Abstract
Hydrogenase enzymes catalyze the rapid and reversible interconversion of H2 with protons and electrons. The active site of the [FeFe] hydrogenase is the H cluster, which consists of a [4Fe–4S]H subcluster linked to an organometallic [2Fe]H subcluster. Understanding the biosynthesis and catalytic mechanism of this structurally unusual active site will aid in the development of synthetic and biological hydrogenase catalysts for applications in solar fuel generation. The [2Fe]H subcluster is synthesized and inserted by three maturase enzymes—HydE, HydF, and HydG—in a complex process that involves inorganic, organometallic, and organic radical chemistry. HydG is a member of the radical S-adenosyl-L-methionine (SAM) family of enzymes and is thought to play a prominent role in [2Fe]H subcluster biosynthesis by converting inorganic Fe2+, L-cysteine (Cys), and L-tyrosine (Tyr) into an organometallic [(Cys)Fe(CO)2(CN)]− intermediate that is eventually incorporated into the [2Fe]H subcluster. In this Forum Article, the mechanism of [2Fe]H subcluster biosynthesis is discussed with a focus on how this key [(Cys)Fe(CO)2(CN)]− species is formed. Particular attention is given to the initial metallocluster composition of HydG, the modes of substrate binding (Fe2+, Cys, Tyr, and SAM), the mechanism of SAM-mediated Tyr cleavage to CO and CN−, and the identification of the final organometallic products of the reaction.
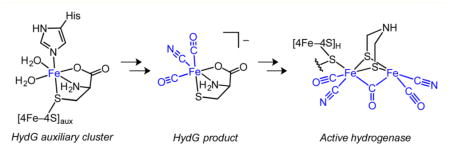
Graphical abstract
INTRODUCTION
The development of artificial photosynthetic technologies for the generation of solar fuels requires the interplay of small molecules with transition-metal clusters that facilitate multielectron catalysis. The only viable source of inexpensive electrons and protons for making reduced fuel is water, and nature has evolved an efficient solar-driven water oxidation machine in the photosystem II (PSII) reaction center with its integral Mn4Ca-oxo catalyst.1–3 On the other hand, various reduced fuels can, in principle, be generated, with the simplest being H2 that results from the combination of the electrons and protons from the water-splitting reaction. Electrical potential can then be generated in a fuel cell with the reoxidation of H2 by the other product of water oxidation, O2. Hydrogenase enzymes employing [FeFe] or [NiFe] active sites4 are efficient catalysts for H2 production. Here we focus on the [FeFe] hydrogenases, for which H2 production turnover frequencies have been reported to be as high as 10000 s−1.5 Their catalytic six-Fe H cluster (Figure 1) contains a unique binuclear Fe subcluster (“[2Fe]H”) with CO and CN− ligands along with a bridging azadithiolate ligand.6–10 The only protein residue that ligates the [2Fe]H subcluster is a cysteine which acts as a bridge to a [4Fe–4S] subcluster (“[4Fe–4S]H”) that comprises the remainder of the H cluster.
Figure 1.
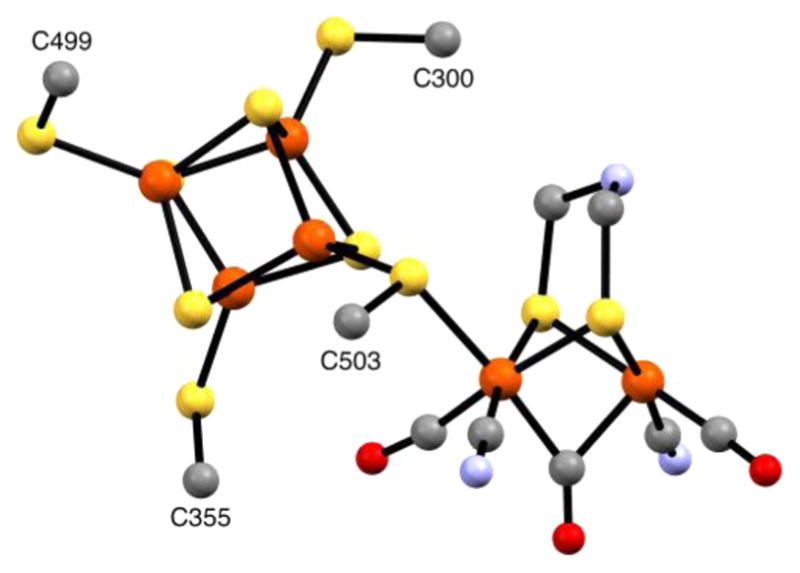
Ball-and-stick representation of the H cluster from the X-ray crystal structure of Clostridium pasteurianum HydA (pdb code 3C8Y),95 with the dithiolate bridging ligand taken as 2-azapropane-1,3-dithiolate. Color code: orange, Fe; yellow, S; gray, C; blue, N; red, O.
Metal-cluster active sites such as those in PSII and the [FeFe] hydrogenase must themselves be assembled, and we can learn much about building artificial catalysts from the natural assembly mechanisms. Interestingly, the inorganic water-splitting catalyst of PSII can be assembled without additional enzymes in a process termed photoactivation, which uses the photooxidation chemistry intrinsic to PSII to oxidize MnII in order to form the Mn4Ca-oxo cluster.11–14 In contrast, assembling the organometallic H cluster of the [FeFe] hydrogenase requires a specific set of Fe–S enzymes—HydE, HydF, and HydG—that perform a series of complex reactions involving elements of inorganic cluster chemistry, organometallic chemistry, and organic radical chemistry. These reactions and their mechanisms are only beginning to be elucidated.
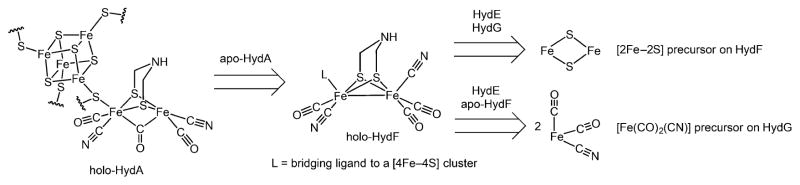
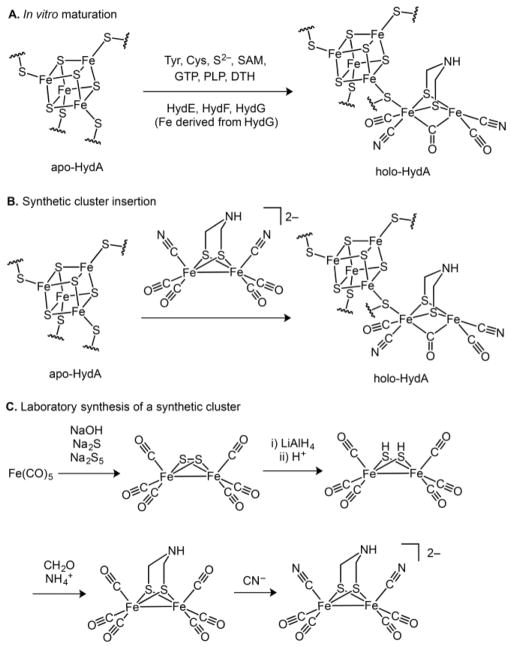
A number of routes can be envisioned for the biosynthesis of the H cluster. Given the complexity of the process, it is often useful to tackle the problem retrosynthetically.15 Working backward, the first established disconnection is between the [2Fe]H and [4Fe–4S]H subclusters (Scheme 1): the [4Fe–4S]H subcluster is synthesized and inserted by the “housekeeping” Fe–S cluster machinery, whereas the HydE, HydG, and HydF “maturase” enzymes are responsible for the biosynthesis of the [2Fe]H subcluster (Scheme 2A).16,17 Thus, hydrogenase (HydA) expressed without coexpression of the maturases harbors only the [4Fe–4S]H subcluster and is therefore referred to as “apo-HydA”.16,17 The [2Fe]H subcluster can be installed using in vitro maturation protocols that employ the individually expressed maturases in conjunction with a cocktail of small-molecule additives (Scheme 2A);18–21 such protocols allow for the individual roles of both the maturases and small molecules to be studied in detail (vide infra) as well as for selective isotopic labeling of the [2Fe]H subcluster.22–25 Alternatively, the [2Fe]H subcluster can be installed into apo-HydA using diiron synthetic precursors (Scheme 2B), a methodology that allows for artificial and isotopically labeled variants to be prepared.10,26–29 These processes take advantage of the stepwise assembly of the H cluster, each employing a late-stage fragment coupling of the [4Fe–4S]H and [2Fe]H subclusters; earlier precedent for this chemical step can be found in the synthesis of a close structural model of the H cluster.30
Scheme 1.
Proposals for Key Synthons in [2Fe]H Subcluster Bioassembly
Scheme 2. Synthesis and Installation of the [2Fe]H Subcluster into apo-HydAa.
aTyr = L-tyrosine; Cys = L-cysteine; SAM = S-adenosyl-L-methionine; GTP = guanosine triphosphate; PLP = pyridoxal phosphate; DTH = dithionite.
