Abstract
Type 2 diabetes (T2D) is a heterogeneous complex disease affecting more than 29 million Americans alone with a rising prevalence trending toward steady increases in the coming decades. Thus, there is a pressing clinical need to improve early prevention and clinical management of T2D and its complications. Clinicians have understood that patients who carry the T2D diagnosis have a variety of phenotypes and susceptibilities to diabetes-related complications. We used a precision medicine approach to characterize the complexity of T2D patient populations based on high-dimensional electronic medical records (EMRs) and genotype data from 11,210 individuals. We successfully identified three distinct subgroups of T2D from topology-based patient-patient networks. Subtype 1 was characterized by T2D complications diabetic nephropathy and diabetic retinopathy; subtype 2 was enriched for cancer malignancy and cardiovascular diseases; and subtype 3 was associated most strongly with cardiovascular diseases, neurological diseases, allergies, and HIV infections. We performed a genetic association analysis of the emergent T2D subtypes to identify subtype-specific genetic markers and identified 1279, 1227, and 1338 single-nucleotide polymorphisms (SNPs) that mapped to 425, 322, and 437 unique genes specific to subtypes 1, 2, and 3, respectively. By assessing the human disease–SNP association for each subtype, the enriched phenotypes and biological functions at the gene level for each subtype matched with the disease comorbidities and clinical differences that we identified through EMRs. Our approach demonstrates the utility of applying the precision medicine paradigm in T2D and the promise of extending the approach to the study of other complex, multi-factorial diseases.
INTRODUCTION
Type 2 diabetes (T2D) is a complex, multifactorial disease that has emerged as an increasing prevalent worldwide health concern associated with high economic and physiological burdens. An estimated 29.1 million Americans (9.3% of the population) were estimated to have some form of diabetes in 2012—up 13% from 2010—with T2D representing up to 95% of all diagnosed cases (1, 2). Risk factors for T2D include obesity, family history of diabetes, physical inactivity, ethnicity, and advanced age (1, 2). Diabetes and its complications now rank among the leading causes of death in the United States (2). In fact, diabetes is the leading cause of nontraumatic foot amputation, adult blindness, and need for kidney dialysis, and multiplies risk for myocardial infarction, peripheral artery disease, and cerebrovascular disease (3–6). The total estimated direct medical cost attributable to diabetes in the United States in 2012 was $176 billion, with an estimated $76 billion attributable to hospital inpatient care alone. There is a great need to improve understanding of T2D and its complex factors to facilitate prevention, early detection, and improvements in clinical management.
A more precise characterization of T2D patient populations can enhance our understanding of T2D pathophysiology (7, 8). Current clinical definitions classify diabetes into three major subtypes: type 1 diabetes (T1D), T2D, and maturity-onset diabetes of the young. Other subtypes based on phenotype bridge the gap between T1D and T2D, for example, latent autoimmune diabetes in adults (LADA) (7) and ketosis-prone T2D. The current categories indicate that the traditional definition of diabetes, especially T2D, might comprise additional subtypes with distinct clinical characteristics. A recent analysis of the longitudinal Whitehall II cohort study demonstrated improved assessment of cardiovascular risks when subgrouping T2D patients according to glucose concentration criteria (9). Genetic association studies reveal that the genetic architecture of T2D is profoundly complex (10–12). Identified T2D-associated risk variants exhibit allelic heterogeneity and directional differentiation among populations (13, 14). The apparent clinical and genetic complexity and heterogeneity of T2D patient populations suggest that there are opportunities to refine the current, predominantly symptom-based, definition of T2D into additional subtypes (7).
Because etiological and pathophysiological differences exist among T2D patients, we hypothesize that a data-driven analysis of a clinical population could identify new T2D subtypes and factors. Here, we develop a data-driven, topology-based approach to (i) map the complexity of patient populations using clinical data from electronic medical records (EMRs) and (ii) identify new, emergent T2D patient subgroups with subtype-specific clinical and genetic characteristics. We apply this approach to a data set comprising matched EMRs and genotype data from more than 11,000 individuals. Topological analysis of these data revealed three distinct T2D subtypes that exhibited distinct patterns of clinical characteristics and disease comorbidities. Further, we identified genetic markers associated with each T2D subtype and performed gene- and pathway-level analysis of subtype genetic associations. Biological and phenotypic features enriched in the genetic analysis corroborated clinical disparities observed among subgroups. Our findings suggest that data-driven, topologic alanalysis of patient cohorts has utility in precision medicine efforts to refine our understanding of T2D toward improving patient care.
RESULTS
T2D-specific patient network
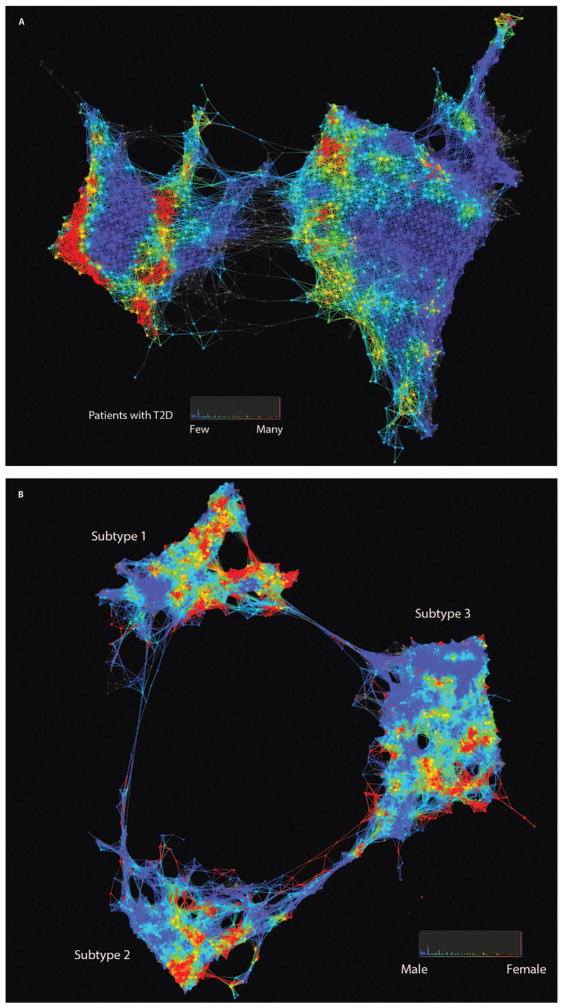
We developed and applied an unsupervised, topology-based approach that uses EMR-derived clinical data to infer a patient-patient similarity network as the computational model to represent a complex patient population. In the resulting patient-patient network, patients (nodes) are connected to one another by edges if they exhibit clinical similarity across many clinical dimensions (for example, laboratory tests). Patients who exhibited very high degrees of similarity were grouped into single nodes (see Materials and Methods). We identified two distinct clusters in the resulting patient-patient network (Fig. 1A) that contained 3889 and 7321 unique patients (the left and right clusters, respectively). The left cluster (n = 3889) was significantly enriched [least absolute shrinkage and selection operator (LASSO), P < 0.05] for endocrine and metabolic diseases, immunity disorders, infectious disease, mental illness, diseases of the circulatory and genitourinary systems, and symptoms/signs/ill-defined conditions and factors that influence health status. The right cluster (n = 7321) was significantly enriched for complications of pregnancy, respiratory diseases, and unclassified E code (external causes of injury) (15). Next, we identified T2D patients in the network to evaluate the heterogeneity of T2D patient groups across the patient-patient topology. We used a previously validated EMRs and genomics (eMERGE) network electronic phenotyping algorithm (16, 17) to define the T2D phenotype (n = 2551) and evaluated the network for topological enrichment of T2D patients. The red areas in Fig. 1A indicate that T2D patients are enriched in that particular location in the network, where the color scheme reflects the P value from hypergeometric enrichment analysis of topological enrichment (see Materials and Methods). We observed multiple distinct clusters or subnetworks of T2D patient enrichment.
Fig. 1. Patient and genotype networks.
(A) Patient-patient network for topology patterns on 11,210 Biobank patients. Each node represents a single or a group of patients with the significant similarity based on their clinical features. Edge connected with nodes indicates the nodes have shared patients. Red color represents the enrichment for patients with T2D diagnosis, and blue color represents the non-enrichment for patients with T2D diagnosis. (B) Patient-patient network for topology patterns on 2551 T2D patients. Each node represents a single or a group of patients with the significant similarity based on their clinical features. Edge connected with nodes indicates the nodes have shared patients. Red color represents the enrichment for patients with females, and blue color represents the enrichment for males.
We then rebuilt the patient-patient network, using the same topology analysis pipeline, with only the 2551 T2D patients identified with the T2D electronic phenotyping algorithm. The filtering step resulted in 73 clinical features that were used for topological inference of the patient-patient similarity network (table S1). From the resulting patient-patient network, we identified three completely segregated clusters with 762 (subtype 1), 617 (subtype 2), and 1096 (subtype 3) patients, respectively (Fig. 1B). We evaluated the network for enrichment of gender and did not observe any elevated enrichment of male or female patients in any of the clusters, suggesting that gender is not an organizing factor in the topology.
To assess the reproducibility of the T2D subtypes identified from the patient-patient network, we examined the performance on random samplings of training and test sets. First, we randomly split the 2551 T2D patients into two groups, with two-thirds as a training set and one-third as a test set. We then rebuilt the patient-patient network using the same 73 clinical features, distance metrics, and filter functions from the topology analysis pipeline. These steps were repeated 10 times. Last, we calculated the average of the precision [positive predictive value (PPV)] and recall (sensitivity) for the 10 tests, for training and test sets individually. The average precisions were 100, 91, and 98%, and the average recalls were 99, 96, and 94% for subtype 1, subtype 2, and subtype 3, respectively, in the training sets. In the test sets, the average precisions were 100, 90, and 97%, and the average recalls were 99, 96, and 93% for subtype 1, subtype 2, and subtype 3, respectively. The overall accuracy was 96% for both the training sets and test sets.
Significant characteristics and clinical features specific to T2D subtypes
We identified 33 clinical variables significantly specific to subtype 1 (n = 761) compared to both of the two other subtypes individually or combined. Three of these variables overlapped with clinical variables that were also specific to subtype 3, resulting in 29 variables unique to subtype 1. In addition, we identified 3 and 11 clinical variables significantly specific to subtype 2 (n = 617) and subtype 3 (n = 1096), respectively, with one shared variable. The only variable the three subtypes had in common was insulin administration (Table 1, A to C).
Table 1. Clinical variables specific to subtypes.
S-1, subtype 1; S-2, subtype 2; S-3, subtype 3; BMI, body mass index.
