Abstract
The molecular effects of total body gamma-irradiation exposure are of critical importance as large populations of people could be exposed either by terrorists, nuclear blast, or medical therapy. In this study, we aimed to identify changes in the urine proteome using a non-human primate model system, Rhesus macaque, in order to characterize effects of acute radiation syndrome following whole body irradiation (Co-60) at 6.7 Gy and 7.4 Gy with a twelve day observation period. The urine proteome is potentially a valuable and non-invasive diagnostic for radiation exposure. Using high-resolution mass spectrometry, we identified 2346 proteins in the urine proteome. We show proteins involved in disease, cell adhesion, and metabolic pathway were significantly changed upon exposure to differing levels and durations of radiation exposure. Cell damage increased at a faster rate at 7.4 Gy compared with 6.7 Gy exposures. We report sets of proteins that are putative biomarkers of time- and dose-dependent radiation exposure. The proteomic study presented here is a comprehensive analysis of the urine proteome following radiation exposure.
Keywords: Rhesus monkey, Quantitative proteomic, Acute radiation syndrome, Urine proteome, Biomarkers
Introduction
The effect of total body gamma-irradiation exposure to large populations of people is of great importance. Whether populations are exposed due to nuclear blast, radiological terrorists’ activities, or medical therapy; the molecular effects on the body need to be identified in order to develop methods for early detection and new therapies for treatment of exposure. In the case of early detection of exposure, one would need a method to rapidly analyze a readily available biospecimen. Such readily available biospecimens include urine, which is the focus of this study. We rationalized that a proteomic analysis of urine following gamma-irradiation exposure could uncover potential biomarkers of exposure as well as provide insight into the in vivo effects of exposure.
Human urine is routinely used for medical diagnostics and is only second to that of plasma [1]. Due to the non-invasive nature of collection and large supply, urine is an attractive source for the study of human pathophysiology [2]. Not only is urine becoming a highly valuable resource for biomarker discovery, but studies are now moving from the discovery phase to validation of those findings in clinical trials for several diseases [3] including steroid resistant nephrotic syndrome [4], polycystic kidney disease [5], acute renal failure [6], prostate cancer [7], bladder cancer [8,9], type 1 diabetes [10], and several others. The urine proteome is derived from the ultrafiltration of plasma in the kidney to eliminate waste products such as proteins, urea and metabolites. Under normal physiologic conditions, the kidney generates a large amount of ultrafiltrate (150-180 L/day) [3,11] in which the majority of components are reabsorbed and only less than 1% is excreted in urine. In addition, serum proteins are filtered by size and charge by passing through the glomeruli and abundant proteins are reabsorbed. Therefore, the protein concentration in normal urine is low (less than 100 mg/L) which corresponds to about 1000-fold less protein concentration compared with other body fluids [3]. Regardless of the lower protein concentrations, urine has become a valuable diagnostic tool due to the fact it can be collected noninvasively in large supply. Kentsis et al. [12] have identified over 2300 proteins in routinely collected urine specimens using high-resolution mass spectrometry based methods. Annotation methods applied to the aforementioned proteome identified possible associations with 27 common and more than 500 rare human diseases giving further weight to the importance and widely useful resource of urine for the study of human pathophysiology.
In this research, we aimed to identify changes in the urine proteome using a non-human primate system, Rhesus macaque, in order to characterize effects of acute radiation syndrome following whole body irradiation (Co-60) with a twelve day observation period. We used high resolution mass spectrometry to compare changes in the proteome indicative of the effects of radiation exposure on Rhesus macaque pathophysiology. We report the most comprehensive proteomic analysis to date of urine from total body gamma-irradiation exposure using this non- human primate model. The urine samples used for this proteomic study are extremely rare and thus the data reported provide some of the first quantitative information on how the urine proteome changes in response to radiation exposure. The development of the Rhesus macaque proteomic model can then be used to analyze data in clinical conditions.
Method
Animals
The Rhesus monkey was selected for this study because the acute radiation syndrome is well characterized and is the most frequently used in radiation research under the US FDA Animal Rule. Procedures involving the care and use of animals in this study were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) prior to conduct. During the study, the care and use of animals were conducted in accordance with the principles outlined in the current Guidelines. The CRO conducting the study was and is accredited by AAALAC. This study was considered as a category of invasiveness D.
The study included a total of twenty-four animals, which were irradiated at Day 0 and monitored for up to twelve days post-radiation as detailed in Table 1. The animal numbers were selected based on planned statistical analyses, assuming that all responses analyzed will be approximately normally distributed. Animals necropsied on day 4 (two positive radiation doses and a sham control) were used to test for a radiation dose-response relationship. Four animals per dose group (2 males and 2 females) will give 70% power at a 5% significance level to detect a linear dose effect, assuming differences of 1 standard deviation in responses between adjacent dose groups. Animals in the two positive radiation dose groups from all three necropsy days were used to assess the time course and its consistency across doses. With 4 animals in each dose group on necropsy days 4 and 7, and 2 animals per group on necropsy day 12, there will be approximately 80% power to detect dose and time effects with a significance level of 5%, assuming differences of 1 standard deviation in responses between adjacent doses and between adjacent necropsy days. The power to detect a dose-by-time interaction will be higher [13].
Table 1.