In vivo activation of apo-HydA is thought to occur by insertion of a preassembled [2Fe]H subcluster that is generated on HydF. This is based, in part, on the finding that HydF, when coexpressed with HydE and HydG, is capable of activating apo-HydA.31 In addition, HydF expressed in this manner harbors a cluster with Fourier transform infrared (FTIR) and X-ray spectroscopic properties similar to those of the H cluster;32–34 however, the precise structure of this H-cluster precursor awaits further characterization. This preassembly process on HydF has parallels in the bioassembly of the nitrogenase FeMo cofactor in which the FeMo cofactor undergoes its final assembly steps on the NifEN complex before being inserted into the NifDK complex.35
Perhaps the most interesting questions pertain to how the [2Fe]H subcluster is generated by the biosynthetic machinery. Significant progress has been made in the synthesis of structural and functional diiron model complexes,36 some of which uncannily predate the structure determination of the [FeFe] hydrogenase.8,9 The preparations of these complexes provide inspiration for mechanistic proposals for [2Fe]H subcluster biosynthesis and also highlight the challenges inherent to the biosynthetic process. To illustrate this, we briefly describe the first synthesis of [(adt)Fe2(CO)4(CN)2]2− (adt = 2-azapropane-1,3-dithiolate; Scheme 2C),37–40 which can be used to activate apo-HydA (Scheme 2B).10,26,28 In this and related syntheses, Fe and the CO ligands are often sourced from the inexpensive and convenient starting material Fe(CO)5, and CN− ligands are often introduced by ligand substitution for CO ligands.41–44 CO substitution chemistry is likewise important in the biosynthetic process, although displacement by CN− is likely not operative (vide infra). For [(adt)Fe2(CO)4(CN)2]2−, the adt ligand is built by elaboration of an Fe2(μ2-SH)2(CO)4 cluster in a series of condensation reactions using formaldehyde and the appropriate amine (NH4+ in this case; Scheme 2C). Although a related reaction sequence has been proposed for the biosynthesis of the adt ligand,45 a number of alternative pathways could be envisioned including some that involve radical chemistry and the maturase HydE.46 Our knowledge concerning the biosynthesis of the azadithiolate ligand is limited, and the reaction promoted by HydE is not known; as such, we will not discuss either of these in detail herein.
Whereas Fe(CO)5 is a useful starting material in the laboratory syntheses of [2Fe]H subcluster model complexes, it is likely not a precursor to the biological [2Fe]H subcluster. This immediately raises the questions of how the diiron core is assembled and what the identities of its key inorganic and organometallic precursors are. It has been suggested that a [2Fe–2S] cluster on HydF could be an inorganic precursor to the [2Fe]H subcluster (Scheme 1),32,47 with HydE and HydG subsequently installing the CO, CN−, and azadithiolate ligands. One attractive feature of this proposal is that [2Fe–2S] clusters are common motifs in biology and their synthesis and installation by the Fe–S cluster housekeeping machinery has been documented.48,49 However, other electron paramagnetic resonance (EPR) spectroscopic studies of HydF are not consistent with the presence of a [2Fe–2S] cluster.21,50–53 Moreover, 57Fe electron–nuclear double resonance spectroscopy (ENDOR) studies show that Fe in the [2Fe]H subcluster is supplied by HydG, which suggests that a [2Fe–2S] cluster on HydF is unlikely to be an inorganic precursor to the [2Fe]H subcluster.23,25 An alternative mechanistic framework invokes a mononuclear Fe(CO)x(CN)y precursor that is first formed on HydG (Scheme 1). In support of such a process, we have reported FTIR spectroscopic evidence for the formation of an organometallic [Fe(CO)2(CN)] precursor to the H cluster (vide infra).23 Given the 57Fe ENDOR and FTIR spectroscopic results, mechanistic proposals for the biosynthesis of the [2Fe]H subcluster should take into account the donation of Fe from HydG and the formation of an [Fe(CO)2(CN)] synthon on HydG.
In this Forum Article, we discuss the spectroscopic characterization of the maturases in the context of their roles in building the [2Fe]H subcluster with an emphasis on the key role of HydG. We describe recent studies that elucidate how the [Fe(CO)2(CN)] synthon is built including the characterization of its inorganic precursor on HydG, new experimental results pertaining to the mode of the substrate binding, the structures of intermediates, and a recent proposal concerning the organometallic product of the HydG reaction and its role in the H-cluster assembly process.
MATERIALS AND METHODS
Materials
Nonisotopically enriched chemicals were purchased from common commercial vendors. Isotopically enriched chemicals were purchased from Cambridge Isotope Laboratories. All additives except for L-tyrosine (Tyr) were dissolved in 50 mM HEPES buffer (pH = 7.5) with 50 mM KCl and adjusted to pH = 7.5 before use. Tyrosine solutions were prepared as previously described.54
Protein Expression and Purification
Shewanella oneidensis (So) wild-type (WT) HydG (“HydG”), HydGXN, and HydGXC (also called “HydGSxxS”) were expressed in Escherichia coli BL21(DE3) ΔiscR::kan cells, purified using a StrepTactin–Sepharose column as previously described,20,21,55 and frozen before the preparation of spectroscopic samples.
Spectroscopic Sample Preparation
EPR samples were prepared in an anaerobic glovebox under an N2 atmosphere (<1 ppm of O2) and frozen using liquid nitrogen before spectroscopic analysis. Unless otherwise indicated, the final concentrations were as follows: freshly thawed HydG, ~200–1000 μM; dithionite (DTH), 3 mM or 10 mM; all other additives, 3 mM. Other than HydG, each component was added as a solution of 10-fold higher concentration than its final concentration. See relevant references for the details of previously reported experiments.
EPR Spectroscopic Methods
X-band continuous-wave (CW) EPR spectra in Figure 6 were collected on a Bruker ELEXSYS E500 spectrometer equipped with a cylindrical TE011-mode resonator (SHQE-W), an ESR-900 liquid-helium cryostat, and an ITC-5 temperature controller (Oxford Instruments). Spectra were recorded at 9.4 GHz and 10 K using 126 μW microwave power and 5.0 G modulation amplitude. Hyperfine sublevel correlation spectroscopy (HYSCORE) spectra in Figure 7 were recorded using a Bruker ELEXSYS E580 spectrometer, a CF935 cryostat (Oxford Instruments), and an ITC-5 temperature controller (Oxford Instruments). Spectra were acquired at 9.7–9.8 GHz and 10 K with a split-ring (MS5) resonator using the pulse sequence π/2–τ–π/2–t1–π–t2–π/2–τ–echo, wherein both the excitation and inversion pulse lengths were identical (16 ns). Values of τ were chosen to suppress 1H nuclear coherences (τ = 128–140 ns). Four-step phase cycling was employed. Time-domain spectra were baseline-corrected (third-order polynomial), apodized with a Hamming window, zero-filled to 8-fold points, and fast-Fourier-transformed to yield the frequency-domain spectra. Spectral simulations were performed with MATLAB release 2015A using the EasySpin 4.5.5 toolbox.56 The simulations in Figure 7B use g = [2.045, 1.963, 1.908] and the zyz Euler angle convention. See relevant references for the details of previously reported experiments.
Figure 6.
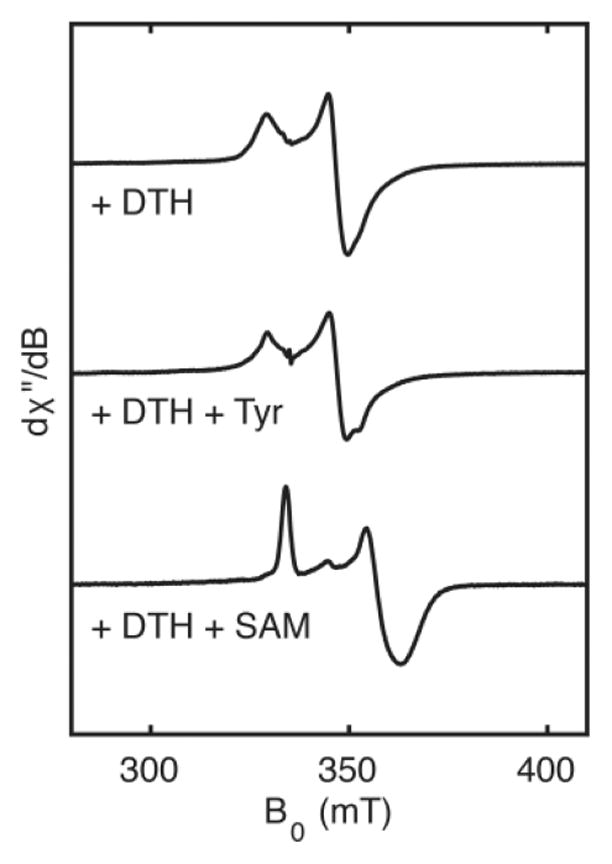
X-band EPR spectroscopic analysis of the interaction between the HydG [4Fe–4S]RS cluster and the substrates, Tyr or SAM.
Figure 7.
X-band HYSCORE spectra of WT HydG treated with DTH and 15N-Tyr (A), HydGXC treated with DTH and 15N-Tyr (B), or WT HydG treated with DTH and 15N-L-phenylalanine (C). Simulations in the bottom of part B are shown in blue, with the data reproduced in red for clarity. The features centered at (3.8, 3.8) MHz that are present in all spectra are assigned to a weakly coupled 14N nucleus.
