Abstract
Carotenoids are not only important to the plants themselves but also are beneficial to human health. Since citrus fruit is a good source of carotenoids for the human diet, it is important to study carotenoid profiles and the accumulation mechanism in citrus fruit. Thus, in the present paper, we describe the diversity in the carotenoid profiles of fruit among citrus genotypes. In regard to carotenoids, such as β-cryptoxanthin, violaxanthin, lycopene, and β-citraurin, the relationship between the carotenoid profile and the expression of carotenoid-biosynthetic genes is discussed. Finally, recent results of quantitative trait locus (QTL) analyses of carotenoid contents and expression levels of carotenoid-biosynthetic genes in citrus fruit are shown.
Keywords: citrus, carotenoid, β-cryptoxanthin, violaxanthin, β-citraurin, gene expression, quantitative trait locus
Introduction
Carotenoids play two key roles in the photosynthetic organisms: they serve as accessory pigments in the photosystems, and they protect the photosynthetic apparatus against toxic reactive oxygen species (Ramel et al. 2013). In higher plants, yellow, orange and red carotenoids accumulate in flowers and fruits to attract pollinators and agents of seed dispersal. In addition, epoxy-carotenoids, violaxanthin, and neoxanthin are precursors for the plant hormone abscisic acid (Rock and Zeevaart 1991).
Carotenoids are also important components of fruit quality and their presence as pigments dictates peel and juice color in fruit. Moreover, carotenoids are beneficial to human health. Some carotenoids are precursors of vitamin A, which is essential to humans, and antioxidants, which reduce the risk of chronic diseases (Männistö et al. 2004, Olson 1989, Yuan et al. 2003). Furthermore, nutritional epidemiologic studies showed that intake of β-cryptoxanthin, a carotenoid plentiful in satsuma mandarin (Citrus unshiu Marcow.) fruit, may reduce the risk of lifestyle-related diseases, such as liver dysfunction (Sugiura et al. 2005), osteoporosis (Sugiura et al. 2011, Sugiura et al. 2012), and metabolic syndrome (Sugiura et al. 2008). These reports indicate that regulating carotenoid content in citrus fruit is important for human health because the fruit is a good major source of carotenoids, especially of β-cryptoxanthin, in the human diet.
In this paper, we explain the diversity in the carotenoid profiles of fruit among citrus genotypes. Moreover, the relationship between the accumulation of carotenoids and the expression of carotenoid-biosynthetic genes is discussed. Finally, the potential for genetic regulation of carotenoid content is shown.
Diversity in carotenoid profiles among citrus genotypes
Citrus fruit is a complex source of carotenoids. Approximately 115 different carotenoids have been reported in citrus, including a large number of isomers (Goodner et al. 2001, Wheaton and Stewart 1973). Thus, several studies have been conducted to differentiate citrus genotypes on the basis of the carotenoid profiles obtained using high-performance liquid chromatography equipped with a photodiode array detector and a C30 column. Goodner et al. (2001) analyzed 12 carotenoids of 32 citrus juices prepared from oranges, mandarins, and their hybrids and demonstrated that these fruits were clearly distinguishable by the difference in their β-cryptoxanthin content. Fanciullino et al. (2006) reported that cis-violaxanthin and β-cryptoxanthin in juice were strong determinants for classification of the 25 citrus genotypes. Matsumoto et al. (2007) developed a highly sensitive liquid chromatography-mass spectrometry (LC-MS) method for the simultaneous quantification of 18 carotenoids and analyzed carotenoid contents in the flavedos and juice sacs of 39 cultivars from October to January. This study also showed that violaxanthin and β-cryptoxanthin are major factors in discriminating citrus genotypes in each fruit tissue. Moreover, the results suggested that citrus cultivars could be roughly divided into three groups: β-cryptoxanthin abundant, violaxanthin abundant, and carotenoid poor. Most mandarin cultivars, including satsuma mandarin and ponkan (C. reticulata Blanco), were classified into the β-cryptoxanthin abundant category in both the flavedo and juice sacs. In contrast, orange cultivars, including common sweet orange (C. sinensis (L.) Osbeck ‘Trovita’) and navel orange (C. sinensis (L.) Osbeck ‘Washington’), were classified into the violaxanthin abundant category in both the flavedo and juice sacs. Other cultivars, including lime (C. aurantifolia (Cristm.) Swingle), lemon (C. limon (L.) Burm. f.), grapefruit (C. paradisi Macfad.), and pummelo (C. grandis (L.) Osbeck), were separated from oranges and mandarins because of the low violaxanthin and β-cryptoxanthin contents both in the flavedo and juice sacs.
The effect of genotypes and environments on carotenoid content was also investigated in citrus juice (Dhuique-Mayer et al. 2009) and juice sacs (Nonaka et al. 2012). On the basis of the carotenoid content of 3 sweet oranges (‘Pera’, ‘Valencia’, and ‘Sanguinelli’) during 3 seasons in Mediterranean conditions, it was demonstrated that annual variations were extremely limited as compared to variations caused by genotype influences in juice (Dhuique-Mayer et al. 2009). In contrast, on the basis of the carotenoid content of sweet orange in highly contrasting geographical conditions, Mediterranean (Corsica), subtropical (New Caledonia), and tropical (Tahiti, Costa Rica, and Cuba), the effect of high variation of geographical conditions on carotenoid was characterized (Dhuique-Mayer et al. 2009). Moreover, it was revealed that the Mediterranean condition amplifies the differentiation among genotypes, particularly by increasing the β-cryptoxanthin and cis-violaxanthin content in sweet oranges and the β-carotene, phytoene, and phytofluene content in mandarins (Dhuique-Mayer et al. 2009). Nonaka et al. (2012) assayed the juice sacs of 48 citrus cultivars and selections cultivated at two locations (Nagasaki and Shizuoka) in Japan. In this study, broad-sense heritabilities of the contents of most carotenoids, such as β-cryptoxanthin and violaxanthin, in the 48 genotypes were high, indicating that the differences among the obtained values for 48 genotypes were mostly derived from genetic variation.
