Abstract
Background
Data are limited on genome-wide association studies (GWAS) for incident coronary heart disease (CHD). Moreover, it is not known whether genetic variants identified to date also associate with risk of CHD in a prospective setting.
Methods
We performed a two-stage GWAS analysis of incident myocardial infarction (MI) and CHD in a total of 64,297 individuals (including 3898 MI cases, 5465 CHD cases). SNPs that passed an arbitrary threshold of 5×10−6 in Stage I were taken to Stage II for further discovery. Furthermore, in an analysis of prognosis, we studied whether known SNPs from former GWAS were associated with total mortality in individuals who experienced MI during follow-up.
Results
In Stage I 15 loci passed the threshold of 5×10−6; 8 loci for MI and 8 loci for CHD, for which one locus overlapped and none were reported in previous GWAS meta-analyses. We took 60 SNPs representing these 15 loci to Stage II of discovery. Four SNPs near QKI showed nominally significant association with MI (p-value<8.8×10−3) and three exceeded the genome-wide significance threshold when Stage I and Stage II results were combined (top SNP rs6941513: p = 6.2×10−9). Despite excellent power, the 9p21 locus SNP (rs1333049) was only modestly associated with MI (HR = 1.09, p-value = 0.02) and marginally with CHD (HR = 1.06, p-value = 0.08). Among an inception cohort of those who experienced MI during follow-up, the risk allele of rs1333049 was associated with a decreased risk of subsequent mortality (HR = 0.90, p-value = 3.2×10−3).
Conclusions
QKI represents a novel locus that may serve as a predictor of incident CHD in prospective studies. The association of the 9p21 locus both with increased risk of first myocardial infarction and longer survival after MI highlights the importance of study design in investigating genetic determinants of complex disorders.
Introduction
There is strong and consistent evidence that coronary heart disease (CHD) is highly heritable and is influenced by a wide range of genetic factors [1, 2]. Recently genome-wide association studies (GWAS) identified common genetic variants involved in cardiovascular disease and its risk factors [3]. The loci reported by the latest and largest GWAS altogether explain around 10% of CHD heritability [4].
To date, GWAS for CHD have been conducted mostly in cross-sectional case-control setting, and this design, which uses prevalent cases, typically oversamples those with long post-event survival times. Although such a design often makes it possible to collect information from a large number of patients, this approach may incorrectly identify factors that are associated with a high or low case-fatality rate. For instance, a factor associated with a low case-fatality will be enriched among surviving cases and may appear to increase the risk of disease when prevalent cases are compared with controls. This bias is known as incidence-prevalence (Neyman) bias [5, 6]. One major advantage of studying incident cases rather than prevalent cases is that incident cases properly represent the fatal cases and persons with only brief post-event survival. To date the strong and reliable evidence for identifying and assessing factors such as LDL-cholesterol and systolic blood pressure that predict future clinical disease are provided by well-designed population-based, prospective cohort studies that collect large number of incident cases [7].
Here we aimed to study genetic variants that affect the incidence of myocardial infarction (MI) and CHD in prospective, population-based cohorts and whether the genetic variants identified to date are also associated with risk of CHD in a prospective setting. Moreover, we investigated whether the known genetic variants are associated with total-mortality after MI. To this end we used the data from the Cohorts for Heart and Aging Research in Genome Epidemiology (CHARGE) Consortium [8] and collaborating prospective studies.
Methods
Study Population
We performed our study in two stages. Stage I studies comprised participants from five prospective cohort studies that form the CHARGE consortium [8]: the Age, Gene Environment Susceptibility Reykjavik Study (AGES) [9]; the Atherosclerosis Risk in Communities (ARIC) Study [10]; the Cardiovascular Health Study (CHS) [11]; the Framingham Heart Study (FHS) [12]; and the Rotterdam Study (RS) [13, 14]. Stage II comprised individuals from: The Health, Aging, and Body Composition (Health ABC) Study; The Health Professionals Follow-Up Study (HPFS); The Nurses’ Health Study (NHS); PROSPER/PHASE Study; the Study of Health in Pomerania (SHIP); The Women’s Genome Health Study (WGHS); the MOnica Risk, Genetics, Archiving and Monograph (MORGAM) Study comprising the Alpha-Tocopherol, Beta-Carotene (ATBC) Study; The FINRISK Study; The PRIME Study (including the PRIME cohorts of Belfast, Lille, Strasbourg and Toulouse); The Northern Sweden Study. Participants in Stages I and II were of European ancestry. Participants with a history of MI or CHD at baseline were excluded. All studies had protocols approved by local institutional review boards. Participants provided written informed consent and gave permission to use their DNA for research purposes. The Supplementary Document provides details about the design and characteristics for these studies.
Case Definitions for MI and CHD
The definitions of incident MI were consistent among the participating studies, including both fatal and non-fatal MI. CHD included fatal or non-fatal MI, and in most studies fatal CHD or sudden death. The definition of MI and CHD for each cohort study is summarized in S1 Table and S2 Table.
Statistical Analysis
The date of entry to the analysis was the date of cohort entry (AGES, ARIC, CHS, RS) or DNA collection (FHS). Within each study, Cox proportional hazards regression models were used to test the association between each SNP and time to incident MI or CHD, while adjusting for sex and baseline age. FHS adjusted for familial correlation by clustering on pedigree. Analyses in CHS and ARIC were adjusted for study site and in FHS, for generation and additionally for ancestry using principal components [15]. The censor date was the time of MI or CHD diagnosis, the time of death, last date of contact, or at the end of follow-up, whichever came first. For each SNP, additive genetic models were used to estimate the regression coefficient for the hazard ratio (HR) for allele dosage and its respective standard error. For each analysis, a genomic control coefficient (λ) was calculated, which estimated the extent of underlying population structure. Further information on the analysis methods can be found in S3 Table and S4 Table.
Information regarding the genotyping and imputation as well as genotype quality control are found in S5 Table and S6 Table. SNPs with a minor allele frequency of less than 1%, imputation quality less than 0.3 or very large regression coefficients (absolute value larger than 5) were excluded from meta-analysis. Results from individual studies were meta-analyzed for a total of 2,543,842 autosomal SNPs based on Phase 2 HapMap. A fixed effects inverse variance weighted meta-analysis approach was implemented in METAL [16] to combine the regression coefficients and their standard errors, producing a summary regression coefficient and standard error from which a p-value was computed. An arbitrary significance threshold for follow-up in Stage II was set at 5.0×10−6. When more than one SNP clustered at a locus, we carried forward four SNPs with smallest p-values in the associated locus for further investigation in Stage II.
In Stage II, three studies provided data both for incident MI and CHD (HABC, MORGAM, and WGHS), two studies provided data only for MI (PROSPER, SHIP), and two others provided data only for CHD (HPFS, NHS). Each Stage II study used the same analytic method as used in Stage I to examine the association of the 60 SNPs with MI or CHD. As in the Stage I meta-analysis, we used inverse-variance weighted fixed effects meta-analysis to evaluate the Stage II results. We applied a Bonferroni correction for 60 SNPs and set 8.3×10−4 as the significance threshold. Finally, results from all studies in Stage I and II were combined using inverse-variance weighted fixed effects meta-analysis.