Description of the non-human primates used in the study.
| Group | Radiation Gy | Necropsy Day 4 | Necropsy Day 7 | Necropsy Day 12 | |||
|---|---|---|---|---|---|---|---|
| Males | Females | Males | Females | Males | Females | ||
| 1 | Sham | 2 | 2 | - | - | - | - |
| 2 | 6.7 (LD70/60) | 2 | 2 | 2 | 2 | 1 | 1 |
| 3 | 7.4 (LD90/60) | 2 | 2 | 2 | 2 | 1 | 1 |
Whole-body irradiation
Historical data from irradiated and control animals including information about individual animals (age, sex, etc.) as well as day of euthanasia were used to determine the most appropriate radiation doses to use. Based on this information, we selected radiation levels of 6.7 Gy and 7.4 Gy to cause significant GI-injury while insuring survival of some animals out to 12 days post-irradiation. Study days 4 and 7 represent the window of major GI-injury while Day 12 is a later endpoint to investigate partial resolution of injury.
Animals were exposed to a single uniform total body dose of gamma radiation from a Co60 source (Theratron1000) at a dose rate of approximately 60 cGy/min for 12 minutes. In order to produce homogenous dose distribution, treatment was divided in two parts. First, the animals received half of the dose by antero-posterior (AP) irradiation and the second half of the dose was delivered by postero-anterior (PA) irradiation. The radiation dose was calibrated using a solid water phantom placed in the same experimental set up used for animal irradiation.
Two dosimeters (Landauer, Inc. Model scanned nanoDot) were placed on each animal during whole body irradiation to quantify the dose. The dosimeters were placed on the mid- plane approximately at the level of the xiphoid process and at the corresponding level in the dorsal area (below the interscapular area). Dosimeters were placed under a gel bolus build-up of approximately 5 mm (superflab) and secured with bandaging.
Urine collection and MS/MS processing
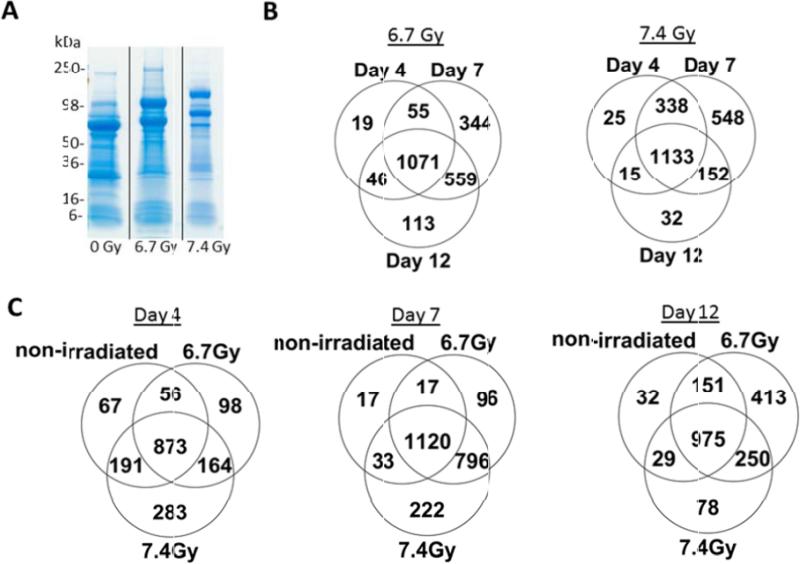
The twenty-four Rhesus macaque urine samples were analyzed by high-resolution mass spectrometric analysis. Non-human primates were gamma-irradiated with 0, 6.7Gy and 7.4Gy and urine was collected at days 4, 7, and 12 post-irradiation (Table 1). The bladder was exposed and urine collected (maximum volume available) with a syringe, via cystocentesis. The urine was aliquoted and stored at −70°C until ready for processing. 500 μL of each urine sample was concentrated with an Amicon Ultra 0.5 mL 3K Ultracel Centrifugal Filtration Unit (Millipore catalog UFC500396) at 14,000 g-force at 4°C until a desired final concentrated volume of 20 μL was reached. The final volume of concentrated urine sample was resolved by 4-20% Tris- Glycine SDS-PAGE (Life Technologies) and urine proteins were visualized by Coomassie- staining (Figure 1A). Each gel lane was sliced into 20 equivalent bands.
Figure 1.
Quantitative proteomic analysis of urine from non-human primates following gamma-irradiation reveals changes in the urine proteome. (A) Urine proteins from non-human primates (days 4, 7 and 12 post-exposure with 0, 6.7 or 7.4 Gy gamma-irradiation) were resolved by SDS-PAGE and visualized by Coomassie-staining. The gel image shows representative samples from non-human primates at 0Gy, 6.7 Gy and 7.4 Gy on day 4 post-irradiation. B) Venn diagram showing the number of total identified proteins that overlap between day 4, 7, and 12 post-irradiation following 6.7 Gy and 7.4 Gy exposures. C) Venn diagram showing total identified proteins from each time point (day 4, 7, 12) for 0 Gy, 6.7 Gy, and 7.4 Gy exposures.
Gel bands were destained (50% methanol, 100 mM ammonium bicarbonate), followed by treatment with 10 mM Tris[2-carboxyethyl] phosphine to reduce protein disulfide bonds and 50 mM iodoacetamide to block reformation of cysteine disulfide bonds. Gel slices were then dehydrated in acetonitrile, followed by addition of 100 ng porcine sequencing grade modified trypsin in 100 mM ammonium bicarbonate and incubated at 37°C for 12-16 hours. Peptide products were then acidified in 0.1% formic acid. Tryptic peptides were separated by reverse phase Jupiter Proteo resin (Phenomenex) on a 100 × 0.075 mm column using a nanoAcquity UPLC system (Waters). Peptides were eluted using a 40 min gradient from 97:3 to 35:65 buffer A:B ratio. [Buffer A=0.1% formic acid, 0.5% acetonitrile; buffer B=0.1% formic acid, 75% acetonitrile]. Eluted peptides were ionized by electrospray (1.9 kV) followed by MS/MS analysis using collision induced dissociation on an LTQ Orbitrap Velos mass spectrometer (Thermo Scientific). MS data were acquired using the FTMS analyzer in profile mode at a resolution of 60,000 over a range of 375 to 1500 m/z. MS/MS data were acquired for the top 15 peaks from each MS scan using the ion trap analyzer in centroid mode and normal mass range with normalized collision energy of 35.0 [14,15].
