Abstract
Mammalian target of rapamycin (mTOR) is a protein serine/threonine kinase that controls a wide range of growth-related cellular processes. In the past several years, many factors have been identified that are involved in controlling mTOR activity. Those factors in turn are regulated by diverse signaling cascades responsive to changes in intracellular and environmental conditions. The molecular connections between mTOR and its regulators form a complex signaling network that governs cellular metabolism, growth and proliferation. In this review, we discuss some key factors in mTOR regulation and mechanisms by which these factors control mTOR activity.
Keywords: mTOR, TSC2, Rheb, FKBP38, PRAS40, hVps34, Rag GTPases, Phosphatidic acid
Introduction
TOR, target of rapamycin, is an atypical serine/threonine kinase that was originally identified in the yeast Saccharomyces cerevisiae and later in human, mouse and other eukaryotic cells [1, 2]. Rapamycin, in complex with a cytosolic protein FKBP12, specifically binds to TOR and interferes with its function [3, 4]. For most of eukaryotic cells, inhibition of TOR by rapamycin results in growth arrest. The drug-affected cells withdraw from cell cycle and become unresponsive to growth factor and nutrient stimulation [5]. However, the sensitivity of each type of cells to rapamycin varies [6]. Lymphocytes and certain types of cancer cells are among those highly susceptible to rapamycin inhibition. This differentiated sensitivity allows the use of rapamycin and its derivatives (rapalogs) to selectively block the growth and proliferation of unwanted cells without affecting normal ones in the human body, leading to the development of rapalogs as immunosuppressive drugs and recently as anti-cancer agents [7]. Hence, ever since its identification, TOR has been at the center of extensive studies driven by our desire to understand fundamental mechanisms governing cell growth and by the clinical applications of TOR inhibitors.
In mammals the homologs of the yeast TOR are collectively referred to as mammalian target of rapamycin (mTOR). Like its counterparts in other organisms, mTOR exists in two distinct complexes termed as mTOR complex 1 (mTORC1) and complex 2 (mTORC2), both of which contain several components that are conserved from yeast to human [8, 9]. Despite the presence of mTOR in both complexes, only mTORC1 is sensitive to rapamycin inhibition [8]. However, prolonged rapamycin treatment still affects mTORC2 function, presumably by preventing new mTORC2 generation [10].
Being the target of rapamycin, mTORC1 has been the main focus in most mTOR-related studies. As discussed below, mTORC1 is regulated by an array of diverse intracellular and environmental cues, including those of growth factors, mitogens, phosphatidic acid (PA), energy, nutrient and stresses (Fig. 1). Upon receiving and integrating upstream signals, mTORC1 in turn controls a wide range of growth-related cellular processes, including translation, transcription, autophagy and hypoxic adaptation [5]. In contrast, mTORC2 is involved in controlling Akt activity and in regulating cell-cycle-dependent organization of actin cytoskeleton [11]. The mechanisms for its regulation have only begun to be revealed. In the following sections, we discuss many recently emerged factors involved in mTOR regulation with emphasis on those directly in contact with mTOR or mTOR complexes.
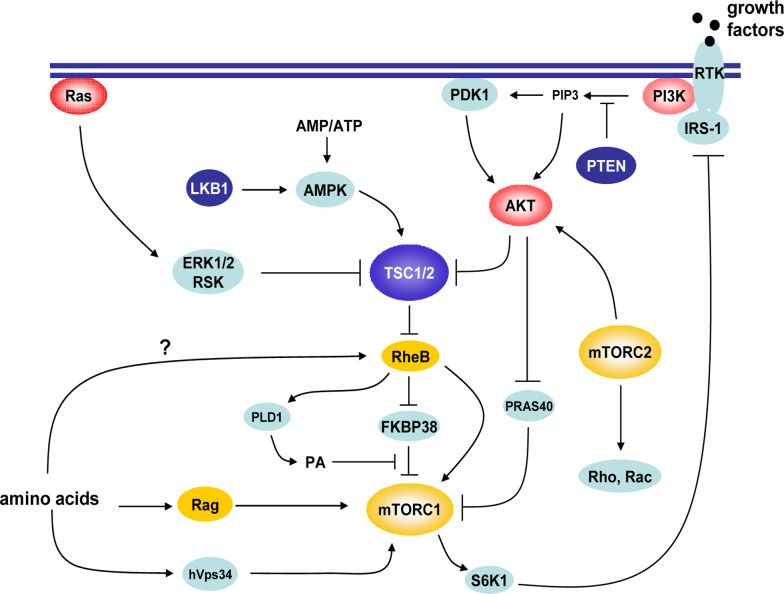
Fig. 1.
The signaling network that controls mTOR. mTOR exists in two distinct complexes, mTORC1 and mTORC2. The function of mTORC1 is regulated by signaling pathways responsive to amino acids sufficiency, growth factor stimulation and changes in energy levels. These pathways converge either on mTORC1 itself or on the TSC1/TSC2 complex, making up a complex signaling network. The proximal regulators of mTORC1 include Rheb and Rag small GTPases as activators and FKBP38 and PRAS40 as inhibitors. The S6K-directed phosphorylation of IRS-1 constitutes a feedback loop that downregulates the signaling activity from insulin receptors to Akt and mTOR. The mechanisms involved in mTORC2 regulation remain elusive. Arrows depict activation and T bars inhibition
mTOR
Mammalian target of rapamycin is responsible for the catalytic activity of both mTORC1 and mTORC2. It is a ~289-kDa multi-domain protein that belongs to the family of the phosphatidylinositol 3-kinase (PI3-K)-related kinases (PIKK). Members of this kinase family are characterized by their large sizes (>2,500 amino acids) and a C-terminally located kinase domain that is structurally related to PI3-K [12]. In addition to the kinase domain, mTOR also contains an N-terminal region that is mainly composed of tandem repeats of the HEAT motif, an α-helix-like structure mediating protein–protein interaction [13]. The middle part of mTOR comprises the FAT domain that is found among all the members of the PIKK family and is believed to be involved in interaction with other proteins [14]. The last ~30 residues of mTOR form the FATC domain, which consists of an α-helix and a disulfide-bonded loop, which can be regulated by cytosolic redox potential [15]. The FKBP12–rapamycin complex binds to a region lying between the FAT and the kinase domains [16]. Binding of the complex to mTOR is thus not expected to directly affect the kinase activity of mTOR (Fig. 2).
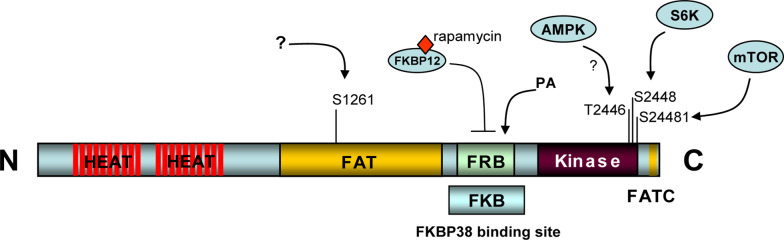
Fig. 2.
A schematic presentation of mTOR structures. mTOR is a large protein kinase containing multiple-functional domains. The kinase domain is located at the C-terminus and is responsible for the catalytic activity of mTORC1. The HEAT motif region located at the N-terminus and the FAT domain in the middle are involved in mediating mTOR interaction with other proteins. The FRB domain is where the FKBP12 and rapamycin complex binds, which is within the region that binds to FKBP38. mTOR is also phosphorylated by several kinases, including itself
Mammalian target of rapamycin is phosphorylated at multiple sites, including Ser2448, Ser2481, Thr2446 and Ser1261. Phosphorylation at Ser2448 is mediated by p70 ribosomal S6 kinase (S6K) and occurs predominantly to mTOR in mTORC1 [17–19]. Ser2481 is an autophosphorylation site. Phosphorylation at this site happens mainly to mTOR in mTORC2 and is insensitive to acute rapamycin treatment [19, 20]. Thr2446 is phosphorylated presumably by cAMP activated protein kinase (AMPK), and the phosphorylation levels increase upon nutrient withdrawal and decrease upon insulin stimulation [21]. Ser1261 was identified recently as a site whose phosphorylation promotes mTORC1 activity and is required for mTOR autophosphorylation at Ser2481 [22]. In all the cases examined, phosphorylation of mTOR appears to alter its kinase activity rather than its association with other components in the mTOR complexes [19, 23].
mTORC1
mTOR complex 1 is sensitive to rapamycin and thus is responsible for all the rapamycin-sensitive processes in mammalian cells. It consists of mTOR, regulatory associated protein of mTOR (raptor) and mLST8 (mammalian lethal with sec13 protein 8, also known as GβL), all of which are essential for the function of the complex (Fig. 3) [8, 24, 25].
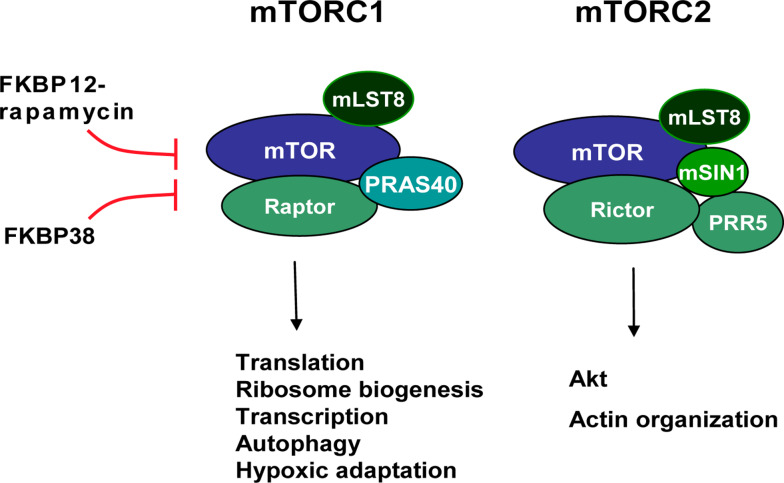
Fig. 3.
Two mTOR complexes and their functions. mTORC1 is composed of mTOR, mLST8, raptor and PRAS40. Its function is involved many growth-related processes and oxygen adaptation and is sensitive to rapamycin. mTORC2 shares mTOR and mLST8 with mTORC1, but contains a few other unique components, including rictor, mSIN1 and PRR5
mLST8 is the only mTOR partner found in both mTORC1 and mTORC2. It is a 36-kDa protein composed entirely of seven WD40 repeats, which are short sequence motifs of ~40 amino acids, often terminating in a tryptophan-aspartic acid (W-D) dipeptide [26]. mLST8 has been found to associate with the kinase domain of mTOR and stimulate mTOR kinase activity [25]. However, it appears to be dispensable for association of mTOR with raptor, and hence may not play a critical role in mTORC1 function [27].
