Abstract
The banana weevil Cosmopolites sordidus is an important and serious insect pest in most banana and plantain-growing areas of the world. In spite of the economic importance of this insect pest very little genomic and transcriptomic information exists for this species. In the present study, we characterized the midgut transcriptome of C. sordidus using massive 454-pyrosequencing. We generated over 590,000 sequencing reads that assembled into 30,840 contigs with more than 400 bp, representing a significant expansion of existing sequences available for this insect pest. Among them, 16,427 contigs contained one or more GO terms. In addition, 15,263 contigs were assigned an EC number. In-depth transcriptome analysis identified genes potentially involved in insecticide resistance, peritrophic membrane biosynthesis, immunity-related function and defense against pathogens, and Bacillus thuringiensis toxins binding proteins as well as multiple enzymes involved with protein digestion. This transcriptome will provide a valuable resource for understanding larval physiology and for identifying novel target sites and management approaches for this important insect pest.
Introduction
The banana weevil Cosmopolites sordidus (Germar) (Coleoptera: Curculionidae) is considered one of the most invasive and destructive pests of banana worldwide [1]. The larvae of C. sordidus are a severe constraint on banana and plantain production in most areas where these crops are cultivated, especially in Africa [2–5] where this insect pest has been associated with rapid plantation decline [6] and with a phenomenon called “yield decline syndrome” in West Africa. The larvae of the banana weevil, which are the most destructive stage of the insect, is responsible for considerable damage of the plant corm, interfering with root initiation, nutrient and water uptake and plant development [6]. When a severe weevil infestation occurs, crop losses of up to 100% have been reported [7]. It is well known that chemical control of this insect pest is not only undesirable but also expensive. Options for biological control are limited and pheromone-based insect trapping results in either low or ineffective captures [8, 9].
Many basic advances have been made by studying the banana weevil, including, studies regarding pest resistance [10], insect resistant germplasm [2, 11, 12], plant antifeedants [13], cultural control practices [14] and biological control [15]. Despite extensive and recent biochemical and physiological studies, limited genomic information exists, especially for important tissues such as the midgut. The availability of transcriptome sequences from insect midgut tissues will facilitate identification of genes that are expressed in the intestinal tract and their respective metabolic and functional roles. It is well known that the curculionids are the largest family of beetles [16], which in general are important plant tissue damaging pests such as the banana weevil C. sordidus[1].
The rapid growth of next-generation DNA sequencing technologies such as 454-based pyrosequencing [17, 18] have allowed the characterization of the transcriptome of many important, non-model insect species [19–23], thus providing valuable and unprecedented opportunities to increase our knowledge of expressed genes, especially in those insect pests where little or no genomic resources exist [24].
In this study, we used a 454-based pyrosequencing platform to sequence the C. sordidus larval midgut transcriptome allowing the characterization of transcripts encoding different genes associated with metabolic functions and potential insecticide targets. Many of these transcripts were protease-like genes from different digestive enzyme families, mainly associated with aminopeptidases, carboxypeptidases, serine proteases and cysteine proteases. The C. sordidus transcriptome represents an important contribution to understanding the biology of this insect pest and for the identification of potential target genes involved in protein digestion and many other metabolic pathways.
Materials and Methods
The experiments were carried out under a standard protocol in the lab and no specific permissions were required for these locations/activities. In addition, these study did not involve any endangered or protected species.
Insect dissection and of midgut RNA extraction
C. sordidus larvae were collected from corms obtained at a plantain field near Manizales, Colombia (1058 m, 5° 4’ 13.2” N, 75° 41’ 7.7” O). Collected larvae were inspected under a stereoscope and the fourth instar larvae were selected based on the size of the head capsule as described by [25] and then used for midgut dissection (Fig 1). Gut tissue was obtained by dissecting in DEPC-treated distilled water. The gut content and peritrophic matrix were removed and the washed midgut tissue was flash-frozen using liquid nitrogen and stored at -80°C. RNA extraction was performed using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. RNA was then purified using the RNeasy MinElute Cleanup Kit (Qiagen, Chatsworth, CA) after removing genomic DNA contamination using the TURBO DNA-free™ Kit (Ambion, Carlsbad, CA) according to manufacturer’s instructions.
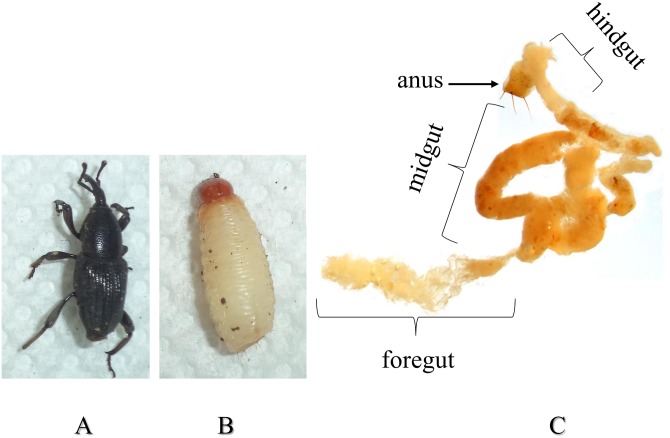
Fig 1.
Cosmopolites sordidus. Adult (A), fourth instar larva (B), digestive tract from a fourth instar larvae (C).
C. sordidus midgut normalized cDNA library preparation
Full-length-enriched double-stranded cDNA was then synthesized using the Mint-2 cDNA synthesis kit (Evrogen, Moscow, Russia/ Cat # SK005). To reduce the prevalence of abundant transcripts, the resulting double-stranded cDNAs were normalized using the Evrogen Trimmer-2 cDNA normalization kit (Evrogen, Moscow, Russia/ Cat # NK003) [26]. The resulting normalized cDNA midgut library was then submitted to 454- high-throughput pyrosequencing.
Sequencing and assembly
For 454 pyrosequencing (Roche Applied Science), 3 μg of normalized cDNAs was sent to the Core for Applied Genomics and Ecology (CAGE) facility at the University of Nebraska-Lincoln. The sequences obtained were preprocessed by filtering reads with low qualities (Q15) that were less than 100 bp as well as trimming SMART adapters and Ns. Finally, processed reads were clustered using the MIRA 3.4.0 assembler.
Homology searches and sequence annotation
Functional annotation of assembled sequences by gene ontology terms (GO; www.geneontology.org), InterPro entries (InterProScan; http://www.ebi.ac.uk/tools/pfa/iprscan/) and enzyme classification codes (EC) was conducted using Blast2Go software suite [27]. For homology analysis, all sequences were searched against the NCBI non-redundant (nr) protein database via BLASTx using an E-value cut-off of 10-25.