We further studied each of the 46 SNPs reported by the CARDIoGRAMplusC4D Consortium [4], for association with incident events in our meta-analysis of longitudinal cohort studies. Moreover, the SNPs were combined into a weighted genetic risk score using beta estimates from the CARDIoGRAMplusC4D Consortium report [4]. The association of each SNP, as well as the score from the combination of all 46 SNPs, was examined with incident MI and CHD using the results of Stage I meta-analysis.
We applied a Cox proportional hazards model adjusted for age and sex to examine the association of the known SNPs with mortality after MI. Five studies including AGES, ARIC, CHS, FHS and the Rotterdam Study provided data for this analysis and in total 2953 individuals were followed after incidence of MI of which 1828 died. The median follow up time ranged from 2.3 years in AGES to 4.7 years in FHS. The baseline characteristics of the study populations for this analysis are presented in S7 Table. Since this analysis was meant to explore potential reasons for weak association or lack of association with incident MI and CHD, we limited the analysis to three SNPs with more than 80% power in Stage I to study its estimated associations with incident MI and CHD.
Results
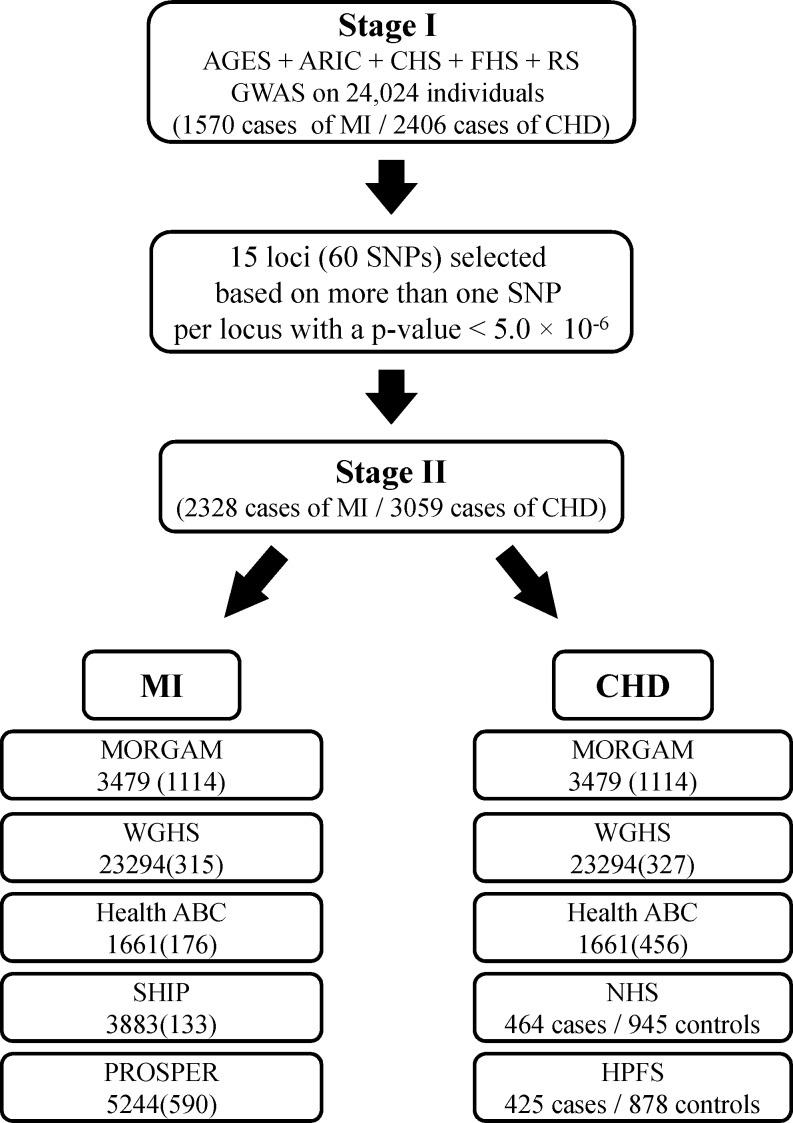
Fig 1 describes Stage I and Stage II of the study. The Stage I panel included five prospective cohort studies comprising a total of 24,024 participants who were free of MI and CHD at baseline. The average age ± standard deviation ranged from 54.1±5.6 in ARIC to 74.6±5.5 in AGES. More than half of the participants (54.5%) were women. The basic characteristics of the participating studies are shown in Table 1. A total of 1570 incident MI events (6.5%) and 2406 incident CHD events (10.0%) occurred over an average of 8.2 years and 8.1 years of follow-up for MI and CHD, respectively. The average age at the time of MI ranged from 65.2 years in ARIC to 80.8 years in CHS.
Fig 1. Study design for identification and validation of SNPs associated with MI and CHD.
Table 1. Baseline characteristics of participants included in incident MI/CHD analysis stratified by cohort.
| Characteristic | AGES | ARIC | CHS | FHS | RS |
|---|---|---|---|---|---|
| Participants, n | 3219 | 7406 | 3291 | 4134 | 5974 |
| Age, years | 76.4 (5.5) | 54.1 (5.57) | 72.3 (5.4) | 64.5 (12.8) | 69.4 (9.1) |
| Women, % | 58.0 | 54.7 | 39.1 | 56.7 | 59.4 |
| Hypertension1, % | 80.6 | 25.7 | 52.8 | 45.3 | 33.4 |
| Diabetes2, % | 11.5 | 7.7 | 11.8 | 10.2 | 10.6 |
| Current smoker3, % | 12.7 | 24.8 | 11.3 | 14.0 | 22.4 |
| Total cholesterol, mg/dL | 217 (45) | 214 (41) | 213 (39) | 203 (40) | 255 (47) |
| HDL cholesterol, mg/dL | 61 (17) | 51 (17) | 55 (16) | 52 (17) | 52 (14) |
| Triglyceride, mg/dL | 107 (59) | 135 (91) | 140 (76) | 144 (127) | NA |
| Body mass index, kg/m2 | 27.1 (4.4) | 27.0 (4.9) | 26.3 (4.5) | 27.7 (5.2) | 26.3 (3.7) |
| Incident MI, N cases | 86 | 486 | 537 | 165 | 296 |
| Mean MI follow-up time | 2.7 | 9.2 | 12.0 | 5.5 | 10.1 |
| Incident CHD, N cases | 209 | 575 | 660 | 201 | 761 |
| Mean CHD follow-up time | 2.6 | 9.1 | 12.0 | 5.5 | 9.9 |
| Incident MI Age, years | 79.1 (5.5) | 65.17 (6.9) | 80.8 (6.2) | 75.2 (12.2) | 80.6 (10.1) |
Numbers in table are Mean (SD) or percentage. AGES = Age, Gene/Environment Study; ARIC = Atherosclerosis Risk in Communities Study; CHS = Cardiovascular Health Study; FHS = Framingham Heart Study; HDL = high density lipoprotein; RS = The Rotterdam Study
1 Hypertension was defined as blood pressure ≥140/90 mmHg or on anti-hypertensive medication
2 Diabetes was defined as fasting blood glucose >125 mg/dL, a random blood glucose of >200 mg/dL, or use of insulin or oral hypoglycemic agents (Rotterdam: diabetes definition: Using anti-diabetic medication or random glucose or oral glucose test more than 200 mg/dl)
3 Current cigarette smoking was defined as self-reported cigarette smoking of at least 1 cigarette per day for a year at any attended exam
The λ coefficient within each cohort was small (≤1.03), suggesting negligible genomic inflation. We combined the results of associations for all SNPs across the five cohorts. S1A Fig and S1B Fig presents the Q-Q plots of combined p-values against the expected p-value distribution for MI and CHD, respectively. The evidence for population admixture was small, both for MI (λ = 1.017) and CHD (λ = 1.022). S2A Fig and S2B Fig illustrates the p-values of the meta-analysis for each of the SNPs across the 22 autosomal chromosomes for MI and CHD, respectively.