The pathway of carotenoid biosynthesis in plants and the expression of carotenoid-biosynthetic genes during maturation of citrus fruit
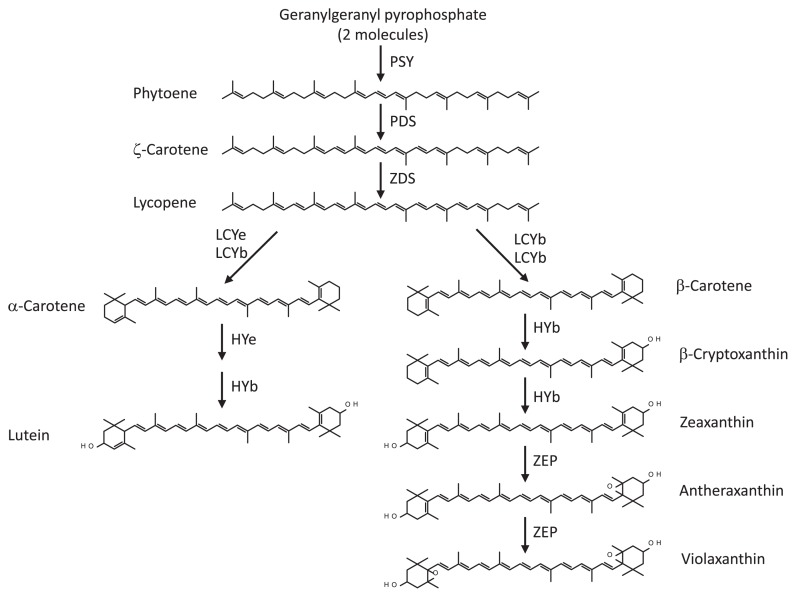
The pathway of carotenoid biosynthesis in plants has been studied extensively (Bouvier et al. 2000, Cunningham et al. 1996, 1998, Park et al. 2002, Ronen et al. 1999, Fig. 1). The first step of carotenoid biosynthesis is condensing two molecules of geranylgeranyl pyrophosphate (C20) to form a colorless phytoene (C40) catalyzed by phytoene synthase (PSY). Phytoene desaturase (PDS) and ζ-carotene desaturase (ZDS) introduce four double bonds into phytoene to produce lycopene. The cyclization of lycopene in the next step is a crucial branching point in this pathway, yielding α-carotene with one ɛ-ring and one β-ring, and β-carotene with two β-rings, in which two cyclases, lycopene β-cyclase (LCYb) and lycopene ɛ-cyclase (LCYe), are responsible for these reactions. The formation of β-carotene is catalyzed by a single cyclase, LCYb, whereas α-carotene is catalyzed by two different cyclases, LCYe and LCYb. β-Carotene is converted to zeaxanthin via β-cryptoxanthin by two-step hydroxylation, which is catalyzed by β-ring hydroxylase (HYb). Furthermore, zeaxanthin is converted to violaxanthin via antheraxanthin by zeaxanthin epoxidase (ZEP). α-Carotene is also converted into lutein by ɛ- and β-ring hydroxylase.
Fig. 1.
Principal pathway of carotenoid biosynthesis in plants. Abbreviations of carotenoid-biosynthetic enzymes: PSY, phytoene synthase; PDS, phytoene desaturase; ZDS, ζ-carotene desaturase; LCYe, lycopene ɛ-cyclase; LCYb, lycopene β-cyclase; HYe, ɛ-ring hydroxylase; HYb, β-ring hydroxylase; ZEP, zeaxanthin epoxidase.
In the initial stage of studying carotenoid-biosynthetic genes in citrus, increases in the expression of each gene, PSY (Ikoma et al. 2001, Kim et al. 2001a), PDS (Kita et al. 2001), and HYb (Kim et al. 2001b), were individually observed in the flavedo and juice sacs during fruit maturation. Subsequently, the simultaneous investigation of the expression of carotenoid-biosynthetic genes, such as PSY, PDS, ZDS, LCYb, LCYe, HYb, and ZEP, was conducted in the flavedo and juice sacs (Fanciullino et al. 2008, Kato et al. 2004, Rodrigo et al. 2004). Kato et al. (2004) showed that with the transition of peel color from green to orange, the change from β,ɛ-carotenoid (α-carotene and lutein) accumulation to β,β-carotenoid (β-carotene, β-cryptoxanthin, zeaxanthin, and violaxanthin) accumulation was observed in the flavedos of satsuma mandarin, ‘Valencia’ sweet orange and ‘Lisbon’ lemon, accompanying the disappearance of LCYe transcripts and the increase in LCYb transcripts. Rodrigo et al. (2004) also indicated transcriptional downregulation of LCYe in the flavedo of ‘Navelate’ navel orange with the transitional stage of peel color. These results suggest that in the flavedo, a decrease in the gene expression of LCYe is predominantly responsible for the pathway changing from β,ɛ-carotenoid synthesis to β,β-carotenoid synthesis with the transition of the flavedo’s color from green to orange. During massive carotenoid accumulation at the orange stage, Kato et al. (2004) showed that a simultaneous increase in the expression of genes (PSY, PDS, ZDS, LCYb, HYb, and ZEP) led to massive β,β-xanthophyll (β-cryptoxanthin, zeaxanthin, and violaxanthin) accumulation in both the flavedo and the juice sacs as fruit maturation progressed in satsuma mandarin and ‘Valencia’ sweet orange. Rodrigo et al. (2004) observed an increase in the expression of PSY, PDS, ZDS, and HYb genes with massive accumulations of carotenoids in the flavedo of ‘Navelate’ navel orange. Fanciullino et al. (2008) indicated that the accumulation of β,β-xanthophylls in the juice sacs of ‘Shamouti’ and ‘Sanguinelli’ sweet oranges was apparently related to increased transcript levels of PSY, PDS, ZDS, HYb, and ZEP. In contrast, Kato et al. (2004) suggested that low levels of gene expression for β,β-xanthophyll synthesis lead to an extremely low concentration of β,β-xanthophyll in the flavedo and juice sacs of ‘Lisbon’ lemon. These results suggest that an increase in the expression of all or most genes related to carotenoid biosynthesis from phytoene to violaxanthin is responsible for the massive accumulation of β,β-xanthophyll. In addition to the expression of carotenoid-biosynthetic genes, Fanciullino et al. (2008) also investigated that of DXS, the deoxyxylulose 5-phosphate synthase that controls the first step of the methylerythritol phosphate (MEP) pathway; they showed that an increase in the gene expression of DXS was responsible for the massive accumulation of β,β-xanthophyll.