| (A) Clinical variables significantly specific to T2D subtype 1
| |||||||
|---|---|---|---|---|---|---|---|
| Clinical variables | Mean or % subtype 1 | Mean or % subtype 2 | Mean or % subtype 3 | P (1 versus 2 + 3) | S-1 | S-2 | S-3 |
| Platelet count (109/liter) | 98.36 ± 17.86 | 228.24 ± 2.90 | 228.61 ± 2.45 | <0.0001 | Y | ||
| Urine protein concentration (mg/dl) | 51.19 ± 14.38 | 152.67 ± 37.21 | 219.98 ± 47.62 | 0.0001 | Y | ||
| Lactate dehydrogenase (U/liter) | 193.35 ± 8.88 | 231.03 ± 8.82 | 251.34 ± 8.17 | <0.0001 | Y | ||
| Age (years) | 59.76 ± 0.45 | 64.25 ± 0.50 | 63.65 ± 0.38 | <0.0001 | Y | ||
| Blood urea nitrogen (mg/dl) | 16.69 ± 0.35 | 19.38 ± 0.59 | 19.52 ± 0.35 | <0.0001 | Y | ||
| Neutrophil count (109/liter) | 2.50 ± 0.58 | 4.78 ± 0.12 | 4.83 ± 0.09 | 0.0024 | Y | ||
| White blood cell count (109/liter) | 5.32 ± 0.57 | 7.28 ± 0.09 | 7.46 ± 0.07 | 0.001 | Y | ||
| Respirations | 16.65 ± 0.16 | 17.50 ± 0.14 | 17.62 ± 0.08 | <0.0001 | Y | ||
| Urine protein-to-creatinine ratio | 0.40 ± 0.09 | 1.19 ± 0.26 | 2.48 ± 0.45 | <0.0001 | Y | Y | |
| Serum creatinine (mg/dl) | 1.00 ± 0.02 | 1.25 ± 0.07 | 1.27 ± 0.04 | <0.0001 | Y | ||
| Eosinophil count (109/liter) | 0.09 ± 0.02 | 0.19 ± 0.01 | 0.20 ± 0.01 | 0.0003 | Y | ||
| Blood protein total (g/dl) | 7.49 ± 0.03 | 7.34 ± 0.04 | 7.14 ± 0.03 | <0.0001 | Y | Y | |
| Serum albumin (g/dl) | 4.27 ± 0.02 | 4.03 ± 0.03 | 4.04 ± 0.02 | <0.0001 | Y | ||
| Serum calcium (mg/dl) | 9.90 ± 0.02 | 9.66 ± 0.03 | 9.60 ± 0.02 | <0.0001 | Y | ||
| CO2 total | 26.60 ± 0.13 | 26.05 ± 0.15 | 26.16 ± 0.09 | 0.0011 | Y | ||
| Mean platelet volume (fl) | 9.97 ± 0.37 | 8.98 ± 0.05 | 8.97 ± 0.04 | 0.008 | Y | ||
| Prothrombin time* (s) | 29.18 ± 3.64 | 14.10 ± 0.33 | 14.13 ± 0.27 | 0.0005 | Y | ||
| INR* | 2.57 ± 0.34 | 1.19 ± 0.03 | 9.32 ± 0.41 | 0.0005 | Y | ||
| BMI | 33.07 ± 0.29 | 31.32 ± 0.30 | 31.19 ± 0.02 | <0.0001 | Y | ||
| Estimated GFR calculation (MDRD, ml/min/1.73 m2) | 74.86 ± 1.47 | 68.40 ± 1.99 | 65.04 ± 1.33 | <0.0001 | Y | ||
| GFR estimate (ml/min/1.73 m2) | 72.26 ± 1.47 | 64.62 ± 1.77 | 63.75 ± 1.22 | <0.0001 | Y | ||
| Glucose* (mg/dl) | 193.69 ± 11.45 | 149.55 ± 4.18 | 158.69 ± 2.90 | 0.0005 | Y | ||
| Insulin | 21.92% | 29.82% | 45.16% | <0.0001 | Y | Y | Y |
| Metformin | 6.43% | 23.01% | 21.17% | <0.0001 | Y | ||
| Loop diuretics | 5.51% | 14.10% | 18.34% | <0.0001 | Y | ||
| DPP4 | 1.05% | 6.48% | 6.39% | <0.0001 | Y | ||
| CCBs | 19.55% | 30.63% | 35.31% | <0.0001 | Y | ||
| β-Blocker | 21.92% | 39.06% | 45.80% | <0.0001 | Y | Y | |
| ARB/ACEI | 48.16% | 57.05% | 62.96% | <0.0001 | Y | Y | |
| Vasodilators | 0.92% | 5.02% | 5.57% | <0.0001 | Y | ||
| Nicotinic acid derivatives | 0.13% | 1.30% | 1.37% | 0.02 | Y | ||
| (B) Clinical variables significantly specific to T2D subtype 2
| |||||||
|---|---|---|---|---|---|---|---|
| Clinical variable | Mean or % subtype 1 | Mean or % subtype 2 | Mean or % subtype 3 | P (2 versus 1 + 3) | S-1 | S-2 | S-3 |
| Weight (kg) | 92.26 ± 1.08 | 85.17 ± 1.14 | 89.16 ± 0.83 | <0.0001 | Y | ||
| Troponin I level (ng/ml) | 0 | 0.03 ± 0.01 | 0.36 ± 0.09 | 0.0003 | Y | Y | |
| Insulin | 21.92% | 29.82% | 45.16% | <0.0001 | Y | Y | Y |
| (C) Clinical variables significantly specific to T2D subtype 3
| |||||||
|---|---|---|---|---|---|---|---|
| Clinical variable | Mean or % subtype 1 | Mean or % subtype 2 | Mean or % subtype 3 | P (3 versus 1 + 2) | S-1 | S-2 | S-3 |
| Blood protein total (g/dl) | 7.49 ± 0.03 | 7.34 ± 0.04 | 7.14 ± 0.03 | 0 | Y | Y | |
| Urine protein-to-creatinine ratio | 0.40 ± 0.09 | 1.19 ± 0.26 | 2.48 ± 0.45 | 0.0006 | Y | Y | |
| Troponin I level (ng/ml) | 0 | 0.03 ± 0.01 | 0.36 ± 0.09 | 0.0003 | Y | Y | |
| Systolic blood pressure (mmHg) | 132.04 ± 0.73 | 132.41 ± 0.92 | 135.7 ± 0.7 | 0.0001 | Y | ||
| Serum chloride level (mEq/liter) | 101.01 ± 0.17 | 101.45 ± 0.18 | 102.03 ± 0.11 | 0 | Y | ||
| HMG-CoA reductase inhibitors (statins) | 42.26% | 45.71% | 56.39% | <0.0001 | Y | ||
| Centrally acting antihypertensives | 1.44% | 1.30% | 4.11% | 0.0001 | Y | ||
| ARB/ACEI | 48.16% | 57.05% | 62.96% | <0.0001 | Y | Y | |
| β-Blocker | 21.92% | 39.06% | 45.80% | <0.0001 | Y | Y | |
| Insulin | 21.92% | 29.82% | 45.16% | <0.0001 | Y | Y | Y |
Point of care.
Patients in subtype 1 were the youngest (59.76 ± 0.45 years) and were notable for features classically associated with T2D, such as the highest BMI (33.07 ± 0.29 kg/m2) and highest serum glucose concentrations at point-of-care testing (POCT) (193.69 ± 11.45 mM). Patients in subtype 1 had the lowest complete blood count, including the lowest white blood cell counts (5.32 ± 0.57 × 109/liter), neutrophil counts (2.50 ± 0.58 × 109/liter), eosinophil counts (0.09 ± 0.02 × 109/liter), and mean platelet volumes (9.97 ± 0.37 fl). In addition, patients in subtype 1 had a considerably lower platelet count, with more than 50% of patients below the reference range (98.36 ± 17.86 × 109/liter). Adding to this curious hematological finding was a prolonged pro-thrombin time at POCT (29.18 ± 3.64 s), which corresponded to an elevated international normalized ratio (INR) (2.57 ± 0.34). Patients in subtype 1 also displayed the highest serum albumin (4.27 ± 0.02 g/dl) and lowest creatinine (1.0 ± 0.02 mg/dl) levels. Although these patients had better kidney function compared to those in the other two subtypes, estimated glomerular filtration rate (GFR) was below the reference range (72.26 ± 1.47 ml/min/1.73 m2; range, 17.3 to 149.7). In addition, patients in subtype 1 had the highest total blood CO2 (26.6 ± 0.13 mmHg) and fewer respirations per minute (16.65 ± 0.16), and lower prescription rates for calcium channel blockers (CCB; 19.55%), angiotensin II receptor blockers and angiotensin-converting enzyme inhibitors (ARB/ACEI, 48.16%) (commonly prescribed for hypertension), dipeptidyl peptidase 4 inhibitor (DPP4, 1.05%), and metformin (MET, 6.43%) (the last two are both prescribed for T2D).
Patients in subtype 2 had the lowest weight (85.17 ± 1.14 kg) compared with those in the other subtypes. Patients in subtype 3 had the highest systolic blood pressure (135.7 ± 0.7 mmHg), serum chloride levels (102.03 ± 0.11 mEq/liter), and troponin I levels (0.36 ± 0.09 μg/liter) and were more often prescribed ARB/ACEI (62.96%) for the treatment of hypertension and statins (56%) for cholesterol reduction. A full list of variables that were significantly specific to each subtype is provided in Table 1(A to C).
Disease comorbidity associated withT2D subtypes
We applied the disease Clinical Classifications Software (CCS; see Materials and Methods) (18) on more than 7000 ICD-9-CM (International Classification of Diseases, Ninth Revision, Clinical Modification) diagnosis codes in our cohort to aggregate the large number of ICD-9-CM codes into a manageable number of either 281 single-level disease categories or 18 level 1 (broader) categories in the multilevel disease categories. By adjusting patient age, gender, and self-reported race, we found that the patients in subtype 1 (n = 762) were more likely to associate with the following ICD-9-CM codes: diseases in the “other upper respiratory infections” [relative risk (RR), mean, 1.68; range, 1.34 to 2.11]; immunization and screening for infectious disease (RR, 1.65; range, 1.32 to 2.06); diabetes mellitus with complications (RR, 1.50; range, 1.22 to 1.84); other skin disorders (RR, 1.41; range, 1.13 to 1.76); and blindness and vision defects (RR, 1.32; range, 1.04 to 1.67), than were the other two subtypes (Table 2A). Patients in subtype 2 (n = 617) were more likely to associate with diseases of cancer of bronchus: lung (RR, 3.76; range, 1.14 to 12.39); malignant neoplasm without specification of site (RR, 3.46; range, 1.23 to 9.70); tuberculosis (RR, 2.93; range, 1.30 to 6.64); coronary atherosclerosis and other heart disease (RR, 1.28; range, 1.01 to 1.61); and other circulatory disease (RR, 1.27; range, 1.02 to 1.58), than were the other two subtypes (Table 2B). Patients in subtype 3 (n = 1096) were more often diagnosed with HIV infection (RR, 1.92; range, 1.30 to 2.85) and were associated with E codes (that is, external causes of injury care) (RR, 1.84; range, 1.41 to 2.39); aortic and peripheral arterial embolism or thrombosis (RR, 1.79; range, 1.18 to 2.71); hypertension with complications and secondary hypertension (RR, 1.66; range, 1.29 to 2.15); coronary atherosclerosis and other heart disease (RR, 1.41; range, 1.15 to 1.72); allergic reactions (RR, 1.42; range, 1.19 to 1.70); deficiency and other anemia (RR, 1.39; range, 1.14 to 1.68); and screening and history of mental health and substance abuse code (RR, 1.30; range, 1.07 to 1.58) (Table 2C).
Table 2.
Significant associated disease categories.MHSA, mental health and substance abuse; LCI, lower confidence interval; UCI, upper confidence interval.
| (A) Significant disease categories associated with T2D subtype 1
| ||||
|---|---|---|---|---|
| Disease category | RR | 95% LCI | 95% UCI | P value |
| Other upper respiratory infections | 1.68 | 1.34 | 2.11 | <0.0001 |
| Immunizations and screening for infectious disease | 1.65 | 1.32 | 2.06 | <0.0001 |
| Diabetes mellitus with complications | 1.50 | 1.22 | 1.84 | 0.0001 |
| Other skin disorders | 1.41 | 1.13 | 1.76 | 0.003 |
| E codes: place of occurrence | 1.38 | 1.08 | 1.77 | 0.01 |
| Blindness and vision defects | 1.32 | 1.04 | 1.67 | 0.02 |
| Other screening for suspected conditions (not mental disorders or infectious diseases) | 1.28 | 1.04 | 1.58 | 0.02 |
| Screening and history of MHSA codes | 0.74 | 0.59 | 0.94 | 0.01 |
| Other circulatory disease | 0.68 | 0.54 | 0.87 | 0.002 |
| Acute and unspecified renal failure | 0.63 | 0.42 | 0.94 | 0.02 |
| Pulmonary heart disease | 0.60 | 0.37 | 0.98 | 0.04 |
| Deficiency and other anemia | 0.57 | 0.45 | 0.71 | <0.0001 |
| E codes: adverse effects of medical care | 0.55 | 0.38 | 0.79 | 0.001 |
| Coronary atherosclerosis and other heart disease | 0.51 | 0.40 | 0.64 | <.0001 |
| Peri-, endo-, and myocarditis; cardiomyopathy (without tuberculosis or sexually transmitted disease) | 0.48 | 0.28 | 0.82 | 0.01 |
| Aortic, peripheral, and visceral artery aneurysms | 0.36 | 0.21 | 0.64 | 0.0004 |
| HIV infection | 0.22 | 0.12 | 0.38 | <0.0001 |
| (B) Significant disease categories associated with T2D subtype 2
| ||||
|---|---|---|---|---|
| Disease category | RR | 95% LCI | 95% UCI | P value |
| Cancer of bronchus: lung | 3.76 | 1.14 | 12.39 | 0.03 |
| Malignant neoplasm without specification of site | 3.46 | 1.23 | 9.70 | 0.02 |
| Tuberculosis | 2.93 | 1.30 | 6.64 | 0.01 |
| Coronary atherosclerosis and other heart disease | 1.28 | 1.01 | 1.61 | 0.04 |
| Other circulatory disease | 1.27 | 1.02 | 1.58 | 0.03 |
| Age | 1.01 | 1.00 | 1.02 | 0.003 |
| Allergic reactions | 0.70 | 0.57 | 0.85 | 0.0004 |
| Other screening for suspected conditions (not mental disorder or infectious disease) | 0.64 | 0.52 | 0.79 | <0.0001 |
| Disorders of lipid metabolism | 0.56 | 0.45 | 0.70 | <0.0001 |
| E codes: struck by; against | 0.41 | 0.18 | 0.92 | 0.03 |
| Peritonitis and intestinal abscess | 0.12 | 0.02 | 0.88 | 0.04 |
| (C) Significant disease categories associated with T2D subtype 3
| ||||
|---|---|---|---|---|
| Disease category | RR | 95% LCI | 95% UCI | P value |
| HIV infection | 1.92 | 1.30 | 2.85 | 0.001 |
| E codes: adverse effects of medical care | 1.84 | 1.41 | 2.39 | <0.0001 |
| Aortic and peripheral arterial embolism or thrombosis | 1.79 | 1.18 | 2.71 | 0.01 |
| Hypertension with complications and secondary hypertension | 1.66 | 1.29 | 2.15 | <0.0001 |
| Coronary atherosclerosis and other heart disease | 1.41 | 1.15 | 1.72 | 0.001 |
| Allergic reactions | 1.42 | 1.19 | 1.70 | 0.0001 |
| Deficiency and other anemia | 1.39 | 1.14 | 1.68 | 0.001 |
| Screening and history of MHSA codes | 1.30 | 1.07 | 1.58 | 0.01 |
| Diabetes mellitus with complications | 0.80 | 0.67 | 0.96 | 0.02 |
| E codes: place of occurrence | 0.71 | 0.56 | 0.89 | 0.003 |
| Other upper respiratory infections | 0.73 | 0.57 | 0.92 | 0.01 |
| Blindness and vision defects | 0.71 | 0.57 | 0.88 | 0.002 |
| Other skin disorders | 0.68 | 0.55 | 0.83 | 0.0003 |
Significant disease–genetic variant enrichments specific to T2D subtypes
We next evaluated the genetic variants significantly associated with each of the three subtypes. Observed genetic associations and gene-level [that is, single-nucleotide polymorphisms (SNPs) mapped to gene-level annotations] enrichments by hypergeometric analysis are considered independent of the clinical phenotype–based network topology, because patient genetic data were not used in the determination of the patient-patient network topology. We identified 1279, 1227, and 1338 genetic variants specific to subtypes 1, 2, and 3, respectively, using a hypergeometric enrichment approach (see Materials and Methods) (significant SNPs are shown in table S3, A to C). After mapping the variants to gene regions, we identified 425, 322, and 437 unique genes specific to subtypes 1, 2, and 3, respectively. We used a comprehensive human disease–SNP association database (VarDi) (19) to assess the agreement between genetic-disease associations and disease comorbidities associated with each subtype. We analyzed the enrichment of phenotypes including both diagnosis (for example, diabetic nephropathy) and laboratory measurements (for example, creatinine levels) associated with the genetic variants at the gene level.