RESULTS AND DISCUSSION
Fe–S Cluster Composition of HydG
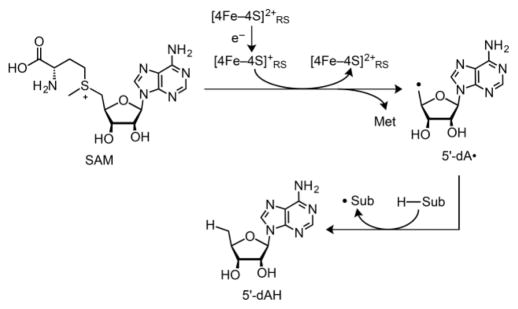
Each of the [FeFe] hydrogenase maturase enzymes contains at least one Fe–S cluster. HydE and HydG are members of the radical S-adenosyl-L-methionine (SAM) family of enzymes,57–60 featuring a [4Fe–4S]RS cluster typically bound by a CX3CX2C motif. The noncysteine-coordinated Fe of the cluster binds SAM as an N/O chelate.61 For many radical SAM enzymes including HydG, reduction of the cluster triggers homolytic SAM cleavage (Scheme 3), producing methionine plus a strongly oxidizing 5′-deoxyadenosyl radical (5′-dA•). 5′-dA• abstracts an H atom from the specific substrate for a given radical SAM enzyme. For HydG, this substrate is Tyr, the small-molecule precursor to the CO and CN ligands of the [2Fe]H subcluster.20,45,62,63 The transformation of Tyr into CO, CN−, and the byproduct p-cresol is a main topic of discussion in this Forum Article.
Scheme 3. General H-Atom Abstraction Reaction Promoted by Radical SAM Enzymesa.
aMet = L-methionine.
HydG contains a second Fe–S cluster, and we have proposed that this “auxiliary” cluster is the site of [Fe-(CO)2(CN)] synthon formation and the eventual source of Fe in the [2Fe]H subcluster.23,25,55,64 As such, a detailed understanding of the structure and reactivity of this intermediate is of paramount importance. When studied in isolation, the Fe–S clusters in radical SAM enzymes can be quite unstable and may decompose during enzyme isolation or manipulation; these clusters are therefore often reconstituted with Fe and S2−.65 Early EPR spectroscopic studies of HydG suggested that this enzyme may66 or may not33 harbor an auxiliary Fe–S cluster. For example, His6-tagged Thermotoga maritima HydG that was expressed in E. coli, isolated using nickel-affinity chromatography, and subsequently reconstituted with iron and sulfide showed a complex EPR spectrum indicative of a [4Fe–4S] cluster, with additional shoulders that could arise from a second [4Fe–4S] cluster.66 Subsequent EPR spectroscopic studies on Fe/S-reconstituted Clostridium acetobutylicum (Ca) HydG compared the EPR spectra of the WT enzyme with those of mutant enzymes containing cysteine deletions near either the N- or C-terminus.67 The EPR results revealed two S = 1/2 EPR signals in reduced samples, with the N-terminal [4Fe–4S] cluster signal showing a characteristic shift upon SAM binding. On the basis of these findings, it was argued that HydG contains two separate [4Fe–4S] clusters: an N-terminal [4Fe–4S] cluster that binds SAM and generates the primary 5′-dA• radical and a C-terminal “auxiliary” [4Fe–4S] cluster that serves another purpose (orienting substrate and ensuring N-terminal cluster reduction were two suggested roles).67
Our HydG EPR spectra,23,54,55,64 obtained from non-reconstituted So HydG expressed in E. coli and isolated using StrepTactin-affinity chromatography, are distinctly different: in addition to the S = 1/2 [4Fe–4S]RS cluster signal, we observe a strong, low-field S = 5/2 signal in spectra of the WT enzyme (Figure 2). The EPR spectrum of the C394/397S, C-terminal Fe–S cluster deletion mutant (called “HydGXC” or “HydGSxxS”) shows only an S = 1/2 [4Fe–4S] cluster signal (Figure 2) that shifts upon the addition of SAM, thereby confirming the assignment of this signal to the [4Fe–4S]RS cluster.54 On the other hand, the EPR spectrum of the C103/107/110S, N-terminal Fe–S cluster deletion mutant (“HydGXN”) shows only an S = 5/2 EPR signal, confirming the assignment of this signal to that of the C-terminal auxiliary cluster (Figure 2).55 S = 5/2 spin systems are unusual for biological Fe–S clusters, and reduced [4Fe–4S] clusters are not known to adopt an S = 5/2 ground state.68,69 In the context of a [4Fe–4S] structural assignment to the C-terminal cluster, one possible model for the S = 5/2 EPR signal is a cuboidal [3Fe–4S] cluster.54 However, the [3Fe–4S] clusters that adopt an S = 5/2 spin state are linear oxidized clusters such as those found in “purple aconitase”70 and related model complexes71 or reduced mixed-metal clusters such as [Zn3Fe–4S]−;72 neither is a satisfying structure for the reduced HydG enzyme. The recent publication of the first X-ray structure of HydG with an intact Fe–S cluster in the C-terminal domain offered a more compelling and intriguing origin of this high-spin EPR signal, revealing that HydG can bind a unique five-Fe cluster in the C-terminal domain.55
Figure 2.
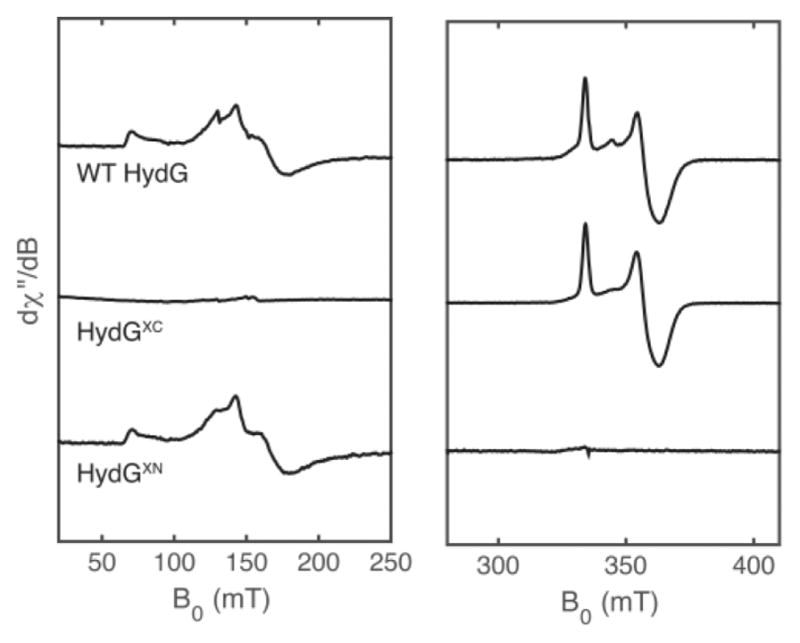
X-band CW EPR spectra of nonreconstituted WT HydG (top), HydGXC (middle), and HydGXN (bottom). All samples were treated with DTH, and the WT HydG and HydGXC samples were further treated with SAM. The S = 5/2 EPR signals in the WT HydG and HydGXN samples (left panel) correspond to that of the C-terminal [4Fe–4S]aux[(Cys)Fe] cluster form. The major S = 1/2 signals in the WT HydG and HydGXC samples (right panel) correspond to that of the SAM-bound form of the N-terminal [4Fe–4S]RS cluster. These WT HydG and HydGXN spectra are representative of typical HydG preparations, wherein the auxiliary cluster is isolated predominantly in its [4Fe–4S]aux[(Cys)Fe] cluster form. Reproduced with permission from ref 64. Copyright 2015 National Academy of Sciences.
Figure 3 shows a schematic of the five-Fe, C-terminal cluster observed in the X-ray structure of Fe/S-reconstituted Thermoanaerobacter italicus (Ti) HydG. In one asymmetric unit (Figure 3A), the C-terminal cluster contains a [4Fe–4S]aux cluster core, with three of the Fe atoms bound by cysteine residues. The fourth Fe of the cubane is linked to the unique fifth Fe (in 73% occupancy) by a bridging sulfide. A conserved histidine (H265) residue provides the single protein ligand to the fifth Fe; the remaining ligands are water molecules and an unidentified nonproteinaceous amino acid. In the other asymmetric unit (Figure 3B), the fifth Fe is missing, which highlights its lability and suggests the possibility that this is the Fe atom that is donated by HydG to build the [2Fe]H subcluster.55 Moreover, the presence of the fifth Fe, assigned to a high-spin Fe2+ center, provides a compelling explanation for our observed S = 5/2 signal in So HydG because this “dangler Fe” contributes an additional S = 2 electron spin center that can couple with unpaired electron spins of the S = 1/2 [4Fe–4S]aux cluster to give the observed signal.25,55,64
Figure 3.