The mechanism of the specific accumulation of β-cryptoxanthin during maturation in citrus juice sacs
β-Cryptoxanthin is an intermediate of a two-step hydroxylation by HYb. Thus, it seems that the massive accumulation of β-cryptoxanthin in juice sacs is controlled by not only transcriptional factors but also other factors, such as enzymatic characteristics. Sun et al. (1996) demonstrated that β-cryptoxanthin, rather than zeaxanthin, was mainly accumulated in Escherichia coli cells carrying the truncated HYb gene. On the basis of this result, they indicated that HYb hydroxylates the β-rings of β-carotene more efficiently than the not-yet-hydroxylated β-ring of β-cryptoxanthin. Li et al. (2010) also analyzed the function of two HYb cDNAs (Zmbch1 and Zmbch2), which were isolated from maize, in E. coli cells that carried their cDNAs without truncation. ZmBCH1 could convert β-carotene into β-cryptoxanthin and zeaxanthin, but ZmBCH2 could form β-cryptoxanthin alone and had lower overall activity than ZmBCH1. Taken together, these observations in truncated HYb and ZmBCH2 suggest that the accumulation of β-cryptoxanthin predominated in two-step hydroxylation by HYb when the enzyme activity was insufficient. Moreover, these observations suggest that HYb preferred the first-step conversion from β-carotene to β-cryptoxanthin rather than the second-step conversion from β-cryptoxanthin to zeaxanthin under low HYb activity and/or excessive β-carotene supply.
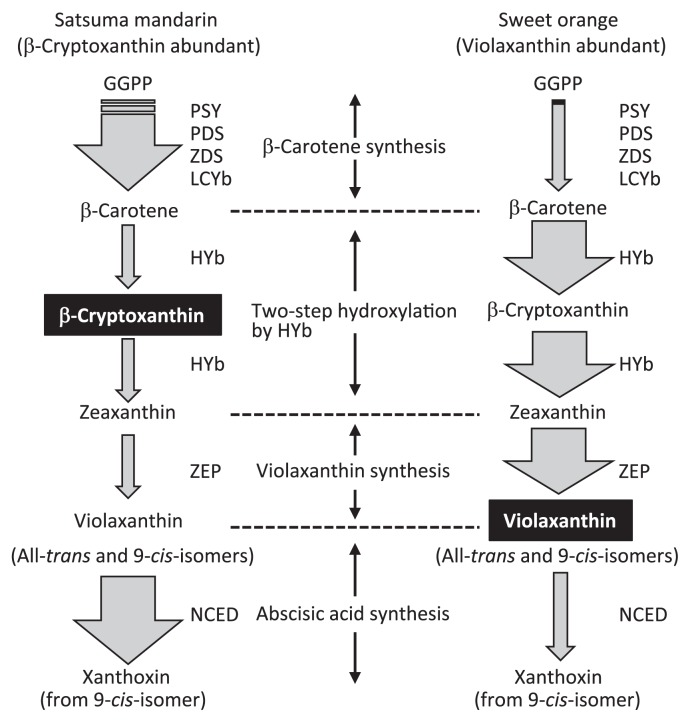
In citrus fruit, the substrate specificity of HYb seems important for regulating β-cryptoxanthin accumulation. Kato et al. (2004) investigated the mechanism causing the diversity of carotenoid profiles in the juice sacs of citrus species, satsuma mandarin (a β-cryptoxanthin abundant species) and ‘Valencia’ orange (a violaxanthin abundant species), on the basis of the gene expression profiles of carotenoid-biosynthetic enzymes (Fig. 2). This study showed that the gene expression of upstream carotene synthesis (PSY, PDS, ZDS, and LCYb) in satsuma mandarin exceeded that in sweet orange, whereas the gene expression of downstream xanthophyll synthesis (HYb and ZEP) in satsuma mandarin was lower than that in sweet orange (Fig. 2). The higher expression of upstream synthesis genes and lower expression of the HYb gene suggested a higher supply of β-carotene and lower HYb activity in the juice sacs of satsuma mandarin than in those of sweet orange. Therefore, Kato et al. (2004) concluded that, amid an equilibrium of a high gene expression of upstream synthesis and low gene expression of HYb (high supply of β-carotene and low HYb activity), HYb predominantly catalyzed the first-step conversion due to its high substrate specificity to β-carotene, leading to a marked accumulation of β-cryptoxanthin in the juice sacs of satsuma mandarin.
Fig. 2.
Comparison of gene expression for carotenoid biosynthesis between the Satsuma mandarin and Valencia orange in juice sacs during massive carotenoid accumulation. The width of the arrows indicates the relative gene expression levels for carotenoid-biosynthetic enzymes. Carotenoids written in white characters were abundant in each citrus species. Abbreviations of carotenoid-biosynthetic enzymes: PSY, phytoene synthase; PDS, phytoene desaturase; ZDS, ζ-carotene desaturase; LCYb, lycopene β-cyclase; HYb, β-ring hydroxylase; ZEP, zeaxanthin epoxidase; NCED, 9-cis-epoxycarotenoid dioxygenase.
In contrast, Kato et al. (2004) suggested that, in the juice sacs of sweet orange, HYb was likely to sufficiently catalyze the reaction to zeaxanthin via β-cryptoxanthin due to the low gene expression of upstream synthesis and high gene expression of HYb (low supply of β-carotene and high HYb activity). Moreover, they suggested that zeaxanthin was rapidly converted to violaxanthin by ZEP in the juice sacs of sweet orange, as the intensity in the gene expression of ZEP was much higher in the juice sacs of sweet orange than in those of satsuma mandarin. Recently, Wei et al. (2014) investigated carotenoid accumulation and gene expression for carotenoid biosynthesis during fruit maturation in ‘Valencia’ sweet orange (common type) and its more deeply colored mutant, ‘Rohde Red Valencia’ sweet orange. This study showed that in the juice sacs, the common type mainly accumulated violaxanthin, but the mutant accumulated β-cryptoxanthin and violaxanthin during orange stages. Moreover, the study showed that several members of upstream genes (PDS, ZDS, and LCYb) were expressed at higher levels in the mutant than in the common type, although distinct differences in the level of gene expression for HYb were not observed among these genotypes. Thus, it seems that the substrate specificity of HYb is also responsible for the high β-cryptoxanthin content of the juice sacs in the mutant, as the gene expression profiles suggest that a supply of β-carotene for HYb is higher in the mutant than in the common type of ‘Valencia’ sweet orange.