Data analysis
Proteins were identified by searching the UniProtKB database (2015_06 release; restricted to Rhesus macaque; 69,970 entries) using Andromeda search engine in MaxQuant (version 1.5.3.8). Search parameters were specified as follows: trypsin digestion with up to three missed cleavages; fixed carbamidomethyl modification of cysteine; variable modification of oxidation on methionine and acetyl on N-terminus; first search 4 ppm precursor ion tolerance and the main search 2 ppm; the number of max modifications of 5; selected label-free quantitation of LFQ with a minimum ratio of 2, minimum number of neighbors of 3 and average number of neighbors of 6; protein minimum ratio count of 2, using unique and razor peptides. A contaminants file was used for the first search. Peptide and protein identifications were validated using Scaffold Q+S (v4.4; Proteome Software). Peptide identifications were accepted at >50.0% probability as determined by the Scaffold Local FDR algorithm. Protein identifications were accepted at >95.0% probability and a minimum of two identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm [16]. Proteins with similar peptides that could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Proteins sharing significant peptide evidence were grouped into clusters.
Quantitative testing was performed in Scaffold Q+S to detect differential presence of a protein between different samples (time dependent: day 0, 4, 7, and 12 for 6.7Gy and 7.4Gy radiation doses; dose dependent: non-irradiated, 6.7 Gy, and 7.4 Gy from days 4, 7, and 12). An intensity-based normalization scheme using a weighted median of the parent ion or MS1 ion intensity values for all the spectra that identify a specific peptide was employed to calculate fold change values as well as statistical testing by Kruskal-Wallis with bonferroni correction. Scaffold not only calculates the fold change ratio values but also calculates a sample wide fold change by subtracting the log2 (secondary sample) minus the log2 (primary sample). The sample wide fold change shows the difference in the log2 fold change for a selected protein in two quantitative samples relative to the differences in the log2 fold change for all other proteins in the same samples. Additionally, the Kruskal-Wallis test was performed for both time and dose dependent comparisons to determine if the evidence that supports the presence or absence of differential abundance is truly conclusive or happens by chance.
Data visualization
The data was visualized using Hierarchical Clustering Explorer (HCE 3.0, [17]) and Search Tool for the Retrieval of Interacting Genes/Proteins (STRING, [18]). Proteins that were significantly differentiated with p-value < 0.05 in a time and dose dependent manner were uploaded into HCE to visually inspect similarities between proteins and samples. The hierarchical clusters were built using the average linkage method with Euclidean distance measure, the distance metric for clustering proteins, in HCE. STRING was employed to investigate protein-protein interactions using the confidence view in order to visualize how the significantly differentiating proteins interact with one another. In addition, STRING identified the enriched KEGG pathways associated with each network of proteins. A KEGG pathway is a collection of manually drawn pathway maps representing our knowledge on the molecular interaction and reaction networks for metabolism, genetic information processing, environmental information processing, cellular processes, organismal systems, human diseases, and drug development.
Results and Discussion
Here, we provide a comprehensive look into the urine proteome from Rhesus macaque and identify changes in the molecular networks and pathways in response to total body gamma- irradiation. Urine was collected from 24 non-human primates under 7 different conditions (0 Gy collected only on day 4 post-irradiation; 6.7 Gy and 7.4 Gy collected on days 4, 7, and 12 post- irradiation) (Table 1). The proteins from the urine samples were resolved by SDS-PAGE and visualized by Coomassie-staining (Figure 1A, Supplemental Figure 1). The gel lanes were sliced into 2 mm bands and analyzed by tandem mass spectrometry using a Thermo LTQ Orbitrap Velos mass spectrometer coupled to a Waters nano-Acquity LC.
Mass spectrometric data were analyzed using an intensity-based approach with MaxQuant and Scaffold Q+S. A total of 2346 proteins (332,607 spectra) were identified at a 95% protein probability threshold, a minimum number of peptides of 2, and 50% peptide probability threshold. Venn diagrams in Figures 1B and 1C show the total number of proteins identified in a time and dose dependent manner, respectively. There are several shared and unique proteins identified among the three time points under two radiation exposures, as well as between the levels of radiation exposures at each time point. When analyzing the proteins in a time-dependent manner, the number of unique proteins filtered in the kidney increases 62% from the lower radiation dose of 6.7 Gy (344) to that of 7.4 Gy (548) by day 7. By day 12, the number of unique proteins is reduced to 113 at 6.7 Gy and 32 at 7.4 Gy. This indicates with the higher radiation exposure damage is occurring faster in the non-human primates and more proteins are being filtered through the blood into the kidneys. The peak in unique proteins identified following 6.7 Gy exposure occurs on day 7 and extends into day 12, while the peak following 7.4 Gy exposure occurs at day 7 (Figure 1B). This suggests day 7 post-exposure could be an optimal time to assay for biomarkers of radiation response at levels of 6.7 Gy and 7.4 Gy exposures, while biomarkers could also be detected in the lower dose even at day 12. A similar trend is found in the dose-dependent analysis (Figure 1C). The number of unique proteins in the lower radiation dose (6.7 Gy) is maximized by day 12 (413), whereas the majority of proteins filtered from the blood at 7.4 Gy are found at days 4 (283) and 7 (222).
