding sites. The latter case was recently observed with Dsx regulation of Fmo-2 expression, in which both a Dsx binding site and nearby enhancers were necessary for the full pattern of sexually dimorphic expression (12).
Female Lgr3-Expressing Neurons Regulate Sexual Behavior.
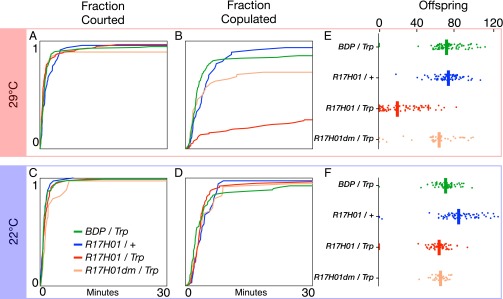
To begin to understand the functional significance of sex-specific Lgr3 regulation, we stimulated Lgr3 neurons using the thermogenetic activator UAS-dTrpA1 and assayed changes in courtship behavior. We placed virgin male and female pairs in courtship chambers and (i) measured latency to courtship initiation by males (indicated by their first wing extension directed at females), (ii) measured latency to successful copulation, and (iii) looked for other obvious changes in behavior. Activation in females of R17H01-GAL4 neurons inhibits female receptivity to male courtship, as indicated by delayed or no copulation during the observation period (Fig. 6). Control females consisted of pBDPGAL4u (which lacks an enhancer insertion) with UAS-dTrpA1 and R17H01-GAL4 without UAS-dTrpA1. Performance of the two controls was not significantly different (P = 0.2 for courtship latency and P = 0.4 for copulation latency, by log-rank test, n = 130 and 47, respectively), and the genotypes were pooled for further comparisons. Although males initiated courtship of R17H01 > dTrpA1 and control females at indistinguishable speeds (P = 0.15, log-rank test) (Fig. 6A), R17H01 > dTrpA1 female receptivity to courtship was significantly reduced at the Trp-activating temperature of 29 °C (P < 0.001, log-rank test, n = 116–177) (Fig. 6B). No difference was seen between the experimental and control groups in courtship or copulation latency at 22 °C, where Trp is inactive (courtship latency P = 0.3, copulation latency P = 0.6, log-rank test, n = 78–115) (Fig. 6 C and D).
Fig. 6.
Activation of R17H01-GAL4 neurons reduces female receptivity and fecundity. Canton-S males were individually placed with females in courtship chambers. Latency to initiation of male courtship, measured by first wing extension, and successful copulation were measured at (A and B) 29 °C and (C and D) 22 °C. Pairs were transferred to a fly vial with food and maintained at (E) 29 °C or (F) 22 °C until offspring were counted at the pharate pupal stage. R17H01-GAL4, UAS-dTrpA1 females had significantly delayed copulation and reduced fecundity at 29 °C. Survival plots in (A–D) show cumulative percentage completion over 30 min. Fecundity plots in (E and F) show each female's number of offspring, along with a bar indicating the average for each group.
After observing their courtship, as described above, we briefly anesthetized the male/female pairs and placed them into food vials to measure their fecundity. Pairs were maintained at 29 °C or 22 °C until offspring were counted at the pharate adult stage. R17H01 > dTrpA1 females displayed significantly lower fecundity than pBDPGAL4u > dTrpA1 or R17H01/+ females at 29 °C (P < 0.001 for both, Kruskal–Wallace test then Dunn post hoc with Bonferroni correction, n = 82–90) (Fig. 6E). This difference was not simply because of a failure to copulate: rather than being a mix of completely infertile and unaffected females, most R17H01 > dTrpA1 females produced fewer offspring, (Fig. 6E). At 22 °C, fecundity was somewhat variable between control genotypes, but all were significantly above R17H01 > dTrpA1 at 29 °C (P < 0.001 for each, Kruskal–Wallace test then Dunn post hoc with Bonferroni correction) (Fig. 6F).
Although the effects of Dsx-binding-site mutations on R17H01dm-GAL4 expression were subtle, we asked whether they were sufficient to alter the phenotype observed in tests of R17H01 > dTrpA1 females (Fig. 6). At 29 °C, R17H01dm > dTrpA1 females were courted as promptly as R17H01 > dTrpA1 females (P = 0.2, log-rank test, n = 48–116) (Fig. 6A) but showed a strong increase in receptivity (P < 0.001, log-rank test) (Fig. 6B). R17H01dm > dTrpA1 female receptivity was statistically indistinguishable from controls (P = 0.3, log-rank test, n = 48–178) (Fig. 6B). At 22 °C there was a slight, but significant, delay in courtship toward R17H01dm > dTrpA1 females (P = 0.043, log-rank test, n = 36–78) (Fig. 6C), but no difference in receptivity (P = 0.3, log-rank test) (Fig. 6D). Similarly, R17H01dm > dTrpA1 females showed a significant recovery in fecundity at 29 °C compared with R17H01 > dTrpA1 (P < 0.001, Kruskal–Wallace test then Dunn post hoc with Bonferroni correction, n = 47–90) (Fig. 6E), but no difference at 22 °C (P = 1, Kruskal–Wallace test then Dunn post hoc with Bonferroni correction, n = 36–69) (Fig. 6 E and F). Thus, although GFP expression was only partially reduced in R17H01dm flies, it appears that the Dsx binding site-dependent effects on R17H01 expression account for most or all of its effects on female receptivity and fecundity.
Other recent studies have also examined the abdominal ganglion neurons necessary for female reproductive behavior in Drosophila (18, 19, 33–35). One question is whether the neurons we report here are the same or different from those previously reported. Gou et al. (33) focused on ascending neurons from the reproductive tract, and both Feng et al. (34) and Bussell et al. (35) reported stimulation of neurons favoring receptivity. In contrast, we observed that stimulation of R17H01-GAL4 neurons inhibits receptivity, suggesting the neurons examined here differ from the ones reported above.