The relationship between carotenoid accumulation and gene expression of 9-cis-epoxycarotenoid dioxygenase in citrus juice sacs
Carotenoid cleavage dioxygenases (CCDs) are a group of enzymes that catalyze the oxidative cleavage of carotenoids (Ryle and Hausinger 2002). In Arabidopsis, the CCD family contains 9 members (CCD1, NCED2, NCED3, CCD4, NCED5, NCED6, CCD7, CCD8, and NCED9); orthologs in other plant species are typically named according to their homology with an Arabidopsis CCD (Huang et al. 2009). In the initial stage of studying CCDs in citrus, 9-cis-epoxycarotenoid dioxygenase (NCED) was isolated and characterized (Kato et al. 2006, Rodrigo et al. 2006). NCED catalyzes the cleavage of 9-cis-violaxanthin or 9′-cis-neoxanthin at the 11–12 position to form C25 epoxy-apocarotenal and xanthoxin (C15), a precursor of the plant hormone abscisic acid (Schwartz et al. 1997, 2001, 2003). It was also demonstrated that citrus NCEDs, which were isolated from satsuma mandarin (Kato et al. 2006), cleaved 9-cis-violaxanthin to form xanthoxin by using their recombinant proteins.
From satsuma mandarin, cDNAs for NCED3 and NCED5 were isolated (Kato et al. 2006). NCED3 and NCED5 were previously described as CitNCED3 and CitNCED2, respectively (Kato et al. 2006). On the basis of these nucleotide sequences, gene expression of NCED3 and NCED5 was investigated in the juice sacs of satsuma mandarin, lemon, and sweet orange. In the juice sacs, increased abscisic acid levels were observed in the satsuma mandarin during fruit maturation. As the abscisic acid accumulated, the gene expressions of NCED3 and NCED5 also increased. In the juice sacs of lemon, the levels of the NCED5 gene expression soared as abscisic acid accumulated during the green stage (from August to October), whereas NCED3 gene expression changed regardless of the abscisic acid level. These results suggest that NCED3 and NCED5 gene expression in satsuma mandarin and NCED5 gene expression in lemon were primarily responsible for the abscisic acid accumulation in their juice sacs. In the juice sacs of sweet orange, the abscisic acid level was much lower than those in satsuma mandarin and lemon. In sweet orange, no noticeable increase in NCED5 gene expression was observed. In addition, NCED3 gene expression changed regardless of the abscisic acid level. This result suggests that, in sweet orange, the extremely low level of NCED5 was primarily responsible for the low level of abscisic acid. Taken together, it seems that in the juice sacs, NCED5 is likely to play a primary role in the cleavage of 9-cis-violaxanthin to form C25 epoxy-apocarotenal and xanthoxin. In contrast, Rodrigo et al. (2006) suggested that in the leaves and flavedos of sweet orange, the cleavage reaction was primarily catalyzed by NCED3. NECD3 was previously described as CsNCED1 (Rodrigo et al. 2006). Thus, NCEDs would be differentially expressed in citrus tissues.
In mature fruit, the juice sacs of satsuma mandarin accumulated a low level of 9-cis-violaxanthin, whereas those of sweet orange accumulated a high level. Kato et al. (2006) explained the mechanism for different 9-cis-violaxanthin content among species. In sweet orange, 9-cis-violaxanthin in the juice sacs was not cleaved efficiently by NCED because NCED5 gene expression remained minimal (Fig. 2). Therefore, in sweet orange, the content of 9-cis-violaxanthin was high in the juice sacs. In satsuma mandarin and lemon, the 9-cis-violaxanthin was cleaved immediately by NCED because an increase in NCED5 gene expression was observed in the juice sacs (Fig. 2). Therefore, the content of 9-cis-violaxanthin in satsuma mandarin and lemon was much lower than that in sweet orange. Overall, it was thought that the oxidative cleavage of 9-cis-violaxanthin catalyzed by NCED affected the 9-cis-violaxanthin content and, consequently, the carotenoid profiles in the juice sacs of the three citrus species during fruit maturation. The oxidative cleavage of 9-cis-violaxanthin by NCED would also be responsible for the specific accumulation of β-cryptoxanthin in the juice sacs of satsuma mandarin.
Accumulation of specific carotenoids, lycopene and β-citraurin, and the expression of genes related to specific carotenoids
Lycopene is an uncommon carotenoid in citrus fruit. Most lycopene-accumulating cultivars are mutant grapefruits, pummelos, and oranges (Xu et al. 2006). Grapefruit have the greatest number of lycopene-accumulating mutants, including famous cultivars ‘Marsh Pink’, ‘Ruby Red’, and ‘Star Ruby’ (Gmitter 1993, Xu et al. 2006). Since characterization of the mutants is a useful experimental system for identifying the molecular mechanism that regulates lycopene accumulation, many studies have been conducted with lycopene-accumulating grapefruit mutants. β-Cyclization of lycopene, catalyzed by LCYb, seems to be a key regulatory step of lycopene accumulation. Thus, two types of LCYb, LCYb1 and LCYb2, have been characterized in grapefruit (Alquézar et al. 2009, Mendes et al. 2011). The function, cyclization of lycopene, of LCYb1 and LCYb2 from satsuma mandarin was confirmed by using their recombinant protein expressed in E. coli cells (Zhang et al. 2012). Mendes et al. (2011) showed that the gene expression of LCYb1 was not significantly different between two grapefruit mutants, ‘Flame’ (lycopene-accumulating mutant) and ‘Marsh’ (non-lycopene-accumulating mutant), was much lower than that of LCYb2, and was nearly constant during fruit maturation. In contrast, they indicated that the gene expression of LCYb2 in ‘Flame’ was significantly lower than that in ‘Marsh’. Alquézar et al. (2009) compared the gene expression of LCYb2 between high lycopene-accumulating ‘Star Ruby’ grapefruit and the non-lycopene-accumulating ‘Washington’ navel orange. This study revealed that the gene expression of LCYb2 in the flavedo and juice sacs was inhibited more in the high lycopene-accumulating grapefruit than in the non-lycopene-accumulating navel orange. Overall, these results suggest that the inhibited expression of the LCYb2 gene is predominantly responsible for lycopene accumulation in the fruit of grapefruit mutants. Moreover, Alquézar et al. (2009) isolated two different alleles of LCYb2, b-LCY2a and b-LCY2b, and demonstrated by functional assays that the LCYb activity of allele b was almost null. In their study, it was noticed that ‘Star Ruby’ grapefruit predominantly expressed the nonfunctional b-LCY2b allele during fruit ripening, whereas navel orange preferably expressed the functional allele. On the other hand, Costa et al. (2012) investigated the expression of PSY, PDS, and ZDS genes in the lycopene-accumulating grapefruit mutant, ‘Flame’, and suggested that the mechanism controlling lycopene accumulation in red grapefruit involved not only the transcriptional downregulation of LCYb2 but also the transcriptional upregulation of PSY, which controls flux into the carotenoid pathway, during fruit ripening in lycopene-accumulating grapefruit. In lycopene-accumulating navel orange mutant, ‘Cara Cara’, and sweet orange mutant, ‘Hong Anliu’, the mechanism for lycopene accumulation has also been studied (Alquézar et al. 2008, Fanciullino et al. 2008, Liu et al. 2007, Tao et al. 2007, Xu et al. 2009, 2010).
