Significance
Calmodulin is essential for sensing intracellular Ca2+ in eukaryotic cells. Calmodulin modulates hundreds of effectors, and it has a highly conserved protein sequence. Humans have three identical copies, but a change in either the protein sequence or the protein expression level of any one of the three copies can cause life-threatening disease. We analyzed calmodulin sequences across eukaryotes and compared biophysical properties and structures to show that all of calmodulin’s four Ca2+-binding sites have conserved properties that distinguish them from one another.
Keywords: calcium signaling, EF-hand, structure, evolution, protein
Abstract
Calmodulin (CaM) is a Ca2+-sensing protein that is highly conserved and ubiquitous in eukaryotes. In humans it is a locus of life-threatening cardiomyopathies. The primary function of CaM is to transduce Ca2+ concentration into cellular signals by binding to a wide range of target proteins in a Ca2+-dependent manner. We do not fully understand how CaM performs its role as a high-fidelity signal transducer for more than 300 target proteins, but diversity among its four Ca2+-binding sites, called EF-hands, may contribute to CaM’s functional versatility. We therefore looked at the conservation of CaM sequences over deep evolutionary time, focusing primarily on the four EF-hand motifs. Expanding on previous work, we found that CaM evolves slowly but that its evolutionary rate is substantially faster in fungi. We also found that the four EF-hands have distinguishing biophysical and structural properties that span eukaryotes. These results suggest that all eukaryotes require CaM to decode Ca2+ signals using four specialized EF-hands, each with specific, conserved traits. In addition, we provide an extensive map of sites associated with target proteins and with human disease and correlate these with evolutionary sequence diversity. Our comprehensive evolutionary analysis provides a basis for understanding the sequence space associated with CaM function and should help guide future work on the relationship between structure, function, and disease.
Eukaryotes use Ca2+ in numerous intracellular signaling pathways. Calmodulin (CaM) is a highly versatile Ca2+ signaling protein that is essential for at least dozens of cellular processes in eukaryotic cells. In humans it binds to more than 300 targets (1–3). Humans have three genes that encode identical CaM proteins, but mutations in just one of the three copies can cause disease (4–8), as can altered gene expression (9). Although CaM has been extensively studied, many details about its function are still poorly understood. The high evolutionary conservation along with the wide range of targets brings up the question of how a single Ca2+-binding protein displays both selectivity and flexibility in the context of its various signaling pathways.
CaM binds Ca2+ at four, nonidentical sites that contain the structural motif called an EF-hand (10, 11), each of which contains an acidic Ca2+-coordinating loop, or “EF-loop” (Fig. 1A). The EF-loop spans 12 amino acids and provides at least six oxygen atoms for coordinating Ca2+ (12). The coordinating oxygen atoms are provided by the side chains at the first, third, fifth, and 12th positions of the EF-loop, and an oxygen from a main chain carbonyl group is provided at the seventh position (10). Water molecules participate in the Ca2 coordination geometry (13). CaM functions as a sensor over a broad range of Ca2+ signals that vary in amplitude, duration, and location. Although biophysical and evolutionary sequence studies have resulted in a general understanding of the bulk properties of EF-hand–binding sites, the implications of differences in Ca2+ affinity among the four EF-hands deserves a thorough investigation.
Fig. 1.
(A) Example of a Ca2+-bound EF-hand structure from PDBID 1CLL. A cartoon of an EF-hand peptide chain threads through a semitransparent representation of its molecular surface. The surface is the interface between molecular atoms and solvent rendered in PyMOL. Only atoms nearest the Ca2+ are shown and are depicted as spheres—green for Ca2+ and red for oxygens. A Ca2+-coordinating water is depicted as a semitransparent red sphere. Helices are gray, and the EF-loop is tan. (B) Maximum likelihood branch lengths of CaM and tubulin constrained to match the species tree in Torruella et al. (40). This tree covers much of eukaryotic diversity. Holozoa and Holomycota include animals and fungi, respectively, and their closely related protist lineages. SARPAE is described in the text. Both proteins are highly constrained, but whereas tubulin’s rate has been fairly consistent across eukaryotes, CaM underwent a dramatic speed-up in Ascomycete fungi, which include the model system S. cerevisiae.
Previous reports showed that the large family of EF-hand proteins likely arose from a founder protein with a single EF-hand in the most recent common ancestor of all extant eukaryotes (11, 14–18). Different EF-hand–containing proteins bind Ca2+ with different affinities, suggesting that a protein with multiple EF-hands, such as CaM, may bind Ca2+ with a different affinity at each site (19–28). It has therefore been suggested that CaM’s four sites display different affinities and perhaps cooperativity (29, 30). We therefore hypothesized that CaM’s four, nonidentical loops may generate some of their functional flexibility by binding Ca2+ using different physical properties and explored whether such differences could be discerned in the evolutionary record.
Evolutionary analyses can provide mechanistic insight into how CaM is used as a Ca2+ sensor across eukaryotes. Prior work showed that the protein sequence of CaM is evolving at a faster pace in fungal species (11, 31–33), reflecting the fact that although CaM is essential in Saccharomyces cerevisiae, the cells can survive with all four EF-hands ablated (34). However, previous evolutionary studies focused on a small subset of eukaryotes, either because few sequences were available at the time of publication or because the study was focused on a particular lineage. The vast expansion of taxonomic coverage in sequence databases, and the recent availability of new NMR and X-ray crystal structures of CaM, therefore demands a more comprehensive analysis. Unfortunately, CaM is a small, ancient, and highly conserved protein and therefore does not contain enough information to infer phylogenetic tree topologies. Kretsinger and Nakayama and coworkers (11, 16, 17, 35), for instance, found little correspondence between phylogenies inferred from protein, DNA, or intron–exon structure.
To overcome this hurdle, we used a variety of techniques to explore sequence and structural conservation in CaM across eukaryotes. Our approach allows us to address several key questions: (i) How fast is CaM diverging in different phyla? (ii) How does the function of a site, or its association with disease, correlate with sequence conservation? (iii) What properties of the EF-hands are conserved over deep evolutionary time, and how might this correspond to functional plasticity?
Results
We searched multiple genomic databanks to compile a list of CaM and CaM-like molecules (Dataset S1). For the purposes of comparing CaM evolution across all eukaryotes, we divide CaM sequences into three major taxonomic groups: Holozoa (animals and closely related protists), Holomycota (fungi and closely related protists), and all other eukaryotes, which we will call SARPAE (Stramenopiles, Alveolates, Rhizaria, Plantae, Amoebozoa, and Excavata). At least one copy of a CaM gene was found from every complete genome, but some genomes have multiple CaM and/or CaM-like genes, as described previously (36–39). Compared with the Holomycota dataset, Holozoa and SARPAE have more CaM genes per organism (see Dataset S1). Although some of the copy number heterogeneity could be from genome assembly errors, most of the genomes containing multiple CaMs assign these genes to different loci. This set of sequences was the basis for all subsequent analyses.
CaM Evolves Faster in Fungi.
To analyze the evolutionary rate of CaM, we mapped an alignment of CaM orthologs onto a well-sampled eukaryotic phylogeny from the literature (40) and estimated the maximum likelihood branch lengths on this tree topology (Fig. 1B). Branch lengths are in units of expected number of substitutions per amino acid site, and a longer branch length therefore corresponds to faster evolution. We compared the maximum likelihood branch lengths of CaM with another highly conserved protein, tubulin α-1 chain (Fig. 1B). Whereas the evolutionary rate of tubulin appears to be relatively stable across the three groups, CaM evolution experienced an acceleration in Holomycota, especially in one sublineage called the Ascomycota, which includes the model yeast S. cerevisiae. Divergence of the CaM protein sequence in yeast has been previously reported (11, 31, 41, 42), but our data show that this increase in evolutionary rate is specific to fungi, especially ascomycetes, and that the increased rate is not a general genomic signature of fungi, as tubulin does not have a similarly elevated rate.
Sequence Conservation Correlates with Target Binding but Not with Disease Association.