Raptor, also known as p150 TOR-scaffold protein, is the unique component of mTORC1. It acts as an adapter protein in mTORC1 by binding and presenting substrates to mTOR [24, 28]. Raptor is a large protein with a M w of ~150 kDa that contains an N-terminal RNC (raptor N-terminal conserved) domain followed by three HEAT repeats and seven WD-40 repeats in the C-terminus of the protein [24, 25]. Analysis of the interaction between the yeast TOR and Kog1, the yeast counterpart of raptor, reveals that the C-terminal WD40 repeats of Kog1 bind to the N-terminal HEAT repeats of TOR, and the RNC domain is in proximity to the TOR kinase domain. The RNC domain is thus likely to function in bringing in substrates into the vicinity of the catalytic region [29]. Raptor binds to a sequence motif, commonly referred to as the TOR signaling motif (TOS), which is found in all known substrates of mTORC1, including S6K, 4E-BP1 and PRAS40 [30, 31]. Mutations in TOS motif of S6K and 4E-BP1 that abolish the binding with raptor also eliminate the ability of these polypeptides to be phosphorylated by mTORC1 in cells. In addition to TOS motif, 4E-BP1 contains another raptor-interacting motif (RAIP) that is required for its binding with raptor and efficient phosphorylation by mTORC1 in cells [32, 33]. Recently, a new raptor interacting motif, SAIN (Shc and IRS-1 NPXY-binding) domain, has been found in insulin receptor substrate-1 (IRS-1), which mediates the mTORC1-dependent phosphorylation of IRS-1 at Ser636 and Ser639 [34].
Besides its function to bring protein substrates to mTOR, raptor itself is also phosphorylated at multiple sites by several kinases, including AMPK, p90 ribosomal S6 kinase (RSK) and mTORC1. AMPK phosphorylates raptor on two well-conserved serine residues, Ser722 and Ser792. The phosphorylation recruits binding of 14-3-3 to raptor, resulting in inhibition of mTORC1 activity [35]. RSK phosphorylates raptor at three sites, including Ser719, Ser721 and Ser722. Phosphorylation at these sites has been shown to promote mTORC1 kinase activity [36]. In addition, a few mTORC1-directed phosphorylation sites have been found in raptor. Mutations in one of the sites, Ser863, attenuate the effect of small GTP-binding protein Ras-homology enriched in brain (Rheb) on mTORC1 activation [37]. Collectively, these findings suggest that raptor is able to integrate various signals for mTOR regulation.
The function of mTORC1 has been implicated in many cellular processes by the nature of their sensitivity to rapamycin inhibition. However, only a few direct substrates of mTORC1 have so far been identified. Among them, 4E-BP1 and S6K are two well-characterized targets. As such, the mTORC1-directed phosphorylation of both proteins is commonly used as an indicator for mTORC1 activity in cells. 4E-BP1 is a translation repressor, which binds and inhibits the translation initiation factor 4E (eIF-4E), a key factor for translation of 5′ capped mRNAs, among which are transcripts encoding growth promoting factors, such as Myc, cyclin D1, vascular endothelial growth factor (VEGF) and signal transducer and activator of transcription 3 (STAT3) [38]. mTORC1 phosphorylates 4E-BP1 at several sites, including Thr37, Thr46, Ser65, Thr70 and Ser83 [39–41]. Phosphorylation at Thr37 and Thr 46 appears to prime 4E-BP1 for additional phosphorylation at the other sites, which leads to its dissociation from eIF-4E [42]. S6K is phosphorylated by mTORC1 at Thr389 within a hydrophobic motif that links its catalytic domain with the C-terminal autoinhibition domain [40, 43, 44]. This phosphorylation permits further phosphorylation by phosphoinoside-dependent kinase 1 (PDK1) at Thr229 in the activation loop of the kinase domain of S6K, resulting in its activation [45, 46]. Once activated, S6K phosphorylates ribosomal protein S6, a protein required for translation of 5′ terminal oligopyrimidine (TOP) mRNAs encoding ribosomal proteins and elongation factors. Although mTORC1 is likely to have many other substrates, 4E-BP1 and S6K are the key factors contributing to the ability of mTORC1 to promote increased cell size [47].
In addition to S6 protein, S6K also phosphorylates several other proteins, including IRS-1, glycogen synthase kinase 3 (GSK3), translation elongation factor 2 (eEF2) kinase and pro-apoptotic protein Bad [48–52]. Of these targets, phosphorylation of IRS-1 is of great importance in mTOR regulation. IRS-1 binds to insulin receptor or insulin-like growth factor-1 (IGF-1) receptor and is required for activation of PI3-K in response to insulin or IGF-1 stimulation [53]. S6K phosphorylates IRS-1 at Ser302, which disrupts its interaction with the receptors, leading to a blockage in insulin signaling to Akt and mTOR [50, 54]. The S6K directed phosphorylation of IRS-1 thus constitutes a negative feedback that downregulates the signaling activity from insulin receptor to mTOR (Fig. 1).
mTORC2
mTOR complex 2 shares mTOR and mLST8 with mTORC1. However, it contains several unique components, including rapamycin-insensitive companion of mTOR (rictor), mammalian stress-activated protein kinase-interacting protein 1 (mSIN1) and proline-rich repeat protein-5 (PRR5) or PRR5-like (also known as protor1 and protor 2) (Fig. 3) [55–60]. The association of rictor to mTOR is critical for mTORC2 kinase activity and requires the presence of mLST8 and mSIN1, which makes the two proteins indispensable for mTORC2 function [27, 55, 60, 61]. PRR5 and its close homolog PRR5L are two newly discovered mTORC2 components. These proteins bind to rictor or mSIN1, but are not essential for mTORC2 stability or kinase activity [56, 58]. In addition to the components of mTORC2, rictor has also been shown to interact with other proteins, such as integrin-linked kinase (ILK), Myo1c and heat shock protein 70 (Hsp70) [62–64]. The significance of these interactions is unclear. It is possible that rictor, through interaction with these proteins, acts as an adaptor to bring mTORC2 to its targets. Alternatively, these proteins may function as regulators to control mTORC2 through rictor.
The FKBP12–rapamycin complex does not bind to mTORC2. Hence, mTORC2 is insensitive to acute rapamycin treatment [8, 57]. However, prolonged treatment with rapamycin has been found to disrupt mTORC2 assembly in some cell lines, resulting in decreased levels of functional mTORC2 [10, 65]. The effect of rapamycin may be mediated by the drug-induced dephosphorylation of rictor and mSIN1 [66, 67]. It has been found that short-term treatment with rapamycin triggers dephosphorylation of rictor and mSIN1 exclusively in the cytoplasm, but does not affect mTORC2 assembly. Prolonged rapamycin treatment leads to complete dephosphorylation and cytoplasmic translocation of nuclear rictor and mSIN1, which correlates with reduction in mTORC2 levels [66, 67].
Two major functions have been ascribed to mTORC2, including regulation of Akt and cell cycle-dependent organization of actin cytoskeleton. mTORC2 phosphorylates Akt at Ser473 in its C-terminal hydrophobic motif, which, in conjunction with PDK1-mediated phosphorylation at Thr308, drives full activation of Akt [61]. mTORC2 regulates actin cytoskeleton through a mechanism that involves the small GTPases Rho and Rac, although the molecular details of the regulation remain elusive [9, 57]. In addition, mTORC2 has been found to phosphorylate PKC and serum- and glucocorticoid-induced protein kinase 1 (SGK1), and may by involved in controlling cell size [68–71]. Thus, mTORC2, like its partner mTORC1, has pleiotropic roles in cell growth control.
TSC1 and TSC2
TSC1 and TSC2 were originally identified as two tumor suppressor genes. Inactivating mutations of the genes cause tuberous sclerosis complex syndrome, an inherited genetic disorder that is manifested by occurrence of tumor in multiple organs [72, 73]. Early studies suggested that the gene products, TSC1 and TSC2, also known as hamartin and tubrin, form a heterodimeric complex that displays a GTPase-activating protein (GAP) activity [74–76]. However, the function of the TSC1/TSC2 complex remained unclear until genetic studies in Drosophila ascribed a role for the complex in cell size control and placed it in a pathway that converges the insulin signaling from Akt to mTOR and S6K [77–79]. Subsequent studies in Drosophila identified small GTPase Rheb as a downstream target of TSC1 and TSC2 and upstream of TOR [80, 81]. Biochemical analyses confirmed Rheb as a bona fide effector of the TSC1/TSC2 complex [82–84]. In mammalian cells, coexpression of TSC1 and TSC2 reduces the levels of GTP-bound Rheb, while insulin stimulation increases the levels in a PI-3K-dependent manner [85]. These seminal studies in Drosophila and mammalian cells established a signaling scheme underlying insulin-stimulated mTOR activation that involves, in a sequential order, PI3-K, Akt, the TSC1/TSC2 complex, Rheb and mTOR (Fig. 1).
TSC2 is a 180-kDa protein that contains a GAP homology domain at its C-terminus. When assayed in vitro, TSC2 stimulates GTP hydrolysis of Rheb, but not other related small GTPases [82, 86]. TSC1 does not possess any GAP activity and is not required for that of TSC2 in vitro. However, in cells TSC1 appears to be essential for TSC2 function, as mutations in TSC1 have been found to reduce the function of the TSC1/TSC2 complex [86, 87]. TSC1 contains a transmembrane (TM) domain and is involved in targeting the TSC1/TSC2 complex to membrane structures [74, 88]. The membrane association allows the complex to directly interact with Rheb and modulate its GTP hydrolysis activity [84].
TSC2 is phosphorylated by multiple kinases involved in various signaling pathways, including Akt, AMPK, RSK and extracellular signal-regulated kinase (ERK). In response to growth factor, such as insulin stimulation, TSC2 is phosphorylated by Akt at multiple sites, including Ser939, Ser981 and Thr1462 [79, 89–91]. A mutant TSC2 lacking these phosphorylation sites acts dominantly in cells to block Akt signaling to mTOR, suggesting that the Akt-directed phosphorylation is to downregulate the function of TSC2. However, the phosphorylation does not appear to affect the GAP activity of TSC2 directly. Instead, it creates binding sites for a cytosolic protein, 14-3-3, which binds to phosphorylated TSC2 and inhibits its function [88]. The mechanism by which 14-3-3 downregulates TSC2 function is not fully understood. Some studies have suggested that binding with 14-3-3 prevents TSC2 from associating with TSC1 and thus reduces the function of the TSC1/TSC2 complex toward Rheb [79, 90], while others show that 14-3-3 may directly inhibit TSC2 function without disrupting the TSC1/TSC2 complex [89, 91].
In addition to being a target of Akt, TSC2 is also phosphorylated at Ser1345 by AMPK. Unlike the Akt-directed phosphorylation, which leads to inhibition of TSC2 function, the AMPK-directed phosphorylation enhances the GAP activity of TSC2 and consequently downregulates of mTORC1 [92]. As part of intracellular energy-sensing pathway, AMPK is activated when intracellular ATP is depleting and AMP levels arise [93]. The AMPK-dependent regulation of TSC2 thus couples intracellular energy levels to mTORC1 activity. The AMPK-dependent phosphorylation at Ser1345 also promotes subsequent phosphorylation at Ser 1337 and Ser1341 by glycogen synthase kinase 3β (GSK3β). These coordinated phosphorylation events lead to full activation of the GAP activity of TSC2 [92, 94]. Since GSK3β is a component in the Wnt signaling pathway, the GSK3β-directed phosphorylation links the pathway to mTORC1 regulation, whereby an activated Wnt signaling pathway, which inhibits GSK3β activity [95], is able to enhance mTORC1 activity by repressing GSK3β-dependent activation of TSC2 [92, 94].