We observed 27 gene-phenotype associations enriched (hypergeometric analysis, P ≤ 0.05) among the genetic variants unique to subtype 1 (Table 3A and Fig. 2). Many of the enriched gene-level phenotype annotations have known associations with T2D, such as increased serum retinol levels (20), increased B cell counts (21), increased albumin-to-creatinine ratios (22), increased diabetes mellitus, increased serum alanine transaminase levels (23), increased diabetic nephropathy (22, 24), increased leptin receptor (a single-transmembrane domain receptor) (25), increased serum levels of mannose-binding lectin (26), increased forced expiratory volume (27), and increased serum vitamin D concentrations (28). A complete list of subtype 1–specific enriched phenotypes is displayed in Table 3A.
Table 3.
Significant phenotypes.
| (A) Significant phenotypes with disease–genetic variant enrichments specific to T2D subtype 1
| ||
|---|---|---|
| Phenotypes | Gene symbol | P |
| Albumin-to-creatinine ratios | ACE | 1.00 × 10−27 |
| Aspartyl phenylalanine levels | ACE | 1.00 × 10−27 |
| B cell count | LAMB4 | 1.00 × 10−27 |
| Chronic heart failure | LEPR | 1.00 × 10−27 |
| Crypt frequency | SEMA3A | 1.00 × 10−27 |
| Dyslexia | CLSTN2 | 1.00 × 10−27 |
| Hypercholesterolemia | BTN2A1 | 1.00 × 10−27 |
| Mannose-binding lectin levels | MBL2 | 1.00 × 10−27 |
| Prominence of right endocanthion | TMTC2 | 1.00 × 10−27 |
| Retinol levels | FFAR4 | 1.00 × 10−27 |
| Phosphorylated τ 181 protein levels | MTUS1, UNC5C | 5.53 × 10−3 |
| Angiotensin-converting enzyme activity | ACE | 1.32 × 10−2 |
| Diabetes mellitus | BTN2A1 | 1.32 × 10−2 |
| Entorhinal cortical volume | F13A1 | 1.32 × 10−2 |
| Multiple system atrophy | SNCA | 1.32 × 10−2 |
| N-acetylornithine levels | ALMS1 | 1.32 × 10−2 |
| Otosclerosis | TGFB1 | 1.32 × 10−2 |
| Pelvic organ prolapse | ZFAT | 1.32 × 10−2 |
| Tanning ability | MC1R | 1.32 × 10−2 |
| Vitamin D concentrations | GC | 1.32 × 10−2 |
| Diabetic retinopathy | PLXDC2, HS6ST3 | 2.32 × 10−2 |
| Alanine transaminase levels | ZNF827 | 3.66 × 10−2 |
| Diabetic nephropathy | ACE | 3.66 × 10−2 |
| Left ventricular wall thickness | GRID1 | 3.66 × 10−2 |
| Leptin receptor | LEPR | 3.66 × 10−2 |
| Forced expiratory volume | ZSCAN31, TNS1 | 5.00 × 10−2 |
| Platelet response to aspirin intervention therapy | ZNF583, GLIS3 | 5.00 × 10−2 |
| (B) Significant phenotypes with disease–genetic variant enrichments specific to T2D subtype 2
| ||
|---|---|---|
| Phenotypes | Gene symbol | P |
| Alcohol and nicotine codependence | PLEKHG1 | 1.00 × 10−27 |
| Bleomycin sensitivity | SAMD12 | 1.00 × 10−27 |
| Epirubicin-induced adverse drug reactions | MCPH1 | 1.00 × 10−27 |
| Follicular lymphoma | SV2B | 1.00 × 10−27 |
| Lactose intolerance | ST5 | 1.00 × 10−27 |
| Pronasale to left alare distance | CACNA2D3 | 1.00 × 10−27 |
| Stem cell transplantation | NLRP3 | 1.00 × 10−27 |
| Geographic atrophy | HTRA1, CFH | 6.57 × 10−4 |
| Brain | CDH4 | 7.58 × 10−3 |
| Left ventricular internal diastolic dimensions | SLC35F1 | 7.58 × 10−3 |
| Mean platelet volume | ARHGEF3 | 7.58 × 10−3 |
| Polypoidal choroidal vasculopathy | CFH | 7.58 × 10−3 |
| Psychosis | ZNF804A | 7.58 × 10−3 |
| Suicidal behavior | GFRA1 | 7.58 × 10−3 |
| Tanning ability | HERC2 | 7.58 × 10−3 |
| Total τ protein levels | CDH4 | 7.58 × 10−3 |
| Meningococcal disease | TMPRSS15, CFHR3, CFH | 7.79 × 10−3 |
| Keratoconus | SOX5, MACROD2 | 1.76 × 10−2 |
| Meningioma | CHN2 | 2.14 × 10−2 |
| Polycystic ovary syndrome | DENND1A | 2.14 × 10−2 |
| Primary sclerosing cholangitis | GAS7 | 2.14 × 10−2 |
| Atrial fibrillation | CAV1, HCN4 | 2.64 × 10−2 |
| Age-related macular degeneration | PLEKHA1, HTRA1, IL8, CFH | 3.09 × 10−2 |
| Open-angle glaucoma | ADAMTSL1, CAV1 | 3.71 × 10−2 |
| Phosphorylated τ 181 protein levels | CHN2 | 4.04 × 10−2 |
| (C) Significant phenotypes with disease–genetic variant enrichments specific to T2D subtype 3
| ||
|---|---|---|
| Phenotypes | Gene symbol | P |
| Gallbladder cancer | CNTN4, DCC | 1.00 × 10−27 |
| Allergy | FHIT | 1.00 × 10−27 |
| B cell chronic lymphocytic leukemia | CD38 | 1.00 × 10−27 |
| Lymphoid interstitial pneumonitis | FGF14 | 1.00 × 10−27 |
| Osteoporosis | ALDH7A1 | 1.00 × 10−27 |
| Peripartum cardiomyopathy | AKAP13 | 1.00 × 10−27 |
| RR interval | GPR133 | 1.00 × 10−27 |
| Spinocerebellar ataxia type 1 | ATXN1 | 1.00 × 10−27 |
| Intraventricular septal thickness | EXT1, CERS6 | 1.65 × 10−3 |
| Endometrial cancer | SLC8A1 | 1.40 × 10−2 |
| HIV-associated neurocognitive disorders | SLC8A1 | 1.40 × 10−2 |
| Response to statin | ASB18 | 1.40 × 10−2 |
| Uterine leiomyoma | TNRC6B | 1.40 × 10−2 |
| Vitamin D concentrations | DAB1 | 1.40 × 10−2 |
| Anxiety disorders | SDK2, FHIT | 2.50 × 10−2 |
| Cognitive decline | CTNND2 | 3.86 × 10−2 |
| Dementia | ABCA1 | 3.86 × 10−2 |
| Estrone levels | ESR1 | 3.86 × 10−2 |
| Impaired play skills | DCC | 3.86 × 10−2 |
| Intelligence | CNTN4 | 3.86 × 10−2 |
| Myopia | MIPEP | 3.86 × 10−2 |
| Plasma progranulin levels | DNAH11 | 3.86 × 10−2 |
| Polycystic ovary syndrome | THADA | 3.86 × 10−2 |
| Renal cell carcinoma | ITPR2 | 3.86 × 10−2 |
| Theta power of electroencephalogram | ST6GALNAC3 | 3.86 × 10−2 |
| Central corneal thickness | COL5A1, FNDC3B | 4.00 × 10−2 |
| Atrial fibrillation | C9orf3, SYNE2 | 5.00 × 10−2 |
| Depression | FHIT, BICC1 | 5.00 × 10−2 |
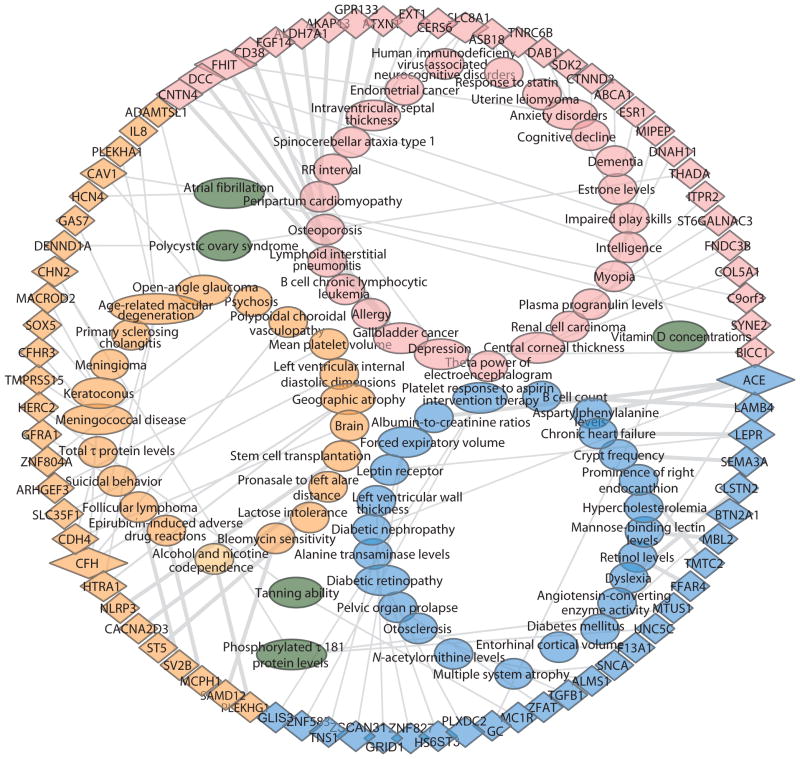
Fig. 2. Genotype-phenotype network for three subtypes in T2D.
The network consists of the significant association between phenotypes and genetic variants at gene level specific to three T2D subtypes (subtype 1 in blue, subtype 2 in orange, and subtype 3 in pink). Phenotypes (oval) and genes (triangle) are connected by gray lines (P value). Oval nodes in dark green indicate the shared phenotypes across subtypes. The edge width reflects the significance of the P value for enrichment. The size of the node reflects the amount of associated genes or phenotypes. This network was visualized using Cytoscape 3.2.0.