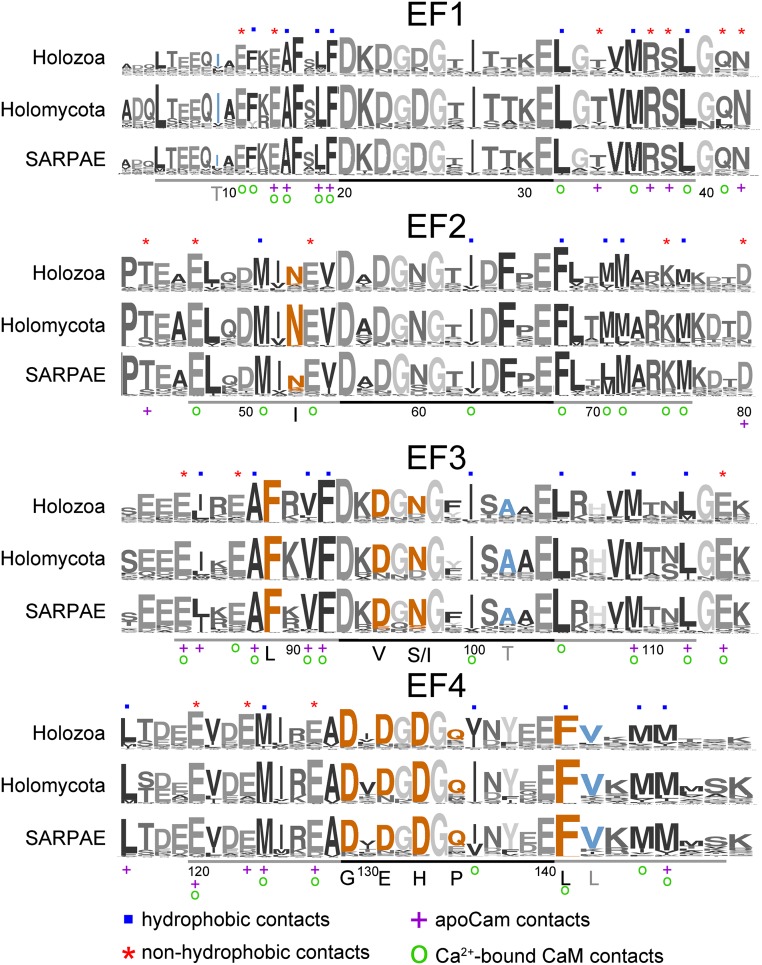
We analyzed CaM sequences from organisms in Holozoa, Holomycota, and SARPAE to see which amino acids were tolerated at different positions and whether these differed among taxonomic groups. CaM sequences from these groups were used to perform three separate group alignments using Multiple Em (Expectation maximization) for Motif Elicitation (MEME) (43), and these data are displayed as sequence logos (44). Table S1 contains consensus sequences for further review. Fig. 2 illustrates amino acid conservation at each residue position. The total height of all of the stacked letters at a single position is proportional to the amount of conservation of the residue (44), and the height of each stacked letter is proportional to the frequency that the amino acid is observed. The main objective of this analysis is to find where substitutions are tolerated in different phyla.
Fig. 2.
Sequence logo of CaM conservation across three phylogenetic groups generated with MEME (32). For visual appeal, amino acids are shown as different shades of gray. The height of the residue stack at a given location represents the relative conservation of that position. Numbering of the amino acids in the protein sequence starts without the methionine. In each row, the EF acid loop is underlined with a black bar, and the helices of the EF-hand are underlined with gray. Orange letters indicate positions that were identified with mutations causing cardiomyopathies in human. Blue letters correspond to mutations found in the ESP (93). Residues that were shown to make contact with target proteins are labeled (46). Blue squares indicate hydrophobic contacts, red asterisks indicate nonhydrophobic contacts, purple crosses indicate apoCaM contacts, and green open circles indicate Ca2+-bound CaM contacts.
Fig. 2 shows that the same sites tend to be conserved in all three eukaryotic supergroups, suggesting that CaM function is at least somewhat conserved across eukaryotes. Seven phenylalanines stand out for having virtually no alternative residues (Table S2 and Dataset S2). The importance of these phenylalanines to cellular viability has been studied using mutagenesis in yeast (45). Mutation of only one of the phenylalanines rarely resulted in an observable growth defect, but nearly all combinations of phenylalanine mutations were strongly deleterious. In humans, mutations at phenylalanine sites result in cardiomyopathies. The mutation F89L has been linked to intraventricular fibrillation, and F141L has been linked to long QT syndrome (5, 6). Leucine is substituted with very low frequencies, 0.005 or 0.010 for F89 and F141, respectively, in other eukaryotes (Dataset S2), implying that leucine substitution is not well tolerated in most organisms. Thus far, most of the other CaM mutations that result in human cardiomyopathies were found in the Ca2+-binding EF-loops; however, not all mutations result in the same cardiac phenotype (4–6), suggesting that different protein pathways are affected by different mutations in CaM.
Many of the most strongly conserved sites in Fig. 2 are either Ca2+-coordinating residues within the EF-loops, which we explore later, or form contacts with other proteins (46). Nearly all CaM residues that make contact with a target protein have higher conservation than the neighboring residue that does not make contact (Fig. 3A). One exception is T34, which has lower conservation than its neighbors but does participate in Ca2+-free interactions (Fig. 2) (46). Fig. 3B shows that the median frequency of residues that bind targets is 0.83; that is, 50% of these residues have the same amino acid in at least 83% of CaM sequences. In contrast, for all other positions in CaM, the median occurs where amino acids are the same in only 72% of the sequences. The elevated conservation in the residues that participate in binding suggests binding targets contribute to the restrictive evolutionary pressure on CaM.
Fig. 3.
(A) Positional frequencies of primary amino acids. Circles are colored based on secondary structure. Filled circles represent random coil, and empty circles represent α-helix. Residues that bind targets are outlined with purple. Red diamonds indicate total frequency where two amino acids, each with a minimum frequency of 0.2, are interchangeable; that is, together they have a combined frequency greater than 0.9. A scheme for the secondary structure based on a crystal structure (PDBID 1CLL) is provided just above the residue number axis. IL, interlobe linker. (B) Comparison of conservation in residues that bind CaM targets, purple circles, with all other residues in CaM, gray circles. The conservation frequency is the frequency of the primary amino acid; each circle represents a position in the CaM sequence (148 total points). The plots are integrated observations going from low frequency to high frequency. Inset shows a box plot summary of the same dataset. For an unpaired, two-tailed, unequal variance t test, the P value is <0.0001.
Of the residues that form binding sites (46), only F141L is associated with a human disease (5): long QT syndrome. In contrast, the human disease mutations N53I and N97S (4) are not in protein interaction sites and appear to have lower conservation than their neighbors (Fig. 2). Other highly conserved sites are most likely not correlated with diseases because the nonsynonymous changes at those loci are so strongly deleterious that they are rarely observed in natural populations.
The Exome Sequencing Project (ESP) data contain three CaM mutations from the general human population with unknown phenotype (47). These mutations include A9I, A102T, and V142L. In the context of evolution, these sites are not strongly conserved (Fig. 2 and Table S2), although these particular substitutions are somewhat rare in other eukaryotes (Dataset S2).
High Conservation Is Found at Residues in Both α-Helices and Loops.
To determine which structural features are preserved from high sequence conservation, we mapped the frequency of an observed residue at each amino acid position onto a previously determined structure of a mammalian CaM as the backbone structure with side chains was omitted (Fig. 4A) (13). The frequency of the dominant amino acid at a given position is represented by thickness and by a color scale where red represents more conserved residues. CaM is a bilobed molecule whose globular domains are joined by α-helices that connect through the interlobe linker (IL in Fig. 4A) (48).
Fig. 4.