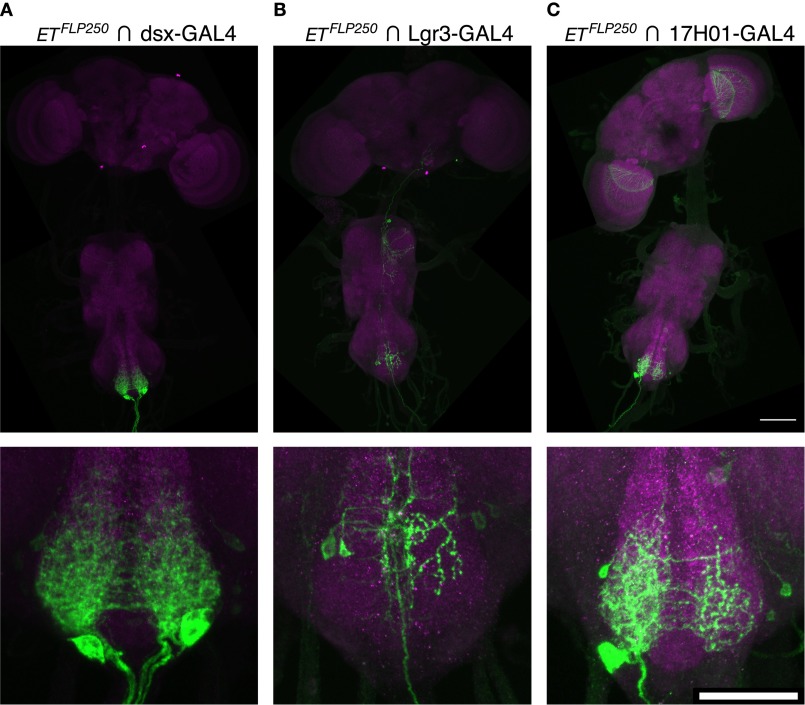
Rezával et al. also identified two populations of Dsx-expressing neurons in the female abdominal ganglion that when activated cause females to become less receptive (18, 19). The relationship between the two populations, labeled by either the ETFLP250 enhancer trap flippase or octopaminergic/tyraminergic Tdc2-GAL4 driver, is not entirely clear, but their overall morphology does not strongly resemble that of Lgr3-GAL4 or R17H01-GAL4. We compared the intersection of ETFLP250 and dsxGAL4(∆2) with that of ETFLP250 and Lgr3-GAL4 or R17H01-GAL4 (Fig. S7). ETFLP250 intersected with R17H01-GAL4 does show a pattern similar to a subset of ETFLP250 intersected with dsxGAL4(∆2), but ETFLP250 intersected with Lgr3-GAL4 does not. Thus, it appears that ETFLP250 coexpresses with separate populations of neurons in the two drivers, despite their otherwise strong correspondence (Fig. 5B). It is also notable that Rezával et al. (18) observed a strong effect of ETFLP250, dsxGAL4, UAS > stop > TrpA1 in females inhibiting the initiation of male courtship, whereas we observed prompt courtship initiation toward R17H01-GAL4, UAS-dTrpA1 females. As a result, although we cannot exclude the possibility that our observed reproductive phenotypes result from effects on the same neurons, it appears unlikely.
Fig. S7.
(Related to Fig. 6.) Intersections with ETFLP250. Full CNSs are shown above, with enlarged abdominal ganglia below. The brain and VNC were imaged in separate 20× tiles and composited. (A) ETFLP250 intersected with dsxGAL4(∆2) and UAS > stop > myrGFP (attP40) in females. (B) ETFLP250 intersected with Lgr3-GAL4 and UAS > stop > myrGFP (attP40) in females. (C) ETFLP250 intersected with R17H01-GAL4 and UAS > stop > myrGFP (attP40) in females. (Scale bars, 100 µm, Upper and 50 µm, Lower).
Discussion
We have shown that DsxF and FruM regulate the expression of Lgr3 in separate populations of Drosophila neurons. FruB inhibits Lgr3 expression in the median bundle in males, whereas DsxF inhibits Lgr3 expression in one population of abdominal ganglion neurons and activates expression in another population. This combined regulation of Lgr3 marks a point of convergence in sex determination after the Dsx/Fru split. Furthermore, we found that activation of Lgr3-expressing neurons inhibits female receptivity toward male courtship and lowers female fecundity, indicating the functional importance of these neurons.
In vitro binding of Fru-DBD-B to R19B09.3A suggests FruM could directly regulate Lgr3 expression in vivo in the median bundle. However, the functional consequences of this regulation remain unclear. FruM-expressing median bundle neurons have previously been shown to play an important role in coordinating steps of male courtship (36). Preventing Lgr3 expression in these neurons may play a role in specifying their male-specific function. Alternatively, Lgr3 median bundle expression could help specify female behavior, with FruM inhibition in males preventing a male-specific side effect.
Expression analysis of Drosophila Lgr3 and Lgr4 at the level of whole tissue was recently reported (37). Although significant differences in expression were reported between males and females, these were outside the central nervous system. The largest reported adult dimorphisms were higher male expression in salivary glands and higher female expression in the fat body. A lack of nervous system changes reported by Van Hiel et al. (37) may not conflict with our results, because only a small number of neurons were affected in our observations, and the changes in the abdominal ganglion were bidirectional.
The distant homology of Lgr3 to mammalian relaxin receptors RXFP1 and RXFP2 presents the possibility of related function, as well as related structure. These receptors and their ligands, relaxin and insulin-like peptide 3, respectively, are necessary for normal reproductive physiology in the male testis and female ovary, along with other functions (reviewed in refs. 23 and 38). This homology has led to the proposal that insulin-like peptides, especially Dilp8, may function as ligands for Lgr3 in Drosophila (39). Dilp8 has been implicated in the regulation of developmental timing and growth in the larva (40, 41), and Lgr3 has recently been found to be necessary for this Dilp8 function (42–44). Lgr3 mutants appear to develop similarly to dilp8 mutants and prevent effects of Dilp8 overexpression, and some evidence has been found for direct interaction between them (44; but see ref. 43).
It is unclear whether Dilp8 signaling is also important for Lgr3’s reproductive function, but it is notable that Dilp8 is highly expressed by the adult female ovary (45). Thus, we hypothesize that the ovaries may signal their state via Dilp8 to Lgr3-expressing neurons in the abdominal ganglion, which then regulate aspects of female reproductive behavior. Regulation of Lgr3 expression by the sex-determination hierarchy may thus pattern the neural components of a sex-specific signaling pathway.
Materials and Methods
Transgenic Drosophila.
Lgr3-GAL4::VP16 was generated by recombination-mediated replacement of the coding portion of the first Lgr3 coding exon with GAL4::VP16 in a bacterial artificial chromosome, which was then inserted into the Drosophila genome at defined attP sites. Other GAL4 reporter stocks were generated by standard methods (46). See SI Materials and Methods for detailed transgenic methods.