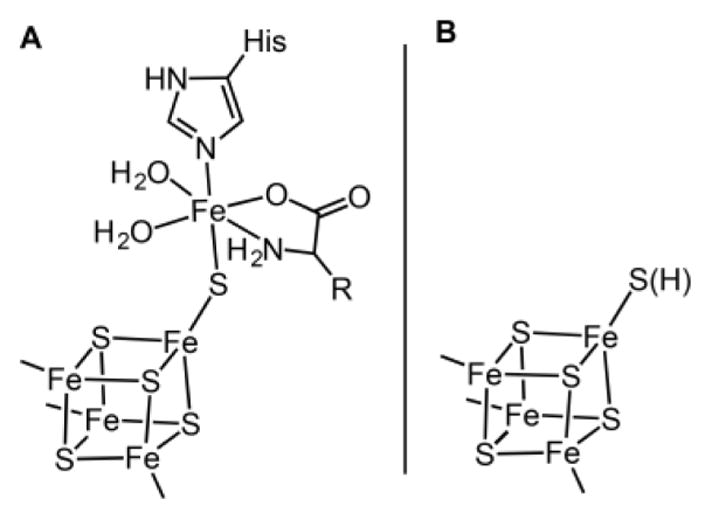
Representations of the two auxiliary cluster forms (A and B) observed in the X-ray crystal structure of Ti HydG.55
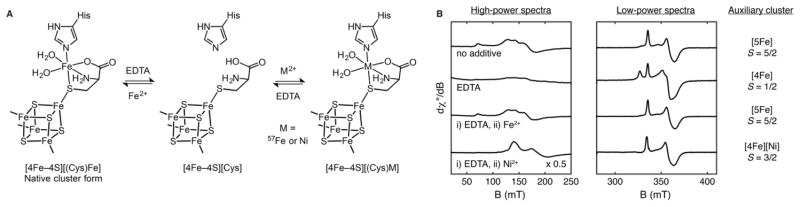
These findings raise the question of why we observe a five-Fe, C-terminal cluster with an S = 5/2 EPR signal, whereas other laboratories have reported a [4Fe–4S]aux cluster with an S = 1/2 EPR signal. In a recent paper, we identified L-cysteine (Cys) as an integral component of the auxiliary cluster.64 Given the presence of the bridging sulfide and the unidentified amino acid ligand in the Ti HydG X-ray structure,55 it seemed possible that Cys provides both structural features in one molecular unit. In addition, Cys is one of the small molecules required for the cell-free synthesis of the H cluster.18,21 We demonstrated Cys binding to the auxiliary cluster using 13C ENDOR spectroscopy and CW EPR spectroscopy of batches of nonreconstituted So HydG samples that show diminished S = 5/2 signal intensity with extra S = 1/2 signal intensity;64 although the reason for why some HydG batches occasionally exhibit weak S = 5/2 signal intensity is not clear at present, such batches provided fortunate test samples for studying reconstitution of the S = 5/2 auxiliary cluster. Treatment of such samples with Cys and Fe2+ quenches the S = 1/2 signal(s) and converts the EPR spectrum to the familiar S = 5/2 form.64 Subsequent treatment with ethylenediaminetetraacetate (EDTA) selectively removes the dangler Fe, resulting in reconversion of the S = 5/2 cluster to an S = 1/2 cluster; this conversion also occurs upon treatment with EDTA of more typical HydG batches that are not deficient in Fe and Cys (Figure 4).64 Using this metal reconstitution procedure, tetranuclear and pentanuclear forms of the auxiliary Fe–S cluster may be interconverted, thereby allowing for the incorporation of either 57Fe or Ni2+ in place of natural-abundance Fe with high selectivity into the dangler Fe site (Figure 4).64 Taken together, these data point to a [4Fe–4S]aux[(Cys)Fe] cluster with a Cys-ligated dangler Fe form as the active state of HydG during both in vitro and in vivo H-cluster biosynthesis. In our view, the expression and purification procedures developed for So HydG may largely preserve this structure, which is why we generally observe the S = 5/2 five-Fe form. On the other hand, purification procedures that involve reconstitution without Cys may not give rise to any appreciable quantity of the S = 5/2 cluster observed in So HydG. Further work toward identifying the factors that result in high levels of Cys and Fe incorporation into HydG is warranted.
Figure 4.
Reversible dangler metal-ion lability at the HydG auxiliary cluster: (A) reaction scheme; (B) X-band EPR spectra of DTH-reduced samples. Adapted with permission from ref 64. Copyright 2015 National Academy of Sciences.
HydG Radical Chemistry
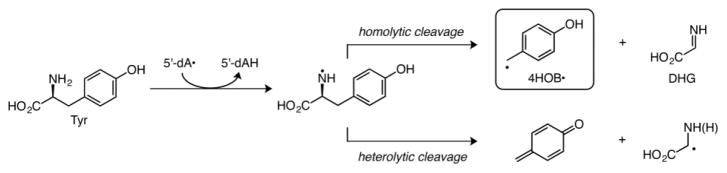
In this article, we have thus-far focused mostly on the auxiliary cluster, which is proposed to be transformed to an [Fe(CO)2(CN)] species.23,25,55,64 We now discuss how this species is formed, beginning with the cleavage of substrate Tyr initiated by 5′-dA• formation at the [4Fe–4S]RS cluster (Scheme 3). Following the addition of SAM, Tyr, and reductant (DTH) to initiate the reaction, a radical intermediate can be trapped and characterized using rapid freeze-quench EPR spectroscopic methods.25,54 The amplitude of this trapped radical EPR signal reaches a maximum at about 30 s, and using a variety of specifically labeled Tyr isotopologues, we were able to determine multiple specific hyperfine interactions through their effects on the EPR line shapes (Figure 5). We compared the observed hyperfine coupling to those predicted for various possible radical species that had been proposed in the literature for HydG and the related radical SAM Tyr lyase ThiH.73–75 We concluded that the radical signal arises from a trapped 4-oxidobenzyl radical (4OB•) or a 4-hydroxybenzyl radical (4HOB•) (Figure 5C) resulting from Cα–Cβ bond scission of a transient Tyr radical produced upon H-atom abstraction of Tyr by the 5′-dA•.54 The other product of the Cα–Cβ bond scission is dehydroglycine (DHG; Scheme 4), which is the source of the CO and CN− ligands of the H cluster.
Figure 5.
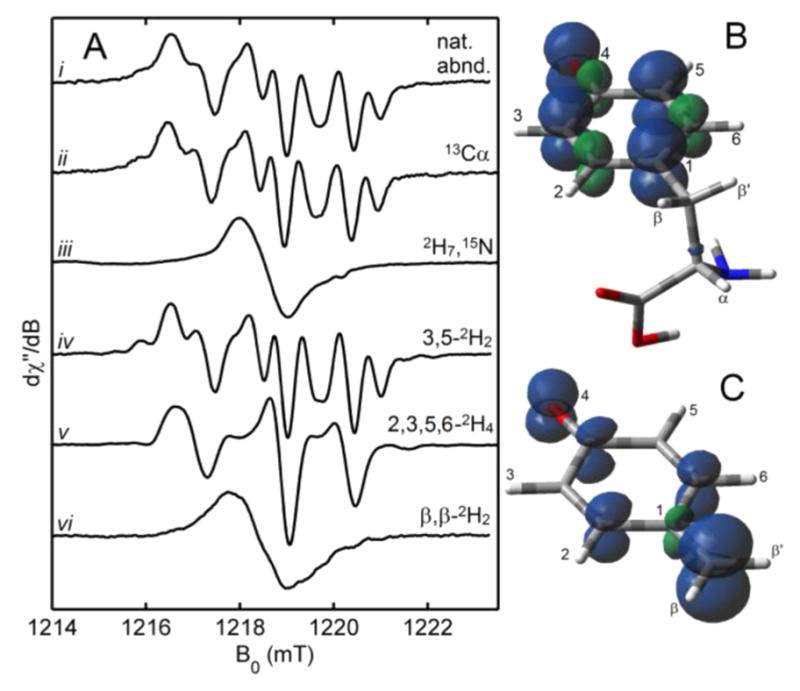
(A) Q-band CW EPR spectra of the HydG radical generated with various Tyr isotopologues. Calculated spin-density plots of a typical O-centered Tyr radical (B) and the 4OB• radical (C). Adapted with permission from 54. Copyright 2013 American Association for the Advancement of Science.
Scheme 4.
HydG-Mediated H-Atom Abstraction of Substrate Tyr Followed by Cα–Cβ Bond Cleavage
Although we have obtained direct evidence for Tyr conversion to the 4(H)OB•—and thus implicated DHG as an intermediate to the CO and CN− ligands of the H cluster—the radical that initially forms upon H-atom abstraction of substrate Tyr by the 5′-dA• has not been observed in HydG or ThiH. Early assumptions were that this transient radical is an O-centered Tyr radical generated by H-atom abstraction of the phenolic H(O) atom.54,73–78 Part of the allure for this proposal was likely the well-documented role of O-centered, neutral Tyr radicals in biology.79–81 However, in general these neutral Tyr radicals are not formed by H-atom abstraction, and moreover they tend to be very stable (e.g., for several hours for the YD• radical of PSII).81 In the formation of these radicals, Tyr oxidation is coupled to H(O) deprotonation, but the transferred electrons and protons typically are vectored to different locations. For example, the PSII Yz• radical is formed upon oxidation by the photogenerated P680+ Chl cation radical as the H(O) proton is transferred across an H bond to an adjacent histidine.81 Upon rereduction of the Yz• radical by an electron from the Mn4Ca oxygen-evolving complex, the H(N) histidine proton hops back to reform a neutral Tyr residue. Such coupled proton- and electron-transfer steps are in contrast to H-atom abstraction of Tyr by 5′-dA• that is proposed for HydG.