Amino acid frequency is plotted and mapped for each residue of CaM. (A) Amino acid frequency mapped onto cartoon putty representation of a CaM crystal structure: PDBID 1CLL. The Inset is a scale for relating thickness and color to amino acid conservation. In both panels, residues that form α-helices are designated from αA–αH in alphabetical order. Red diamonds indicate where two residues have a combined frequency greater than 0.9, but the lowest frequency of the pair is at least 0.2. With frequency in parentheses, these include amino acids K (0.67) or R (0.24) at position 13 in the N-lobe and I (0.57) or L (0.33) at 85, R (0.58) or K (0.33) at 90, T (0.69) or S (0.25) at 117, and I (0.54) or V (0.43) at 136 in the C-lobe. The added values of both primary and alternate are then mapped where there are asterisks in B with transparency at these locations. Close-up views of the non–Ca2+-binding loops that link EF1 to EF2 and EF3 to EF4 are shown in B and C, respectively. (D) Atomic representation of CaM residues close to position 90. Positive-charged residues are blue, and negative-charged residues are red. Other residues are gray.
The most conserved residues are confined to the globular domains, or lobes, of CaM. Both the N-terminal lobe (N-lobe) and the C-terminal lobe (C-lobe) have two EF-hands for binding Ca2+. Each EF-hand is comprised of two α-helices that are joined by an acidic EF-loop (49). The α-helices in both lobes contain many conserved residues. We can infer some roles for these residues by referring to Fig. 2. As suggested in Fig. 2, many of the residues in the α-helices are also residues that participate in binding to targets. For example, several phenylalanines and methionines are recognized by targets (50, 51). In contrast, a few conserved residues do not reside at the protein interaction sites, so these might instead be conserved for structural roles. These include residues F16, L48, L69, and F89. Their side chains do not line the exposed hydrophobic binding pockets (13, 48), but they do provide support to the pocket, suggesting a role for protein stability.
Flexible amino acids also appear to be crucial to CaM’s structure or function. High conservation is seen in the non–Ca2+-binding loops that connect EF1 to EF2 and EF3 to EF4 (Fig. 4 B and C). Residues that are in exposed random coil or turn regions are typically more variable during protein evolution, unless the loop plays a vital functional or structural role (52). G40, G113, and P43 are among the most highly conserved residues in CaM (Fig. 4 B and C and Table S2). Both loops also end with highly conserved glutamates at residues 47 and 120 that initiate the α-helix into the second and fourth EF-hands, respectively. E47 and E120 have been postulated to play a role in forming molecular contacts (46), so in addition to a structural role, these loops may also contribute to molecular recognition. To summarize our structural analysis, highly conserved residues are found throughout the protein architecture, and many residues were under strong purifying selection with clear roles with either binding or structure.
Until now, we have focused on residues that have very low frequencies of alternative amino acids, which is to say they are under strong purifying selection. Another class of residue positions is clearly under purifying selection but often toggles between two amino acids. We define this class as a position that has another amino acid that is present in more than 20% of the CaMs, but adding the top two amino acid frequencies at a residue position accounts for greater than 90% of the observations (Figs. 3 and 4 denoted by diamonds). It can be inferred that the two amino acids share a property that is necessary at this location. For example, position 90 has either a positively charged lysine or arginine (Fig. 2). In structures of CaM bound to targets, these residues face the solvent and usually away from direct contact. The high conservation at position 90 implies that the role of a positive charge is to provide a countercharge to the large number of acidic residues in CaM, whereas the placement indicates that the effect is meant to be local. Many CaM targets have net positive charges (53). Nearby to R/K 90 are glutamate residues E84 and E87 that participate in binding (46) and that have conserved negative charges (Dataset S2), but they line another face of the molecule (Fig. 4D). The conserved residue placement on the structure suggests that positive charges may orient the molecule so that the right face of CaM can bind the target.
Each EF-Loop Has a Unique Selection of Residues That Are Highly Conserved.
Each Ca2+-binding site of CaM may act individually or depend on allosteric coupling with other EF-hands. With our wide sampling of organismal diversity, we are able to study the differences between each EF-hand of CaM in detail. A putty representation of all of the EF-loops beside their sequence logos is provided in Fig. 5. Each EF-loop has 12 residues, the approximate positions of which are shown in Fig. 5A. Conservation was highly variable both within and between loops (Fig. 5B). This indicates that each EF-loop has experienced different evolutionary constraints that involve different sites.
Fig. 5.
Comparing conservation of structural positions and sequence of Ca2+-binding loops from each EF-hand. (A) A schematic that shows the location of each residue position in the loop structure. The putty scale for conservation is the same as in Fig. 4. (B) EF-loops are all oriented to the model scheme so that residue locations can be compared. The peptide backbone is represented by a putty cartoon that shows frequency of most observed amino acids at each position. (C) Sequence logos that correspond to the structures are shown on the right side of the panel. The position coloring of the sequence is identical to the structure. A red diamond represents a position that has two amino acids with a combined high frequency, isoleucine and valine.
Four Different EF-Loop Structures Are Conserved Across Eukaryota.
Early phylogenetic studies suggested that CaM’s four EF-hands could have evolved from two rounds of internal duplications from a precursor protein containing a single EF-hand (16), similar to four-domain ion channels (54). Support for this hypothesis was low due to the depth of evolutionary time under consideration (12). To see whether the hypothesis of two rounds of duplication was supported by structural comparisons, we compared crystallographic or NMR structures of EF-loops bound to Ca2+ modeled from distantly related species including an animal, a protist, a plant, and a fungus (Fig. 6). Each EF-loop is more similar to the same EF-loop from four distantly related eukaryotes than with other EF-loops of the same species (with the exception of EF4 in yeast, which does not bind Ca2+ and was not considered here) (55–57).
Fig. 6.
A Ca2+-bound EF-loop is similar across eukaryotes but not similar to other loops in the same protein. All loops were aligned to the same arbitrary orientation by minimizing the all-atom root-mean-square distance. The solvent-accessible surface for each EF-loop is shown. The electrostatic potentials were determined using the APBS software built into PyMOL. Ca2+ was included for these calculations. Colors are red for more negative potentials, blue for positive, and white for neutral. The following CaMs are represented (PDBID): paramecium (1EXR), potato (1RFJ), vertebrate CaM (1CLL), and a mutant with a deletion of a nonfunctional fourth EF-hand from S. cerivisiae (2LHH). Each loop comprises 12 residues: EF1, residues 20–31; EF2, 56–67; EF3, 93–104; EF4, 129–140. All coordinates were determined by X-ray crystallography except for 2LHH, which was determined by NMR. A distance tree derived from root-mean-square analysis of EF-hand amino acid composition is provided in the lower right-hand corner.
Under the hypothesis of two rounds of internal duplication, each followed by evolutionary divergence, EF1 and EF3 should resemble each other, as should EF2 and EF4. Early studies found weak phylogenetic support for this (12), so we computed a tree based on the distances between the alignments of all of the four EF-loops (Fig. 6, Inset). The ratio of divergence within each EF-loop (external branches) to shared divergence after the first round of duplication (internal branch) is clearly very high. However, the pattern appears in the structure as well (Fig. 6), supporting the hypothesis of two rounds of internal duplication.
EF-Loops Are Distinguished by Their Biophysical Attributes.
