Abstract
Lagerstroemia (crape myrtle) is an important plant genus used in ornamental horticulture in temperate regions worldwide. As such, numerous hybrids have been developed. However, DNA sequence resources and genome information for Lagerstroemia are limited, hindering evolutionary inferences regarding interspecific relationships. We report the complete plastid genome of Lagerstroemia fauriei. To our knowledge, this is the first reported whole plastid genome within Lythraceae. This genome is 152,440 bp in length with 38% GC content and consists of two single-copy regions separated by a pair of 25,793 bp inverted repeats. The large single copy and the small single copy regions span 83,921 bp and 16,933 bp, respectively. The genome contains 129 genes, including 17 located in each inverted repeat. Phylogenetic analysis of genera sampled from Geraniaceae, Myrtaceae, and Onagraceae corroborated the sister relationship between Lythraceae and Onagraceae. The plastid genomes of L. fauriei and several other Lythraceae species lack the rpl2 intron, which indicating an early loss of this intron within the Lythraceae lineage. The plastid genome of L. fauriei provides a much needed genetic resource for further phylogenetic research in Lagerstroemia and Lythraceae. Highly variable markers were identified for application in phylogenetic, barcoding and conservation genetic applications.
Introduction
The Lythraceae include approximately 620 species in 31 genera; most are herbs, with some trees and shrubs adapted to a wide variety of habitats. The four largest genera (Cuphea, Diplusodon, Lagerstroemia, and Nesaea) include three-fourths of all species in Lythraceae [1]. The family has been traditionally classified in the order Myrtales and closely allied with the Onagraceae based on morphological, anatomical, and embryological evidence [2,3]. Within the Lythraceae, Lagerstroemia (“crape myrtle”) is the most economically important and well-known genus. Lagerstroemia comprises about 55 species [4–6] and its center of diversity is in southeast Asia and Australia [7], mainly in tropical and sub-tropical habitats of southern China, Japan, and northeast Australia. Most Lagerstroemia species are easily propagated, resistant to multiple pathogens, grow rapidly, and have colorful flowers that open from summer to fall [8]. Given the importance of Lagerstroemia as an ornamental, more than 260 cultivars have been created and registered (http://www.usna.usda.gov/Research/Herbarium/Lagerstroemia/index.html). Due to the ornamental and economic value of Lagerstroemia, research programs have been initiated to develop hybrid cultivars, study the genetic diversity of cultivars, and evaluate germplasm [9–13]. Molecular tools have been employed to identify Lagerstroemia cultivars and interspecific hybrids [14,15]. Despite the development of microsatellite markers and subsequent research in Lagerstroemia, no complete chloroplast (plastid) genomes have been described from Lythraceae.
Phylogenomic-related research in Lythraceae is limited. Within the Myrtales, Lythraceae was resolved as sister to Onagraceae using the plastid gene rbcL [16]. Within Lythraceae, Lagerstroemia and Duabanga are supported as sister groups based on atpB-rbcL, psaA-ycf3, rbcL, trnK-matK, trnL-trnF, and ITS (internal transcribed spacer region of the nuclear genome) data [1,17]. Phylogenetic inferences within Lagerstroemia and the Lythraceae could be improved if plastid genomes are made available, potentially providing dozens of valuable molecular markers for further research.
In contrast to huge nuclear genomes, the plastid genome, with uniparental inheritance, has a highly conserved circular DNA arrangement ranging from115 to 165 kb [18,19], and the gene content and gene order are conserved across most land plants [20]. With the development of next-generation sequencing approaches, sequencing whole plastid genomes has become cheaper and faster [21]. To date, more than 900 land-plant species’ completed plastomes can be accessed through the National Center for Biotechnology Information (NCBI) public database [22]. Such genetic resources have provided a useful set of tools for researchers interested in species identification by using DNA barcoding [23], genetic data used for plastid transformation [24], and designing molecular makers for systematic and population studies [25,26]. All of these research areas have benefitted from the conserved sequences and structure as well as the lack of recombination found in plastid genomes to simplify analyses. For example, plastids maintain a positive homologous recombination system [27–30], which enables precise transgene targeting into a specific genome region during transformation. Different plastid loci have been used for evaluating phylogenetic relationships at different taxonomic levels, including the interspecific and intraspecific levels [31]. Recently phylogenomic approaches [32] to study plant relationships have employed complete-plastid-genome sequences for studying phylogenetic relationships.
In an effort to comprehensively understand the organization of the Lagerstroemia plastid genome, we present the first complete plastid genome sequence of L. fauriei, which was generated using Illumina sequencing. The three aims of our study are to: deepen our understanding of the structural diversity of the complete L. fauriei plastid genome, compare molecular evolutionary patterns of the L. fauriei plastid genome with other plastid genomes in the Myrtales, and provide a set of genetic resources for future research in Lagerstroemia and the Lythraceae.
Materials and Methods
Plant materials, DNA extraction and sequencing
Leaves of L. fauriei were obtained from the nursery of Zhejiang Agriculture and Forestry University (Hangzhou, Zhejiang, China) and preserved in silica gel. Total genomic DNA was extracted from leaves using a cetyl-trimethyl-ammonium-bromide DNA-extraction protocol [33]. Total genomic DNA was used to construct a sequence library following the manufacturer's instructions (Illumina Inc., San Diego, CA). Paired-end (PE) sequencing libraries with an insert size of approximately 300 bp were sequenced on an Illumina HiSeq 2000 sequencer at the Beijing Genomics Institute (BGI) and 30,887,628 clean reads were obtained, each with a read length of 100 bp.
Plastid genome assembly and annotation
The raw Illumina reads were demultiplexed, trimmed and filtered by quality score with Trimmomatic v0.3 [34] using the following settings: leading: 3, trailing: 3, sliding window: 4:15 and minlen: 50. Then the CLC Genomics Workbench v7 (CLCbio; http://www.clcbio.com) was used to conduct de novo assembly of reads from L. fauriei with the default parameters. The following three separate de novo assemblies were made: PE reads, single-end forward reads and single-end reverse reads [22]. These three separate assemblies were then combined into a single assembly. Assembled contigs (≥0.5 kb) with > 100× coverage from the complete CLC assembly were compared to several Myrtales species with completed plastid genomes, including Oenothera argillicola (Onagraceae; NC_010358), Syzygium cumini (Myrtaceae; GQ870669), and Eucalyptus aromaphloia (Myrtaceae; NC_022396). Local BlastN [35] searches were used to match the contigs from the plastid genomes. Based on the conserved features of the plastid genome [19,22], the mapped contigs were orientated onto the related plastid genomes [36] and those separate contigs were connected into a single contig to construct the circular map of the genome using Informax Vector NTI Contig Express 2003 (Invitrogen, Carlsbad, CA). Seven short gaps (≤100 bp) were filled by aligning individual Illumina sequence reads that overlapped at the contig ends. Longer gaps (>100 bp) between contigs were filled by designing primers in flanking regions, conducting PCR amplifications, and closing the gap regions by adding sequence data generated from Sanger sequencing (by BGI).
We designed additional primers (S1 Table) to test for correct sequence assembly. PCR was conducted in 40μl volumes containing 4 μl 10× Taq buffer, 0.8 μl dNTP (10 mM), 0.4μl Taq polymerase (5 U/μl), 0.5ul each primer (20 pmol/ul; all from Sangong Biotech (Shanghai, China)), 0.5 ul DNA template, and 33.3 μl ddH2O. The amplification program consisted of an initial heating at 94°C for 5 min, then 32 cycles including denaturation at 94°C for 45 s, annealing at 55°C for 45 s, elongation at 72°C for 2 min, and a final elongation at 72°C for 10 min. After incorporation of the Sanger results, the finished plastid genomes were applied as the reference to map the previously unincorporated short reads in order to iteratively refine the assembly based on evenness of sequence coverage.
DOGMA v1.2 [37] was employed for genome annotation of the protein-coding genes, transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs). To accurately confirm the start and stop codons and the exon-intron boundaries of genes, the draft annotation was subsequently inspected and adjusted manually based on plastomes from a related species, Syzygium cumini [36], from the NCBI database. Additionally, both tRNA and rRNA genes were identified by BLASTN searches against the same database of plastomes. Finally, tRNAscan-SE v1.21 [38] was also used to further verify the tRNA genes. The schematic diagram of the plastid genome map was generated using OGDraw [39].