In addition, it has been shown that, during the course of the HydG reaction, SAM abstracts a solvent-exchangeable H atom,54,78 which is consistent with H(O)-atom abstraction. One piece of evidence that is not readily rationalized by H(O)-atom abstraction is the finding that H-atom abstraction by 5′-dA• is a reversible process: when run in 2H2O, up to three 2H atoms can be incorporated into 5′-deoxyadenosine (5′-dAH).54,78 This finding casts doubt on phenolic H(O)-atom abstraction because a Tyr O–H bond is significantly weaker than the primary C–H bond of 5′-dAH, making multiple reversible H-atom abstractions unlikely (though not impossible). Thus, it is the stability of the O-centered Tyr radical that renders it an unlikely intermediate in this process.
The other solvent-exchangeable candidate for H-atom abstraction is an amino H(N) atom. The first proposal that this is the site of H-atom abstraction during the HydG reaction was based on the crystal structure of the related radical SAM enzyme NosL,82 which cleaves the Cα–Cβ bond in substrate L-tryptophan (Trp). In the NosL structure, Trp is positioned such that the NH2 group—rather than the indolyl NH moiety—is 3.8 Å from the 5′-C of S-adenosyl-L-homocysteine (an unreactive structural analog of SAM). This finding strongly suggests that the amino H(N) atom of Trp is abstracted by 5′-dA•, and it was proposed that such regiochemistry may be generalized to related amino acid lyases including HydG and ThiH.82 The strong amino N–H bond strength is better matched with that of the 5′-dAH C–H bond and is therefore more consistent with the experimentally observed reversibility of H-atom abstraction. An analogous X-ray crystal structure for HydG with bound Tyr and SAM (or a structural analogue of SAM) has not been reported. However, both reported crystal structures of HydG were accompanied by computational modeling that shows Tyr and SAM binding with the amino group oriented for H(N)-atom abstraction,55,83 consistent with the NosL structure.84
One reason we had also originally favored H(O)-atom abstraction was because we observed that the S = 5/2 auxiliary cluster signal was diminished in a sample that contained reductant and Tyr without SAM.54 We interpreted this as evidence for Tyr interacting with the auxiliary cluster likely via its carboxylate and amino groups,54 both of which end up directly bound to the auxiliary cluster as CO and CN−, respectively. Tyr binding in this manner could leave the phenolic H(O) atom pointed toward the [4Fe–4S]RS cluster and primed for H-atom abstraction by the 5′-dA•. One problem with this model is that it requires the radical SAM and auxiliary Fe–S clusters to be relatively close together,58 a feature that has not been borne out in the crystal structures of HydG55,83 and is not consistent with modeling studies that predated the crystal structures.45,85,86 Thus, the proposal that Tyr binds to the auxiliary cluster is in disagreement with the currently accepted model of H-atom abstraction (Scheme 4), a point that we will further address below.
In principle, the structure of HydG with bound Tyr and SAM could be studied in detail using orientation-selective EPR spectroscopic techniques such as ENDOR and electron spin echo envelope modulation (ESEEM). Such an experiment entails measuring the hyperfine coupling between the electron spin on an S = 1/2 [4Fe–4S] cluster and the nuclear spins (e.g., 15N and 13C) of labeled substrates. However, it requires that the [4Fe–4S] cluster be in its reduced form, which is difficult to observe in the presence of both SAM and substrate because SAM is rapidly reduced to generate 5′-dA• (which initiates the substrate H-atom abstraction). One approach to overcoming this problem is to use a SAM analogue that generates a more stable allylic radical;87 this radical can then serve as the electron spin to which substrate nuclear hyperfine couplings can be measured.88 Such an experiment has not yet been reported for HydG.
To help address the question of how Tyr binds, we generated HydG samples with a reduced, S = 1/2 [4Fe–4S]RS cluster in the presence of Tyr and the absence of SAM and report their EPR spectroscopic characterization herein. Since our initial finding that the addition of Tyr to HydG reduces the S = 5/2 signal intensity of the auxiliary cluster (vide supra),54 we have found that Tyr generally has no effect on the auxiliary cluster EPR signal; we suspect that the original Tyr-containing sample was prepared with a batch of HydG that was deficient in Cys and/or Fe and therefore had less of the S = 5/2 signal, which we now know corresponds to a [4Fe–4S]aux[(Cys)Fe] cluster (vide supra).64 As such, our original interpretation—that Tyr interacts with the auxiliary cluster—was incorrect, and we now report that the addition of Tyr to HydG in the absence of SAM induces a slight sharpening of the [4Fe–4S]RS cluster (Figure 6). This finding is broadly in line with the report that the S = 1/2 EPR signal(s) in Ca HydG are subtly perturbed upon the addition of Tyr, although the presence of S = 1/2 forms of both the [4Fe–4S]RS cluster and the auxiliary cluster precluded assigning which cluster signal(s) are perturbed by Tyr.67
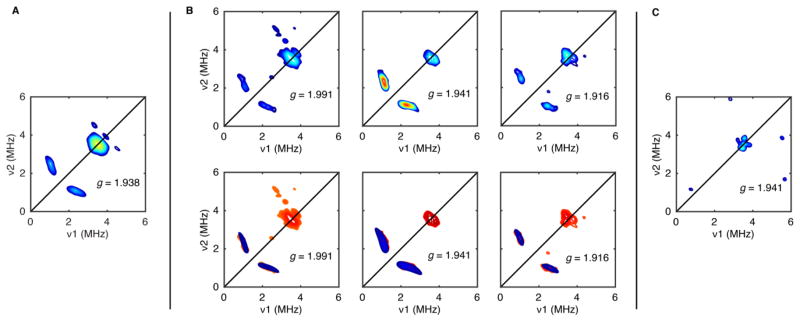
The observation that Tyr induces sharpening of the [4Fe–4S]RS signal with little effect on the g values suggests that Tyr does bind to HydG but not directly to the [4Fe–4S]RS cluster (e.g., in place of SAM). In order to examine this effect more closely using pulse EPR spectroscopy, we prepared samples with isotopically labeled Tyr; doing so allows for measurement of the hyperfine coupling between Tyr nuclei and the [4Fe–4S]RS cluster electron spin. The X-band HYSCORE spectrum of a HydG sample containing DTH and 15N-Tyr shows a pair of correlation ridges centered at the 15N Larmor frequency (Figure 7A); these spectral features are absent in samples either lacking Tyr or using different isotopologues of Tyr (not shown). Similar 15N hyperfine coupling is also observed in samples of the C-terminal cluster deletion mutant (HydGXC), which contains no auxiliary cluster (Figure 7B); thus, the Tyr hyperfine coupling may be definitively assigned to the [4Fe–4S]RS cluster. In order to gain more insight into the nature of Tyr binding, we recorded HYSCORE spectra of the HydGXC + 15N-Tyr + DTH sample at two additional field positions. The observed orientation-dependent 15N hyperfine coupling may be simulated using A(15N) = ±[0.4, 0.7, 3.7] MHz with corresponding Euler angles of [0°, 40°, 0°]. Rewritten as A(15N) = aiso + Tdip[−(1 + ρ), −(1 − ρ), 2], this gives aiso = ±1.6 MHz, Tdip = ±1.05 MHz, and ρ = 0.15 MHz.
The magnitude of aiso is small but nonnegligible. For reference, the 15N hyperfine tensor for bound 15N-SAM is expected to be approximately 6–9 MHz.61,89–91 Thus, the small magnitude of aiso is consistent with Tyr not binding directly to the [4Fe–4S]RS cluster. However, because aiso is indicative of the electron spin density at the 15N nucleus, the observed value of aiso = ±1.6 MHz indicates that there is some bonding interaction between Tyr and the [4Fe–4S]RS cluster; a through-space interaction is expected to give rise to purely dipolar coupling with aiso = 0. Interestingly, ESEEM spectra of [2Fe–2S] clusters have shown similar aiso values [aiso(14N) ≈ 1 MHz, which corresponds to aiso(15N) = 1.4 MHz] that have been attributed to peptide amides engaged in NH–S H bonding to a sulfide of the [2Fe–2S] cluster.92,93 As such, we suggest that Tyr binding in the absence of SAM may be partially mediated by an H-bonding interaction between the Tyr amino group and either one of the [4Fe–4S]RS cluster sulfides (Figure 8) or one of the three Cys ligands to the [4Fe–4S]RS cluster. The precise orientation of Tyr with respect to the [4Fe–4S]RS cluster will require further investigation, and this binding mode is unlikely to correspond exactly to the Tyr binding mode when SAM is present (which is thought not to involve H bonding between Tyr and the [4Fe–4S]RS cluster).55,83 However, these data do suggest that Tyr can bind with its amino group in close proximity to the [4Fe–4S]RS cluster. Interestingly, preliminary experiments suggest that Tyr binding is specific: no 15N correlation ridges are observed in the X-band HYSCORE spectra of samples containing DTH and 15N-L-phenylalanine, an amino acid with a structure similar to that of Tyr (Figure 7C). These results are consistent with a substrate binding preequilibrium in the absence of SAM that is specific for Tyr. However, the functional relevance of this Tyr adduct to HydG activity is unclear.
Figure 8.