Immunohistochemistry, in Situ Hybridization, and Microscopy.
For immunohistochemistry, adult fly central nervous systems were dissected in Schneider’s insect medium and fixed in 4% (wt/vol) paraformaldehyde. Primary and secondary antibodies were applied to label neuropil and neuronal membranes. For in situ hybridization, pharate pupal flies were frozen in OCT compound, cryostat sectioned, and transferred to adhesive slides. They were exposed to DIG-labeled sense and antisense probes from Lgr3 and processed with the PerkinElmer TSA Plus Cyanine 3 System. GFP was then labeled by standard immunohistochemical methods. All samples were imaged on a Zeiss LSM 710 and processed using Fiji and Computational Morphometry Toolkit. See SI Materials and Methods for a detailed description.
Protein Expression and EMSA.
Fru A, B, and C DNA-binding domains were obtained by RT-PCR from wild-type flies and expressed in bacteria. EMSA was performed as described previously (5). See SI Materials and Methods for further description.
Courtship and Fecundity Assays.
Four- to 7-d-old flies were aspirated into standard courtship chambers and allowed to recover. The divider between males and females was removed, and the latency until courtship initiation and successful copulation were measured. After filming, flies were CO2-anesthetized and moved into standard food vials. Numbers of pupal offspring were counted one week later. See SI Materials and Methods for a full description of behavioral methods.
SI Materials and Methods
Fly Stocks.
Flies were raised on standard corn meal molasses food. The stocks used in this paper included the following: R19B09-GAL4, R17G11-GAL4, R17H01-GAL4, and their LexAop2::p65 versions (30, 47); UAS-Stinger-GFP (48); LexAop > tdTomato (27); LexAop-Stinger-GFP (49); UAS-RedStinger (50); pJFRC106-13XLexAop2-IVS-nlstdTomato (VK22) (gift of Barret D. Pfeiffer, Rubin Lab, Janelia Research Campus, Ashburn, VA); fruLexA (27); dsxGAL4(∆2) (51); fru4-40 (52); pJFRC79-8XLexAop2-FlpL (attP40) (53); pJFRC41-10XUAS > STOP > myrGFP (su(Hw)attP1) (47); dsx683-7058 (Df(3R)f00683-d07058) (6, 54); dsxM+R15 (55); dsx1 (56); pJFRC12-10XUAS-myrGFP (attP2, attP40, su(Hw)attP5) (47); UAS-dTrpA1 (57); Canton-S A (gift of Jeff Hall, Brandeis University, Waltham, MA); pBDPGAL4u (attP2) (46); fru∆A, fru∆A2, fru∆B, fru∆B2, fru∆C (15, 31); pJFRC21-10XUAS-IVS-mCD8::RFP (attP18) (58); pJFRC15-13XLexAop2-mCD8::GFP (su(Hw)attP8) (47, 58); and ETFLP250 (18).
Deficiency dsx1649-1098 (Df(3R)f01649-e01098) was generated by FLP/FRT-mediated, transchromosomal recombination between the FRT elements in PBac{RB}e01098 and PBac{WH}dsxf01649, following the method of Parks et al. (59). This deficiency specifically deletes all dsx-coding sequences but leaves the final male-specific exon (corresponding to the male 3′UTR), as well as all neighboring genes intact. This deficiency is marked with a single miniwhite (mw) transgene. R17H01dm-GAL4 was generated by PCR mutagenesis of R17H01-GAL4 to replace Dsx binding site GCAACATTGAAGTT with GCAgtgccaggGT and site GTTACATTGTTGC with GTTACgccaccGC.
Lgr3-GAL4::VP16 (VK33 and attP40) was generated by replacement of the first Lgr3 coding exon with GAL4::VP16. Generic landing-site cassette pGKU was created by flanking the RpsL-kana (60) selectable marker with ∼500-bp 5′ and 3′ homology arms from the GAL4 gene, including HSP70 termination sequences, amplified from pBPGUw (46). An Lgr3-specific cassette was created by amplifying the generic cassette from pGKU using primers that added 50-base gene-specific homology arms to the GAL4-RpsL::kana cassette. The upstream primer comprised the 50 bases up to (not including) the translation-initiating ATG of Lgr3 (lowercase below), followed by GAL4 annealing sequences, starting with the “ATG” (capitalized); the 3′ arm comprised the reverse complement of the 50 bases immediately following the end of the first coding exon, followed by sequences from the cassette terminator; Lgr3-F: agtaattagataagcgagcgtgcaaaacaggagcaaaccgataaatcgccATGAAGCTACTGTCTTCTATCGAACAAGC; Lgr3-R: taggtttgcgaatgatggaattttgaaaggacatcactctaaggactcacGATCTAAACGAGTTTTTAAGCAAACTCACTCCC.
The cassette was amplified with Phusion proofreading polymerase (New England Biolabs), gel-purified, and recombined into BAC clone CH322-155A11 (61) in SW102 cells using methods similar to those described previously (62). Recombinants were selected on kanamycin medium. Putative clones were analyzed by colony PCR to ensure that proper recombination had occurred. Full-length GAL4::VP16, amplified from pBPGAL4.2::VP16Uw, a gift from Gerald Rubin, Janelia Research Campus, Ashburn, VA (Addgene plasmid #26228) (47), was recombined into the modified BAC across the 500-bp homology arms added earlier. Potential recombinants were identified by faster growth on streptomycin medium. Clones were analyzed by colony PCR, and putative recombinant BACs were sequenced at the recombined regions. Correct BACs were isolated and electroporated into EPI300 cells (Epicentre), and EPI300 clones were shipped to Genetic Services or Rainbow Transgenic Flies as agar stabs for integration. Integrant flies were analyzed by PCR to ensure proper integration of the transgene.
Immunohistochemistry.
Zero- to 3-d-old flies were anesthetized with CO2 then placed on ice until dissection in cold Schneider’s (S2) insect medium (Sigma). Isolated CNSs were fixed in 4% (wt/vol) paraformaldehyde in S2 medium for 30 min at room temperature on a rotator, and incubated in PAT (0.5% BSA, 0.5% Triton X-100, 1× PBS) at 4 °C overnight.