TSC2 is also phosphorylated by ERK and RSK in response to mitogen stimulation and Ras activation. Activated ERK phosphorylates TSC2 at Ser664, which causes dissociation of TSC2 from TSC1 and impairs its function [96, 97]. RSK phosphorylates RSC2 at Ser1798, resulting in inhibition of TSC2 function. The mechanism for the inhibition is not clear. However, it has been found to be additive to the inhibitory effect of Akt-directed phosphorylation [98, 99].
The ability to receive phosphorylation by multiple kinases and alter its GAP activity accordingly provides a mechanism for TSC2 to integrate signals from various signaling pathways for mTOR regulation.
Rheb
Ras-homology enriched in brain was identified as a gene whose expression was induced in rat brain by synaptic activity and growth factor stimulation [100, 101]. Rheb is a small GTPase that is structurally more close to Ras than to other small GTPases. The regions in Rheb involved in GTP binding are highly similar to those in Ras. Particularly, the core effector domain of Rheb, the region that involves in targeting and binding to downstream effectors, shares six out of nine identical and two similar amino acid residues with that of Ras. It is thus believed that the action mechanism of Rheb is similar to that of Ras, which involves binding to its effector through the effector domain in a GTP-dependent manner [102, 103]. One striking difference, however, is that Rheb contains an arginine residue at position 15 rather than a glycine residue that is present at the corresponding site in Ras. This difference results in a low intrinsic GTPase activity in Rheb, which correlates with a relatively high level of GTP-bound Rheb in cells [102, 104, 105].
The activity of the small GTPases of Ras family depends on their nucleotide-binding states, which are regulated reciprocally by their cognate GAPs and guanine nucleotide exchange factors (GEFs) [102]. The GAP for Rheb has been shown to be the TSC1/TSC2 complex, which binds GTP-bound Rheb and stimulates GTP hydrolysis [82–84, 86]. However, the identity of the Rheb GEF remains elusive, although recent studies have suggested that the translationally controlled tumor protein (TCTP) may fill this role [106, 107].
Because the GAP activity of the TSC1/TSC2 complex is regulated by Akt- and AMPK-dependent phosphorylation of TSC2, the nucleotide-binding state of Rheb is expected to be controlled by these kinases, which couples, respectively, growth factor and energy signals to the TSC1/TSC2 complex. In support of this notion, insulin has been shown to regulate the nucleotide-binding states of Rheb through TSC2 [85, 108, 109]. However, whether Rheb is regulated by amino acids is controversial. Studies have shown that starving cells for amino acids reduces the levels of GTP-bound Rheb, while re-addition of amino acids increases the levels in the cells, suggesting that Rheb is regulated by amino acid conditions [109]. In contrast, two other studies have failed to detect any significant changes in the nucleotide-binding states of Rheb upon re-addition of amino acids to amino acid-starved cells [83, 110]. It is worth noting that in the latter cases, the GTP/GDP loading assays were done against overexpressed recombinant Rheb, while in the first cases, the assays were against endogenous Rheb or recombinant Rheb expressed at levels similar to those of the endogenous protein [108, 109]. When overexpressed in cells, Rheb stays largely in GTP-bound state, and its activity for mTOR activation becomes insensitive to amino acids [104, 108, 111], which raises a possibility that the overproduced Rheb rendered it unresponsive to changes in amino acid conditions in these studies. Since there is no evidence suggesting that the function of the TSC1/TSC2 complex is affected by amino acid conditions. If amino acids regulate Rheb, they may do so through TSC-independent mechanisms. One possibility is that amino acids control Rheb through a putative GEF of Rheb. Importantly, while Rheb is largely GTP-bound in tsc2 null cells, starving the cells for amino acids still results in mTORC1 inactivation, which suggests that amino acids are able to control mTORC1 activity independent of Rheb [108]. Although this finding does not answer whether Rheb is regulated by amino acids, it makes the issue less significant in mTOR regulation.
Overexpression of Rheb has been shown to activate mTORC1, and the activation is dominant over the effect of serum or amino acid withdrawal, suggesting that Rheb is a proximal activator of mTORC1 [80, 82]. Indeed, recombinant Rheb has been found to associate with mTOR, and this association is stimulated by amino acids [112]. However, the association does not appear to be GTP-dependent. It has been shown that Rheb mutants devoid of nucleotide-binding associate more strongly with mTOR than wild-type or active Rheb. Despite this, the kinase activity of mTOR has been found to be higher when it is copurified with recombinant wild-type Rheb than with inactive mutants, suggesting that the ability of Rheb to stimulate mTOR relies on its activity. Consistent with the notion, it has been found that GTP-loaded Rheb, when added to partially purified mTORC1 in vitro, increases its kinase activity [113, 114]. These findings strongly support that Rheb is able to bind to mTOR and stimulate its activity. However, evidence is lacking showing that Rheb directly binds with mTOR [112, 114, 115].
FKBP38
FKBP38, also known as FKBP8, belongs to the peptidyl prolyl cis/trans isomerase (PPIase) family of FK506-binding protein (FKBP) [116, 117]. Members of this protein family share a PPIase or PPIase-like domain, commonly referred to as the FKBP-C domain, and function as chaperones to facilitate protein folding and functions [118, 119]. The prominent member is FKBP12, which consists of entirely the FKBP-C domain and serves as the receptor for rapamycin [120, 121]. FKBP38 contains a N-terminally located FKBP-C domain, a tetratricopeptide repeat (TPR) motif sitting in the middle of the protein, a putative Ca2+/calmodulin (CaM)-binding site lying immediately adjacent to a TM domain at the very C-terminus. The TPR motif is found in other members of the FKBP family proteins and is believed to be involved in mediating protein–protein interaction. The Ca2+/CaM-binding site is responsible for binding with CaM in a Ca2+ dependent manner. The TM domain in FKBP38 is unique among all the FKBP proteins, which is required for anchoring FKBP38 to mitochondria, where it functions [122–125].
FKBP38 has been suggested to be the endogenous inhibitor of mTOR [126]. In cells, the expression levels of FKBP38 have been found to correlate inversely with the activity of mTORC1 [114, 127, 128]. In vitro, recombinant FKBP38 has been shown to inhibit mTORC1 kinase activity [126, 128]. FKBP38 inhibits mTORC1 by directly binding to mTOR. The FKBP-C domain of FKBP38, the region that is highly similar to FKBP12, is responsible for the binding. On the other hand, the minimal region in mTOR that is capable of binding with FKBP38 comprises amino acids 1967–2191, which embraces the FKBP12–rapamycin-binding (FRB) domain (amino acids 2015–2114). These attributes suggest that FKBP38 acts in the same way as the FKBP12–rapamycin complex to interfere with mTOR function. In accordance with this notion, it has been found that FKBP38, like the FKBP12–rapamycin complex, does not associate with mTORC2 [126].
The interaction of FKBP38 with mTOR is regulated by Rheb, which binds directly to FKBP38 and prevents it from interaction with mTOR [126]. Unlike its association with mTOR, the binding of Rheb with FKBP38 is GTP-dependent. It has been shown that Rheb binds to FKBP38 strongly when loaded with GTP, but weakly when loaded with GDP in vitro [126]. Rheb interacts with FKBP38 through its effector domain within the switch I region. Mutations in the effector domain impair the ability of Rheb to interact with FKBP38 and eliminate the ability of Rheb to stimulate mTORC1 [129]. This effector domain-mediated and GTP-dependent binding is a common mechanism for other members of the Ras family of small GTPases to interact with their targets [102], indicating that Rheb elicits its signaling activity as do by other members of the Ras family and that FKBP38 is a bona fide effector of Rheb [129].
Rheb binds to the FKBP-C domain in FKBP38, the same region that interacts with mTOR. However, the interaction of FKBP38 with Rheb and that with mTOR are not mutually exclusive. A dominant inactive mutant of Rheb, D60K, has been shown to bind to FKBP38 without displacing mTOR [126]. It is likely that Rheb controls FKBP38 allosterically in a GTP-dependent manner, resulting in the release of mTOR from its inhibition.
The binding of Rheb with FKBP38 in cells is regulated by growth factor and amino acid conditions. In cells starved for serum or amino acids, Rheb exhibits a weak binding with FKBP38, which is strongly stimulated upon re-addition of the omitted ingredients. Accompanied with the increased binding of Rheb with FKBP38 is a reduction in the association of FKBP38 with mTOR. These observations suggest that growth factors and amino acids control the association of FKBP38 with mTOR through Rheb, presumably by modulating its nucleotide-binding states [126]. The same observations also support a signaling model in which Rheb activates mTORC1 by antagonizing the inhibitory effect of FKBP38 on mTOR (Fig. 4). Consistent with this scheme, it has been shown that addition of recombinant GTP-loaded Rheb blocks the inhibitory effect of FKBP38 on mTORC1 activity in vitro [128].
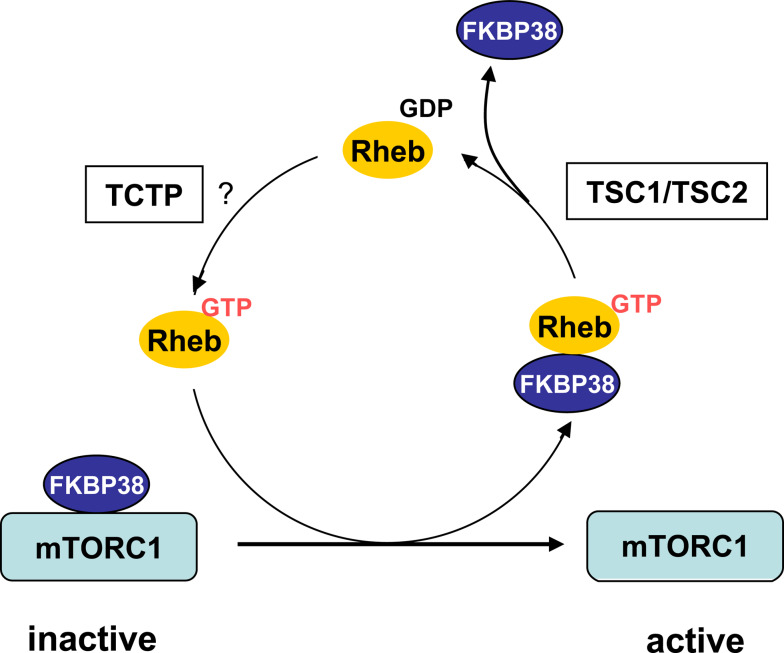
Fig. 4.
A model for the role of FKBP38 in mTORC1 regulation. FKBP38 inhibits mTOR activity through direct binding to mTOR. In GTP-bound form, Rheb interacts with FKBP38 and liberates mTORC1 for activation. The action of Rheb is terminated by the TSC1/TSC2 complex, which promotes the hydrolysis of bounded GTP to GDP. Reloading of Rheb with GTP by a putative guanine exchange factor repeats the cycle
Although the model provides a logical explanation for the role of Rheb and FKBP38 in mTOR regulation, controversies exist. One major concern is that the inhibitory effect of FKBP38 overexpression on mTORC1 activity has been found to be modest [114, 127, 130], which has led to the suggestion that FKBP38 is not the sole mediator of Rheb in mTORC1 regulation and that Rheb may control mTORC1 through both FKBP38-dependent and independent mechanisms [131].