Comparative plastid genomic analysis
Expansion and contraction of four junction regions
Genome-size variation among different photosynthetic species is generally caused by different junctions between the two inverted-repeat regions (IRA and IRB) and the two single-copy regions (LSC and SSC) [36]. There are four junctions (JLA, JLB, JSA, and JSB) in the plastid genome between the two single copy (LSC and SSC) regions and the two IRs (IRA and IRB) [40]. The detailed IR border positions and the adjacent genes among seven Myrtales species plastomes (Lagerstroemia fauriei, Oenothera argillicola, Angophora costata, Corymbia eximia, Eucalyptus aromaphloia, Stockwellia quadrifida, and Syzygium cumini) were compared in this study.
Survey for loss of the rpl2 intron
In the process of annotation and comparison with other species in the Myrtales, we found that the intron of rpl2 is absent in the plastome of L. fauriei. In order to infer the history of this intron loss, we designed a pair of primers (Forward-CAAAACTTCTACCCCAAGCA; Reverse-TCTTCTTCCAAGTGCAGGAT) to amplify the whole rpl2 region and then applied them to 11 Lagerstroemia species and three species (Cuphea hyssopifolia, Punica granatum, and Lythrum salicaria) from other Lythraceae genera, as well as the outgroups Oenothera albicaulus and Catha edulis. In L. fauriei, the target rpl2 fragment without the intron is about 750 bp, whereas it is about 1,400 bp in species containing the intact intron. PCR was used to amplify the rpl2 region and the amplicons were run out on 1% agarose gels. Fragment sizes were determined by comparison to DNA size standards [41]. Sanger sequencing of forward and reverse sequence of gene rpl2 was done for Cuphea hyssopifolia, Punica granatum, L. salicaria, L. fauirei, L. limii and Oenothera albicaulus at the Proteomics and Metabolomics Facility of Colorado State University.
Repetitive sequence analysis
Repetitive elements were investigated using two different approaches. In order to avoid redundancy, repeat-sequence analysis was only carried out using just one IR region [42]. Tandem Repeat Finder [43] was used with the minimum-alignment score and maximum-period size set at 50 and 500, respectively, with default parameters for all other search criteria to find small tandem repeats from 15 to 30 bp in length. The numbers of forward, reverse, complementary and palindromic repeats were quantified using the REPuter [44], setting Hamming distance equal to three and minimum repeat size ≥30 bp. Overlapping repeats were merged into one repeat motif where possible. Microsatellites (SSRs) were detected using SSR Hunter v1.3 [45]. We identified SSRs as mononucleotides with ≥ 8 repeats, dinucleotides ≥ 4, trinucleotides ≥ 3, and tetranucleotides and pentanucleotides both ≥ 3.
Dot-plot analysis
We compared plastomes of the other six Myrtales species to L. fauriei with dot-plot analysis using Perl scripts to visualize arrangement recurrences and structural differences in two-dimensional plots (S1 Fig).
Informative variables analysis from coding and non-coding regions
To identify divergent regions that may be highly informative for phylogenetic analyses, each region, including CDS (coding regions), introns, and IGS (intergenic regions) from seven Myrtales plastid genomes was individually examined. For the longer genes (>1500 bp), we employed the sliding window method to divide the gene into shorter fragments to detect the most informative portions by using a 1000 bp sliding window and 500 bp increments. These regions were aligned using Clustal X 2.0 [46] and adjusted manually using the similarity criterion [47]. The aligned sequences were analyzed using parsimony in PAUP*4.0b10 [48] with tree-bisection-reconnection branch-swapping. The ensemble retention index (RI) [49] was calculated for each of the 78 coding regions and 128 non-coding regions. The 10 coding and 10 non-coding regions with the highest percentages of parsimony-informative characters were then selected as candidates for phylogenetic markers.
Phylogenetic analysis
The 73 shared protein-coding genes from the plastid genomes in the seven Myrtales species and the three Geraniaceae outgroup species were aligned in Clustal X using the default settings, followed by manual adjustment to preserve the reading frames. The data matrix is posted as S1 Matrix. Three phylogenetic-inference methods were used to infer trees from these 73 concatenated genes. Parsimony analysis was implemented in PAUP* 4.0b10 [48], maximum likelihood (ML) in PHYML v 2.4.5 [50], and Bayesian inference (BI) in MrBayes 3.1.2 [51] using the settings from [22].
Results and Discussion
Sequencing, assembly and annotation
The whole plastid genome for Lagerstroemia fauriei was found to be 152,440 bp in length after combining the Sanger and Illumina sequence data. Through mapping the paired reads onto the finished genome, we verified our assembled length for the finished plastid genome with 1,473,293 (5% of the total reads) mapped reads across the whole genome with at least 951 reads per position. Based on this number of reads we consider the assembled genome to be of high-quality. Our annotated plastid genome of L. fauriei is available from GenBank (KT358807).
Plastid genome features
In most land plants, the plastid genome is a single circular structure of 115–165 kb in length that consists of one large single-copy (LSC) region, one small single-copy (SSC) region, and a pair of inverted repeats (IRs). Although gene order and content are highly conserved in plastid genomes, they differ in the extent of gene duplication, size of intergenic spacers, presence or absence of introns, as well as the length and number of small repeats [52]. Such differences not only leave molecular patterns that allow for the inference of evolutionary history, but can also influence the molecular functioning of the cell as a whole (e.g., [20, 32]).
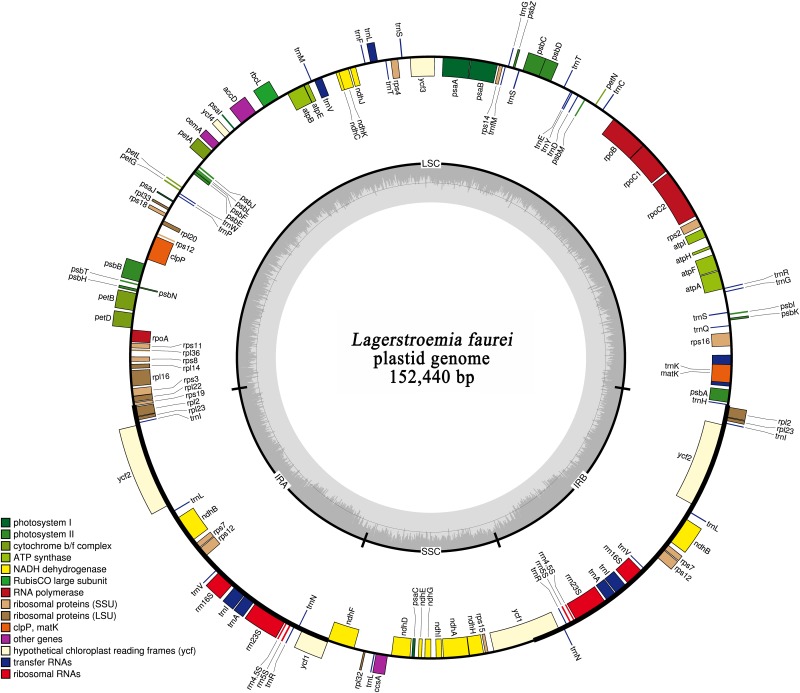
The plastid genome of L. fauriei is composed of two single-copy regions separated by a pair of 25,793 bp IRs (Fig 1, Table 1), which account for 34% of the whole plastid genome. The LSC and SSC regions span 83,921 bp and 16,933 bp, respectively. The proportion of LSC and SSC length in the total plastid genome is 55% and 11%, respectively (Table 1). The L. fauriei plastid genome consists of protein coding genes, transfer RNA (tRNA), ribosomal RNA (rRNA), intronic and intergenic regions (Table 2). 81,412 bp (53%) of the whole L. fauriei plastid genome are non-coding DNA, 68,655 bp (45%) are protein-coding exons, 2,373 bp (2%) are tRNA, 4,517 bp (3%) are rRNA, 14,503 bp (10%) are intronic regions, and 62,570 bp (41%) are intergenic regions (Table 2).
