Abstract
Citrus is one of the most cultivated fruits in the world, and satsuma mandarin (Citrus unshiu Marc.) is a major cultivated citrus in Japan. Many excellent cultivars derived from satsuma mandarin have been released through the improvement of mandarins using a conventional breeding method. The citrus breeding program is a lengthy process owing to the long juvenility, and it is predicted that marker-assisted selection (MAS) will overcome the obstacle and improve the efficiency of conventional breeding methods. To promote citrus molecular breeding in Japan, a genetic mapping was initiated in 1987, and the experimental tools and resources necessary for citrus functional genomics have been developed in relation to the physiological analysis of satsuma mandarin. In this paper, we review the progress of citrus breeding and genome researches in Japan and report the studies on genetic mapping, expression sequence tag cataloguing, and molecular characterization of breeding characteristics, mainly in terms of the metabolism of bio-functional substances as well as factors relating to, for example, fruit quality, disease resistance, polyembryony, and flowering.
Keywords: Citrus, genome, breeding, EST, functional genomics
Introduction
Citrus is one of the most cultivated fruits in the world, and the total citrus fruit production for 2012 has been estimated at 131.3 million tons (8.8 kha) (FAO 2014: http://faostat3.fao.org/; accessed 08/05/2015), which was 20.0% of total fruit production. Among citrus production, sweet oranges account for 52.6%, followed by mandarins (21.1%), lemons and limes (11.2%), and pummelos and grapefruits (6.2%). There are wide variations in citrus in terms of, for example, fruit shape, quality, embryo types, inflorescence, tree growth and habit, and adaptability. Mainly owing to sexual compatibility between Citrus species, a wide diversity has been generated during a long history of cultivation and wide dispersion (Nicolosi et al. 2000); this has led to a complicated system of citrus taxonomy and nomenclature over a long period. Taxonomy and phylogeny have been based mainly on morphological data and geographical data, and two different classification systems are commonly accepted for the citrus taxonomy: the system of Swingle (1943, 1967) and that of Tanaka (1954, 1961) (Nicolosi 2007). The major difference between the two systems is the number of recognized species: Swingle recognized 16 species in the genus Citrus, whereas Tanaka recognized 162 species. The latter system has been essentially accepted in most Japanese citrus researches.
Japan is located to the east of the natural distribution location of citrus, and there are two endemic species: tachibana (Citrus tachibana Tanaka) and shiikuwasha (C. depressa Hayata). During prehistoric ages and historical ages, many types of citrus have been introduced, or naturally migrated to Japan, or occurred as natural hybrids. These have been cultivated widely in the coastal region of southwest Japan, including satsuma mandarin, kinokuni (C. kinokuni hort. ex Tanaka), kunenbo (C. nobilis Lour.), iyo (C. iyo hort. ex Tanaka), natsudaidai (C. natsudaidai Hayata), hassaku (C. hassaku hort. ex Tanaka), hyuga-natsu (C. tamurana hort. ex Tanaka) and yuzu (C. junos Sieb. ex Tanaka). Using these genetic resources, citrus industry has developed through breeding by cross hybridization and selection, as well as improvement of culture management. At present, predominantly satsuma mandarin is cultivated, and accounted for 62.5% (45.5 kha) of all citrus acreage (72.6 kha) in 2014 in Japan (http://www.maff.go.jp/j/tokei/kouhyou/sakumotu/sakkyou_kazyu/index.html), followed by ‘Shiranuhi’ (3,139 ha), iyo (2,854 ha), yuzu (2,216ha), ponkan (2,056 ha), natsudaidai (2,023 ha), hassaku (1,747ha) and ‘Kiyomi’ (1,019 ha), among others. The first artificial hybrid ‘Tanigawa-buntan’ was released in the Taisho period. A large scale breeding program was began in 1937 in Japan, and thereafter numerous promising varieties have been developed by the citrus breeding program of the national institute, so the citrus industry has benefited from conventional breeding methods (Nesumi and Matsumoto 2003). ‘Shiranuhi’ and ‘Kiyomi’ have also been developed through the breeding program and their cultivated acreages are ranked within the top ten varieties. Conventional citrus breeding is a long-term process, requiring approximately 20 years until a new variety is developed and cultivated for commercial shipment. The efficiency of breeding is affected by long-juvenility, heterozygosis, gametophytic cross-incompatibility, male sterility, apomixis, seedlessness and unstability of characteristics under different environmental conditions. To produce new attractive and promising cultivars in the future, it is expected that the use of marker-assisted selection (MAS) will overcome these obstacles and improve the efficiency of conventional breeding programs. To promote citrus molecular breeding, a genome mapping program was initiated in Japan in 1987 and numerous efforts have been conducted to develop the experimental tools and resources necessary for citrus functional genomics. On the one hand, the International Citrus Genome Consortium (ICGC, composed of researchers from Australia, Brazil, China, France, Israel, Italy, Japan, Spain, and the USA) was inaugurated in 2003 to sequence the genome of sweet orange and Clementine mandarin. The genomes of sweet orange (diploid) and mandarin (haploid) were sequenced (Wu et al. 2014), and their draft sequences are available to view from public databases (http://phytozome.jgi.doe.gov/pz/portal.html). This progress in genome research has extended to molecular breeding and the isolation of agronomically important genes, resulting in many worldwide reviews and publications on Citrus genomics, genetics and breeding (Gmitter et al. 2007, 2012, Khan 2007, Talon and Gmitter 2008). In this review, we introduce the current research activities relating to citrus breeding in Japan, along with the progress of genome researches in genetic mapping and EST analysis, which have focused on the molecular characterization of breeding characteristics in Japan, mainly bio-functional substance metabolism in citrus. Such information may contribute not only to the advance of citrus molecular breeding and functional genomics but also to botanical science because Citrus species have various unique characteristics such as polyembryony, oil gland and juice sac development in fruit, and terpenoid metabolites.
Citrus breeding in Japan
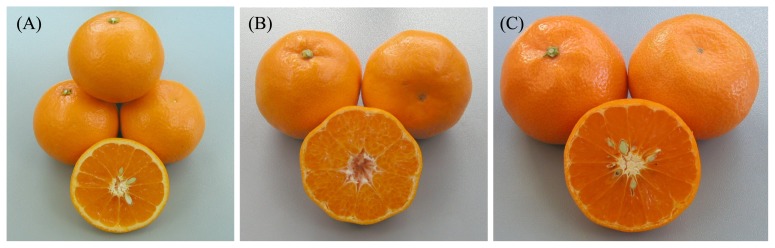
Before modern ages, the introduced or migrated citrus and their hybrids have been selected and cultivated. Since the national institute of Horticultural Research Station (present organization: the NARO Institute of Fruit Tree Science (NIFTS)) was founded in 1902, citrus germplasm has been collected from a number of countries as well as from Japan, and the breeding program was begun by the cross breeding and screening of mutations (Nishiura 1964). The satsuma mandarin is suited to the Japanese climate and has been predominantly cultivated for more than 100 years in Japan because this citrus fruit offers many favorable characteristics that facilitate its cultivation and consumption, such as seedlessness, easy peelability, early maturing, disease resistance, and high productivity. Within the citrus breeding program, the primary objectives have been improving the satsuma mandarin to add more excellent fruit quality, fragrance, stable production and stress resistance as the most important breeding objects. To generate a new cultivar with desirable traits and harvesting season from August to early spring, satsuma mandarin and the other mandarins were pollinated with sweet orange, pummelo, other citrus species and their hybrids (Nesumi and Matsumoto 2003). However, the many mandarins and the derived hybrids produce polyembryonic seeds which arise when the nucellar tissue surrounding the embryo sac generates somatic embryos so that that seed contains more than one embryo. Because most embryos in polyembryonic seeds originate from maternal tissue; this is a serious barrier to conventional breeding in addition to the prolonged juvenile periods until flowering from seedling. Several efforts have been conducted to mitigate serious limitations imposed by polyembryony and juvenility. Iwamasa et al. (1967) researched the heredity of polyembryony among 220 hybrids, and found that polyembryony was likely controlled by single dominant gene. Okudai et al. (1980) established the top-grafting method to promote and flowering and fruiting in the seedlings, and succeeded to shorten the breeding cycle. By using a culture of separate embryos obtained from ‘Miyagawa wase’ (the most popular satsuma mandarin cultivar) pollinated by ‘Trovita’ orange (sweet orange), a hybrid was developed and launched as ‘Kiyomi’ (tangor) in 1979. ‘Kiyomi’ is also a superior seed parent cultivar owing to monoembryonic (strictly zygotic) seed and male sterility, so it has contributed to the production of many promising cultivars with excellent quality such as ‘Shiranuhi’, ‘Harumi’, ‘Setoka’ and ‘Harehime’. Over the past 25 years, 49 cultivars have been released as progeny cultivars of ‘Kiyomi’ through the breeding program (Fig. 1). The cultivation of ‘Harumi’, ‘Shiranuhi’ and their progeny cultivars has been replacing the cultivation of traditional mid and late ripening citrus varieties.
Fig. 1.
‘Kiyomi’ and its progeny cultivars released from the NARO Institute of Fruit Tree Science (NIFTS). (A): ‘Kiyomi’ (‘Miyagawa wase’ (C. unshiu Marc.) × ‘Trovita’ orange (C. sinensis (L.) Osbeck).) (B): ‘Harumi’(‘Kiyomi’ × ponkan (C. reticulata Blanco)) (C): ‘Tamami’ (‘Kiyomi’ × ‘Wilking’(C. deliciosa Ten. × ‘King’ (C. nobilis Lour.))
In addition to the breeding program in national institute, more than 200 cultivars including kumquat (Fortunella spp.) and trifoliate orange (Poncirus trifoliata (L.) Raf.) have been discovered/selected and registered for cultivar license since 1981 http://www.hinsyu.maff.go.jp/vips/CMM/apCMM110.aspx?MOSS=1). Among them, bud sports and nucellar seedlings of satsuma mandarin accounted for 90 cultivars, which varied in fruit maturity season, fruit shape, rind color and texture, reduced degree of rind puffing, and sugar content. Somatic hybridization, ploidy breeding and graft chimera techniques also contributed to the development of 6, 9 and 17 cultivars, respectively.
From breeding activities aimed at further improvement, many excellent cultivars have been developed and launched, for example ‘Tamami’, which is rich in β-cryptoxanthin, ‘Chuukanbohon Nou-5’ (Nou-5) and ‘Chuukanbohon Nou-6’ (Nou-6) exhibiting seedlessness, and ‘Chuukanbohon Nou-8’ (Nou-8) incorporating the citrus tristeza virus (CTV) resistant gene from Poncirus. The present breeding program has 2 major processes for seedling selection. In the primary selection, 30–40 combinations of parental cultivars are cross-hybridized, and approximately 3,000 seeds are disseminated. Seedlings are subjected to grafting onto trifoliate orange rootstock, and are selected through the evaluation for tree habits, fruit quality and disease tolerance. The primary selection of seedlings generally takes 9–10 years and the population size varies depending on feasibility of cross-hybridization among parent varieties, and target of breeding traits. In the secondary selection, the evaluation of selected individuals is carried out at multiple research stations in different environments, and is mainly focused on fruit quality, cultivation, and stable fruit production. Secondary selection of individual generally takes 7–8 years, and the promising seedling with high evaluation progress to the process of the cultivar registration. The following characteristics remain important breeding targets: attractive fruit color, excellent flavor and aroma, rich health-promoting substances, easy-peelability, seedlessness, parthenocarpy, resistance to biotic and non-biotic stress, and a stable crop yield and tree habit. Since the cross-hybridization program was started, the current fifth- and sixth-generation cultivars by intercross between the improved cultivars and lines have piled on the favorable genetic traits. Present breeding cultivars and populations have been developed from the inter-crossing among the limited number of progenitor cultivars, and the accumulation of favorable genes has progressed in breeding populations in each successive generation. Recent demands for new cultivars enriched with various health-promoting substances in addition to seedless fruit with high quality and stable flower generation requires the need to identify suitable germplasm as well as the application of new strategies to single out individuals with favorable genes using genomic and marker information.
