Abstract
Xanthomonas citri subsp. citri causes citrus canker disease, which is characterized by the formation of water-soaked lesions, white or yellow spongy pustules and brown corky canker. In this work, we report the contribution of extracellular endoglucanase to canker development during infection. The ectopic expression of nine putative cellulases in Escherichia coli indicated that two endoglucanases, BglC3 and EngXCA, show carboxymethyl cellulase activity. Both bglC3 and engXCA genes were transcribed in X. citri subsp. citri, however, only BglC3 protein was detected outside the cell in western blot analysis. The deletion of bglC3 gene resulted in complete loss of extracellular carboxymethyl cellulase activity and delayed the onset of canker symptoms in both infiltration- and wound-inoculation assays. When growing in plant tissue, the cell density of bglC3 mutant was lower than that of the wild type. Our data demonstrated that BglC3 is an extracellular endoglucanase required for the full virulence of X. citri subsp. citri.
Introduction
Plant phytobacteria produce several extracellular enzymes that digest host plant primary cell walls, thus playing roles in bacterial pathogenicity. Extracellular enzymes have been well characterized in Pectobacterium and Dickeya species because of their essential roles in soft rot symptom production [1]. The Pectobacterium and Dickeya species produce pectinase, cellulase and protease, leading to the maceration of plant tissues [2,3]. The production of these extracellular enzymes is controlled by a few transcriptional factors. A two-component system, GacA/GacS, has a positive effect on extracellular enzyme activities, while a lysR-like protein HexA and an IclI-like protein KdgR negatively regulate cellulase production [4–6]. In Xanthomonads, the endoglucanase engXCA and engXCB double mutant of X. campestris pv. campestris show a five-fold reduction in cellulose-degradation suggesting additional candidates contribute to cellulase production [7]. A mutation in the engXCB homolog in X. oryzae pv. oryzae results in an 87% reduction in leaf lesion lengths, representing a remarkable reduction in virulence [8].
In most bacteria, cellulases are enzyme complexes containing roughly five different enzymatic subunits namely: endocellulases, exocellulases, cellobiases, oxidative cellulases and cellulose phosphorylases. Among the five subunits, only the endocellulases and cellobiases participate in the actual hydrolysis of cellulose and some related polysaccharides [9]. The endoglucanases (EC 3.2.1.4) break down internal β-1,4-glycosidic bonds, causing the exposure of cellulose polysaccharide chains. Exo-1,4-cellobiohydrolases (EC 3.2.1.91) and β-glucosidases (EC 3.2.1.21) degrade oligos and dimers, releasing mainly cellobiose and glucose, respectively [9]. For plant bacterial pathogens, researchers have mainly focused on the cellulase subunits, which are secreted outside the cells and contribute to pathogenicity [3,8].
X. citri subsp. citri is the causal agent of citrus canker type A, a widespread and severe bacterial disease attacking all citrus species [10]. This pathogen executes several mechanisms to colonize host plants. In addition to the type III secretion system that delivers pathogenicity effector proteins [11–13], the type II secretion system (T2SS) is proposed to be involved in canker development [14–16]. A mutation in the T2SS blocks extracellular enzyme secretions, including those of cellulase, protease and amylase [16]. However, the exact role of cellulase during infection has not been experimentally confirmed. In this work, we identified bglC3 as an extracellular endoglucanase in X. citri subsp. citri. The deletion of bglC3 gene led to complete loss of extracellular carboxymethyl cellulase activity, reduced bacterial growth in plant tissues and delayed canker symptom emergence.
Materials and Methods
Bacterial strains, plasmids and culture conditions
The bacterial strains and plasmids used in this study are listed in S1 Table. The X. citri subsp. citri strain 29–1 (Xcc 29–1) was cultivated in nutrient broth medium or nutrient broth supplemented with 1.5% agar at 28°C [15]. Nutrient agar with or without 10% sucrose was used to cultivate X. citri. subsp. citri during mutant construction [17]. E. coli strains were cultured in Luria-Bertani medium at 37°C. Antibiotics were applied at the following concentrations: kanamycin (Km), 50 μg ml–1 and gentamycin (Gm), 10 μg ml–1.
Extracellular carboxymethyl cellulase activity analysis
Carboxymethyl cellulose (0.5%) was incorporated into agar plates to test carboxymethyl cellulase activity. For ectopic expression, nine cellulase genes were amplified from Xcc 29–1 gDNA and cloned into pET41a(+) (S1 and S2 Tables). The recombinant constructs were transformed into BL21(DE3) to test the extracellular cellulase activity on Luria-Bertani solid medium supplemented with 1.0 mM isopropyl-β-D-thiogalactopyranoside [18]. The cellulase activity of X. citri subsp. citri was tested on nutrient agar medium. For each strain, 1.5 μl cells (OD600 1.0) were spotted on plates. The halos surrounding the colonies indicated the extracellular cellulase activity [17]. The hydrolytic activities were calculated by subtracting colony diameters from the halos. The tests were repeated three times, and the data shown in the figures are mean values.