We observed 25 gene-phenotype associations significantly enriched among the genetic variants unique to subtype 2. The four enriched gene-level phenotype annotations for subtype 2 were related to either cancer or treatment of cancer including bleomycin sensitivity, epirubicin-induced adverse drug reactions, stem cell transplantation, and follicular lymphoma. In addition, we identified two cardiovascular phenotypes, left ventricular internal diastolic dimensions and atrial fibrillation. The enriched gene-level phenotypes matched with patient comorbidities associated with subtype 2 (Table 3B and Fig. 2), suggesting a possible link between observed disease comorbidities and underlying subtype genetics.
We observed 28 gene-phenotype associations significantly enriched among the genetic variants unique to subtype 3 (Table 3C and Fig. 2). Ten phenotypes were related to mental and neurological diseases, including spinocerebellar ataxia type 1, intraventricular septal thickness, anxiety disorders, cognitive decline, dementia, impaired play skills, intelligence, depression, θ power of electroencephalogram, and HIV-associated neurocognitive disorders. Three were related to the cardiovascular system, including heart rate interval (RR), peripartum cardiomyopathy, and atrial fibrillation. Increased serum vitamin D concentrations (28) were recently implicated as a risk factor for T2D and also were enriched in subtype 1. Furthermore, two phenotypes, allergy and response to statins, were enriched for genetic variants that matched with the identified clinical variables and phenotype comorbidities specific to subtype 3, including cardiovascular disease and mental illness. Disease comorbidities and clinical variables associated with subtype 3 matched particularly well with the gene-level phenotype enrichments. A complete list of enriched phenotypes for subtype 3 is shown in Table 3C.
The network of genetic variants in gene-level and associated phenotypes for the three T2D subtypes is shown in Fig. 2 (produced with Cytoscape 3.2.0) (29).
Significant pathway and toxicity functions specific to T2D subtypes
We assessed the toxicity functions and signaling pathways for gene-level enrichments unique to each subtype (425, 322, and 437 gene-level enrichments specific to subtypes 1, 2, and 3, respectively) using Qiagen’s Ingenuity Pathway Analysis (IPA) program. Canonical pathways include metabolic and cell signaling pathways that have been curated from the literature by IPA. We identified five, two, and six canonical pathways to subtypes 1, 2, and 3, respectively (P < 0.01), by Fisher’s exact test right-tailed for enrichment.
Pathways that were enriched in subtype 1 were fatty acid β-oxidation III, which is increased in diabetic liver disease (30), acetate conversion to acetyl-CoA, which is involved in the metabolism of carbon sugars (31–33), and cAMP (adenosine 3′,5′-monophosphate)–mediated signaling, which normalizes glucose-stimulated insulin secretion in uncoupling protein 2–overexpressing pancreatic β cells (34). Two pathways were associated with disease comorbidities for subtype 1, including netrin signaling, which acts in a protective role during diabetic nephropathy (35), and GABA (γ-aminobutyric acid) receptor signaling, which can often be detected early in the course of diabetic retinopathy (36, 37).
Pathways enriched in subtype 2 include those involved in pattern recognition receptors in the recognition of bacteria and viruses, which might explain why patients in subtype 2 had an increased prevalence of tuberculosis. We also found an enrichment for thrombopoietin signaling, which activates a number of secondary messengers that promote cell survival, proliferation, and differentiation (38). Increased thrombopoietin levels might contribute to the development and progression of coronary artery disease (39, 40).
Pathways enriched in subtype 3 include α-adrenergic signaling, which is implicated in diverse physiological functions, in particular those of the cardiovascular and central nervous systems (41, 42); synaptic long-term depression (43); CREB (cAMP response element–binding protein) signaling in neurons, which has a well-documented role in neuronal plasticity and long-term memory formation in the brain (44) as well as therapeutic potential for patients who have Alzheimer’s disease (45); glutamate receptor signaling, which has been implicated in brain pathologies in neurological diseases (46); hepatic fibrosis and hepatic stellate cell activation; and sperm motility. The complete list of pathways and their related genes for all subtypes are shown in Table 4.
Table 4.
Canonical pathways at gene level for each T2D subtype.ns, not significant.
| Canonical pathway | Subtype 1 | Subtype 2 | Subtype 3 | Genes |
|---|---|---|---|---|
| Fatty acid β-oxidation III | 1.1 × 10−3 | ns | ns | ECI1, ECI2 |
| Acetate conversion to acetyl-CoA | 3.5 × 10−3 | ns | ns | ACSL1, ACSL2 |
| Netrin signaling | 6.2 × 10−3 | ns | ns | ABLIM1, PRKG1, UNC5B, UNC5C |
| GABA receptor signaling | 8.8 × 10−3 | ns | ns | ADCY8, ALDH5A1, GABBR1, GABRR2, GPHN |
| cAMP-mediated signaling | 9.2 × 10−3 | ns | 2.0 × 10−2 | Subtype 1: ADCY8, AKAP12, CAMK1D, CNGB1, CNGB3, GABBR1, MC1R, PDE3A, PKIA, RGS7 Subtype 3: AKAP13, CAMK4, CHRM5, GNAI3, HTR1D, PDE4B, PDE6A, PRKAR2B, RAF1 |
| Role of pattern recognition receptors in recognition of bacteria and viruses | ns | 1.8 × 10−3 | ns | CXCL8, MAPK10, NLRP3, OAS1, OAS3, PRKCD, PRKCH |
| Thrombopoietin signaling | ns | 6.8 × 10−3 | ns | GAB2, PRKCD, PRKCH, SOS1 |
| α-Adrenergic signaling | ns | ns | 1.2 × 10−3 | CAMK4, GNAI3, GYS1, ITPR2, PRKAR2B, RAF1, SLC8A1 |
| Synaptic long-term depression | ns | ns | 1.4 × 10−3 | GNA11, GNAI3, GRID2, GRM1, ITPR2, PLA2G4C, PLA2R1, PPP2R5B, RAF1, |
| CREB signaling in neurons | ns | ns | 1.4 × 10−3 | CAMK4, GNA11, GNAI3, GRID2, GRIK4, GRM1, ITPR2, POLR2I, PRKAR2B, RAF1 |
| Glutamate receptor signaling | ns | ns | 4.2 × 10−3 | CAMK4, GRID2, GRIK4, GRM1, PICK1 |
| Hepatic fibrosis/hepatic stellate cell activation | 3.0 × 10−2 | ns | 4.0 × 10−3 | Subtype 1: BCL2, COL19A1, COL28A1, IGF1R, IL1RAP, LEPR, TGFB1, TGFB2 Subtype 3: BAX, COL15A1, COL25A1, COL4A4, COL5A1, COL5A3,COL9A3, FGF2, KLF12, MYH7B |
| Sperm motility | ns | ns | 7.3 × 10−3 | CAMK4, ITPR2, PDE4B, PLA2G4C, PLA2R1, PRKAR2B, SLC12A2 |
Enriched toxicity functions included hepatotoxicity, nephrotoxicity, cardiovascular toxicity, and clinical pathology endpoints. We identified nine, three, and three toxicity functions enriched in subtypes 1, 2, and 3, respectively (P < 0.01). In subtype 1, four of the nine functions are related to renal dysfunction, including glomerular injury, renal hypertrophy, renal proliferation, and renal degeneration, suggesting that diabetic nephropathy exists in the subtype 1 cohort (47, 48). The remaining five functions are related to liver dysfunction, which match the two liver enzymes, alanine transaminase levels and aspartyl phenylalanine levels, identified by VarDi (19). Surprisingly, subtypes 2 and 3 were both associated with cardiac arteriopathy, even though they were associated with different sets of genes. Most toxicity functions that are related to cardiovascular disorders and liver fibrosis match the findings that both cohorts have high risk for cardiovascular diseases, as deduced on the basis of disease comorbidities from the EMRs and genetic variant associations by VarDi (19). The complete list of enriched toxicity functions for all subtypes and their related genes are listed in Table 5.
Table 5.
Toxicity functions at the gene level for each T2D subtype.
| Toxicity functions | Subtype1 | Subtype2 | Subtype3 | Genes |
|---|---|---|---|---|
| Biliary hyperplasia | 3.5 × 10−3 | ns | ns | CFTR, PKHD1 |
| Glutathione depletion in liver | 3.5 × 10−3 | ns | ns | LEPR, TGFB1 |
| Liver fibrosis | 3.5 × 10−3 | ns | ns | TGFB1, LEPR, TGFB2, PKHD1 |
| Glomerular injury | 4.7 × 10−3 | ns | ns | FYN, TGFB1, LEPR, RARA, TNS1, PKN1, PTGER1, BCL2 |
| Renal hypertrophy | 4.7 × 10−3 | ns | ns | TGFB1, LEPR, RARA, BCL2 |
| Liver damage | 5.1 × 10−3 | ns | ns | SLC10A1, TGFB1, IGF1R, GABBR1, SERPINA1, CD274, PARK2, PTGER1 |
| Liver inflammation/hepatitis | 5.1 × 10−3 | ns | ns | AKAP12, SLC10A1, TGFB1, PDE3A, IGF1R, GABBR1, CD274, PARK2 |
| Renal proliferation | 7.6 × 10−3 | ns | ns | PRKG1, TGFB1, UNC5B, TTLL4, CRK, ZNF512B, DLC1, BCL2, UNC5C, AFF1 |
| Renal degeneration | 8.0 × 10−3 | ns | ns | TGFB1, TNS1, BCL2 |
| Cardiac arrhythmia | ns | 1.0 × 10−3 | ns | KCND3, HCN4, KCNG2, KCNQ1, CNTN5 |
| Bradycardia | ns | 4.9 × 10−3 | ns | HCN4, KCNQ1 |
| Cardiac arteriopathy | ns | 9.3 × 10−3 | 4.8 × 10−6 | Subtype 2: SAMD12, KALRN, ITGA8, PDE5A, DOCK4, CNTN6, PRKCH, CSMD2, CPEB3, CNTN5 Subtype 3: CERS6, CLIC5, ZMYM2, CDCP1, ABCG1, FRMD4A, PDE4B, PTPRM, ABCA1, F2, SPATA5, AKAP13, MCF2L, PBX3, CNTNAP5, FMN2, CACNA2D1, SLC8A1, ESR2 |
| Liver fibrosis | ns | ns | 3.3 × 10−3 | FGF2, PLAUR, BMP7, CC2D2A, F2, HSPB1 |
| Congenital heart anomaly | ns | ns | 5.8 × 10−3 | DNAH11, BICC1, PDS5B, INVS |
Together, these results suggest that the current clinical definition of T2D subsumes more nuanced subtypes whose definition and recognition might inform important clinical distinctions. Furthermore, the genetic findings suggest that these differences between T2D subtypes are potentially rooted in biological differences that relate to the observed clinical differences, and these biological differences might suggest new opportunities for biomarker discovery or improving our understanding of disease mechanisms.
DISCUSSION
Previous efforts to analyze or mine large clinical populations with associated genome-wide genotyping information have largely focused on replicating known clinical genotype-phenotype correlations, or discovering new correlations from more narrowly defined clinical phenotypes that can be extracted from EMRs (49, 50). Previous efforts to develop and apply phenome-wide association study (PheWAS) approaches represent a new approach in which data from EMRs are integrated and used for systematic discovery of new clinical genotype-phenotype correlations (51). However, the goal of PheWAS is to discover new pleiotropic genotype-phenotype associations—that is, to identify many clinical phenotypes linked to a single genetic locus. The goal of our study was to develop a precision medicine approach to characterize the complexity of T2D patient populations through data-driven, topological analysis of patient-patient similarity across clinical phenotype traits. Our approach is distinct from previous efforts in that we developed and applied a patient-centric clinical phenotype similarity network and then used the topology of the resulting patient-patient similarity network to define patient subgroups, which were subsequently used as the basis of clinical and genotype risk factor associations.
We hypothesized that topological analysis of patient populations in high-dimensional clinical phenotype space may identify meaningful subpopulations of T2D patients. We focused our analysis on T2D patients, who are of high clinical importance and the most prevalent disease group in the population. We identified 2551 T2D patients in our outpatient cohort as determined by the eMERGE T2D electronic phenotyping algorithm (16, 17). Using our data-driven, topology-based approach, we identified three distinct subtypes of T2D. Subtype 1 comprises ~30% (n = 761) of the overall T2D cases and was enriched for diabetic nephropathy and diabetic retinopathy, both microvascular complications. Subtype 2 comprises ~24% (n = 617) of all T2D cases and was enriched for cancer malignancy and cardiovascular diseases. Subtype 3 comprises ~43% (n = 1096) of all T2D cases and associated most strongly with cardiovascular diseases, neurological diseases, allergies, and HIV infections. Macrovascular complications are generally best averted by stringent control of blood pressure and low-density lipoprotein. We identified 1279, 1227, and 1338 SNPs, which mapped to 425, 322, and 437 genes, specific to subtypes 1, 2, and 3, respectively. The enriched phenotypes and biological functions defined at the gene level for each subtype matched with the disease comorbidities and clinical differences that we identified through EMR-based topology data analysis (TDA). This observed agreement is likely meaningful mechanistically because the genetic data were not used to inform patient subgroup topology.