In Stage I, 27 SNPs in 8 loci reached our arbitrary threshold of 5×10−6 for MI and 29 SNPs in 8 loci reached this threshold for CHD (Table 2). The most significant association with MI was seen for rs6941513 located on chromosome 6 upstream of QKI (Hazard Ratio = 1.22 [95% Confidence Interval: 1.13, 1.31], p-value = 2.0×10−7). For CHD, rs986080, a SNP located on chromosome 1 between two genes (SNX7 and PAP2D) showed the strongest association (HR = 1.19 [95%CI: 1.12, 1.27], p-value = 6.6×10−8).
Table 2. Description and association of SNPs of the top loci associated with incident MI and CHD in Stage I.
| Phenotype | SNP | Band | Alleles* | HR (95%CI) | P-value | Gene | Location |
|---|---|---|---|---|---|---|---|
| MI | rs6941513 | 6q26 | G/A | 1.22 (1.13–1.31) | 2.0×10−7 | QKI | Closest gene |
| rs13139636 | 4q35.1 | T/A | 1.44 (1.25–1.66) | 6.4×10−7 | ODZ3 | Intron | |
| rs217597 | 7p21.2 | T/C | 1.26 (1.15–1.38) | 1.3×10−6 | DGKB | Intron | |
| rs9923194 | 16q24.1 | C/T | 1.88 (1.44–2.44) | 1.9×10−6 | FOXL1 | Closest gene | |
| rs6504582 | 17q21.32 | A/G | 1.20 (1.11–1.30) | 2.1×10−6 | CALCOCO2 | Closest gene | |
| rs7591615 | 2q35 | T/C | 1.21 (1.12–1.32) | 3.3×10−6 | BARD1 | Intron | |
| rs17777478 | 3q12.1 | T/A | 1.73 (1.37–2.20) | 4.4×10−6 | COL8A1 | Intron | |
| rs2299063 | 6p22.3 | A/C | 1.24 (1.13–1.37) | 4.8×10−6 | ATXN1 | Intron | |
| CHD | rs986080 | 1p21.3 | C/T | 1.19 (1.12–1.27) | 6.6×10−8 | PAP2D | Intron |
| rs16945166 | 13q31.3 | G/A | 1.34 (1.20–1.51) | 4.7×10−7 | GPC5 | Closest gene | |
| rs10740220 | 10q21.3 | G/T | 1.20 (1.11–1.29) | 1.0×10−6 | CTNNA3 | Intron | |
| rs10922855 | 1p22.2 | T/G | 1.22 (1.12–1.33) | 1.9×10−6 | BARHL2 | Closest gene | |
| rs7794677 | 7p12.3 | T/C | 1.20 (1.11–1.29) | 2.3×10−6 | IGFBP3 | Closest gene | |
| rs2916260 | 6p21.2 | T/C | 1.21 (1.12–1.31) | 2.8×10−6 | LRFN2 | Intron | |
| rs3812189 | 6p22.3 | C/T | 1.21 (1.12–1.31) | 3.1×10−6 | ATXN1 | Intron | |
| rs356228 | 4q22.1 | G/C | 1.16 (1.09–1.23) | 3.2×10−6 | SNCA | Closest gene |
*Coded/non-coded allele
HR = Hazard Ratio; CI = Confidence Interval
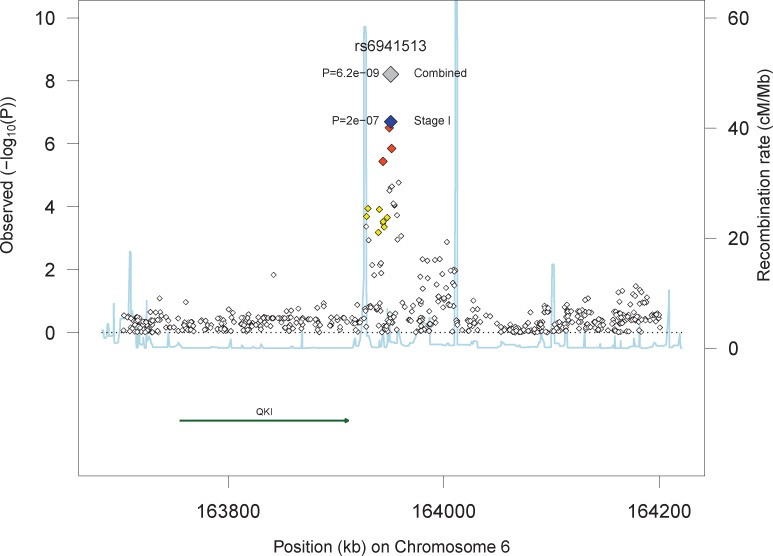
In Stage II, we sought additional evidence for associations in eight loci for MI (QKI, ODZ3, DGKB, FOXL1, CALCOCO2, BARD1, COL8A1, ATXN1) and eight loci for CHD (PAP2D, GPC5, CTNNA3, BARHL2, IGFBP3, LRFN2, ATXN1, SNCA) using four SNPs per locus, for a total of 60 SNPs in 15 loci (ATXN1 was associated with both MI and CHD). Baseline characteristics of the participants of Stages II are shown in S8 and S9 Tables. The results for all 60 SNPs are presented in S10 Table and S11 Table, for MI and CHD, respectively. None of the SNPs passed the Bonferroni adjusted threshold of 8.3×10−4. The results for the best association in each locus are shown in Table 3. Four SNPs located upstream of QKI showed nominal evidence in Stage II for association with MI. The analysis of the combined Stage I and Stage II yielded genome-wide significant associations for three SNPs close to QKI, (rs6941513: HR = 1.21 [95%CI: 1.13, 1.28], p-value = 6.2×10−9). Fig 2 presents the linkage disequilibrium (LD) and p-values of regional markers for this locus. We tested for evidence of replication of this association in 8201 African American individuals including 546 incident cases from the PAGE Study [17], however, rs6941513 was not significantly associated with risk of MI in this population (p = 0.49).