TCTP
Activity of the Ras family of small GTPases is normally regulated by their cognate GAPs and nucleotide exchange factors (GEFs) [102]. With the identification of the TSC1/TSC2 complex as the GAP for Rheb, the search for its GEF has drawn extensive attention. Recently, TCTP, a highly conserved protein in eukaryotes, has been suggested to be the long-sought GEF for Rheb [106]. TCTP, also called histamine-releasing factor (HRF), is a key protein in the process of tumor reversion and well conserved through phylogeny as a pro-survival, growth-stimulating and anti-apoptotic factor [132]. Hsu et al. [106] showed that in Drosophila, dTCTP acts upstream of dS6K, and down-regulation of dTCTP reduced cell size, cell number and organ size. Importantly, they found that dTCTP displayed a guanine nucleotide exchange activity toward Rheb in cells and in vitro. Consistent with these findings, Dong et al. [107] demonstrated that human TCTP accelerated its GDP release in vitro and activated the mTORC1 pathway in cells. However, the identity of TCTP as Rheb GEF has been challenged by several other studies in mammalian cells showing that TCTP is unable to interact with Rheb in vitro and is largely ineffective in regulating S6K phosphorylation in cells [130, 133, 134]. The reason for these discrepancies is not clear. Further independent studies are needed to clarify the role of TCTP in controlling mTORC1 signaling.
PRAS40
PRAS40 was originally identified as a proline-rich Akt substrate that is capable of binding to 14-3-3 upon phosphorylation by Akt [135]. It was later recognized as a component of the mTORC1 complex by mass spectrometry analysis of proteins associated with mTOR [136]. PRAS40 has been shown to associate with raptor and inhibit mTORC1 kinase activity both in vitro and in cells [58, 113, 137, 138]. Furthermore, the association of PRAS40 with mTORC1 increases under conditions that inhibit mTOR signaling, such as nutrient or serum deprivation or mitochondrial metabolic inhibition [136]. These findings suggest that PRAS40 is an inhibitor of mTORC1. Intriguingly, while overexpression of PRAS40 inhibits mTORC1 activity, knocking down the expression of PRAS40 also reduces the insulin-stimulated S6K phosphorylation. These findings suggest that the role of PRAS40 in mTORC1 function is not simply inhibitory [136, 139].
PRAS40 contains a TOS motif that is involved in binding with raptor. This motif is commonly found in mTORC1 substrates, such as S6K and 4E-BP1, indicating that PRAS40 is a substrate of mTORC1. Hence, it has been suggested that PRAS40 inhibits mTORC1-directed phosphorylation of S6K and 4E-BP1 by competing with these proteins for raptor binding [137, 138]. PRAS40 is phosphorylated by mTORC1 at several sites, including Ser183, Ser212 and Ser221 [138–140]. Phosphorylation at Ser183 and Ser221 appears to reduce the association of PRAS40 with raptor and relieve its inhibition on mTORC1 [138, 139], which may constitute a positive feed-forward mechanism for mTORC1 activation, such that a suboptimal mTORC1 signaling activity potentiates its further activation by reducing PRAS40 association.
PRAS40 is also phosphorylated at Thr246 by Akt in response to insulin stimulation. The Akt-directed phosphorylation creates a docking site for 14-3-3, which binds to PRAS40 and interferes with its association with mTORC1 [113, 137]. This Akt-PRAS40 connection counts for the second mechanism, in addition to Akt-TSC2-Rheb signaling axis, whereby Akt activates mTORC1 and explains why in Drosophila Akt is able to maintain a normal development in the absence of TSC2 [141]. In addition, Thr246 has also been found to be phosphorylated by PIM1, a serine/threonine kinase that has diverse biological roles in cell survival, proliferation and differentiation [142]. Like the Akt-directed phosphorylation, the phosphorylation by PIM1 also reduces the association of PRAS40 with mTORC1 and increases the mTORC1-directed phosphorylation of 4E-BP1 and S6K [143].
IKK
Inhibitor of nuclear factor κB (NFκB) kinase (IKK) complex is a key component in the tumor necrosis factor α (TNFα)/NF-κB signaling pathway that is involved in systemic inflammation [144]. It consists of three tightly associated subunits, IKKα, IKKβ and IKKγ, of which IKKα and IKKβ serve as the catalytic subunits and IKKγ is the regulatory subunit [145]. IKK is activated by proinflammatory cytokines, such as TNFα and lipopolysaccharide (LPS) [146]. In addition to activation of IKK, TNFα has been known to stimulate mTORC1 activity [147]. However, the underlying mechanism remained unclear until a recent finding that IKKβ associates with TSC1 [148]. Lee et al. [148] have found that in response to TNFα treatment, IKKβ interacts with TSC1 and phosphorylates it at Ser487 and Ser511. The phosphorylation represses the function of the TSC1/TSC2 complex, resulting in mTORC1 activation. In addition, the IKK-directed phosphorylation has been found to correlate with TNFα-induced VEGF production in multiple tumor types, indicating that TNFα promotes VEGF expression and angiogenesis through IKK-dependent mTORC1 activation.
Moreover, studies from Baldwin’s [149, 150] group have revealed an association between IKKα and mTORC1 in PTEN-deficient cancer cells. The association is regulated by Akt and is required for Akt-stimulated mTORC1 activation. Overexpression of IKKα has been found to enhance mTORC1 kinase activity in vitro, suggesting that IKKα is a positive regulator of mTORC1. It has thus been proposed that IKKα may activate mTORC1 by phosphorylating mTOR or components in mTORC1.
Phosphatidic acid and phospholipase D
Phosphatidic acid is a phospholipid metabolite that serves as a second messenger in mitogenic signaling pathways [151–153]. An elevated PA level in cells, either by serum stimulation or treatment with exogenous PA, results in an increase in mTORC1 activity. In contrast, inhibition of PA accumulation by treatment with 1-butanol represses mTORC1 activity [154]. The action of PA on mTORC1 is mediated through a direct binding of PA to mTOR to a region within the FKBP12-rapamcyin binding (FRB) domain. Residue Arg2109 in this domain has been found to be critical for the binding. Mutations at this site reduce the interaction of PA with mTOR and repress mTORC1 signaling [154]. How binding of PA leads to mTORC1 activation is not fully understood. Direct activation of mTORC1 kinase activity has not been observed. However, two mechanisms have been suggested that may mediate the effect of PA on mTORC1. Sun et al. recently reported that PA was able to reduce the association of FKBP38 with mTOR both in vitro and in cells, which suggests that PA may activate mTORC1 by alleviating the inhibitory of FKBP38 [131, 155]. On the other hand, Toschi et al. [156] showed that suppression of PA production blocked the association of mTOR with raptor, suggesting that PA binding is required for the integrity of mTORC1. These two mechanisms are not necessarily mutually exclusive. Given the role of FKBP38 in mTORC1 regulation, it is possible that an enhanced binding of FKBP38 with mTOR upon PA depletion alters the association of mTOR with raptor.
Mitogen and growth factor stimulate PA production by activating phospholipase D (PLD), which catalyzes the hydrolysis of membrane phosphatidylcholine to produce PA and choline [152]. In mammalian cells, two isoforms of PLD, PLD1 and PLD2, have been identified. Consistent with their role in PA production, overexpression of either protein has been shown to activate mTORC1 activity and knockdown of their expression to reduce mitogen-stimulated mTORC1 activity [32, 157–160]. The activity of PLD1 in cells is regulated by protein kinase C and several small GTPases, including Arf, Cdc42, Rac1 and RhoA [161]. Cdc42 and RhoA have been shown to interact directly with PLD1 and stimulate its activity. Interestingly, while overexpression of Cdc42 increases mTORC1 activity, overexpression of RhoA does not [162]. The reason for this dilemma is not clear. However, we have recently observed that RhoA is able to associate with mTORC1 and downregulates its kinase activity (Y. Lai and Y. Jiang, unpublished observation), suggesting that RhoA, in addition to stimulating PA production through PLD1 activation, may have a negative effect that directly acts on mTORC1. In addition to the Rho family of small GTPases, PLD1 has been found to be activated by Rheb. Sun et al. [160] have shown that Rheb binds to PLD1 in a GTP-dependent manner and stimulates its activity. Downregulation of Rheb, either by siRNA knockdown or TSC2 overexpression, reduces mitogen-stimulated PLD1 activation. Importantly, downregulation of PLD1 or inhibition of PA production blocks Rheb-stimulated mTORC1 activation, suggesting that the activation effect of Rheb on mTORC1 requires PLD1-dependent PA production [160]. In addition to PA production, PLD2 may also regulate mTORC1 through physical interaction. PLD2 has been found to interact with raptor, and the interaction appears to be important for PLD2-stimulated mTORC1 activation [158].
The Rag small GTPases
The Rag small GTPases are a group of unique small GTPases remotely related to Ras [163, 164]. Four members of this type of GTPase have been identified in mammalian cells, including RagA, RagB, RagC and RagD [164, 165]. RagA and RagB are closely related. RagC and RagD differ significantly from RagA and RagB, but share high similarity with each other. The Rag small GTPases function as heterodimers containing one subunit from either RagA or RagB and the other from either RagC or RagD [165–167]. Like other small GTPases, the activity of the Rag proteins is regulated by their nucleotide-binding states. However, a functional configuration of the dimer requires a GTP-bound RagA or RagB in complex with a GDP-bound RagC or RagD [168]. The activity of the complex is believed to be determined mainly by the GTP-bound subunit, but its stability requires the other subunit in GDP-bound state [169].
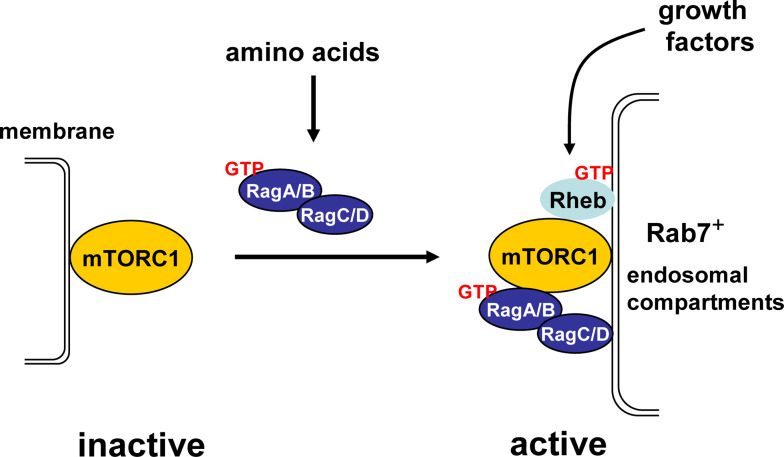
In mammalian cells, the Rag small GTPases have recently been found to associate with mTORC1 as heterodimers [168]. It has been found that GTP-bound RagA or RagB (RagA/BGTP), when complex with GDP-bound RagC or RagD (RagC/DGDP), associates with mTORC1, likely through a direct binding to raptor. The association is regulated by amino acid conditions; re-addition of amino acids to amino acid-starved cells stimulates the association. Since amino acids also stimulate the GTP-loading of RagA/B, it is likely that amino acids promote the association by controlling the GTP-loading of RagA/B. Importantly, in cells expressing the RagA/BGTP–RagC/DGDP heterodimer, the mTORC1 activity becomes insensitive to deprivation of amino acids, but remains responsive to other conditions known to regulate mTORC1, including oxidative stress, mitochondrial inhibition, serum and energy deprivation [168]. This observation suggests that the Rag GTPases mediates mTORC1 activity in response exclusively to amino acid availability and that expression of RagA/BGTP mimics amino acids sufficiency. However, despite the ability to activate mTORC1 in cells, the Rag heterodimers are ineffective in stimulating mTORC1 kinase activity in vitro, suggesting that the GTPases controls mTORC1 activity by means other than increasing its kinase activity [168].