The patient-patient network representation was constructed using cosine distance metric with two filter functions to assess the similarity of the clinical variables from EMRs. The clinical data set comprises more than 500 clinical variables represented in the EMRs, including patient demographics, laboratory tests, and medication orders.
The observed differences in comorbidity and genetic associations between T2D subtypes might serve as useful features for informing the clinical characterization of T2D patients. We found several notable associations between disease diagnosis categories and T2D subtypes. We used CCS developed by the U.S. Agency for Healthcare Research and Quality (AHRQ) (18) to narrow down more than 7000 ICD-9-CM diagnosis codes in our cohort to higher-order single-level disease categories (n = 281) that include exclusively mental health and substance abuse (CCS-MHSA) general categories, which were more useful for presenting data at a descriptive statistical categorical level than using individual ICD-9-CM codes. Patients in subtype 1 associated most with prototypical microvascular diabetic complications, namely, diabetic nephropathy and diabetic retinopathy, which was supported by both clinical data and genotype data independently. In support of a genetic etiology for subtype 1 phenotype manifestation, the ACE gene, which encodes angiotensin I converting enzyme and was specifically associated with this cohort (Table 3A and Fig. 2), has been implicated in diabetic nephropathy (52, 53) and also in platelet aggregation (53). Accordingly, this association could reasonably suggest a mechanism to explain the lower platelet counts observed in subtype 1 patients (54). In addition, we extracted hemoglobin A1c (HbA1c) levels from our EMRs and found that patients in subtype 1 had the highest HbA1c levels compared with other two groups (7.68 ± 1.75, 7.45 ± 1.87, and 7.47 ± 1.78 in subtypes 1, 2, and 3, respectively, P < 0.05), which confirmed that subtype 1 was most likely enriched with microvascular diabetic complications best prevented by glycemic control (55).
Patients in subtype 2 were more likely to associate with cancer of the bronchus and lung (RR, 3.76; range, 1.14 to 12.39) and malignant neoplasm without specification of site (RR, 3.46; range, 1.23 to 9.7). Epidemiological studies have demonstrated an association between T2D and cancer (56). To try to unravel a putatively causal ordering for this disease link, we compared the first diagnosis dates for both diseases in our cohort to determine whether one more often predated the other. We identified 40% patients who were diagnosed with T2D before any instance of cancer and 60% of patients who were diagnosed with a cancer before T2D. This pattern indicates that T2D can be either the risk factor for or consequence of many forms of cancer (56, 57). Patients in subtype 3 were most likely to be associated with cardiovascular diseases and mental illness according to clinical data and genotype data independently. These patients were more often prescribed the top psychiatric medications to treat anxiety and depression (58), with 3.4% (P = 0.01) and 8.3% (P = 0.02), compared with other two subtypes from χ2 tests, respectively, as well as insulin treatment (45%, P < 0.0001). The 61 patients diagnosed with HIV infection could have a poorer response to therapy for diabetes because antiretroviral agents and chronic inflammation could adversely affect glycemic control (59). To address any potential bias from HIV infection or treatment, we removed these HIV patients from the cohort and reanalyzed the data using the LASSO algorithm (60). Except for allergies, disease comorbidities remained the same, dismissing the possibility of HIV infection bias and exhibiting the robustness of our methodology. Furthermore, the FHIT gene, which encodes the fragile histidine triad protein and was specifically associated with the subtype 3 cohort, has been associated with allergy and neurological disorders, including anxiety and depression (Table 3C and Fig. 2) (61–63), indicating that FHIT could be a driver for these conditions and could explain why patients who had allergies also had an increased rate of suicide (64–67). Although patients in subtypes 2 and 3 had significantly lower BMIs than those in subtype 1 (P < 0.0001, Table 1B), both were enriched for cardiovascular morbidity, whereas patients in subtype 1 were not. A recent study showed that weight loss does not reduce the rate of cardiovascular events in obese adults with T2D (68, 69). These data suggest that the cardiovascular morbidity seen in patients in subtypes 2 and 3 might be independent from obesity and potentially driven by genetic variants. Another interesting finding along these lines is our observation that hypertensive macrovascular variants were associated with subtypes 2 and 3, whereas hyperglycemic microvascular variants were associated with subtype 1.
Our study has several potential limitations. We identified 2551 T2D patients on the basis of an eMERGE algorithm (16, 17) from an 11,210 genotyped outpatient cohort. The sample size is relatively modest for identifying risk variants from a genome-wide association study (GWAS) point of view. Given that we investigated 38 million variants, it was a great challenge to control for false discovery rate. In our study, however, we derived our genetic data from more than ~10,000 published GWAS at the P < 1 × 10−6 significance level. The stringency of this inclusion criterion adds a measure of control to the procedure because subtype enrichments were identified using these disease-associated variants.
Another limitation is the lack of a deep consideration for the temporal aspects of disease trajectories. In analyzing the EMRs in Mount Sinai Medical Center (MSMC), we cannot always be clear when and where the first diagnosis of disease took place. Specifically, we cannot determine whether the patient had been diagnosed beforehand in other hospitals and, if so, how long the patient had the diagnosed disease before his or her first observed ICD-9-CM diagnosis. One possible solution is to explore the integration of insurance claims data. We will explore an extension of our analytical framework that incorporates temporal analysis in future studies.
In addition, T2D inclusion and exclusion criteria were precisely refined by the eMERGE algorithm (16, 17), and the other disease categories developed by AHRQ were all based on the current ICD-9-CM diagnosis code. Furthermore, CCS developed by AHRQ (18) only assigns one disease classification of a disease. As of now, only 20 phenotypes have been validated by eMERGE (70) using iteratively refined phenotype algorithms incorporating both structured and unstructured data to achieve high PPVs to identify true cases and controls from EMRs.
Our approach combines imputed variant information from the whole genome with high-dimensional EMRs, which facilitates pinpointing the differences between clinical and genetic factors specific to each subtype. This provides a tractable framework that enables initial steps toward the T2D redefinition informed by genetic markers. Our genetic analysis used the imputed variants from the 1000 Genome Projects, not limiting the variants in the genotyping arrays. This strategy offers better coverage on the intergenic and noncoding regions when investigating the associations between variants and phenotypes. The Encyclopedia of DNA Elements (ENCODE) project has shown that ~95% of known variants within sequenced genomes and 88% of those variants from GWAS fall outside of coding regions (71), and a functional SNP most strongly supported by experimental evidence is an SNP in the linkage disequilibrium region (72). The technique of imputation uses information of haplotypes from a more comprehensive whole-genome sequencing study (the 1000 Genome Projects) to infer variants that were not profiled by the original technology (73). With the information on variants from the whole genome, we were able to identify more variants associated with subtypes as well as to achieve better mapping of the identified variants to published GWAS.
Our study offers several important conclusions for translational research. First, our approach demonstrates the utility and promise of applying the precision medicine paradigm in T2D, and can be extended toward the study of other complex, multifactorial diseases. Next, our study demonstrates the utility of using higher-dimensional clinical data to first define the complex topology of a clinical phenotype before genetic marker discovery. This stands in contrast with previous precision medicine efforts that begin with molecular stratification and rely on established clinical phenotype definitions. Furthermore, the subtype-specific genetic factors identified by this study can be further explored through additional population genetic and experimental work to evaluate their utility for identifying subtype-specific biomarkers or to improve understanding of T2D disease mechanisms. Last, incorporation of the temporal dimension in future development of our topology-based approach might provide additional insight into the complexity of T2D patient populations along the natural history of disease and inform disease prevention efforts.
MATERIALS AND METHODS
Study design
The aim of our study was to develop a precision medicine approach to better understand and to characterize the complexity of T2D patient populations through data-driven, topological analysis of patient-patient similarity across clinical phenotype traits. We performed topological analysis for the data set, which comprises EMRs and genotype data from 11,210 individuals from MSMC’s large outpatient population. T2D and non-T2D control phenotypes were defined by the eMERGE phenotyping algorithm (16, 17). We assessed the disease comorbidities and human disease–SNP association for each subtype in T2D, as well as the enriched phenotypes and biological functions at gene level for each subtype.
Patient population
We recruited and analyzed 11,210 unique patients who are consented participants in the Mount Sinai BioMe Biobank Program, an ongoing, EMR-linked bio- and data repository. The data set comprises adult patients recruited nonselectively from MSMC’s large outpatient population. Participants are predominantly recruited from local diverse communities in New York with 46% Hispanic, 32% African American, 20% European white, and 2% others as self-reported. The data were composed of 6857 (61%) females and 4350 (39%) males, and the average age is 55.5 years for overall, female, and male populations (fig. S1). The overall characteristics of 11,210 Biobank patients are shown in table S2. The individuals represented in the clinical data set are drawn from diverse racial, ethnic, and socioeconomic backgrounds. The EMR data are deidentified, and this study was governed by institutional review board approval and informed consent.
Genotype data processing and identification of genetic variants and genes
A total of 11,210 unique patients were genotyped for genome-wide Illumina OmniExpress and Illumina Human Exome BeadChip arrays. We used a default GenCall score cutoff of 0.15 in GenomeStudio (v2011.1) as recommended by Illumina. Quality control was performed by zCall (74) for SNP quality. SNPs were removed if they had (i) a call rate of <95%, (ii) no minor alleles, (iii) Hardy-Weinberg equilibrium within population (P < 5 × 10−5), and (iv) removed A/T and G/C SNPs and any SNPs that deviate from 1 kg (<40% versus >60% and vice versa). After quality control for call quality and population equilibrium, the genotype data were phased by ShapeIt v2 r644 (75), yielding 850,067 SNPs, and then imputed by IMPUTE2.3 (73) using the 1000 Genomes Project (76) version 3 and integrated variant set (August 2012) as the reference panel, resulting in 38,068,758 variants. A complete list of the number of variants, in coding regions, and genes in both original genotype and the imputed data using genome build GRCh37/hg19 is shown in table S4. The rationale for using the 1000 Genomes Project as reference panel for imputation is that it contains the largest sample size of most diverse ethnicity background. Given the diversity in the Mount Sinai Biobank patients, using the 1000 Genomes Project allows us to identify the closest individuals for each patient and impute for genotypes that were not profiled in the original array. We mapped the imputed variants to gene regions by SnpEff v2 r644 (77) and AILUN [(78); http://ailun.stanford.edu] using human genome assembly (GRCh37/hg19) reference genome (UCSC Genome Browser, http://genome.ucsc.edu). The imputed variants data covering variants originally profiled by the genotyping arrays as well as variants observed in the 1000 Genomes Projects were then used for association analysis.
Clinical phenotype data
We generated a pseudo cross-sectional data set from our deidentified patient records using the following phenotypic logic scheme. Using the initial enrollment date into the BioMe program (D1) as an anchor, we populated all (first) laboratory values, vitals, and specified medications ±30 days from D1. We collected the last laboratory/vital/medication date (D2) where the upper bound of the D2 date was constrained to D1 +30 days, and the lower bound constrained to D2 = D1. In most cases, D2 = D1. We then populated all ICD-9-CM codes for patients, where ICD-9-CM date ≤ D2 date. We then populated all medication orders for patient, where medication orders date ≤ D2 date. The data set also includes self-reported demographic data collected at D1.
T2D and non-T2D control phenotypes were defined by an electronic phenotyping algorithm that was developed by the eMERGE network (16, 17) based on ICD-9-CM diagnosis codes, laboratory tests (LONIC), prescribed medications (RxNorm), physician notes (natural language processing), and family history. Interim results were vetted by subject matter experts (SMEs) to verify that the queries were capturing the specified data appropriately. Adjustments to the queries were implemented iteratively as per the feedback received. Once the SMEs were satisfied with the algorithm components, the separate queries were packaged into a single job flow and executed against the base population datamart, resulting in the identification of cases and controls. We randomly selected samples of 100 cases and 100 controls for manual chart review by clinical experts from the endocrinology division at Mount Sinai Hospital and performance statistics generated. The algorithm achieved a PPV of 96% for cases and 100% for controls.
The processed data were then assembled into a data matrix of n patients by P clinical variables. The data set used for analysis represented 11,210 individual patients, 505 clinical variables (480 of which were clinical laboratory measures), and 7097 unique ICD-9-CM codes (1 to 218 per patient). On average, there were 64 clinical variables collected per patient (range, 25 to 212). To avoid overfitting, we selected the clinical variables with at least 50% of patients who had the values, resulting in 73 variables to perform the analysis (table S1).