Fig 1. Map of the L. fauriei plastid genome.
Genes shown outside the outer circle are transcribed clockwise and genes inside the outer circle are transcribed counterclockwise. Genes in different functional groups are color coded. The shaded area inside the inner circle indicates the GC content, with dark shading indicating percent CG.
Table 1. Comparison of plastid genome size among seven Myrtales species.
| Region | L. fauriei | O. argillicola | A. costata | C. eximia | E. aromaphloia | S. quadrifida | S. cumini |
|---|---|---|---|---|---|---|---|
| LSC | |||||||
| Length (bp) | 83,923 | 88,511 | 88,768 | 88,522 | 88,925 | 88,247 | 89,081 |
| GC content (%) | 36 | 37 | 35 | 35 | 35 | 35 | 35 |
| Length percentage (%) | 55 | 54 | 55 | 55 | 56 | 55 | 56 |
| SSC | |||||||
| Length (bp) | 16,933 | 19,000 | 18,772 | 18,672 | 18,468 | 18,544 | 18,508 |
| GC Content (%) | 31 | 35 | 30 | 31 | 31 | 31 | 31 |
| Length Percentage (%) | 11 | 12 | 12 | 12 | 12 | 12 | 12 |
| IR | |||||||
| Length (bp) | 25,792 | 28,772 | 26,392 | 26,409 | 26,378 | 26,385 | 26,392 |
| GC Content (%) | 43 | 43 | 43 | 43 | 43 | 43 | 43 |
| Length Percentage (%) | 34 | 35 | 33 | 33 | 33 | 33 | 33 |
| Total | |||||||
| Length (bp) | 152,440 | 165,055 | 160,326 | 160,012 | 160,149 | 159,561 | 160,373 |
| GC Content (%) | 38 | 39 | 37 | 37 | 37 | 37 | 37 |
Table 2. Comparison of coding and non-coding region size among seven Myrtales species.
| Region | Species | L. fauriei | O. argillicola | A. costata | C. eximia | E. aromaphloia | S. quadrifida | S. cumini |
|---|---|---|---|---|---|---|---|---|
| Protein coding | Length (bp) | 68,477 | 70,706 | 68,257 | 68,889 | 68,085 | 68,746 | 68,448 |
| GC content (%) | 45 | 43 | 43 | 43 | 43 | 43 | 43 | |
| Length percentage (%) | 38 | 40 | 37 | 37 | 37 | 37 | 38 | |
| tRNA | length (bp) | 2,373 | 2,303 | 3,184 | 2,199 | 2,270 | 2,387 | 2,310 |
| GC content (%) | 54 | 53 | 49 | 53 | 53 | 52 | 53 | |
| Length percentage (%) | 2 | 1 | 2 | 1 | 1 | 2 | 1 | |
| rRNA | length (bp) | 4,517 | 4,551 | 4,510 | 4,528 | 4,528 | 4,528 | 4,525 |
| GC content (%) | 56 | 55 | 55 | 55 | 55 | 55 | 55 | |
| Length percentage (%) | 3 | 3 | 3 | 3 | 3 | 3 | 3 | |
| Intron | length (bp) | 14,503 | 13,311 | 15,514 | 15,499 | 14,720 | 15,465 | 15,496 |
| GC content (%) | 36 | 38 | 35 | 36 | 36 | 36 | 36 | |
| Length percentage (%) | 10 | 8 | 10 | 10 | 9 | 10 | 10 | |
| Intergenic | length (bp) | 62,570 | 70,706 | 68,861 | 68,897 | 70,546 | 68,435 | 69,594 |
| GC content (%) | 36 | 37 | 35 | 35 | 35 | 35 | 35 | |
| Length percentage (%) | 41 | 43 | 43 | 43 | 44 | 43 | 43 |
The plastid genome of L. fauriei contains 129 coding genes, including 84 protein-coding genes, 37 tRNA genes, and eight rRNA genes. Among the 129 genes, 4 rRNA genes, 7 tRNA genes and 6 coding genes are duplicated in the two IR regions (Fig 1; Table 3). Of the 112 unique genes, 82 are located in the LSC region (60 protein-coding genes, 22 tRNA genes), 13 in the SSC region (12 protein-coding genes, 1 tRNA gene), and 17 in both IR regions (6 coding genes, 4 rRNA genes, 7 tRNA genes). The following four genes span regional plastid boundaries: ycf1 spans the SSC and IRB regions, rps12 spans the LSC and two IR regions (5’ end exon was in LSC and two 3’end exons were duplicated in IR regions), ndhF spans the IRA and SSC regions and rps19 spans the LSC and IRA region (Fig 1). In the whole plastid genome, 17 genes contain introns, including eight protein-coding genes with a single intron each (atpF, ndhA, ndhB, petB, petD, rpl16, rpoC1, rps16), five tRNA genes with a single intron each (trnAGUC, trnGUCC, trnIGAU, trnKUUU, trnLUAA, trnVUAC), and three protein coding genes with two introns each (clpP, rps12 and ycf3). Among the 17 genes with introns, 13 genes are located in LSC, one in SSC, and three in both IRs (S2 Table). The rps12 gene is a trans-spliced gene with a 5’ end exon in the LSC region and two duplicated 3’-end exons in IR regions. The 2,497 bp intron of trnKUUU is the longest, but 1491 bp of it codes for the matK gene.
Table 3. List of genes in the L. fauriei plastid genome.
| Gene category | Group of genes | Name of genes |
|---|---|---|
| Self-replication | Transfer RNA genes | trnA-UGCa,b trnC-GCA trnD-GUC trnE-UUC trnF-GAA trnfM-CAU trnG-UCC trnG-GCC trnH-GUG trnI-CAUb trnI-GAUa,b trnK-UUUa trnL-CAAb trnL-UAAa trnL-UAG trnM-CAU trnN-GUUb trnP-UGG trnQ-UUG trnR-ACGb trnR-UCU trnS-GCU trnS-GGA trnS-UGA trnT-GGU trnT-UGU trnV-GACb trnV-UACa trnW-CCA trnY-GUA |
| Small subunit of ribosome | rps2 rps3 rps4 rps7b rps8 rps11 rps12a,b rps14 rps15 rps16* rps18 rps19 | |
| Ribosomal RNA genes | rrn16b rrn23b rrn4.5b rrn5b | |
| Large subunit of ribosome | rpl2b rpl14 rpl16a rpl20 rpl22 rpl23b rpl32 rpl33 rpl36 | |
| DNA dependent RNA polymerase | rpoA rpoB rpoC1a rpoC2 | |
| Photosynthesis | Subunits of photosystem I | psaA psaB psaC psaI psaJ |
| Subunits of photosystem II | psbA psbB psbC psbD psbE psbF psbHpsbI psbJ psbK psbL psbM psbN psbT psbZ | |
| Subunits of cytochrome | petA petBa petDa petG petL petN | |
| Subunits of ATP synthase | atpA atpB atpE atpFa atpH atpI | |
| ATP-dependent protease subunit p gene | clpPa | |
| Large subunit of Rubisco | rbcL | |
| Subunits of NADH dehydrogenase | ndhAa ndhBa,b ndhC ndhD ndhE ndhF ndhG ndhH ndhI ndhJ ndhK | |
| Other genes | Maturase | matK |
| Envelop membrane protein | cemA | |
| Subunit of acetyl-CoA-carboxylase | accD | |
| c-type cytochrome synthesis gene | ccsA | |
| Genes of unknown function | Conserved open reading frames | ycf1 ycf2b ycf3a ycf4 |
a: Genes containing introns;
b: Duplicated gene (Genes present in the IR regions).