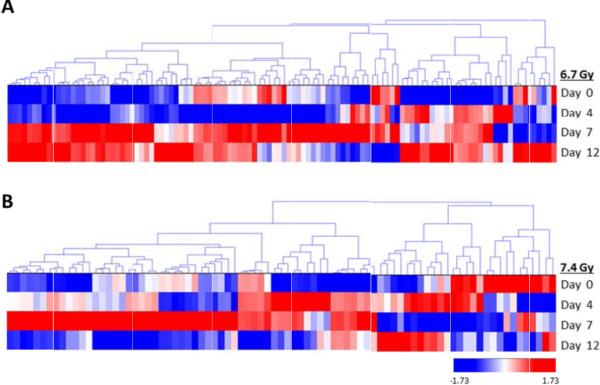
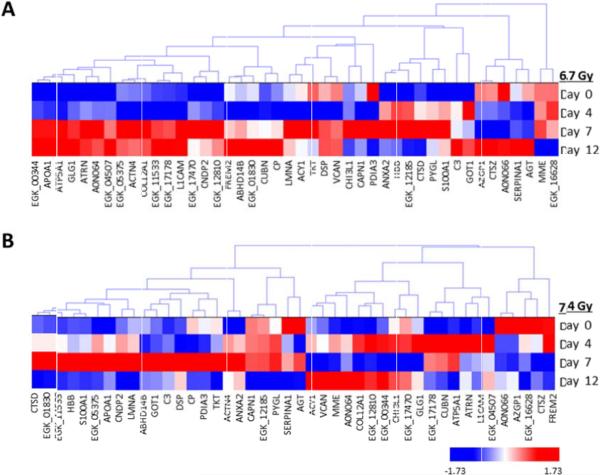
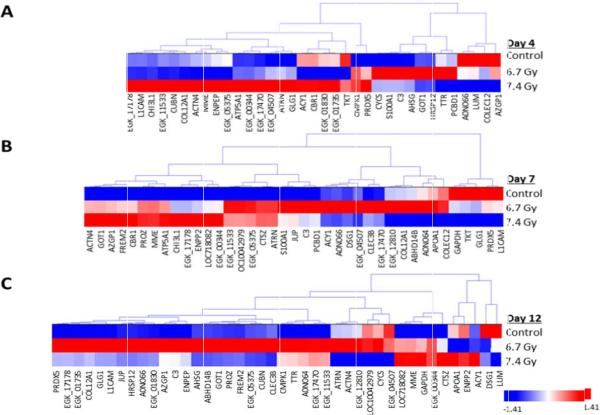
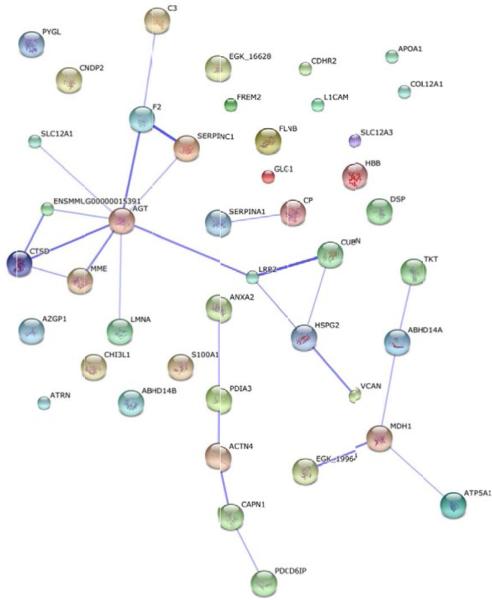
Non-irradiated samples on day 4 were utilized as the 0 Gy baseline control samples and were compared with both 6.7 Gy and 7.4 Gy gamma-irradiated specimens for three post- irradiation time points (days 4, 7, and 12). Non-irradiated Rhesus macaque urine collected on day 4 was used as the control specimen for data analysis. We also compared 6.7 Gy with 7.4 Gy radiation exposures at each time point. Proteins with a p-value<0.05 by the Kruskal-Wallis test were considered to be significant. The log2 normalized intensity values for each significant protein was used to generate a hierarchical cluster to visualize the monkey specimens and the protein expression in a time- and dose-dependent manner (Figures 2 and 3, Figures 5 and 6; Supplemental Figures 2 and 3, Supplemental Tables 1 to 5). There are clear differences in protein intensity values in each comparison. The data in Figure 2 shows a large change in the abundance of proteins at day 7 and extending into day 12 for 6.7 Gy, which is similar to the trend observed in Figure 1B. For 7.4 Gy exposures, the large change in the abundance of proteins occurs at days 4 and 7, which is also consistent with the data in Figure 1B. Thus, the increased level of radiation exposure more rapidly increases the amount of proteins filtered from the blood stream. Proteins that were identified as significant between time points in both the 6.7 Gy and 7.4 Gy radiation induced monkeys were re-clustered in order to identify putative biomarkers of radiation exposure that significantly change in urine abundance over time (Figure 3, Table 2). The proteins showing an increased level (i.e., the more red color in Figure 3) in urine over time are the time-dependent putative biomarkers of exposure. Significantly differentiated proteins from Figure 3 were visualized by STRING using the confidence view (Search Tool for the Retrieval of Interacting Genes/Proteins, version 10, http://string-db.org) in order to easily identify protein-protein associations (Figure 4). STRING provides a critical assessment and integration of protein-protein interactions, including direct or physical as well as indirect or functional associations [19,20]. In addition, STRING also links the proteins to KEGG pathways. KEGG pathways with p-value of <0.05 are listed in Table 3 for the time-dependent significant proteins, respectively. KEGG analysis revealed changes in several disease, cell adhesion, and metabolic pathways. It is interesting that over time several disease pathways and cell adhesion molecules pathways are significantly changed possibly leading to less cell to cell contact and potentially indicating cell damage following exposure.