Disease classification
Each individual patient had at least one ICD-9-CM code diagnosis at the time his or her DNA sample was collected. CCS is a tool that was developed at AHRQ for clustering patient diagnoses and procedures into a manageable number of clinically meaningful categories (18). The single level of CCS is used to classify all diagnoses and procedures into unique groups based on the patient’s ICD-9-CM codes. The multilevel characterization of CCS is used to group single-level CCS categories into broader body systems or condition categories (for example, “Diseases of the Circulatory System,” “Mental Disorders”). The multilevel system has four levels of groupings for diagnoses, and we use the highest, most broad level to examine and assess general groupings for the disease category (18). In our study, we used 281 mutually exclusive single-level and 18 multilevel categories (broadest level) from CCS to map the disease categories based on their ICD-9-CM codes.
TDA pipeline
We developed a novel TDA-based approach to perform unsupervised clustering of patients using various clinical features to produce a patient-patient network organized according to the high-dimensional clinical phenotype similarity among patients. We use Ayasdi 3.0 (79, 80) (http://ayasdi.com, Ayasdi Inc.) to perform the TDA analysis. We used TDA pipeline for overall patients, random samplings of training and test data sets. A cosine distance metric was used to assess the similarity of the data points based on clinical variables (Eq. 1). Two filter functions, L-infinity centrality and principal metric singular value decomposition (SVD1), were used to generate the patient-patient network based on clinical variables. L-infinity centrality is defined for each data point y to be the maximum distance from y to any other data point in the data set. It produces a more detailed and succinct description of the data set than a typical scatter plots display (80). Large values of this function correspond to points that are far from the center of the data set. SVD1 also was used in the data matrix to obtain subspaces within the column space, and dimensionality reduction is accomplished by projection on these subspaces (80). This is done with standard linear algebraic techniques when possible, and when the number of points is too large, numerical optimization techniques are used.
| (1) |
where D1 and D2 represent two individual data points.
Statistical analysis
We used Ayasdi 3.0 (79, 80) (http://ayasdi.com, Ayasdi Inc.) to perform TDA for generating the patient-patient network. We used Qiagen’s IPA program version 24390178 (IPA, Qiagen, http://qiagen.com/ingenuity) to assess the toxicity functions and pathways for significant genes associated with each subtype. For imputed SNPs, we performed hypergeometric analysis to identify the significant SNPs associated with each subtypes based on their allele frequency and then examined the disease enrichment associated with the genes mapped from SNPs. The goal of performing hypergeometric tests is to identify genes that are highly associated with each subtype, which would lead to distinct phenotypes associated with each subtype. Such analysis is by nature different from traditional GWAS, where the goal is to identify disease-causing variants. Therefore, the hypergeometric test P values were used as an association measure instead of the evaluation of significance for individual SNPs. Similar analysis can also be seen in gene set–based gene expression analysis such as gene set enrichment analysis (81). We used our curated VarDi (19) to assess the significance of the genotype-phenotype enrichment. VarDi (19) is composed of 24,435 variants mapped to 3694 unique genes in 904 distinct phenotypes with a significant level (P < 1 × 10−6) from over ~13,000 GWAS, and we used P < 1 × 10−6 to identify variants from VarDi (19). LASSO provides stability and robustness statistics, which are used to inform consistency and sparsity. LASSO seeks a model that not only fits well but also is “simple” to avoid large variation, which occurs in estimating complex models (60). We used the LASSO algorithm with corrected Akaike information criterion statistic (AICC) (Eq. 2) (82) for feature selection and logistic regression for RR estimate of disease comorbidities based on CCS disease classification. We used analysis of variance (ANOVA), two-tailed t test, or χ2 tests to compare multiple or two-class continuous or categorical clinical variables. Data were presented as means ± SE. Statistical analyses and random samplings were carried out using SAS 9.3.2 (SAS Institute) and R 2.15.1 (83). We used Cytoscape 3.2.0 (29) to visualize the networks for the significant genotype-phenotype association identified from VarDi (19) specific to each of the T2D subtypes.
| (2) |
where k is the number of parameters in the model, and n is the sample size.
Acknowledgments
We thank D. Ruderfer for insights on Biobank data; M. Menon for helpful comments and suggestions; and the IT group in Icahn School of Medicine at Mount Sinai for Hadoop computing and database support.
Funding: This study was supported by funding from the NIH National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) (R01DK098242) and National Cancer Institute (NCI) (U54CA189201). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Author contributions: Conceived and designed the study: L.L. and J.T.D. Performed the TDA and statistical analysis: L.L. Analyzed the EMRs: L.L. Analyzed the genotyping data: L.L. and W.-Y.C. Contributed VarDi analysis tools: R.C. Contributed Biobank genotyping data: J.T.D., O.G., and E.P.B. Contributed clinical interpretation: L.L. and R.T. Wrote and edited the paper: L.L., W.-Y.C., J.T.D., B.S.G., O.G., and R.T.
Competing interests: The authors declare that they have no competing interests.
SUPPLEMENTARY MATERIALS
www.sciencetranslationalmedicine.org/cgi/content/full/7/311/311ra174/DC1
Fig. S1. Age distributions for overall, female, and male populations.
Table S1. Clinical features.
Table S2. Patient characteristics across entire Biobank cohort.
Table S3. Significant SNPs specific for each T2D subtype.
Table S4. Genes and variants.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIDDK, NCI, or NIH.
REFERENCES AND NOTES
- 1.Centers for Disease Control and Prevention. National Diabetes Statistics Report: Estimates of Diabetes and Its Burden in the United States, 2014. U.S. Department of Health and Human Services; Atlanta, GA: 2014. [Google Scholar]
- 2.American Diabetes Association. Standards of medical care in diabetes—2009. Diabetes Care. 2009;32(Suppl 1):S13–S61. doi: 10.2337/dc09-S013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fong DS, Aiello LP, Ferris FL, III, Klein R. Diabetic retinopathy. Diabetes Care. 2004;27:2540–2553. doi: 10.2337/diacare.27.10.2540. [DOI] [PubMed] [Google Scholar]
- 4.Lehto S, Rönnemaa T, Pyörälä K, Laakso M. Predictors of stroke in middle-aged patients with non–insulin-dependent diabetes. Stroke. 1996;27:63–68. doi: 10.1161/01.str.27.1.63. [DOI] [PubMed] [Google Scholar]
- 5.Beckman JA, Creager MA, Libby P. Diabetes and atherosclerosis: Epidemiology, pathophysiology, and management. JAMA. 2002;287:2570–2581. doi: 10.1001/jama.287.19.2570. [DOI] [PubMed] [Google Scholar]
- 6.Boulton AJM, Vinik AI, Arezzo JC, Bril V, Feldman EL, Freeman R, Malik RA, Maser RE, Sosenko JM, Ziegler D A. American Diabetes. Diabetic neuropathies: A statement by the American Diabetes Association. Diabetes Care. 2005;28:956–962. doi: 10.2337/diacare.28.4.956. [DOI] [PubMed] [Google Scholar]
- 7.American Diabetes Association. Diagnosis and classification of diabetes mellitus. Diabetes Care. 2010;33(Suppl 1):S62–S69. doi: 10.2337/dc10-S062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.National Research Council Committee on a Framework for Developing a New Taxonomy of Disease. Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease. The National Academies Press; Washington, DC: 2011. [PubMed] [Google Scholar]
- 9.Færch K, Witte DR, Tabák AG, Perreault L, Herder C, Brunner EJ, Kivimäki M, Vistisen D. Trajectories of cardiometabolic risk factors before diagnosis of three subtypes of type 2 diabetes: A post-hoc analysis of the longitudinal Whitehall II cohort study. Lancet Diabetes Endocrinol. 2013;1:43–51. doi: 10.1016/S2213-8587(13)70008-1. [DOI] [PubMed] [Google Scholar]
- 10.Morris AP, Voight BF, Teslovich TM, Ferreira T, Segrè AV, Steinthorsdottir V, Strawbridge RJ, Khan H, Grallert H, Mahajan A, Prokopenko I, Min Kang H, Dina C, Esko T, Fraser RM, Kanoni S, Kumar A, Lagou V, Langenberg C, Luan J, Lindgren CM, Müller-Nurasyid M, Pechlivanis S, William Rayner N, Scott LJ, Wiltshire S, Yengo L, Kinnunen L, Rossin EJ, Raychaudhuri S, Johnson AD, Dimas AS, Loos RJF, Vedantam S, Chen H, Florez JC, Fox C, Liu C-T, Rybin D, Couper DJ, Kao WHL, Li M, Cornelis MC, Kraft P, Sun Q, van Dam RM, Stringham HM, Chines PS, Fischer K, Fontanillas P, Holmen OL, Hunt SE, Jackson AU, Kong A, Lawrence R, Meyer J, Perry JRB, Platou CGP, Potter S, Rehnberg E, Robertson N, Sivapalaratnam S, Stančáková A, Stirrups K, Thorleifsson G, Tikkanen E, Wood AR, Almgren P, Atalay M, Benediktsson R, Bonnycastle LL, Burtt N, Carey J, Charpentier G, Crenshaw AT, Doney ASF, Dorkhan M, Edkins S, Emilsson V, Eury E, Forsen T, Gertow K, Gigante B, Grant GB, Groves CJ, Guiducci C, Herder C, Hreidarsson AB, Hui J, James A, Jonsson A, Rathmann W, Klopp N, Kravic J, Krjutškov K, Langford C, Leander K, Lindholm E, Lobbens S, Männistö S, Mirza G, Mühleisen TW, Musk B, Parkin M, Rallidis L, Saramies J, Sennblad B, Shah S, Sigurđsson G, Silveira A, Steinbach G, Thorand B, Trakalo J, Veglia F, Wennauer R, Winckler W, Zabaneh D, Campbell H, van Duijn C, Uitterlinden AG, Hofman A, Sijbrands E, Abecasis GR, Owen KR, Zeggini E, Trip MD, Forouhi NG, Syvänen A-C, Eriksson JG, Peltonen L, Nöthen MM, Balkau B, Palmer CNA, Lyssenko V, Tuomi T, Isomaa B, Hunter DJ, Qi L, Shuldiner AR, Roden M, Barroso I, Wilsgaard T, Beilby J, Hovingh K, Price JF, Wilson JF, Rauramaa R, Lakka TA, Lind L, Dedoussis G, Njølstad I, Pedersen NL, Khaw K-T, Wareham NJ, Keinanen-Kiukaanniemi SM, Saaristo TE, Korpi-Hyövälti E, Saltevo J, Laakso M, Kuusisto J, Metspalu A, Collins FS, Mohlke KL, Bergman RN, Tuomilehto J, Boehm BO, Gieger C, Hveem K, Cauchi S, Froguel P, Baldassarre D, Tremoli E, Humphries SE, Saleheen D, Danesh J, Ingelsson E, Ripatti S, Salomaa V, Erbel R, Jöckel K-H, Moebus S, Peters A, Illig T, de Faire U, Hamsten A, Morris AD, Donnelly PJ, Frayling TM, Hattersley AT, Boerwinkle E, Melander O, Kathiresan S, Nilsson PM, Deloukas P, Thorsteinsdottir U, Groop LC, Stefansson K, Hu F, Pankow JS, Dupuis J, Meigs JB, Altshuler D, Boehnke M Wellcome Trust Case Control Consortium, Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) Investigators, Genetic Investigation of ANthropometric Traits (GIANT) Consortium, Asian Genetic Epidemiology Network–Type 2 Diabetes (AGEN-T2D) Consortium, South Asian Type 2 Diabetes (SAT2D) Consortium, Mark I McCarthy for the DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malandrino N, Smith RJ. Personalized medicine in diabetes. Clin Chem. 2011;57:231–240. doi: 10.1373/clinchem.2010.156901. [DOI] [PubMed] [Google Scholar]