Table 3. Description and association of top SNPs with incident MI and CHD in Stage II and their combined results with Stage I.
| Phenotype | SNPID | Alleles* | Stage II | Combined | Closest Gene | ||
|---|---|---|---|---|---|---|---|
| HR (95%CI) | P-value | HR (95%CI) | P-value | ||||
| MI | rs6941513 | G/A | 1.16 (1.04–1.31) | 8.8×10−3 | 1.2 (1.13–1.28) | 6.2×10−9 | QKI |
| rs7692395 | T/G | 1.03 (0.75–1.40) | 0.86 | 0.69 (0.59–0.79) | 2.5×10−7 | ODZ3 | |
| rs4721377 | T/G | 0.98 (0.87–1.10) | 0.73 | 1.26 (1.14–1.38) | 1.9×10−6 | DGKB | |
| rs9923194 | C/T | 1.04 (0.69–1.58) | 0.84 | 1.58 (1.27–1.97) | 4.3×10−5 | FOXL1 | |
| rs6504582 | A/G | 1.03 (0.91–1.15) | 0.65 | 1.15 (1.08–1.22) | 1.8×10−5 | CALCOCO2 | |
| rs7591615 | T/C | 0.89 (0.78–1.02) | 0.09 | 1.11 (1.03–1.19) | 3.8×10−3 | BARD1 | |
| rs17777478 | T/A | 1.15 (0.80–1.64) | 0.45 | 1.54 (1.26–1.88) | 1.6×10−5 | COL8A1 | |
| rs2299063 | A/C | 0.93 (0.81–1.07) | 0.29 | 1.13 (1.05–1.23) | 1.6×10−3 | ATXN1 | |
| CHD | rs986080 | C/T | 0.97 (0.91–1.04) | 0.39 | 0.97 (0.91–1.04) | 1.1×10−3 | PAP2D |
| rs16945166 | G/A | 1.02 (0.92–1.14) | 0.69 | 1.02 (0.92–1.14) | 2.2×10−4 | GPC5 | |
| rs10509258 | C/T | 1.02 (0.96–1.09) | 0.50 | 1.02 (0.96–1.09) | 3.0×10−4 | CTNNA3 | |
| rs12031583 | G/A | 0.89 (0.79–1.00) | 0.05 | 0.89 (0.79–1.00) | 7.8×10−3 | BARHL2 | |
| rs1551837 | A/G | 1.03 (0.94–1.12) | 0.58 | 1.03 (0.94–1.12) | 2.3×10−4 | IGFBP3 | |
| rs6925172 | C/T | 1.12 (1.01–1.25) | 0.04 | 1.12 (1.01–1.25) | 3.3×10−6 | LRFN2 | |
| rs9297015 | T/A | 0.93 (0.86–1.01) | 0.07 | 0.93 (0.86–1.01) | 0.12 | ATXN1 | |
| rs356228 | G/C | 1.02 (0.96–1.09) | 0.50 | 1.02 (0.96–1.09) | 1.7×10−4 | SNCA | |
*Coded/non-coded allele
Chr. = Chromosome; HR = Hazard Ratio; CI = Confidence Interval
Fig 2. Regional plots for the association of SNPs with MI in the region of QKI.
We sought evidence for the association of 46 SNPs recently reported in the largest GWAS to date for coronary artery disease [4] with the incidence of MI and CHD (Table 4). Despite excellent power, we found only modest evidence for replication of the association with 9p21 locus (CDKN2A/B), the most established finding from previous cross-sectional case-control GWAS. The most replicated SNP at 9p21 locus, rs1333049, was nominally associated with MI (HR: 1.09 [95%CI: 1.01, 1.18], p-value = 0.02) and marginally with CHD (HR = 1.06 [95%CI: 0.99, 1.13], p-value = 0.08). The most significant association with MI was found for rs15563, a SNP in UBE2Z (HR: 1.12 [95%CI: 1.04, 1.20], p-value = 1.9×10−3) and the most significant association with CHD was found for rs10947789, a SNP within the KCNK5 locus (HR: 1.13 [95%CI: 1.05, 1.22], p-value = 5.6×10−4). We found nominally significant associations (p<0.05) with SNPs annotated to CDKN2A/B for MI, LIPA for CHD and COL4A2, TCF21, PDGFD, KCNK5, VAMP8, MRAS, UBE2Z and TCF21 for both MI and CHD (Table 4). A weighted genetic risk score composed of these 46 SNPs was associated with MI (p-value = 1.3×10−3) and CHD (p-value = 1.2×10−3) in the Stage I meta-analysis.
Table 4. Association of the known SNPs for coronary artery disease with incident MI and CHD in Stage I.
| SNP | Chr. | Freq. | Alleles* | Reported GWAS | GWAS on Incident MI | GWAS on Incident CHD | Gene | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | P-value | Power | HR (95% CI) | P-value | Power | HR (95% CI) | P-value | |||||
| rs3217992 | 9 | 0.38 | A/G | 1.16 | 7.8×10−57 | 0.98 | 1.07 (0.99–1.15) | 0.10 | 0.99 | 1.04 (0.98–1.11) | 0.21 | CDKN2A/B |
| rs1333049 | 9 | 0.47 | C/G | 1.23 | 1.4×10−52 | 0.99 | 1.09 (1.01–1.18) | 0.02 | 0.99 | 1.06 (0.99–1.13) | 0.08 | CDKN2A/B |
| rs602633 | 1 | 0.77 | C/A | 1.12 | 1.5×10−25 | 0.73 | 1.08 (0.99–1.17) | 0.10 | 0.89 | 1.05 (0.97–1.12) | 0.21 | PSRC1 |
| rs9369640 | 6 | 0.65 | A/C | 1.09 | 7.5×10−22 | 0.62 | 1.05 (0.98–1.13) | 0.17 | 0.78 | 1.03 (0.97–1.10) | 0.27 | PHACTR1 |
| rs11556924 | 7 | 0.65 | C/T | 1.09 | 6.7×10−17 | 0.62 | 1.05 (0.97–1.14) | 0.26 | 0.78 | 1.06 (0.99–1.14) | 0.07 | ZC3HC1 |
| rs9982601 | 21 | 0.13 | T/C | 1.13 | 7.7×10−17 | 0.63 | 0.98 (0.88–1.09) | 0.68 | 0.79 | 1.01 (0.93–1.10) | 0.81 | MRPS6 |
| rs6725887 | 2 | 0.11 | C/T | 1.12 | 1.2×10−15 | 0.51 | 1.06 (0.95–1.18) | 0.29 | 0.67 | 0.99 (0.90–1.08) | 0.78 | WDR12 |
| rs1122608 | 19 | 0.76 | G/T | 1.10 | 6.3×10−14 | 0.62 | 1.07 (0.98–1.16) | 0.13 | 0.77 | 1.04 (0.97–1.12) | 0.23 | SMARCA4 |