Construction of non-polar deletion mutants
The non-polar mutants of ΔbglC3, ΔengXCA and the double mutant ΔengXCAΔbglC3 were constructed using the suicide vector pKMS1 through a double homologous recombination strategy [17]. The primer pairs 0028.1.F/0028.1.R and 0028.2.F/0028.2.R were used to PCR amplify two DNA fragments flanking bglC3 gene (S2 Table) while the primer pairs 0612.1.F/0612.1.R and 0612.2.F/0612.2.R were used to amplify the two DNA fragments flanking engXCA (S2 Table). For each gene, two flanking fragments were ligated together and inserted into pKMS1 vector at the BamHI and SacI sites, resulting in pKMS-0028 and pKMS-0612, respectively (S1 Table). Each plasmid was individually introduced into wild-type Xcc 29–1 by electroporation. Integration events were selected on nutrient agar without 10% sucrose containing kanamycin and then transferred to nutrient agar supplemented with 10% sucrose to select for crossover events that resulted in the loss of the sacB gene [17]. The engXCA bglC3 double mutant was then generated by introducing the recombinant plasmid pKMS-0028 into the mutant ΔengXCA genetic background. Sucrose-resistant colonies were screened by using PCR, and positive colonies containing the desired deletion were confirmed by sequencing [17].
Complementation analysis
The DNA fragments containing the entire bglC3 or engXCA genes were amplified using PCR with primers C0028.F/C0028.R and C0612.F/C0612.R, respectively (S2 Table). To constitutively express the genes in the mutants, the promoter region of wxacO was cloned into the vector pBBR1MCS-5 at the KpnI and XhoI sites using the primers wxaco.p.F and wxaco.p.R (S2 Table) [19]. The bglC3 gene was inserted into the XhoI and HindIII sites, resulting in pC0028. The engXCA gene was inserted into the XbaI and SacI sites, resulting in pC0612. For the double mutant, the bglC3 and engXCA genes were inserted into pBBR1MCS-5 together, resulting in pC0028-0612. The recombinant constructs were introduced into the corresponding mutant strains for complementation analysis by electroporation.
Pathogenicity assays
The X. citri. subsp. citri strains were cultured and re-suspended in sterile water to a final concentration of 108 CFU ml–1 (OD600 0.3). Bacterial suspensions were injected into fully expanded citrus leaves with needleless syringes [19]. In parallel experiments for each strain, five wounds were produced near the inoculation sites with a needle. The cell suspensions were then placed on the wounds. To maintain a humidity level that facilitates bacterial infection, ~3.5 cm2 of plastic wrap was used to cover the inoculation zones for 2 d. Disease symptoms were scored and photographed 4 and 10 d post inoculation. Each test was repeated at least three times.
RNA isolation and RT-PCR
To measure the transcription of cellulase genes, RNA was extracted from Xcc 29–1 cells grown in NA medium and in citrus plant tissue. Cell samples were prepared as described previously [15]. The total RNA was extracted from wild-type Xcc 29–1 using Trizol reagent as recommended by the manufacturer (Invitrogen, Shanghai, China) [15]. Potential traces of genomic DNA were removed using RNase-free DNase I (Takara, Dalian, China) before first-strand cDNA synthesis. All primers used for RT-PCR are listed in S2 Table. The conditions for semi-quantitative RT-PCR were as follows: initial denaturation at 95°C for 30 s; 31 cycles of 95°C for 35 s, 52°C for 30 s, and 72°C for 35 s; followed by 72°C for 5 min. The expression of 16S rRNA was used as the internal control to verify whether there was significant variation of the cDNA level. RT-PCR experiments were performed in triplicate.
Western blot
To generate c-Myc-tagged fusion proteins, bglC3 and engXCA genes were PCR amplified using primers 0028.S.F/0028.S.R and 0612.S.F/0612.S.R, respectively. The PCR products were cloned into pUFR034Myc at the SacI and KpnI sites in-frame with a c-Myc epitope-encoding sequence, generating p0028Myc and p0612Myc (S1 Table) [20]. The western blot was performed following a previously described procedure [20].