Protein sequence alignment and phylogenetic analysis
The protein sequence of insect carboxypeptidases were aligned with ClustalW program (http://www.ebi.ac.uk/clustalw/). The evolutionary relationship among carboxypeptidases was determined using phylogenetic analysis based on protein sequences and carried out using the Neighbor-joining method using MEGA 6.0 software.
Semi-quantitative RT-PCR
One microgram of total RNA was used as template for synthesis of the first strand of cDNA with an oligo-(dT) primer and the Maxima H Minus cDNA synthesis kit (Thermo Scientific, kat # K1681)). The cDNA was employed as a template for amplification and detection of Carboxypeptidase (CsoCp), Chitin Synthase (CsoChs), and Aminopeptidase (CsoAp) transcripts in five larval development and pupae stages of C. sordidus. The expression level of these transcripts was evaluated using a specific set of primers as follow: CsoCp sequence (forward primer 5'-CCGAACCTTGCTCTGATACC-3', and reverse primer 5'-CGTACCCCCATGGATACAAC-3'), CsoChs sequence (forward primer 5'-CCATTTACCCCGAAGATCAA-3', and reverse primer 5'-TGGATAAACATGCAAATACATTG-3'), and CsoAp sequence (forward primer 5'-TTCCTGAATGAGGGATTTGC-3', and reverse primer 5'-GGTGCTTGAAGTGCTTGTGA-3'). C. sordidus β-actin gene was also amplified by PCR using the following set of primers: forward primer 5'-AAGACATCAGGGCGTAATGG-3', and reverse primer 5'-GAAGGTGTGGTGCCAGATTT-3'. The PCR reaction was carried out in a 10 μl final volume. PCR conditions were: 95°C for 3 min, 60°C for 30 s, 72°C for 30 s followed by 32 cycles, and 5-min extension at 72°C. All PCR products were resolved by electrophoresis on 1% agarose gels.
Results
Pyrosequencing, assembly, and annotation
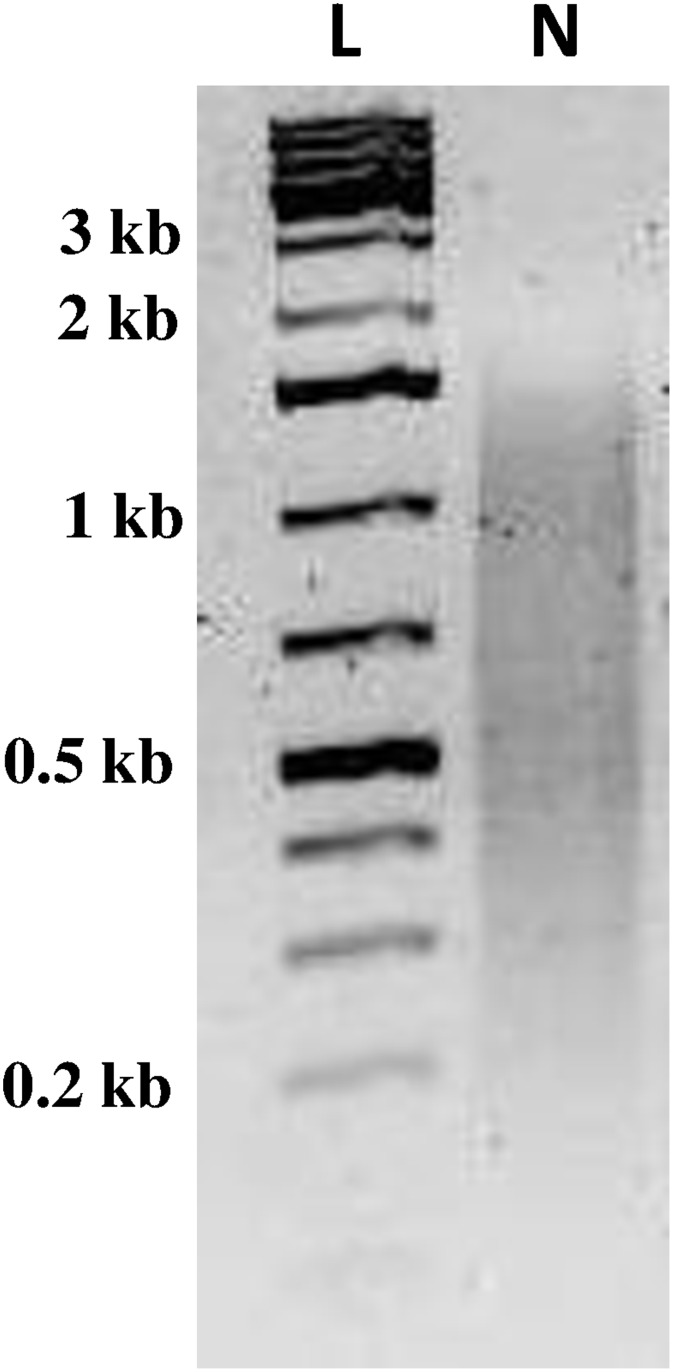
Normalization of the C. sordidus midgut cDNA library resulted in an even distribution of transcripts ranging from 0.2 to 1.5 kb in length (Fig 2). 454-pyrosequencing of the normalized library from C. sordidus midgut transcriptome generated a total of 596,389 sequencing reads with an average length of 491 bp (Table 1). After filtering reads with low quality (Q15) and less than 100 bp in length as well as trimming SMART adapters and Ns, 425,605 reads were assembled using the MIRA 3.4.0 assembler. The assembly resulted in 47,729 contigs and 139,600 singletons that did not assemble into a contig. The average contig length was 491 bp (100–4270 bp) with N50 of 505 bp (Table 1). These data were deposited in NIH Short Read Archive with accession number SRP061782. It was found that almost 35% of all contigs returned at least one blast hit and one GO term (Table 1). In addition, 13.5% of these contigs (6,457) received an EC number, which assigned a known enzymatic function.
Fig 2. Electrophoresis of normalized cDNA from C. sordidus midgut.
Normalized cDNA library containing transcripts ranging from 0.2 to 1.5 kb in size were subject to 454-mediated pyrosequencing. L, molecular mass markers. N, normalized cDNA library.