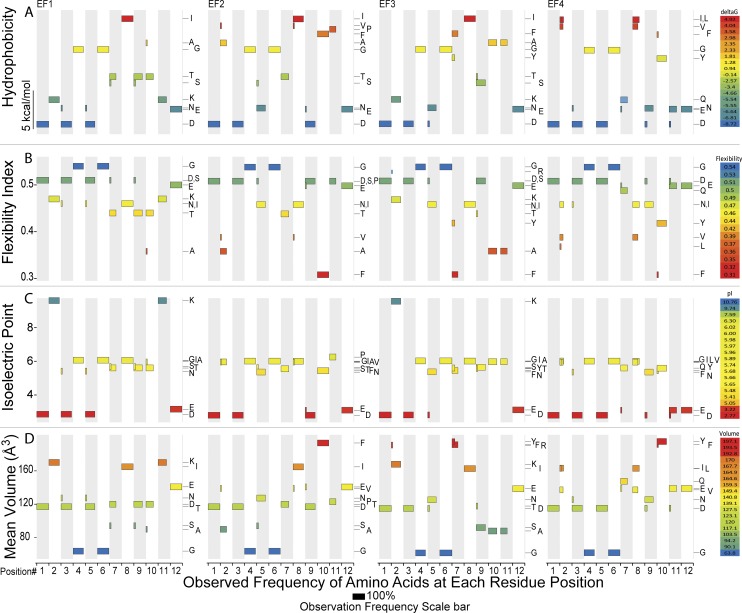
Because the level of sequence divergence within the EF-loops is quite high (Fig. 6, Inset), but the structures retained some signal from the two rounds of internal gene duplication (Fig. 6), we wanted to determine whether the EF-loops were distinguished from one another in terms of their biophysical properties—a kind of midpoint between raw sequence and structure. We aligned all four EF-loops from each sampled species and recoded each amino acid according to one of four biophysical properties: hydrophobicity (58), positional flexibility (59), isoelectric point (60), and amino acid volume when packed in a protein (61). Hydrophobicity, or solvation energy, strongly distinguishes charged from nonpolar R-groups of amino acids. Backbone flexibility is an empirical value determined from the comparisons of numerous protein structures. Amino acids have an index value according to their overall association with rigid or flexible domains. To distinguish oppositely charged amino acids, the isoelectric point (pI in log units) is best suited, although uncharged amino acids have nearly indistinguishable values along the pH axis. The mean volume of an amino acid buried in a protein can vary from 64 (glycine) to 232 (tryptophan) cubic angstroms. The full distributions of these properties within the four EF-hands are plotted in Fig. S1.
Fig. S1.
Conservation of physical properties of each residue of each Ca2+-binding site. Each panel corresponds to an EF-loop and to a physical property. A panel is further divided into 12 EF-loop positions depicted by alternating gray and white vertical bands. All amino acids with a frequency greater than 0.10 are plotted as horizontal bars for each position. Each bar has a horizontal length that is proportional to the frequency of an amino acid, normalized to the total number of CaM sequences. The vertical position of the bar plots the value for the physical parameter. Color coding is provided to assist the reader in seeing patterns across EF-loops. The physical parameters plotted are as follows: (A) hydrophobicity, plotted as solvation energy in the first row of panels, (B) the flexibility index in the second row, (C) isoelectric point of the free amino acid in the third row, and (D) residue volume in angstroms in the fourth row.
We then used principle component analysis (PCA) on the biophysical properties of the EF-loop residues to determine whether the four EF-hands had evolutionary conserved differences in the four biophysical properties. The input data are provided in Table S3, and the code we used to implement PCA is provided in Script S1. The output data points, each corresponding to one EF-hand from one CaM, are plotted in the space of the first two principal components. These first two components capture much of the variance in the data (Fig. 7 A–D). Proximity of points to one another in this space indicates similarity of biophysical properties. Our implementation of PCA also calculates the loadings of the 12 original variables (biophysical properties of each site) onto the first two principal components (Fig. 7 E–H). These loadings show how each amino acid position correlates with the two principal components and therefore how it contributes to the distribution of the points in this space (Table S4).
Fig. 7.
PCA of the EF-loops for four different biophysical parameters. In A–D, each EF-hand from each species is plotted on the first two principal components, which describe much of the variance in the data. The four EF-loops are clearly distinguished from one another for both hydrophobicity and flexibility. EF4 and EF3 are distinguished for isoelectric point and volume, respectively. The loadings for each amino acid position on the first two principal components are plotted in E–H. Each loading vector (arrow) is determined by the two coordinates to which it points, and the coordinates are proportional to that site’s contribution to variance on the two principle components. The loadings therefore give a sense of which EF-hand sites are driving the patterns in A–D.
With the full power of PCA analysis, conclusions can be drawn as to what biophysical parameters are important for CaM’s identity as a Ca2+ sensor. PCA tells how EF-loops are different in a way that sequence alignments and static structural comparisons cannot show. All four EF-loops are clearly distinguishable from one another in both hydrophobicity and flexibility (Fig. 7 A and B). The patterns are more complex for the other biophysical parameters, with EF4 being clearly distinguished from others by isoelectric point and EF3 by volume (Fig. 7 C and D). The four EF-loops therefore have biophysical differences that are generally conserved across eukaryotes. We next take a closer look at some of the residues that contribute to each EF-loop’s identity.
Five Positions Have the Same Residues in All Four EF-Loops.
Our analyses of the conservation of sequence, structure, and biophysical qualities of the EF-loops gives us insight into selective pressures that have acted on CaM’s Ca2+-binding activities across more than a billion years of eukaryotic evolution and how these pressures determine the sequence space available to CaM. This information is summarized in Fig. 8. Most CaMs have the same amino acids at positions 1, 3, 4, 6, and 12 in all four of CaM’s EF-loops (Fig. S1 and Dataset S2). The PCA loadings associated with these positions are the smallest vectors because they do not contribute variance to the data (Fig. 7 E, F, and H). Positions 1, 3, 5, and 12 all provide oxygen atoms from their side chains to coordinate Ca2+ (Fig. 1A), and the backbone carbonyl of position 7 also provides an oxygen atom (13). Consistent with earlier studies that used a reduced dataset (16), the only Ca2+-coordinating residues that are usually found in all four EF-hands of CaM are aspartates at positions 1 and 3 and a glutamate at position 12 (Fig. 5B). The flexibility in the Ca2+ site is determined in large part by two glycines that are located at positions 4 and 6 (Fig. S1B). The position 6 glycines in particular are highly conserved with a frequency greater than 0.94 for each EF-loop (Table S2).
Fig. 8.
Comparison of positions that are identical in different EF-loops. Residues that appear at a high frequency in multiple EF-loops are connected by a line. Positions that are unique to only one EF-loop are filled circles. Positions that appear to have only one alternate residue across multiple EF-loops are connected by a dashed line. Residues are colored as follows: black, polar; blue, basic; gray, nonpolar; green, glycine; red, acidic.
Different Residue Positions Distinguish Each Ca2+-Binding Site.
The remaining seven EF-loop positions are more variable. Although each EF-loop has at least two positions with a unique amino acid, EF1 shares the most properties with other loops and EF4 is the most different. We briefly highlight two sites that appear to be key players in distinguishing the four EF-loops from one another. The lysine in position 2 strongly distinguishes EF1 and EF3 from EF2 and EF4 with regards to hydrophobicity, flexibility, and volume (Figs. 7 E, F, and H and 8) but not for isoelectric point (Fig. 7G). Lysines provide flexibility and a positive charge, which appears to be important for EF1 and EF3 (Fig. 5B) but not EF2 and EF4, where that site has lower conservation (Fig. S1B). At position 5, a Ca2+-coordinating residue, EF1 is similar to EF4 and EF2 is similar to EF3 (see Fig. 7 for loading vectors at this site). Although EF1 and EF4 have a highly conserved aspartate at this site, the most frequent residue in EF2 and EF3 is an asparagine (Fig. S1). A glutamate or a glutamine, which is just one carbon atom longer in side chain length, is quite rare (Dataset S2), indicating that the short branch length of an aspartate or an asparagine is optimal for Ca2+ binding. The requirement for a short side chain length is further supported by the finding that a human cardiomyopathy results when a glutamate is substituted for an aspartate in EF4 (7).
Using our results, specific details can be inferred about the conservation of biophysical properties at certain residue positions. As an example, position 9 in EF1 and EF3 both share the characteristic of being small and polar (Fig. S1 A and D and Fig. 7 E and H); however, the threonine in EF1 and serine in EF3 differ in flexibility, which leads to different EF-loop pairs being distinguished by different attributes of these similar residues (Fig. S1B and Fig. 7F). Further examination of our data at other sites will provide equally useful insight into the conserved biophysical properties of each EF-hand.