Bacterial growth in citrus plants
The cultured X. citri subsp. citri cells were adjusted to a final concentration of 108 CFU ml-1, and infiltrated into citrus leaves [15]. At 2, 4 and 6 d post inoculation, 1.0 cm2 leaf planchets were cut using a cork borer. After being surface sterilized twice with 75% ethanol, the planchets were ground completely and diluted with 1 ml of sterile double-distilled water, and then plated to determine the cell number. The number of bacteria represented the approximate colony-forming units per cm2 of the leaf area. Standard deviation was calculated using colony counts from the three triplicate spots from each of the three samples per time point per inoculum [15]. Experiments were repeated three times.
Results
Hydrolysis activity of carboxymethyl cellulase from nine putative cellulases
The Xcc 29–1 genome has been completely sequenced (GenBank No. PRJNA193774). Like the first sequenced citrus canker A strain 306 from Brazil, Xcc 29–1 contains one chromosome and two plasmids. The nine putative cellulase genes, including five putative endoglucanases and four cellulases, were all localized in the chromosome (S3 Table). The three bglC genes (bglC1, bglC2 and bglC3) were closely linked in the chromosome. The endoglucanase Egl2 and EngXCA were encoded by XAC29_12820 and XAC29_03125, respectively. The later one is homologous with an extracellular cellulase in Pseudomonas syringae. XAC29_08905 gene product was an 81 kDa protein CelA. The remaining three cellulases included one degenerate cellulase, XAC29_01790, and two truncated cellulase precursors. All nine genes were found in the citrus canker type A strains 306 and Aw, as well as X. fuscans subsp. fuscans (S3 Table).
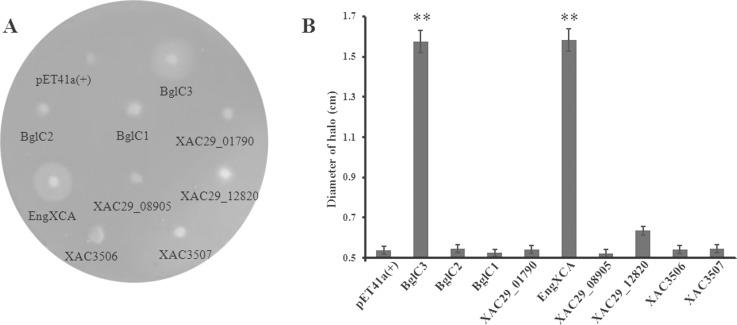
The E. coli BL21(DE3) possesses type II secretion system and does not have cellulase activity, which allowed us to characterize the putative extracellular cellulases by ectopic expression. All nine cellulase genes were cloned in-frame into the pET41a(+) vector, and their expression was induced in E. coli BL21(DE3) by isopropyl-β-D-thiogalactopyranoside (S1 Fig). BglC1, BglC2, XAC29_12820 and the four cellulases did not show extracellular cellulase activities. Their hydrolytic halos showed no distinct differences from that of the empty vector control. By contrast, clear and large hydrolysis halos were found surrounding the colonies expressing BglC3 and EngXCA. The halos were greater than 1.5 cm, which was nearly 3-fold greater than that of the empty vector control (Fig 1).
Fig 1. Ectopic expression of nine cellulases in Escherichia coli BL21(DE3).
(A) Degradation halos on Luria-Bertani plates supplemented with 0.5% carboxymethyl cellulose and 1.0 mM isopropyl-β-D-thiogalactopyranoside. (B) Diameters of the degradation halos. The tests were repeated three times; the data in the figure are the mean values. Two asterisks in the data column indicated significant differences at P = 0.01 by Student’s t test.