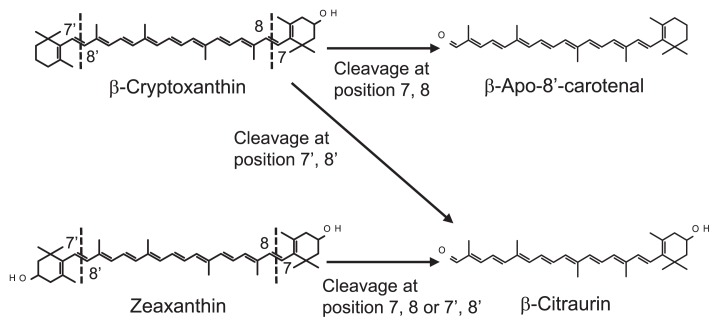
β-Citraurin, a C30 apocarotenoid, is an uncommon carotenoid that accumulates specifically in the peel of some citrus cultivars (Oberholster et al. 2001). The color is responsible for the reddish color of citrus peel (Farin et al. 1983, Oberholster et al. 2001). Although β-citraurin was presumed to be a degradation product of β-cryptoxanthin or zeaxanthin (Oberholster et al. 2001, Ríos et al. 2010, Rodrigo et al. 2004) on the basis of the similarity in the structures among these carotenoids, the biosynthetic pathway could not be determined until recently. In 2013, two groups independently elucidated that CCD4, a carotenoid cleavage dioxygenase, was responsible for β-citraurin biosynthesis (Ma et al. 2013, Rodrigo et al. 2013). In these studies, functional analyses in vitro showed that the recombinant protein of CCD4 cleaved zeaxanthin at its 7, 8 or 7′, 8′ position, leading to β-citraurin production (Ma et al. 2013, Rodrigo et al. 2013, Fig. 3). These analyses also demonstrated that the recombinant protein of CCD4 cleaved β-cryptoxanthin at its 7, 8 or 7′, 8′ position, leading to β-apo-8′-carotenal production and β-citraurin production, respectively (Ma et al. 2013, Rodrigo et al. 2013, Fig. 3). Moreover, the gene expression of CCD4 was investigated in two satsuma mandarin cultivars of ‘Yamashitabeni-wase’, which accumulates β-citraurin predominantly, and ‘Miyagawa-wase’, which does not accumulate β-citraurin (Ma et al. 2013). In this study, increased expression of CCD4 was consistent with accumulation of β-citraurin in the flavedo of ‘Yamashitabeni-wase’. In contrast, the expression of CCD4 remained at an extremely low level during the ripening process in the flavedo of ‘Miyagawa-wase’, resulting in the absence of β-citraurin. Rodrigo et al. (2013) also observed that in the flavedos of 3 citrus cultivars, the increase in β-citraurin content was consistent with upregulation of CCD4 gene expression. Zheng et al. (2015) revealed that in the flavedos of some citrus cultivars, there was a positive correlation between CCD4 expression levels and the presence of β-citraurin. Taken together, these observations in functional analyses of CCD4 and the gene expression of CCD4 in flavedos suggest that the accumulation of β-citraurin is controlled by the cleavage of β-cryptoxanthin and zeaxanthin by CCD4.
Fig. 3.
The pathway from β-cryptoxanthin and zeaxanthin to β-citraurin catalyzed by carotenoid cleavage dioxygenase 4 (CCD4) in citrus fruit. Dotted lines indicate the positions cleaved by CCD4.
Quantitative trait locus analysis for carotenoid content and expression levels of carotenoid-biosynthetic genes in citrus fruit
An increase in carotenoids, especially the β-cryptoxanthin content, is an important breeding objective for citrus in Japan. However, there have been few detailed genetic analyses of carotenoid content in citrus until recently. Analysis of quantitative trait locus (QTL) using a segregating population is an effective means of obtaining genetic information on agronomically important traits. This method allows the identification of genetic regions associated with certain quantitative traits on linkage groups, whereupon genetically linked selection markers can be obtained for breeding.
QTL analyses of carotenoid content have been reported for the tomato (Blauth et al. 1998) and carrot (Santos and Simon 2002). In maize kernels, two QTLs were detected, and their locations on the genetic map were associated with the loci of phytoene synthase and ζ-carotene desaturase genes (Wong et al. 2004). In the cauliflower, Lu et al. (2006) showed that the Or gene, which regulates the synthesis of a chaperone protein of the DnaJ cysteine-rich domain, regulates β-carotene accumulation. Thus, these previous studies detected QTLs related to the accumulation of various carotenoids such as β-carotene and lycopene. However, QTL analysis had not been conducted on a plant with high β-cryptoxanthin content.
Sugiyama et al. (2011) performed QTL analysis to identify loci related to carotenoid content in the juice sacs of citrus fruit. In their study, QTL of a mapping population derived from a cross between two citrus parents, ‘Okitsu-46’ and ‘Nou-5’, were investigated. The study showed that the strongest QTL for β-cryptoxanthin content was broadly located from 4.1 to 7.0 cM on linkage group 6 of Nou-5, including the Gn0005 locus. The study also demonstrated that the mean of the β-cryptoxanthin content in the juice sacs was 1.3 mg/100 g for homozygous progenies genotyped with the Gn0005 marker, whereas that for heterozygous genotypes was 0.9 mg/100 g. When comparing Gn0005 marker genotypes, a significant difference in the means of the β-cryptoxanthin content of the juice sacs was detected.