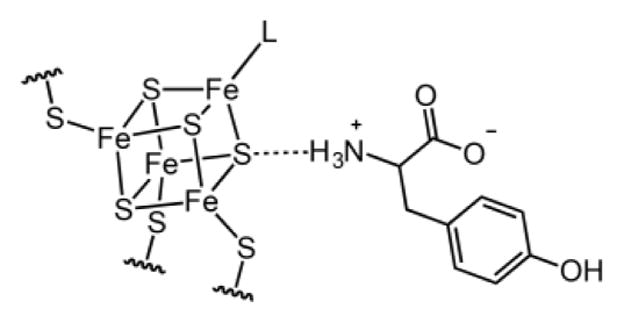
Representation of a possible H-bonding interaction between the substrate Tyr and the [4Fe–4S]RS cluster in the absence of SAM.
CO and CN− Binding to the Auxiliary Fe–S Cluster
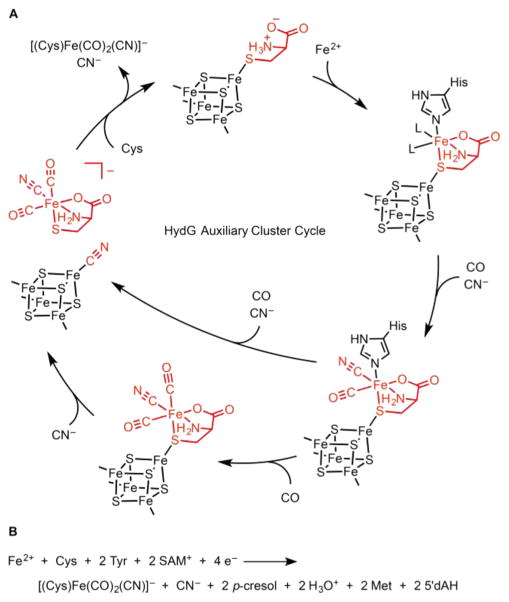
The Tyr-derived 4HOB• radical starts to decay about 30 s after reaction initiation. On the same time scale of the radical decay, an [Fe(CO)(CN)] species may be observed in parallel stopped-flow FTIR (SF-FTIR) experiments.23 In the context of the recent dangler Fe model for the auxiliary cluster,55 we assign this species as the product of the addition of CO and CN− (derived from DHG) to the dangler Fe (Scheme 5A). At longer time scales, the SF-FTIR spectra reveal a second CO ligand to the dangler Fe, verified by isotopic shift effects on the FTIR line shapes observed in experiments carried out with mixed Tyr isotopologues.23 Because CO and CN− are produced in a 1:1 ratio from Tyr cleavage, a second CN− should form concurrently with this second CO ligand, but we have no SF-FTIR spectroscopic evidence of a second CN− ligand to Fe.
Scheme 5. (A) Mechanistic Proposal for the HydG Auxiliary Cluster Reactiona and (B) Proposed Net HydG Reaction.
aKey steps include the following: (1) the initial binding of Cys and Fe2+ to the [4Fe–4S]aux cluster either in a stepwise fashion (as shown here) or by binding of a preassembled (Cys)Fe complex; (2) binding of Tyr-derived CO and CN−; (3) binding of a second equivalent of Tyr-derived CO at the dangler Fe and CN− at the [4Fe–4S]aux cluster; (4) substitution of CN− with Cys at the [4Fe–4S]aux cluster and release of the [(Cys)Fe(CO)2(CN)]− product. The His residue depicted is that observed in the Ti HydG X-ray crystal structure.55.
However, on the same 20 min time scale in which we observe the [Fe(CN) (CO)2] moiety using SF-FTIR spectroscopy, the EPR spectrum of the reaction mixture shows a component with g values of [2.09, 1.94, 1.93],64 identical with what we had previously observed in HydG samples that are chemically treated with exogenous KCN.55 For KCN-treated samples, we verified that this EPR signal corresponds to a [4Fe–4S]aux[CN] cluster by recording the 13C hyperfine interaction of a K13CN-treated sample using HYSCORE spectroscopy and noting its similarity to that of the previously studied 13CN-bound [4Fe–4S] cluster of Pyrococcus furiosus ferredoxin;94 the g values are also nearly identical.
We next sought to evaluate whether this [4Fe–4S]aux[CN] species is indeed generated during the reaction with Tyr, SAM, and DTH as the substrates (without added KCN). Because the CN− ligands of the [2Fe]H subcluster are derived from the N and 2-C atoms of Tyr, we used either 2-13C-Tyr or 15N-Tyr as substrates during HydG turnover to isotopically label any CN−-containing intermediates with high selectivity.64 HYSCORE spectra of these samples correspond precisely to those of the [4Fe–4S]aux[CN] cluster as generated by the addition of K13CN or KC15N,64 thus demonstrating that an S = 1/2 [4Fe–4S]aux[CN] cluster builds up during the reaction with Tyr, SAM, and DTH.64 Given this finding, we proposed a possible fate of the second Tyr-derived CN−: binding to the [4Fe-4S]aux cluster, with CN− acting as a nucleophile to displace a [(Cys)Fe(CO)2(CN)]− species from the [4Fe–4S]aux cluster (Scheme 5A).64 Although further experiments are required to determine whether a [4Fe–4S]aux[CN] species is a certified intermediate in the HydG mechanism, this discovery provides an interesting new clue as to how HydG may release the [(Cys)Fe(CO)2(CN)]− species to its downstream acceptor. Finally, we also showed using CW EPR spectroscopy that treatment of the [4Fe–4S]aux[CN] cluster with Cys allows for regeneration of the Cys-bound form (Scheme 5A).64 Because subsequent remetalation with Fe2+ is facile (Figure 4),64 this reactivity points to the possibility that [(Cys)Fe(CO)2(CN)]− production by HydG could be catalytic, although further experiments are required to assess this possibility.
CONCLUSION
These results provide experimental support for the formation and interconversions of several intermediates during the HydG reaction (Scheme 5A), and they also support the proposal that the identity of the [Fe(CO)2(CN)] synthon is a [(Cys)Fe-(CO)2(CN)]− intermediate to the [2Fe]H subcluster.64 It is unclear at this time whether the [(Cys)Fe(CO)2(CN)]− species detected by FTIR is bound to the [4Fe–4S]aux cluster, unbound from the [4Fe–4S]aux cluster yet within the protein environment, or released from HydG altogether. Regardless, the finding that Cys is involved in the HydG reaction allows for a complete reaction to be proposed (Scheme 5B)64 and should inspire new testable hypotheses. Of course, many questions remain: (i) Can we reconcile the sulfide-linked five-Fe auxiliary cluster observed in the crystal structure of Ti HydG55 and the spectroscopically supported model with the integral Cys ligand?64 (ii) Can the structure of the [(Cys)Fe(CO)2(CN)]− species be fully elucidated? (iii) What is the fate of the Cys ligand in the [(Cys)Fe(CO)2(CN)]− species? (iv) How is the [(Cys)Fe(CO)2(CN)]− species transferred to its acceptor, and how is it subsequently modified by the other maturase enzymes? Answers to these and other questions await further experimentation, and further surprising discoveries will surely be made.
Acknowledgments
We acknowledge the National Institute of General Medical Sciences of the National Institutes of Health for funding (Grant F32GM111025 to D.L.M.S and Grant R01GM104543 to R.D.B.) and the Division of Materials Science and Engineering of the Department of Energy for funding of discussions, protein preparations, and manuscript writing (Grant DE-FG02-09ER46632 to J.R.S.).
Footnotes
Notes
The authors declare no competing financial interest.