Discussion
CaM is the primary Ca2+ sensor of eukaryotes. It is a versatile model protein that, for several decades, has been studied broadly in multiple disciplines including both experimental and theoretical research. CaM is one of the most highly conserved eukaryotic proteins (31). However, most of the protein sequence varies across deep evolutionary divergences (11). Our results show that variation in CaM sequences occurs at similar sites in Holomycota, Holozoa, and SARPAE (Fig. 2). The fact that some organisms in Holozoa and SARPAE have multiple, differentiated copies of CaM whereas others, including humans, have multiple identical copies (17) suggests that CaM evolution is shaped by changes in the coding sequence and by changes in gene expression but that these two modes are more or less important in different lineages. Because all vertebrates have multiple identical copies, it seems likely that the coding sequence is under very strong purifying selection. In contrast, organisms within Holomycota usually have just one single, essential copy of CaM. S. cerevisiae has a single copy of CaM in its entire genome, sharing about 60% sequence identity with vertebrate CaM (62). But all three examined lineages have very similar patterns of conserved and unconserved sites, suggesting deep similarities in CaM function across eukaryotes. The fact that the sites associated with protein–protein interactions appear conserved across all eukaryotes suggests three testable hypotheses: (i) Protein–protein interactions are conserved over deep evolutionary time, (ii) the same sites play roles in binding many different proteins, or (iii) these sites also play a structural role in CaM itself.
CaM sequence evolves much faster within fungi and especially within ascomycetes (Fig. 1B). This correlates with some existing experimental evidence that yeast are more robust to perturbation in CaM sequence than are vertebrates. For instance, although CaM is essential in yeast, CaM gene knockout can be rescued with a vertebrate CaM, which is only 60% identical (62, 63). Additionally, it appears that yeast can survive with Ca2+ binding knocked out in all four EF-hands (34), whereas single mutations in just one of the three identical copies of CaM present in humans can cause major diseases (4–6). Yeast is therefore more robust to changes in CaM sequence than vertebrates, and this robustness probably translates into the higher evolutionary rate within ascomycetes. Why exactly yeast is more robust remains a major question. One possibility is that CaM has fewer target proteins in yeast. Fungi are known, for instance, to have lost many of the ion channel genes present in the common ancestor of fungi and animals (64), but further investigation of this phenomenon is required.
We have shown that differences in both sequence and the biophysical properties clearly distinguish EF-loops from one another and that these differences are maintained over the large evolutionary distances comprised by our dataset. Each EF-hand likely has a unique physiological purpose that contributes to CaM’s enormous functional plasticity. Numerous NMR and X-ray crystal structures show CaM in a myriad of conformations and stoichiometries when it is bound to Ca2+ and to different targets (65, 66). Adding to the complexity, a single target may bind CaM with different conformations and stoichiometries that are Ca2+-dependent (67). Protein targets of CaM affect the apparent Ca2+ affinity (68, 69), and the protein complex is tuned to diverse physiological roles through evolution (70–72). How do the four EF-loops contribute to these diverse roles? Many attempts have been made to measure the Ca2+ affinity of CaM, but simplified models, such as the Adair and Hill equations, are often used to fit data. A recent study shows that the methods used to measure parameters from standard binding curves do not have uniquely identifiable solutions (73), so prior studies that determined the Ca2+ affinity of CaM are called into question. Our dataset suggests a need for a thorough investigation of each EF-loop’s contribution to CaM’s deeply conserved functions and provides a starting point by highlighting key residues that differentiate the four EF-loops. Our analysis strongly suggests that Ca2+ binding should not be assumed to be equal at all four sites or at the paired sites within the two lobes.
The high conservation in the non–Ca2+-binding loops suggests important roles for CaM function that also merit further investigation. Allosteric coupling between EF-hands has been studied for a long time (23), but the discussion is usually limited to the β-strands and α-helices that are at the interface between Ca2+ sites. The distance of the non-Ca2+ loops from the Ca2+ sites may have deterred consideration for a role in Ca2+ binding, but a recent review discusses how allosteric coupling can occur over large distances within proteins (74). Perhaps the non–Ca2+-binding loops play such a role. Alternatively, the non-Ca2+ loop may play a role in target protein interactions, but not all of its most conserved residues are involved with target contacts in known crystal structures (46).
Evolutionary studies of CaM provide a wealth of information that can help prioritize future functional analyses. This work used a variety of methods that helped glean mechanistic insights from sequence data for which traditional phylogenetic approaches were infeasible. Our dataset presents a map of CaM as seen by well over a billion years of eukaryotic evolution. It is our hope that this map will serve as a reference for guiding future experimental work on this widely studied molecule.
Methods
Sequence Blasts and Alignments.
The entire genomes of each organism were searched, one at a time, to identify CaM genes. Several gene databanks were searched, including National Center for Biotechnology Information (blast.ncbi.nlm.nih.gov/Blast.cgi), The Genome Portal of the Department of Energy Joint Genome Institute (JGI) (75), Compagen (76), The UniProt Consortium (77), and The Origins of Multicellularity Sequencing Project, Broad Institute of Harvard and MIT (www.broadinstitute.org/). Query sequences used to search genomes for CaM were vertebrate (NP_001008160.1), brown rot fungus (JGI Genome, Pospl1Protein Id 117693), or a diatom (XP_002295755.1). All hits were reciprocally blasted into the originating genome for the query, and only proteins that matched CaM in the reciprocal blast were retained. This process produced a list of all putative CaMs, including the one-to-one orthologs used for evolutionary rate analysis, and all putative paralogs. In 12 out of 237 sequences, centrin, troponin C, or myosin regulatory light chain cdc4 were returned with a greater sequence identity than CaM, and in each case, close inspection of the level of amino acid identity confirmed that those 12 genes are obviously not CaM, and they were eliminated from our dataset. Alignments were performed using the Guidance web server running PRANK (PRobabilistic AligNment Kit) (78, 79). Because the N-terminal methionine of CaM is removed in most organisms (80), our CaM residue numbering system assumes the methionine is cleaved. CaM is part of a large EF-hand–bearing protein family, so we used best reciprocal BLAST searches as described in Methods to identify true CaM sequences. The reciprocal BLASTs were necessary to ensure that we only analyzed protein sequences that match with CaM better than with any other protein.
Evolutionary Rate Analysis.
CaM sequences were too highly conserved to estimate a phylogenetic tree, so we used the topology from Torruella et al. to guide our evolutionary rate analysis (40). This tree was chosen because it had a wide sampling of eukaryotic diversity and used robust phylogenetic analyses. Only the sequence from each organism that had the highest identity with CaM used in the initial sequence search was used in this analysis. We aligned one-to-one CaM orthologs using Mafft’s l-ins-i algorithm (81). Maximum likelihood branch lengths were then estimated on the fixed topology using Garli (82).
We used the Whelan and Goldman or “WAG” model of amino substitution with estimated equilibrium frequencies and 10 discrete gamma distributed rate categories (83).
Sequence Logo.
We collated CaM from our three groups and used MEME[20] (43) to perform motif discovery on each EF-hand region shown in Fig. 2. As CaM is highly conserved, these motifs correspond directly to those other amino acids that exist in CaM from multiple species at each position of CaM. We found that performing motif discovery separately, by group, was more useful than performing motif discovery simultaneously and allows us to see a finer resolution of the consensus sequence for CaM within each group.
Structural Evaluation of Evolution Data.
Amino acid frequency at each residue position was determined using counting functions in Excel 2007 (Microsoft). Frequencies were manually added to the B-factor column of the Protein Data Bank (PDB) file for vertebrate CaM, PDBID (PDB ID code) 1CLL. The cartoon putty and space filling structures of CaM were rendered using The PyMOL Molecular Graphics System, Version 0.99rc6 (Schrödinger, LLC). Space filling and solvent-accessible surface areas were also rendered using PyMOL. Electrostatic surface potential was determined with Ca2+ ions included using the PDB2PQR and APBS (Adaptive Poisson-Boltzmann Solver) plugins for PyMOL (84, 85). The following CaMs are represented (PDB ID code): vertebrate (vert.) CaM [1CLL (13)], paramecium (param.) [1EXR (86)], potato [1RFJ (87)], and a yeast deletion mutant of a nonfunctional fourth EF-hand [2LHH (88)].
Distance Tree.
Mean-squared distances between alignments of each EF-hand were computed by calculating the frequency vectors over all 20 amino acids for each of the 12 sites in each alignment and then summing over and averaging the distance between each value for each site. These distances were used as input for a heuristic distance tree search using the program PAUP* (Phylogenetic Analysis Using Parsimony) (89).