Cytological information and genome analysis
The basic chromosome number of Citrus has been recognized as x = 9 since the studies conducted in the 1920s (Frost 1925, Nakamura 1929), and this largely applies to species from the Rutaceae family. The genome size was estimated as 0.76 or 0.82 μg DNA/2C, equivalent to 367 or 396 Mb/1C, in C. sinensis L. (Arumuganathan and Earle 1991), but varied among species, ranging from the largest citron (C. medica L., average value 398 Mb/haploid genome) to the smallest mandarins (C. reticulata Blanco, 360 Mb). The pummelo (C. grandis L.) had an intermediate genome size of 383 Mb (Ollitrault et al. 1994).
The fluorescent staining of chromosomes (Guerra 1993) showed karyotype variation in the size and distribution of chromomycin A+ (CMA+) bands (Befu et al. 2000, Miranda et al. 1997). Proximal heterochromatin with CMA+ was highly variable among various Citrus species (Befu et al. 2000, 2001). Variations were also observed between possible homologous chromosomes. Fluorescence in situ hybridization (FISH) detection of ribosomal DNA (rDNA) sites also showed variations in signals between homologous chromosome pairs in sweet orange (Matsuyama et al. 1996, Roose et al. 1998), suggesting that commercial citrus cultivars such as C. sinensis originated from hybridization from basic species and are characterized by high heterozygosity (Herrero et al. 1996).
The establishment of reference whole-genome sequences involved the identification and development of the haploid or the doubled haploid from anther culture (Hidaka et al. 1979, 1982, Xu et al. 2013), interploid hybridization (Germanà and Chiancone 2001, Oiyama and Kobayashi 1993), or pollination with irradiated pollen (Aleza et al. 2009). A reference genetic map of Clementine mandarin was developed (Ollitrault et al. 2012) and sequence of haploid sweet orange and haploid Clementine were released (Wu et al. 2014, Xu et al. 2013). Recent comparative genome sequencing has revealed that species of possible interspecific hybrid origin, such as sweet orange (C. sinensis L.), sour orange (C. aurantium L.), lemon (C. limon L.) and grapefruit (C. paradisi Macf.), have mosaic genomes with large DNA fragments inherited from the basic taxa of citron (C. medica L.), pummelo (C. grandis L.) and mandarin (C. reticulata Blanco) and the others.
cDNA cataloguing and expression profiling
The availability of EST and genome sequences of Citrus species from public databases was quite limited in 1990, and the random sequencing of EST clones from various fruit tissues of the satsuma mandarin such as pulp, albedo, ovary and young seed has been promoted since 1994 (Fujii et al. 2003, Hisada et al. 1996, 1997, 1999, Kita et al. 2000a, 2000b, Moriguchi et al. 1998a, Shimada et al. 2003) (Table 1). To date, 21,976 independent sequences have been registered in the DDBJ database (the DNA Data Bank of Japan). In addition to the utilization of the ESTs for genetic mapping, they were used for the physiological and molecular analysis of flower- and fruit-related traits, including the metabolism of bio-functional substances.
Table 1.
Citrus EST catalogs and its application in Japan
| Library | NO of EST analyzed | ESTs-gene homologs used for functional analysis (example) | Reference | No of frame loci (AGI map)a | ||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Name | Source cultivar | Latin name | Tissue (organ) | Stage | ||||
| VSS | Valencia orange | C. sinensis (L.) Osbeck | Young seed | 1 month after anthesis | 577 | CuMFT1 promoter, etc. | Hisada et al. 1996 | 17 |
| FRI | Miyagawa wase | C. unshiu Marc. | Fruit pulp | Fruit developing | 1,051 | Hisada et al. 1997 | 11 | |
| FRM | Miyagawa wase | C. unshiu Marc. | Fruit pulp | Maturation | 385 | CiFT, CitMT45 promoter, CitPAP, CitCHS, etc. | Moriguchi et al. 1998a, 1998b | 21 |
| ALM | Miyagawa wase | C. unshiu Marc. | Albedo | Maturation | 623 | CitLGT, etc. | Kita et al. 2000 | 57 |
| OVA | Miyagawa wase | C. unshiu Marc. | Ovary | Flowering | 827 | CitSERK, CitMADS, CuSEP, CuSEP, CuFUL | Shimada et al. 2003 | 34 |
| ALP | Miyagawa wase | C. unshiu Marc. | Alvedo | Initiation stage of rind peeling | 941 | Fujii et al. 2003a | 26 | |
| WFY | Miyagawa wase | C. unshiu Marc. | Whole fruit | Young (1 month after flowering) | 1,689 | Direct submission on DDBJ | 40 | |
| BFC | Miyagawa wase | C. unshiu Marc. | Fuit rind | Rind coloring | 1,650 | CuMTSE2 promoter, monoterpene synthases, etc. | Nishikawa et al. 2009b, Shimada et al. 2004, etc. | 54 |
| FBI | Miyagawa wase | C. unshiu Marc. | Flower bud | 1 month prior to anthesis | 2,367 | 57 | ||
| GSA | Miyagawa wase | C. unshiu Marc. | Seed | 4 days after imbibition | 1,920 | 1 | ||
| RGP | Miyagawa wase | C. unshiu Marc. | Root | 2 week seedling | 960 | |||
| SLG | Miyagawa wase | C. unshiu Marc. | Shoot | 2 week seedling | 1,920 | |||
| YJS | Miyagawa wase | C. unshiu Marc. | Juice sac | 60 days after flowering | 1,926 | 2 | ||
| PCC | Miyagawa wase | C. unshiu Marc. | Callus | Culture for proliferation | 960 | 1 | ||
| EIC | Miyagawa wase | C. unshiu Marc. | Callus | Culture for embryogenesis | 1,152 | |||
| STG | Miyagawa wase | C. unshiu Marc. | Stigma | Flowering | 3,552 | 21 | ||
| ANT | Miyagawa wase | C. unshiu Marc. | Anther | Flowering | 2,600 | 6 | ||
| LLL | Lisbon lemon | C. limon (L.) Burm. f. | Leaf | Young (before full expansion) | 2,016 | 10 | ||
| EGJ | Kinokuni | C. kinokuni hort. ex Tanaka | Ovule | 60–70 days after flowering | 2,112 | 14 | ||
|
| ||||||||
| Total | 29,228 | 372 | ||||||
Refer to Shimada et al. (2014a).
A microarray analysis is one of the most powerful tools in functional genomics for addressing transcriptional profiling for a huge number of genes in one experiment. In terms of the first generation of microarray systems, where cDNA inserts were spotted onto glass slides, Shimada et al. (2005a) reported a cDNA microarray containing 2,213 cDNA probes which were independent genes from 8,333-cDNA collection. At the same time, the Spanish citrus genomic consortium generated 25 cDNA libraries covering different tissues, developmental stages and stress conditions and developed a cDNA microarray containing 12,672 cDNA probes, corresponding to 6,875 putative unigenes of a 22,635-EST collection (Forment et al. 2005). In terms of the second generation of microarray systems, where short oligonucleotide sequences were printed onto glass slides, Affymetrix (CA, USA) developed and released a GeneChip citrus array in 2006. The sequence information for this array was selected from Citrus HarvEST http://harvest-web.org/hweb/hmain.wc?versid=19) and cDNA clustering database. This array contains 30,171 probe sets representing up to 33,879 citrus transcripts. Moreover, a citrus 22K oligoarray comprising 21,495 independent EST probes derived from citrus species was developed on an Agilent oligoarray system (Fujii et al. 2007).
Using these microarray platforms, various researches have been reported. Cercós et al. (2006) identified more than 2,200 putative unique genes that exhibited significant changes of expression level during fruit development and these included genes in the metabolism of carbohydrates, acids, secondary metabolites, the genes in cell expansion and transcription regulatory genes. Fujii et al. (2007, 2008a) characterized ethylene and GA-responsive genes in satsuma mandarin mature fruit using a citrus 22K oligoarray and found that carotenoid biosynthetic genes show reverse responses to ethylene and GA treatments compared with chlorophyll biosynthetic genes. In addition, a type-II ethylene receptor (ETR2) plays a major role perception sensing the ethylene signal in citrus mature fruit.
DNA marker development and application
Up to the present, numerous DNA markers, such as restriction fragment length polymorphism (RFLP), randomly amplified polymorphic DNA (RAPD) and cleaved amplified polymorphic sequences (CAPS) were developed and applied to cultivar identification (Matsuyama et al. 1992, Omura et al. 2003, Sugawara et al. 2002, Ueda et al. 2003). Numerous EST-SSR and genomic SSR markers have been developed from citrus genome sequences and assigned to the genetic map of the Clementine mandarin (Luro et al. 2008, Ollitrault et al. 2012). Among these markers, CAPS markers are generally less polymorphic than the other molecular markers but represent a convenient method that does not require special analysis instruments for genetic diagnosis. Therefore, CAPS markers were utilized as the principal DNA marker for cultivar identification and for phylogenetic and linkage analyses in Japan. To the present, 3,562 primer sets have been tested for DNA marker construction, and 708 CAPS markers were found to be eligible for genetic analysis (Shimada et al. 2014a). Using CAPS markers, cultivar identification was carried out using 33 representative Japanese cultivars (Ninomiya et al. 2015) and a database was created for the genotypes of the examined CAPS markers. Based on “MinimalMarker”, which is a computer program for the identification of minimal sets of discriminating DNA markers for efficient cultivar identification (Fujii et al. 2013a), nine CAPS markers were sufficient to discriminate examined 33 cultivars. Fujii et al. (2008b, 2010) also developed the computer programs “MarkerToolkit” and “MARCO” which conduct various calculations automatically and estimate parentage based on the genotypes of DNA markers. Application of these DNA markers and computer programs enables us to evaluate the parentage of cultivars and resolve several areas of confusion regarding certain cultivars, and to secure rights for the citrus breeders. CAPS markers were also applied to evaluate the diversity of 103 various mandarin and related species using chloroplast DNA polymorphisms (Yamamoto et al. 2013), dividing them among seven groups.
Fujii et al. (2013b) developed a single nucleotide polymorphism (SNP) genotyping array using Illumina’s GoldenGate assay system. Among 1,497 SNPs candidates, 384 SNPs were selected for a high-throughput genotyping array based on the physical parameters of Illumina’s BeadArray criteria and applied to genotyping for a hybrid population of 88 progenies and 103 citrus accessions, for breeding in Japan. Genome-wide genotyping of citrus cultivars indicated that the chromosome transmission to progeny tended to be inherited in the large linkage blocks from both of the parent cultivars (Omura et al. 2003) and suggested that the frequency of recombination in a chromosome was low. To accumulate and combine the favorable genes distributed through-over the genome in cross-breeding procedure, detailed genotyping of breeding resources using recent SNP analysis (Fujii et al. 2013b) might provide the genetic information needed to select suitable parental candidate lines and to perform genomic selection in the near future for breeding purposes.