Two 15-min PAT washes at room temperature were followed by blocking for 1 h in PAT with 3% (vol/vol) normal goat serum (NGS) at room temperature. Two additional PAT washes were followed by the addition of primary antibodies in PAT+NGS at room temperature for 3 h, then 12–36 h at 4 °C. Primary antibodies consisted of either mouse anti-Bruchpilot nc82 at 1:30 (Developmental Studies Hybridoma Bank) and rabbit anti-GFP at 1:1,000 (Life Technologies A-11122) or rabbit anti-RFP at 1:500 (Life Technologies R10367) and chicken anti-GFP at 1:1,000 (Life Technologies A-10262).
Three additional 15-min room temperature PAT washes were followed by the addition of secondary antibodies in PAT+NGS at room temperature for 3 h then 48–96 h at 4 °C. Secondary antibodies consisted of either Alexa Fluor 568 goat anti-mouse at 1:200 (Life Technologies A-11031) and Alexa Fluor 488 goat anti-rabbit at 1:1,000 (Life Technologies A-11034), or Alexa Fluor 568 goat anti-rabbit at 1:500 (Life Technologies A-11036) and Alexa Fluor 488 goat anti-chicken at 1:500 (Life Technologies A-11039). After four final 15-min room temperature washes in PAT, samples were mounted in VECTASHIELD (Vector Laboratories) on glass slides with coverslip spacers, and sealed with nail polish (Ted Pella). Samples were imaged using a Zeiss LSM 710 confocal microscope at 20× using ZEN software and MultiTime macros. Images were processed with Fiji software (63). Unless otherwise specified (see registration description, below), all images are full z-projections through a sample.
Registration and Line Scans.
To compare expression in the abdominal ganglion, VNCs were registered based on nc82 labeling using the Computational Morphometry Toolkit (64). A nc82 template was created by iterative shape averaging of 20 dorsal-up VNCs. Five to seven VNCs for each group (Fig. 5 G and H) were aligned to the template using Computational Morphometry Toolkit nonrigid registration. The VNCs for each group were averaged together in three dimensions using Fiji software. A z-projection was made of each averaged group from slice 27 (a short distance into the neuropil from the dorsal side) to the end of the image (slice 109). An intensity profile was measured along one of two lines across either the terminal abdominal ganglion or the metathoracic neuromere.
In Situ Hybridization.
Probes were designed to tile across the Lgr3 transcript. For each probe, DNA was PCR-amplified from FlyBase clone RE38148 (FlyBase ID: FBcl0230445, provided by the Drosophila Genomics Resource Center). T7 and T3 sites were added to the forward and reverse primers, respectively, for the production of sense and antisense transcripts. DIG-labeled RNA probes were produced using DIG RNA Labeling Mix (Roche). Initially two to three probes were pooled together in three groups and examined in females, where Lgr3 expression is stronger. We focused on probe 3 (primers TAATACGACTCACTATAGGGtggccgccgtgatattctacttga and AATTAACCCTCACTAAAGGGcagctccaggcgttccaggtga), targeting an early 571-bp section of the Lgr3 coding region, based on strong labeling and low background. The sense control was a pool of sense probe 2 (TAATACGACTCACTATAGGGcgaacgcgtttggtccttttgtcc and AATTAACCCTCACTAAAGGGagaatatcacggcggccagcagaa) and sense probe 3 (TAATACGACTCACTATAGGGtggccgccgtgatattctacttga and AATTAACCCTCACTAAAGGGcagctccaggcgttccaggtga).
Pharate pupae were sorted by sex and embedded in Tissue-Tek OCT compound (Sakura Finetek) then quickly frozen on dry ice and stored at −20 °C to −80 °C until sectioning. Next, 10-μm horizontal sections were cut at −30 °C on a cryostat and transferred to adhesive slides using the CryoJane Tape-Transfer System (Leica Biosystems).
Sections were fixed for 1 h in RNase free 4% (wt/vol) paraformaldehyde and washed twice for 5 min in RNase-free PBST (PBS with 0.1% Triton X-100). Sections were passed through the following series of RNase-free methanol solutions for 2 min each: 25%, 50%, 75%, 100%, 75%, 50%, and 25% (vol/vol) methanol. After two more PBST washes, sections were acetylated by 5 min of 50 mL 0.1 M TEA (triethanolamine, HCl) followed by the addition of 125 µL acetic anhydride for 10 min and two more PBST washes. Sections were prehybridized for 1 h at 65 °C in hybridization buffer [25% (vol/vol) formamide, 5× SSC, 5× Denhardts, 0.25 mg/mL yeast tRNA, 0.5 mg/mL herring sperm DNA, 0.05 mg/mL heparin sodium, 2.5 mM EDTA, 0.1% Tween 20, and 2.5 mg/mL CHAPS].
DIG-labeled probes diluted 1:10,000 in hybridization buffer were denatured for 5 min at 80 °C, chilled on ice, pipetted onto the sections, and coverslipped using Hybrislip plastic coverslips. Slides were hybridized horizontally in slide mailers in a humidified box at 65 °C for 16–18 h. Coverslips were removed by soaking slides in 5× SSC at 65 °C, followed by three 20 min washes in 0.2× SSC at 65 °C. Sections were rinsed in 5 µg/mL RNase A in 2× SSC for 15 min at room temperature, then equilibrated in TN (100 mM Tris, 150 mM NaCl) buffer for 5 min and blocked in TN buffer plus 0.5% Blocking Reagent (PerkinElmer) for 30 min.
Antidigoxigenin-POD (Roche) at 1:500 in blocking buffer was applied, followed by coverslips and a 30-min incubation. Sections were washed in TNT (TN buffer plus 0.05% Tween 20) three times for 5 min. Cy3-Tyramide (PerkinElmer TSA Plus Cyanine 3 System) at 1:50 in TSA Amplification Reagent was applied to sections, followed by coverslips, a 10-min incubation, then three 5-min washes in TNT. To label GFP, sections were blocked in 0.1% Triton, 2% (wt/vol) BSA, 1× PBS for 30 min, then chicken anti-GFP (Aves Labs) at 1:1,000 in blocking buffer was applied overnight at 4 °C. After additional washes in blocking buffer, goat anti-chicken at 1:500 in blocking buffer was added for 2 h at room temperature. After final washes, VECTASHIELD with DAPI was added and sections were coverslipped. Sections were imaged at 20× on a Zeiss LSM 710 confocal microscope, with variable gain between samples.