Table 1. Summary statistics for C. sordidus midgut transcriptome after assembly and annotation.
| Assembly | |
| Total number of reads (before filtering) | 596,389 |
| Number of reads that entered assembly (after filtering) | 425,600 |
| Total base pairs that entered assembly | 178,143,660 |
| Average contig length (range) | 491 (100–4270) |
| N50 length (bp) | 505 |
| Number of contigs >400 bps | 30,840 |
| Annotation | |
| % contigs with at least one GO term | 34.4% |
| % contigs with at least one blast hit | 35.4% |
| % contigs with at least one InterPro cross ref | 34.5% |
| % contigs with an EC number | 13.5% |
Functional classifications, homology searches and Gene Ontology Analysis
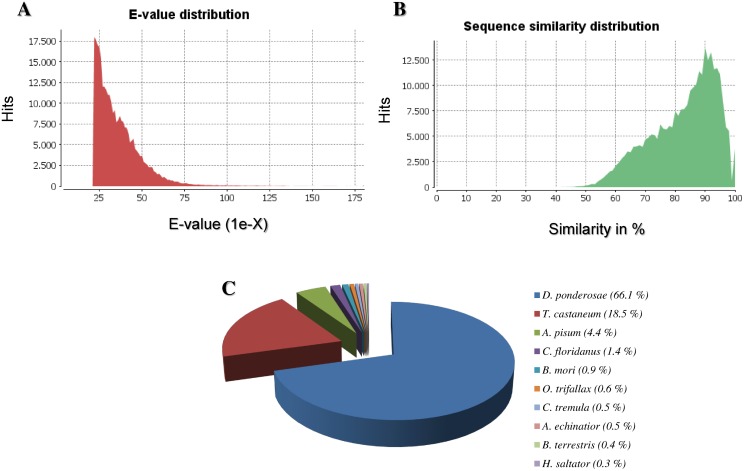
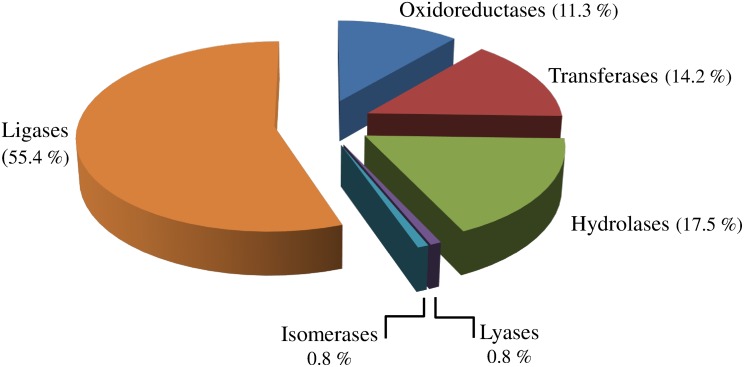
After read assembly, contigs were submitted to BLASTx similarity search against NCBI non-redundant protein database (nr) to assess their putative function. The similarity distributions and E-value of the C. sordidus BLAST hits against the non-redundant database are presented in Fig 3. Most of the BLAST hits are to the bark beetle Dendroctonus ponderosae (66%) and to the model coleopteran, Tribolium castaneum genomes (18.5%) (Fig 3C), which is one of the few beetle genomes that has been fully sequenced so far. Enzyme classification (EC) was used to classify the predicted C. sordidus midgut proteins. Enzyme classification shows that ligases account for the largest proportion of C. sordidus enzymes (55.4%), followed by hydrolases (17.5%), transferases (14.2%) and oxidoreductases (11.3%) (Fig 4). In addition to enzyme classification, gene ontology (GO) assignments were used to classify the functions of the predicted proteins, producing 37,982 terms for biological process categories, 16,457 terms for cellular component categories and 22,870 terms for molecular function categories.
Fig 3. Summary of homology searches (BLAST) of C. sordidus midgut 454-pyrosequencing data against the non-redundant (nr) database.
(A) E-value distribution (Cut-off 10–20). (B) Similarity distribution of the top BLAST hit. (C) Species distribution of the top BLAST hit. Note that the majority of top hits are to the beetles D. ponderosae (66.1%) and T. castaneum (18.5%).
Fig 4. General Enzyme Classification (EC) terms for the contigs of C. sordidus midgut transcriptome.
Oxidoreductases (EC: 1.x.x), Transferases (EC: 2.x.x), Hydrolases (EC: 3.x.x), Lyases (EC: 4.x.x), Isomerases (EC: 5.x.x), and Ligases (EC: 6.x.x).
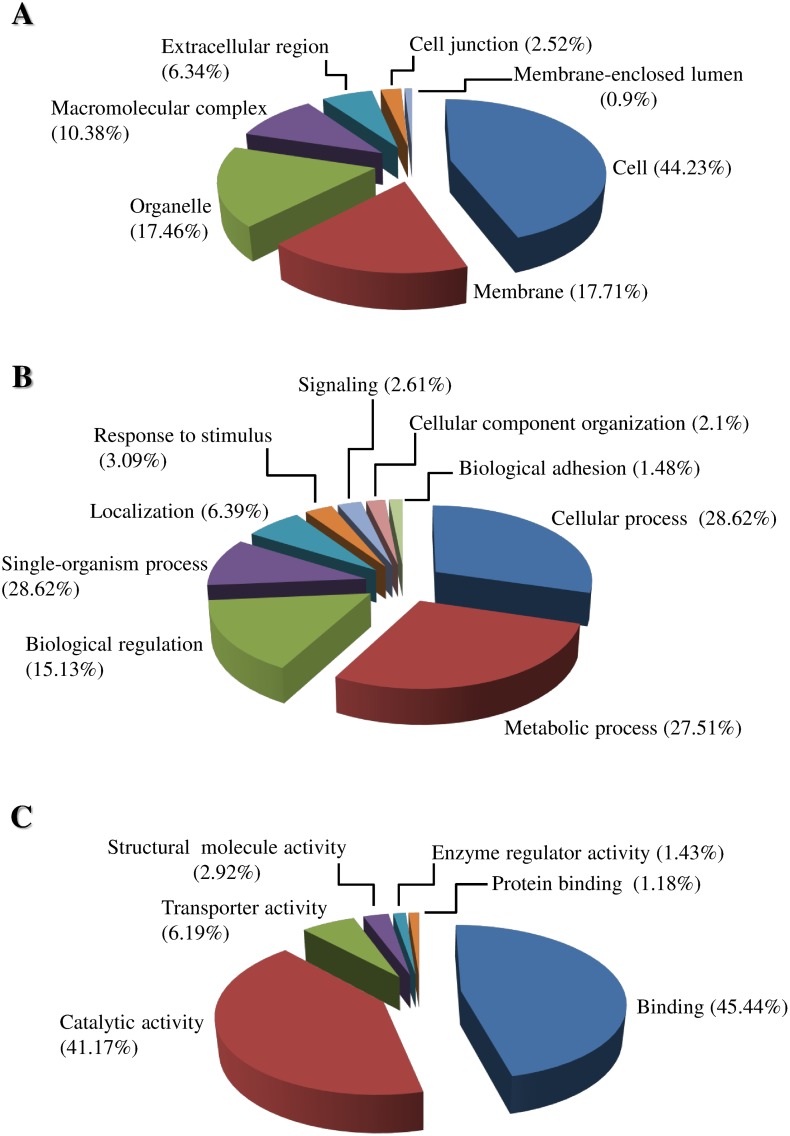
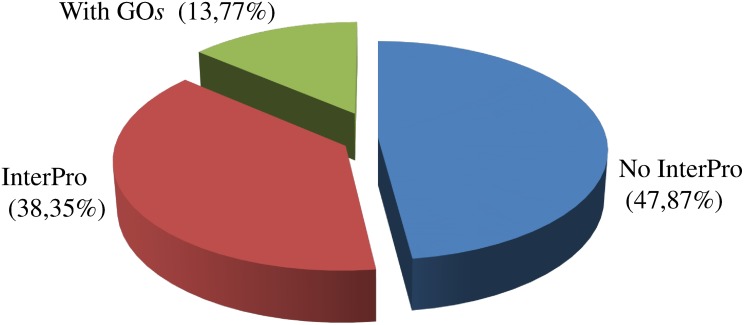
Most of the cellular component GO terms (Fig 5A) were associated with the cell (44.23%) followed by the membrane (17.71%) and organelle (17.46%). Metabolic (27.51%) and cellular processes (28.62%) were involved with more than half of the biological process GO terms followed by biological regulation (15.13%) (Fig 5B). Most of the molecular function GO terms were associated with binding (45.44%) followed by catalytic activity (41.17%) and transporter activity (6.19%) (Fig 5C). The InterPro analysis was also used in addition to enzyme classification and GO assignments and identified that almost 14% of predicted proteins received a GO assignment and almost 48% of the predicted C. sordidus proteins did not have an InterPro assignment (Fig 6).
Fig 5. Gene ontology (GO) assignments for the C. sordidus midgut transcriptome.
Panel A represents cellular components, while panel B represents biological processes and panel C is for molecular function at level 2.
Fig 6. Summary information for InterProScan results in the C. sordidus midgut transcriptome.
Genes of interest related to midgut metabolic functions and xenobiotic metabolism