Moreover, to identify the regulatory factors of gene expression for carotenoid-biosynthetic enzymes, Sugiyama et al. (2014) investigated the expression quantitative trait locus (eQTL) by using the above-mentioned population. In their study, significant eQTLs for PSY, PDS, ZDS, HYb, and ZEP were detected. The results indicated that the expression levels of HYb and ZEP were influenced by cis-elements in their upstream regions, since the eQTLs for HYb and ZEP were located on their responsible gene loci. In contrast, the results showed that the eQTLs for PDS and ZDS were located in different loci from those of the responsible genes, indicating that the loci were trans-regulating factors. In addition, the results suggested that the expression levels of PDS and ZDS were regulated by a common transcription factor because their eQTLs were co-localized. Thus, it was thought that eQTL analysis was a powerful tool for identifying cis- or trans-regulation for carotenoid-biosynthetic genes. Conclusively, it seems that information regarding QTL and eQTL is useful for the development of DNA markers to select progeny with higher carotenoid content. However, the results in these studies (Sugiyama et al. 2011, 2014) were preliminary because they obtained data in only one season and from a limited number of individuals (n = 51).
Conclusion
In this paper, the diversity of carotenoid profiles among citrus genotypes was discussed. The mechanism of carotenoid accumulation in citrus fruit was also explained by comparing the expression of genes related to carotenoid biosynthesis and catabolism among citrus species and mutants. Moreover, we showed genetic information for regulating carotenoid accumulation by QTL and eQTL analyses. We hope that in the future, the information reported in this paper will aid further technical development toward controlling carotenoid content in citrus and boosting its quality.
Literature Cited
- Alquézar, B., Rodrigo, M.J. and Zacarías, L. (2008) Regulation of carotenoid biosynthesis during fruit maturation in the red-fleshed orange mutant Cara Cara. Phytochemistry 69: 1997–2007. [DOI] [PubMed] [Google Scholar]
- Alquézar, B., Zacarías, L. and Rodrigo, M.J. (2009) Molecular and functional characterization of a novel chromoplast-specific lycopene β-cyclase from Citrus and its relation to lycopene accumulation. J. Exp. Bot. 60: 1783–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blauth, S.L., Churchill, G.A. and Mutschler, M.A. (1998) Identification of quantitative trait loci associated with acylsugar accumulation using intraspecific populations of the wild tomato, Lycopersicon pennellii. Theor. Appl. Genet. 96: 458–467. [DOI] [PubMed] [Google Scholar]
- Bouvier, F., D’Harlingue, A., Backhaus, R.A., Kumagai, M.H. and Camara, B. (2000) Identification of neoxanthin synthase as a carotenoid cyclase paralog. Eur. J. Biochem. 267: 6346–6352. [DOI] [PubMed] [Google Scholar]
- Costa, M.G.C., Moreira, C.D., Melton, J.R., Otoni, W.C. and Moore, G.A. (2012) Characterization and developmental expression of genes encoding the early carotenoid biosynthetic enzymes in Citrus paradisi Macf. Mol. Biol. Rep. 39: 895–902. [DOI] [PubMed] [Google Scholar]
- Cunningham, F.X., Pogson, B., Sun, Z., McDonald, K.A., DellaPenna, D. and Gantt, E. (1996) Functional analysis of the β and ɛ lycopene cyclase enzymes of Arabidopsis reveals a mechanism for control of cyclic carotenoid formation. Plant Cell 8: 1613–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham, F.X. and Gantt, E. (1998) Genes and enzymes of carotenoid biosynthesis in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49: 557–583. [DOI] [PubMed] [Google Scholar]
- Dhuique-Mayer, C., Fanciullino, A.L., Dubois, C. and Ollitrault, P. (2009) Effect of genotype and environment on citrus juice carotenoid content. J. Agric. Food Chem. 57: 9160–9168. [DOI] [PubMed] [Google Scholar]
- Fanciullino, A.L., Dhuique-Mayer, C., Luro, F., Casanova, J., Morillon, R. and Ollitrault, P. (2006) Carotenoid diversity in cultivated citrus is highly influenced by genetic factors. J. Agric. Food Chem. 54: 4397–4406. [DOI] [PubMed] [Google Scholar]
- Fanciullino, A.L., Cerćos, M., Dhique-Mayer, C., Froelicher, Y., Talón, M., Ollitrault, P. and Morillon, R. (2008) Changes in carotenoid content and biosynthetic gene expression in juice sacs of four orange varieties (Citrus sinensis) differing in flesh fruit color. J. Agric. Food Chem. 56: 3628–3638. [DOI] [PubMed] [Google Scholar]
- Farin, D., Ikan, R. and Gross, J. (1983) The carotenoid pigments in the juice and flavedo of a mandarin hybrid (Citrus reticulata) cv Michal during ripening. Phytochemistry 22: 403–408. [Google Scholar]
- Gmitter, F.G.Jr. (1993) “Marsh” grapefruit”. Fruit Var. J. 47: 130–133. [Google Scholar]
- Goodner, K.L., Rouseff, R.L. and Hofsommer, H.J. (2001) Orange, mandarin, and hybrid classification using multivariate statistics based on carotenoid profiles. J. Agric. Food Chem. 49: 1146–1150. [DOI] [PubMed] [Google Scholar]
- Huang, F.C., Molnár, P. and Schwab, W. (2009) Cloning and functional characterization of carotenoid cleavage dioxygenase 4 genes. J. Exp. Bot. 60: 3011–3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikoma, Y., Komatsu, A., Kita, M., Ogawa, K., Omura, M., Yano, M. and Moriguchi, T. (2001) Expression of a phytoene synthase gene and characteristic carotenoid accumulation during citrus fruit development. Physiol. Plant. 111: 232–238. [Google Scholar]