Linkage mapping and marker-assisted breeding
Genetic maps are useful tools for breeding program to select hybrids with economically important traits, and high density genetic maps provide information on quantitative trait loci (QTLs) associated with pathogen resistance and fruit quality. Numerous molecular markers have been developed and used to construct citrus genetic maps over the past decade. The initial citrus genetic maps were constructed from RFLPs and isozymes (Durham et al. 1992, Jarrell et al. 1992). Subsequently, SSR markers were adopted for mapping (Kijas et al. 1997). Thereafter, genetic maps have been extended to localize important traits, such as cold-acclimation (Cai et al. 1994), citrus tristeza virus (CTV) resistance (Cristofani et al. 1999, Fang et al. 1998, Gmitter et al. 1996), fruit acidity (Fang et al. 1997), apomixis (Garcia et al. 1999, Kepiro and Roose 2010), nematode resistance (Ling et al. 2000) and Phytophthora gummosis resistance (Siviero et al. 2006). These individual linkage maps provide us with useful information, but they are somewhat arbitrary and are difficult to compare. Recently, reference genetic maps have been developed. Ollitrault et al. (2012) constructed a genetic map of the Clementine mandarin using a SNP and EST-SSR marker, and Shimada et al. (2014a) constructed a framework genetic map of mandarin progenies using CAPS and SNP markers (Table 2). These maps were constructed using co-dominant genetic markers associated with a rather conserved and functional genome-coding region, and include functional annotation and positional information about the Clementine genome sequence. Therefore, framework genetic maps represent a useful resource for structural, functional and evolutionary studies in Citrus species. Using these framework genetic maps, Ohta et al. (2015) conducted graphical genotyping of pseudo-backcrossed F2 (BC2) progeny to introduce a CTV resistance from trifoliate orange into Citrus via introgression breeding, and developed CTV linkage markers (Ohta et al. 2011).
Table 2.
Mapping of breeding objective traits in Japan
| Parents of population | Population size | Trait | Linkage group | Reference |
|---|---|---|---|---|
| Kiyomi × Miyagawa wase | 125 | Seed number (QTL) | 6 | Omura et al. 2003 |
| Kiyomi × Miyagawa wase | 125 | Polyembryony | 1 | Nakano et al. 2012 |
| Kiyomi × Okitsu41 | 92 | Male sterility | 8 | Nakano et al. 2003 |
| Nou8 × Shiamese acidless | 92 | CTV resistance | 2 | Ohta et al. 2011 |
| Okitsu46 × Nou5 | 87 | Seedlessness | 9 | Shimada et al. 2014a |
| Okitsu46 × Nou5 | 87 | Carotenoids (QTL) | 6 | Sugiyama et al. 2011 |
‘Kiyomi’is derived from the cross between ‘Miyagawa wase’ (C. unshiu Marc.) and ‘Trovita’ orange (C. sinensis (L.) Osbeck).
Okistu-41 is selected breeding line derived from the cross between ‘Kiyomi’ and × ‘Wilking’(‘King’ tangor (C. nobilis Lour.) × ‘Willowleaf’ mandarin (C. deliciosa Ten.).
Okistu-46 is selected breeding line derived from the cross between ‘Sweet spring’ (‘Ueda unshiu’ (C. unshiu Marc.) × Hassaku (C. hassaku hort. ex Tanaka)) and ‘Trovita’ orange.
Nou-8 is a CTV resistant line derived from the cross between ‘Kiyomi’ and ‘H·FD-1’ (Hassaku × ‘Hiryu’ (P. trifoliata var. monstrosa)).
‘Siamese acidless’ is acidless variety of pummelo (Citrus grandis [L.] Osb.).
Nou-5 is a seedless line derived from the cross between ‘Lee’ (Clementine (C. clementina hort. ex Tanaka) and ‘Orland’ (‘Dancy tangerine’ × ‘Duncan’ grapefruit)) and ‘Mukaku kishiu’ (C. kinokuni hort. ex Tanaka).
Only the QTL showing the highest LOD score.
As an approach to breeding a seedless cultivar in Japan, the male sterility derived from ‘Kiyomi’ was genetically analyzed (Nakano et al. 2001) and mapped on linkage group 8 (Nakano et al. 2003). The seedlessness derived from ‘Mukaku kishu’ (Yamasaki et al. 2007a) was mapped on linkage group 9 (Shimada et al. 2014a). Nakano et al. (2008) also mapped a polyembryony locus and constructed bacterial artificial chromosome (BAC) contigs covering this region. Nakano et al. (2012) characterized genomic sequences for 380 kbps of BAC contigs and elucidated that the polyembryony locus consisted of 70 predicted open reading frames (ORFs), developing DNA markers that discriminated polyembryony from monoembryony. Recently, QTL mapping and expression QTL (eQTL) mapping were carried out to identify DNA markers that could be used to increase the amount of β-cryptoxanthin within a cultivar (Sugiyama et al. 2011, 2014).
Flowering
Citrus has a long juvenile period, for example, sweet orange needs more than 10 years until growing up to adult and flowering from seed. This juvenile period has hampered conventional breeding and genetic studies and flowering control is required to reduce juvenile period. In plants, flowering genes such as APETALA1 (AP1), LEAFY (LFY) and FLOWERING TIME (FT) promote flower development, and MADS-box genes are involved in floral organ development and/or flower meristem identification (Kobayashi et al. 1999, Wigge et al. 2005). Several flowering genes have been functionally characterized using transgenic plants to understand the processes regulating the phase transition from vegetative to reproductive growth in citrus. Peña et al. (2001) reported that constitutive expression of AtLFY or AtAP1 genes derived from Arabidopsis dramatically reduced their generation time in transgenic citranges (hybrids between sweet orange and trifoliate orange). Endo et al. (2005) reported that the 35S::CiFT1 trifoliate orange exhibited extremely early flowering. These results suggest that FT, LFY and AP1, which are key genes promoting flowering in Arabidopsis, may regulate the phase transition from the juvenile to the adult phase in citrus and its relatives. Pillitteri et al. (2004) reported that juvenility in sweet orange was positively correlated with the transcription accumulation of TERMINAL FLOWER LOCUS (CsTFL), a key negative regulator of floral timing. Endo et al. (2006) characterized five MADS-box cDNA clones (CitMADS1, CitMADS3, CitMADS5, CitMADS6 and CitMADS8) from the fruit tissues of satsuma mandarin. Most of them, except CitMADS1 and CitMADS3, were expressed in the seedlings before the phase transition from vegetative to reproductive growth. In 35S::CiFT1 trifoliate orange, the transcripts for some of these genes were found to be more significantly accumulated than in normal plants (Nishikawa et al. 2010). These results indicate that CiFT plays an important role in flowering induction and is more closely involved in seasonal periodicity than LFY and AP1.
CiFT shows a seasonal increase during the floral induction period in satsuma mandarin, and low temperature promotes floral induction via the activation of CiFT transcription in adult satsuma mandarin trees (Nishikawa et al. 2007). The endogenous expression of flowering-related genes has been investigated in satsuma mandarin and trifoliate orange, which are evergreen and deciduous, respectively (Nishikawa et al. 2009a). In satsuma mandarin, in which floral induction is triggered by low temperatures, mRNA levels of CiFT increased during fall and winter, corresponding to the floral induction period, and mRNA levels of CsLFY and SEPALLATA homologs (CuSEP) increased during early spring just before blooming. CsAP1 and FRUITFULL homologs (CuFUL) do not show a significant association with seasonal flowering. In trifoliate orange, in which floral induction and flower bud development occur during early summer, as in many deciduous trees, expression of CiFT, CsLFY, CsAP1, CuSEPs and CuFUL increases during early summer, corresponding to the period of floral induction and flower bud development. The CuSEPs expression peaks again during early spring just before blooming. In both species, CsTFL shows low transcript levels during the period of floral induction and flower bud development. Thus, despite the difference in flowering season, in both species transcriptional changes in CiFT, CsLFY, CsTFL and CuSEPs are correlated with seasonal flowering. In contrast, the correspondence between CsAP1 and CuFUL expression and seasonal flowering differs between species. Similar results have been observed in kumquat (genus Fortunella), a close relative of species in the genus Citrus and Poncirus (Nishikawa et al. 2011). Elucidation of the flowering mechanism is important to obtain a new insight into resolving the problem of stable fruit production due to the alternate bearing of flower and fruit.
Sugar and acid metabolism
High sugar content and low organic acid content in fruits is one of the most important traits of fruit quality in citrus breeding. A high ratio of sugars to organic acids in fruit is a kind index of taste. Generally, the sugar (sucrose, glucose and fructose) content of the fruit increases during fruit maturation while the content of organic acids (citric and malic) decreases. These storage components in fruit originate from sucrose as a main translocation form of photoassimilate in leaves (Goldschmidt and Koch 1996) and mainly accumulate in the vacuoles of juice sac cells. Sucrose synthase activity in juice sac tissues plays an important role during the early stages of fruit development, when cell division, cell wall synthesis and respiration rate are occurring at a maximum rate (Lowell et al. 1989). Sucrose synthase activity is high in the early stages and decreases until mid-developmental stage, then rapidly increases during the maturation stage (Komatsu et al. 2002). The increase in sucrose synthase activity directly promotes sucrose accumulation during fruit maturation. The three sucrose synthase genes (CitSUS1, CitSUS2 and CitSUSA) have been isolated, and their gene expression patterns correspond with changes in sucrose content during fruit development and maturation. Komatsu et al. (1996) also characterized sucrose-phosphate synthase (SPS), which is another key enzyme in the synthetic pathway of sucrose. SPS is involved in many processes relating to important agronomic traits such as the growth and yield of plants, and their transcripts accumulated in flower, leaves and mature fruits in citrus. Katz et al. (2011) reported the results of a metabolite analysis of citrus fruit during fruit development using label-free shotgun proteomics. In all of these studies, the protein change responsible for sugar metabolism shows a similar pattern. The invertases, an important family of proteins responsible for sucrose degradation to glucose and fructose, did not change in the proteomics analysis. The transcription activity of two acidic and one neutral invertase genes showed peaks at the early stage of fruit development and then decreased in the later stages (Kubo et al. 2001). Interestingly, an invertase inhibitor protein, responsible for the reduction in invertase activity, was up-regulated in the later stages of fruit development.
Citrus fruit accumulate large amounts of organic acids, mainly citric acid, in juice sac cells. Citric acid is produced through the TCA cycles and is an intermediate metabolite in energy production, accumulating in the vacuole during early-mid fruit development. Toward fruit maturation, citric acid is released from vacuole to cytosol to assist with a vacuolar citrate/H+ symporter (Shimada et al. 2006). In the cytosol, the released citric acid is catabolized by cytosolic aconitase, to be converted to amino acids (Cercós et al. 2006, Sadka et al. 2000). In terms of the consumption of citrus fruits, a high citric acid content reduces fruit quality, but the mechanisms regulating citric acid accumulation and degradation are still unknown. Degu et al. (2011) reported that inhibition of aconitase activity by citramalate and oxalomalate were effective at reducing citric acid metabolism. Citramalate, as an endogenous inhibitor, inhibited mitochondrial aconitase activity and resulted in an increase in citric acid accumulation, although it did not inhibit cytosolic aconitase activity. In addition, oxalomalate inhibited cytosolic aconitase activity and resulted in a metabolic shift towards amino acid biosynthesis by inducing the gamma-aminobutyric acid (GABA) shunt. The usage of citramalate and oxalomalate is one of the manipulations to control fruit acidity in citrus.