| rs12190287 | 6 | 0.59 | C/G | 1.07 | 4.9×10−13 | 0.45 | 1.13 (1.04–1.23) | 4.4×10−3 | 0.60 | 1.09 (1.01–1.16) | 0.02 | TCF21 |
| rs7173743 | 15 | 0.58 | T/C | 1.07 | 6.7×10−13 | 0.45 | 1.05 (0.97–1.13) | 0.23 | 0.60 | 1.04 (0.98–1.10) | 0.23 | MORF4L1 |
| rs17114036 | 1 | 0.91 | A/G | 1.11 | 5.8×10−12 | 0.39 | 1.12 (0.97–1.28) | 0.11 | 0.52 | 1.06 (0.95–1.19) | 0.27 | PPAP2B |
| rs9515203 | 13 | 0.74 | T/C | 1.08 | 5.9×10−12 | 0.46 | 1.06 (0.95–1.17) | 0.31 | 0.61 | 1.07 (0.99–1.17) | 0.09 | COL4A2 |
| rs2505083 | 10 | 0.42 | C/T | 1.06 | 1.4×10−11 | 0.35 | 1.05 (0.97–1.13) | 0.19 | 0.47 | 1.03 (0.97–1.10) | 0.36 | KIAA1462 |
| rs4773144 | 13 | 0.42 | G/A | 1.07 | 1.4×10−11 | 0.45 | 1.09 (1.01–1.18) | 0.03 | 0.60 | 1.09 (1.02–1.16) | 0.01 | COL4A2 |
| rs7692387 | 4 | 0.81 | G/A | 1.06 | 2.7×10−11 | 0.39 | 1.00 (0.91–1.09) | 0.95 | 0.52 | 1.01 (0.93–1.09) | 0.83 | GUCY1A3 |
| rs974819 | 11 | 0.29 | A/G | 1.07 | 3.6×10−11 | 0.39 | 0.91 (0.83–0.98) | 0.02 | 0.53 | 0.93 (0.87–1.00) | 0.04 | PDGFD |
| rs3184504 | 12 | 0.40 | T/C | 1.07 | 5.4×10−11 | 0.44 | 1.03 (0.95–1.11) | 0.47 | 0.60 | 1.04 (0.98–1.11) | 0.22 | SH2B3 |
| rs2075650 | 19 | 0.14 | G/A | 1.11 | 5.9×10−11 | 0.53 | 1.03 (0.91–1.16) | 0.67 | 0.68 | 1.01 (0.91–1.13) | 0.79 | TOMM40 |
| rs2048327 | 6 | 0.35 | G/A | 1.06 | 6.9×10−11 | 0.33 | 1.00 (0.93–1.08) | 0.98 | 0.45 | 1.01 (0.95–1.08) | 0.65 | SLC22A3 |
| rs9319428 | 13 | 0.32 | A/G | 1.05 | 7.3×10−11 | 0.32 | 0.99 (0.91–1.09) | 0.91 | 0.43 | 1.03 (0.96–1.11) | 0.44 | FLT1 |
| rs17514846 | 15 | 0.44 | A/C | 1.05 | 9.3×10−11 | 0.45 | 1.00 (0.93–1.08) | 0.94 | 0.60 | 1.02 (0.96–1.09) | 0.52 | FURIN |
| rs1561198 | 2 | 0.45 | A/G | 1.05 | 1.2×10−10 | 0.35 | 1.09 (1.01–1.17) | 0.02 | 0.48 | 1.09 (1.02–1.15) | 6.9×10−3 | VAMP8 |
| rs515135 | 2 | 0.83 | G/A | 1.08 | 2.6×10−10 | 0.29 | 1.09 (0.98–1.20) | 0.10 | 0.40 | 1.03 (0.95–1.12) | 0.52 | APOB |
| rs4845625 | 1 | 0.47 | T/C | 1.04 | 3.6×10−10 | 0.36 | 1.01 (0.94–1.08) | 0.86 | 0.48 | 1.03 (0.97–1.10) | 0.28 | IL6R |
| rs2895811 | 14 | 0.43 | C/T | 1.06 | 4.1×10−10 | 0.35 | 1.03 (0.95–1.11) | 0.44 | 0.48 | 1.03 (0.96–1.10) | 0.39 | KIAA1822 |
| rs4252120 | 6 | 0.73 | T/C | 1.06 | 4.9×10−10 | 0.38 | 0.99 (0.92–1.07) | 0.82 | 0.51 | 0.99 (0.93–1.06) | 0.74 | PLG |
| rs273909 | 5 | 0.14 | C/T | 1.09 | 9.6×10−10 | 0.26 | 1.04 (0.91–1.17) | 0.59 | 0.35 | 1.01 (0.91–1.12) | 0.84 | SLC22A4 |
| rs12936587 | 17 | 0.59 | G/A | 1.06 | 1.2×10−9 | 0.35 | 1.00 (0.92–1.08) | 0.94 | 0.47 | 0.97 (0.91–1.04) | 0.40 | RAI1 |
| rs2047009 | 10 | 0.48 | C/A | 1.05 | 1.6×10−9 | 0.27 | 1.02 (0.94–1.09) | 0.68 | 0.36 | 1.03 (0.97–1.1) | 0.30 | CXCL12 |
| rs501120 | 10 | 0.83 | A/G | 1.07 | 1.8×10−9 | 0.29 | 1.08 (0.96–1.21) | 0.18 | 0.39 | 1.00 (0.91–1.09) | 0.96 | CXCL12 |
| rs9818870 | 3 | 0.14 | T/C | 1.07 | 2.6×10−9 | 0.26 | 1.10 (1.00–1.21) | 0.05 | 0.35 | 1.09 (1.01–1.18) | 0.02 | MRAS |
| rs264 | 8 | 0.86 | G/A | 1.05 | 2.9×10−9 | 0.53 | 0.99 (0.89–1.10) | 0.83 | 0.68 | 1.05 (0.96–1.14) | 0.31 | LPL |
| rs2281727 | 17 | 0.36 | C/T | 1.05 | 7.8×10−9 | 0.25 | 1.07 (0.99–1.15) | 0.09 | 0.34 | 1.02 (0.96–1.09) | 0.50 | SMG6 |
| rs445925 | 19 | 0.9 | C/T | 1.13 | 8.8×10−9 | 0.54 | 1.16 (0.97–1.39) | 0.10 | 0.56 | 1.05 (0.92–1.19) | 0.46 | APOC1 |
| rs10947789 | 6 | 0.76 | T/C | 1.06 | 9.8×10−9 | 0.36 | 1.12 (1.02–1.22) | 0.01 | 0.48 | 1.13 (1.05–1.22) | 5.6×10−4 | KCNK5 |
| rs579459 | 9 | 0.21 | C/T | 1.07 | 2.7×10−8 | 0.33 | 1.06 (0.97–1.17) | 0.20 | 0.45 | 1.03 (0.96–1.12) | 0.39 | ABO |
| rs2252641 | 2 | 0.46 | G/A | 1.04 | 5.3×10−8 | 0.36 | 1.00 (0.92–1.07) | 0.91 | 0.48 | 0.97 (0.91–1.03) | 0.34 | ZEB2 |
| rs12413409 | 10 | 0.89 | G/A | 1.10 | 6.3×10−8 | 0.38 | 1.05 (0.92–1.20) | 0.49 | 0.52 | 1.01 (0.90–1.13) | 0.91 | CNNM2 |
| rs9326246 | 11 | 0.10 | C/G | 1.09 | 1.5×10−7 | 0.51 | 0.93 (0.79–1.08) | 0.32 | 0.41 | 0.93 (0.82–1.06) | 0.26 | BUD13 |
| rs11203042 | 10 | 0.44 | T/C | 1.04 | 6.1×10−6 | 0.19 | 1.05 (0.98–1.13) | 0.15 | 0.25 | 1.07 (1.01–1.14) | 0.03 | LIPA |
| rs15563 | 17 | 0.52 | C/T | 1.04 | 9.4×10−6 | 0.19 | 1.12 (1.04–1.20) | 1.9×10−3 | 0.25 | 1.07 (1.01–1.13) | 0.03 | UBE2Z |
| rs2246833 | 10 | 0.38 | T/C | 1.06 | 9.5×10−6 | 0.34 | 1.07 (1.00–1.16) | 0.06 | 0.46 | 1.06 (1.00–1.13) | 0.05 | LIPA |
| rs11206510 | 1 | 0.84 | T/C | 1.06 | 1.8×10−5 | 0.22 | 1.02 (0.92–1.14) | 0.67 | 0.30 | 1.01 (0.93–1.10) | 0.80 | PCSK9 |
| rs12205331 | 6 | 0.81 | C/T | 1.04 | 4.2×10−5 | 0.14 | 1.03 (0.93–1.13) | 0.58 | 0.17 | 1.05 (0.97–1.13) | 0.24 | ANKS1A |