BglC3 is an extracellular carboxymethyl cellulase in X. citri subsp. citri
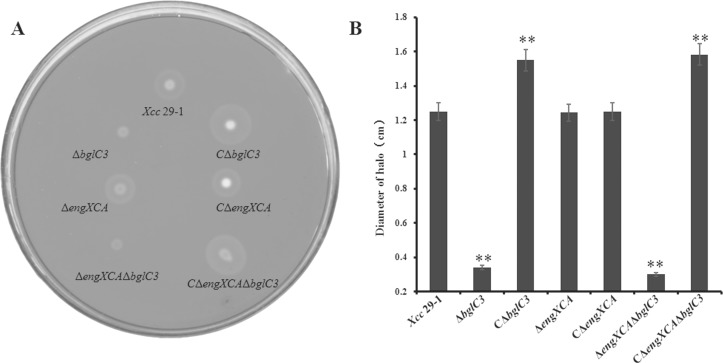
Because BglC3 and EngXCA showed carboxymethyl cellulose hydrolysis activities, we were interested in finding out whether they contributed to extracellular cellulase activities in X. citri subsp. citri. The deletion mutants ΔbglC3 and ΔengXCA were constructed using two-step homologous recombination. The deletions of the coding regions were confirmed using PCR, and the products were sequenced for verification (S2 Fig). On the nutrient agar plates supplemented with carboxymethyl cellulose, the extracellular cellulase activity of the mutant ΔbglC3 was completely impaired. No hydrolysis halos were formed around the bacterial colonies (Fig 2A). By contrast, the mutant ΔengXCA retained its extracellular cellulase activity. The diameters of hydrolysis halos were the same as those of the wild-type Xcc 29–1, suggesting that the EngXCA was not secreted outside the cells. To confirm this hypothesis, we also deleted bglC3 gene in the ΔengXCA genetic background to produce engXCA bglC3 double mutant (S2 Fig). As expected, ΔengXCAΔbglC3 double mutant did not show extracellular cellulase activity. For complementation analysis, bglC3 and engXCA were constitutively expressed under control of the wxac promoter. The complemented strain CΔengXCA, expressing the engXCA gene, did not have increased extracellular cellulase activity. Meanwhile, CΔbglC3 and CΔengXCAΔbglC3, expressing the bglC3 gene, restored the extracellular cellulase activity, and the diameter of their hydrolysis halos were 24% and 27% larger than those of the wild type, respectively (Fig 2B).
Fig 2. Testing the extracellulase activity of deficient mutants.
(A) Extracellular cellulase activity on a nutrient agar medium supplemented with carboxymethyl cellulose. (B) Degradation halo diameters. The tests were repeated three times; the data in the figure are the mean values. Two asterisks in the data column indicated significant differences at P = 0.01 by Student’s t test.
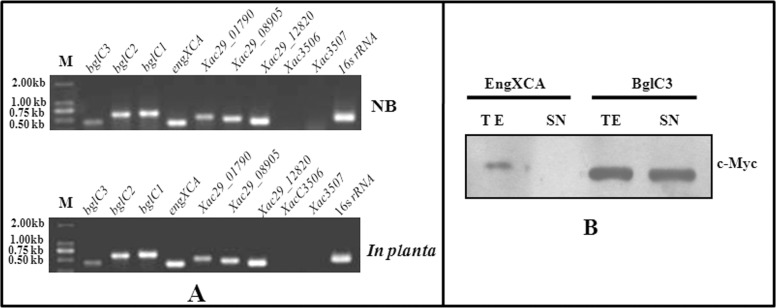
A semi-quantitative RT-PCR was used to detect the expression of nine cellulases in NB broth medium and in citrus plants. Except for two cellulase precursors, 7 genes were transcribed in cells either during culturing in NA broth medium or when growing in citrus plants (Fig 3A and 3B). To study the secretion traits, engXCA and bglC3 were expressed as C-terminal c-Myc epitope-tagged derivatives in plasmid pUFR034Myc and introduced into wild-type strain Xcc 29–1. Immunoblotting revealed that both endoglucanases were present in the total-cell extract (TE). In the culture supernatant (SN), BglC3 showed obvious hybridization bands, whereas no hybridization signal was found using engXCA samples (Fig 3C). This suggested that the engXCA gene was transcribed in X. citri subsp. citri, but the protein was not secreted out of the cells.
Fig 3. Expression of cellulase genes in X. citri subsp. citri.
(A) RT-PCR analysis of gene transcription in X. citri subsp. citri culturing in nutrient broth medium (NB) and growing in citrus. M, DL2000 marker. (B) Detection of BglC3 and EngXCA secretion using immunoblotting. Protein samples were analyzed by SDS-PAGE and immunoblotted using anti-c-Myc antibodies. TE, Total protein extract; SE, Culture supernatant.