Comparison of the plastid genomes with six other Myrtales
We compared the plastid genome of L. fauriei (Lythraceae) to six other species in the Myrtales with dot-plot analysis. The plastid genomes in these species possess identical gene order with the exception of O. argillicola, which contains a large inversion of about 56 kb in the LSC region (S1 Fig) [53,54]. These results further verified the conserved feature of the plant plastid genome and partial lineage-specific variation [19]. The seven plastid genomes vary in length from 152,440 to 165,055 bp. From the comparative results (Table 1), the plastid genome of O. argillicola is the longest of the seven species, which is explained partly by expansion of intergenic regions in the SSC and IR regions. However, the plastome of L. fauriei is the shortest because of reduction of intergenic regions, which only occupy 41% of the genome (Table 2). These comparisons demonstrate that the dynamic variation of the intergenic regions is the main cause of length differences between plastid genomes [19, 22].
The GC content of the plastid genome is stable across most land plants [19]. The GC content of the entire L. fauriei plastid genome is 38%, with 36% GC content in the LSC region, 31% in the SSC region and 43% in the IR regions. These percentages are generally similar to other plastid genomes [55]. The overall GC contents in seven Myrtales plastid genomes ranged from 37% to 39%, with O. argillicola having the highest GC content and A. costata having the lowest (Table 1). The GC content of protein-coding regions in the seven Myrtales species range from 37% to 40%, of which O. argillicola has the highest and C. eximia has the lowest (Table 1).
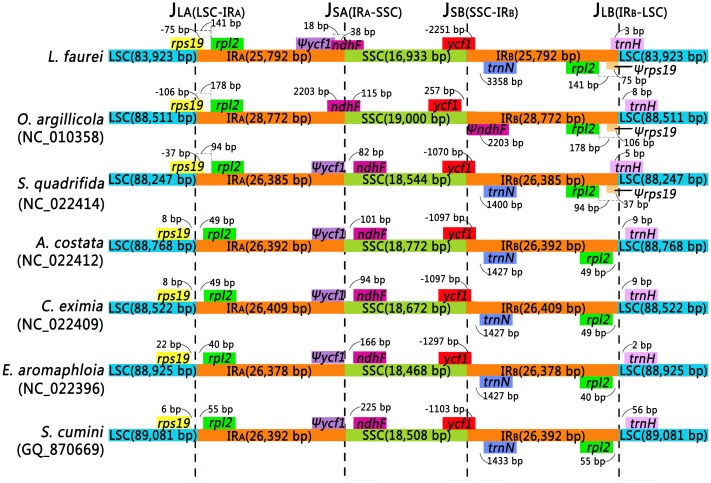
From these cross-species comparisons, we verified that the Myrtales plastid genomes are highly conserved in genome content, gene order and overall genomic structure relative to L. fauriei. They have similar gene orders at the IR-SSC and IR-LSC borders, with the exception of ψycf1 (pseudogene ycf1), which is absent from the border of IRA and SSC in O. argillicola. Instead O. argillicola has a ψndhF (pseudogene ndhF) on the border of SSC and IRB (Fig 2).
Fig 2. Comparison of junctions between the LSC, SSC, and two IR regions among seven Myrtales species.
ψ means pseudogene; distance in the figure is not to scale.
Expansion and contraction of four junction regions
The typical quadripartite structure of plastomes includes two single-copy regions and two inverted repeat regions, though length of the IRs differ between plant species because of contraction and expansion in these regions [19]. We examined the four junctions (JLA, JLB, JSA, and JSB) across the seven Myrtales species to assess the junction variation between the IRs and single-copy regions following Wang [40] and Wu [22].
The length of the IRs ranged from 25,792 to 28,772 bp, and the positions of all four IR boundaries (JLA, JLB, JSA, and JSB) varied (Fig 2) [56]. The LSC/IRA junctions in plastid genomes of L. fauriei, O. argillicola, and S. quadrifida were located in the coding region of rps19, which extended into the IRB region 75 bp, 106 bp, and 37 bp, respectively. In the other four species the LSC includes an intact rps19 gene together with 8 bp (A. costata, C. eximia), 22 bp (E. aromaphloia), or 6 bp (S. cumini) of non-coding region beyond the LSC/IRA border. The IRB/LSC border in these four species is located in the intergenic spacer between rpl2 and trnH. The trnH gene of S. cumini is 56 bp away from the IRB/SSC border, whereas in L. fauriei and S. quadrifida the trnH gene extends into the IRB by 3 bp and 5 bp respectively. In the other four species the trnH gene is 2–9 bp away from the IRB/SSC border.
In O. argillicola, the ycf1 gene does not extend into the IRB region at the border of SSC/IRA. Rather, in contrast to the other six species wherein ycf1 extends across the border, ycf1 in O. argillicola is separated by 257 bp. Hence the SSC/IRB junction resulted in the duplication of the 3’ end region of ycf1 in these six species, and consequently a pseudogene with variable length at the IRA/SSC border (Fig 2) [49].
Variable gene composition was found at the IRA/SSC border. In O. argillicola the ψycf1 gene is absent, and instead the IRA/SSC border was positioned in the ndhF gene, which had 115 bp in the SSC region and 2,203 bp in the IRA region. Similarly, ndhF extends 38 bp into the IRA region in L. fauriei, which also has 20 bp overlap with ψycf1. The entire ndhF gene is located in the SSC region in the other five species and is separated by 82–225 bp from the IRA/SSC border. The IR/LSC border region has been used extensively for phylogenetic studies in Eucalyptus [36,57] and given the variation we observed, this region could be similarly useful for resolving the relationships between L. fauriei and its relatives.
Loss of the rpl2 intron from Lagerstroemia and Lythraceae
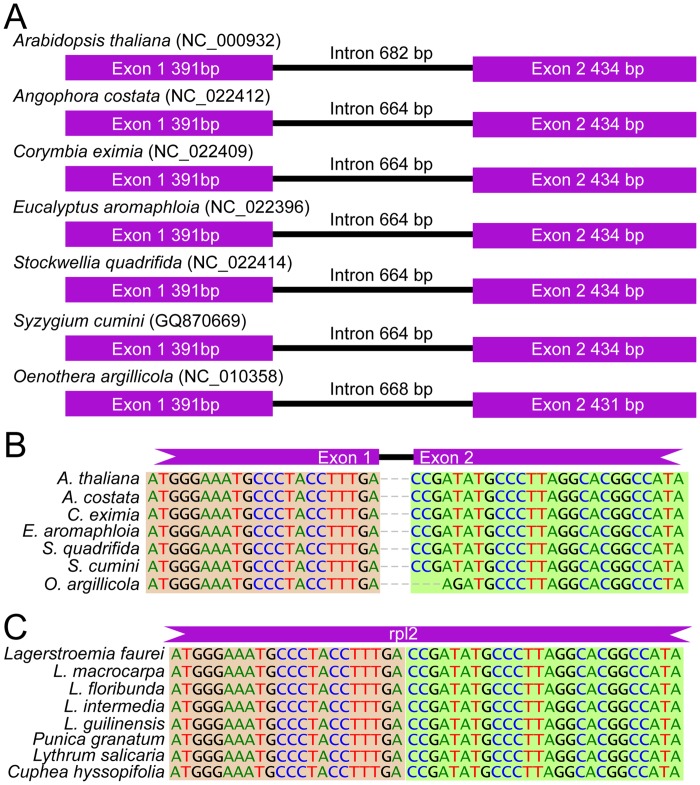
The distribution and number of introns in the L. fauriei plastid genome are similar to other Myrtales plastid genomes (S2 Table), with the exception of the intron of rpl2. The structure and the length of the intron for rpl2 is conserved across all other Myrtales and also present in the more distant Arabidopsis thaliana (NC_000932; Fig 3A). The length of this intron is approximately 660 bp in the other sampled six Myrtales species and the two exons are also highly conserved. To verify the loss of the rpl2 intron in the whole Lagerstroemia or even broadly within Lythraceae as a whole, we designed a pair of primers in the flanking exons to amplify and sequence the region spanning the intron among different species. From the rpl2 gene alignment, the intron was absent among all 14 Lythraceae species sampled (S2 and S3 Figs), but the intron was present in Oenothera albicaulus (Fig 3B; from the arrow of S2A Fig). From the PCR amplification test (S3 Fig), the rpl2 amplicon is about 750 bp in 14 samples species of Lythraceae, whereas in the amplicons from the outgroups O. albicalus and Catha edulis were about 1,400 bp (S3 Fig). These results indicate that the intron was lost after the divergence of the Lythraceae from the Onagraceae (S2B and S3 Figs) but prior to the divergence of the four Lythraceae genera sampled.