A list of C. sordidus larval genes related to general digestion, peritrophic membrane biosynthesis, degradation and remodeling as well as detoxification and protease-like related genes are presented in Table 2. A total of 51 detoxification related contigs were identified in the C. sordidus midgut transcriptome. Of these, 22 corresponded to cytochrome P450 genes, 11 to glutathione-S-transferases, 13 to carboxylesterases and 5 to superoxide dismutases (SOD). Contigs related to peritrophic membrane biosynthesis, degradation and remodeling include chitinase (15), chitin synthase (6), and chitin deacetylase (5). Contigs associated with general digestion from this pyrosequencing analysis include cysteine proteinases (143), serine proteinases (61), aminopeptidases (38), carboxypeptidases (22), Dipeptidyl peptidases (8), lipases (3), and β-glucosidases (7). An additional 40 contigs related to immunity and defense against pathogens were identified from the C. sordidus midgut transcriptome. Among these contigs, lectin (17) and serine protease inhibitors (serpin) (11) were the most abundant.
Table 2. Selection of C. sordidus larval genes related to midgut metabolic functions and “Detox” related.
| EC number | Total number of contigs | |
|---|---|---|
| “Detox” related | ||
| Cytochrome P450 | - | 22 |
| Glutathione-S-transferase | 2.5.1.18 | 11 |
| Carboxylesterase | 3.1.1.1 | 13 |
| Superoxide dismutase | 1.15.1.1 | 5 |
| Peritrophic membrane biosynthesis, degradation, and remodeling | ||
| Chitinase | 3.2.1.14 | 15 |
| Chitin synthase | 2.4.1.16 | 6 |
| Chitin deacetylase | 3.1.5.41 | 5 |
| General digestion | ||
| Cysteine proteinase all types | - | 143 |
| Serine proteinase all types | - | 61 |
| Aminopeptidase all types | - | 38 |
| Carboxypeptidase all types | - | 22 |
| Dipeptidyl peptidase all types | - | 8 |
| Lipase | 3.1.1.3 | 3 |
| β-glucosidase | 3.2.1.21 | 7 |
| Immunity-related and defense against pathogens | ||
| Peptidoglycan recognition protein | - | 5 |
| C-type lectins | - | 17 |
| Defensin-like | - | 6 |
| Lysozyme | - | 1 |
| Serin protease inhibitors (Serpin) | - | 11 |
Protein alignment of protease-like enzymes and phylogenetic analysis
A carboxypeptidase predicted protein (AFH35127.1), which was recently submitted to the GenBank from our group, shows 45–58% amino acid identity to other coleopteran carboxypeptidase-like proteins. Amino acid alignment of the predicted carboxypeptidase CsoCP1 with insect protease-like proteins is shown in the supplementary materials (S1 Fig). To determine the relatedness of the predicted proteinase-like proteins from the C. sordidus midgut transcriptome with other insect digestive enzymes, phylogenetic trees were constructed based on protein sequence. It was found that the carboxypeptidase predicted protein from C. sordidus (AFH35127.1) clustered together with two carboxypeptidase-like proteins from D. ponderosae (Scolytidae) (AEE63523 and AEE62416) (Fig 7).
Fig 7. Phylogenetic analysis of carboxypeptidases from C. sordidus and other insect species (accession numbers are given).
Phylogenetic analysis was conducted in MEGA6.0 using the Neighbor-joining method. Positions containing alignment gaps and missing data were eliminated only with pairwise deletion. Amino acid sequences of carboxypeptidases used for this analysis were C. sordidus (CsoCP), D. ponderosae (DpoCP), Eupolyphaga sinensis (EsiCP), Nasonia vitripennis (NviCP), Coptotermes formosanus (CfoCP), Bombus terrestris (BteCP), Bombus impatiens (BimCP), T.castaneum (TcaCP), Apis mellifera (AmeCP), A. dorsata (AdoCP), Phlebotomus papatasi (PhpaCP), Aedes polynesiensis (ApoCP), Aedes aegypti (AaeCP), Lutzomyia longipalpis (LloCP), Culex quinquefasciatus (CquCP), N. vitripennis (NviCP), Bombyx mori (BmoCP), Acromyrmex echinatior (AecCP), Ochlerotatus epactius (OepCP), Musca domestica (MdoCP), Harpegnathos saltator (HsaCP), Trichoplusia ni (TniCP).