Aroma production and diversity
The molecular identification and characterization of genes related to chemical constituents of citrus is very informative on genetic and physiological improvement of the target traits involved in fruit quality. Most aerial citrus tissues and organs have oil glands that contain and emit a wide diversity of volatile terpenoids, such as hemiterpenes, monoterpenes and sesquiterpenes, including their alcohol, ester and acetate derivatives as fragrance components (Sawamura 2000, Vekiari et al. 2002). Monoterpenes and sesquiterpenes have roles in communication between plants and insects, and plants and pathogens, and in attracting pollinators (Pichersky and Gershenzon 2002). Plant terpene synthases comprise a large gene family, of which 49 putative members have been estimated in Citrus germplasm through the use of in silico characterization of ESTs (Dornelas and Mazzafera 2007). Several citrus cDNAs encoding monoterpene and sesquiterpene synthase genes have been functionally characterized (Lücker et al. 2002, Murayama et al. 2001, Sharon-Asa et al. 2003, Shimada et al. 2004, 2005a, 2005b, 2012, 2014b). Most of these genes encode cyclic monoterpene and sesquiterpene synthases, and they share the common structural features of a tandem arginine (RR) motif and a DDXXD motif. In addition, most produce a specific main product accompanied by significant amounts of by-products. This enzymatic feature may contribute the complex factor in aroma breeding programs as a quantitative trait. In rough lemon (C. jambhiri Lush), in situ hybridization revealed that the RlemTPS4 (delta-elemene synthase gene) was specifically expressed in specialized epithelial cells surrounding the oil secretory cavities of leaf tissues (Yuya et al. 2015).
Certain citrus volatiles, including the most predominant monoterpenes of fruit peels (d-limonene, α-pinene, β-pinene and myrcene), have stimulatory effects on host recognition, germination and growth of Penicillium digitatum and P. italicum (Droby et al. 2008). The essential oils of ponkan have been proposed as a promising source of fungicidal substances because they show antifungal activity in vitro against several diseases caused by, for example, P. italicum and P. digitatum (Chutia et al. 2009, Tao et al. 2014). In rough lemon leaves, monoterpene volatiles likely play an important role in defense against microbes and herbivores (Yamasaki et al. 2007b). In studies of a transgenic orange with downregulated d-limonene biosynthesis, the aroma components of the transgenic fruits were significantly altered (i.e., levels of certain oxygenated monoterpenes strongly increased) to generate broad-spectrum resistance against pathogens (Rodríguez et al. 2011, 2014). Thus, citrus aroma components from leaves and fruit peels have clear biological roles in plant resistance and susceptibility responses to pests and pathogens, but the activity of each aroma component and particularly that of specific oxygenated monoterpenes in such biological interactions has not yet been clarified in detail. Shimada et al. (2014b) reported that linalool, which is a major oxygenated monoterpene in citrus, showed anti-bacterial and antifungal activities against the citrus major diseases of canker (Xanthomonas citri subsp. citri) and molds (P. digitatum and P. italicum). The biosynthetic genes of linalool were transcriptionally induced by wounding and elicitor treatments [bacteria, fungi and jasmonic acid (JA)]. Ponkan, one of the breeding materials with field resistance against cankers and molds, contains a high level of linalool in its leaves and mature fruit. In rice, JA signaling plays an important role in resistance to rice bacterial blight caused by Xanthomonas oryzae pv. oryzae (Xoo), and is accompanied with linalool accumulation (Taniguchi et al. 2014). In transgenic orange fruits harboring bacterial and mold resistances, cis-(+)-12-oxophytodienoic acid (OPDA), as a precursor of JA biosynthesis, has been found to be highly accumulated and to produce extraordinary amounts of JA after elicitor treatments compared with normal orange (Rodríguez et al. 2014). From these findings, we will gain a new insight into elucidating the molecular mechanisms involved in the field resistance of ponkan.
Limonoid metabolism
Limonoids are highly oxygenated triterpenes present in Rutaceae spp. and a few other plants such as Meliaceae spp.; 36 limonoids have been identified in Citrus and its related species (Berhow et al. 2000). Limonoids have several biological functions linked to chemically induced carcinogenesis in mice, hamsters and human breast cancer cell growth (Lam and Hasegawa 1989), which may offer commercial benefits. However, limonoids in fruits cause a gradual bitterness called delayed bitterness in navel orange after juice processing, and reduce the quality of the juice. In contrast, mature fruits of satsuma mandarin cause much less delayed limonoid bitterness than sweet orange; probably because the concentration of limonoid aglycones in the juice from satsuma mandarin is low. Limonoid metabolism is characterized by radioactive tracers (Hasegawa et al. 1984, Ou et al. 1988) but many of the corresponding enzymes and biosynthetic genes have been not isolated. Kita et al. (2000c) isolated cDNA clones encoding limonoid UDP-glucosyltransferase (LGT) that regulated the conversion of limonoid aglycones such as limonin bitter compound to their nonbitter glucosides. Satsuma mandarin has 2 LGT allelic genes (CitLGT1 and CitLGT2), and their expression patterns were found to be different among fruit development (Kita et al. 2003). CitLGT2 is the gene responsible for converting limonoid aglycones to their nonbitter glucosides throughout fruit development. The delayed bitterness in sweet orange was presumed to be caused by a lack of CitLGT2. Endo et al. (2002) performed a study involving genetic engineering to maximize the formation of limonoid glucosides in a suspension cell culture system through Agrobacterium transformation of the functional coding region of CitLGT1. The suspension cell culture system study attempted to evaluate limonoid metabolism; genetic modification resulted in an accumulation of limonoid glucoside in transgenic cells but the quantity was very low due to other unexpected factors. Thus, limonoid metabolism is still not fully understood, and further researches are required to isolate a series of biosynthetic genes and elucidate the regulators of limonoid metabolism.
Flavonoid metabolism
Flavonoids are a group of pigments contained in plants and are responsible for flower and fruit coloration, and for bitterness of citrus fruit. Approximately 60 flavonoids have been identified in citrus and categorized into six classes: flavones, flavanones, flavonols, isoflavones, anthocyanidins and flavanols (or catechins), according to their molecular structures (Peterson et al. 1998). The yellow or orange color of general citrus fruits originates from an accumulation of carotenoid pigments, while the red color of blood orange predominantly originates from an accumulation of anthocyanin pigments. Citrus species accumulate large amounts of flavonols, especially flavanone glycosides (Horowitz 1964). Flavanone, which is synthesized from naringenin chalcone by the catalysis of chalcone isomerase, is modified stepwise to various derivatives by hydroxylation, methylation and glycosylation, then rhamnosylation (Lewinsohn et al. 1989). The most common glycosidic group attached to the flavonoids in citrus is rhamnoglucose diglycoside. Flavanone neohesperidosides mainly accumulate in citrus species related to the pummelo, such as grapefruit, sour orange and natsudaidai, and lend a bitter taste to citrus fruit (Horowitz 1964, Rouseff 1980). Other species related to citron and mandarin mainly accumulate the tasteless flavanone rutinosides narirutin and hesperidin (Nishiura et al. 1971). In addition to the bitter property of citrus flavonoids, pharmacological investigations have shown that several flavonoids such as naringenin and hesperetin function to prevent atherosclerosis and cancer (Tripoli et al. 2007). Moreover, citrus peels contain several polymethoxylated flavonoids (PMFs) tangeretin and nobiletin that are enriched in orange and tangerine peel (Kurowska and Manthey 2004). They are O-methylated flavones and are reported to have several anticancer, anti-inflammatory, anti-atherogenic and anti-diabetic properties (Miyata et al. 2011).
Moriguchi et al. (2001) isolated and characterized major flavonoid biosynthetic genes (CitCHS1, CitCHS2, CitCHI and CitF3H) and reported that flavonoid accumulation and transcription of the related genes were correlated and abundant in the young tissue of leaves and fruits. Frydman et al. (2004) isolated and characterized the gene encoding 1,2 rhamnosyltransferase, a key enzyme in the biosynthesis of the bitter flavonoids of citrus. However, the other metabolic genes responsible for hydroxylation, methylation and glycosylation of flavonoids are still not fully known.
Recently, the mechanism involved in the accumulation of anthocyanin pigments in blood orange has been elucidated. The transcription level of the gene that specifies dihydroflavonol 4-reductase (DRF), which is a key gene in anthocyanin biosynthesis and catalyzes the conversion of dihydroflavonols to leucoanthocyanins, is lower in non-red orange cultivars than in blood orange cultivars (Lo piero et al. 2006). Anthocyanin biosynthesis is regulated mainly at a transcriptional level in plants controlled by a regulatory complex that is composed of proteins of the Myb, bHLH and WD-repeat family. Butelli et al. (2012) isolated R2R3-type Myb gene (Ruby) which is responsible for anthocyanin biosynthesis, and the insertion of a Copia-like retrotransposon adjacent to Ruby activated the production of anthocyanin in blood orange. The cultivation of blood orange is becoming more widespread in Japan, but the poor blood coloration of both rind and flesh is a principal issue owing to physiological and climatic factors. Further research would be valuable in the creation of a new citrus cultivar with a different color.
Transgenic research on gene function relating to citrus breeding traits
Transgenic research is considerably important to progress the functional genomics of Citrus. Transformation efficiency is generally low in mandarins (Hidaka et al. 1990) and the varieties suitable for transformation are limited to fruits such as trifoliate orange and sweet orange (Kaneyoshi et al. 1994, Peña et al. 2008). The reduction in generation time by genetic engineering could accelerate the research in terms of understanding the complex regulatory mechanisms involved in biological and agronomical traits as well as in manipulating the nutritional value and quality of fruits. Several efforts have been made to achieve a reduction in long juvenile periods in citrus using flowering genes. Peña et al. (2001) succeeded in reducing the generation time of citrange by the introduction of either APETALA1 (AP1) or LEAFY (LFY). Endo et al. (2005) introduced citrus FLOWERING LOCUS T (CiFT) to trifoliate orange and succeeded in developing a precocious flowering phenotype. The precocious flowering phenotypes in 35S::CiFT trifoliate orange may result primarily from the conversion of a vegetative shoot apical meristem into an inflorescence meristem rather than by the transition from the juvenile to adult phases. Early flowering phenotype caused by CiFT could be inherited in F1 progenies obtained by crossing ‘Kiyomi’ tangor (C. unshiu × C. sinensis) with pollen from a transgenic line. Mendel’s segregating ratio (1:1) was fitted to the segregation of the transgene, resulting in the complete co-segregation of the transgene and the early flowering phenotype. Interestingly, the null segregant, which lost the transgene in the progress to the next generation, showed no early flowering phenotype. In maize, a seed production technology (SPT) process was developed to improve the efficiency of hybrid seed production by using the segregation-out of transgene. In Europe and the United States, the SPT seeds are outside of the regulation governing genetically modified organisms. Because the introduced CiFT in a transformed trifoliate orange could be also segregated out, CiFT would be useful to accelerate the cycles of breeding generations of backcrossing with citrus and produce non-GMO citrus hybrids. In addition, Endo et al. (2009) developed a CiFT co-expression binary vector, which drives the target gene under El2-Ω-cauliflower mosaic virus P35S (El2-35S) promoter with CiFT. Using this vector, they succeeded in modifying aroma components in transgenic trifoliate orange to repress the biosynthesis of d-limonene using antisense transgenes of d-limonene synthase genes. Transgenic trifoliate orange formed normal flowers and fruits within 18 months of infection by Agrobacterium. Thus, the functional analysis of transgenic citrus facilitated evaluate the mature traits of flowers and fruits, unlike in past research. The utilization of flowering genes contributes to overcome problems relating to long juvenile periods in transgenic research.