Fig 3. The structure and sequence variation for rpl2 gene with and without intron.
(A) The structural components of rpl2 gene from Arabidopsis thaliana (NC_000932) and the other six Myrtales species. (B) The boundary sequences of the two exons: the dashed lines represent the intron sequences; the sequence from the maple shade is from first exon and from the green shade is from the second intron. (C) Exons borders of rpl2 sequences from genus Lagerstroemia and other species from Lythraceae: the maple and green shades mean the sequence from exon 1 and 2.
Plastid introns have been lost numerous times in other species, such as those reported from the legume tribe Desmodieae [58,59], and have been documented in both monocots and dicots [60]. Specifically, rpl2 intron loss has been reported from five other lineages of dicotyledons: Saxifragaceae, Convolvulaceae, Menyanthaceae, two genera of Geraniaceae, and one genus of Droseraceae [59]. The discovery of this intron loss indicates a structural difference between Lythraceae and the six other Myrtales families sampled. And we could confirm that many times instances of independent intron loss have happened in the history of plastid genome evolution. Two different theories had been proposed to explain loss of the rpl2 intron [61,62]. First, through the homologous recombination, the full rpl2 transcript (cDNA) could replace the rpl2 gene by the reverse-transcriptase mediated mechanism to precisely delete the entire intron. Alternatively, rpl2 intron loss could be caused by unknown processes involving intron removal by DNA-level deletion or gene conversion between an intron-containing gene and its spliced transcript. In near future, by combining the density samplings within Lythraceae and Onagraceae, and by employing the data from RNA and DNA could answer this intron loss history around this family.
Long repetitive sequences
Long repetitive sequences have an important role in structural variation in plastid genomes via recombination and rearrangement [63]. Tandem repeats (≥15 bp), and forward and palindromic repeats (≥30 bp) were compared across the seven Myrtales species (S4B Fig). Most of these repeats are located in intergenic spacers, except for some that are distributed in the shared coding regions of ycf2 and psaB. L. faurei has the fewest (22) repeats, which is consistent with the small genome size of L. fauriei compared with the six other Myrtales species sampled (S4B Fig).
Repeated sequences have been demonstrated to affect genome length [64]. Our data are consistent with these findings given that the length and number of repeat in O. argillicola and L. fauriei (S4 Fig) are correlated with their genome size. Forward-repeat sequences are often associated with transposons [65], which can proliferate during episodes of cellular stress [66, 67]. The origins and proliferation of large tandem repeats are not as well understood as interspersed repetitive sequences [68]. Forward repeats can cause genomic reconfiguration, and therefore have potential to be useful markers in phylogenetic studies.
Plastid SSRs
Simple sequence repeats (SSRs) in the plastid genome can be highly variable at the intraspecific level, and therefore valuable markers for population-genetic studies [56]. We identified 204 SSRs in the plastid genome of L. fauriei, of which 132 are located in non-coding regions and 72 in coding regions. These SSRs include 115 mononucleotide SSRs (homopolymers; 56%), 35 dinucleotide SSRs (17%), 46 trinucleotide SSRs (23%), seven tetranucleotide (3%), and one pentanucleotide SSR (1%). Of the 204 SSRs, 143 are in the LSC region, 35 in SSC, and 26 in IRA region accounting for 70%, 17%, and 13% of the total SSRs, respectively. Among the 115 homopolymer SSRs, 113 (98%) are the A/T type with a repeat number from 8 to 14. Among the coding regions, ycf2 was found to possess 13 SSRs, followed by ycf1 with eight SSRs. This result is consistent with previous studies which found that these genes are highly variable in other species [67, 68, 69]. From this result ycf1and ycf2 are potential candidates for species-level DNA barcoding[70].
Among the seven Myrtales species sampled, L.faurei has the fewest SSRs (S4C Fig). The total length of SSRs in these species does not have a strong overall correlation to genome size. However L. fauriei has the shortest chloroplast genome and had the smallest contribution from SSRs. Thus, reduction in the size and presence of SSR’s may contribute somewhat to the short chloroplast genome of L. fauriei [71].
Highly informative regions and potential markers for phylogenetic analysis
Identifying highly variable gene regions provides an important resource for phylogenetic analyses and DNA barcoding [72]. Regions such as atpB, atpB-rbcL, matK, ndhF, rbcL, rpl16, rps4-trnS, rps16, trnH-psbA, trnL-F, and trnS-G have been extensively employed for phylogenetic reconstructions [73–75] and barcoding applications [76,77]. Using complete plastid genomes, we identified additional informative loci for use within the Myrtales, including Lagerstroemia.
We aligned all coding and non-coding regions ≥ 200 bp separately to identify regions with the highest percentage of parsimony-informative sites, and the highest ensemble retention index, among the seven Myrtales species sampled (Table 4, S3 Table). Among the coding regions, rpoA and matK have the highest percentage of parsimony-informative characters (7% and 6%, respectively). Among non-coding regions, trnRUCU-atpA and trnKUUU-rps16 have the highest percentages (20% and 14%, respectively). These non-coding regions should be particularly informative for DNA barcoding and species-level phylogenetic analyses within the Myrtales given the high percentage of variable sites (S3 Table). In order to better understand the variation from the longer genes (>1500 bp) and make them usable in practical applications, we employed the sliding-window method (S4 Table). By applying this method, we identified the most variable regions within each gene that would be valuable as molecular makers in phylogeny or for marker-assisted breeding analysis. For example, the most variable region of ycf1, which is over 7000 bp in length, is located from 5 to 6 kb downstream from the start.
Table 4. Top ten coding regions ordered with respect to their potential phylogenetic signal.
| No. | Region | Length (bp) a | Aligned length (bp) b | Conserved sites | Pars. Inf. c | Pars. Inf.% d | RIe |
|---|---|---|---|---|---|---|---|
| 1 | rpoA | 1002 | 1101 | 989 | 77 | 6.99 | 0.96 |
| 2 | matK | 1500 | 1593 | 1295 | 98 | 6.15 | 0.92 |
| 3 | rps15 | 273 | 297 | 253 | 15 | 5.05 | 0.86 |
| 4 | rpl22 | 471 | 486 | 396 | 24 | 4.94 | 0.82 |
| 5 | rpl32 | 174 | 306 | 271 | 15 | 4.90 | 0.77 |
| 6 | ccsA | 960 | 966 | 827 | 47 | 4.87 | 0.80 |
| 7 | ndhF | 2244 | 2349 | 1967 | 113 | 4.81 | 0.82 |
| 8 | ycf1 | 5613 | 7356 | 5102 | 350 | 4.76 | 0.70 |
| 9 | ndhG | 531 | 555 | 496 | 24 | 4.32 | 0.88 |
| 10 | petL | 96 | 96 | 91 | 4 | 4.17 | 0.75 |
a: Length: refers to sequence length in L.fauriei;
b: Aligned length: refers to the alignment of seven Myrtales species considered in the comparative analysis (see Materials and Methods);
c: Number of parsimony informative sites;
d: Percentage of parsimony informative sites;
e: RI-Ensemble retention index.
Shaw [25,78] evaluated the phylogenetic utility of noncoding plastid regions and found that those that are most commonly used for phylogenetic analyses (e.g., trnL intron, trnL-trnF spacer) are among the least variable. Thus, our identification of ten more variable noncoding regions provides a valuable resource for future phylogenetic studies within Myrtales, including our focal genus, Lagerstroemia.