Figure 2.
Hierarchical cluster of proteins significantly differentiated in a time dependent manner. A) Proteins significantly differentiated at differing time points at 6.7 Gy exposures. B) Proteins significantly differentiated at differing time points at 7.4 Gy exposures. Proteins were clustered using the average linkage method and Euclidean distance metric. Proteins were considered significant at a p-value<0.05.
Figure 3.
Hierarchical cluster of proteins identified as significantly differentiated at both levels of radiation exposure in a time dependent manner. A) Proteins significantly differentiated at differing time points at 6.7 Gy exposures. B) Proteins significantly differentiated at differing time points at 7.4 Gy exposures. Proteins were clustered using the average linkage method and Euclidean distance metric. Proteins were considered significant at a p-value<0.05.
Figure 5.
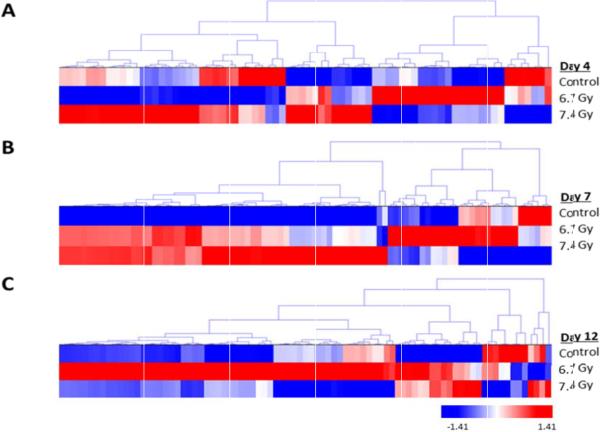
Hierarchical cluster of proteins significantly differentiated in a dose-dependent manner. Proteins significantly differentiated at differing levels of radiation exposure for days A) 4, B) 7, and C) 12. Proteins were clustered using the average linkage method and Euclidean distance metric. Proteins were considered significant at a p-value<0.05.
Figure 6.
Hierarchial cluster of proteins identified as significantly differentiated in a dose-dependent manner at each time point. Shown are the proteins identified at all three time points: A) day 4, B) day 7, and C) day 12. Proteins were clustered using the average linkage method and Euclidean distance metric. Proteins were considered significant at a p-value<0.05.
Table 2.
Time-dependent significant proteins.
| Identified Proteins | Accession Number | Gene symbol |
|---|---|---|
| Complement C3 | H9EXI6 | C3 |
| Putative uncharacterized protein | G7N891 | EGK_04507 |
| Putative uncharacterized protein | G7ML00 | EGK_11533 |
| Thiazide-sensitive sodium-chloride co-transporter | G7NPT3 | EGK_12810 |
| Uncharacterized protein (Fragment) | F7GN59 | CUBN |
| Uncharacterized protein | F7HN19 | FREM2 |
| Annexin | F7H0C7 | ANXA2 |
| Putative uncharacterized protein | G7MX91 | EGK_17470 |
| Alpha-1-antitrypsin | F6SEN6 | SERPINA1 |
| Transketolase | I0FJ64 | TKT |
| Putative uncharacterized protein (Fragment) | G7MHF2 | EGK_00344 |
| Putative uncharacterized protein | G7MU29 | EGK_17178 |
| Chitinase-3-like protein 1 | F7E1U6 | CHI3L1 |
| Putative uncharacterized protein (Fragment) | G7MF07 | EGK_01830 |
| Prelamin-A/C isoform 1 | F7GLE9 | LMNA |
| Apolipoprotein A-I | APOA1 | APOA1 |
| Malate dehydrogenase | G7NA62 | EGK_05375 |
| Alpha-actinin-4 | F7HU82 | ACTN4 |
| Neural cell adhesion molecule L1 isoform 1 | H9FUA5 | L1CAM |
| Uncharacterized protein | F7GFB4 | CP |
| Cathepsin D preproprotein | F7H7Y3 | CTSD |
| Uncharacterized protein | F6SIX4 | COL12A1 |
| Aminoacylase-1 | H9EWL7 | ACY1 |
| Uncharacterized protein | F6YWS9 | AZGP1 |
| Antithrombin III | A0N066 | A0N066 |
| Abhydrolase domain-containing protein 14B | F7CU68 | ABHD14B |
| Alpha-1,4 glucan phosphorylase | F7FLT9 | PYGL |
| Neprilysin | F7H3Y6 | MME |
| Golgi apparatus protein 1 isoform 1 (Fragment) | H9F9K8 | GLG1 |
| Uncharacterized protein | F6X826 | ATRN |
| Protein disulfide-isomerase | F6W5I4 | PDIA3 |
| Uncharacterized protein | F6WDG6 | DSP |
| Uncharacterized protein | F6TLR3 | AGT |
| Putative uncharacterized protein | G7MK64 | EGK_12185 |
| Calpain-1 catalytic subunit | H9G0C4 | CAPN1 |
| Aspartate aminotransferase | H9EVG6 | GOT1 |
| Hemoglobin beta chain | F7AV81 | HBB |
| Prothrombin | A0N064 | A0N064 |
| Cathepsin Z | G7N4B3 | CTSZ |
| Protein S100 | F7HGE0 | S100A1 |
| Uncharacterized protein | F7C5T5 | VCAN |
| Betaine-homocysteine S-methyltransferase 1 | G7MUU6 | EGK_16628 |
| ATP synthase subunit alpha | F7ETD0 | ATP5A1 |
| Cytosolic non-specific dipeptidase isoform 1 | F7EW08 | CNDP2 |
Figure 4.