Semi-quantitative RT-PCR
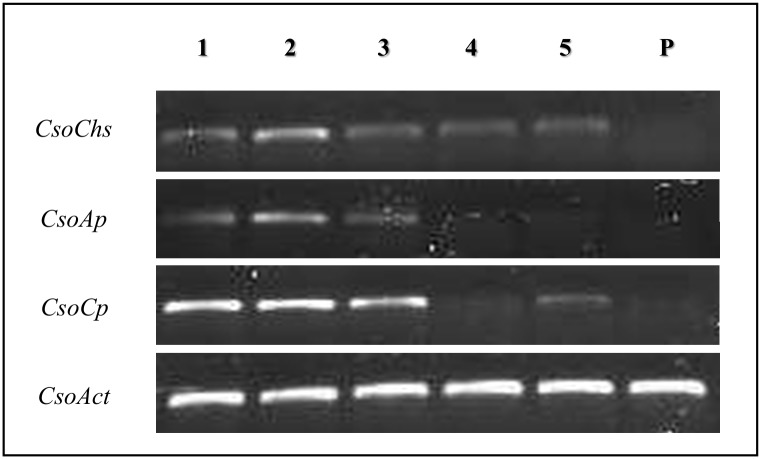
RT-PCR expression analysis was carried out to determine the expression profiles of specific enzyme-like transcripts in larval and pupae stages of C. sordidus (Fig 8). Results shows that expression of all protease-like transcripts were clearly visible through the first three larval stages. However, it was found that Chitin Synthase (CsoChs) transcript is expressing in all five larval development stages but not in pupae stage. In addition, none of evaluated transcripts was expressed in pupae stage. Just the control actin housekeeping gene (CsoAct) was clearly visible at all insect developmental stages.
Fig 8. Semi-quantitative RT-PCR expression analysis of Carboxypeptidase (CsoCp), Chitin Synthase (CsoChs), Aminopeptidase (CsoAp) transcripts in five larval development and pupae stages of C. sordidus.
Expression of the Actin Housekeeping gene (CsoAct) is shown as control. 1–5 represent first, second, third, fourth and fifth instar larvae while P is for pupae stage.
Discussion
Despite the enormous economic impact of the banana weevil C. sordidus on plantain crops worldwide [1], there is a general lack of transcriptome sequence data for this insect pest that could be used to examine traits of biological relevance that might be exploited for developing novel control methods. By using 454-based pyrosequencing, we obtained extensive sequence data providing an unprecedented opportunity for genomic research in an insect pest for which little genomic information is currently available. For example, transcriptome analysis in insects using 454-based pyrosequencing technologies have contributed significantly to the discovery of insect molecular markers (SNPs)[28], Bt receptors [20], immune responses [29, 30], insecticide targets and detoxifying enzymes [19, 31].
The transcriptomic data of the banana weevil C. sordidus that is presented here dramatically increases the number of C. sordidus midgut ESTs. For instance, the number of reported nucleotide sequences related to C. sordidus that were previously available represents only six genes (GenBank, February 15, 2016). This insect EST midgut collection provided by this study will allow the characterization of different genes not only for those closely related to other insect pests, but for many other coleopterans. A good example of this statement is found in the study of the molecular evolution of glycoside hydrolase (GH) genes in the western corn rootworm Diabrotica virgifera virgifera [32]. Results from this study have revealed the presence of three GH family genes (GH45, GH48, and GH28), which are found almost exclusively in Chrysomeloidea and Curculionoidea superfamilies, indicating the possibility of their acquisitions by horizontal gene transfer rather than simple vertical transmission. The transcriptomic analysis of the C. sordidus midgut provides an opportunity to identify genes unique to the C. sordidus midgut, thus providing an unprecedented opportunity for future insect specific management approaches. The 454-based deep pyrosequencing of the C. sordidus midgut transcriptome allowed the identification of contigs encoding proteins with functions strongly related not only to peritrophic membrane biosynthesis, membrane degradation and remodeling, detoxification, and immunity-related genes as well as defense against pathogens, but also to key digestive proteases involved with midgut physiology among many others (Table 2). Importantly, such genes could be targeted by using RNA interference (RNAi) which has been proposed as a novel control technology for other coleopterans. The banana weevil C. sordidus, like many other most insect species, can metabolize not only secondary plant chemicals but also insecticide-like chemicals, a metabolic process that includes a pool of detoxification enzymes such as cytochrome P450s, glutathione-S-transferase (EC 2.5.1.18), carboxylesterase (EC 3.1.1.1) and superoxide dismutase (EC 1.15.1.1) [19]. Transcripts encoding proteins linked to these detoxification enzyme families were found in the C. sordidus normalized midgut transcriptome. In total, 51 contigs were associated with xenobiotic metabolism. It has been reported that P450s represent a large superfamily of heme-containing monooxygenases that catalyze the metabolisms of exogenous and endogenous compounds [33]. A454-based transcriptomic analysis of greenhouse whitefly Trialeurodes vaporariorum identified 57 putative P450s [19]. However, it is possible that the number of these detoxification-related transcripts in the C. sordidus midgut transcriptome could be greater, especially the great variation of total number of P450 genes identified in different insect species [34] and availability of full length of some genes related to detoxification in this database, which could be a valuable prospect to be explored in future, this in turn will facilitate a better understanding of the role of these genes in xenobiotic metabolism and to evaluate the possibility of targeting some of them by using RNAi silencing technology.
As presented in Table 2, the most abundant uncovered protease-like transcripts in the C. sordidus midgut transcriptome are cysteine proteinases, serine proteinases, aminopeptidases and carboxypeptidases, indicating the widespread distribution of these protease-like genes in the C. sordidus midgut. It is well known that proteases are hydrolytic enzymes that are involved in many important roles in insect physiology from protein digestion to polyphenol oxidase activation [35]. The abundance of protease-like transcripts in the C. sordidus midgut transcriptome, as well as the expression of some of these specific transcripts as presented in results session of this manuscript, shows that the development of the banana weevil is extremely dependent on proteolytic enzymes indicating that those genes could represent a good target for RNAi-based technologies. In addition, the finding of the expression of the specific Chitin Synthase (CsoChs) transcript through all five larval development stages represent a strong evidence of the importance of these remodeling-like genes in insect metabolism. It is well known that chitin is not only the principal component of the arthropod cuticle, but also an integral part of peritrophic matrices [36], thus chitin synthesis is essential for insect development and survival and a potential target for RNA-based silencing technology (RNAi). In this context, previous research works have showed that RNAi-mediated down-regulation of T. castaneum CHS genes results in the reduction of chitin content [37].