In amino acid-starved cells, mTOR is distributed throughout cytoplasm on punctate membrane structures resembling endosomal compartments [168]. Upon restimulation with amino acids, mTOR, as well as raptor, accumulate to late endosomal or lysosomal compartments that are enriched with Rab7. This relocation appears to be dependent on the Rag small GTPases, since it does not occur in cells when expression of the Rag proteins is downregulated by siRNA [168]. Interestingly, it has been previously shown that the Rab7-enriched compartments are also the place where Rheb resides [170, 171]. This colocalization thus promotes a model which explains the role of amino acids and Rag proteins in mTORC1 regulation. In this model, amino acids stimulate the activity of the Rag GTPases, which in turn binds to mTORC1 and recruits it to late endosomal and lysosomal compartments, where it is activated by Rheb (Fig. 5). Accordingly, it can be envisaged that the role of growth factors in mTORC1 activation is to activate Rheb and that of amino acids is to present mTORC1 to an activated Rheb. This explains why mTORC1 cannot be activated in the absence of either amino acids or growth factors.
Fig. 5.
A model for the role of the Rag GTPases in mTORC1 regulation. In cells starved for amino acids, mTORC1 resides on membrane structures distributed throughout the cytoplasm. Re-stimulation with amino acids activates the heterodimers of the Rag small GTPases, which causes mTORC1 redistribution to Rab7-enriched endosomal compartments, where it is activated by Rheb
hVps34 and calcium
Vps34 was identified in yeast as an unconventional (Class III) PI3-K that converts phosphatidylinositol to phosphatidylinositol 3-phosphate (PI3P), a bioactive messenger [172]. It is structurally highly similar to the catalytic subunit of classic (Class I) PI3-K [the 110-kilodalton (p110) subunit of PI 3-kinase] [173]. In yeast, Vps34 is involved in protein sorting to the yeast lysosome-like vacuole [174].
Nobukuni et al. [111] observed that wortmanin, a potent inhibitor of PI3-K, prevented amino acid-stimulated mTORC1 activation. However, downregulation of class I PI3-K by siRNA, while blocks insulin-induced mTORC1 activation, had no effect on amino acid-stimulated mTORC1 activation. This observation suggests that the effect of wortmanin on amino acid-induced mTORC1 activation is mediated by another PI3-K. In support of this notion, it was found that addition of amino acids to amino acid-starved cells induced a sharp increase in the intracellular levels of PI3P, which coincided with an increase in the kinase activity of human Vps34 (hVps34), supporting a role for hVps34 in amino acid-mediated mTORC1 activation [111]. Consistent with this notion, overexpression of hVps34 has been shown to activate mTORC1, while downregulation of hVps34, either by siRNA or antibodies specifically against hVps34, to reduce amino acid-induced mTORC1 activation. In addition, siRNA knockdown of hVps15, a protein kinase that activates hVps34, also inhibits amino acid-induced mTORC1 activation [111, 175]. These studies demonstrate that hVpd34 mediates the effect of amino acids for mTORC1 activation.
How does hVps34 activate mTORC1 in response to amino acid stimulation? Gulati et al. [176] showed that amino acid-stimulated mTORC1 activation was accompanied by a rapid rise in intracellular [Ca2+]. Treating cells with the endoplasmic reticulum Ca2+-ATPase inhibitor thapsigargin (Tg), which increased [Ca2+] in cytosol, was able to activate mTORC1 in the absence of amino acids. Conversely, depletion of Ca2+ suppressed amino acid-induced mTORC1 activation. These results suggest that a rise in [Ca2+] is required and sufficient for mTORC1 activation. Interestingly, the Ca2+-induced mTORC1 activation is attenuated when hVp34 expression levels are reduced by siRNA knockdown, suggesting that Ca2+ mediated mTORC1 activation requires hVps34 [176]. Since hVps34 contains a putative Ca2+/CaM binding domain, it is possible that Ca2+ and CaM may bind to hVps34 and regulate its activity. Indeed, Gulati et al. [176] showed that CaM bound directly to hVps34 in the presence of Ca2+, and the binding of CaM was required for the lipid kinase activity of hVps34. Collectively, these results support a model whereby amino acids stimulate mTORC1 activity through Ca2+-dependent activation of hVps34, which results in an increase in PI3P in cells. However, it remains to be determined how PI3P activates mTORC1. In this regard, it is interesting to note that hVps34 is co-localized with Rab7 on late endosomal compartments, where the Rag small GTPases bring mTORC1 for activation by Rheb [168, 177]. A potential connection among PI3P, Rheb and Rag GTPases is thus worth exploring.
Conclusion
Recent advances in mTOR study have uncovered a complex regulatory network that governs mTOR activity. As discussed above, mTOR is regulated at multiple levels, including direct phosphorylation, oligomerization and interactions with its regulators, which in turn are controlled by an array of signaling cascades that are responsive to changes in intracellular and environmental conditions. Such a complex regulation network ensures a fine tuning of mTOR activity in response to numerous conditions and fits the pivotal role of mTOR in cell growth control.
Despite these advances, one important level of regulation remains largely unexplored, that is, the spatial control. mTOR has been found to associate with various intracellular compartments, including endosomal membranes, the Golgi complex, mitochondria and shuttling between cytoplasm and nucleus [67, 178–182]. This wide distribution pattern of mTOR suggests that localization is likely to play a critical role in mTOR function. This notion is highlighted by the recent finding that the Rag small GTPases control mTORC1 by targeting it to late endosomal compartments for activation by Rheb [168]. Other issues pertaining to spatial control of mTOR include how mTOR function is affected by its localization and whether mTOR localized at various places is regulated differently. In this regard, the use of S6K and 4E-BP1 phosphorylation as a surrogate for mTORC1 activity carries a risk of overlooking regulatory mechanisms that control mTORC1 for functions other than translation. In future studies, incorporation of spatial control in mTOR regulation may be the key for answering the fundamental question in mTOR study, that is, how does mTOR integrate signals of different origins to control cell growth?
Acknowledgments
The authors thank their colleagues for stimulating discussion and critical reading of this manuscript. This work was supported by NIH grants CA129821 and GM068832 to YJ.
Contributor Information
Xiaochun Bai, Phone: +86-20-61648207, Email: xiaochunbai@yahoo.com.cn.
Yu Jiang, Phone: +1-412-6483390, FAX: +1-412-6481945, Email: yuj5@pitt.edu.
References
- 1.De Virgilio C, Loewith R. Cell growth control: little eukaryotes make big contributions. Oncogene. 2006;25:6392–6415. doi: 10.1038/sj.onc.1209884. [DOI] [PubMed] [Google Scholar]
- 2.Jacinto E, Hall MN. Tor signalling in bugs, brain and brawn. Nat Rev Mol Cell Biol. 2003;4:117–126. doi: 10.1038/nrm1018. [DOI] [PubMed] [Google Scholar]
- 3.Heitman J, Movva NR, Hiestand PC, Hall MN. FK 506-binding protein proline rotamase is a target for the immunosuppressive agent FK 506 in Saccharomyces cerevisiae . Proc Natl Acad Sci USA. 1991;88:1948–1952. doi: 10.1073/pnas.88.5.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koltin Y, Faucette L, Bergsma DJ, Levy MA, Cafferkey R, Koser PL, Johnson RK, Livi GP. Rapamycin sensitivity in Saccharomyces cerevisiae is mediated by a peptidyl-prolyl cis–trans isomerase related to human FK506-binding protein. Mol Cell Biol. 1991;11:1718–1723. doi: 10.1128/mcb.11.3.1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 6.Huang S, Bjornsti MA, Houghton PJ. Rapamycins: mechanism of action and cellular resistance. Cancer Biol Ther. 2003;2:222–232. doi: 10.4161/cbt.2.3.360. [DOI] [PubMed] [Google Scholar]
- 7.Easton JB, Houghton PJ. Therapeutic potential of target of rapamycin inhibitors. Expert Opin Ther Targets. 2004;8:551–564. doi: 10.1517/14728222.8.6.551. [DOI] [PubMed] [Google Scholar]
- 8.Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P, Hall MN. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell. 2002;10:457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 9.Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, Hall A, Hall MN. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol. 2004;6:1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- 10.Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 11.Bhaskar PT, Hay N. The two TORCs and Akt. Dev Cell. 2007;12:487–502. doi: 10.1016/j.devcel.2007.03.020. [DOI] [PubMed] [Google Scholar]
- 12.Keith CT, Schreiber SL. PIK-related kinases: DNA repair, recombination, and cell cycle checkpoints. Science. 1995;270:50–51. doi: 10.1126/science.270.5233.50. [DOI] [PubMed] [Google Scholar]
- 13.Andrade MA, Bork P. HEAT repeats in the Huntington’s disease protein. Nat Genet. 1995;11:115–116. doi: 10.1038/ng1095-115. [DOI] [PubMed] [Google Scholar]
- 14.Bosotti R, Isacchi A, Sonnhammer EL. FAT: a novel domain in PIK-related kinases. Trends Biochem Sci. 2000;25:225–227. doi: 10.1016/s0968-0004(00)01563-2. [DOI] [PubMed] [Google Scholar]
- 15.Dames SA, Mulet JM, Rathgeb-Szabo K, Hall MN, Grzesiek S. The solution structure of the FATC domain of the protein kinase target of rapamycin suggests a role for redox-dependent structural and cellular stability. J Biol Chem. 2005;280:20558–20564. doi: 10.1074/jbc.M501116200. [DOI] [PubMed] [Google Scholar]
- 16.Chen J, Zheng XF, Brown EJ, Schreiber SL. Identification of an 11-kDa FKBP12-rapamycin-binding domain within the 289-kDa FKBP12-rapamycin-associated protein and characterization of a critical serine residue. Proc Natl Acad Sci USA. 1995;92:4947–4951. doi: 10.1073/pnas.92.11.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiang GG, Abraham RT. Phosphorylation of mammalian target of rapamycin (mTOR) at Ser-2448 is mediated by p70S6 kinase. J Biol Chem. 2005;280:25485–25490. doi: 10.1074/jbc.M501707200. [DOI] [PubMed] [Google Scholar]