| rs17464857 | 1 | 0.87 | T/G | 1.05 | 6.1×10−5 | 0.15 | 1.11 (0.97–1.27) | 0.13 | 0.19 | 1.09 (0.97–1.22) | 0.14 | TAF1A |
| rs12539895 | 7 | 0.19 | A/C | 1.08 | 5.3×10−4 | 0.39 | 1.03 (0.94–1.12) | 0.57 | 0.52 | 0.99 (0.92–1.07) | 0.87 | GPR22 |
Chr. = Chromosome; Freq. = Frequency; OR = Odds Ratio; HR = Hazard Ratio; CI = Confidence Interval
*Coded / Non-coded allele
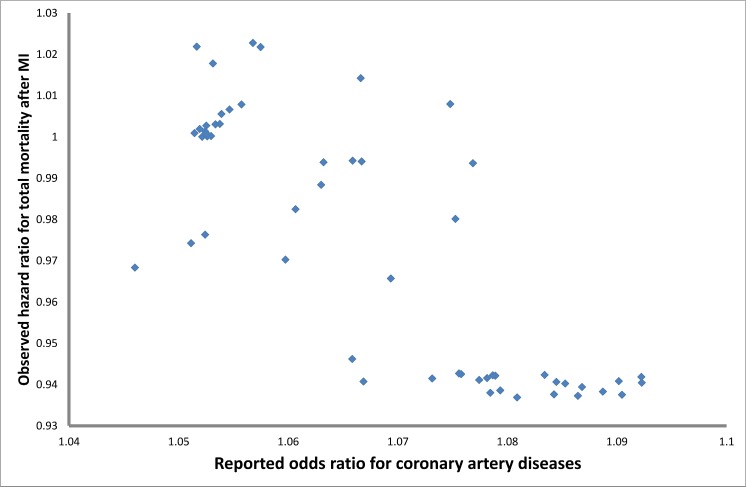
Among individuals who experienced MI during follow-up, the risk allele of rs1333049 was associated with a significantly decreased risk of mortality (HR: 0.90 [95% CI: 0.84, 0.97], p-value = 5.5×10−3) (Table 5). In both SNPs at 9p21 locus the “risk allele” from cross-sectional case-control GWAS was associated with longer survival after MI and would have been enriched in surviving prevalent cases. Fig 3 illustrates the inverse association of 78 top SNPs at the 9p21 locus as reported by CARDIoGRAMplusC4D Consortium [4] with survival after MI. We also examined the association of rs6941513 with mortality after MI, however, the association was not significant.
Table 5. Association of the known SNPs for coronary artery disease with mortality after MI.
| SNP | Closest Gene | Alleles | HR(95%CI) | P-value |
|---|---|---|---|---|
| rs1333049 | CDKN2A/B | C/G | 0.90 (0.84–0.97) | 5.5×10−3 |
| rs3217992 | CDKN2A/B | A/G | 0.94 (0.88–1.01) | 0.10 |
| rs602633 | PSRC1 | C/A | 1.01 (0.93–1.09) | 0.93 |
Fig 3. The association of top 79 SNPs with coronary artery disease as reported by CardiogramplusC4D for 9p21 locus and their association with total mortality after MI.
Discussion
We performed a GWAS on incident MI and CHD and examined whether the gene variants identified to date are also associated with risk of CHD in a prospective setting. In a two-stage design, involving 37,561 participants with 2,328 cases of incident MI, we identified a novel genome-wide significant locus, QKI, associated with incident MI. This finding requires further replication. The results also highlighted the difference between the genes identified in prospective versus cross-sectional case-control studies. The 9p21 locus was associated with both an increased risk of incident MI and, during follow-up post-MI, a decreased risk of total mortality, indicating that genetic variants may operate differently in an alternative setting.
In this two-stage design, we found evidence for MI-associated genetic variants nearby QKI (KH domain containing, RNA binding). The combined p-value for three out of four genetic variants that were examined in the region exceeded genome-wide significant threshold. Although these data provide evidence for an association between the QKI locus and incident MI, this finding should be confirmed by further studies since these variants attained conventional levels of genome-wide significant p-value only in the combined meta-analysis.
If confirmed, the QKI finding may represent a novel pathway in developing CHD. QKI is known to be involved in cell cycle regulation, a pathway for which there is emerging evidence for a key role in developing atherosclerotic plaques and cardiovascular disease [18, 19]. A functional study has reported that QKI is a central regulator of vascular smooth muscle cell phenotypic plasticity and that intervention in QKI activity can improve pathogenic fibro-proliferative responses to vascular injury [20]. Moreover, a recent paper shows that the RNA-binding properties of QKI play a critical role in regulating human monocyte to macrophage differentiation [21]. de Bruin and co-workers identified that the conversion of monocytes to both pro- and anti-inflammatory macrophages with GM-CSF or M-CSF, respectively, markedly increased expression of the QKI, which all were readily detected in CD68+ macrophages of fibrous cap atheromata and atherosclerotic lesions with intraplaque hemorrahage. Furthermore, reduced expression of QKI in monocytes delayed their differentiation into macrophages, perturbed their capacity to become lipid-engorged foam cells, and led to a reduction in monocyte infiltration in atherosclerotic lesions [21]. Altogether we propose that QKI is involved in inflammatory responses to injury and could be a potential thrapeutic target to prevent cardiovascular disease. Further functional investigation is needed to robustly identify mechanisms involved for this locus.