STRING protein network analysis of proteins identified as significantly differentiated at both levels of radiation exposure in a time dependent manner. Proteins were considered significant at a p-value<0.05. The thicker the connecting lines in between proteins indicates a stronger the protein-protein association.
Table 3.
KEGG pathway for time-dependent significantly differentiating proteins.
| GO ID | Term | Number of Genes | p-value |
|---|---|---|---|
| 4610 | Complement of coagulation cascades | 3 | 0.0246 |
| 5412 | Arrhythmogenic right ventricular cardiomyopathy (ARVC) | 3 | 2.83E-04 |
| 4614 | Renin-angiotensin system | 2 | 5.02E-04 |
| 4514 | Cell adhesion molecules (CAMs) | 3 | 2.31E-03 |
| 270 | Cysteine and methionine metabolism | 2 | 2.53E-03 |
| 5010 | Alzheimer's disease | 3 | 5.62E-03 |
| 4974 | Protein digestion and absorption | 2 | 1.05E-02 |
| 4141 | Protein processing in endoplasmic reticulum | 2 | 3.67E-02 |
| 4810 | Regulation of actin cytoskeleton | 2 | 5.41E-02 |
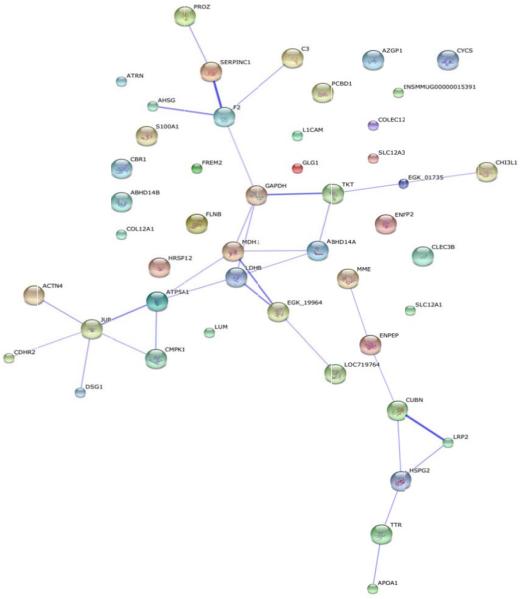
The same trend is shown in the dose-dependent analysis (Figures 5 and 6) relative to the time-dependent analysis (Figures 2 and 3). At day 4 (Figure 5), there is a small cluster of proteins that are elevated at 6.7 Gy compared to a larger cluster of proteins at 7.4 Gy radiation exposure. By day 7, there are more elevated proteins at 7.4 Gy than at the 6.7 Gy. By day 12, the majority of the elevated proteins are now found in non-human primates exposed to 6.7 Gy. Those proteins significantly changing as a function of dose in Figure 5 were re-clustered in Figure 6 (listed in Table 4). The proteins showing an increased level (i.e., the more red color in Figure 6) in urine as a function of dose are the dose-dependent putative biomarkers of exposure. As was done for the time-dependent analysis in Figure 3, the proteins showing a significant dose-response at all three time points were analyzed by STRING (Figure 7) and categorized by KEGG pathway (Table 5). More metabolic changes are indicated to be significant in the proteins significantly differentiated in a dose- relative to time-dependent manner (Tables 3 and 5). This is to be expected since the urine proteome contains filtered proteins from the blood circulation [1,21,22]. The proteins that change due to longer exposure of gamma-irradiation or differing levels of radiation are interesting targets for future study.
Table 4.
Dose-dependent significant proteins identified at all three time points.
| Identified Proteins | Accession Number | Gene Symbol |
|---|---|---|
| Putative uncharacterized protein | G7ML00 | EGK_11533 |
| Thiazide-sensitive sodium-chloride cotransporter | G7NPT3 | EGK_12810 |
| Complement C3 | H9EXI6 | C3 |
| Alpha-actinin-4 | F7HU82 | ACTN4 |
| Chitinase-3-like protein 1 | F7E1U6 | CHI3L1 |
| Putative uncharacterized protein | G7N891 | EGK_04507 |
| Putative uncharacterized protein | G7MX91 | EGK_17470 |
| Putative uncharacterized protein (Fragment) | G7MHF2 | EGK_00344 |
| Antithrombin III | A0N066 | A0N066 |
| Uncharacterized protein | F7HN19 | FREM2 |
| Neprilysin | F7H3Y6 | MME |
| Peroxiredoxin-5, mitochondrial isoform a | H9Z7D9 | PRDX5 |
| Malate dehydrogenase | G7NA62 | EGK_05375 |
| Uncharacterized protein | F6YWS9 | AZGP1 |
| Uncharacterized protein (Fragment) | F7D1C5 | CBR1 |
| Uncharacterized protein | F6SIX4 | COL12A1 |
| Golgi apparatus protein 1 isoform 1 (Fragment) | H9F9K8 | GLG1 |
| Aminoacylase-1 | H9EWL7 | ACY1 |
| Uncharacterized protein | F6SAZ0 | PROZ |
| Uncharacterized protein | F6X826 | ATRN |
| ATP synthase subunit alpha | F7ETD0 | ATP5A1 |
| Uncharacterized protein (Fragment) | F6TSU2 | LOC100429793 |
| Desmoplakin-3 | F7H7V0 | JUP |
| Aspartate aminotransferase | H9EVG6 | GOT1 |
| Prothrombin | A0N064 | A0N064 |
| Tetranectin | F6T0W5 | CLEC3B |
| Putative uncharacterized protein | G7MU29 | EGK_17178 |
| Uncharacterized protein (Fragment) | F6RUR9 | COLEC12 |
| Neural cell adhesion molecule L1 isoform 1 | H9FUA5 | L1CAM |
| Apolipoprotein A-I | APOA1 | APOA1 |
| L-lactate dehydrogenase | F7HKA5 | LOC718082 |
| Ectonucleotide pyrophosphatase/phosphodiesterase family member 2 isoform 2 preproprotein | F7CJD1 | ENPP2 |
| Transketolase | I0FJ64 | TKT |
| Uncharacterized protein | F7GSK8 | DSG1 |
| Abhydrolase domain-containing protein 14B | F7CU68 | ABHD14B |
| Protein S100 | F7HGE0 | S100A1 |
Figure 7.