- 18.Holz MK, Blenis J. Identification of S6 kinase 1 as a novel mammalian target of rapamycin (mTOR)-phosphorylating kinase. J Biol Chem. 2005;280:26089–26093. doi: 10.1074/jbc.M504045200. [DOI] [PubMed] [Google Scholar]
- 19.Copp J, Manning G, Hunter T. TORC-specific phosphorylation of mammalian target of rapamycin (mTOR): phospho-Ser2481 is a marker for intact mTOR signaling complex 2. Cancer Res. 2009;69:1821–1827. doi: 10.1158/0008-5472.CAN-08-3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peterson RT, Beal PA, Comb MJ, Schreiber SL. FKBP12-rapamycin-associated protein (FRAP) autophosphorylates at serine 2481 under translationally repressive conditions. J Biol Chem. 2000;275:7416–7423. doi: 10.1074/jbc.275.10.7416. [DOI] [PubMed] [Google Scholar]
- 21.Cheng SW, Fryer LG, Carling D, Shepherd PR. Thr2446 is a novel mammalian target of rapamycin (mTOR) phosphorylation site regulated by nutrient status. J Biol Chem. 2004;279:15719–15722. doi: 10.1074/jbc.C300534200. [DOI] [PubMed] [Google Scholar]
- 22.Acosta-Jaquez HA, Keller JA, Foster KG, Ekim B, Soliman GA, Feener EP, Ballif BA, Fingar DC. Site-specific mTOR phosphorylation promotes mTORC1-mediated signaling and cell growth. Mol Cell Biol. 2009;29:4308–4324. doi: 10.1128/MCB.01665-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosner M, Siegel N, Valli A, Fuchs C, Hengstschlager M (2009) mTOR phosphorylated at S2448 binds to raptor and rictor. Amino Acids [Epub ahead of print] [DOI] [PubMed]
- 24.Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 25.Kim DH, Sarbassov DD, Ali SM, Latek RR, Guntur KV, Erdjument-Bromage H, Tempst P, Sabatini DM. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol Cell. 2003;11:895–904. doi: 10.1016/s1097-2765(03)00114-x. [DOI] [PubMed] [Google Scholar]
- 26.Smith TF, Gaitatzes C, Saxena K, Neer EJ. The WD repeat: a common architecture for diverse functions. Trends Biochem Sci. 1999;24:181–185. doi: 10.1016/s0968-0004(99)01384-5. [DOI] [PubMed] [Google Scholar]
- 27.Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, Brown M, Fitzgerald KJ, Sabatini DM. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 28.Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, Tokunaga C, Avruch J, Yonezawa K. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–189. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- 29.Adami A, Garcia-Alvarez B, Arias-Palomo E, Barford D, Llorca O. Structure of TOR and its complex with KOG1. Mol Cell. 2007;27:509–516. doi: 10.1016/j.molcel.2007.05.040. [DOI] [PubMed] [Google Scholar]
- 30.Nojima H, Tokunaga C, Eguchi S, Oshiro N, Hidayat S, Yoshino K, Hara K, Tanaka N, Avruch J, Yonezawa K. The mammalian target of rapamycin (mTOR) partner, raptor, binds the mTOR substrates p70 S6 kinase and 4E-BP1 through their TOR signaling (TOS) motif. J Biol Chem. 2003;278:15461–15464. doi: 10.1074/jbc.C200665200. [DOI] [PubMed] [Google Scholar]
- 31.Schalm SS, Fingar DC, Sabatini DM, Blenis J. TOS motif-mediated raptor binding regulates 4E-BP1 multisite phosphorylation and function. Curr Biol. 2003;13:797–806. doi: 10.1016/s0960-9822(03)00329-4. [DOI] [PubMed] [Google Scholar]
- 32.Hornberger TA, Chu WK, Mak YW, Hsiung JW, Huang SA, Chien S. The role of phospholipase D and phosphatidic acid in the mechanical activation of mTOR signaling in skeletal muscle. Proc Natl Acad Sci USA. 2006;103:4741–4746. doi: 10.1073/pnas.0600678103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee VH, Healy T, Fonseca BD, Hayashi A, Proud CG. Analysis of the regulatory motifs in eukaryotic initiation factor 4E-binding protein 1. Febs J. 2008;275:2185–2199. doi: 10.1111/j.1742-4658.2008.06372.x. [DOI] [PubMed] [Google Scholar]
- 34.Tzatsos A. Raptor binds the SAIN (Shc and IRS-1 NPXY-binding) domain of IRS-1 and regulates the phosphorylation of IRS-1 at Ser636/639 by mTOR. J Biol Chem. 2009;284(34):22525–22534. doi: 10.1074/jbc.M109.027748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carriere A, Cargnello M, Julien LA, Gao H, Bonneil E, Thibault P, Roux PP. Oncogenic MAPK signaling stimulates mTORC1 activity by promoting RSK-mediated raptor phosphorylation. Curr Biol. 2008;18:1269–1277. doi: 10.1016/j.cub.2008.07.078. [DOI] [PubMed] [Google Scholar]
- 37.Wang L, Lawrence JC, Jr, Sturgill TW, Harris TE. Mammalian target of rapamycin complex 1 (mTORC1) activity is associated with phosphorylation of raptor by mTOR. J Biol Chem. 2009;284:14693–14697. doi: 10.1074/jbc.C109.002907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 39.Brunn GJ, Hudson CC, Sekulic A, Williams JM, Hosoi H, Houghton PJ, Lawrence JC, Jr, Abraham RT. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science. 1997;277:99–101. doi: 10.1126/science.277.5322.99. [DOI] [PubMed] [Google Scholar]
- 40.Burnett PE, Barrow RK, Cohen NA, Snyder SH, Sabatini DM. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc Natl Acad Sci USA. 1998;95:1432–1437. doi: 10.1073/pnas.95.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, Aebersold R, Sonenberg N. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 1999;13:1422–1437. doi: 10.1101/gad.13.11.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 43.Pearson RB, Dennis PB, Han JW, Williamson NA, Kozma SC, Wettenhall RE, Thomas G. The principal target of rapamycin-induced p70s6k inactivation is a novel phosphorylation site within a conserved hydrophobic domain. EMBO J. 1995;14:5279–5287. doi: 10.1002/j.1460-2075.1995.tb00212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brown EJ, Beal PA, Keith CT, Chen J, Shin TB, Schreiber SL. Control of p70 s6 kinase by kinase activity of FRAP in vivo. Nature. 1995;377:441–446. doi: 10.1038/377441a0. [DOI] [PubMed] [Google Scholar]
- 45.Pullen N, Dennis PB, Andjelkovic M, Dufner A, Kozma SC, Hemmings BA, Thomas G. Phosphorylation and activation of p70s6k by PDK1. Science. 1998;279:707–710. doi: 10.1126/science.279.5351.707. [DOI] [PubMed] [Google Scholar]
- 46.Alessi DR, Kozlowski MT, Weng QP, Morrice N, Avruch J. 3-Phosphoinositide-dependent protein kinase 1 (PDK1) phosphorylates and activates the p70 S6 kinase in vivo and in vitro. Curr Biol. 1998;8:69–81. doi: 10.1016/s0960-9822(98)70037-5. [DOI] [PubMed] [Google Scholar]
- 47.Fingar DC, Salama S, Tsou C, Harlow E, Blenis J. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 2002;16:1472–1487. doi: 10.1101/gad.995802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harada H, Andersen JS, Mann M, Terada N, Korsmeyer SJ. p70S6 kinase signals cell survival as well as growth, inactivating the pro-apoptotic molecule BAD. Proc Natl Acad Sci USA. 2001;98:9666–9670. doi: 10.1073/pnas.171301998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tremblay F, Marette A. Amino acid and insulin signaling via the mTOR/p70 S6 kinase pathway. A negative feedback mechanism leading to insulin resistance in skeletal muscle cells. J Biol Chem. 2001;276:38052–38060. doi: 10.1074/jbc.M106703200. [DOI] [PubMed] [Google Scholar]
- 50.Harrington LS, Findlay GM, Gray A, Tolkacheva T, Wigfield S, Rebholz H, Barnett J, Leslie NR, Cheng S, Shepherd PR, Gout I, Downes CP, Lamb RF. The TSC1-2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. J Cell Biol. 2004;166:213–223. doi: 10.1083/jcb.200403069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang X, Li W, Williams M, Terada N, Alessi DR, Proud CG. Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase. EMBO J. 2001;20:4370–4379. doi: 10.1093/emboj/20.16.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang HH, Lipovsky AI, Dibble CC, Sahin M, Manning BD. S6K1 regulates GSK3 under conditions of mTOR-dependent feedback inhibition of Akt. Mol Cell. 2006;24:185–197. doi: 10.1016/j.molcel.2006.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun XJ, Rothenberg P, Kahn CR, Backer JM, Araki E, Wilden PA, Cahill DA, Goldstein BJ, White MF. Structure of the insulin receptor substrate IRS-1 defines a unique signal transduction protein. Nature. 1991;352:73–77. doi: 10.1038/352073a0. [DOI] [PubMed] [Google Scholar]
- 54.Craparo A, Freund R, Gustafson TA. 14-3-3 (epsilon) interacts with the insulin-like growth factor I receptor and insulin receptor substrate I in a phosphoserine-dependent manner. J Biol Chem. 1997;272:11663–11669. doi: 10.1074/jbc.272.17.11663. [DOI] [PubMed] [Google Scholar]
- 55.Jacinto E, Facchinetti V, Liu D, Soto N, Wei S, Jung SY, Huang Q, Qin J, Su B. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 2006;127:125–137. doi: 10.1016/j.cell.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 56.Pearce LR, Huang X, Boudeau J, Pawlowski R, Wullschleger S, Deak M, Ibrahim AF, Gourlay R, Magnuson MA, Alessi DR. Identification of protor as a novel rictor-binding component of mTOR complex-2. Biochem J. 2007;405:513–522. doi: 10.1042/BJ20070540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 58.Thedieck K, Polak P, Kim ML, Molle KD, Cohen A, Jeno P, Arrieumerlou C, Hall MN. PRAS40 and PRR5-like protein are new mTOR interactors that regulate apoptosis. PLoS One. 2007;2:e1217. doi: 10.1371/journal.pone.0001217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Woo SY, Kim DH, Jun CB, Kim YM, Haar EV, Lee SI, Hegg JW, Bandhakavi S, Griffin TJ, Kim DH. PRR5, a novel component of mTOR complex 2, regulates platelet-derived growth factor receptor beta expression and signaling. J Biol Chem. 2007;282:25604–25612. doi: 10.1074/jbc.M704343200. [DOI] [PubMed] [Google Scholar]