Cysteine proteinases (EC 3.4.22) are digestive enzymes that have been isolated and partially characterized and which are widely distributed among many coleopteran species [38, 39]. Despite their importance in insect digestion, many of these protease-like enzymes remain poorly understood for their molecular functions. It is well known that the study of insect digestive enzymes has often focused on aminopeptidase-like enzymes due to the fact that this group of digestive proteases may act as natural receptors for Bt endotoxins [40, 41]. In fact, the insect midgut has become the primary target for both Bt-derived insecticides and a Bt alternative for pest control of Chrysomela tremulae [20]. The physiological role of these digestive-like enzymes in herbivorous insects like C. sordidus is to participate actively in the digestion of proteins. These enzymes cleave single amino acid residue from the N-terminus of proteins, which represents one of the most abundant compounds that are currently found in plant tissues. It is known that the expression level of proteases in insect guts depends on the protein content of the plant tissue that the insect uses as the main food source [35]. It is also important to point out that carboxypeptidases, with 22 contigs found in the C. sordidus transcriptome, represents an important group of peptidases in the banana weevil midgut. The lack of nucleotide sequences in the GenBank for this specific group of insect digestive enzymes (1 from our transcriptome data (AFH35127.1), as well as for many other gene sequences, will facilitate future research approaches that focus on C. sordidus peptidases and proteases. In addition, genes encoding proteinase inhibitors (PI) can represent a valuable alternative for control of insect pests when considering their inclusion into plant genomes using transgenic approaches [42]. Serine proteases are a group of digestive enzymes that are widely distributed in animals and microorganisms [43], playing key roles in many biological processes. As in C. sordidus, it has been also reported that many other insect species contains serine-type proteinases in their intestinal tract, allowing the insect to digest proteins that are naturally found in their food [44]. It has been observed that insects with alkaline midgut pH usually show higher serine proteinase activity [45], which are more active at neutral to alkaline pH, the condition of many lepidopteran insects. However, coleopteran insects that have a more acid pH in the digestive tract rely on cysteine or aspartic proteinases, which have better enzymatic activity at acidic pH [45].
Transcriptomic analysis of the C. sordidus genes involved in the insect immune and defense response led to the identification of C-type lectins, a protein family that has diverse functions, such as pathogen recognition and neutralization [46]. In the C. sordidus larval midgut EST database, the most abundant contig associated with immune response are C-type lectins followed by putative serine proteinase inhibitors or serpins. Similar results were found in the Plutella xylostella larval midgut transcriptome [47]. Results presented in this report represent the first transcriptomic analysis of the banana weevil C. sordidus, the most invasive and destructive pest of banana and plantain worldwide. This analysis has not only dramatically increased the number of known genes for this insect pest but it has also allowed the identification of novel gene sequences that are expressed in the intestinal tract providing a valuable source of information for understanding larval physiology and for identifying potential targets and management approaches for this insect pest or even as an important source of cDNAs in genome annotation. In addition, this transcriptome data adds to other research work focused on insect genome sequencing projects [48–50].
Sequence Submission
The raw data obtained by 454-based pyrosequencing was submitted to the Short Read Archive database at NCBI (http://www.ncbi.nlm.nih.gov/guide/howto/submit-data/) (Accession SRP#: SRP061782).
Supporting Information
An asterisk (*) indicates identical residues, semicolon (:) indicates highly conserved substitutions and a period (.) indicates semi-conserved substitutions. Dashes represent gaps introduced to preserve alignment. Species and accession numbers included in the alignment were C. sordidus (AFH35127), D. ponderosae (AEE63523), P. papatasi (ABV44754), and L. longipalpis (ABV60312).
(TIF)
Acknowledgments
The authors thank the Universidad de Caldas (Manizales, Colombia) and the Entomology Department at the University of Nebraska (Lincoln, USA) for support of this research. Authors also thank the Colombian National Fund for Science, Technology and Innovation, Francisco Jose de Caldas-COLCIENCIAS (Colombia).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Funding came from the Universidad de Caldas (Manizales, Colombia) grant # 1659114 and from the National Bank for Reconstruction and Development (BIRF) (Contract No. FP44842-242-2015) administered by Colombian National Fund for Science, Technology and Innovation, Francisco Jose de Caldas-COLCIENCIAS.
References
- 1.Waterhouse DF. Biological control: Pacific prospects. Melbourne: Inkata Press; 1987. viii, 454 p. p. [Google Scholar]
- 2.Gold CS, Pena JE, Karamura EB. Biology and integrated pest management for the banana weevil Cosmopolites sordidus (Germar) (Coleoptera: Curculionidae). Integrated Pest Management Reviews. 2001;6:79–155. [Google Scholar]
- 3.Gold CS, Messiaen S. The banana weevil Cosmopolites sordidus. Musa Pest INIBAP Fact Sheet. 2000;No 4.
- 4.Forgain R, Messiaen S, Foure E. Studies on the banana borer weevil in Cameroon. 2002.