BglC3 is required for full virulence in canker development
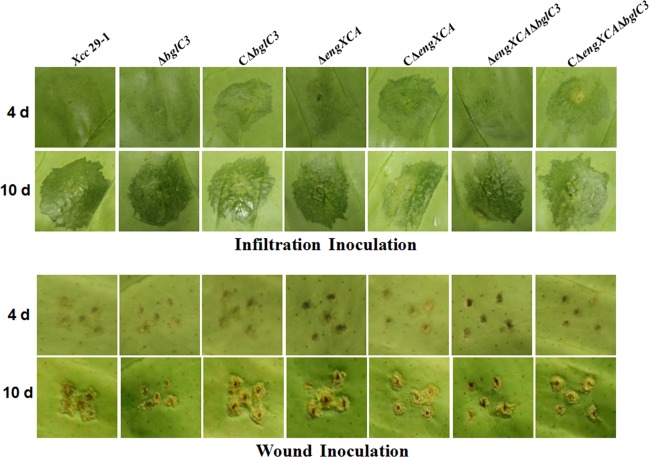
Infiltration- and wound-inoculation approaches were used to examine the pathogenicity of bglC3- and engXCA -deficient mutants. At 4 d post infiltration, water-soaked symptoms were clearly visible at the inoculation site of wild type, and slightly spongy pustules began to appear. The mutant ΔengXCA showed a similar phenotype. However, the ΔbglC3 and ΔengXCAΔbglC3 mutants caused slight water-soaked symptoms, and the symptoms caused by the double mutant were extremely weak. At 10 d post infiltration, wild type and ΔengXCA caused fully spongy pustules, while the ΔbglC3 and ΔengXCAΔbglC3 mutants maintained only water-soaked symptoms (Fig 4). In the wound-inoculation experiments, ΔbglC3 and ΔengXCAΔbglC3 mutants caused a delayed canker symptom similar with the phenotype in infiltration experiments. The complemented strains restored pathogenicity to wild type, indicating that the extracellular endoglucanase was required for optimum canker development during infection (Fig 4).
Fig 4. The pathogenicity of extracellular endoglucanase-deficient mutants in citrus plants.
(a) Symptoms in citrus plants in an infiltration-inoculation assay. (b) Symptoms in citrus plants in a wound-inoculation assay.
In planta growth of endoglucanase mutants
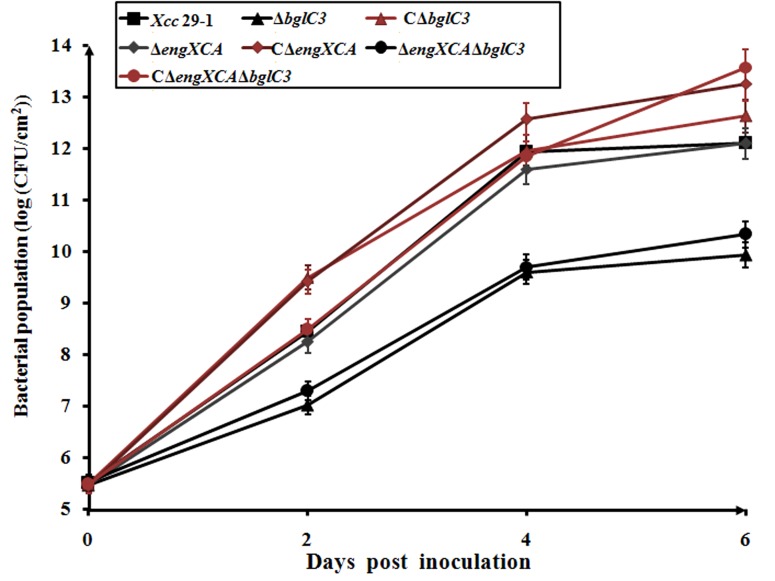
To determine whether extracellular endoglucanase was required for bacterial growth in plant tissues or not, the cultured cells were infiltrated into citrus leaves. At 2, 4 and 6 d post inoculation, 1 cm2 of leaves was punched and ground completely to release the X. citri subsp. citri cells. At 2 d post inoculation, the cell density of wild type had reached 108 colony-forming units per cm2. The deletion of endoglucanase bglC3 resulted in over a 10-fold reduction in cell numbers. At 4 and 6 d post inoculation, the cell densities of ΔbglC3 and ΔengXCAΔbglC3 were reduced 100- and 1000-fold, respectively. The constitutively expressed bglC3 gene in the mutant strains increased the cell density to slightly higher than wild type. By contrast, the deletion of the engXCA gene had no distinct effect on bacterial growth (Fig 5).
Fig 5. Bacterial growth in citrus tissue.
Leaf planchets were cut from each infiltration area at 2-d intervals post inoculation to analyze bacterial growth. The values shown are means of three technical repeats with standard deviations.
Discussion
In this work, we identified BglC3 and EngXCA as endoglucanases showing carboxymethyl cellulase activity. Although both genes were transcribed inside the cells, only BglC3 was externally secreted. The deletion mutation of the bglC3 gene resulted in complete loss of extracellular carboxymethyl hydrolysis activity, reduction in cell growth in citrus plants and delayed canker symptom emergence.