Phylogenetic analysis
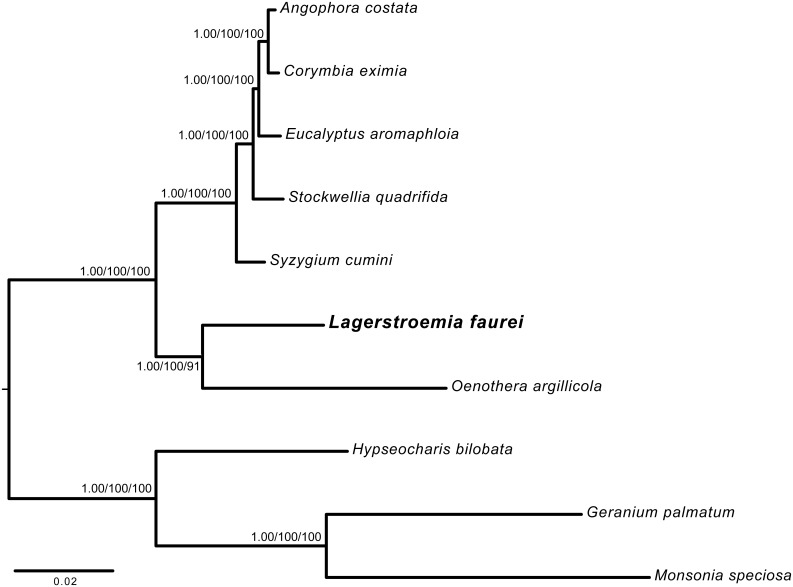
Phylogenetic analysis using plastid sequences have resolved numerous lineages within the angiosperms [79,80]. Furthermore, atpF-atpH, matK, psbK-psbI, rbcL and trnH-psbA have been used successfully as species-level barcodes [76,81,82]. Phylogenetic relationships within Lythraceae have been inferred using morphology and DNA sequences from the rbcL gene, the trnL-F region, and the psaA-ycf3 intergenic spacer from the plastid genome, together with ITS from the nuclear genome [1,17]. Our phylogenetic analyses included seven Myrtales species together with three outgroups from Geraniaceae. These analyses all corroborated the sister relationship between Lythraceae and Onagraceae based on 73 shared protein-coding genes (Fig 4). From the branch-length differences between the two main Myrtales clades, we infer that both Lythraceae and Onagraceae have undergone a more rapid rate of nucleotide substitution than their Myrtaceae sister group. This more rapid nucleotide-substitution rate was also accompanied by more structural differences in the Onagraceae and Lythraceae.
Fig 4. Phylogenetic tree inferred by Bayesian inference, maximum likelihood, and parsimony using 73 shared protein-coding genes among 10 plastid genomes (1 Lythraceae, 1 Onagraceae, 5 Myrtaceae, 3 Geraniaceae).
Numbers above nodes indicate posterior probability followed by bootstrap values.
Supporting Information
(TIF)
(A) The boundary sequences of two exons: the dash lines represents the elliptical intron sequences; the sequence from the maple shade is from first exon and from the green shade is from the second intron. The Sanger sequencing chromatograms with first exon and intron regions was from O. albicaulus. (B) The joints of two exons of rpl2 sequences from genus Lagerstroemia and other species from Lythraceae: the maple and green shades mean the sequence from exon 1 and 2. The Sanger sequencing chromatograms from five species from Lythraceae show the loss of intron.
(TIF)
MA = L. macrocarpa, FL = L. floribunda, INT = L. intermedia, GU = L. guilinensis, FA = L. fauriei, VE = L. venusa, CAU = L. caudata, LI = L. limii, SUB = L. subcostata, IND = L. indica, PA = L. parvifolia, PU = Punica granatum, LY = Lythrum salicaria, CU = Cuphea hyssopifolia, OEN = Oenothera albicaulus, CATHA = Catha edulis, C = negative control.
(TIF)
A. Plastid genome size comparison among seven Myrtales species (1 = Lagerstroemia fauriei, 2 = Oenothera argillicola, 3 = Angophora costata, 4 = Corymbia eximia, 5 = Eucalyptus aromaphloia, 6 = Stockwellia quadrifida, 7 = Syzygium cumini, with species listed according to their distance). B. All repeat sequences, tandem repeats (≥15 bp), and forward and palindromic repeats (≥30 bp) for each of seven Myrtales species. Bars indicate total length of each type of repeat. C. Total length contribution from SSRs for each of seven Myrtales species, separated by motif type.
(TIF)
(NEX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by The National Natural Science Fund (NO. 31300581), Open Fund of the most important disciplines of Forestry, Zhejiang Agriculture and Forestry University (KF201322), and Lagerstroemia indica Breeding and Industrialization Development (NO. 2012C12909–5) sponsored by Science Technology Department of Zhejiang Province.
References
- 1.Graham SA, Hall J, Sytsma K, Shi S (2005) Phylogenetic analysis of the Lythraceae based on four gene regions and morphology. Int J Plant Sci 166: 995–1017. [Google Scholar]
- 2.Dahlgren R, Thorne RF (1984) The Order Myrtales: Circumscription, variation, and relationships. Ann Missouri Bot Gard 71: 633–699. [Google Scholar]
- 3.Johnson LA., Briggs BG (1984) Myrtales and Myrtaceae -A phylogenetic analysis. Ann Missouri Bot Gard 71: 700–756. [Google Scholar]
- 4.Qin H, Graham SA (2007) Lagerstroemia Flora of China pp. 277–281. Available: http://www.bioone.org/doi/abs/10.3100/1043-4534-13.2.301
- 5.Koehne E (1883) Botanische Jahrbücher für Systematik. Pflanzengeschichte und Pflanzengeographie 4: 252–270. [Google Scholar]
- 6.Furtado CX, Montien Srisuko (1969) A revision of Lagerstroemia L. (Lythraceae). Gard Bull 24: 185–335. [Google Scholar]
- 7.Egolf DR, Andrick AO (1978) The Lagerstroemia Handbook/Checklist. American Association of Botanical Gardens and Arboreta.
- 8.Wang X, Wadl PA, Pounders C, Trigiano RN, Cabrera RI, Scheffler BE, et al. (2011) Evaluation of genetic diversity and pedigree within crapemyrtle cultivars using simple sequence repeat markers. J Amer Soc Hort Sci 136: 116–128. [Google Scholar]
- 9.Unno T, Sugimoto A, Kakuda T (2004) Xanthine oxidase inhibitors from the leaves of Lagerstroemia speciosa (L.) Pers. J Ethnopharmacol 93: 391–395. 10.1016/j.jep.2004.04.012 [DOI] [PubMed] [Google Scholar]
- 10.Ichikawa H, Yagi H, Tanaka T, Cyong JC, Masaki T (2010) Lagerstroemia speciosa extract inhibit TNF-induced activation of nuclear factor-kB in rat cardiomyocyte H9c2 cells. J Ethnopharmacol 128: 254–256. 10.1016/j.jep.2009.12.033 [DOI] [PubMed] [Google Scholar]
- 11.Cai M, Pan HT, Wang XF, He D, Wang XY, Wang XJ, et al. (2011) Development of novel microsatellites in Lagerstroemia indica and DNA fingerprinting in Chinese Lagerstroemia cultivars. Sci Hortic (Amsterdam). 131: 88–94. 10.1016/j.scienta.2011.09.031 [DOI] [Google Scholar]
- 12.Pan HT, He D, Liu Y, Cai M, Zhang QX, Wang XY, et al. (2012) Genetic diversity of Lagerstroemia (Lythraceae) species assessed by simple sequence repeat markers. Genet Mol Res 11: 3522–3533. 10.4238/2012.September.26.9 [DOI] [PubMed] [Google Scholar]
- 13.He D, Liu Y, Cai M, Pan H, Zhang Q (2014) The first genetic linkage map of crape myrtle (Lagerstroemia) based on amplification fragment length polymorphisms and simple sequence repeats markers. Plant Breed 133: 138–144. 10.1111/pbr.12100 [DOI] [Google Scholar]
- 14.Pooler MR (2003) Molecular genetic diversity among 12 clones of Lagerstroemia fauriei revealed by AFLP and RAPD markers. HortScience 38: 256–259. [Google Scholar]
- 15.Pounders C, Rinehart T, Sakhanokho H (2007) Evaluation of interspecific hybrids between Lagerstroemia indica and L. speciosa. HortScience 42: 1317–1322. [Google Scholar]
- 16.Conti E, Litt A, Wilson P, Graham S (1997) Interfamilial relationships in Myrtales: molecular phylogeny and patterns of morphological evolution. Syst Bot 22: 629–647. Available: http://www.jstor.org/stable/10.2307/2419432 [Google Scholar]
- 17.Huang YL, Shi SH (2002) Phylogenetics of Lythraceae sensu lato: a preliminary analysis based on chloroplast rbcL gene, psaA—ycf3 spacer, and nuclear rDNA internal transcribed spacer (ITS). Int J Plant Sci 163: 215–225. [Google Scholar]
- 18.Palmer JD (1985) Comparative organization of chloroplast genomes. Annu Rev Genet 19: 325–354. 10.1146/annurev.genet.19.1.325 [DOI] [PubMed] [Google Scholar]
- 19.Ravi V, Khurana JP, Tyagi AK, Khurana P (2008) An update on chloroplast genomes. Plant Syst Evol 271: 101–122. 10.1007/s00606-007-0608-0 [DOI] [Google Scholar]