- 60.Yang Q, Inoki K, Ikenoue T, Guan KL. Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes Dev. 2006;20:2820–2832. doi: 10.1101/gad.1461206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 62.Hagan GN, Lin Y, Magnuson MA, Avruch J, Czech MP. A rictor-myo1c complex participates in dynamic cortical actin events in 3T3-L1 adipocytes. Mol Cell Biol. 2008;28:4215–4226. doi: 10.1128/MCB.00867-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martin J, Masri J, Bernath A, Nishimura RN, Gera J. Hsp70 associates with rictor and is required for mTORC2 formation and activity. Biochem Biophys Res Commun. 2008;372:578–583. doi: 10.1016/j.bbrc.2008.05.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McDonald PC, Oloumi A, Mills J, Dobreva I, Maidan M, Gray V, Wederell ED, Bally MB, Foster LJ, Dedhar S. Rictor and integrin-linked kinase interact and regulate Akt phosphorylation and cancer cell survival. Cancer Res. 2008;68:1618–1624. doi: 10.1158/0008-5472.CAN-07-5869. [DOI] [PubMed] [Google Scholar]
- 65.Zeng Z, Sarbassov dos D, Samudio IJ, Yee KW, Munsell MF, Ellen Jackson C, Giles FJ, Sabatini DM, Andreeff M, Konopleva M. Rapamycin derivatives reduce mTORC2 signaling and inhibit AKT activation in AML. Blood. 2007;109:3509–3512. doi: 10.1182/blood-2006-06-030833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Akcakanat A, Singh G, Hung MC, Meric-Bernstam F. Rapamycin regulates the phosphorylation of rictor. Biochem Biophys Res Commun. 2007;362:330–333. doi: 10.1016/j.bbrc.2007.07.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rosner M, Hengstschlager M. Cytoplasmic and nuclear distribution of the protein complexes mTORC1 and mTORC2: rapamycin triggers dephosphorylation and delocalization of the mTORC2 components rictor and sin1. Hum Mol Genet. 2008;17:2934–2948. doi: 10.1093/hmg/ddn192. [DOI] [PubMed] [Google Scholar]
- 68.Facchinetti V, Ouyang W, Wei H, Soto N, Lazorchak A, Gould C, Lowry C, Newton AC, Mao Y, Miao RQ, Sessa WC, Qin J, Zhang P, Su B, Jacinto E. The mammalian target of rapamycin complex 2 controls folding and stability of Akt and protein kinase C. EMBO J. 2008;27:1932–1943. doi: 10.1038/emboj.2008.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ikenoue T, Inoki K, Yang Q, Zhou X, Guan KL. Essential function of TORC2 in PKC and Akt turn motif phosphorylation, maturation and signalling. EMBO J. 2008;27:1919–1931. doi: 10.1038/emboj.2008.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Garcia-Martinez JM, Alessi DR. mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1) Biochem J. 2008;416:375–385. doi: 10.1042/BJ20081668. [DOI] [PubMed] [Google Scholar]
- 71.Rosner M, Fuchs C, Siegel N, Valli A, Hengstschlager M. Functional interaction of mTOR complexes in regulating mammalian cell size and cell cycle. Hum Mol Genet. 2009;18(17):3298–3310. doi: 10.1093/hmg/ddp271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Leung AK, Robson WL. Tuberous sclerosis complex: a review. J Pediatr Health Care. 2007;21:108–114. doi: 10.1016/j.pedhc.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 73.Mak BC, Yeung RS. The tuberous sclerosis complex genes in tumor development. Cancer Invest. 2004;22:588–603. doi: 10.1081/cnv-200027144. [DOI] [PubMed] [Google Scholar]
- 74.Plank TL, Yeung RS, Henske EP. Hamartin, the product of the tuberous sclerosis 1 (TSC1) gene, interacts with tuberin and appears to be localized to cytoplasmic vesicles. Cancer Res. 1998;58:4766–4770. [PubMed] [Google Scholar]
- 75.van Slegtenhorst M, Nellist M, Nagelkerken B, Cheadle J, Snell R, van den Ouweland A, Reuser A, Sampson J, Halley D, van der Sluijs P. Interaction between hamartin and tuberin, the TSC1 and TSC2 gene products. Hum Mol Genet. 1998;7:1053–1057. doi: 10.1093/hmg/7.6.1053. [DOI] [PubMed] [Google Scholar]
- 76.Wienecke R, Konig A, DeClue JE. Identification of tuberin, the tuberous sclerosis-2 product. Tuberin possesses specific Rap1GAP activity. J Biol Chem. 1995;270:16409–16414. doi: 10.1074/jbc.270.27.16409. [DOI] [PubMed] [Google Scholar]
- 77.Tapon N, Ito N, Dickson BJ, Treisman JE, Hariharan IK. The Drosophila tuberous sclerosis complex gene homologs restrict cell growth and cell proliferation. Cell. 2001;105:345–355. doi: 10.1016/s0092-8674(01)00332-4. [DOI] [PubMed] [Google Scholar]
- 78.Gao X, Pan D. TSC1 and TSC2 tumor suppressors antagonize insulin signaling in cell growth. Genes Dev. 2001;15:1383–1392. doi: 10.1101/gad.901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Potter CJ, Pedraza LG, Xu T. Akt regulates growth by directly phosphorylating Tsc2. Nat Cell Biol. 2002;4:658–665. doi: 10.1038/ncb840. [DOI] [PubMed] [Google Scholar]
- 80.Saucedo LJ, Gao X, Chiarelli DA, Li L, Pan D, Edgar BA. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat Cell Biol. 2003;5:566–571. doi: 10.1038/ncb996. [DOI] [PubMed] [Google Scholar]
- 81.Stocker H, Radimerski T, Schindelholz B, Wittwer F, Belawat P, Daram P, Breuer S, Thomas G, Hafen E. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat Cell Biol. 2003;5:559–565. doi: 10.1038/ncb995. [DOI] [PubMed] [Google Scholar]
- 82.Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang Y, Gao X, Saucedo LJ, Ru B, Edgar BA, Pan D. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat Cell Biol. 2003;5:578–581. doi: 10.1038/ncb999. [DOI] [PubMed] [Google Scholar]
- 84.Castro AF, Rebhun JF, Clark GJ, Quilliam LA. Rheb binds tuberous sclerosis complex 2 (TSC2) and promotes S6 kinase activation in a rapamycin- and farnesylation-dependent manner. J Biol Chem. 2003;278:32493–32496. doi: 10.1074/jbc.C300226200. [DOI] [PubMed] [Google Scholar]
- 85.Garami A, Zwartkruis FJ, Nobukuni T, Joaquin M, Roccio M, Stocker H, Kozma SC, Hafen E, Bos JL, Thomas G. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol Cell. 2003;11:1457–1466. doi: 10.1016/s1097-2765(03)00220-x. [DOI] [PubMed] [Google Scholar]
- 86.Li Y, Inoki K, Guan KL. Biochemical and functional characterizations of small GTPase Rheb and TSC2 GAP activity. Mol Cell Biol. 2004;24:7965–7975. doi: 10.1128/MCB.24.18.7965-7975.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nellist M, Sancak O, Goedbloed MA, Rohe C, van Netten D, Mayer K, Tucker-Williams A, van den Ouweland AM, Halley DJ. Distinct effects of single amino-acid changes to tuberin on the function of the tuberin-hamartin complex. Eur J Hum Genet. 2005;13:59–68. doi: 10.1038/sj.ejhg.5201276. [DOI] [PubMed] [Google Scholar]
- 88.Cai SL, Tee AR, Short JD, Bergeron JM, Kim J, Shen J, Guo R, Johnson CL, Kiguchi K, Walker CL. Activity of TSC2 is inhibited by AKT-mediated phosphorylation and membrane partitioning. J Cell Biol. 2006;173:279–289. doi: 10.1083/jcb.200507119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dan HC, Sun M, Yang L, Feldman RI, Sui XM, Ou CC, Nellist M, Yeung RS, Halley DJ, Nicosia SV, Pledger WJ, Cheng JQ. Phosphatidylinositol 3-kinase/Akt pathway regulates tuberous sclerosis tumor suppressor complex by phosphorylation of tuberin. J Biol Chem. 2002;277:35364–35370. doi: 10.1074/jbc.M205838200. [DOI] [PubMed] [Google Scholar]
- 90.Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 91.Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol Cell. 2002;10:151–162. doi: 10.1016/s1097-2765(02)00568-3. [DOI] [PubMed] [Google Scholar]
- 92.Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 93.Kahn BB, Alquier T, Carling D, Hardie DG. AMP-activated protein kinase: ancient energy gauge provides clues to modern understanding of metabolism. Cell Metab. 2005;1:15–25. doi: 10.1016/j.cmet.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 94.Inoki K, Ouyang H, Zhu T, Lindvall C, Wang Y, Zhang X, Yang Q, Bennett C, Harada Y, Stankunas K, Wang CY, He X, MacDougald OA, You M, Williams BO, Guan KL. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell. 2006;126:955–968. doi: 10.1016/j.cell.2006.06.055. [DOI] [PubMed] [Google Scholar]
- 95.Moon RT (2005) Wnt/beta-catenin pathway. Sci STKE (271):cm1. Accessed 15 Feb 2005 [DOI] [PubMed]
- 96.Ma L, Chen Z, Erdjument-Bromage H, Tempst P, Pandolfi PP. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell. 2005;121:179–193. doi: 10.1016/j.cell.2005.02.031. [DOI] [PubMed] [Google Scholar]
- 97.Ma L, Teruya-Feldstein J, Bonner P, Bernardi R, Franz DN, Witte D, Cordon-Cardo C, Pandolfi PP. Identification of S664 TSC2 phosphorylation as a marker for extracellular signal-regulated kinase mediated mTOR activation in tuberous sclerosis and human cancer. Cancer Res. 2007;67:7106–7112. doi: 10.1158/0008-5472.CAN-06-4798. [DOI] [PubMed] [Google Scholar]
- 98.Roux PP, Ballif BA, Anjum R, Gygi SP, Blenis J. Tumor-promoting phorbol esters and activated Ras inactivate the tuberous sclerosis tumor suppressor complex via p90 ribosomal S6 kinase. Proc Natl Acad Sci USA. 2004;101:13489–13494. doi: 10.1073/pnas.0405659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ballif BA, Roux PP, Gerber SA, MacKeigan JP, Blenis J, Gygi SP. Quantitative phosphorylation profiling of the ERK/p90 ribosomal S6 kinase-signaling cassette and its targets, the tuberous sclerosis tumor suppressors. Proc Natl Acad Sci USA. 2005;102:667–672. doi: 10.1073/pnas.0409143102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yamagata K, Sanders LK, Kaufmann WE, Yee W, Barnes CA, Nathans D, Worley PF. rheb, a growth factor- and synaptic activity-regulated gene, encodes a novel Ras-related protein. J Biol Chem. 1994;269:16333–16339. [PubMed] [Google Scholar]
- 101.Kinouchi H, Arai S, Kamii H, Izaki K, Kunizuka H, Mizoi K, Yoshimoto T. Induction of Rheb mRNA following middle cerebral artery occlusion in the rat. Neuroreport. 1999;10:1055–1059. doi: 10.1097/00001756-199904060-00029. [DOI] [PubMed] [Google Scholar]
- 102.Bos JL. Ras-like GTPases. Biochim Biophys Acta. 1997;1333:M19–M31. doi: 10.1016/s0304-419x(97)00015-2. [DOI] [PubMed] [Google Scholar]
- 103.Aspuria PJ, Tamanoi F. The Rheb family of GTP-binding proteins. Cell Signal. 2004;16:1105–1112. doi: 10.1016/j.cellsig.2004.03.019. [DOI] [PubMed] [Google Scholar]
- 104.Im E, von Lintig FC, Chen J, Zhuang S, Qui W, Chowdhury S, Worley PF, Boss GR, Pilz RB. Rheb is in a high activation state and inhibits B-Raf kinase in mammalian cells. Oncogene. 2002;21:6356–6365. doi: 10.1038/sj.onc.1205792. [DOI] [PubMed] [Google Scholar]
- 105.Yu Y, Li S, Xu X, Li Y, Guan K, Arnold E, Ding J. Structural basis for the unique biological function of small GTPase RHEB. J Biol Chem. 2005;280:17093–17100. doi: 10.1074/jbc.M501253200. [DOI] [PubMed] [Google Scholar]