To colonize host plants, a number of plant pathogenic bacteria possess genes that encode cellulase complexes for cellulose hydrolysis, and thus destroy plant cell structures [2], [21–24]. However, only a few cellulase subunits are secreted out of the cell [7, 22, 25]. Among the nine cellulase candidates from Xcc 29–1, BglC3 and EngXCA showed carboxymethyl cellulose hydrolysis activity when ectopically expressed in E. coli BL21(DE3). The transcripts of bglC3 and engXCA were detected in RT-PCR experiments, but only BglC3 was secreted out of the cell. Furthermore, deletion of bglC3 resulted in a complete loss of extracellular carboxymethyl hydrolysis activity. It appeared that BglC3 was an extracellular endoglucanase facilitating bacterial infection in X. citri subsp. citri. However, the biological function of the engXCA gene remains unclear. Further studies are necessary to explore the role of engXCA in X. citri subsp. citri.
The typical symptoms of citrus canker include water-soaked lesions, white or yellow spongy pustules and brown corky canker [26]. The leaf-spot symptom is quite different from soft rot which is characterized by a massive degradation of the plant cell wall caused by sets of extracellular hydrolysis enzymes secreted by other organisms [27, 28]. In addition to the extracellular enzyme deficiency, a mutation in X. citri subsp. citri T2SS delays the appearance of canker symptoms [14–16]. Because X. citri subsp. citri can produce several extracellular enzymes, such as cellulases, proteases and amylases, it is difficult to assess the exact roles of cellulases in pathogenicity [15]. In this work, we constructed deletion mutants of extracellular endoglucanases and reported their roles in pathogenicity on host plants. The loss of extracellular endoglucanase activity caused a delay in the appearance of canker symptoms, similar to the phenotypes of T2SS mutants. It is possible that extracellular endoglucanases affect bacterial growth during the colonization stage. This is the first report confirming the role of an extracellular endoglucanase in the pathogenicity in X. citri subsp. citri.
The T2SS is composed of 12–16 proteins and enables bacteria to secrete exoproteins from the periplasm to the outer environment [29]. The proteins are secreted into the extracellular medium by a two-step process in which the proteins are exported across the cytoplasmic membrane and released into the periplasm before being transported across the outer membrane [30]. Although T2SS has been widely studied in plant and animal pathogenic bacteria, the common secretion signal of the secreted proteins has not been well characterized. The key steps of this secretion occur when putative substrates interact with the outer membrane component GspD, which plays a chaperon-like role [31, 32]. A XAC29_03125 homolog EngXCA, is a major cellulase in X. campestris pv. campestris [7]. In this study, EngXCA in Xcc 29–1 showed endoglucanase activity but was not secreted into the medium. Even though there was lack of direct evidence, we postulated that differences in secretion manner in these two bacteria may stem from whether EngXCA interacts with the T2SS component GspD, since X. citri subsp. citri and X. campestris pv campestris both possess EngXCA protein but with different amino acid sequences.
Supporting Information
Bacteria were cultured in LB medium at 37°C to OD600 0.5. The recombinant proteins were induced for 3 h by supplementationwith 0.5 mM IPTG. Cells were harvested, washed in PBS, and then resuspended in 10 mM PBS (pH 7.5, 500 mM NaCl). After several freeze/thaw cycles, the cell suspension was sonicated for 3 min with an interval of 4 s between pulses, and then centrifuged at 5000 g for 10 min at 37°C. Twenty microliter supernatant samples were analysed by 12% SDS-PAGE.
(PDF)
The size differences of PCR products from wild-type Xcc 29–1, ΔbglC3, ΔengXCA and ΔengXCAΔbglC3 were revealed using primer sets 0028.1.F/0028.2.R and 0612.1.F/0612.2.R. PCR products were sequenced to confirm that the target genes were deleted from chromosome. 1, PCR product produced by primer set 0028.1.F/0028.2.R; 2, PCR product produced by primer set 0612.1.F/0612.2.R.
(PDF)
(DOCX)
(DOCX)
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the National Natural Science Foundation of China (31171832) to HZ. The funder had no role in the study design, data analysis, decision to publish, or preparation of the manuscript.