Prior GWAS which included extremely large sample sizes did not report QKI though they should have had enough statistical power to detect a locus with such an effect. However rs6941513 was not associated with CAD in the Cardiogram plusC4D GWAS (OR = 1.01, p-value = 0.45). In contrast to former GWAS, we have used a prospective, longitudinal cohort design to examine genetic association with incident cases of MI and CHD. It is possible that the magnitude of the effect with prevalent cases is smaller than with incident cases; thus the locus was not detected by previously published GWAS that primarily use a case-control design.
Although CHD includes MI events by definition, the loci we found for MI and CHD overlapped only for one locus (ATX1). One reason could be differences in mechanisms involved in the restrictive diagnosis of MI versus the broader diagnosis of CHD. However, unstable effect estimates and p-values due to lack of statistical power could have contributed to this observation as well.
Despite excellent statistical power, we identified only a modest signal at the 9p21 locus. This locus, initially identified by GWAS, has been validated by numerous studies in different geographic and ethnic subgroups. However, our study is not the first study to report a weak signal or lack of association at this locus. In fact, prominent differences have been observed between cross-sectional case-control versus longitudinal studies. For instance, in a meta-analysis by Chan et al [22], cross-sectional analyses of angiographically defined cases and controls show a strong per allele association with 9p21 (OR: 1.31, 95% CI: 1.20, 1.43). However, in a meta-analysis of follow-up studies by Patel et al [23], the per allele hazard ratio of the 9p21 variants for fatal and non-fatal adjudicated MI was 1.09 (95% CI: 1.03–1.16). The latter is the same as what we report in this study, though the meta-analysis includes earlier reports from some of our studies. One explanation for this inconsistency is the incidence-prevalence bias. Most GWAS for coronary artery disease to date have consisted of cross-sectional case-control studies, a design that over represents patients who survived their MI or CHD event. Using data from five population based cohort studies we found that the reported risk alleles for this locus are associated with longer survival after MI. This finding that was previously reported as well [23–25] supports the conjecture. Thus, the high prevalence of the risk allele in various types of cross-sectional analyses may not be due entirely to a high risk of experiencing MI or CHD, but also to an improved chance of survival after MI.
The molecular biology behind the protective effect of the risk alleles at 9p21 is yet unclear, however, there is a growing body of evidence to show that 9p21 locus is only increasing the risk of CHD for the first event and not for the subsequent events. For instance, Patel et al found no association with subsequent CHD events in a recent meta-analysis of 25,163 individuals with established CHD [23]. Thus, it could be concluded that 9p21 locus is contributing to the formation and progression of plaques and not to their instability prior to events; therefore, the association is merely observed in early stages of the disease. This is in agreement with the report by Palomaki [26] that suggests a diminished effect of 9p21 locus by age, a finding that is confirmed by Patel et al for secondary events. It should be noted that the mean age of participants was more than 70 years old in two and more than 60 years old in four of the participating cohorts. In this context, the older mean age of our population could be another reason why our findings do not replicate known loci such as 9p21.Our study is the largest collection of population-based prospective GWAS on incident MI and CHD and includes high quality genotyping and phenotyping data from well-known cohort studies in the field of cardiovascular disease. Moreover, similar case definitions for MI and CHD, comparable quality control for genotyped data, harmonized imputation strategies and collaboratively designed analysis plans are further strengths of our study. Despite these strengths, there are several limitations that merit discussion. First, nearly all studies who contributed to our GWAS are also members of the CARDIoGRAMPlusC4D Consortium [27], however, they have used only their prevalent cases in CARDIoGRAMPlusC4D project and therefore there is no overlap between the two GWAS. Second, since our sample size was limited, further susceptibility variants of weaker effects may have been missed in our study. Third, we have tried to use consistent definitions for MI, however, slight differences exist between the definitions for CHD. This might have introduced heterogeneity in our case definition. Finally, our findings may not be directly generalizable to non-European populations.
A potential clinical application of risk alleles identified from GWAS is the prospective prediction of cardiovascular disease. To date, the totality of evidence from prospective studies suggests that there is only modest, independent prediction of increased cardiovascular disease risk using genetic information with small to modest incremental reclassification for prediction beyond the known clinical CVD risk scores [28]. This lack of success has been attributed to the small percentage of variance explained by known genetic factors. However, our results also suggest that genetic risk prediction needs to consider differences in genetic variants that predict the risk of cardiovascular disease in prospective and cross-sectional settings.
In summary, using the largest collection of population- based prospective genome-wide association studies we have identified QKI as a potential locus for incident myocardial infarction. Furthermore, we have shown that the genes associated with risk of cardiovascular disease may differ in effect size when studied in a cross-sectional case-control versus cohort settings. The role of 9p21 locus may be complex, increasing the risk of incident MI and decreasing mortality among those with CHD. This highlights the importance of examining longitudinal cohort studies in the study of etiology even for genetic factors. These findings may have implications for application of genetic variants in risk estimation for cardiovascular disease, an effort that so far has not provided strong evidence for incremental risk prediction by genetic markers.
Supporting Information
(DOCX)
a. QQ Plot for Discovery GWAS MI. b. QQ Plot for Discovery GWAS CHD.
(ZIP)
a. Log-plot for Discovery GWAS MI. b. Log-plot for Discovery GWAS CHD.
(ZIP)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability
Data are available from dbGaP with Study Accession: phs000930.v1.p1.
Funding Statement
There were no current funding sources for this study. Merck Sharp & Dohme Corp. provided support in the form of salary for author Cynthia J. Girman, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific role of this author is articulated in the ‘author contributions’ section. Novartis Institutes for Biomedical Research provided support in the form of salary for author Ying A. Wang, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific role of this author is articulated in the ‘author contributions’ section.