STRING protein network analysis of proteins identified as significantly differentiated at all three time points in a dose-dependent manner. Proteins were considered significant at a p-value<0.05. The thicker the connecting lines in between proteins indicates a stronger the protein-protein association.
Table 5.
KEGG pathway for dose-dependent significantly differentiating proteins.
| GO ID | Term | Number of Genes | p-value |
|---|---|---|---|
| 4614 | Renin-angiotensin system | 2 | 6.00E-04 |
| 5010 | Alzheimer's disease | 4 | 6.48E-04 |
| 270 | Cysteine and methionine metabolism | 2 | 3.02E-03 |
| 620 | Pryruvate metabolism | 2 | 4.77E-03 |
| 1120 | Microbial metabolism in diverse environments | 3 | 4.99E-03 |
| 4610 | Complement of coagulation cascades | 2 | 7.85E-03 |
| 5412 | Arrhythmogenic right ventricular cardiomyopathy (ARVC) | 2 | 8.60E-03 |
| 5205 | Proteoglycans in cancer | 3 | 9.68E-03 |
| 1100 | Metabolic pathways | 7 | 1.01E-02 |
| 10 | Glycolysis/Gluconeogenesis | 2 | 1.07E-02 |
| 4974 | Protein digestion and absorption | 2 | 1.25E-02 |
| 1200 | Carbon metabolism | 2 | 2.29E-02 |
| 4514 | Cell adhesion molecules (CAMs) | 2 | 3.36E-02 |
| 5161 | Hepatitis B | 2 | 3.78E-02 |
| 5012 | Parkinson's disease | 2 | 4.84E-02 |
Consistent with a previous analysis of radiation-induced proteins in human breast cancer MDA-MB-231 cells [23], we also identified significant changes in fibronection I (FN1), Cathepsin D preproprotein (CTSD), and peroxiredoxin 5 (PRDX5) in varying concentrations of radiation exposure and at differing time points. Previous reports have shown ionizing radiation leads to an increase in FN1 expression and poor prognosis [23-26]. Interaction analysis indicates potential interactions of FN1 and CTSD with cluster of differentiation 44 (CD44), integrin alpha 4 (ITGA4), and integrin alpha 5 (ITGA5), which are involved in cell-cell adhesion and immune response [23]. In our data, we show the highest expression of FN1 at day 7 post-irradiation We also found CTSD over expression in both 6.7Gy and 7.4Gy radiation treatments at day 7. It is possible that the change in FN1 expression from day 4 to day 7 also increases CTSD expression leading to poor prognosis. FN1 is associated with radiation resistance and cell motility and in various human cancers. CTSD has been shown to be overexpressed and plays a role in apoptosis, invasion, migration, and angiogenesis [27-29]. Further studies will be needed to investigate this possible correlation.
Taken together our data suggests that cell damage occurs faster with higher levels of radiation. Cells then lose their cell to cell contact and cell membranes are disrupted allowing the intracellular proteins to be released into the blood stream, filtered in the kidney and excreted into the urine. Thus the urine proteome is altered due to the effects of total body gamma- irradiation and is a valuable and non-invasive resource for diagnostics. The proteins showing elevated levels in Figures 3 and 6 are the candidate urine biomarkers of radiation exposure that will require extensive follow-up studies beyond the scope of this initial, discovery-phase proteomic survey. Eventually these proteins could be assayed to measure both the dose and duration of whole-body gamma radiation exposure in humans.
Supplementary Material
Acknowledgement
This project has been partially funded with Federal funds from the Office of the ASPR, BARDA under contract # HHSO100201100045C. The ideas presented are the authors and not those of the Government. Additional funding for this work has come from NIH grants R01GM106024, R33CA173264, R21ES025268, UL1TR000039, P20GM103625, P20GM103429, and P20GM109005.