References
- 1.Matsumoto H, Jitareerat P, Baba Y, Tsuyumu S. Comparative study of regulatory mechanisms for pectinase production by Erwinia carotovora subsp. carotovora and Erwinia chrysanthemi. Mol Plant Microbe Interact. 2003;16: 226–237. [DOI] [PubMed] [Google Scholar]
- 2.Barras F, van Gijsegem F, Chatterjee AK. Extracellular enzymes and pathogenesis of soft-rot Erwinia. Annu Rev Phytopathol. 1994; 32: 201–234. [Google Scholar]
- 3.Mäe A, Heikinheimo R, Palva ET. Structure and regulation of the Erwinia carotovora subspecies carotovora SCC3193 cellulase gene celV1 and the role of cellulase in phytopathogenicity. Mol Gen Genet. 1995;247: 17–26. [DOI] [PubMed] [Google Scholar]
- 4.Cui YY, Chatterjee A, Chatterjee AK. Effects of the two-component system comprising GacA and GacS of Erwinia carotovora subsp. carotovora on the production of global regulatory rsmB RNA, extracellular enzymes, and HarpinEcc. Mol Plant Microbe Interact. 2001;14: 516–526. [DOI] [PubMed] [Google Scholar]
- 5.Mukherjee A, Cui YY, Ma WL, Liu Y, Chatterjee AK. hexA of Erwinia carotovora subsp. carotovora strain Ecc71 negatively regulates production of RpoS and rsmB RNA, a global regulator of extracellular proteins, plant virulence and the quorum sensing signal, N-(3-oxohexanoyl)-L-homoserine lactone. Environ Microbiol. 2000;2: 203–215. [DOI] [PubMed] [Google Scholar]
- 6.Schell M. Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol. 1993;47: 597–626. [DOI] [PubMed] [Google Scholar]
- 7.Schröter K, Flaschel E, Pühler A, Becker A. Xanthomonas campestris pv. campestris secretes the endoglucanases ENGXCA and ENGXCB: construction of an endoglucanase-deficient mutant for industrial xanthan production. Appl Microbiol Biotechnol. 2001;55: 727–733. [DOI] [PubMed] [Google Scholar]
- 8.Hu J, Qian W, He CZ. The Xanthomonas oryzae pv. oryzae eglXoB endoglucanase gene is required for virulence to rice. FEMS Microbiol Lett. 2007;269: 273–279. [DOI] [PubMed] [Google Scholar]
- 9.Bayer EA, Chanzy H, Lamed R, Shoham Y. Cellulose, cellulases and cellulosomes. Curr Opin Struct Biol. 1998;8: 548–557. [DOI] [PubMed] [Google Scholar]
- 10.Vauterin L, Hoste B, Kersters K, Swings J. Reclassification of Xanthomonas. Internat J System Evolut Bacteriol. 1995;45: 472–489. [Google Scholar]
- 11.Dunger G, Arabolaza AL, Gottig N, Orellano EG, Ottado J. Participation of Xanthomonas axonopodis pv. citri hrp cluster in citrus canker and nonhost plant responses. Plant Pathol. 2005;54: 781–788. [Google Scholar]
- 12.Dunger G, Garofalo CG, Gottig N, Garavaglia BS, Rosa MCP, Farah CS, et al. Analysis of three Xanthomonas axonopodis pv. citri effector proteins in pathogenicity and their interactions with host plant proteins. Mol Plant Pathol. 2012;13: 865–876. 10.1111/j.1364-3703.2012.00797.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sgro GG, Ficarra FA, Dunger G, Scarpec TE, Valle EM, Cortadi A, et al. Contribution of a harpin protein from Xanthomonas axonopodis pv. citri to pathogen virulence. Mol Plant Pathol. 2012;13: 1047–1059. 10.1111/j.1364-3703.2012.00814.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baptista JC, Machado MA, Homem RA, Torres PS, Vojnov AA, do Amaral AM. Mutation in the xpsD gene of Xanthomonas axonopodis pv. citri affects cellulose degradation and virulence. Genet Mol Biol. 2010;33: 146–153. 10.1590/S1415-47572009005000110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song X, Guo J, Ma WX, Ji ZY, Zou LF, Chen GY, et al. Identification of seven novel virulence genes from Xanthomonas citri subsp. citri by Tn5-based random mutagenesis. J Microbiol. 2015;53: 330–336. 10.1007/s12275-015-4589-3 [DOI] [PubMed] [Google Scholar]
- 16.Yan Q, Wang N. High-throughput screening and analysis of genes of Xanthomonas citri subsp. citri involved in citrus canker symptom development. Mol Plant Microbe Interact. 2012;25: 69–84. 10.1094/MPMI-05-11-0121 [DOI] [PubMed] [Google Scholar]
- 17.Guo J, Song X, Zou LF, Zou HS, Chen GY. The small and large subunits of carbamoyl-phosphate synthase exhibit diverse contributions to pathogenicity in Xanthomonas citri subsp. citri. J Integr Agr. 2015;14: 1338–1347. [Google Scholar]