- 12.Muller G. Personalized prognosis and diagnosis of type 2 diabetes—Vision or fiction? Pharmacology. 2010;85:168–187. doi: 10.1159/000283780. [DOI] [PubMed] [Google Scholar]
- 13.Ng MCY, Shriner D, Chen BH, Li J, Chen W-M, Guo X, Liu J, Bielinski SJ, Yanek LR, Nalls MA, Comeau ME, Rasmussen-Torvik LJ, Jensen RA, Evans DS, Sun YV, An P, Patel SR, Lu Y, Long J, Armstrong LL, Wagenknecht L, Yang L, Snively BM, Palmer ND, Mudgal P, Langefeld CD, Keene KL, Freedman BI, Mychaleckyj JC, Nayak U, Raffel LJ, Goodarzi MO, Chen Y-DI, Taylor HA, Jr, Correa A, Sims M, Couper D, Pankow JS, Boerwinkle E, Adeyemo A, Doumatey A, Chen G, Mathias RA, Vaidya D, Singleton AB, Zonderman AB, Igo RP, Jr, Sedor JR, Kabagambe EK, Siscovick DS, McKnight B, Rice K, Liu Y, Hsueh W-C, Zhao W, Bielak LF, Kraja A, Province MA, Bottinger EP, Gottesman O, Cai Q, Zheng W, Blot WJ, Lowe WL, Pacheco JA, Crawford DC, Grundberg E, Rich SS, Hayes MG, Shu X-O, Loos RJF, Borecki IB, Peyser PA, Cummings SR, Psaty BM, Fornage M, Iyengar SK, Evans MK, Becker DM, Linda Kao WH, Wilson JG, Rotter JI, Sale MM, Liu S, Rotimi CN, Bowden DW eMERGE Consortium; DIAGRAM Consortium, MuTHER Consortium, FIND Consortium, MEta-analysis of type 2 DIabetes in African Americans Consortium. Meta-analysis of genome-wide association studies in African Americans provides insights into the genetic architecture of type 2 diabetes. PLOS Genet. 2014;10:e1004517. doi: 10.1371/journal.pgen.1004517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen R, Corona E, Sikora M, Dudley JT, Morgan AA, Moreno-Estrada A, Nilsen GB, Ruau D, Lincoln SE, Bustamante CD, Butte AJ. Type 2 diabetes risk alleles demonstrate extreme directional differentiation among human populations, compared to other diseases. PLOS Genet. 2012;8:e1002621. doi: 10.1371/journal.pgen.1002621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li L, Dudley J. Mining topological patterns in electronic medical records for clinical population discovery. Proceedings of 2nd International Workshop on Pattern Recognition for Healthcare Analytics; Stockholm, Sweden: International Conference for Pattern Recognition; 2014. [Google Scholar]
- 16.Wei W-Q, Leibson CL, Ransom JE, Kho AN, Caraballo PJ, Chai HS, Yawn BP, Pacheco JA, Chute CG. Impact of data fragmentation across healthcare centers on the accuracy of a high-throughput clinical phenotyping algorithm for specifying subjects with type 2 diabetes mellitus. J Am Med Inform Assoc. 2012;19:219–224. doi: 10.1136/amiajnl-2011-000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kho AN, Hayes MG, Rasmussen-Torvik L, Pacheco JA, Thompson WK, Armstrong LL, Denny JC, Peissig PL, Miller AW, Wei W-Q, Bielinski SJ, Chute CG, Leibson CL, Jarvik GP, Crosslin DR, Carlson CS, Newton KM, Wolf WA, Chisholm RL, Lowe WL. Use of diverse electronic medical record systems to identify genetic risk for type 2 diabetes within a genome-wide association study. J Am Med Inform Assoc. 2012;19:212–218. doi: 10.1136/amiajnl-2011-000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cowen ME, Dusseau DJ, Toth BG, Guisinger C, Zodet MW, Shyr Y. Casemix adjustment of managed care claims data using the clinical classification for health policy research method. Med Care. 1998;36:1108–1113. doi: 10.1097/00005650-199807000-00016. [DOI] [PubMed] [Google Scholar]
- 19.Glicksberg BS, Li L, Cheng W-Y, Shameer K, Hakenberg J, Castellanos R, Ma M, Shi L, Shah H, Dudley JT, Chen R. An integrative pipeline for multi-modal discovery of disease relationships. Pac Symp Biocomput. 2015:407–418. [PMC free article] [PubMed] [Google Scholar]
- 20.Aubertin J. Developmental aspects of diabetic retinopathy (retinographic study) Diabete. 1965;13:105–113. [PubMed] [Google Scholar]
- 21.DeFuria J, Belkina AC, Jagannathan-Bogdan M, Snyder-Cappione J, Carr JD, Nersesova YR, Markham D, Strissel KJ, Watkins AA, Zhu M, Allen J, Bouchard J, Toraldo G, Jasuja R, Obin MS, McDonnell ME, Apovian C, Denis GV, Nikolajczyk BS. B cells promote inflammation in obesity and type 2 diabetes through regulation of T-cell function and an inflammatory cytokine profile. Proc Natl Acad Sci USA. 2013;110:5133–5138. doi: 10.1073/pnas.1215840110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raile K, Galler A, Hofer S, Herbst A, Dunstheimer D, Busch P, Holl RW. Diabetic nephropathy in 27,805 children, adolescents, and adults with type 1 diabetes: Effect of diabetes duration, A1C, hypertension, dyslipidemia, diabetes onset, and sex. Diabetes Care. 2007;30:2523–2528. doi: 10.2337/dc07-0282. [DOI] [PubMed] [Google Scholar]
- 23.Wang C-S, Chang T-T, Yao W-J, Wang S-T, Chou P. Impact of increasing alanine aminotransferase levels within normal range on incident diabetes. J Formos Med Assoc. 2012;111:201–208. doi: 10.1016/j.jfma.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 24.Krolewski AS, Warram JH, Freire MBS. Epidemiology of late diabetic complications. A basis for the development and evaluation of preventive programs. Endocrinol Metab Clin North Am. 1996;25:217–242. doi: 10.1016/s0889-8529(05)70322-4. [DOI] [PubMed] [Google Scholar]
- 25.Chen H, Charlat O, Tartaglia LA, Woolf EA, Weng X, Ellis SJ, Lakey ND, Culpepper J, Moore KJ, Breitbart RE, Duyk GM, Tepper RI, Morgenstern JP. Evidence that the diabetes gene encodes the leptin receptor: Identification of a mutation in the leptin receptor gene in db/db mice. Cell. 1996;84:491–495. doi: 10.1016/s0092-8674(00)81294-5. [DOI] [PubMed] [Google Scholar]
- 26.Hansen TK, Gall M-A, Tarnow L, Thiel S, Stehouwer CD, Schalkwijk CG, Parving H-H, Flyvbjerg A. Mannose-binding lectin and mortality in type 2 diabetes. Arch Intern Med. 2006;166:2007–2013. doi: 10.1001/archinte.166.18.2007. [DOI] [PubMed] [Google Scholar]
- 27.Klein OL, Meltzer D, Carnethon M, Krishnan JA. Type II diabetes mellitus is associated with decreased measures of lung function in a clinical setting. Respir Med. 2011;105:1095–1098. doi: 10.1016/j.rmed.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 28.Song Y, Wang L, Pittas AG, Del Gobbo LC, Zhang C, Manson JE, Hu FB. Blood 25-hydroxy vitamin D levels and incident type 2 diabetes: A meta-analysis of prospective studies. Diabetes Care. 2013;36:1422–1428. doi: 10.2337/dc12-0962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Asayama K, Sandhir R, Sheikh FG, Hayashibe H, Nakane T, Singh I. Increased peroxisomal fatty acid β-oxidation and enhanced expression of peroxisome proliferator-activated receptor-α in diabetic rat liver. Mol Cell Biochem. 1999;194:227–234. doi: 10.1023/a:1006930513476. [DOI] [PubMed] [Google Scholar]
- 31.Kakuda H, Shiroishi K, Hosono K, Ichihara S. Construction of Pta-Ack pathway deletion mutants of Escherichia coli and characteristic growth profiles of the mutants in a rich medium. Biosci Biotechnol Biochem. 1994;58:2232–2235. doi: 10.1271/bbb.58.2232. [DOI] [PubMed] [Google Scholar]
- 32.Diaz-Ricci JC, Regan L, Bailey JE. Effect of alteration of the acetic acid synthesis pathway on the fermentation pattern of Escherichia coli. Biotechnol Bioeng. 1991;38:1318–1324. doi: 10.1002/bit.260381109. [DOI] [PubMed] [Google Scholar]
- 33.Person S, Snipes W, Krasin F. Mutation production from tritium decay: A local effect for [3H]2a-adenosine and [3H]6-thymidine decays. Mutat Res. 1976;34:327–332. doi: 10.1016/0027-5107(76)90137-8. [DOI] [PubMed] [Google Scholar]
- 34.McQuaid TS, Saleh MC, Joseph JW, Gyulkhandanyan A, Manning-Fox JE, MacLellan JD, Wheeler MB, Chan CB. cAMP-mediated signaling normalizes glucose-stimulated insulin secretion in uncoupling protein-2 overexpressing β-cells. J Endocrinol. 2006;190:669–680. doi: 10.1677/joe.1.06723. [DOI] [PubMed] [Google Scholar]
- 35.Tak E, Ridyard D, Badulak A, Giebler A, Shabeka U, Werner T, Clambey E, Moldovan R, Zimmerman MA, Eltzschig HK, Grenz A. Protective role for netrin-1 during diabetic nephropathy. J Mol Med. 2013;91:1071–1080. doi: 10.1007/s00109-013-1041-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramsey DJ, Ripps H, Qian H. Streptozotocin-induced diabetes modulates GABA receptor activity of rat retinal neurons. Exp Eye Res. 2007;85:413–422. doi: 10.1016/j.exer.2007.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramsey DJ, Ripps H, Qian H. An electrophysiological study of retinal function in the diabetic female rat. Invest Ophthalmol Vis Sci. 2006;47:5116–5124. doi: 10.1167/iovs.06-0364. [DOI] [PubMed] [Google Scholar]
- 38.Kaushansky K. Molecular mechanisms of thrombopoietin signaling. J Thromb Haemost. 2009;7(Suppl 1):235–238. doi: 10.1111/j.1538-7836.2009.03419.x. [DOI] [PubMed] [Google Scholar]
- 39.Lupia E, Goffi A, Bosco O, Montrucchio G. Thrombopoietin as biomarker and mediator of cardiovascular damage in critical diseases. Mediators Inflamm. 2012;2012:390892. doi: 10.1155/2012/390892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Şenaran H, Ileri M, Altinbaş A, Koşar A, Yetkin E, Oztürk M, Karaaslan Y, Kirazli Ş. Thrombopoietin and mean platelet volume in coronary artery disease. Clin Cardiol. 2001;24:405–408. doi: 10.1002/clc.4960240511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schramm NL, McDonald MP, Limbird LE. The α2A-adrenergic receptor plays a protective role in mouse behavioral models of depression and anxiety. J Neurosci. 2001;21:4875–4882. doi: 10.1523/JNEUROSCI.21-13-04875.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kable JW, Murrin LC, Bylund DB. In vivo gene modification elucidates subtype-specific functions of α2-adrenergic receptors. J Pharmacol Exp Ther. 2000;293:1–7. [PubMed] [Google Scholar]
- 43.Linden DJ, Connor JA. Long-term synaptic depression. Annu Rev Neurosci. 1995;18:319–357. doi: 10.1146/annurev.ne.18.030195.001535. [DOI] [PubMed] [Google Scholar]
- 44.Mantamadiotis T, Lemberger T, Bleckmann SC, Kern H, Kretz O, Martin Villalba A, Tronche F, Kellendonk C, Gau D, Kapfhammer J, Otto C, Schmid W, Schütz G. Disruption of CREB function in brain leads to neurodegeneration. Nat Genet. 2002;31:47–54. doi: 10.1038/ng882. [DOI] [PubMed] [Google Scholar]
- 45.Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- 46.Ribeiro FM, Paquet M, Cregan SP, Ferguson SSG. Group I metabotropic glutamate receptor signalling and its implication in neurological disease. CNS Neurol Disord Drug Targets. 2010;9:574–595. doi: 10.2174/187152710793361612. [DOI] [PubMed] [Google Scholar]
- 47.Wolf G. Cell cycle regulation in diabetic nephropathy. Kidney Int Suppl. 2000;77:S59–S66. doi: 10.1046/j.1523-1755.2000.07710.x. [DOI] [PubMed] [Google Scholar]
- 48.Berkman J, Rifkin H. Unilateral nodular diabetic glomerulosclerosis (Kimmelstiel-Wilson): Report of a case. Metabolism. 1973;22:715–722. doi: 10.1016/0026-0495(73)90243-6. [DOI] [PubMed] [Google Scholar]