R19B09 Fragments.
R19B09 was subdivided by PCR into overlapping thirds in two rounds, to create R19B09.1, R19B09.2, and R19B09.3, then R19B09.3A, R19B09.3B, and R19B09.3C. R19B09.3D included R19B09.3A and R19B09.3B, whereas R19B09.3E included R19B09.3B and R19B09.3C. Primers used were as follows: 1-F: CCATGGACGAGCCTTTGCGAATCT, 1-R: CCGTGTTGCTACACTTGGCATACGA, 2-F: GAGCTGCAAATCCTTTACGCCTGT, 2-R: CCCATCGCAAGTGCGTCAGAATCA, 3-F: GATAAACCTGTTCGATCATTGGAGAT, 3-R: ACTCCTTAATGGGATTACCGACA, 3a-R: GATCCTACCCGGCGGCCAAATGA, 3b-F: GGCCGGATATTTATGAAGTTATTAAG, 3b-R: GTTTCCTTTCAAACTCTGCGAGAA, 3c-F: AGCCGAATAACTTTCTCCACA.
fru microRNAs.
To target the male-specific fru transcript, we used a strategy outlined in Chen et al., which makes use of the Drosophila miRNA miR6.1 stem-loop precursor and its flanking sequences, allowing for subsequent processing by Drosha and Dicer (65). We annealed two primers that were complementary to specific regions of the male-specific fru transcript (PCR1: see specific primers below). In a second PCR (PCR2), the PCR1 product was then amplified using primers Mir6 5′ EcoRI/BglII (5′ GGCGAATTCCGCCAGATCTTTTAAAGTCCACAACTC ATCAAGGAAAATGAAAGTCAAAGTTGGCAGCTTACTTAAACTTA), and Mir6 3′ BamHI/NotI (5′GGCCGCGGCCGCACGGATCCAAAACGGCATGGTTATTCGTGTGC CAAAAAA AAAAAAAATTAAATAATGATGTTAGGCAC). This entire concatenated sequence was then digested with BglII and NotI and ligated into pMUH (44). This construct was then injected into Drosophila embryos and integrated into attP2. Three lines of flies were generated: fru mi-1, fru mi-2 and fru mi 1 SCR. fru mi-1 consists of sequences that are complementary to the UTR and ExonA (UTRa and ExonA). fru mi-2 consists of sequences complementary to another region of the UTR and the same region of ExonA as in fru mi-1 (UTRb and ExonA). fru mi 1 SCR consists of sequences that are scrambled for the fru mi-1 UTR and ExonA (scrambled UTRa and scrambled ExonA).
PCR1, general strategy.
Forward Primer: GGCAGCTTACTTAAACTTAATCACAGCCTTTAATGT SEQUENCE TAAGTTAATATACCATATC.
Reverse Primer: AATAATGATGTTAGGCACTTTAGGTAC SEQUENCE TAGATATGGTATATTAACTTAGATT (should be complementary to last 4 mers of target sequence).
Specific primers.
UTRa-F: GGCAGCTTACTTAAACTTAATCACAGCCTTTAATGT CCACAACATTCCAATTCAAATCTAAGTTAATATACCATATC.
UTRa-R: AATAATGATGTTAGGCACTTTAGGTACCCACAACATTCCAATTCAAATCTAGATATGGTATATTAACTTAGATT.
ExonA-F: GGCAGCTTACTTAAACTTAATCACAGCCTTTAATGT GATGGCGACGTCACAGGATTATTAAGTTAATATACCATATC.
ExonA-R: AATAATGATGTTAGGCACTTTAGGTACGATGGCGACGTCACAGGATTAT TAGATATGGTATATTAACTTAATAA.
UTRb-F: GGCAGCTTACTTAAACTTAATCACAGCCTTTAATGT GAAACGCCAATTAGCCAAACATTAAGTTAATATACCATATC.
UTRb-R: AATAATGATGTTAGGCACTTTAGGTACGAAACGCCAATTAGCCAAACAT TAGATATGGTATATTAACTTAATGT.
UTRaScr-F: GGCAGCTTACTTAAACTTAATCACAGCCTTTAATGT TATCCCAACACTCCATAATCAATAAGTTAATATACCATATC.
UTRaScr-R: AATAATGATGTTAGGCACTTTAGGTACTATCCCAACACTCCATAATCAA TAGATATGGTATATTAACTTAATTG.
ExonAScr-F: GGCAGCTTACTTAAACTTAATCACAGCCTTTAATGT CGTATCATTCGGGCAGTGAAGATAAGTTAATATACCATATC.
ExonAScr-R: AATAATGATGTTAGGCACTTTAGGTACCGTATCATTCGGGCAGTGAAGA TAGATATGGTATATTAACTTATCTT.
Zn A-F: GGCAGCTTACTTAAACTTAATCACAGCCTTTAATGTGTTCAGTCT CGGTTTCAATAGCTAAGTTAATATACCATATC.
Zn A-R: AATAATGATGTTAGGCACTTTAGGTACGTTCAGTCTCGGTTTCA ATAGCTAGATATGGTATATTAACTTA.
Zn A scr-F: GGCAGCTTACTTAAACTTAATCACAGCCTTTAATGTTGTTAG TCCAACCTATTGGGCTTAAGTTAATATACCATATC.
Zn A scr-R: AATAATGATGTTAGGCACTTTAGGTACTGTTAGTCCAACCTAT TGGGCTTAGATATGGTATATTAACTTA.
Zn B-F: GGCAGCTTACTTAAACTTAATCACAGCCTTTAATGTCCTGGCA CATGCGGCTAACTTTTAAGTTAATATACCATATC.
Zn B-R: AATAATGATGTTAGGCACTTTAGGTACCCTGGCACATG CGGCTAACTTTTAGATATGGTATATTAACTTA.
Zn B scr-F: GGCAGCTTACTTAAACTTAATCACAGCCTTTAATGTCCTCTCGC AGGTTCATGCGAATTAAGTTAATATACCATATC.
Zn B scr-R: AATAATGATGTTAGGCACTTTAGGTACCCTCTCGCAGGTT CATGCGAATTAGATATGGTATATTAACTTA.
Zn C-F: GGCAGCTTACTTAAACTTAATCACAGCCTTTAATGTC CTGTCGGCGGCCCTGAAACGCTAAGTTAATATACCATATC.