- 18.Zou HS, Song X, Zou LF, Yuan L, Li YR, Guo W, et al. EcpA, an extracellular protease, is a specific virulence factor required by Xanthomonas oryzae pv. oryzicola but not by X. oryzae pv. oryzae in rice. Microbiology 2012;158: 2372–2383. 10.1099/mic.0.059964-0 [DOI] [PubMed] [Google Scholar]
- 19.Li JY, Wang N. The wxacO gene of Xanthomonas citri ssp. citri encodes a protein with a role in lipopolysaccharide biosynthesis, biofilm formation, stress tolerance and virulence. Mol Plant Pathol. 2011;12: 381–396. 10.1111/j.1364-3703.2010.00681.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xue XB, Zou LF, Ma WX, Liu ZY, Chen GY. Identification of 17 HrpX-regulated proteins including two novel type III effectors, XOC_3956 and XOC_1550, in Xanthomonas oryzae pv. oryzicola. PLoS ONE. 2014;9: e93205 10.1371/journal.pone.0093205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bateman DF, Miilar KL. Pectic enzymes in tissue degradation. Ann Rev Phytopathol. 1966;4: 119–146. [Google Scholar]
- 22.Roberts DP, Denny TP, Schell MA. Cloning of the egl gene of Pseudomonas solanacearum and analysis of its role in phytopathogenicity. J Bacteriol. 1988;170: 1445–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang JL, Liu YN, Barber CE, Dow JM, Wootton JC, Daniels MJ. Genetic and molecular analysis of a cluster of rpf genes involved in positive regulation of synthesis of extracellular enzymes and polysaccharide in Xanthomonas campestris pathovar campestris. Mol Gen Genet. 1991; 226: 409–417. [DOI] [PubMed] [Google Scholar]
- 24.Thowthampitak J, Shaffer BT, Prathuangwong S, Loper JE. Role of rpfF in virulence and exoenzyme production of Xanthomonas axonopodis pv. glycines the causal agent of bacterial pustule of soybean. Phytopathology. 2008;98: 1252–1260. 10.1094/PHYTO-98-12-1252 [DOI] [PubMed] [Google Scholar]
- 25.Py B, Salmond GPC, Chippaux M, Barras F. Secretion of cellulases in Erwinia chrysanthemi and E. carotovora is species specific. FEMS Microbiol Lett. 1991;79: 315–322. [Google Scholar]
- 26.Brunings AM, Gabriel DW. Xanthomonas citri: breaking the surface. Mol Plant Pathol. 2003;4: 141–157. 10.1046/j.1364-3703.2003.00163.x [DOI] [PubMed] [Google Scholar]
- 27.Collmer A, Keen NT. The role of pectic enzymes in plant pathogenesis. Annu Rev Phytopathol. 1986;24: 383–409. [Google Scholar]
- 28.Pérombélon MCM, Kelman A. Ecology of the soft rot Erwinias. Annu Rev Phytopathol. 1980;18: 361–387. [Google Scholar]
- 29.Cianciotto NP. Type II secretion: a protein secretion system for all seasons. Trends Microbiol. 2005;13: 581–588. [DOI] [PubMed] [Google Scholar]
- 30.Korotkov KV, Sandkvist M, Hol WG. The type II secretion system: biogenesis, molecular architecture and mechanism. Nat Rev Microbiol. 2012;10: 336–351. 10.1038/nrmicro2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu NT, Hung MN, Chen DC, Tsai RT. Insertion mutagenesis of XpsD, an outer-membrane protein involved in extracellular protein secretion in Xanthomonas campestris pv. campestris. Microbiology. 1998;144: 1479–1486. [DOI] [PubMed] [Google Scholar]
- 32.Shevchik VE, Robert-Baudouy J, Condemine G. Specific interaction between OutD, an Erwinia chrysanthemi outer membrane protein of the general pathway, and secreted proteins. EMBO J. 1997;16: 3007–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bacteria were cultured in LB medium at 37°C to OD600 0.5. The recombinant proteins were induced for 3 h by supplementationwith 0.5 mM IPTG. Cells were harvested, washed in PBS, and then resuspended in 10 mM PBS (pH 7.5, 500 mM NaCl). After several freeze/thaw cycles, the cell suspension was sonicated for 3 min with an interval of 4 s between pulses, and then centrifuged at 5000 g for 10 min at 37°C. Twenty microliter supernatant samples were analysed by 12% SDS-PAGE.
(PDF)
The size differences of PCR products from wild-type Xcc 29–1, ΔbglC3, ΔengXCA and ΔengXCAΔbglC3 were revealed using primer sets 0028.1.F/0028.2.R and 0612.1.F/0612.2.R. PCR products were sequenced to confirm that the target genes were deleted from chromosome. 1, PCR product produced by primer set 0028.1.F/0028.2.R; 2, PCR product produced by primer set 0612.1.F/0612.2.R.
(PDF)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.