Meanwhile, additional efforts should be made to develop collections of tissue-specific and inducible promoters for the control of incorporated gene expression. To the present, several promoters controlling tissue-specific expression have been reported in citrus. Endo et al. (2007) developed a CitMT45 promoter that was derived from genomic clones of type 3 metallothionein gene, which was exclusively expressed in the fruit tissue of citrus (Moriguchi et al. 1998a). Nishikawa et al. (2008) developed a CuMFT1 promoter that was derived from a FT/TFL1 homolog (CuMFT1), which may be useful in regulating seed-specific expression in transgenic plants in Citrus. Nishikawa et al. (2009b) developed a CuMTSE2 promoter that was derived from d-limonene synthase gene 2 with peel-specific expression in mature fruit.
The establishment of transgenic technology combined with the technical improvement in gene function assays could accelerate the identification of genes related to breeding factors such as fruit quality and bio-functional substance production.
Conclusion and future prospects
The development of experimental tools and resources for citrus genome analysis has contributed to elucidating the genetic composition and molecular mechanism of agronomically important traits and to advancing the isolation of such genes. In breeding programs, DNA marker-based selection methods can be further developed and applied to the selection of seedlings with new favorable traits such as CTV resistance and seedlessness. In addition to these traditionally important breeding traits, the breeding of new cultivars enriched with health-promoting substances has progressed. Research into the mechanisms involved in the metabolism of monoterpenes, limonoids, flavonoids and carotenoids has advanced, and the development of DNA markers is under way as part of the practice of selection in target of β-cryptoxanthin, aurapten and nobiletin (Table 3).
Table 3.
Target of functional metabolites aimed to enrichment in fruit
| Compounds | Breeding resource (Latin name) | Target gene or metabolite |
|---|---|---|
| Limonoids | Satsuma mandarin (C. unshiu Marc.) | UDP-gulcose: limonoid glucosyltransferase |
| Polymethoxyflavonoids | Nou-6 (‘King’ (C. nobilis Lour.) × ‘Mukaku kishu’ (C. kinokuni hort. ex Tanaka)), King (C. nobilis Lour.) | Flavonoid O methyltransferases (nobiletin and tangertine from sinensetine) |
| Hesperisin | Satsuma mandarin (C. unshiu Marc.), Hassaku (C. hassaku hort. ex Tanaka) | Conversion of rhamoside |
| Auraptene | ‘Aura star’ (‘H·FD-1’ (Hassaku (C. hassaku hort. ex Tanaka) × ‘Hiryu’ (P. trifoliata var. monstrosa)), ‘Banpeiyu’ (C. grandis Osbeck)) | Conversion of geranyloxy group |
| β-cryptoxanthin | ‘Tamami’ (‘Kiyomi’ (‘Miyagawa wase’ (C. unshiu Marc.) × ‘Trovita’ orange (C. sinensis (L.) Osbeck)) × ‘Wilking’ (‘King’ tangor (C. nobilis Lour.) × ‘Willowleaf’ mandarin (C. deliciosa Ten.)) | Carotenoid biosynthetic genes |
| Monoterpenes | Ponkan (C. reticulata Blanco), sweet orange (C. sinensis (L.) Osbeck) | Monoterpene biosynthetic genes |
The use of molecular technologies will help to overcome the obstacles of juvenility and polyembryony in citrus breeding. The utilization of CiFT will succeed in reducing the time needed to evaluate fruit traits in transgenic plants and will shorten the cycles of breeding generation. Furthermore, researches on the genes involved in flower bud formation will contribute to improving breeding technology as well as regulating the development of flower buds so as to avoid alternate fruit bearing in cultures. Seedlessness is perhaps one of the most important traits relating to citrus fruit breeding in the future, so the isolation and identification of genes involved in seed and embryo development, as well as male sterility, are vitally important. The genes regulating polyembryony have been investigated through a positional cloning procedure combined with expression analysis (Nakano et al. 2012). The isolation of a gene corresponding to the locus is likely to be achieved in the near future. This will provide a new strategy for the production of seeds from heterozygous plants and a practical application for MAS.
In citrus, most of the important breeding objects such as fruit quality and fruit yield are quantitatively inherited, and the development of the QTL marker is urgently needed (Table 2). The established experimental tools and resources used in genome analysis are expected to accelerate the discovery of the loci associated with various quantitative traits and contribute to the design of breeding programs supported by MAS, including genomic selection in citrus breeding.
Acknowledgement
We thank Mr. Terutaka Yoshioka for providing photographs of citrus cultivars and information on citrus breeding.
Literature Cited
- Aleza, P., Juárez, J., Hernández, M., Pina, J.A., Ollitrault, P. and Navarro, L. (2009) Recovery and characterization of a Citrus clementina Hort. ex Tan. ‘Clemenules’ haploid plant selected to establish the reference whole Citrus genome sequence. BMC Plant Biol. 9: 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumuganathan, K. and Earle, E.D. (1991) Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9: 208–218. [Google Scholar]
- Befu, M., Kitajima, A., Yang, X.L. and Hasegawa, K. (2000) Classification of ‘Tosa-Buntan’ pummelo (Citrus grandis [L.] Osb.), ‘Washington’ navel orange (C. sinensis [L.] Osb.) and trifoliate orange (Poncirus trifoliata [L.] Raf.) chromosomes using young leaves. J. Japan. Soc. Hort. Sci. 69: 22–28. [Google Scholar]
- Befu, M., Kitajima, A. and Hasegawa, K. (2001) Chromosome composition of some citrus species and cultivars based on chromomycin A3 (CMA) banding patterns [in Japanese with English summary]. J. Jpn. Soc. Hort. Sci. 70: 83–88. [Google Scholar]
- Berhow, M.A., Hasegawa, S., Kwan, K. and Bennett, R.D. (2000) Limonoids and the chemotaxonomy of Citrus and the Rutaceae family. In Citrus limonoids. Functional chemicals in agriculrue and foods. (Ed.) Berhow M.A., Hasegawa S. and Manners G.D.. ACS Symposium series 758 pp. 212–229. [Google Scholar]
- Butelli, E., Licciardello, C., Zhang, Y., Liu, J., Mackay, S., Bailey, P., Reforgiato-Recupero, G. and Martin, C. (2012) Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell 24: 1242–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, Q., Guy, C.G. and Moore, G.A. (1994) Extension of the linkage map in Citrus using random amplified polymorphic DNA (RAPD) markers and RFLP mapping of cold-acclimation-responsive loci. Theor. Appl. Genet. 89: 606–614. [DOI] [PubMed] [Google Scholar]
- Cercós, M., Soler, G., Iglesias, D.J., Gadea, J., Forment, J. and Talón, M. (2006) Global analysis of gene expression during development and ripening of citrus fruit flesh. A Proposed Mechanism for Citric Acid Utilization. Plant Mol. Biol. 62: 513–527. [DOI] [PubMed] [Google Scholar]
- Chutia, M., Bhuyan, P.D., Pathak, M.G., Sarma, T.C. and Boruah, P. (2009) Antifungal activity and chemical composition of Citrus reticulata Blanco essential oil against phytopathogens from North East India. LMW-Food Sci. Technol. 42: 777–780. [Google Scholar]
- Cristofani, M., Machado, M.A. and Grattapaglia, D. (1999) Genetic linkage maps of Citrus sunki Hort. Ex. Tan. and Poncirus trifoliata (L.) Raf. and mapping of citrus tristeza virus resistance gene. Euphytica 109: 25–32. [Google Scholar]
- Degu, A., Hatew, B., Nubes-Nesi, A., Shlizerman, L., Zur, N., Katz, E., Fernie, A.R. and Blumwald, E. (2011) Inhibition of aconitase in citrus fruit callus results in a metabolic shift towards amino acid biosynthesis. Planta 234: 501–513. [DOI] [PubMed] [Google Scholar]
- Dornelas, M.C. and Mazzafera, P. (2007) A genomic approach to characterization of the Citrus terpene synthase gene family. Genet. Mol. Biol. 30: 832–840. [Google Scholar]
- Droby, S., Eick, A., Macarisin, D., Cohen, L., Rafael, G., Stange, R., McColum, G., Dudai, N., Nasser, A., Wisniewski, M.et al. (2008) Role of citrus volatiles in host recognition, germination and growth of Penicillium digitatum and Penicillium italicum. Postharvest Biol. Technol. 49: 386–396. [Google Scholar]
- Durham, R.E., Liou, P.C., Gmitter, F.G.Jr. and Moore, G.A. (1992) Linkage of restriction fragment length polymorphisms and isozymes in Citrus. Theor. Appl. Genet. 84: 39–48. [DOI] [PubMed] [Google Scholar]
- Endo, T., Kita, M., Shimada, T., Moriguchi, T., Hidaka, T., Matsumoto, R., Hasegawa, S. and Omura, M. (2002) Modification of limonoid metabolism in suspension cell culture of citrus. Plant Biotechnol. 19: 397–403. [Google Scholar]
- Endo, T., Shimada, T., Fujii, H., Kobayashi, Y., Araki, T. and Omura, M. (2005) Ectopic expression of an FT homolog from Citrus confers an early flowering phenotype on trifoliate orange (Poncirus trifoliata L. Raf.). Transgenic Res. 14: 703–712. [DOI] [PubMed] [Google Scholar]
- Endo, T., Shimada, T., Fujii, H. and Omura, M. (2006) Cloning and characterization of 5 MADS-box cDNAs isolated from citrus fruit tissue. Sci. Hortic. 109: 315–321. [Google Scholar]
- Endo, T., Shimada, T., Fujii, H., Moriguchi, T. and Omura, M. (2007) Promoter analysis of a type 3 metallothionein-like gene abundant in Satsuma mandarin (Citrus unshiu Marc.) fruit. Sci. Hortic. 112: 207–214. [Google Scholar]
- Endo, T., Shimada, T., Fujii, H., Nishikawa, F., Sugiyama, A., Nakano, M., Shimizu, T., Kobayashi, Y., Araki, T., Peña, L.et al. (2009) Development of a CiFT co-expression system for functional analysis of genes in citrus flowers and fruit. J. Japan. Soc. Hort. Sci. 78: 74–83. [Google Scholar]
- Fang, D.Q., Federici, C.T. and Roose, M.L. (1997) Development of molecular markers linked to a gene controlling fruit acidity in citrus. Genome 40: 841–849. [DOI] [PubMed] [Google Scholar]