References
- 1.Decramer S, Gonzalez de Peredo A, Breuil B, Mischak H, Monsarrat B, et al. Urine in clinical proteomics. Mol Cell Proteomics. 2008;7:1850–1862. doi: 10.1074/mcp.R800001-MCP200. [DOI] [PubMed] [Google Scholar]
- 2.Nagaraj N, Mann M. Quantitative Analysis of the Intra- and Inter-Individual Variability of the Normal Urinary Proteome. J Proteome Res. 2011;10:637–645. doi: 10.1021/pr100835s. [DOI] [PubMed] [Google Scholar]
- 3.Adachi J, Kumar C, Zhang Y, Olsen JV, Mann M. The human urinary proteome contains more than 1500 proteins, including a large proportion of membrane proteins. Genome Biol. 2006;7:R80. doi: 10.1186/gb-2006-7-9-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Traum AZ. Urine proteomic profiling to identify biomarkers of steroid resistance in pediatric nephrotic syndrome. Expert Rev Proteomics. 2008;5:715–719. doi: 10.1586/14789450.5.5.715. [DOI] [PubMed] [Google Scholar]
- 5.Hogan MC, Manganelli L, Woollard JR, Masyuk AI, Masyuk TV, et al. Characterization of PKD protein-positive exosome-like vesicles. J Am Soc Nephrol. 2009;20:278–288. doi: 10.1681/ASN.2008060564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ho J, Lucy M, Krokhin O, Hayglass K, Pascoe E, et al. Mass Spectrometry–Based Proteomic Analysis of Urine in Acute Kidney Injury Following Cardiopulmonary Bypass: A Nested Case-Control Study. Am J Kidney Dis. 2009;53:584–595. doi: 10.1053/j.ajkd.2008.10.037. [DOI] [PubMed] [Google Scholar]
- 7.Rehman I, Azzouzi AR, Catto JW, Allen S, Cross SS, et al. Proteomic analysis of voided urine after prostatic massage from patients with prostate cancer: a pilot study. Urology. 2004;64:1238–1243. doi: 10.1016/j.urology.2004.06.063. [DOI] [PubMed] [Google Scholar]
- 8.Celis JE, Wolf H, Ostergaard M. Bladder squamous cell carcinoma biomarkers derived from proteomics. Electrophoresis. 2000;21:2115–2121. doi: 10.1002/1522-2683(20000601)21:11<2115::AID-ELPS2115>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 9.Rasmussen HH, Orntoft TF, Wolf H, Celis JE. Towards a comprehensive database of proteins from the urine of patients with bladder cancer. J Urol. 1996;155:2113–2119. [PubMed] [Google Scholar]
- 10.Meier JJ, Bhushan A, Butler AE, Rizza RA, Butler PC. Sustained beta cell apoptosis in patients with long-standing type 1 diabetes: indirect evidence for islet regeneration? Diabetologia. 2005;48:2221–2228. doi: 10.1007/s00125-005-1949-2. [DOI] [PubMed] [Google Scholar]
- 11.Brunzel NA. Fundamentals of urine and body fluid analysis. 3rd Edn Vol. 201. Elsevier; St. Louis: 2013. [Google Scholar]
- 12.Kentsis A, Monigatti F, Dorff K, Campagne F, Bachur R, et al. Urine proteomics for profiling of human disease using high accuracy mass spectrometry. Proteomics Clin Appl. 2009;3:1052–1061. doi: 10.1002/prca.200900008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Landes RD, Lensing SY, Kodell RL, Hauer-Jensen M. Practical Advice on Calculating Confidence Intervals for Radioprotection Effects and Reducing Animal Numbers in Radiation Countermeasure Experiments. Radiat Res. 2013;180:567–574. doi: 10.1667/RR13429.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Byrum S, Avaritt NL, Mackintosh SG, Munkberg JM, Badgwell BD, et al. A quantitative proteomic analysis of FFPE melanoma. J Cutan Pathol. 2011;38:933–936. doi: 10.1111/j.1600-0560.2011.01761.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Byrum SD, Taverna SD, Tackett AJ. Quantitative analysis of histone exchange for transcriptionally active chromatin. J Clin Bioinforma. 2011;1:17. doi: 10.1186/2043-9113-1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 17.Hierarchical Clustering Explorer.
- 18.STRING.
- 19.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, et al. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma M, Halligan BD, Wakim BT, Savin VJ, Cohen EP, et al. The urine proteome for radiation biodosimetry: effect of total body vs. local kidney irradiation. Health Phys. 2010;98:186–195. doi: 10.1097/HP.0b013e3181b17cbd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mann M. Functional and quantitative proteomics using SILAC. Nat Rev Mol Cell Biol. 2006;7:952–958. doi: 10.1038/nrm2067. [DOI] [PubMed] [Google Scholar]
- 23.Kim MH, Jung SY, Ahn J, Hwang SG, Woo HJ, et al. Quantitative proteomic analysis of single or fractionated radiation-induced proteins in human breast cancer MDA-MB-231 cells. Cell Biosci. 2015;5:2. doi: 10.1186/2045-3701-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bae YK, Kim A, Kim MK, Choi JE, Kang SH, et al. Fibronectin expression in carcinoma cells correlates with tumor aggressiveness and poor clinical outcome in patients with invasive breast cancer. Hum Pathol. 2013;44:2028–2037. doi: 10.1016/j.humpath.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 25.Jerhammar F, Ceder R, Garvin S, Grénman R, Grafström RC, et al. Fibronectin 1 is a potential biomarker for radioresistance in head and neck squamous cell carcinoma. Cancer Biol Ther. 2010;10:1244–1251. doi: 10.4161/cbt.10.12.13432. [DOI] [PubMed] [Google Scholar]
- 26.Park S, Ahn JY, Lim MJ, Kim MH, Yun YS, et al. Sustained expression of NADPH oxidase 4 by p38 MAPK-Akt signaling potentiates radiation-induced differentiation of lung fibroblasts. J Mol Med (Berl) 2010;88:807–816. doi: 10.1007/s00109-010-0622-5. [DOI] [PubMed] [Google Scholar]
- 27.Wolf M, Clark-Lewis I, Buri C, Langen H, Lis M, et al. Cathepsin D specifically cleaves the chemokines macrophage inflammatory protein-1 alpha, macrophage inflammatory protein-1 beta, and SLC that are expressed in human breast cancer. Am J Pathol. 2003;162:1183–1190. doi: 10.1016/s0002-9440(10)63914-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benes P, Vetvicka V, Fusek M. Cathepsin D-many functions of one aspartic protease. Crit Rev Oncol Hematol. 2008;68:12–28. doi: 10.1016/j.critrevonc.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masson O, Bach AS, Derocq D, Prébois C, Laurent-Matha V, et al. Pathophysiological functions of cathepsin D: Targeting its catalytic activity versus its protein binding activity? Biochimie. 2010;92:1635–1643. doi: 10.1016/j.biochi.2010.05.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.