Principal Components Analysis.
Our implementation of PCA reduces the dimensions by using standardized linear combinations through single-value decomposition of the scaled data (90). For PCA, we aligned each EF-loop from each species separately, removed sequences with gaps, and converted these alignments to values of four different biophysical parameters for each amino acid (58–61). The results were four tables, one for each parameter, in which each row was one of the four EF-loops from one species and each column was one of the 12 sites in the EF-loops. Each entry was therefore a biophysical parameter value for a given amino acid at a given position in the EF-loop for a given species. These tables were analyzed in R using the prcomp function in the core package (91) after normalization and plotted using ggbiplot (92). Each cluster is partially bounded by an oval that circumscribes ∼68% of the data.
Analysis of the Biophysical Properties of Residues.
A matrix of frequencies at each residue position and published values for physical parameters were created (58–61). Frequencies were calculated and put into a Microsoft Excel spreadsheet. Plots were created using IGOR Pro-5.05A (Wavemetrics, Inc.) and colored using Adobe Illustrator from CS4 suite (Adobe Systems Incorporated).
Supplementary Material
Acknowledgments
We thank Jennifer Eldridge, Tom Middendorf, Margaux Miller, and Jenni Bernier-Greeson for helpful discussions. The research was supported by the National Institutes of Health under Ruth L. Kirschstein National Research Service Awards F32GM93626-02 and 1F32GM112504-01A1 from the National Institute of General Medical Sciences and by National Institutes of Health Grant NS077821.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1600385113/-/DCSupplemental.
References
- 1.Yap KL, et al. Calmodulin target database. J Struct Funct Genomics. 2000;1(1):8–14. doi: 10.1023/a:1011320027914. [DOI] [PubMed] [Google Scholar]
- 2.Shen X, Valencia CA, Szostak JW, Dong B, Liu R. Scanning the human proteome for calmodulin-binding proteins. Proc Natl Acad Sci USA. 2005;102(17):5969–5974. doi: 10.1073/pnas.0407928102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Connell DJ, et al. Integrated protein array screening and high throughput validation of 70 novel neural calmodulin-binding proteins. Mol Cell Proteomics. 2010;9(6):1118–1132. doi: 10.1074/mcp.M900324-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nyegaard M, et al. Mutations in calmodulin cause ventricular tachycardia and sudden cardiac death. Am J Hum Genet. 2012;91(4):703–712. doi: 10.1016/j.ajhg.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crotti L, et al. Calmodulin mutations associated with recurrent cardiac arrest in infants. Circulation. 2013;127(9):1009–1017. doi: 10.1161/CIRCULATIONAHA.112.001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marsman RF, et al. A mutation in CALM1 encoding calmodulin in familial idiopathic ventricular fibrillation in childhood and adolescence. J Am Coll Cardiol. 2014;63(3):259–266. doi: 10.1016/j.jacc.2013.07.091. [DOI] [PubMed] [Google Scholar]
- 7.Makita N, et al. Novel calmodulin mutations associated with congenital arrhythmia susceptibility. Circ Cardiovasc Genet. 2014;7(4):466–474. doi: 10.1161/CIRCGENETICS.113.000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reed GJ, Boczek NJ, Etheridge SP, Ackerman MJ. CALM3 mutation associated with long QT syndrome. Heart Rhythm. 2015;12(2):419–422. doi: 10.1016/j.hrthm.2014.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedrich FW, et al. EUROGENE Heart Failure Project A new polymorphism in human calmodulin III gene promoter is a potential modifier gene for familial hypertrophic cardiomyopathy. Eur Heart J. 2009;30(13):1648–1655. doi: 10.1093/eurheartj/ehp153. [DOI] [PubMed] [Google Scholar]
- 10.Kretsinger RH, Nockolds CE. Carp muscle calcium-binding protein. II. Structure determination and general description. J Biol Chem. 1973;248(9):3313–3326. [PubMed] [Google Scholar]
- 11.Moncrief ND, Kretsinger RH, Goodman M. Evolution of EF-hand calcium-modulated proteins. I. Relationships based on amino acid sequences. J Mol Evol. 1990;30(6):522–562. doi: 10.1007/BF02101108. [DOI] [PubMed] [Google Scholar]
- 12.Kawasaki H, Nakayama S, Kretsinger RH. Classification and evolution of EF-hand proteins. Biometals. 1998;11(4):277–295. doi: 10.1023/a:1009282307967. [DOI] [PubMed] [Google Scholar]
- 13.Chattopadhyaya R, Meador WE, Means AR, Quiocho FA. Calmodulin structure refined at 1.7 A resolution. J Mol Biol. 1992;228(4):1177–1192. doi: 10.1016/0022-2836(92)90324-d. [DOI] [PubMed] [Google Scholar]
- 14.Baba ML, Goodman M, Berger-Cohn J, Demaille JG, Matsuda G. The early adaptive evolution of calmodulin. Mol Biol Evol. 1984;1(6):442–455. doi: 10.1093/oxfordjournals.molbev.a040330. [DOI] [PubMed] [Google Scholar]
- 15.Marsden BJ, Shaw GS, Sykes BD. Calcium binding proteins. Elucidating the contributions to calcium affinity from an analysis of species variants and peptide fragments. Biochem Cell Biol. 1990;68(3):587–601. doi: 10.1139/o90-084. [DOI] [PubMed] [Google Scholar]
- 16.Nakayama S, Moncrief ND, Kretsinger RH. Evolution of EF-hand calcium-modulated proteins. II. Domains of several subfamilies have diverse evolutionary histories. J Mol Evol. 1992;34(5):416–448. doi: 10.1007/BF00162998. [DOI] [PubMed] [Google Scholar]
- 17.Nakayama S, Kretsinger RH. Evolution of EF-hand calcium-modulated proteins. III. Exon sequences confirm most dendrograms based on protein sequences: Calmodulin dendrograms show significant lack of parallelism. J Mol Evol. 1993;36(5):458–476. doi: 10.1007/BF02406722. [DOI] [PubMed] [Google Scholar]
- 18.Kretsinger RH, Nakayama S. Evolution of EF-hand calcium-modulated proteins. IV. Exon shuffling did not determine the domain compositions of EF-hand proteins. J Mol Evol. 1993;36(5):477–488. doi: 10.1007/BF02406723. [DOI] [PubMed] [Google Scholar]
- 19.Hofmann T, et al. Site-site interactions in EF-hand calcium-binding proteins. Laser-excited europium luminescence studies of 9-kDa calbindin, the pig intestinal calcium-binding protein. Eur J Biochem. 1988;172(2):307–313. doi: 10.1111/j.1432-1033.1988.tb13888.x. [DOI] [PubMed] [Google Scholar]
- 20.Linse S, Helmersson A, Forsén S. Calcium binding to calmodulin and its globular domains. J Biol Chem. 1991;266(13):8050–8054. [PubMed] [Google Scholar]
- 21.Falke JJ, Snyder EE, Thatcher KC, Voertler CS. Quantitating and engineering the ion specificity of an EF-hand-like Ca2+ binding. Biochemistry. 1991;30(35):8690–8697. doi: 10.1021/bi00099a029. [DOI] [PubMed] [Google Scholar]
- 22.Kilhoffer MC, Kubina M, Travers F, Haiech J. Use of engineered proteins with internal tryptophan reporter groups and pertubation techniques to probe the mechanism of ligand-protein interactions: Investigation of the mechanism of calcium binding to calmodulin. Biochemistry. 1992;31(34):8098–8106. doi: 10.1021/bi00149a046. [DOI] [PubMed] [Google Scholar]
- 23.Falke JJ, Drake SK, Hazard AL, Peersen OB. Molecular tuning of ion binding to calcium signaling proteins. Q Rev Biophys. 1994;27(3):219–290. doi: 10.1017/s0033583500003012. [DOI] [PubMed] [Google Scholar]