Zn C-R: AATAATGATGTTAGGCACTTTAGGTACCCTGTCGGC GGCCCTGAAACGCTAGATATGGTATATTAACTTA.
Zn C scr-F: GGCAGCTTACTTAAACTTAATCACAGCCTTTAATGT CCTCGCTGGGGCAATCGCCACGTAAGTTAATATACCATATC.
Zn C scr-R: AATAATGATGTTAGGCACTTTAGGTACCCTCGCTGG GGCAATCGCCACGTAGATATGGTATATTAACTTA.
Protein Expression and EMSA.
The coding sequence for the different Fru DNA-binding domains (A, B, C) were obtained by RT-PCR from wild-type Canton-S fly RNA and cloned into the bacterial expression vector Pet15I-mOrange (gift from Loren Looger, Janelia Research Campus, Ashburn, VA), between the EcoRI and HindIII sites. The primers used for RT-PCR were (capital letters indicate added sequence for restriction sites):
FruA-F: CCAGAATTCatgtttgcaggtcgcgtcaagtgttttaac;
FruA-R: TCGAAGCTTtcacatatgtacatagtggctaaagaatc;
FruB-F: CCAGAATTCatgcggctaactttcgagcgcctctc;
FruB-R: TCGAAGCTTtcattcacttgtggcattgtgctgctg;
FruC-F: CCAGAATTCatgcacctgtcggcggccctgaaac;
FruC-R: TCGAAGCTTttaaatggatgagttcagcttgagca.
Fusion proteins were purified using Ni-NTA beads (Qiagen) according to the manufacturer’s protocol. EMSA was performed using biotin-labeled probes as described previously (8). For the competition assay, 10-fold of nonlabeled DNA (with various mutations) were added as competitors.
Mutation of Putative FruA and FruB Binding Sites.
Mutation of each site was introduced into the R19B09.3A sequence by PCR mutagenesis. The resulting sequence was confirmed by sequencing to verify each mutation. All GAL4 transgenes were integrated at the attP2 site.
Courtship and Fecundity Assays.
Flies were raised at 18–22 °C until eclosion, and maintained at 22 °C on a 12:12 light:dark cycle until testing. Four- to 7-d-old flies were aspirated into half of a standard 10-mm diameter, 6-mm high cylindrical courtship chamber, and allowed to recover for 30 min either at 22 °C for controls, or 29 °C for Trp activation. After recovery, the divider between chamber halves was removed, and courtship and copulation were filmed. Courtship latency was scored as the time from introduction to the first unilateral wing extension. Copulation latency was scored as the time from introduction to successful copulation.
For fecundity assays, after filming flies were anesthetized with CO2 and moved to standard fly vials, where they remained at 22 °C or 29 °C as appropriate, until the first offspring developed into pharate pupae (7–8 d at 29 °C or 11–12 d at 22 °C). All pupae in each vial were then counted.
Statistics.
Cell counts were compared by Student’s t tests or ANOVA with Tukey post hoc tests when normally distributed, or by Mann–Whitney u tests otherwise, with Bonferroni corrections for multiple comparisons. For courtship assays, survival analysis was used to analyze the total number of flies showing each behavior, and their latency. The log-rank test was used to compare survival curves. Fecundity was nonnormally distributed, and was analyzed using a Kruskal–Wallis rank sum test, followed by a post hoc Dunn test between pairs, with Bonferroni correction for multiple comparisons. Statistical analysis used JMP and R software.
Acknowledgments
We thank Dave Mellert and Kevin McGowan for help planning in situ experiments; Xiaorong Zhang for in situ probes; Susan Michael for sectioning and in situ hybridization; Heather Dionne and Gerald Rubin for GAL4 and LexA lines and constructs; Barret Pfeiffer and Gerald Rubin for providing pJFRC106; members of the B.S.B. laboratory, David Mellert, and Troy Shirangi for feedback; and Alison Howard for administrative support. The Drosophila Genomics Resource Center, supported by National Institutes of Health Grant 2P40OD010949-10A1, is thanked for supplying cDNA clones. This study was supported in part by the Howard Hughes Medical Institute for funding.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1600241113/-/DCSupplemental.
References
- 1.Sturtevant A. Experiments on sex recognition and the problem of sexual selection in Drosophila. J Anim Behav. 1915;5(5):351–366. [Google Scholar]
- 2.Erickson JW, Quintero JJ. Indirect effects of ploidy suggest X chromosome dose, not the X:A ratio, signals sex in Drosophila. PLoS Biol. 2007;5(12):e332. doi: 10.1371/journal.pbio.0050332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamamoto D, Koganezawa M. Genes and circuits of courtship behaviour in Drosophila males. Nat Rev Neurosci. 2013;14(10):681–692. doi: 10.1038/nrn3567. [DOI] [PubMed] [Google Scholar]
- 4.Cline TW, Meyer BJ. Vive la différence: Males vs females in flies vs worms. Annu Rev Genet. 1996;30:637–702. doi: 10.1146/annurev.genet.30.1.637. [DOI] [PubMed] [Google Scholar]
- 5.Luo SD, Shi GW, Baker BS. Direct targets of the D. melanogaster DSXF protein and the evolution of sexual development. Development. 2011;138(13):2761–2771. doi: 10.1242/dev.065227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chatterjee SS, Uppendahl LD, Chowdhury MA, Ip P-L, Siegal ML. The female-specific doublesex isoform regulates pleiotropic transcription factors to pattern genital development in Drosophila. Development. 2011;138(6):1099–1109. doi: 10.1242/dev.055731. [DOI] [PubMed] [Google Scholar]
- 7.Burtis KC, Coschigano KT, Baker BS, Wensink PC. The doublesex proteins of Drosophila melanogaster bind directly to a sex-specific yolk protein gene enhancer. EMBO J. 1991;10(9):2577–2582. doi: 10.1002/j.1460-2075.1991.tb07798.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coschigano KT, Wensink PC. Sex-specific transcriptional regulation by the male and female doublesex proteins of Drosophila. Genes Dev. 1993;7(1):42–54. doi: 10.1101/gad.7.1.42. [DOI] [PubMed] [Google Scholar]
- 9.Hutson SF, Bownes M. The regulation of yp3 expression in the Drosophila melanogaster fat body. Dev Genes Evol. 2003;213(1):1–8. doi: 10.1007/s00427-002-0286-4. [DOI] [PubMed] [Google Scholar]