- Fang, D.Q., Federici, C.T. and Roose, M.L. (1998) A high-resolution linkage map of the citrus tristeza virus resistance gene region in Poncirus trifoliata (L.) Raf. Genetics 150: 883–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forment, J., Gadea, J., Huerta, L., Abizanda, L., Agusti, J., Alamar, S., Alos, E., Andres, F., Arribas, R., Beltran, J.P.et al. (2005) Development of a citrus genome-wide EST collection and cDNA microarray as resources for genomic studies. Plant Mol. Biol. 57: 375–391. [DOI] [PubMed] [Google Scholar]
- Frost, H.B. (1925) Tetraploidy in Citrus. Proc. Natl. Acad. Sci. USA 11: 535–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydman, A., Weisshaus, O., Bar-Peled, M., Huhman, D.V., Sumner, L.W., Marin, F.R., Lewinsohn, E., Fluhr, R., Gressel, J. and Eyal, Y. (2004) Citrus fruit bitter flavors: isolation and functional characterization of the gene Cm1,2RhaT encoding a 1,2 rhamnosyltransferase, a key enzyme in the biosynthesis of the bitter flavonoids of citrus. Plant J. 40: 88–100. [DOI] [PubMed] [Google Scholar]
- Fujii, H., Kita, M., Shimada, T., Endo, T. and Omura, M. (2003) Expressed sequence tags from citrus albedo at the initiation stage of rind peeling. Bull. Natl. Inst. Fruit Tree Sci. 2: 127–143. [Google Scholar]
- Fujii, H., Shimada, T., Sugiyama, A., Nishikawa, F., Endo, T., Nakano, M., Ikoma, Y., Shimizu, T. and Omura, M. (2007) Profiling ethylene-responsive genes in mature mandarin fruit using a citrus 22K oligoarray. Plant Sci. 173: 340–348. [Google Scholar]
- Fujii, H., Shimada, T., Sugiyama, A., Endo, T., Nishikawa, F., Nakano, M., Ikoma, Y., Shimizu, T. and Omura, M. (2008a) Profiling gibberellin (GA3)-responsive genes in mature mandarin fruit using a citrus 22K oligoarray. Sci. Hortic. 116: 291–298. [Google Scholar]
- Fujii, H., Yamashita, H., Shimada, T., Endo, T., Shimizu, T. and Yamamoto, T. (2008b) MakerToolKit: an analysis program for data sets consist of DNA maker types obtained from various varieties. DNA Polymorphism 16: 103–107. [Google Scholar]
- Fujii, H., Yamashita, H., Hosaka, F., Terakami, S. and Yamamoto, T. (2010) Development of a software to presume the parent-child relationship using the result of DNA marker typing. J. Japan. Soc. Hort. Sci. abst. 9: 34. [Google Scholar]
- Fujii, H., Ogata, T., Shimada, T., Endo, T., Iketani, H., Shimizu, T., Yamamoto, T. and Omura, M. (2013a) Minimal marker: an algorithm and computer program for the identification of minimal sets of discriminating DNA markers for efficient variety identification. J. Bioinform. Comput. Biol. 11: 1250022. [DOI] [PubMed] [Google Scholar]
- Fujii, H., Shimada, T., Nonaka, K., Kita, M., Kuniga, T., Endo, T., Ikoma, Y. and Omura, M. (2013b) High-throughput genotyping in citrus accessions using an SNP genotyping array. Tree Genet. Genomes 9: 145–153. [Google Scholar]
- Garcia, R., Asins, M.J., Forner, J. and Carbonell, E.A. (1999) Genetic analysis of apomixis in Citrus and Poncirus by molecular markers. Theor. Appl. Genet. 99: 511–518. [DOI] [PubMed] [Google Scholar]
- Germana, M.A. and Chiancone, B. (2001) Gynogenetic haploids of citrus after in vitro pollination with triploid pollen grains. Plant Cell Tissue Organ Cult. 66: 59–66. [Google Scholar]
- Gmitter, F.G.Jr, Xiao, S.Y., Huang, S., Hu, X.L., Garnsey, S.M. and Deng, Z. (1996) A localized linkage map of the citrus tristeza virus resistance gene region. Theor. Appl. Genet. 92: 688–695. [DOI] [PubMed] [Google Scholar]
- Gmitter, F.G.Jr, Chen, C., Rao, M.N. and Soneji, J.R. (2007) Citrus fruits. In: Kole, C. (ed.) Genome mapping and meolecular breeding in plants vol. 4 Fruit and Nut Springer, pp. 265–279. [Google Scholar]
- Gmitter, F.G.Jr., Chen, C., Machado, M.A., Alves de Souza, A., Ollitrault, P., Froehlicher, Y. and Shimizu, T. (2012) Citrus genomics. Tree Genet. Genomes 8: 611–626. [Google Scholar]
- Goldschmidt, E.E. and Koch, K.E. (1996) In: Zamski, E. and Schaeffer A.A. (eds.) Photoassimilate distribution in plants and crops: Source sink relationships. Marcel Dekker Inc, New York. [Google Scholar]
- Guerra, M. (1993) Cytogenetics of Rutaceae. V. High chromosomal variability in Citrus species revealed by CMA/DAPI staining. Heredity 71: 234–241. [Google Scholar]
- Hasegawa, S., Benett, R.D. and Maier, V.P. (1984) Biosynthesis of limonoids in citrus seedlings. Phytochemistry 23: 1601–1603. [Google Scholar]
- Herrero, R., Asins, M.J., Carbonell, E.A. and Navarro, L. (1996) Genetic diversity in the orange subfamily Aurantioideae. I. Intraspecies and intragenus genetic variability. Theor. Appl. Genet. 92: 599–609. [DOI] [PubMed] [Google Scholar]
- Hidaka, T., Yamada, Y. and Shichijo, T. (1979) In vitro differentiation of haploid plants by anther culture in Poncirus trifoliata (L.) RAF. Japan. J. Breed. 29: 248–254. [Google Scholar]
- Hidaka, T., Yamada, Y. and Shichijo, T. (1982) Plantlet formation by anther culture of Citrus aurantium L. Japan. J. Breed. 32: 247–252. [Google Scholar]
- Hidaka, T., Omura, M., Ugaki, M., Tomiyama, M., Kato, A., Ohshima, M. and Motoyoshi, F. (1990) Agrobacterium-mediated transformation and regeneration of Citrus Spp. from suspension cells. Japan. J. Breed. 40: 199–207. [Google Scholar]
- Hisada, S., Moriguchi, T., Hidaka, T., Koltunow, A.M., Akihama, T. and Omura, M. (1996) Random sequencing of sweet orange (Citrus sinensis Osbeck) cDNA library derived from young seeds. J. Japan. Soc. Hort. Sci. 65: 487–495. [Google Scholar]
- Hisada, S., Akihama, T., Endo, T., Moriguchi, T. and Omura, M. (1997) Expressed sequence tags of Citrus fruit during rapid cell development phase. J. Amer. Soc. Hort. Sci. 122: 808–812. [Google Scholar]
- Hisada, S., Kita, M., Endo-Inagaki, T., Omura, M. and Moriguchi, T. (1999) Refinement of cDNA clone expression analysis in random sequencing from the rapid cell development phase of citrus fruit. J. Plant Physiol. 155: 699–705. [Google Scholar]
- Horowitz, R.M. (1964) Relations between the taste and structure of some phenolic glycosides. In: Harborne, J.B. (ed.) Biochemistry of Phenolic Compounds. Academic Press, New York, NY, pp. 545–571. [Google Scholar]
- Iwamasa, M., Ueno, I. and Nishiura, M. (1967) Inheritance of nucellar embryony in citrus. Bull. Hort. Res. Stn. B7: 1–10. [Google Scholar]
- Jarrell, D.C., Roose, M.L., Traugh, S.N. and Kupper, R.S. (1992) A genetic map of citrus based on the segregation of isozymes and RFLPs in an intergeneric cross. Theor. Appl. Genet. 84: 49–56. [DOI] [PubMed] [Google Scholar]
- Kaneyoshi (Hiramatsu), J., Kobayashi, S., Nakamura, Y., Shigemoto, N. and Doi, Y. (1994) A simple and efficient gene transfer system of trifoliate orange (Poncirus trifoliata Raf.). Plant Cell Reports 13: 541–545. [DOI] [PubMed] [Google Scholar]
- Katz, E., Boo, K.H., Kim, H.Y., Eigenheer, R.A., Phinney, B.S., Shulaev, V., Negre-Zakharov, F., Sadka, A. and Blumwald, E. (2011) Label-free shotgun proteomics and metabolite analysis reveal a significant metabolic shift during citrus fruit development. J. Exp. Bot. 62: 5367–5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepiro, J.L. and Roose, M.L. (2010) AFLP markers closely linked to a major gene essential for nucellar embryony (apomixis) in Citrus maxima × Poncirus trifoliata. Tree Genet Genomes 6: 1–11. [Google Scholar]
- Khan, I.A. (2007) Citrus genetics, breeding and biotechnology. CAB International, Wallingford, pp. 141–150. [Google Scholar]
- Kijas, J.M.H., Thomas, M.R., Fowler, J.S.C. and Roose, M.L. (1997) Integration of trinucleotide microsatellites into a linkage map of Citrus. Theor. Appl. Genet. 94: 701–706. [Google Scholar]
- Kita, M., Endo-Inagaki, T., Moriguchi, T. and Omura, M. (2000a) cDNA catalogs expressed in albedo of citrus fruit: a comparative analysis of cDNA libraries from pulp and albedo of Satsuma mandarin (Citrus unshiu Marc.). Acta. Hortic. 521: 179–183. [Google Scholar]
- Kita, M., Hisada, S., Endo-Inagaki, T., Omura, M. and Moriguchi, T. (2000b) Changes in the levels of mRNAs for putative cell growth-related genes in the albedo and flavedo during citrus fruit development. Plant Cell Reports 19: 582–587. [DOI] [PubMed] [Google Scholar]
- Kita, M., Hirata, Y., Moriguchi, T., Endo-Inagaki, T., Matsumoto, R., Hasegawa, S., Suhayda, C.G. and Omura, M. (2000c) Molecular cloning and characterization of a novel gene encoding limonoid UDP-glucosyltransferase in Citrus. FEBS Lett. 469: 173–178. [DOI] [PubMed] [Google Scholar]
- Kita, M., Endo, T., Shimada, T., Moriguchi, T., Hirata, Y., Hasegawa, S. and Omura, M. (2003) Allelic structures of UDP-glucose: limonoid glucosyltransferase affect limonoid bitterness in Citrus unshiu and C. sinensis. Euphytica 132: 87–94. [Google Scholar]
- Kobayashi, Y., Kaya, H., Goto, K., Iwabuchi, M. and Araki, T. (1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962. [DOI] [PubMed] [Google Scholar]
- Komatsu, A., Takanokura, Y., Omura, M. and Akihama, T. (1996) Cloning and molecular analysis of cDNAs encoding three sucrose phosphate synthase isoforms from a citrus fruit (Citrus unshiu Marc.). Mol. Gen. Genet. 252: 346–351. [DOI] [PubMed] [Google Scholar]
- Komatsu, A., Moriguchi, T., Koyama, K., Omura, M. and Akihama, T. (2002) Analysis of sucrose synthase genes in citrus suggests different roles and phylogenetic relationships. J. Exp. Bot. 53: 61–71. [PubMed] [Google Scholar]
- Kubo, T., Honjo, I. and Hiratsuka, S. (2001) Sucrose accumulation and its related enzyme activities in the juice sacs of satsuma mandarin fruit from trees with different crop loads. Sci. Hortic. 91: 215–225. [Google Scholar]
- Kurowska, E.M. and Manthey, A.J. (2004) Hypolipidemic effects and absorption of citrus polymethoxylated flavones in hamsters with diet-induced hypercholesterolemia. J. Agric. Food Chem. 52: 2879–2886. [DOI] [PubMed] [Google Scholar]