- 20.Wicke S, Schneeweiss GM, DePamphilis CW, Müller KF, Quandt D (2011) The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol 76: 273–297. 10.1007/s11103-011-9762-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soltis DE, Gitzendanner M, Stull G, Chester M, Chanderbali A, Jordon-Thaden I, et al. (2013) The potential of genomics in plant systematics. Taxon 62: 886–898. 10.12705/625.13 [DOI] [Google Scholar]
- 22.Wu Z, Tembrock LR, Ge S (2015) Are differences in genomic data sets due to true biological variants or errors in genome assembly: an example from two chloroplast genomes. PLoS One 10: 1–14. 10.1371/journal.pone.0118019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.CBOL (2009) A DNA barcode for land plants. Proc Natl Acad Sci 106: 12794–12797. 10.1073/pnas.0905845106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Day A, Goldschmidt-Clermont M (2011) The chloroplast transformation toolbox: selectable markers and marker removal. Plant Biotechnol J 9: 540–553. 10.1111/j.1467-7652.2011.00604.x [DOI] [PubMed] [Google Scholar]
- 25.Shaw J, Lickey EB, Beck JT, Farmer SB, Liu W, Miller J, et al. (2005) The tortoise and the hare II: Relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am J Bot 92: 142–166. 10.3732/ajb.92.1.142 [DOI] [PubMed] [Google Scholar]
- 26.Wu ZQ, Ge S (2012) The phylogeny of the BEP clade in grasses revisited: Evidence from the whole-genome sequences of chloroplasts. Mol Phylogenet Evol 62: 573–578. 10.1016/j.ympev.2011.10.019 [DOI] [PubMed] [Google Scholar]
- 27.Cerutti H, Johnson AM, Boynton JE, Gillham NW (1995) Inhibition of chloroplast DNA recombination and repair by dominant negative mutants of Escherichia coli RecA. Mol Cell Biol 15: 3003–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Svab Z, Maliga P (1993) High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc Natl Acad Sci U S A 90: 913–917. 10.1073/pnas.90.3.913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pal M, Jeffrey S, Helaine C, Ivan Kanevski ZS (1994) Homologous recombination and integration of foreign DNA in plastids of higher plants. Paszkowski J, editor. Amsterdam: Kluwer Academic [Google Scholar]
- 30.Maliga P (2004) Plastid transformation in higher plants. Annu Rev Plant Biol 55: 289–313. 10.1146/annurev.arplant.55.031903.141633 [DOI] [PubMed] [Google Scholar]
- 31.Yang JB, Li DZ, Li HT (2014) Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol Ecol Resour 14: 1024–1031. 10.1111/1755-0998.12251 [DOI] [PubMed] [Google Scholar]
- 32.O’Brien SJ, Stanyon R (1999) Phylogenomics: ancestral primate viewed. Nature 402: 365–366. 10.1038/46450 [DOI] [PubMed] [Google Scholar]
- 33.Doyle JJ, Doyle J (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 11–15. [Google Scholar]
- 34.Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. (2009) BLAST+: architecture and applications. BMC Bioinformatics 10: 421 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Asif H, Khan A, Iqbal A, Khan IA, Heinze B, Azim MK (2013) The chloroplast genome sequence of Syzygium cumini (L.) and its relationship with other angiosperms. Tree Genet Genomes 9: 867–877. 10.1007/s11295-013-0604-1 [DOI] [Google Scholar]
- 37.Wyman SK, Jansen RK, Boore J (2004) Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20: 3252–3255. 10.1093/bioinformatics/bth352 [DOI] [PubMed] [Google Scholar]
- 38.Schattner P, Brooks AN, Lowe TM (2005) The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res 33: 686–689. 10.1093/nar/gki366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lohse M, Drechsel O, Bock R (2007) OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet 52: 267–274. 10.1007/s00294-007-0161-y [DOI] [PubMed] [Google Scholar]
- 40.Wang RJ, Cheng CL, Chang CC, Wu CL, Su TM, Chaw SM (2008) Dynamics and evolution of the inverted repeat-large single copy junctions in the chloroplast genomes of monocots. BMC Evol Biol 8: 36 10.1186/1471-2148-8-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jansen RK, Wojciechowski MF, Sanniyasi E, Lee SB, Daniell H (2008) Complete plastid genome sequence of the chickpea (Cicer arietinum) and the phylogenetic distribution of rps12 and clpP intron losses among legumes (Leguminosae). Mol Phylogenet Evol 48: 1204–1217. 10.1016/j.ympev.2008.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang H, Shi C, Liu Y, Mao SY, Gao LZ (2014) Thirteen Camellia chloroplast genome sequences determined by high-throughput sequencing: genome structure and phylogenetic relationships. BMC Evol Biol 14: 151 10.1186/1471-2148-14-151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benson G (1999) Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27: 573–580. 10.1093/nar/27.2.573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R (2001) REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res 29: 4633–4642. 10.1093/nar/29.22.4633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Q, Wan JM (2005) SSRHunter: development of a local searching software for SSR sites. Yi Chuan 27: 808–810. [PubMed] [Google Scholar]
- 46.Larkin MA, Blackshields G, Brown NP, Chenna R, Mcgettigan PA, Mcwilliam H, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 47.Simmons MP (2004) Independence of alignment and tree search. Mol Phylogenet Evol 31: 874–879. 10.1016/j.ympev.2003.10.008 [DOI] [PubMed] [Google Scholar]
- 48.Swofford DL (2002) Paup*: Phylogenetic analysis using parsimony (and other methods) pp. 1–142. 10.1007/BF02198856 [DOI] [Google Scholar]
- 49.Farris JS (1989) The retention index and the rescaled consistency index. Cladistics. 417–419. [DOI] [PubMed] [Google Scholar]
- 50.Guindon S, Dufayard JF, Lefort V, Anisimova M (2010) New alogrithms and methods to estimate maximum—likelihoods phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- 51.Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, et al. (2012) Mrbayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61: 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Green BR. Chloroplast genomes of photosynthetic eukaryotes. Plant J. 2011:66: 34–44. 10.1111/j.1365-313X.2011.04541.x [DOI] [PubMed] [Google Scholar]
- 53.Sytsma KJ, Hahn WJ, Smith JF, Wagner WL (1993) Characterisation and phylogenetic utility of a large inversion in the chloroplast genome of some species in Oenothera (Onagraceae). Am J Bot 80:79. [Google Scholar]
- 54.Greiner S, Wang X, Rauwolf U, Silber MV, Mayer K, Meurer J, et al. (2008) The complete nucleotide sequences of the five genetically distinct plastid genomes of Oenothera, subsection Oenothera: I. Sequence evaluation and plastome evolution. Nucleic Acids Res 36: 2366–2378. 10.1093/nar/gkn081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raubeson LA, Peery R, Chumley TW, Dziubek C (2007) Comparative chloroplast genomics: analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. BMC Genomics 8: 174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nguyen PA, Kim JS, Kim JH (2015) The complete chloroplast genome of colchicine plants (Colchicum autumnale L. and Gloriosa superba L.) and its application for identifying the genus. Planta 242: 223–37. 10.1007/s00425-015-2303-7 [DOI] [PubMed] [Google Scholar]
- 57.Bayly MJ, Rigault P, Spokevicius A, Ladiges PY, Ades PK, Anderson C, et al. (2013) Chloroplast genome analysis of Australian eucalypts—Eucalyptus, Corymbia, Angophora, Allosyncarpia and Stockwellia (Myrtaceae). Mol Phylogenet Evol 69: 704–716. 10.1016/j.ympev.2013.07.006 [DOI] [PubMed] [Google Scholar]