- 10.Williams TM, et al. The regulation and evolution of a genetic switch controlling sexually dimorphic traits in Drosophila. Cell. 2008;134(4):610–623. doi: 10.1016/j.cell.2008.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shirangi TR, Dufour HD, Williams TM, Carroll SB. Rapid evolution of sex pheromone-producing enzyme expression in Drosophila. PLoS Biol. 2009;7(8):e1000168. doi: 10.1371/journal.pbio.1000168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo SD, Baker BS. Constraints on the evolution of a doublesex target gene arising from doublesex’s pleiotropic deployment. Proc Natl Acad Sci USA. 2015;112(8):E852–E861. doi: 10.1073/pnas.1501192112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clough E, et al. Sex- and tissue-specific functions of Drosophila doublesex transcription factor target genes. Dev Cell. 2014;31(6):761–773. doi: 10.1016/j.devcel.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ito H, et al. Fruitless recruits two antagonistic chromatin factors to establish single-neuron sexual dimorphism. Cell. 2012;149(6):1327–1338. doi: 10.1016/j.cell.2012.04.025. [DOI] [PubMed] [Google Scholar]
- 15.Dalton JE, et al. Male-specific Fruitless isoforms have different regulatory roles conferred by distinct zinc finger DNA binding domains. BMC Genomics. 2013;14(1):659. doi: 10.1186/1471-2164-14-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neville MC, et al. Male-specific fruitless isoforms target neurodevelopmental genes to specify a sexually dimorphic nervous system. Curr Biol. 2014;24(3):229–241. doi: 10.1016/j.cub.2013.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vernes SC. Genome wide identification of fruitless targets suggests a role in upregulating genes important for neural circuit formation. Sci Rep. 2014;4(4412):4412. doi: 10.1038/srep04412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rezával C, et al. Neural circuitry underlying Drosophila female postmating behavioral responses. Curr Biol. 2012;22(13):1155–1165. doi: 10.1016/j.cub.2012.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rezával C, Nojima T, Neville MC, Lin AC, Goodwin SF. Sexually dimorphic octopaminergic neurons modulate female postmating behaviors in Drosophila. Curr Biol. 2014;24(7):725–730. doi: 10.1016/j.cub.2013.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou C, Pan Y, Robinett CC, Meissner GW, Baker BS. Central brain neurons expressing doublesex regulate female receptivity in Drosophila. Neuron. 2014;83(1):149–163. doi: 10.1016/j.neuron.2014.05.038. [DOI] [PubMed] [Google Scholar]
- 21.Dolan J, et al. The extracellular leucine-rich repeat superfamily; a comparative survey and analysis of evolutionary relationships and expression patterns. BMC Genomics. 2007;8(1):320. doi: 10.1186/1471-2164-8-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo C-W, et al. Bursicon, the insect cuticle-hardening hormone, is a heterodimeric cystine knot protein that activates G protein-coupled receptor LGR2. Proc Natl Acad Sci USA. 2005;102(8):2820–2825. doi: 10.1073/pnas.0409916102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anand-Ivell R, Ivell R. Regulation of the reproductive cycle and early pregnancy by relaxin family peptides. Mol Cell Endocrinol. 2014;382(1):472–479. doi: 10.1016/j.mce.2013.08.010. [DOI] [PubMed] [Google Scholar]
- 24.Robinett CC, Vaughan AG, Knapp J-M, Baker BS. Sex and the single cell. II. There is a time and place for sex. PLoS Biol. 2010;8(5):e1000365. doi: 10.1371/journal.pbio.1000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rideout EJ, Dornan AJ, Neville MC, Eadie S, Goodwin SF. Control of sexual differentiation and behavior by the doublesex gene in Drosophila melanogaster. Nat Neurosci. 2010;13(4):458–466. doi: 10.1038/nn.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee G, et al. Spatial, temporal, and sexually dimorphic expression patterns of the fruitless gene in the Drosophila central nervous system. J Neurobiol. 2000;43(4):404–426. doi: 10.1002/1097-4695(20000615)43:4<404::aid-neu8>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 27.Mellert DJ, Knapp J-M, Manoli DS, Meissner GW, Baker BS. Midline crossing by gustatory receptor neuron axons is regulated by fruitless, doublesex and the Roundabout receptors. Development. 2010;137(2):323–332. doi: 10.1242/dev.045047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cachero S, Ostrovsky AD, Yu JY, Dickson BJ, Jefferis GSXE. Sexual dimorphism in the fly brain. Curr Biol. 2010;20(18):1589–1601. doi: 10.1016/j.cub.2010.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu JY, Kanai MI, Demir E, Jefferis GSXE, Dickson BJ. Cellular organization of the neural circuit that drives Drosophila courtship behavior. Curr Biol. 2010;20(18):1602–1614. doi: 10.1016/j.cub.2010.08.025. [DOI] [PubMed] [Google Scholar]
- 30.Jenett A, et al. A GAL4-driver line resource for Drosophila neurobiology. Cell Reports. 2012;2(4):991–1001. doi: 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Billeter J-C, et al. Isoform-specific control of male neuronal differentiation and behavior in Drosophila by the fruitless gene. Curr Biol. 2006;16(11):1063–1076. doi: 10.1016/j.cub.2006.04.039. [DOI] [PubMed] [Google Scholar]
- 32.Erdman SE, Chen HJ, Burtis KC. Functional and genetic characterization of the oligomerization and DNA binding properties of the Drosophila doublesex proteins. Genetics. 1996;144(4):1639–1652. doi: 10.1093/genetics/144.4.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gou B, Liu Y, Guntur AR, Stern U, Yang C-H. Mechanosensitive neurons on the internal reproductive tract contribute to egg-laying-induced acetic acid attraction in Drosophila. Cell Reports. 2014;9(2):522–530. doi: 10.1016/j.celrep.2014.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng K, Palfreyman MT, Häsemeyer M, Talsma A, Dickson BJ. Ascending SAG neurons control sexual receptivity of Drosophila females. Neuron. 2014;83(1):135–148. doi: 10.1016/j.neuron.2014.05.017. [DOI] [PubMed] [Google Scholar]
- 35.Bussell JJ, Yapici N, Zhang SX, Dickson BJ, Vosshall LB. Abdominal-B neurons control Drosophila virgin female receptivity. Curr Biol. 2014;24(14):1584–1595. doi: 10.1016/j.cub.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manoli DS, Baker BS. Median bundle neurons coordinate behaviours during Drosophila male courtship. Nature. 2004;430(6999):564–569. doi: 10.1038/nature02713. [DOI] [PubMed] [Google Scholar]