References
- 1.Ferreira KN, Iverson TM, Maghlaoui K, Barber J. Science. 2004;303:1831–1838. doi: 10.1126/science.1093087. [DOI] [PubMed] [Google Scholar]
- 2.Umena Y, Kawakami K, Shen JR, Kamiya N. Nature. 2011;473:55–60. doi: 10.1038/nature09913. [DOI] [PubMed] [Google Scholar]
- 3.Suga M, Akita F, Hirata K, Ueno G, Murakami H, Nakajima Y, Shimizu T, Yamashita K, Yamamoto M, Ago H, Shen JR. Nature. 2014;517:99–103. doi: 10.1038/nature13991. [DOI] [PubMed] [Google Scholar]
- 4.Vignais PM, Billoud B. Chem Rev. 2007;107:4206–4272. doi: 10.1021/cr050196r. [DOI] [PubMed] [Google Scholar]
- 5.Vincent KA, Parkin A, Armstrong FA. Chem Rev. 2007;107:4366–4413. doi: 10.1021/cr050191u. [DOI] [PubMed] [Google Scholar]
- 6.Wang G, Benecky MJ, Huynh BH, Cline JF, Adams M, Mortenson LE, Hoffman BM, Münck E. J Biol Chem. 1984;259:4328–4331. [PubMed] [Google Scholar]
- 7.Van Der Spek TM, Arendsen AF, Happe RP, Yun SY, Bagley KA, Stufkens DJ, Hagen WR, Albracht S. Eur J Biochem. 1996;237:629–634. doi: 10.1111/j.1432-1033.1996.0629p.x. [DOI] [PubMed] [Google Scholar]
- 8.Peters JW, Lanzilotta WN, Lemon BJ, Seefeldt LC. Science. 1998;282:1853–1858. doi: 10.1126/science.282.5395.1853. [DOI] [PubMed] [Google Scholar]
- 9.Nicolet Y, Piras C, Legrand P, Hatchikian CE, Fontecilla-Camps JC. Structure. 1999;7:13–23. doi: 10.1016/s0969-2126(99)80005-7. [DOI] [PubMed] [Google Scholar]
- 10.Berggren G, Adamska A, Lambertz C, Simmons TR, Esselborn J, Atta M, Gambarelli S, Mouesca JM, Reijerse E, Lubitz W, Happe T, Artero V, Fontecave M. Nature. 2013;499:66–69. doi: 10.1038/nature12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheniae GM, Martin IF. Biochim Biophys Acta, Bioenerg. 1971;253:167–181. doi: 10.1016/0005-2728(71)90242-8. [DOI] [PubMed] [Google Scholar]
- 12.Ananyev GM, Dismukes GC. Biochemistry. 1996;35:14608–14617. doi: 10.1021/bi960894t. [DOI] [PubMed] [Google Scholar]
- 13.Cinco RM, McFarlane Holman KL, Robblee JH, Yano J, Pizarro SA, Bellacchio E, Sauer K, Yachandra VK. Biochemistry. 2002;41:12928–12933. doi: 10.1021/bi026569p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burnap RL. Phys Chem Chem Phys. 2004;6:4803. [Google Scholar]
- 15.Corey EJ, Cheng X-M. The Logic of Chemical Synthesis. Wiley; New York: 1989. [Google Scholar]
- 16.Mulder DW, Ortillo DO, Gardenghi DJ, Naumov AV, Ruebush SS, Szilagyi RK, Huynh B, Broderick JB, Peters JW. Biochemistry. 2009;48:6240–6248. doi: 10.1021/bi9000563. [DOI] [PubMed] [Google Scholar]
- 17.Mulder DW, Boyd ES, Sarma R, Lange RK, Endrizzi JA, Broderick JB, Peters JW. Nature. 2010;465:248–251. doi: 10.1038/nature08993. [DOI] [PubMed] [Google Scholar]
- 18.Kuchenreuther JM, Stapleton JA, Swartz JR. PLoS One. 2009;4:e7565. doi: 10.1371/journal.pone.0007565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuchenreuther JM, Grady-Smith CS, Bingham AS, George SJ, Cramer SP, Swartz JR. PLoS One. 2010;5:e15491. doi: 10.1371/journal.pone.0015491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuchenreuther JM, George SJ, Grady-Smith CS, Cramer SP, Swartz JR. PLoS One. 2011;6:e20346. doi: 10.1371/journal.pone.0020346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuchenreuther JM, Britt RD, Swartz JR. PLoS One. 2012;7:e45850. doi: 10.1371/journal.pone.0045850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuchenreuther JM, Guo Y, Wang H, Myers WK, George SJ, Boyke CA, Yoda Y, Alp EE, Zhao J, Britt RD, Swartz JR, Cramer SP. Biochemistry. 2013;52:818–826. doi: 10.1021/bi301336r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuchenreuther JM, Myers WK, Suess DLM, Stich TA, Pelmenschikov V, Shiigi S, Cramer SP, Swartz JR, Britt RD, George SJ. Science. 2014;343:424–427. doi: 10.1126/science.1246572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Myers WK, Stich TA, Suess DLM, Kuchenreuther JM, Swartz JR, Britt RD. J Am Chem Soc. 2014;136:12237–12240. doi: 10.1021/ja507046w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suess DLM, Britt RD. Top Catal. 2015;58:699–707. doi: 10.1007/s11244-015-0412-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Esselborn J, Lambertz C, Adamska-Venkatesh A, Simmons T, Berggren G, Noth J, Siebel J, Hemschemeier A, Artero V, Reijerse E, Fontecave M, Lubitz W, Happe T. Nat Chem Biol. 2013;9:607–609. doi: 10.1038/nchembio.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adamska-Venkatesh A, Simmons TR, Siebel JF, Artero V, Fontecave M, Reijerse E, Lubitz W. Phys Chem Chem Phys. 2015;17:5421–5430. doi: 10.1039/c4cp05426a. [DOI] [PubMed] [Google Scholar]
- 28.Gilbert-Wilson R, Siebel JF, Adamska-Venkatesh A, Pham CC, Reijerse E, Wang H, Cramer SP, Lubitz W, Rauchfuss TB. J Am Chem Soc. 2015;137:8998–9005. doi: 10.1021/jacs.5b03270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adamska-Venkatesh A, Roy S, Siebel JF, Simmons TR, Fontecave M, Artero V, Reijerse E, Lubitz W. J Am Chem Soc. 2015;137:12744–12747. doi: 10.1021/jacs.5b06240. [DOI] [PubMed] [Google Scholar]
- 30.Tard C, Liu X, Ibrahim SK, Bruschi M, De Gioia L, Davies SC, Yang X, Wang LS, Sawers G, Pickett CJ. Nature. 2005;433:610–613. doi: 10.1038/nature03298. [DOI] [PubMed] [Google Scholar]
- 31.McGlynn SE, Shepard EM, Winslow MA, Naumov AV, Duschene KS, Posewitz MC, Broderick WE, Broderick JB, Peters JW. FEBS Lett. 2008;582:2183–2187. doi: 10.1016/j.febslet.2008.04.063. [DOI] [PubMed] [Google Scholar]
- 32.Shepard EM, McGlynn SE, Bueling AL, Grady-Smith CS, George SJ, Winslow MA, Cramer SP, Peters JW, Broderick JB. Proc Natl Acad Sci U S A. 2010;107:10448–10453. doi: 10.1073/pnas.1001937107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Czech I, Silakov A, Lubitz W, Happe T. FEBS Lett. 2010;584:638–642. doi: 10.1016/j.febslet.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 34.Czech I, Stripp S, Sanganas O, Leidel N, Happe T, Haumann M. FEBS Lett. 2011;585:225–230. doi: 10.1016/j.febslet.2010.11.052. [DOI] [PubMed] [Google Scholar]
- 35.Hu Y, Corbett MC, Fay AW, Webber JA, Hodgson KO, Hedman B, Ribbe MW. Proc Natl Acad Sci U S A. 2006;103:17119–17124. doi: 10.1073/pnas.0602647103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tard C, Pickett CJ. Chem Rev. 2009;109:2245–2274. doi: 10.1021/cr800542q. [DOI] [PubMed] [Google Scholar]
- 37.Hieber W, Gruber J. Z Anorg Allg Chem. 1958;296:91–103. [Google Scholar]
- 38.Seyferth D, Henderson RS. J Am Chem Soc. 1979;101:508–509. [Google Scholar]
- 39.Seyferth D, Henderson RS. J Organomet Chem. 1981;218:C34–C36. [Google Scholar]
- 40.Li H, Rauchfuss TB. J Am Chem Soc. 2002;124:726–727. doi: 10.1021/ja016964n. [DOI] [PubMed] [Google Scholar]
- 41.Cloirec AL, Davies SC, Evans DJ, Hughes DL, Pickett CJ, Best SP, Borg S. Chem Commun. 1999:2285–2286. [Google Scholar]
- 42.Lyon EJ, Georgakaki IP, Reibenspies JH, Darensbourg MY. Angew Chem, Int Ed. 1999;38:3178–3180. [PubMed] [Google Scholar]
- 43.Lyon EJ, Georgakaki IP, Reibenspies JH, Darensbourg MY. J Am Chem Soc. 2001;123:3268–3278. doi: 10.1021/ja003147z. [DOI] [PubMed] [Google Scholar]
- 44.George SJ, Cui Z, Razavet M, Pickett CJ. Chem - Eur J. 2002;8:4037–4046. doi: 10.1002/1521-3765(20020902)8:17<4037::AID-CHEM4037>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 45.Pilet E, Nicolet Y, Mathevon C, Douki T, Fontecilla-Camps JC, Fontecave M. FEBS Lett. 2009;583:506–511. doi: 10.1016/j.febslet.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 46.Betz JN, Boswell NW, Fugate CJ, Holliday GL, Akiva E, Scott AG, Babbitt PC, Peters JW, Shepard EM, Broderick JB. Biochemistry. 2015;54:1807–1818. doi: 10.1021/bi501205e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Joshi N, Shepard EM, Byer AS, Swanson KD, Broderick JB, Peters JW. FEBS Lett. 2012;586:3939–3943. doi: 10.1016/j.febslet.2012.09.028. [DOI] [PubMed] [Google Scholar]