- 5.Onwueme IC, Sinha TD, Technical Centre for Agricultural and Rural Cooperation (Ede Netherlands). Field crop production in tropical Africa: principles and practice. Ede, Netherlands: CTA; 1991. v, 480 p. p. [Google Scholar]
- 6.Gold CS, Rukazambuga NDTM, Karamura EB, Nemeye P, Night G. Recent advances in banana weevil biology, population dynamics and pest status with emphasis on East Africa: International Network for the Improvement of Banana and Plantain (INIBAP), Montpellier (France); 1999. [Google Scholar]
- 7.Sengooba T. Survey of banana pest problem complex in Rakai and Masaka districts. Uganda: Ministry of Agriculture, Namulonge Research Station. [Google Scholar]
- 8.Reddy GVP, Cruz ZT, Guerrero A. Development of an Efficient Pheromone-Based Trapping Method for the Banana Root Borer Cosmopolites sordidus. J Chem Ecol. 2009;35(1):111–7. 10.1007/s10886-008-9580-6. WOS:000262928000013. [DOI] [PubMed] [Google Scholar]
- 9.Tinzaara W, Gold CS, Kagezi GH, Dicke M, Van Huis A, Nankinga CM, et al. Effects of two pheromone trap densities against banana weevil, Cosmopolites sordidus, populations and their impact on plant damage in Uganda. J Appl Entomol. 2005;129(5):265–71. 10.1111/j.1439-0418.2005.00962.x. WOS:000229580800006. [DOI] [Google Scholar]
- 10.Collins PJ, Treverrow NL, Lambkin TM. Organophosphorus Insecticide Resistance and Its Management in the Banana Weevil Borer, Cosmopolites-Sordidus (Germar) (Coleoptera, Curculionidae), in Australia. Crop Prot. 1991;10(3):215–21. [Google Scholar]
- 11.Kiggundu A, Gold CS, Labuschagne MT, Vuylsteke D, Louw S. Levels of host plant resistance to banana weevil, Cosmopolites sordidus (Germar) (Coleoptera: Curculionidae), in Ugandan Musa germplasm. Euphytica. 2003;133(3):267–77. [Google Scholar]
- 12.Seshu-Reddy KV, Lubega MC. Evaluation of banana cultivars for resistance to tolerance of the weevil Cosmopolites sordidus Germar Breeding Banana and Plantain for Resistance to Diseases and Pests: CIRAD/INIBAP, Montpellier, France; 1993. p. 143–8. [Google Scholar]
- 13.Kiggundu A. Host-plant interactions and resistance mechanisms to banana weevil Cosmopolites sordidus (Germar) in Ugandan Musa germplasm: University of the Orange Free State, Bloemfontein, South Africa; 2000. [Google Scholar]
- 14.Gold CS, Ogenga Latigo MW, Tushemereirwe W., Kashaija I., Nankinga C.M. Farmer perceptions of banana pest constraints in Uganda. Gold CS, Gemmil B., editor. Cotonou, Benin, 12–14 November, 1991: International Institute of Tropical Agriculture; 1991. [Google Scholar]
- 15.Lopes RB, Michereff M, Tigano MS, Neves PMOJ, Lopez EL, Fancelli M, et al. Virulence and horizontal transmission of selected Brazilian strains of Beauveria bassiana against Cosmopolites sordidus under laboratory conditions. B Insectol. 2011;64(2):201–8. WOS:000298339400008. [Google Scholar]
- 16.Hunt T, Bergsten J, Levkanicova Z, Papadopoulou A, John OS, Wild R, et al. A comprehensive phylogeny of beetles reveals the evolutionary origins of a superradiation. Science. 2007;318(5858):1913–6. 10.1126/science.1146954. WOS:000251786600056. [DOI] [PubMed] [Google Scholar]
- 17.Metzker ML. Applications of Next-Generation Sequencing Sequencing Technologies—the Next Generation. Nat Rev Genet. 2010;11(1):31–46. [DOI] [PubMed] [Google Scholar]
- 18.Shendure J, Ji HL. Next-generation DNA sequencing. Nat Biotechnol. 2008;26(10):1135–45. 10.1038/Nbt1486. WOS:000259926000028. [DOI] [PubMed] [Google Scholar]
- 19.Karatolos N, Pauchet Y, Wilkinson P, Chauhan R, Denholm I, Gorman K, et al. Pyrosequencing the transcriptome of the greenhouse whitefly, Trialeurodes vaporariorum reveals multiple transcripts encoding insecticide targets and detoxifying enzymes. BMC genomics. 2011;12 Artn 56 10.1186/1471-2164-12-56. WOS:000287201900001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pauchet Y, Wilkinson P, van Munster M, Augustin S, Pauron D, Ffrench-Constant RH. Pyrosequencing of the midgut transcriptome of the poplar leaf beetle Chrysomela tremulae reveals new gene families in Coleoptera. Insect Biochem Molec. 2009;39(5–6):403–13. 10.1016/j.ibmb.2009.04.001. WOS:000267505200011. [DOI] [PubMed] [Google Scholar]
- 21.Peng X, Zha W, He R, Lu T, Zhu L, Han B, et al. Pyrosequencing the midgut transcriptome of the brown planthopper, Nilaparvata lugens. Insect molecular biology. 2011;20(6):745–62. 10.1111/j.1365-2583.2011.01104.x. WOS:000297289000006. [DOI] [PubMed] [Google Scholar]
- 22.Zagrobelny M, Scheibye-Alsing K, Jensen NB, Moller BL, Gorodkin J, Bak S. 454 pyrosequencing based transcriptome analysis of Zygaena filipendulae with focus on genes involved in biosynthesis of cyanogenic glucosides. BMC genomics. 2009;10 Artn 574 10.1186/1471-2164-10-574. WOS:000272804700002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang FJ, Guo HY, Zheng HJ, Zhou T, Zhou YJ, Wang SY, et al. Massively parallel pyrosequencing-based transcriptome analyses of small brown planthopper (Laodelphax striatellus), a vector insect transmitting rice stripe virus (RSV). BMC genomics. 2010;11 Artn 303 10.1186/1471-2164-11-303. WOS:000279862300001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gibbons JG, Janson EM, Hittinger CT, Johnston M, Abbot P, Rokas A. Benchmarking Next-Generation Transcriptome Sequencing for Functional and Evolutionary Genomics. Mol Biol Evol. 2009;26(12):2731–44. 10.1093/molbev/msp188. WOS:000271818500008. [DOI] [PubMed] [Google Scholar]
- 25.Pantoja A, Salazar A, Macchiavelli R. Recognition of instars and adult trap catches of Cosmopolites sordidus (Coleoptera: Curculionidae) from plantains in Puerto Rico. Ann Entomol Soc Am. 2006;99(5):875–8. [Google Scholar]
- 26.Zhulidov PA, Bogdanova EA, Shcheglov AS, Vagner LL, Khaspekov GL, Kozhemyako VB, et al. Simple cDNA normalization using kamchatka crab duplex-specific nuclease. Nucleic Acids Res. 2004;32(3). ARTN e37 10.1093/nar/gnh031. WOS:000220490400054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–6. 10.1093/bioinformatics/bti610. WOS:000231694600016. [DOI] [PubMed] [Google Scholar]