References
- 1.Lloyd-Jones DM, Nam BH, D'Agostino RB Sr., Levy D, Murabito JM, Wang TJ, et al. Parental cardiovascular disease as a risk factor for cardiovascular disease in middle-aged adults: a prospective study of parents and offspring. JAMA. 2004;291(18):2204–11. Epub 2004/05/13. 10.1001/jama.291.18.2204 291/18/2204 [pii]. . [DOI] [PubMed] [Google Scholar]
- 2.Marenberg ME, Risch N, Berkman LF, Floderus B, de Faire U. Genetic susceptibility to death from coronary heart disease in a study of twins. N Engl J Med. 1994;330(15):1041–6. Epub 1994/04/14. 10.1056/NEJM199404143301503 . [DOI] [PubMed] [Google Scholar]
- 3.O'Donnell CJ, Nabel EG. Genomics of cardiovascular disease. N Engl J Med. 2011;365(22):2098–109. Epub 2011/12/02. 10.1056/NEJMra1105239 . [DOI] [PubMed] [Google Scholar]
- 4.Consortium CAD, Deloukas P, Kanoni S, Willenborg C, Farrall M, Assimes TL, et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45(1):25–33. Epub 2012/12/04. ng.2480 [pii] 10.1038/ng.2480 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hill G, Connelly J, Hebert R, Lindsay J, Millar W. Neyman's bias re-visited. J Clin Epidemiol. 2003;56(4):293–6. Epub 2003/05/28. S0895435602005711 [pii]. . [DOI] [PubMed] [Google Scholar]
- 6.Neyman J. Statistics; servant of all sciences. Science. 1955;122(3166):401–6. Epub 1955/09/02. . [DOI] [PubMed] [Google Scholar]
- 7.Wilson PW. Risk scores for prediction of coronary heart disease: an update. Endocrinol Metab Clin North Am. 2009;38(1):33–44. Epub 2009/02/17. S0889-8529(08)00084-4 [pii] 10.1016/j.ecl.2008.11.001 . [DOI] [PubMed] [Google Scholar]
- 8.Psaty BM, O'Donnell CJ, Gudnason V, Lunetta KL, Folsom AR, Rotter JI, et al. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: Design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet. 2009;2(1):73–80. Epub 2009/12/25. 2/1/73 [pii] 10.1161/CIRCGENETICS.108.829747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris TB, Launer LJ, Eiriksdottir G, Kjartansson O, Jonsson PV, Sigurdsson G, et al. Age, Gene/Environment Susceptibility-Reykjavik Study: multidisciplinary applied phenomics. Am J Epidemiol. 2007;165(9):1076–87. Epub 2007/03/14. kwk115 [pii] 10.1093/aje/kwk115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. The ARIC investigators. Am J Epidemiol. 1989;129(4):687–702. Epub 1989/04/01. . [PubMed] [Google Scholar]
- 11.Fried LP, Borhani NO, Enright P, Furberg CD, Gardin JM, Kronmal RA, et al. The Cardiovascular Health Study: design and rationale. Ann Epidemiol. 1991;1(3):263–76. Epub 1991/02/01. . [DOI] [PubMed] [Google Scholar]
- 12.Feinleib M, Kannel WB, Garrison RJ, McNamara PM, Castelli WP. The Framingham Offspring Study. Design and preliminary data. Prev Med. 1975;4(4):518–25. Epub 1975/12/01. . [DOI] [PubMed] [Google Scholar]
- 13.Hofman A, Grobbee DE, de Jong PT, van den Ouweland FA. Determinants of disease and disability in the elderly: the Rotterdam Elderly Study. Eur J Epidemiol. 1991;7(4):403–22. . [DOI] [PubMed] [Google Scholar]
- 14.Hofman A, van Duijn CM, Franco OH, Ikram MA, Janssen HL, Klaver CC, et al. The Rotterdam Study: 2012 objectives and design update. Eur J Epidemiol. 2011;26(8):657–86. 10.1007/s10654-011-9610-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–9. Epub 2006/07/25. ng1847 [pii] 10.1038/ng1847 . [DOI] [PubMed] [Google Scholar]
- 16.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–1. Epub 2010/07/10. btq340 [pii] 10.1093/bioinformatics/btq340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matise TC, Ambite JL, Buyske S, Carlson CS, Cole SA, Crawford DC, et al. The Next PAGE in understanding complex traits: design for the analysis of Population Architecture Using Genetics and Epidemiology (PAGE) Study. Am J Epidemiol. 2011;174(7):849–59. Epub 2011/08/13. kwr160 [pii] 10.1093/aje/kwr160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316(5830):1491–3. Epub 2007/05/05. 1142842 [pii] 10.1126/science.1142842 . [DOI] [PubMed] [Google Scholar]
- 19.McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316(5830):1488–91. Epub 2007/05/05. 1142447 [pii] 10.1126/science.1142447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van der Veer E, de Bruin RG, Kraaijeveld A, de Vries MR, Bot I, Pera T, et al. The RNA-Binding Protein Quaking is a Critical Regulator of Vascular Smooth Muscle Cell Phenotype. Circ Res. 2013. Epub 2013/08/22. CIRCRESAHA.113.301302 [pii] . [DOI] [PubMed] [Google Scholar]
- 21.de Bruin RG, Shiue L, Prins J, de Boer HC, Singh A, Fagg W.S., et al. Quaking post-transcriptionally promotes monocyte differentiation into pro-atherogenic macrophages by controlling pre-mRNA splicing and gene expression. Nat Commun. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan K, Patel RS, Newcombe P, Nelson CP, Qasim A, Epstein SE, et al. Association between the chromosome 9p21 locus and angiographic coronary artery disease burden: a collaborative meta-analysis. J Am Coll Cardiol. 2013;61(9):957–70. Epub 2013/01/29. S0735-1097(12)05975-X [pii] 10.1016/j.jacc.2012.10.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patel RS, Asselbergs FW, Quyyumi AA, Palmer TM, Finan CI, Tragante V, et al. Genetic variants at chromosome 9p21 and risk of first versus subsequent coronary heart disease events: a systematic review and meta-analysis. J Am Coll Cardiol. 2014;63(21):2234–45. Epub 2014/03/13. S0735-1097(14)01383-7 [pii] 10.1016/j.jacc.2014.01.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szpakowicz A, Kiliszek M, Pepinski W, Waszkiewicz E, Franaszczyk M, Skawronska M, et al. Polymorphism of 9p21.3 locus is associated with 5-year survival in high-risk patients with myocardial infarction. PLoS One. 2014;9(8):e104635 Epub 2014/08/12. 10.1371/journal.pone.0104635 PONE-D-14-14817 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gong Y, Beitelshees AL, Cooper-DeHoff RM, Lobmeyer MT, Langaee TY, Wu J, et al. Chromosome 9p21 haplotypes and prognosis in white and black patients with coronary artery disease. Circ Cardiovasc Genet. 2011;4(2):169–78. Epub 2011/03/05. CIRCGENETICS.110.959296 [pii] 10.1161/CIRCGENETICS.110.959296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palomaki GE, Melillo S, Bradley LA. Association between 9p21 genomic markers and heart disease: a meta-analysis. JAMA. 2010;303(7):648–56. Epub 2010/02/18. 303/7/648 [pii] 10.1001/jama.2010.118 . [DOI] [PubMed] [Google Scholar]
- 27.Deloukas P, Kanoni S, Willenborg C, Farrall M, Assimes TL, Thompson JR, et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2012. Epub 2012/12/04. ng.2480 [pii] 10.1038/ng.2480 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Vries PS, Kavousi M, Ligthart S, Uitterlinden AG, Hofman A, Franco OH, et al. Incremental predictive value of 152 single nucleotide polymorphisms in the 10-year risk prediction of incident coronary heart disease: the Rotterdam Study. Int J Epidemiol. 2015;44(2):682–8. Epub 2015/05/09. dyv070 [pii] 10.1093/ije/dyv070 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
a. QQ Plot for Discovery GWAS MI. b. QQ Plot for Discovery GWAS CHD.
(ZIP)
a. Log-plot for Discovery GWAS MI. b. Log-plot for Discovery GWAS CHD.
(ZIP)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
Data are available from dbGaP with Study Accession: phs000930.v1.p1.