- 24.Linse S, Chazin WJ. Quantitative measurements of the cooperativity in an EF-hand protein with sequential calcium binding. Protein Sci. 1995;4(6):1038–1044. doi: 10.1002/pro.5560040602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye Y, et al. Metal binding affinity and structural properties of an isolated EF-loop in a scaffold protein. Protein Eng. 2001;14(12):1001–1013. doi: 10.1093/protein/14.12.1001. [DOI] [PubMed] [Google Scholar]
- 26.VanScyoc WS, et al. Calcium binding to calmodulin mutants monitored by domain-specific intrinsic phenylalanine and tyrosine fluorescence. Biophys J. 2002;83(5):2767–2780. doi: 10.1016/S0006-3495(02)75286-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ye Y, Lee H-W, Yang W, Shealy S, Yang JJ. Probing site-specific calmodulin calcium and lanthanide affinity by grafting. J Am Chem Soc. 2005;127(11):3743–3750. doi: 10.1021/ja042786x. [DOI] [PubMed] [Google Scholar]
- 28.Beccia MR, et al. Thermodynamics of calcium binding to the calmodulin N-terminal domain to evaluate site-specific affinity constants and cooperativity. J Biol Inorg Chem. 2015;20(5):905–919. doi: 10.1007/s00775-015-1275-1. [DOI] [PubMed] [Google Scholar]
- 29.Tsalkova TN, Privalov PL. Thermodynamic study of domain organization in troponin C and calmodulin. J Mol Biol. 1985;181(4):533–544. doi: 10.1016/0022-2836(85)90425-5. [DOI] [PubMed] [Google Scholar]
- 30.Waltersson Y, Linse S, Brodin P, Grundström T. Mutational effects on the cooperativity of Ca2+ binding in calmodulin. Biochemistry. 1993;32(31):7866–7871. doi: 10.1021/bi00082a005. [DOI] [PubMed] [Google Scholar]
- 31.Copley RR, Schultz J, Ponting CP, Bork P. Protein families in multicellular organisms. Curr Opin Struct Biol. 1999;9(3):408–415. doi: 10.1016/S0959-440X(99)80055-4. [DOI] [PubMed] [Google Scholar]
- 32.Wang L, Zhuang WY. Phylogenetic analyses of penicillia based on partial calmodulin gene sequences. Biosystems. 2007;88(1-2):113–126. doi: 10.1016/j.biosystems.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 33.Romeo O, Scordino F, Criseo G. New insight into molecular phylogeny and epidemiology of Sporothrix schenckii species complex based on calmodulin-encoding gene analysis of Italian isolates. Mycopathologia. 2011;172(3):179–186. doi: 10.1007/s11046-011-9420-z. [DOI] [PubMed] [Google Scholar]
- 34.Geiser JR, van Tuinen D, Brockerhoff SE, Neff MM, Davis TN. Can calmodulin function without binding calcium? Cell. 1991;65(6):949–959. doi: 10.1016/0092-8674(91)90547-c. [DOI] [PubMed] [Google Scholar]
- 35.Kretsinger R, Nakayama S. Evolution of EF-hand calcium-modulated proteins. IV. Exon shuffling did not determine the domain compositions of EF-hand proteins. J Mol Evol. 1993;36(5):477–488. doi: 10.1007/BF02406723. [DOI] [PubMed] [Google Scholar]
- 36.Braam J, Davis RW. Rain-, wind-, and touch-induced expression of calmodulin and calmodulin-related genes in Arabidopsis. Cell. 1990;60(3):357–364. doi: 10.1016/0092-8674(90)90587-5. [DOI] [PubMed] [Google Scholar]
- 37.Xiao C, Xin H, Dong A, Sun C, Cao K. A novel calmodulin-like protein gene in rice which has an unusual prolonged C-terminal sequence carrying a putative prenylation site. DNA Res. 1999;6(3):179–181. doi: 10.1093/dnares/6.3.179. [DOI] [PubMed] [Google Scholar]
- 38.Karabinos A, Bhattacharya D. Molecular evolution of calmodulin and calmodulin-like genes in the cephalochordate Branchiostoma. J Mol Evol. 2000;51(2):141–148. doi: 10.1007/s002390010074. [DOI] [PubMed] [Google Scholar]
- 39.Simpson RJ, Wilding CS, Grahame J. Intron analyses reveal multiple calmodulin copies in Littorina. J Mol Evol. 2005;60(4):505–512. doi: 10.1007/s00239-004-0232-3. [DOI] [PubMed] [Google Scholar]
- 40.Torruella G, et al. Phylogenetic relationships within the Opisthokonta based on phylogenomic analyses of conserved single-copy protein domains. Mol Biol Evol. 2012;29(2):531–544. doi: 10.1093/molbev/msr185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davis TN, Urdea MS, Masiarz FR, Thorner J. Isolation of the yeast calmodulin gene: Calmodulin is an essential protein. Cell. 1986;47(3):423–431. doi: 10.1016/0092-8674(86)90599-4. [DOI] [PubMed] [Google Scholar]
- 42.Friedberg F, Rhoads AR. Evolutionary aspects of calmodulin. IUBMB Life. 2001;51(4):215–221. doi: 10.1080/152165401753311753. [DOI] [PubMed] [Google Scholar]
- 43.Bailey TL, et al. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009;37(Web Server issue) suppl 2:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneider TD, Stephens RM. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 1990;18(20):6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohya Y, Botstein D. Structure-based systematic isolation of conditional-lethal mutations in the single yeast calmodulin gene. Genetics. 1994;138(4):1041–1054. doi: 10.1093/genetics/138.4.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Villarroel A, et al. The ever changing moods of calmodulin: How structural plasticity entails transductional adaptability. J Mol Biol. 2014;426(15):2717–2735. doi: 10.1016/j.jmb.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 47.Sorensen AB, Søndergaard MT, Overgaard MT. Calmodulin in a heartbeat. FEBS J. 2013;280(21):5511–5532. doi: 10.1111/febs.12337. [DOI] [PubMed] [Google Scholar]
- 48.Babu YS, et al. Three-dimensional structure of calmodulin. Nature. 1985;315(6014):37–40. doi: 10.1038/315037a0. [DOI] [PubMed] [Google Scholar]
- 49.Kretsinger RH. Gene triplication deduced from the tertiary structure of a muscle calcium binding protein. Nat New Biol. 1972;240(98):85–88. doi: 10.1038/newbio240085a0. [DOI] [PubMed] [Google Scholar]
- 50.Meador WE, Means AR, Quiocho FA. Target enzyme recognition by calmodulin: 2.4 A structure of a calmodulin-peptide complex. Science. 1992;257(5074):1251–1255. doi: 10.1126/science.1519061. [DOI] [PubMed] [Google Scholar]
- 51.Ikura M, et al. Solution structure of a calmodulin-target peptide complex by multidimensional NMR. Science. 1992;256(5057):632–638. doi: 10.1126/science.1585175. [DOI] [PubMed] [Google Scholar]
- 52.Brown CJ, Johnson AK, Daughdrill GW. Comparing models of evolution for ordered and disordered proteins. Mol Biol Evol. 2010;27(3):609–621. doi: 10.1093/molbev/msp277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mruk K, Farley BM, Ritacco AW, Kobertz WR. Calmodulation meta-analysis: Predicting calmodulin binding via canonical motif clustering. J Gen Physiol. 2014;144(1):105–114. doi: 10.1085/jgp.201311140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Strong M, Chandy KG, Gutman GA. Molecular evolution of voltage-sensitive ion channel genes: On the origins of electrical excitability. Mol Biol Evol. 1993;10(1):221–242. doi: 10.1093/oxfordjournals.molbev.a039986. [DOI] [PubMed] [Google Scholar]
- 55.Ogura K, et al. Solution structures of yeast Saccharomyces cerevisiae calmodulin in calcium- and target peptide-bound states reveal similarities and differences to vertebrate calmodulin. Genes Cells. 2012;17(3):159–172. doi: 10.1111/j.1365-2443.2012.01580.x. [DOI] [PubMed] [Google Scholar]
- 56.Matsuura I, et al. A site-directed mutagenesis study of yeast calmodulin. J Biochem. 1991;109(1):190–197. doi: 10.1093/oxfordjournals.jbchem.a123344. [DOI] [PubMed] [Google Scholar]
- 57.Starovasnik MA, Davis TN, Klevit RE. Similarities and differences between yeast and vertebrate calmodulin: An examination of the calcium-binding and structural properties of calmodulin from the yeast Saccharomyces cerevisiae. Biochemistry. 1993;32(13):3261–3270. doi: 10.1021/bi00064a008. [DOI] [PubMed] [Google Scholar]