- 28.O'Neil ST, Dzurisin JDK, Carmichael RD, Lobo NF, Emrich SJ, Hellmann JJ. Population-level transcriptome sequencing of nonmodel organisms Erynnis propertius and Papilio zelicaon. BMC genomics. 2010;11 Artn 310 10.1186/1471-2164-11-310. WOS:000279862800001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jaworski DC, Zou Z, Bowen CJ, Wasala NB, Madden R, Wang Y, et al. Pyrosequencing and characterization of immune response genes from the American dog tick, Dermacentor variabilis (L.). Insect molecular biology. 2010;19(5):617–30. 10.1111/j.1365-2583.2010.01037.x. WOS:000281796000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zou Z, Najar F, Wang Y, Roe B, Jiang HB. Pyrosequence analysis of expressed sequence tags for Manduca sexta hemolymph proteins involved in immune responses. Insect Biochem Molec. 2008;38(6):677–82. 10.1016/j.ibmb.2008.03.009. WOS:000257002900008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang XW, Luan JB, Li JM, Bao YY, Zhang CX, Liu SS. De novo characterization of a whitefly transcriptome and analysis of its gene expression during development. BMC genomics. 2010;11 Artn 400 10.1186/1471-2164-11-400. WOS:000279870400001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eyun SI, Wang HC, Pauchet Y, Ffrench-Constant RH, Benson AK, Valencia-Jimenez A, et al. Molecular Evolution of Glycoside Hydrolase Genes in the Western Corn Rootworm (Diabrotica virgifera virgifera). Plos One. 2014;9(4). ARTN e94052 10.1371/journal.pone.0094052. WOS:000334339000073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hlavica P. Insect cytochromes P450: Topology of structural elements predicted to govern catalytic versatility. J Inorg Biochem. 2011;105(10):1354–64. 10.1016/j.jinorgbio.2011.05.003. WOS:000295423700013. [DOI] [PubMed] [Google Scholar]
- 34.Oakeshott JG, Johnson RM, Berenbaum MR, Ranson H, Cristino AS, Claudianos C. Metabolic enzymes associated with xenobiotic and chemosensory responses in Nasonia vitripennis. Insect molecular biology. 2010;19:147–63. 10.1111/j.1365-2583.2009.00961.x. WOS:000273947400015. [DOI] [PubMed] [Google Scholar]
- 35.Neurath H. Evolution of Proteolytic-Enzymes. Science. 1984;224(4647):350–7. [DOI] [PubMed] [Google Scholar]
- 36.Merzendorfer H. Insect chitin synthases: a review. J Comp Physiol B. 2006;176(1):1–15. 10.1007/s00360-005-0005-3. WOS:000235309200001. [DOI] [PubMed] [Google Scholar]
- 37.Arakane Y, Muthukrishnan S, Beeman RW, Kanost MR, Kramer KJ. Laccase 2 is the phenoloxidase gene required for beetle cuticle tanning. P Natl Acad Sci USA. 2005;102(32):11337–42. 10.1073/pnas.0504982102. WOS:000231253400039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murdock LL, Brookhart G, Dunn PE, Foard DE, Kelley S, Kitch L, et al. Cysteine Digestive Proteinases in Coleoptera. Comp Biochem Phys B. 1987;87(4):783–7. 10.1016/0305-0491(87)90388-9. WOS:A1987J493000021. [DOI] [Google Scholar]
- 39.Thie NMR, Houseman JG. Identification of Cathepsin-B, Cathepsin-D and Cathepsin-H in the Larval Midgut of Colorado Potato Beetle, Leptinotarsa-Decemlineata Say (Coleoptera, Chrysomelidae). Insect Biochem. 1990;20(3):313–8. [Google Scholar]
- 40.Gill M, Ellar D. Transgenic Drosophila reveals a functional in vivo receptor for the Bacillus thuringiensis toxin Cry1Ac1. Insect molecular biology. 2002;11(6):619–25. 10.1046/j.1365-2583.2002.00373.x. WOS:000179068300012. [DOI] [PubMed] [Google Scholar]
- 41.Rajagopal R, Sivakumar S, Agrawal N, Malhotra P, Bhatnagar RK. Silencing of midgut aminopeptidase N of Spodoptera litura by double-stranded RNA establishes its role as bacillus thuringiensis toxin receptor. J Biol Chem. 2002;277(49):46849–51. 10.1074/jbc.C200523200. WOS:000179663700003. [DOI] [PubMed] [Google Scholar]
- 42.Reeck GR, Kramer K. J., Baker J. E., Kanost M. R., Fabrick J. A., and Behnke C. A. Advances in Insect Control: The Role of Transgenic Plants In: Carozzi NaK M. G., editor. Proteinase inhibitors and resistance of transgenic plants to insects. London: Taylor and Francis; 1997. p. 157–83. [Google Scholar]
- 43.Joanitti GA, Freitas S.M., Silva L.P. Proteinaceous Protease Inhibitors: Structural Features and Multiple Functional Faces. Curr Enzyme Inhibition 2 2006:199–217. [Google Scholar]
- 44.Masoud SA, Johnson LB, White FF, Reeck GR. Expression of a cysteine proteinase inhibitor (oryzacystatin-I) in transgenic tobacco plants. Plant Mol Biol. 1993;21(4):655–63. . [DOI] [PubMed] [Google Scholar]
- 45.Johnson KS, Rabosky D. Phylogenetic distribution of cysteine proteinases in beetles: evidence for an evolutionary shift to an alkaline digestive strategy in Cerambycidae, Comp. Comp Biochem Phys A. 2000;(126):10. [DOI] [PubMed] [Google Scholar]
- 46.Weis WI, Taylor ME, Drickamer K. The C-type lectin superfamily in the immune system. Immunol Rev. 1998;163:19–34. 10.1111/j.1600-065X.1998.tb01185.x. WOS:000074973900003. [DOI] [PubMed] [Google Scholar]
- 47.Xie W, Lei YY, Fu W, Yang ZX, Zhu X, Guo ZJ, et al. Tissue-Specific Transcriptome Profiling of Plutella Xylostella Third Instar Larval Midgut. Int J Biol Sci. 2012;8(8):1142–55. 10.7150/Ijbs.4588. WOS:000310055600006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Richards S, Gibbs RA, Weinstock GM, Brown SJ, Denell R, Beeman RW, et al. The genome of the model beetle and pest Tribolium castaneum. Nature. 2008;452(7190):949–55. 10.1038/Nature06784. WOS:000255208600030. [DOI] [PubMed] [Google Scholar]
- 49.Richards S, Liu Y, Bettencourt BR, Hradecky P, Letovsky S, Nielsen R, et al. Comparative genome sequencing of Drosophila pseudoobscura: Chromosomal, gene, and cis-element evolution. Genome Res. 2005;15(1):1–18. 10.1101/Gr.3059305. WOS:000226170900001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weinstock GM, Robinson GE, Gibbs RA, Worley KC, Evans JD, Maleszka R, et al. Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 2006;443(7114):931–49. 10.1038/Nature05260. WOS:000241523400044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
An asterisk (*) indicates identical residues, semicolon (:) indicates highly conserved substitutions and a period (.) indicates semi-conserved substitutions. Dashes represent gaps introduced to preserve alignment. Species and accession numbers included in the alignment were C. sordidus (AFH35127), D. ponderosae (AEE63523), P. papatasi (ABV44754), and L. longipalpis (ABV60312).
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.