- Kato, M., Ikoma, Y., Matsumoto, H., Sugiura, M., Hyodo, H. and Yano, M. (2004) Accumulation of carotenoids and expression of carotenoid biosynthetic genes during maturation in citrus fruit. Plant Physiol. 134: 824–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, M., Matsumoto, H., Ikoma, Y., Okuda, H. and Yano, M. (2006) The role of carotenoid cleavage dioxygenases in the regulation of carotenoid profiles during maturation in citrus fruit. J. Exp. Bot. 57: 2153–2164. [DOI] [PubMed] [Google Scholar]
- Kim, I.J., Ko, K.C., Kim, C.S. and Chung, W.I. (2001a) Isolation and expression patterns of a cDNA encoding phytoene synthase in Citrus. J. Plant Physiol. 158: 795–800. [Google Scholar]
- Kim, I.J., Ko, K.C., Kim, C.S. and Chung, W.I. (2001b) Isolation and characterization of cDNAs encoding β-carotene hydroxylase in Citrus. Plant Sci. 161: 1005–1010. [Google Scholar]
- Kita, M., Komatsu, A., Omura, M., Yano, M., Ikoma, Y. and Moriguchi, T. (2001) Cloning and expression of CitPDS1, a gene encoding phytoene desaturase in citrus. Biosci. Biotechnol. Biochem. 65: 1424–1428. [DOI] [PubMed] [Google Scholar]
- Li, Q., Farre, G., Naqvi, S., Breitenbach, J., Sanahuja, G., Bai, C., Sandmann, G., Capell, T., Christou, P. and Zhu, C. (2010) Cloning and functional characterization of the maize carotenoid isomerase and β-carotene hydroxylase genes and their regulation during endosperm maturation. Transgenic Res. 19: 1053–1068. [DOI] [PubMed] [Google Scholar]
- Liu, Q., Xu, J., Liu, Y., Zhao, X., Deng, X., Guo, L. and Gu, J. (2007) A novel bud mutation that confers abnormal patterns of lycopene accumulation in sweet orange fruit (Citrus sinensis L. Osbeck). J. Exp. Bot. 58: 4161–4171. [DOI] [PubMed] [Google Scholar]
- Lu, S., Eck, J.V., Zhou, X., Lopez, A.B., O’Halloran, D.M., Cosman, K.M., Conlin, B.J., Paolillo, D.J., Garvin, D.F., Vrebalov, J.et al. (2006) The cauliflower Or gene encodes a DnaJ cysteine-rich domain-containing protein that mediates high levels of β-carotene accumulation. Plant Cell 18: 3594–3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, G., Zhang, L., Matsuta, A., Matsutani, K., Yamawaki, K., Yahata, M., Wahyudi, A., Motohashi, R. and Kato, M. (2013) Enzymatic formation of β-citraurin from β-cryptoxanthin and zeaxanthin by carotenoid cleavage dioxygenase4 in the flavedo of citrus fruit. Plant Physiol. 163: 682–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Männistö, S., Smith-Warner, S.A., Spiegelman, D., Albanes, D., Anderson, K., Brandt, P.A., Cerhan, J.R., Colditz, G., Feskanich, D., Freudenheim, J.L.et al. (2004) Dietary carotenoids and risk of lung cancer in a pooled analysis of seven cohort studies. Cancer Epidemiol. Biomark. Prev. 13: 40–48. [DOI] [PubMed] [Google Scholar]
- Matsumoto, H., Ikoma, Y., Kato, M., Kuniga, T., Nakajima, N. and Yoshida, T. (2007) Quantification of carotenoids in citrus fruit by LC-MS and comparison of patterns of seasonal changes for carotenoids among citrus varieties. J. Agric. Food Chem. 55: 2356–2368. [DOI] [PubMed] [Google Scholar]
- Mendes, A.F.S., Chen, C., Gmitter, F.G.Jr., Moore, G.A. and Costa, M.G.C. (2011) Expression and phylogenetic analysis of two new lycopene β-cyclases from Citrus paradisi. Physiol. Plant. 141: 1–10. [DOI] [PubMed] [Google Scholar]
- Nonaka, K., Kita, M., Ikoma, Y., Fukamachi, H., Imai, A., Yoshioka, T. and Yamada, M. (2012) Genetic differences and environmental variations in carotenoid contents of fruit flesh in parental population used in citrus breeding in Japan. J. Amer. Soc. Hort. Sci. 137: 243–249. [Google Scholar]
- Oberholster, R., Cowan, A.K., Molnár, P. and Tóth, G. (2001) Biochemical basis of color as an aesthetic quality in Citrus sinensis. J. Agric. Food Chem. 49: 303–307. [DOI] [PubMed] [Google Scholar]
- Olson, J.A. (1989) Provitamin A function of carotenoids: the conversion of β-carotene into vitamin A. J. Nutr. 119: 105–108. [DOI] [PubMed] [Google Scholar]
- Park, H., Kreunen, S.S., Cuttriss, A.J., DellaPenna, D. and Pogson, B.J. (2002) Identification of the carotenoid isomerase provides insight into carotenoid biosynthesis, prolamellar body formation, and photomorphogenesis. Plant Cell 14: 321–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramel, F., Mialoundama, A.S. and Havaux, M. (2013) Nonenzymic carotenoid oxidation and photooxidative stress signalling in plants. J. Exp. Bot. 64: 799–805. [DOI] [PubMed] [Google Scholar]
- Ríos, G., Naranjo, M.A., Rodrigo, M.J., Alós, E., Zacarías, L., Cercós, M. and Talón, M. (2010) Identification of a GCC transcription factor responding to fruit colour change events in citrus through the transcriptomic analyses of two mutants. BMC Plant Biol. 10: 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock, C.D. and Zeevaart, J.A. (1991) The aba mutant of Arabidopsis thaliana is impaired in epoxy-carotenoid biosynthesis. Proc. Natl. Acad. Sci. USA 88: 7496–7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo, M.J., Marcos, J.F. and Zacarías, L. (2004) Biochemical and molecular analysis of carotenoid biosynthesis in flavedo of orange (Citrus sinensis L.) during fruit development and maturation. J. Agric. Food Chem. 52: 6724–6731. [DOI] [PubMed] [Google Scholar]
- Rodrigo, M.J., Alquézar, B. and Zacarías, L. (2006) Cloning and characterization of two 9-cis-epoxycarotenoid dioxygenase genes, differentially regulated during fruit maturation and under stress conditions, from orange (Citrus sinensis L. Osbeck). J. Exp. Bot. 57: 633–643. [DOI] [PubMed] [Google Scholar]
- Rodrigo, M.J., Alquézar, B., Alós, E., Medina, V., Carmona, L., Bruno, M., Al-Babili, S. and Zacarías, L. (2013) A novel carotenoid cleavage activity involved in the biosynthesis of Citrus fruit-specific apocarotenoid pigments. J. Exp. Bot. 64: 4461–4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronen, G., Cohen, M., Zamir, D. and Hirschberg, J. (1999) Regulation of carotenoid biosynthesis during tomato fruit development: Expression of the gene for lycopene epsilon-cyclase is down-regulated during ripening and is elevated in the mutant. Delta. Plant J. 17: 341–351. [DOI] [PubMed] [Google Scholar]