- 58.Bailey CD, Doyle JJ, Kajita T, Nemoto T, Bailey CD, Doyle JJ (1997) The chloroplast rpl2 intron and ORF184 as phylogenetic markers in the Legume Tribe Desmodieae. Syst Bot 22: 133–138. [Google Scholar]
- 59.Downie SR, Olmstead RG, Zurawski G, Soltis DE, Soltis S, Watson JC, et al. (1991) Six independent losses of the chloroplast DNA rpl2 intron in Dicotyledons : molecular and phylogenetic implications. Evolution 45: 1245–1259. [DOI] [PubMed] [Google Scholar]
- 60.Jansen RK, Raubeson LA, Boore JL, DePamphilis CW, Chumley TW, Haberle RC, et al. (2005) Methods for obtaining and analyzing whole chloroplast genome sequences. Methods Enzymol 395: 348–384. 10.1016/S0076-6879(05)95020-9 [DOI] [PubMed] [Google Scholar]
- 61.Fink GR (1987) Pseudogenes in yeast? Cell 49: 5–6. 10.1016/0092-8674(87)90746-X [DOI] [PubMed] [Google Scholar]
- 62.Dujon B (1989) Group I introns as mobile genetic elements: facts and mechanistic speculations—a review. Gene 82: 91–114. 10.1016/0378-1119(89)90034-6 [DOI] [PubMed] [Google Scholar]
- 63.Cavalier-Smith T (2002) Chloroplast evolution: secondary symbiogenesis and multiple losses. Curr Biol 12: 62–64. 10.1016/S0960-9822(01)00675-3 [DOI] [PubMed] [Google Scholar]
- 64.Rubinsztein DC, Amos W, Leggo J, Goodburn S, Jain S, Li SH, et al. (1995) Microsatellite evolution—evidence for directionality and variation in rate between species. Nat Genet 10: 337–343. 10.1038/ng0795-337 [DOI] [PubMed] [Google Scholar]
- 65.Gemayel R, Cho J, Boeynaems S, Verstrepen KJ (2012) Beyond junk-variable tandem repeats as facilitators of rapid evolution of regulatory and coding sequences. Genes (Basel) 3: 461–480. 10.3390/genes3030461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Voronova A, Belevich V, Jansons A, Rungis D (2014) Stress-induced transcriptional activation of retrotransposon-like sequences in the Scots pine (Pinus sylvestris L.) genome. Tree Genet Genomes 10: 937–951. 10.1007/s11295-014-0733-1 [DOI] [Google Scholar]
- 67.Grassi F, Labra M, Scienza A, Imazio S (2002) Chloroplast SSR markers to assess DNA diversity in wild and cultivated grapevines. Vitis 41: 157–158. [Google Scholar]
- 68.Timme RE, Kuehl JV, Boore JL, Jansen RK (2007) A comparative analysis of the Lactuca and Helianthus (Asteraceae) plastid genomes: Identification of divergent regions and categorization of shared repeats. Am J Bot 94: 302–312. 10.3732/ajb.94.3.302 [DOI] [PubMed] [Google Scholar]
- 69.Curci PL, De Paola D, Danzi D, Vendramin GG, Sonnante G (2015) Complete Chloroplast genome of the multifunctional crop globe artichoke and Comparison with Other Asteraceae. PLoS One 10: e0120589 10.1371/journal.pone.0120589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kumar S, Hahn FM, McMahan CM, Cornish K, Whalen MC (2009) Comparative analysis of the complete sequence of the plastid genome of Parthenium argentatum and identification of DNA barcodes to differentiate Parthenium species and lines. BMC Plant Biol 9: 131 10.1186/1471-2229-9-131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.George B, Bhatt BS, Awasthi M, George B, Singh AK (2015) Comparative analysis of microsatellites in chloroplast genomes of lower and higher plants. Curr Genet. 10.1007/s00294-015-0495-9 [DOI] [PubMed] [Google Scholar]
- 72.Dong W, Liu J, Yu J, Wang L, Zhou S (2012) Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS One 7: e35071 10.1371/journal.pone.0035071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim KJ, Jansen RK (1995) ndhF sequence evolution and the major clades in the sunflower family. Proc Natl Acad Sci U S A 92: 10379–10383. 10.1073/pnas.92.22.10379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hilu KW, Black C, Diouf D, Burleigh JG (2008) Phylogenetic signal in matK vs. trnK: a case study in early diverging eudicots (angiosperms). Mol Phylogenet Evol 48: 1120–1130. 10.1016/j.ympev.2008.05.021 [DOI] [PubMed] [Google Scholar]
- 75.Li J (2008) Phylogeny of Catalpa (Bignoniaceae) inferred from sequences of chloroplast ndhF and nuclear ribosomal DNA. J Syst Evol 46: 341–348. 10.3724/SP.J.1002.2008.08025 [DOI] [Google Scholar]
- 76.Kress WJ, Erickson DL (2007) A two-locus global DNA barcode for land plants: the coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS One 2 10.1371/journal.pone.0000508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li X, Yang Y, Henry RJ, Rossetto M, Wang Y, Chen S (2014) Plant DNA barcoding: from gene to genome. Biol Rev 90: 157–166. 10.1111/brv.12104 [DOI] [PubMed] [Google Scholar]
- 78.Shaw J, Lickey EB, Schilling EE, Small RL (2007) Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: The Tortoise and the hare III. Am J Bot 94: 275–288. 10.3732/ajb.94.3.275 [DOI] [PubMed] [Google Scholar]
- 79.Jansen RK, Cai Z, Raubeson LA, Daniell H, Depamphilis CW, Leebens-Mack J, et al. (2007) Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci U S A 104: 19369–19374. 10.1073/pnas.0709121104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Moore MJ, Bell CD, Soltis PS, Soltis DE (2007) Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. Proc Natl Acad Sci U S A 104: 19363–19368. 10.1073/pnas.0708072104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pennisi E (2007) Wanted: a barcode for plants. Science 318: 190–191. [DOI] [PubMed] [Google Scholar]
- 82.Li DZ, Gao LM, Li HT, Wang H, Ge XJ, Liu JQ, et al. (2011) Comparative comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc Natl Acad Sci U S A 108: 19641–19646. 10.1073/pnas.1104551108 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(A) The boundary sequences of two exons: the dash lines represents the elliptical intron sequences; the sequence from the maple shade is from first exon and from the green shade is from the second intron. The Sanger sequencing chromatograms with first exon and intron regions was from O. albicaulus. (B) The joints of two exons of rpl2 sequences from genus Lagerstroemia and other species from Lythraceae: the maple and green shades mean the sequence from exon 1 and 2. The Sanger sequencing chromatograms from five species from Lythraceae show the loss of intron.
(TIF)
MA = L. macrocarpa, FL = L. floribunda, INT = L. intermedia, GU = L. guilinensis, FA = L. fauriei, VE = L. venusa, CAU = L. caudata, LI = L. limii, SUB = L. subcostata, IND = L. indica, PA = L. parvifolia, PU = Punica granatum, LY = Lythrum salicaria, CU = Cuphea hyssopifolia, OEN = Oenothera albicaulus, CATHA = Catha edulis, C = negative control.
(TIF)
A. Plastid genome size comparison among seven Myrtales species (1 = Lagerstroemia fauriei, 2 = Oenothera argillicola, 3 = Angophora costata, 4 = Corymbia eximia, 5 = Eucalyptus aromaphloia, 6 = Stockwellia quadrifida, 7 = Syzygium cumini, with species listed according to their distance). B. All repeat sequences, tandem repeats (≥15 bp), and forward and palindromic repeats (≥30 bp) for each of seven Myrtales species. Bars indicate total length of each type of repeat. C. Total length contribution from SSRs for each of seven Myrtales species, separated by motif type.
(TIF)
(NEX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.