- Lam, L.K.T. and Hasegawa, S. (1989) Inhibition of benzo[α] pyrene-induced forestomach neoplasia in mice by citrus limonids. Nutr. Cancer 12: 43–47. [DOI] [PubMed] [Google Scholar]
- Lewinsohn, E., Britsch, L., Mazur, Y. and Gressel, J. (1989) Flavanone glycoside biosynthesis in Citrus: Chalcone synthase, UDP-glucose: flavanone-7-O-glucosyl-transferase and -rhamnosyl-transferase activities in cell-free extracts. Plant Physiol. 91: 1323–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling, P., Duncan, L.W., Deng, Z., Dunn, D., Hu, X., Huang, S. and Gmitter, F.G.Jr. (2000) Inheritance of citrus nematode resistance and its linkage with molecular markers. Theor. Appl. Genet. 100: 1010–1017. [Google Scholar]
- Lo Piero, A.R., Puglisi, I. and Petrone, G. (2006) Gene characterization, analysis of expression and in vitro synthesis of dihydroflavonol 4-reductase from [Citrus sinensis (L.) Osbeck]. Phytochemistry 67: 684–695. [DOI] [PubMed] [Google Scholar]
- Lowell, C.A., Tomlinson, P.T. and Koch, K.E. (1989) Sucrose-metabolizing enzymes in transport tissues and adjacent sink structures in developing citrus fruit. Plant Physiol. 90: 1394–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lücker, J., El Tamer, M.K., Schwab, W., Verstappen, F.W.A., van der Plas, L.H.W., Bouwmeester, H.J. and Verhoeven, H.A. (2002) Monoterpene biosynthesis in lemon (Citrus limon): cDNA isolation and functional analysis of four monoterpene synthases. Eur. J. Biochem. 269: 3160–3171. [DOI] [PubMed] [Google Scholar]
- Luro, F.L., Costantino, G., Terol, J., Argout, X., Allario, T., Wincker, P., Talon, M., Ollitrault, P. and Morillon, R. (2008) Transferability of the EST-SSRs developed on Nules Clementine (Citrus clementina Hort ex Tan) to other Citrus species and their effectiveness for genetic mapping. BMC Genomics 9: 287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama, T., Motohashi, R., Akihama, T. and Omura, M. (1992) DNA Fingerprinting in Citrus Cultivars. Breed Sci. 42: 155–159. [Google Scholar]
- Matsuyama, T., Akihama, T., Ito, Y., Omura, M. and Fukui, K. (1996) Characterization of heterochromatic regions in ‘Trovita’ orange (Citrus sinensis Osbeck) chromosomes by the fluorescent staining and FISH methods. Genome 39: 941–945. [DOI] [PubMed] [Google Scholar]
- Miranda, M., Ikeda, F., Endo, T., Moriguchi, T. and Omura, M. (1997) Comparative analysis on the distribution of heterochromatin in Citrus, Poncirus and Fortunella chromosomes. Chromosome Res. 5: 86–92. [DOI] [PubMed] [Google Scholar]
- Miyata, Y., Tanaka, H., Shimada, A., Sato, T., Ito, A., Yamanouchi, T. and Kosano, H. (2011) Regulation of adipocytokine secretion and adipocyte hypertrophy by polymethoxyflavonoids, nobiletin and tangeretin. Life Sci. 88: 613–618. [DOI] [PubMed] [Google Scholar]
- Moriguchi, T., Kita, M., Hisada, S., Endo-Inagaki, T. and Omura, M. (1998a) Characterization of gene repertories at mature stage of citrus fruit through random sequencing and analysis of redundant metallothionein-like genes expressed during fruit development. Gene 211: 221–227. [DOI] [PubMed] [Google Scholar]
- Moriguchi, T., Kita, M., Endo-Inagaki, T., Ikoma, Y. and Omura, M. (1998b) Characterization of a cDNA homologous to carotenoid-associated protein in citrus fruits. Biochim. Biophys. Acta 1442: 334–338. [DOI] [PubMed] [Google Scholar]
- Moriguchi, T., Kita, M., Tomono, Y., Endo-Inagaki, T. and Omura, M. (2001) Gene expression in flavonoid biosynthesis: Correlation with flavonoid accumulation in developing citrus fruit. Physiol. Plant. 111: 66–74. [Google Scholar]
- Nakamura, M. (1929) Cytological studies in the genus Citrus. I. On the Wase satsuma originated through bud variation. Stud. Citrol. 3: 1–14. [Google Scholar]
- Nakano, M., Nesumi, H., Yoshioka, T. and Yoshida, T. (2001) Segregation of plants with undeveloped anthers among hybrids derived from the seed parent, ‘Kiyomi’ (Citrus unshiu × C. sinensis). J. Japan. Soc. Hort. Sci. 70: 539–545. [Google Scholar]
- Nakano, M., Nesumi, H., Yoshioka, T., Omura, M. and Yoshida, T. (2003) Linkage analysis between male sterility of citrus and STS markers. Proc. Intl. Soc. Citricultut. IX Ccngr. 2000, pp. 179–180. [Google Scholar]
- Nakano, M., Shimizu, T., Fujii, H., Shimada, T., Endo, T., Nesumi, H., Kuniga, T. and Omura, M. (2008) Marker enrichment and construction of haplotype-specific BAC contigs for polyembryony genomic region in Citrus. Breed. Sci. 58: 375–383. [Google Scholar]
- Nakano, M., Shimada, T., Endo, T., Fujii, H., Nesumi, H., Kita, M., Ebina, M., Shimizu, T. and Omura, M. (2012) Characterization of genomic sequence showing strong association with polyembryony among diverse Citrus species and cultivars, and its synteny with Vitis and Populus. Plant Sci. 183: 131–142. [DOI] [PubMed] [Google Scholar]
- Nesumi, H. and Matsumoto, R. (2003) Improvement of citrus scion cultivars by crossbreeding in Japan. Proc. Intl. Soc. Citricul. IX Congr. 2000, pp. 46–47. [Google Scholar]
- Nicolosi, E. (2007) Origin and taxonomy. In: Khan, I.A. (ed.) Citrus Genetics, Breeding and Biotechnology. CAB International, Wallingford, UK, pp. 19–44. [Google Scholar]
- Nicolosi, E.Z., Deng, N., Gentile, A., La Malfa, S., Continella, G. and Tribulato, E. (2000) Citrus phylogeny and genetic origin of important species as investigated by molecular markers. Theor. Appl. Genet. 100: 1155–1166. [Google Scholar]
- Ninomiya, T., Shimada, T., Endo, T., Nonaka, K., Omura, M. and Fujii, H. (2015) Development of citrus cultivar identification by CAPS markers and parentage analysis. Hort. Res. (Japan) 14: 127–133. [Google Scholar]
- Nishikawa, F., Endo, T., Shimada, T., Fujii, H., Shimizu, T., Omura, M. and Ikoma, Y. (2007) Increased CiFT abundance in the stem correlates with floral induction by low temperature in Satsuma mandarin (Citrus unshiu Marc.). J. Exp. Bot. 58: 3915–3927. [DOI] [PubMed] [Google Scholar]
- Nishikawa, F., Endo, T., Shimada, T., Fujii, H., Shimizu, T. and Omura, M. (2008) Isolation and characterization of a Citrus FT/TFL1 homologue (CuMFT1), which shows quantitatively preferential expression in Citrus seeds. J. Japan. Soc. Hort. Sci. 77: 38–46. [Google Scholar]
- Nishikawa, F., Endo, T., Shimada, T., Fujii, H., Shimizu, T. and Omura, M. (2009a) Differences in seasonal expression of flowering genes between deciduous trifoliate orange and evergreen Satsuma mandarin. Tree Physiol. 29: 921–926. [DOI] [PubMed] [Google Scholar]
- Nishikawa, F., Endo, T., Shimada, T., Fujii, H., Shimizu, T. and Omura, M. (2009b) Characterization of the 5′ flanking region of the citrus d-limonene synthase gene, which shows a quantitatively preferential expression in peel. J. Japan. Soc. Hort. Sci. 78: 84–89. [Google Scholar]
- Nishikawa, F., Endo, T., Shimada, T., Fujii, H., Shimizu, T., Kobayashi, Y., Araki, T. and Omura, M. (2010) Transcriptional changes in CiFT-introduced transgenic trifoliate orange (Poncirus trifoliata L. Raf.). Tree Physiol. 30: 431–439. [DOI] [PubMed] [Google Scholar]
- Nishikawa, F., Iwasaki, M., Fukamachi, H., Nokana, K., Imai, A. and Endo, T. (2011) Seasonal changes of citrus flowering locus T gene expression in kumquat. Bull. Natl. Inst. Fruit Tree Sci. 12: 27–32. [Google Scholar]
- Nishiura, M. (1964) Citrus breeding in Japan. Florida State Horticultural Society, pp. 79–83. [Google Scholar]
- Nishiura, M., Kamiya, S. and Esaki, S. (1971) Flavonoids in citrus and related genera. Part III. Flavonoid pattern and citrus taxonomy. Agric. Biol. Chem. 35: 1691–1971. [Google Scholar]
- Ohta, S., Endo, T., Shimada, T., Fujii, H., Shimizu, T., Kuniga, T., Yoshioka, T., Nesumi, H., Yoshida, T. and Omura, M. (2011) PCR primers for marker assisted backcrossing to introduce a CTV resistance gene from Poncirus trifoliata (L.) Raf. into Citrus. J. Japan. Soc. Hort. Sci. 80: 295–307. [Google Scholar]
- Ohta, S., Endo, T., Shimada, T., Fujii, H., Shimizu, T., Kita, M., Kuniga, T., Yoshioka, T., Nesumi, H., Yoshida, T.et al. (2015) Construction of genetic linkage map and graphical genotyping of pseudo-backcrossed F2 (BC2) progeny to introduce a CTV resistance from Poncirus trifoliata (L.) Raf. into Citrus by introgression breeding. Tree Genet Genomes 11: 797. [Google Scholar]
- Oiyama, I. and Kobayashi, S. (1993) Haploids obtained from diploid × triploid crosses of citrus. J. Japan Soc. Hort. Sci. 62: 89–93. [Google Scholar]
- Okudai, N., Yoshinaga, K., Takahara, T., Ishiuchi, D. and Oiyama, I. (1980) Studies on the promotion of flowering and fruiting of juvenile citrus seedlings. II. Effect of grafting. Bull. Fruit Tree Res. Stn. D2: 15–28. [Google Scholar]
- Ollitrault, P., Dambier, D., Luro, F. and Duperray, C. (1994) Nuclear genome size variations in Citrus. Fruits 49: 390–393. [Google Scholar]
- Ollitrault, P., Terol, J., Chen, C., Federici, C.T., Lotfy, S., Hippolyte, I., Ollitrault, F., Berard, A., Chauveau, A., Cuenca, J.et al. (2012) A reference genetic map of C. clementina hort. ex Tan.; citrus evolution inferences from comparative mapping. BMC Genomics 13: 593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omura, M., Ueda, T., Shimada, T., Endo, T., Fujii, H., Nesumi, H. and Yoshida, T. (2003) Graphical genotype of citrus cultivars by codominant CAPS markers. Abst. Plant & Animal genome XI Conference p.22 Jan. 11–15, San Diego, CA. [Google Scholar]
- Ou, P., Hasegawa, S., Herman, Z. and Fong, C.H. (1988) Limonoid biosynthesis in the stem of Citrus limon. Photochemistry 27: 115–118. [Google Scholar]
- Peña, L., Martín-Trillo, M., Juárez, J., Pina, J.A., Navarro, L. and Martínez-Zapater, J.M. (2001) Constitutive expression of Arabidopsis LEAFY or APETALA1 genes in citrus reduces their generation time. Nat. Biotechnol. 19: 263–267. [DOI] [PubMed] [Google Scholar]