- Ryle, M.J. and Hausinger, R.P. (2002) Non-heme iron oxygenases. Curr. Opin. Chem. Biol. 6: 193–201. [DOI] [PubMed] [Google Scholar]
- Santos, C.A.F. and Simon, P.W. (2002) QTL analyses reveal clustered loci for accumulation of major provitamin A carotenes and lycopene in carrot roots. Mol. Genet. Genomics 268: 122–129. [DOI] [PubMed] [Google Scholar]
- Schwartz, S.H., Tan, B.C., Gage, D.A., Zeevaart, J.A.D. and McCarty, D.R. (1997) Specific oxidative cleavage of carotenoids by VP14 of maize. Science 276: 1872–1874. [DOI] [PubMed] [Google Scholar]
- Schwartz, S.H., Qin, X. and Zeevaart, J.A.D. (2001) Characterization of a novel carotenoid cleavage dioxygenase from plants. J. Biol. Chem. 276: 25208–25211. [DOI] [PubMed] [Google Scholar]
- Schwartz, S.H., Qin, X. and Zeevaart, J.A.D. (2003) Elucidation of the indirect pathway of abscisic acid biosynthesis by mutants, genes, and enzymes. Plant Physiol. 131: 1591–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura, M., Nakamura, M., Ikoma, Y., Yano, M., Ogawa, K., Matsumoto, H., Kato, M., Ohshima, M. and Nagao, A. (2005) High serum carotenoids are inversely associated with serum gamma-glutamyltransferase in alcohol drinkers within normal liver function. J. Epidemiol. 15: 180–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura, M., Nakamura, M., Ogawa, K., Ikoma, Y., Matsumoto, H., Ando, F., Shimokata, H. and Yano, M. (2008) Associations of serum carotenoid concentrations with the metabolic syndrome: Interaction with smoking. Br. J. Nutr. 100: 1297–1306. [DOI] [PubMed] [Google Scholar]
- Sugiura, M., Nakamura, M., Ogawa, K., Ikoma, Y., Ando, F., Shimokata, H. and Yano, M. (2011) Dietary patterns of antioxidant vitamin and carotenoid intake associated with bone mineral density: Findings from post-menopausal Japanese female subjects. Osteoporos. Int. 22: 143–152. [DOI] [PubMed] [Google Scholar]
- Sugiura, M., Nakamura, M., Ogawa, K., Ikoma, Y. and Yano, M. (2012) High serum carotenoids associated with lower risk for bone loss and osteoporosis in post-menopausal Japanese female subjects: Prospective cohort study. PLoS ONE 7: e52643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama, A., Omura, M., Matsumoto, H., Shimada, T., Fujii, H., Endo, T., Shimizu, T., Nesumi, H. and Ikoma, Y. (2011) Quantitative trait loci (QTL) analysis of carotenoid content in Citrus fruit. J. Japan. Soc. Hort. Sci. 80: 136–144. [Google Scholar]
- Sugiyama, A., Omura, M., Shimada, T., Fujii, H., Endo, T., Shimizu, T., Nesumi, H., Nonaka, K. and Ikoma, Y. (2014) Expression quantitative trait loci analysis of carotenoid metabolism-related genes in citrus. J. Japan. Soc. Hort. Sci. 83: 32–43. [Google Scholar]
- Sun, Z., Gantt, E. and Cunningham, F.X.Jr. (1996) Cloning and functional analysis of the β-carotene hydroxylase of Arabidopsis thaliana. J. Biol. Chem. 271: 24349–24352. [DOI] [PubMed] [Google Scholar]
- Tao, N., Hu, Z., Liu, Q., Xu, J., Cheng, Y., Guo, L., Guo, W. and Deng, X. (2007) Expression of phytoene synthase gene (Psy) is enhanced during fruit ripening of Cara Cara navel orange (Citrus sinensis Osbeck). Plant Cell Rep. 26: 837–843. [DOI] [PubMed] [Google Scholar]
- Wei, X., Chen, C., Yu, Q., Gady, A., Yu, Y., Liang, G. and Gmitter, F.G.Jr. (2014) Comparison of carotenoid accumulation and biosynthetic gene expression between Valencia and Rohde Red Valencia sweet oranges. Plant Sci. 227: 28–36. [DOI] [PubMed] [Google Scholar]
- Wheaton, T.A. and Stewart, I. (1973) Optimum temperature and ethylene concentrations for postharvest development of carotenoid pigments in citrus. J. Amer. Soc. Hort. Sci. 98: 337–340. [Google Scholar]
- Wong, J.C., Lambert, R.J., Wurtzel, E.T. and Rocheford, T.R. (2004) QTL and candidate genes phytoene synthase and ζ-carotene desaturase associated with the accumulation of carotenoids in maize. Theor. Appl. Genet. 108: 349–359. [DOI] [PubMed] [Google Scholar]
- Xu, C.J., Fraser, P.D., Wang, W.J. and Bramley, P.M. (2006) Differences in the carotenoid content of ordinary citrus and lycopene-accumulating mutants. J. Agric. Food Chem. 54: 5474–5481. [DOI] [PubMed] [Google Scholar]
- Xu, Q., Yu, K., Zhu, A., Ye, J., Liu, Q., Zhang, J. and Deng, X. (2009) Comparative transcripts profiling reveals new insight into molecular processes regulating lycopene accumulation in a sweet orange (Citrus sinensis) red-flesh mutant. BMC Genomics 10: 540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Q., Liu, Y., Zhu, A., Wu, X., Ye, J., Yu, K., Guo, W. and Deng, X. (2010) Discovery and comparative profiling of microRNAs in a sweet orange red-flesh mutant and its wild type. BMC Genomics 11: 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, J.M., Stram, D.O., Arakawa, K., Lee, H. and Yu, M.C. (2003) Dietary cryptoxanthin and reduced risk of lung cancer: The Singapore Chinese health study. Cancer Epidemiol. Biomark. Prev. 12: 890–898. [PubMed] [Google Scholar]
- Zhang, L., Ma, G., Shirai, Y., Kato, M., Yamawaki, K., Ikoma, Y. and Matsumoto, H. (2012) Expression and functional analysis of two lycopene β-cyclases from citrus fruits. Planta 236: 1315–1325. [DOI] [PubMed] [Google Scholar]
- Zheng, X., Xie, Z., Zhu, K., Xu, Q., Deng, X. and Pan, Z. (2015) Isolation and characterization of carotenoid cleavage dioxygenase 4 genes from different citrus species. Mol. Genet. Genomics 290: 1589–1603. [DOI] [PubMed] [Google Scholar]