- 48.Lill R. Nature. 2009;460:831–838. doi: 10.1038/nature08301. [DOI] [PubMed] [Google Scholar]
- 49.Shakamuri P, Zhang B, Johnson MK. J Am Chem Soc. 2012;134:15213–15216. doi: 10.1021/ja306061x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brazzolotto X, Rubach JK, Gaillard J, Gambarelli S, Atta M, Fontecave M. J Biol Chem. 2006;281:769–774. doi: 10.1074/jbc.M510310200. [DOI] [PubMed] [Google Scholar]
- 51.Albertini M, Vallese F, Di Valentin M, Berto P, Giacometti GM, Costantini P, Carbonera D. Int J Hydrogen Energy. 2014;39:18574–18582. [Google Scholar]
- 52.Albertini M, Galazzo L, Maso L, Vallese F, Berto P, De Rosa E, Di Valentin M, Costantini P, Carbonera D. Top Catal. 2015;58:708–718. [Google Scholar]
- 53.Berggren G, Garcia-Serres R, Brazzolotto X, Clemancey M, Gambarelli S, Atta M, Latour JM, Hernandez HL, Subramanian S, Johnson MK, Fontecave M. JBIC, J Biol Inorg Chem. 2014;19:75–84. doi: 10.1007/s00775-013-1062-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuchenreuther JM, Myers WK, Stich TA, George SJ, Nejatyjahromy Y, Swartz JR, Britt RD. Science. 2013;342:472–475. doi: 10.1126/science.1241859. [DOI] [PubMed] [Google Scholar]
- 55.Dinis P, Suess DLM, Fox SJ, Harmer JE, Driesener RC, De La Paz L, Swartz JR, Essex JW, Britt RD, Roach PL. Proc Natl Acad Sci U S A. 2015;112:1362–1367. doi: 10.1073/pnas.1417252112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stoll S, Schweiger A. J Magn Reson. 2006;178:42–55. doi: 10.1016/j.jmr.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 57.Frey PA, Hegeman AD, Ruzicka FJ. Crit Rev Biochem Mol Biol. 2008;43:63–88. doi: 10.1080/10409230701829169. [DOI] [PubMed] [Google Scholar]
- 58.Vey JL, Drennan CL. Chem Rev. 2011;111:2487–2506. doi: 10.1021/cr9002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stich TA, Myers WK, Britt RD. Acc Chem Res. 2014;47:2235–2243. doi: 10.1021/ar400235n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Broderick JB, Duffus BR, Duschene KS, Shepard EM. Chem Rev. 2014;114:4229–4317. doi: 10.1021/cr4004709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Walsby CJ, Ortillo D, Broderick WE, Broderick JB, Hoffman BM. J Am Chem Soc. 2002;124:11270–11271. doi: 10.1021/ja027078v. [DOI] [PubMed] [Google Scholar]
- 62.Driesener RC, Challand MR, McGlynn SE, Shepard EM, Boyd ES, Broderick JB, Peters JW, Roach PL. Angew Chem, Int Ed. 2010;49:1687–1690. doi: 10.1002/anie.200907047. [DOI] [PubMed] [Google Scholar]
- 63.Shepard EM, Duffus BR, George SJ, McGlynn SE, Challand MR, Swanson KD, Roach PL, Cramer SP, Peters JW, Broderick JB. J Am Chem Soc. 2010;132:9247–9249. doi: 10.1021/ja1012273. [DOI] [PubMed] [Google Scholar]
- 64.Suess DLM, Bürstel I, De La Paz L, Kuchenreuther JM, Pham CC, Cramer SP, Swartz JR, Britt RD. Proc Natl Acad Sci U S A. 2015;112:11455–11460. doi: 10.1073/pnas.1508440112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cheek J, Broderick JB. JBIC, J Biol Inorg Chem. 2001;6:209–226. doi: 10.1007/s007750100210. [DOI] [PubMed] [Google Scholar]
- 66.Rubach JK, Brazzolotto X, Gaillard J, Fontecave M. FEBS Lett. 2005;579:5055–5060. doi: 10.1016/j.febslet.2005.07.092. [DOI] [PubMed] [Google Scholar]
- 67.Driesener RC, Duffus BR, Shepard EM, Bruzas IR, Duschene KS, Coleman NJR, Marrison APG, Salvadori E, Kay CWM, Peters JW, Broderick JB, Roach PL. Biochemistry. 2013;52:8696–8707. doi: 10.1021/bi401143s. [DOI] [PubMed] [Google Scholar]
- 68.Onate YA, Vollmer SJ, Switzer RL, Johnson MK. J Biol Chem. 1989;264:18386–18391. [PubMed] [Google Scholar]
- 69.Boll M, Fuchs G, Tilley G, Armstrong FA, Lowe DJ. Biochemistry. 2000;39:4929–4938. doi: 10.1021/bi9927890. [DOI] [PubMed] [Google Scholar]
- 70.Kennedy MC, Kent TA, Emptage M, Merkle H, Beinert H, Münck E. J Biol Chem. 1984;259:14463–14471. [PubMed] [Google Scholar]
- 71.Richards AJM, Thomson AJ, Holm RH, Hagen KS. Spectrochimica Acta Part A. 1990;46:987–993. [Google Scholar]
- 72.Surerus KK, Münck E, Moura I, Moura JJG, LeGall J. J Am Chem Soc. 1987;109:3805–3807. [Google Scholar]
- 73.Kriek M, Martins F, Challand MR, Croft A, Roach PL. Angew Chem, Int Ed. 2007;46:9223–9226. doi: 10.1002/anie.200702554. [DOI] [PubMed] [Google Scholar]
- 74.Kriek M, Martins F, Leonardi R, Fairhurst SA, Lowe DJ, Roach PL. J Biol Chem. 2007;282:17413–17423. doi: 10.1074/jbc.M700782200. [DOI] [PubMed] [Google Scholar]
- 75.Nicolet Y, Martin L, Tron C, Fontecilla-Camps JC. FEBS Lett. 2010;584:4197–4202. doi: 10.1016/j.febslet.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 76.Challand MR, Martins FT, Roach PL. J Biol Chem. 2010;285:5240–5248. doi: 10.1074/jbc.M109.056606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Roach PL. Curr Opin Chem Biol. 2011;15:267–275. doi: 10.1016/j.cbpa.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 78.Duffus BR, Ghose S, Peters JW, Broderick JB. J Am Chem Soc. 2014;136:13086–13089. doi: 10.1021/ja504618y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stubbe J, van der Donk WA. Chem Rev. 1998;98:705–762. doi: 10.1021/cr9400875. [DOI] [PubMed] [Google Scholar]
- 80.Stubbe J, Riggs-Gelasco P. Trends Biochem Sci. 1998;23:438–443. doi: 10.1016/s0968-0004(98)01296-1. [DOI] [PubMed] [Google Scholar]
- 81.Diner BA, Britt RD. Photosystem II. Vol. 22. Springer; Dordrecht, The Netherlands: 2005. pp. 207–233. [Google Scholar]
- 82.Nicolet Y, Zeppieri L, Amara P, Fontecilla-Camps JC. Angew Chem, Int Ed. 2014;53:11840–11844. doi: 10.1002/anie.201407320. [DOI] [PubMed] [Google Scholar]
- 83.Nicolet Y, Pagnier A, Zeppieri L, Martin L, Amara P, Fontecilla-Camps JC. Chem Bio Chem. 2015;16:397–402. doi: 10.1002/cbic.201402661. [DOI] [PubMed] [Google Scholar]
- 84.Nicolet Y, Rohac R, Martin L, Fontecilla-Camps JC. Proc Natl Acad Sci U S A. 2013;110:7188–7192. doi: 10.1073/pnas.1302388110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shepard EM, Mus F, Betz JN, Byer AS, Duffus BR, Peters JW, Broderick JB. Biochemistry. 2014;53:4090–4104. doi: 10.1021/bi500210x. [DOI] [PubMed] [Google Scholar]
- 86.Tron C, Cherrier MV, Amara P, Martin L, Fauth F, Fraga E, Correard M, Fontecave M, Nicolet Y, Fontecilla-Camps JC. Eur J Inorg Chem. 2011;2011:1121–1127. [Google Scholar]
- 87.Magnusson OT, Reed GH, Frey PA. J Am Chem Soc. 1999;121:9764–9765. [Google Scholar]
- 88.Horitani M, Byer AS, Shisler KA, Chandra T, Broderick JB, Hoffman BM. J Am Chem Soc. 2015;137:7111–7121. doi: 10.1021/jacs.5b00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen D, Walsby C, Hoffman BM, Frey PA. J Am Chem Soc. 2003;125:11788–11789. doi: 10.1021/ja036120z. [DOI] [PubMed] [Google Scholar]
- 90.Gambarelli S, Luttringer F, Padovani D, Mulliez E, Fontecave M. Chem Bio Chem. 2005;6:1960–1962. doi: 10.1002/cbic.200500182. [DOI] [PubMed] [Google Scholar]
- 91.Walsby CJ, Ortillo D, Yang J, Nnyepi MR, Broderick WE, Hoffman BM, Broderick JB. Inorg Chem. 2005;44:727–741. doi: 10.1021/ic0484811. [DOI] [PubMed] [Google Scholar]
- 92.Cammack R, Chapman A, McCracken J, Cornelius JB, Peisach J, Weiner JH. Biochim Biophys Acta, Protein Struct Mol Enzymol. 1988;956:307–312. doi: 10.1016/0167-4838(88)90148-3. [DOI] [PubMed] [Google Scholar]
- 93.Cammack R, Gay E, Shergill JK. Coord Chem Rev. 1999;190–192:1003. [Google Scholar]
- 94.Telser J, Smith ET, Adams MWW, Conover RC, Johnson MK, Hoffman BM. J Am Chem Soc. 1995;117:5133–5140. [Google Scholar]
- 95.Pandey AS, Harris TV, Giles LJ, Peters JW, Szilagyi RK. J Am Chem Soc. 2008;130:4533–4540. doi: 10.1021/ja711187e. [DOI] [PubMed] [Google Scholar]