- Peña, L., Cervera, M., Fagoaga, C., Romero, J., Ballester, A., Soler, N., Pons, E., Rodríguez, A., Peris, J., Juárez, J.et al. (2008) Citrus. In: Kole, C. and Hall T.C. (eds.) A compendium of transgenic crop plants. vol. 5, Wilkey-Blackwell, Malden, Mass, USA. [Google Scholar]
- Peterson, J.M.S., Dwyer, J. and Dsc, R.D. (1998) Flavonoids: Dietary occurrence and biochemical activity. Nutr. Res. 18: 1995–2018. [Google Scholar]
- Pichersky, E. and Gershenzon, J. (2002) The formation and function of plant volatiles: Perfumes for pollinator attraction and defense. Curr. Opin. Plant Biol. 5: 237–243. [DOI] [PubMed] [Google Scholar]
- Pillitteri, L.J., Lovatt, C.J. and Walling, L.L. (2004) Isolation and characterization of a TERMINAL FLOWER homolog and its correlation with juvenility in citrus. Plant Physiol. 135: 1540–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez, A., San Andrés, V., Cervera, M., Redondo, A., Alquézar, B., Shimada, T., Gadea, J., Rodrigo, M.J., Zacarías, L., Palou, L.et al. (2011) Terpene down-regulation in orange reveals the role of fruit aromas in mediating interactions with insect herbivores and pathogens. Plant Physiol. 156: 793–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez, A., Shimada, T., Cervera, M., Alquézar, B., Gadea, J., Gómez-Cadenas, A., De Ollas, C.J., Rodrigo, M.J., Zacarías, L. and Peña, L. (2014) Terpene down-regulation triggers defense responses in transgenic orange leading to resistance against fungal pathogens. Plant Physiol. 164: 321–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roose, M.L., Schwarzacher, T. and Heslop-Harrison, J.S. (1998) The chromosomes of Citrus and Poncirus species and hybrids: identification of characteristic chromosomes and physical mapping of rDNA loci using in situ hybridization and fluorochrome banding. J. Hered. 89: 83–86. [DOI] [PubMed] [Google Scholar]
- Rouseff, R.L. (1980) Flavonoids and Citrus quality. In: Nagy, S. and Attaway J. (eds.) Citrus Nutrition and Quality, Symposium Series Vol. 143 American Chemical Society, Washington, DC, pp. 83–108. [Google Scholar]
- Sadka, A., Dahan, E., Cohen, L. and Marsh, K.B. (2000) Aconitase activity and expression during the development of lemon fruit. Physiol. Plant. 108: 255–262. [Google Scholar]
- Sawamura, M. (2000) Volatile components of essential oils of the Citrus genus. Recent Res. Devel. Agric. Food Chem. 4: 131–164. [Google Scholar]
- Sharon-Asa, L., Shalit, M., Frydman, A., Bar, E., Holland, D., Or, E., Lavi, U., Lewinsohn, E. and Eyal, Y. (2003) Citrus fruit flavor and aroma biosynthesis: isolation, functional characterization, and developmental regulation of Cstps1, a key gene in the production of the sesquiterpene aroma compound valencene. Plant J. 36: 664–674. [DOI] [PubMed] [Google Scholar]
- Shimada, T., Kita, M., Endo, T., Fujii, H., Ueda, T., Moriguchi, T. and Omura, M. (2003) Expressed sequence tags of ovary tissue cDNA library in Citrus unshiu Marc. Plant Sci. 165: 167–168. [Google Scholar]
- Shimada, T., Endo, T., Fujii, H., Hara, M., Ueda, T., Kita, M. and Omura, M. (2004) Molecular cloning and functional characterization of four monoterpene synthase genes from Citrus unshiu Marc. Plant Sci. 166: 49–58. [Google Scholar]
- Shimada, T., Fuiii, H., Endo, T., Yazaki, J., Kishimoto, N., Shimbo, K., Kikuchi, S. and Omura, M. (2005a) Toward comprehensive expression profiling by microarray analysis in citrus: monitoring the expression profiles of 2213 genes during fruit development. Plant Sci. 168: 1383–1385. [Google Scholar]
- Shimada, T., Endo, T., Fujii, H., Hara, M. and Omura, M. (2005b) Isolation and characterization of (E)-beta-ocimene and 1,8 cineole synthases in Citrus unshiu Marc. Plant Sci. 168: 987–995. [Google Scholar]
- Shimada, T., Endo, T., Fujii, H., Hara, M. and Omura, M. (2005c) Isolation and characterization of a new d-limonene synthase gene with a different expression pattern in Citrus unshiu Marc. Sci. Hortic. 105: 507–512. [Google Scholar]
- Shimada, T., Nakano, R., Shulaev, V., Sadka, A. and Blumwald, E. (2006) Vacuolar citrate/H+ symporter of citrus juice cells. Planta 224: 472–480. [DOI] [PubMed] [Google Scholar]
- Shimada, T., Endo, T., Rodríguez, A., Fujii, H., Nakano, M., Sugiyama, A., Shimizu, T., Peña, L. and Omura, M. (2012) Isolation and characterization of germacrene A synthases gene in Citrus unshiu Marc. Sci. Hortic. 145: 102–108. [Google Scholar]
- Shimada, T., Fujii, H., Endo, T., Ueda, T., Sugiyama, A., Nakano, M., Kita, M., Yoshioka, T., Shimizu, T., Nesumi, H.et al. (2014a) Construction of a citrus framework genetic map anchored by 708 gene-based markers. Tree Genet. Genomes 10: 1001–1013. [Google Scholar]
- Shimada, T., Endo, T., Fujii, H., Rodríguez, A., Peña, L. and Omura, M. (2014b) Characterization of three linalool synthase genes from Citrus unshiu Marc. and analysis of linalool-mediated resistance against Xanthomonas citri subsp. citri and Penicilium italicum in citrus leaves and fruits. Plant Sci. 229: 154–166. [DOI] [PubMed] [Google Scholar]
- Siviero, A., Cristofani, M., Furtado, E.L., Garcia, A.A.F., Coelho, A.S.G. and Machado, M.A. (2006) Identification of QTLs associated with citrus resistance to Phytophthora gummosis. J. Appl. Genet. 47: 23–28. [DOI] [PubMed] [Google Scholar]
- Sugawara, K., Wakizuka, T., Oowada, A., Moriguchi, T. and Omura, M. (2002) Histogenic identification by RAPD analysis of leaves and fruit of newly synthesized chimeric citrus. J. Amer. Soc. Hort. Sci. 127: 104–107. [Google Scholar]
- Sugiyama, A., Omura, M., Matsumoto, H., Shimada, T., Fujii, H., Endo, T., Shimizu, T., Nesumi, H. and Ikoma, Y. (2011) Quantitative trait loci (QTL) analysis of carotenoid content in citrus fruit. J. Japan. Soc. Hort. Sci. 80: 136–144. [Google Scholar]
- Sugiyama, A., Omura, M., Shimada, T., Fujii, H., Endo, T., Shimizu, T., Nesumi, H., Nonaka, K. and Ikoma, Y. (2014) Expression quantitative trait loci analysis of carotenoid metabolism-related genes in Citrus. J. Japan. Soc. Hort. Sci. 83: 32–43. [Google Scholar]
- Swingle, W.T. (1943) The botany of citrus and its wild relatives of the orange subfamily. In: Webber, H.J. and Batchelor L.D. (eds.) The Citrus Industry. University of California Press, California, pp. 190–430. [Google Scholar]
- Swingle, W.T. (1967) The botany of citrus and its wild relatives. In: Reuther, W., Webber H.J. and Batchelor L.D. (eds.) The Citrus Industry, vol. 1 University of California Press, Berkeley, CA, USA, pp. 389–390. [Google Scholar]
- Talon, M. and Gmitter, F.G.Jr. (2008) Citrus genomics. Intl. J. Plant Genomics. Article ID 528361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, T. (1954) Species problem in Citrus (Revisio aurantiaceraum, IX). Japan Soci Prom. Sci., Veno, Tokyo. [Google Scholar]
- Tanaka, T. (1961) Contribution to the knowledge of citrus classification. Reports Citrologia 107–114. [Google Scholar]
- Taniguchi, S., Hosokawa-Shinonaga, Y., Tamaoki, D., Yamada, S., Akimitsu, K. and Gomi, K. (2014) Jasmonate induction of the monoterpene linalool confers resistance to rice bacterial blight and its biosynthesis is regulated by JAZ protein in rice. Plant Cell Environ. 37: 451–461. [DOI] [PubMed] [Google Scholar]
- Tao, N., Jia, L. and Zhou, H. (2014) Anti-fungal activity of Citrus reticulata Blanco essential oil against Penicillium italicum and Penicillium digitatum. Food Chem. 153: 265–271. [DOI] [PubMed] [Google Scholar]
- Tripoli, E., La Guardia, M., Giammanco, S., Majo, D.D. and Giammanco, M. (2007) Citrus flavonoids: Molecular structure, biological activity and nutritional properties: A review. Food Chem. 104: 466–479. [Google Scholar]
- Ueda, T., Ikeda, F., Kita, M., Shimada, T., Endo, T. and Omura, M. (2003) Evaluation of a CAPS method based on ESTs in Citrus. Proc. Intl. Soc. Citricult. IX Congr. 2000, pp. 116–117. [Google Scholar]
- Vekiari, S.A., Protopapadakis, E.E., Papadopoulou, P., Papanicolaou, D., Panou, C. and Vamvakias, M. (2002) Composition and seasonal variation of the essential oil from leaves and peel of a Cretan lemon variety. J. Agric. Food Chem. 50: 147–153. [DOI] [PubMed] [Google Scholar]
- Wigge, P.A., Kim, M.C., Jaeger, K.E., Busch, W., Schmid, M., Lohmann, J.U. and Weigel, D. (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309: 1056–1059. [DOI] [PubMed] [Google Scholar]
- Wu, G.A., Prochnik, S., Jenkins, J., Salse, J., Hellsten, U., Murat, F., Perrier, X., Ruiz, M., Scalabrin, S., Terol, J.et al. (2014) Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat. Biotechnol. 32: 656–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Q., Chen, L.L., Ruan, X., Chen, D., Zhu, A., Chen, C., Bertrand, D., Jiao, W.B., Hao, B.H., Lyon, M.P.et al. (2013) The draft genome of sweet orange (Citrus sinensis). Nat. Genet. 45: 59–66. [DOI] [PubMed] [Google Scholar]
- Yamamoto, M., Tsuchimochi, Y., Ninomiya, T., Koga, T., Kitajima, A., Yamasaki, A., Inafuku-Teramoto, S., Yang, X., Yang, X., Zhong, G.et al. (2013) Diversity of chloroplast DNA in various mandarins (Citrus spp.) and other citrus demonstrated by CAPS analysis. J. Japan. Soc. Hort. Sci. 82: 106–113. [Google Scholar]
- Yamasaki, A., Kitajima, A., Ohara, N., Tanaka, M. and Hasegawa, K. (2007a) Histological study on expression of seedlessness in Citrus kinokuni ‘Mukaku Kishu’ and its progenies. J. Amer. Soc. Hort. Sci. 132: 869–875. [Google Scholar]
- Yamasaki, Y., Kunoh, H., Yamamoto, H. and Akimitsu, K. (2007b) Biological roles of monoterpene volatiles derived from rough lemon (Citrus jambhiri Lush) in citrus defense. J. Gen. Plant Pathol. 73: 168–179. [Google Scholar]
- Yuya, U., Ozawa, R., Shishido, H., Taniguchi, S., Takabayashi, J., Akimitsu, K. and Gomi, K. (2015) Isolation of a sesquiterpene synthase expressing in specialized epithelial cells surrounding the secretory cavities in rough lemon (Citrus jambhiri) J. Plant Physiol. 180: 67–71. [DOI] [PubMed] [Google Scholar]