- 49.Denny JC, Bastarache L, Ritchie MD, Carroll RJ, Zink R, Mosley JD, Field JR, Pulley JM, Ramirez AH, Bowton E, Basford MA, Carrell DS, Peissig PL, Kho AN, Pacheco JA, Rasmussen LV, Crosslin DR, Crane PK, Pathak J, Bielinski SJ, Pendergrass SA, Xu H, Hindorff LA, Li R, Manolio TA, Chute CG, Chisholm RL, Larson EB, Jarvik GP, Brilliant MH, McCarty CA, Kullo IJ, Haines JL, Crawford DC, Masys DR, Roden DM. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013;31:1102–1110. doi: 10.1038/nbt.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kvale MN, Hesselson S, Hoffmann TJ, Cao Y, Chan D, Connell S, Croen LA, Dispensa BP, Eshragh J, Finn A, Gollub J, Iribarren C, Jorgenson E, Kushi LH, Lao R, Lu Y, Ludwig D, Mathauda GK, McGuire WB, Mei G, Miles S, Mittman M, Patil M, Quesenberry CP, Jr, Ranatunga D, Rowell S, Sadler M, Sakoda LC, Shapero M, Shen L, Shenoy T, Smethurst D, Somkin CP, Van Den Eeden SK, Walter L, Wan E, Webster T, Whitmer RA, Wong S, Zau C, Zhan Y, Schaefer C, Kwok P-Y, Risch N. Genotyping informatics and quality control for 100,000 subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA) Cohort. Genetics. 2015;200:1051–1060. doi: 10.1534/genetics.115.178905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Denny JC, Ritchie MD, Basford MA, Pulley JM, Bastarache L, Brown-Gentry K, Wang D, Masys DR, Roden DM, Crawford DC. PheWAS: Demonstrating the feasibility of a phenome-wide scan to discover gene–disease associations. Bioinformatics. 2010;26:1205–1210. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Naresh VV, Reddy ALK, Sivaramakrishna G, Sharma PVGK, Vardhan RV, Kumar VS. Angiotensin converting enzyme gene polymorphism in type II diabetics with nephropathy. Indian J Nephrol. 2009;19:145–148. doi: 10.4103/0971-4065.59335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wiwanitkit V. Angiotensin-converting enzyme gene polymorphism: I and D alleles from some different countries. Clin Appl Thromb Hemost. 2004;10:179–182. doi: 10.1177/107602960401000209. [DOI] [PubMed] [Google Scholar]
- 54.Würtz M, Hvas A-M, Kristensen SD, Grove EL. Platelet aggregation is dependent on platelet count in patients with coronary artery disease. Thromb Res. 2012;129:56–61. doi: 10.1016/j.thromres.2011.08.019. [DOI] [PubMed] [Google Scholar]
- 55.Vijan S, Hofer TP, Hayward RA. Estimated benefits of glycemic control in microvascular complications in type 2 diabetes. Ann Intern Med. 1997;127:788–795. doi: 10.7326/0003-4819-127-9-199711010-00003. [DOI] [PubMed] [Google Scholar]
- 56.Giovannucci E, Harlan DM, Archer MC, Bergenstal RM, Gapstur SM, Habel LA, Pollak M, Regensteiner JG, Yee D. Diabetes and cancer: A consensus report. Diabetes Care. 2010;33:1674–1685. doi: 10.2337/dc10-0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cannata D, Fierz Y, Vijayakumar A, LeRoith D. Type 2 diabetes and cancer: What is the connection? Mt Sinai J Med. 2010;77:197–213. doi: 10.1002/msj.20167. [DOI] [PubMed] [Google Scholar]
- 58.Grohol J. Top 25 Psychiatric Medication Prescriptions for 2013. Psych Central; Newburyport, MA: 2014. [Google Scholar]
- 59.Han JH, Crane HM, Bellamy SL, Frank I, Cardillo S, Bisson GP Centers for AIDS Research Network of Integrated Clinical Systems. HIV infection and glycemic response to newly initiated diabetic medical therapy. AIDS. 2012;26:2087–2095. doi: 10.1097/QAD.0b013e328359a8e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tibshirani R. Regression shrinkage and selection via the lasso. J R Stat Soc Ser B. 1996;58:267–288. [Google Scholar]
- 61.Luciano M, Huffman JE, Arias-Vásquez A, Vinkhuyzen AA, Middeldorp CM, Giegling I, Payton A, Davies G, Zgaga L, Janzing J, Ke X, Galesloot T, Hartmann AM, Ollier W, Tenesa A, Hayward C, Verhagen M, Montgomery GW, Hottenga J-J, Konte B, Starr JM, Vitart V, Vos PE, Madden PAF, Willemsen G, Konnerth H, Horan MA, Porteous DJ, Campbell H, Vermeulen SH, Heath AC, Wright A, Polasek O, Kovacevic SB, Hastie ND, Franke B, Boomsma DI, Martin NG, Rujescu D, Wilson JF, Buitelaar J, Pendleton N, Rudan I, Deary IJ. Genome-wide association uncovers shared genetic effects among personality traits and mood states. Am J Med Genet B Neuropsychiatr Genet. 2012;159B:684–695. doi: 10.1002/ajmg.b.32072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium. Ripke S, Wray NR, Lewis CM, Hamilton SP, Weissman MM, Breen G, Byrne EM, Blackwood DH, Boomsma DI, Cichon S, Heath AC, Holsboer F, Lucae S, Madden PA, Martin NG, McGuffin P, Muglia P, Noethen MM, Penninx BP, Pergadia ML, Potash JB, Rietschel M, Lin D, Müller-Myhsok B, Shi J, Steinberg S, Grabe HJ, Lichtenstein P, Magnusson P, Perlis RH, Preisig M, Smoller JW, Stefansson K, Uher R, Kutalik Z, Tansey KE, Teumer A, Viktorin A, Barnes MR, Bettecken T, Binder EB, Breuer R, Castro VM, Churchill SE, Coryell WH, Craddock N, Craig IW, Czamara D, De Geus EJ, Degenhardt F, Farmer AE, Fava M, Frank J, Gainer VS, Gallagher PJ, Gordon SD, Goryachev S, Gross M, Guipponi M, Henders AK, Herms S, Hickie IB, Hoefels S, Hoogendijk W, Hottenga JJ, Iosifescu DV, Ising M, Jones I, Jones L, Jung-Ying T, Knowles JA, Kohane IS, Kohli MA, Korszun A, Landen M, Lawson WB, Lewis G, Macintyre D, Maier W, Mattheisen M, McGrath PJ, McIntosh A, McLean A, Middeldorp CM, Middleton L, Montgomery GM, Murphy SN, Nauck M, Nolen WA, Nyholt DR, O’Donovan M, Oskarsson H, Pedersen N, Scheftner WA, Schulz A, Schulze TG, Shyn SI, Sigurdsson E, Slager SL, Smit JH, Stefansson H, Steffens M, Thorgeirsson T, Tozzi F, Treutlein J, Uhr M, van den Oord EJ, Van Grootheest G, Volzke H, Weilburg JB, Willemsen G, Zitman FG, Neale B, Daly M, Levinson DF, Sullivan PF. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry. 2013;18:497–511. doi: 10.1038/mp.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McCormack M, Urban TJ, Shianna KV, Walley N, Pandolfo M, Depondt C, Chaila E, O’Conner GD, Kasperavičiūtė D, Radtke RA, Heinzen EL, Sisodiya SM, Delanty N, Cavalleri GL. Genome-wide mapping for clinically relevant predictors of lamotrigine- and phenytoin-induced hypersensitivity reactions. Pharmacogenomics. 2012;13:399–405. doi: 10.2217/pgs.11.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Postolache TT, Komarow H, Tonelli LH. Allergy: A risk factor for suicide? Curr Treat Options Neurol. 2008;10:363–376. doi: 10.1007/s11940-008-0039-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Timonen M, Jokelainen J, Hakko H, Silvennoinen-Kassinen S, Meyer-Rochow VB, Herva A, Rasanen P. Atopy and depression: Results from the Northern Finland 1966 Birth Cohort Study. Mol Psychiatry. 2003;8:738–744. doi: 10.1038/sj.mp.4001274. [DOI] [PubMed] [Google Scholar]
- 66.Timonen M, Jokelainen J, Herva A, Zitting P, Meyer-Rochow VB, Räsänen P. Presence of atopy in first-degree relatives as a predictor of a female proband’s depression: Results from the Northern Finland 1966 Birth Cohort. J Allergy Clin Immunol. 2003;111:1249–1254. doi: 10.1067/mai.2003.1546. [DOI] [PubMed] [Google Scholar]
- 67.Wamboldt MZ, Hewitt JK, Schmitz S, Wamboldt FS, Räsänen M, Koskenvuo M, Romanov K, Varjonen J, Kaprio J. Familial association between allergic disorders and depression in adult Finnish twins. Am J Med Genet. 2000;96:146–153. doi: 10.1002/(sici)1096-8628(20000403)96:2<146::aid-ajmg4>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 68.Espeland MA, Glick HA, Bertoni A, Brancati FL, Bray GA, Clark JM, Curtis JM, Egan C, Evans M, Foreyt JP, Ghazarian S, Gregg EW, Hazuda HP, Hill JO, Hire D, Horton ES, Hubbard VS, Jakicic JM, Jeffery RW, Johnson KC, Kahn SE, Killean T, Kitabchi AE, Knowler WC, Kriska A, Lewis CE, Miller M, Montez MG, Murillo A, Nathan DM, Nyenwe E, Patricio J, Peters AL, Pi-Sunyer X, Pownall H, Redmon JB, Rushing J, Ryan DH, Safford M, Tsai AG, Wadden TA, Wing RR, Yanovski SZ, Zhang P Look AHEAD Research Group. Impact of an intensive lifestyle intervention on use and cost of medical services among overweight and obese adults with type 2 diabetes: The action for health in diabetes. Diabetes Care. 2014;37:2548–2556. doi: 10.2337/dc14-0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Look AHEAD Research Group. Wing RR, Bolin P, Brancati FL, Bray GA, Clark JM, Coday M, Crow RS, Curtis JM, Egan CM, Espeland MA, Evans M, Foreyt JP, Ghazarian S, Gregg EW, Harrison B, Hazuda HP, Hill JO, Horton ES, Hubbard VS, Jakicic JM, Jeffery RW, Johnson KC, Kahn SE, Kitabchi AE, Knowler WC, Lewis CE, Maschak-Carey BJ, Montez MG, Murillo A, Nathan DM, Patricio J, Peters A, Pi-Sunyer X, Pownall H, Reboussin D, Regensteiner JG, Rickman AD, Ryan DH, Safford M, Wadden TA, Wagenknecht LE, West DS, Williamson DF, Yanovski SZ. Cardiovascular effects of intensive lifestyle intervention in type 2 diabetes. N Engl J Med. 2013;369:145–154. doi: 10.1056/NEJMoa1212914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Newton KM, Peissig PL, Kho AN, Bielinski SJ, Berg RL, Choudhary V, Basford M, Chute CG, Kullo IJ, Li R, Pacheco JA, Rasmussen LV, Spangler L, Denny JC. Validation of electronic medical record-based phenotyping algorithms: Results and lessons learned from the eMERGE network. J Am Med Inform Assoc. 2013;20:e147–e154. doi: 10.1136/amiajnl-2012-000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, Karczewski KJ, Park J, Hitz BC, Weng S, Cherry JM, Snyder M. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schaub MA, Boyle AP, Kundaje A, Batzoglou S, Snyder M. Linking disease associations with regulatory information in the human genome. Genome Res. 2012;22:1748–1759. doi: 10.1101/gr.136127.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLOS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Goldstein JI, Crenshaw A, Carey J, Grant GB, Maguire J, Fromer M, O’Dushlaine C, Moran JL, Chambert K, Stevens C, Sklar P, Hultman CM, Purcell S, McCarroll SA, Sullivan PF, Daly MJ, Neale BM Swedish Schizophrenia Consortium, ARRA Autism Sequencing Consortium. zCall: A rare variant caller for array-based genotyping: Genetics and population analysis. Bioinformatics. 2012;28:2543–2545. doi: 10.1093/bioinformatics/bts479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.O’Connell J, Gurdasani D, Delaneau O, Pirastu N, Ulivi S, Cocca M, Traglia M, Huang J, Huffman JE, Rudan I, McQuillan R, Fraser RM, Campbell H, Polasek O, Asiki G, Ekoru K, Hayward C, Wright AF, Vitart V, Navarro P, Zagury J-F, Wilson JF, Toniolo D, Gasparini P, Soranzo N, Sandhu MS, Marchini J. A general approach for haplotype phasing across the full spectrum of relatedness. PLOS Genet. 2014;10:e1004234. doi: 10.1371/journal.pgen.1004234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.1000 Genomes Project Consortium. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Reumers J, Maurer-Stroh S, Schymkowitz J, Rousseau F. SNPeffect v2.0: A new step in investigating the molecular phenotypic effects of human non-synonymous SNPs. Bioinformatics. 2006;22:2183–2185. doi: 10.1093/bioinformatics/btl348. [DOI] [PubMed] [Google Scholar]
- 78.Chen R, Li L, Butte AJ. AILUN: Reannotating gene expression data automatically. Nat Methods. 2007;4:879. doi: 10.1038/nmeth1107-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Carlsson G. Topology and data. Bull Amer Math Soc. 2009;46:255–308. [Google Scholar]
- 80.Lum PY, Singh G, Lehman A, Ishkanov T, Vejdemo-Johansson M, Alagappan M, Carlsson J, Carlsson G. Extracting insights from the shape of complex data using topology. Sci Rep. 2013;3:1236. doi: 10.1038/srep01236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hurvich CM, Tsai C-L. A corrected Akaike information criterion for vector autoregressive model selection. J Time Ser Anal. 2008;14:271–279. [Google Scholar]
- 83.Ihaka R, Gentleman R. R: A language for data analysis and graphics. J Comput Graph Stat. 1996;5:299–314. [Google Scholar]