- 58.Fersht A. Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding. W. H. Freeman and Company; New York: 1999. Forces between molecules, and binding energies; pp. 324–348. [Google Scholar]
- 59.Bhaskaran R, Ponnuswamy PK. Positional flexibilities of amino-acid residues in globular-proteins. Int J Pept Protein Res. 1988;32(4):241–255. doi: 10.1111/j.1399-3011.1984.tb00944.x. [DOI] [PubMed] [Google Scholar]
- 60.Zimmerman JM, Eliezer N, Simha R. The characterization of amino acid sequences in proteins by statistical methods. J Theor Biol. 1968;21(2):170–201. doi: 10.1016/0022-5193(68)90069-6. [DOI] [PubMed] [Google Scholar]
- 61.Harpaz Y, Gerstein M, Chothia C. Volume changes on protein folding. Structure. 1994;2(7):641–649. doi: 10.1016/s0969-2126(00)00065-4. [DOI] [PubMed] [Google Scholar]
- 62.Davis TN, Thorner J. Vertebrate and yeast calmodulin, despite significant sequence divergence, are functionally interchangeable. Proc Natl Acad Sci USA. 1989;86(20):7909–7913. doi: 10.1073/pnas.86.20.7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ohya Y, Anraku Y. Functional expression of chicken calmodulin in yeast. Biochem Biophys Res Commun. 1989;158(2):541–547. doi: 10.1016/s0006-291x(89)80083-x. [DOI] [PubMed] [Google Scholar]
- 64.Liebeskind BJ, Hillis DM, Zakon HH. Convergence of ion channel genome content in early animal evolution. Proc Natl Acad Sci USA. 2015;112(8):E846–E851. doi: 10.1073/pnas.1501195112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kursula P. The many structural faces of calmodulin: A multitasking molecular jackknife. Amino Acids. 2014;46(10):2295–2304. doi: 10.1007/s00726-014-1795-y. [DOI] [PubMed] [Google Scholar]
- 66.Halling DB, Aracena-Parks P, Hamilton SL. Regulation of voltage-gated Ca2+ channels by calmodulin. Sci STKE. 2005;2005(315):re15. doi: 10.1126/stke.3152005re15. [DOI] [PubMed] [Google Scholar]
- 67.Halling DB, Kenrick SA, Riggs AF, Aldrich RW. Calcium-dependent stoichiometries of the KCa2.2 (SK) intracellular domain/calmodulin complex in solution. J Gen Physiol. 2014;143(2):231–252. doi: 10.1085/jgp.201311007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gaertner TR, Putkey JA, Waxham MN. RC3/neurogranin and Ca2+/calmodulin-dependent protein kinase II produce opposing effects on the affinity of calmodulin for calcium. J Biol Chem. 2004;279(38):39374–39382. doi: 10.1074/jbc.M405352200. [DOI] [PubMed] [Google Scholar]
- 69.Peersen OB, Madsen TS, Falke JJ. Intermolecular tuning of calmodulin by target peptides and proteins: Differential effects on Ca2+ binding and implications for kinase activation. Protein Sci. 1997;6(4):794–807. doi: 10.1002/pro.5560060406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tadross MR, Dick IE, Yue DT. Mechanism of local and global Ca2+ sensing by calmodulin in complex with a Ca2+ channel. Cell. 2008;133(7):1228–1240. doi: 10.1016/j.cell.2008.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liang H, et al. Unified mechanisms of Ca2+ regulation across the Ca2+ channel family. Neuron. 2003;39(6):951–960. doi: 10.1016/s0896-6273(03)00560-9. [DOI] [PubMed] [Google Scholar]
- 72.Slavov N, Carey J, Linse S. Calmodulin transduces Ca2+ oscillations into differential regulation of its target proteins. ACS Chem Neurosci. 2013;4(4):601–612. doi: 10.1021/cn300218d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hines KE, Middendorf TR, Aldrich RW. Determination of parameter identifiability in nonlinear biophysical models: A Bayesian approach. J Gen Physiol. 2014;143(3):401–416. doi: 10.1085/jgp.201311116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Motlagh HN, Wrabl JO, Li J, Hilser VJ. The ensemble nature of allostery. Nature. 2014;508(7496):331–339. doi: 10.1038/nature13001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Grigoriev IV, et al. The genome portal of the Department of Energy Joint Genome Institute. Nucleic Acids Res. 2012;40(Database issue) D1:D26–D32. doi: 10.1093/nar/gkr947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hemmrich G, Bosch TCG. Compagen, a comparative genomics platform for early branching metazoan animals, reveals early origins of genes regulating stem-cell differentiation. BioEssays. 2008;30(10):1010–1018. doi: 10.1002/bies.20813. [DOI] [PubMed] [Google Scholar]
- 77.Consortium TU. UniProt Consortium Activities at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2014;42(Database issue) D1:D191–D198. doi: 10.1093/nar/gkt1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Löytynoja A, Goldman N. An algorithm for progressive multiple alignment of sequences with insertions. Proc Natl Acad Sci USA. 2005;102(30):10557–10562. doi: 10.1073/pnas.0409137102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Penn O, et al. GUIDANCE: A web server for assessing alignment confidence scores. Nucleic Acids Res. 2010;38(Web Server issue):W23–W28. doi: 10.1093/nar/gkq443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Frottin F, et al. The proteomics of N-terminal methionine cleavage. Mol Cell Proteomics. 2006;5(12):2336–2349. doi: 10.1074/mcp.M600225-MCP200. [DOI] [PubMed] [Google Scholar]
- 81.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33(2):511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zwickl DJ. Genetic Algorithm Approaches for the Phylogenetic Analysis of Large Biological Sequence Datasets Under the Maximum Likelihood Criterion. The University of Texas at Austin; Austin, TX: 2006. [Google Scholar]
- 83.Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001;18(5):691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 84.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98(18):10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. PDB2PQR: An automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32(Web Server issue) suppl 2:W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wilson MA, Brunger AT. The 1.0 A crystal structure of Ca(2+)-bound calmodulin: An analysis of disorder and implications for functionally relevant plasticity. J Mol Biol. 2000;301(5):1237–1256. doi: 10.1006/jmbi.2000.4029. [DOI] [PubMed] [Google Scholar]
- 87.Yun C-H, et al. Structure of potato calmodulin PCM6: The first report of the three-dimensional structure of a plant calmodulin. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 7):1214–1219. doi: 10.1107/S0907444904009771. [DOI] [PubMed] [Google Scholar]
- 88.Nakashima K, Ishida H, Nakatomi A, Yazawa M. Specific conformation and Ca(2+)-binding mode of yeast calmodulin: Insight into evolutionary development. J Biochem. 2012;152(1):27–35. doi: 10.1093/jb/mvs048. [DOI] [PubMed] [Google Scholar]
- 89.Swofford DL. 2003 PAUP*. Phylogenetic Analysis Using Parsimony (*and other methods). Version 4 (Sinauer Associates, Sunderland, MA). Available at paup.csit.fsu.edu/about.html.
- 90.Afifi A, May S, Clark VA. Practical Multivariate Analysis. 5th Ed Taylor & Francis Group, LLC; Boca Raton, FL: 2011. [Google Scholar]
- 91.R_Core_Team . R: A Language and Environment for Statistical Computing. Foundation for Statistical Computing; Vienna, Austria: 2013. https://www.r-project.org/. Accessed April 16, 2014. [Google Scholar]
- 92.Vu VQ. 2011. ggbiplot: A ggplot2 based biplot. R Package Version 0.55, https://github.com/vqv/ggbiplot. Accessed April 16, 2014.
- 93.National Heart, Lung, and Blood Institute 2015. Exome Variant Server in NHLBI GO ESP, evs.gs.washington.edu/EVS/. Accessed July 3, 2015.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.