- 37.Van Hiel MB, Vandersmissen HP, Proost P, Vanden Broeck J. Cloning, constitutive activity and expression profiling of two receptors related to relaxin receptors in Drosophila melanogaster. Peptides. 2015;68:83–90. doi: 10.1016/j.peptides.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 38.Ivell R, Kotula-Balak M, Glynn D, Heng K, Anand-Ivell R. Relaxin family peptides in the male reproductive system—A critical appraisal. Mol Hum Reprod. 2011;17(2):71–84. doi: 10.1093/molehr/gaq086. [DOI] [PubMed] [Google Scholar]
- 39.Veenstra JA. The contribution of the genomes of a termite and a locust to our understanding of insect neuropeptides and neurohormones. Front Physiol. 2014;5:454. doi: 10.3389/fphys.2014.00454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Colombani J, Andersen DS, Léopold P. Secreted peptide Dilp8 coordinates Drosophila tissue growth with developmental timing. Science. 2012;336(6081):582–585. doi: 10.1126/science.1216689. [DOI] [PubMed] [Google Scholar]
- 41.Garelli A, Gontijo AM, Miguela V, Caparros E, Dominguez M. Imaginal discs secrete insulin-like peptide 8 to mediate plasticity of growth and maturation. Science. 2012;336(6081):579–582. doi: 10.1126/science.1216735. [DOI] [PubMed] [Google Scholar]
- 42.Colombani J, et al. Drosophila Lgr3 couples organ growth with maturation and ensures developmental stability. Curr Biol. 2015;25(20):2723–2729. doi: 10.1016/j.cub.2015.09.020. [DOI] [PubMed] [Google Scholar]
- 43.Garelli A, et al. Dilp8 requires the neuronal relaxin receptor Lgr3 to couple growth to developmental timing. Nat Commun. 2015;6:8732. doi: 10.1038/ncomms9732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vallejo DM, Juarez-Carreño S, Bolivar J, Morante J, Dominguez M. A brain circuit that synchronizes growth and maturation revealed through Dilp8 binding to Lgr3. Science. 2015;350(6262):aac6767. doi: 10.1126/science.aac6767. [DOI] [PubMed] [Google Scholar]
- 45.Chintapalli VR, Wang J, Dow JAT. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39(6):715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- 46.Pfeiffer BD, et al. Tools for neuroanatomy and neurogenetics in Drosophila. Proc Natl Acad Sci USA. 2008;105(28):9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pfeiffer BD, et al. Refinement of tools for targeted gene expression in Drosophila. Genetics. 2010;186(2):735–755. doi: 10.1534/genetics.110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barolo S, Carver LA, Posakony JW. GFP and beta-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. Biotechniques. 2000;29(4):726, 728, 730, 732. doi: 10.2144/00294bm10. [DOI] [PubMed] [Google Scholar]
- 49.Meissner GW, et al. Functional dissection of the neural substrates for sexual behaviors in Drosophila melanogaster. Genetics. 2011;189(1):195–211. doi: 10.1534/genetics.111.129940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barolo S, Castro B, Posakony JW. New Drosophila transgenic reporters: Insulated P-element vectors expressing fast-maturing RFP. Biotechniques. 2004;36(3):436–440, 442. doi: 10.2144/04363ST03. [DOI] [PubMed] [Google Scholar]
- 51.Pan Y, Robinett CC, Baker BS. Turning males on: Activation of male courtship behavior in Drosophila melanogaster. PLoS One. 2011;6(6):e21144. doi: 10.1371/journal.pone.0021144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anand A, et al. Molecular genetic dissection of the sex-specific and vital functions of the Drosophila melanogaster sex determination gene fruitless. Genetics. 2001;158(4):1569–1595. doi: 10.1093/genetics/158.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pan Y, Meissner GW, Baker BS. Joint control of Drosophila male courtship behavior by motion cues and activation of male-specific P1 neurons. Proc Natl Acad Sci USA. 2012;109(25):10065–10070. doi: 10.1073/pnas.1207107109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mellert DJ, Robinett CC, Baker BS. doublesex functions early and late in gustatory sense organ development. PLoS One. 2012;7(12):e51489. doi: 10.1371/journal.pone.0051489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baker BS, Hoff G, Kaufman TC, Wolfner MF, Hazelrigg T. The doublesex locus of Drosophila melanogaster and its flanking regions: A cytogenetic analysis. Genetics. 1991;127(1):125–138. doi: 10.1093/genetics/127.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hildreth PE. doublesex, a recessive gene that tranforms both males and females of Drosophila into intersexes. Genetics. 1965;51(4):659–678. doi: 10.1093/genetics/51.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hamada FN, et al. An internal thermal sensor controlling temperature preference in Drosophila. Nature. 2008;454(7201):217–220. doi: 10.1038/nature07001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu C, et al. A subset of dopamine neurons signals reward for odour memory in Drosophila. Nature. 2012;488(7412):512–516. doi: 10.1038/nature11304. [DOI] [PubMed] [Google Scholar]
- 59.Parks AL, et al. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat Genet. 2004;36(3):288–292. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- 60.Wang S, Zhao Y, Leiby M, Zhu J. A new positive/negative selection scheme for precise BAC recombineering. Mol Biotechnol. 2009;42(1):110–116. doi: 10.1007/s12033-009-9142-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Venken KJT, et al. Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat Methods. 2009;6(6):431–434. doi: 10.1038/nmeth.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33(4):e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schindelin J, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rohlfing T, Maurer CR., Jr Nonrigid image registration in shared-memory multiprocessor environments with application to brains, breasts, and bees. IEEE Trans Inf Technol Biomed. 2003;7(1):16–25. doi: 10.1109/titb.2003.808506. [DOI] [PubMed] [Google Scholar]
- 65.Chen CH, et al. A synthetic maternal-effect selfish genetic element drives population replacement in Drosophila. Science. 2007;316(5824):597–600. doi: 10.1126/science.1138595. [DOI] [PubMed] [Google Scholar]