- 106.Hsu YC, Chern JJ, Cai Y, Liu M, Choi KW. Drosophila TCTP is essential for growth and proliferation through regulation of dRheb GTPase. Nature. 2007;445:785–788. doi: 10.1038/nature05528. [DOI] [PubMed] [Google Scholar]
- 107.Dong X, Yang B, Li Y, Zhong C, Ding J. Molecular basis of the acceleration of the GDP-GTP exchange of human Rheb by human TCTP. J Biol Chem. 2009;284(35):23754–23764. doi: 10.1074/jbc.M109.012823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Smith EM, Finn SG, Tee AR, Browne GJ, Proud CG. The tuberous sclerosis protein TSC2 is not required for the regulation of the mammalian target of rapamycin by amino acids and certain cellular stresses. J Biol Chem. 2005;280:18717–18727. doi: 10.1074/jbc.M414499200. [DOI] [PubMed] [Google Scholar]
- 109.Roccio M, Bos JL, Zwartkruis FJ. Regulation of the small GTPase Rheb by amino acids. Oncogene. 2006;25:657–664. doi: 10.1038/sj.onc.1209106. [DOI] [PubMed] [Google Scholar]
- 110.Long X, Ortiz-Vega S, Lin Y, Avruch J. Rheb binding to mammalian target of rapamycin (mTOR) is regulated by amino acid sufficiency. J Biol Chem. 2005;280:23433–23436. doi: 10.1074/jbc.C500169200. [DOI] [PubMed] [Google Scholar]
- 111.Nobukuni T, Joaquin M, Roccio M, Dann SG, Kim SY, Gulati P, Byfield MP, Backer JM, Natt F, Bos JL, Zwartkruis FJ, Thomas G. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc Natl Acad Sci USA. 2005;102:14238–14243. doi: 10.1073/pnas.0506925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Long X, Lin Y, Ortiz-Vega S, Yonezawa K, Avruch J. Rheb binds and regulates the mTOR kinase. Curr Biol. 2005;15:702–713. doi: 10.1016/j.cub.2005.02.053. [DOI] [PubMed] [Google Scholar]
- 113.Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, Carr SA, Sabatini DM. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell. 2007;25:903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 114.Sato T, Nakashima A, Guo L, Tamanoi F. Specific activation of mTORC1 by Rheb G-protein in vitro involves enhanced recruitment of its substrate protein. J Biol Chem. 2009;284:12783–12791. doi: 10.1074/jbc.M809207200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tee AR, Blenis J, Proud CG. Analysis of mTOR signaling by the small G-proteins, Rheb and RhebL1. FEBS Lett. 2005;579:4763–4768. doi: 10.1016/j.febslet.2005.07.054. [DOI] [PubMed] [Google Scholar]
- 116.Marks AR. Cellular functions of immunophilins. Physiol Rev. 1996;76:631–649. doi: 10.1152/physrev.1996.76.3.631. [DOI] [PubMed] [Google Scholar]
- 117.Nielsen JV, Mitchelmore C, Pedersen KM, Kjaerulff KM, Finsen B, Jensen NA. Fkbp8: novel isoforms, genomic organization, and characterization of a forebrain promoter in transgenic mice. Genomics. 2004;83:181–192. doi: 10.1016/j.ygeno.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 118.Fischer G, Aumuller T. Regulation of peptide bond cis/trans isomerization by enzyme catalysis and its implication in physiological processes. Rev Physiol Biochem Pharmacol. 2003;148:105–150. doi: 10.1007/s10254-003-0011-3. [DOI] [PubMed] [Google Scholar]
- 119.Ratajczak T, Ward BK, Minchin RF. Immunophilin chaperones in steroid receptor signalling. Curr Top Med Chem. 2003;3:1348–1357. doi: 10.2174/1568026033451934. [DOI] [PubMed] [Google Scholar]
- 120.Griffith JP, Kim JL, Kim EE, Sintchak MD, Thomson JA, Fitzgibbon MJ, Fleming MA, Caron PR, Hsiao K, Navia MA. X-ray structure of calcineurin inhibited by the immunophilin-immunosuppressant FKBP12-FK506 complex. Cell. 1995;82:507–522. doi: 10.1016/0092-8674(95)90439-5. [DOI] [PubMed] [Google Scholar]
- 121.Kissinger CR, Parge HE, Knighton DR, Lewis CT, Pelletier LA, Tempczyk A, Kalish VJ, Tucker KD, Showalter RE, Moomaw EW, et al. Crystal structures of human calcineurin and the human FKBP12-FK506-calcineurin complex. Nature. 1995;378:641–644. doi: 10.1038/378641a0. [DOI] [PubMed] [Google Scholar]
- 122.Maestre-Martinez M, Edlich F, Jarczowski F, Weiwad M, Fischer G, Lucke C. Solution structure of the FK506-binding domain of human FKBP38. J Biomol NMR. 2006;34:197–202. doi: 10.1007/s10858-006-0018-6. [DOI] [PubMed] [Google Scholar]
- 123.Pedersen KM, Finsen B, Celis JE, Jensen NA. muFKBP38: a novel murine immunophilin homolog differentially expressed in Schwannoma cells and central nervous system neurons in vivo. Electrophoresis. 1999;20:249–255. doi: 10.1002/(SICI)1522-2683(19990201)20:2<249::AID-ELPS249>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 124.Shirane M, Nakayama KI. Inherent calcineurin inhibitor FKBP38 targets Bcl-2 to mitochondria and inhibits apoptosis. Nat Cell Biol. 2003;5:28–37. doi: 10.1038/ncb894. [DOI] [PubMed] [Google Scholar]
- 125.Edlich F, Weiwad M, Erdmann F, Fanghanel J, Jarczowski F, Rahfeld JU, Fischer G. Bcl-2 regulator FKBP38 is activated by Ca2+/calmodulin. EMBO J. 2005;24:2688–2699. doi: 10.1038/sj.emboj.7600739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bai X, Ma D, Liu A, Shen X, Wang QJ, Liu Y, Jiang Y. Rheb activates mTOR by antagonizing its endogenous inhibitor, FKBP38. Science. 2007;318:977–980. doi: 10.1126/science.1147379. [DOI] [PubMed] [Google Scholar]
- 127.Uhlenbrock K, Weiwad M, Wetzker R, Fischer G, Wittinghofer A, Rubio I. Reassessment of the role of FKBP38 in the Rheb/mTORC1 pathway. FEBS Lett. 2009;583:965–970. doi: 10.1016/j.febslet.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 128.Dunlop EA, Dodd KM, Seymour LA, Tee AR. Mammalian target of rapamycin complex 1-mediated phosphorylation of eukaryotic initiation factor 4E-binding protein 1 requires multiple protein–protein interactions for substrate recognition. Cell Signal. 2009;21:1073–1084. doi: 10.1016/j.cellsig.2009.02.024. [DOI] [PubMed] [Google Scholar]
- 129.Ma D, Bai X, Guo S, Jiang Y. The switch I region of Rheb is critical for its interaction with FKBP38. J Biol Chem. 2008;283:25963–25970. doi: 10.1074/jbc.M802356200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wang X, Fonseca BD, Tang H, Liu R, Elia A, Clemens MJ, Bommer UA, Proud CG. Re-evaluating the roles of proposed modulators of mammalian target of rapamycin complex 1 (mTORC1) signaling. J Biol Chem. 2008;283:30482–30492. doi: 10.1074/jbc.M803348200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Avruch J, Long X, Ortiz-Vega S, Rapley J, Papageorgiou A, Dai N. Amino acid regulation of TOR complex 1. Am J Physiol Endocrinol Metab. 2009;296:E592–E602. doi: 10.1152/ajpendo.90645.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Telerman A, Amson R. The molecular programme of tumour reversion: the steps beyond malignant transformation. Nat Rev Cancer. 2009;9:206–216. doi: 10.1038/nrc2589. [DOI] [PubMed] [Google Scholar]
- 133.Chen SH, Wu PS, Chou CH, Yan YT, Liu H, Weng SY, Yang-Yen HF. A knockout mouse approach reveals that TCTP functions as an essential factor for cell proliferation and survival in a tissue- or cell type-specific manner. Mol Biol Cell. 2007;18:2525–2532. doi: 10.1091/mbc.E07-02-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rehmann H, Bruning M, Berghaus C, Schwarten M, Kohler K, Stocker H, Stoll R, Zwartkruis FJ, Wittinghofer A. Biochemical characterisation of TCTP questions its function as a guanine nucleotide exchange factor for Rheb. FEBS Lett. 2008;582:3005–3010. doi: 10.1016/j.febslet.2008.07.057. [DOI] [PubMed] [Google Scholar]
- 135.Kovacina KS, Park GY, Bae SS, Guzzetta AW, Schaefer E, Birnbaum MJ, Roth RA. Identification of a proline-rich Akt substrate as a 14-3-3 binding partner. J Biol Chem. 2003;278:10189–10194. doi: 10.1074/jbc.M210837200. [DOI] [PubMed] [Google Scholar]
- 136.Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol. 2007;9:316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- 137.Wang L, Harris TE, Roth RA, Lawrence JC., Jr PRAS40 regulates mTORC1 kinase activity by functioning as a direct inhibitor of substrate binding. J Biol Chem. 2007;282:20036–20044. doi: 10.1074/jbc.M702376200. [DOI] [PubMed] [Google Scholar]
- 138.Oshiro N, Takahashi R, Yoshino K, Tanimura K, Nakashima A, Eguchi S, Miyamoto T, Hara K, Takehana K, Avruch J, Kikkawa U, Yonezawa K. The proline-rich Akt substrate of 40 kDa (PRAS40) is a physiological substrate of mammalian target of rapamycin complex 1. J Biol Chem. 2007;282:20329–20339. doi: 10.1074/jbc.M702636200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang L, Harris TE, Lawrence JC., Jr Regulation of proline-rich Akt substrate of 40 kDa (PRAS40) function by mammalian target of rapamycin complex 1 (mTORC1)-mediated phosphorylation. J Biol Chem. 2008;283:15619–15627. doi: 10.1074/jbc.M800723200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Fonseca BD, Smith EM, Lee VH, MacKintosh C, Proud CG. PRAS40 is a target for mammalian target of rapamycin complex 1 and is required for signaling downstream of this complex. J Biol Chem. 2007;282:24514–24524. doi: 10.1074/jbc.M704406200. [DOI] [PubMed] [Google Scholar]
- 141.Dong J, Pan D. Tsc2 is not a critical target of Akt during normal Drosophila development. Genes Dev. 2004;18:2479–2484. doi: 10.1101/gad.1240504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Shah N, Pang B, Yeoh KG, Thorn S, Chen CS, Lilly MB, Salto-Tellez M. Potential roles for the PIM1 kinase in human cancer—a molecular and therapeutic appraisal. Eur J Cancer. 2008;44:2144–2151. doi: 10.1016/j.ejca.2008.06.044. [DOI] [PubMed] [Google Scholar]
- 143.Zhang F, Beharry ZM, Harris TE, Lilly MB, Smith CD, Mahajan S, Kraft AS. PIM1 protein kinase regulates PRAS40 phosphorylation and mTOR activity in FDCP1 cells. Cancer Biol Ther. 2009;8:846–853. doi: 10.4161/cbt.8.9.8210. [DOI] [PubMed] [Google Scholar]
- 144.Li H, Lin X. Positive and negative signaling components involved in TNFalpha-induced NF-kappaB activation. Cytokine. 2008;41:1–8. doi: 10.1016/j.cyto.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 145.Häcker H, Karin M (2006) Regulation and function of IKK and IKK-related kinases. Sci STKE (357):re13. Accessed 17 Oct 2006 [DOI] [PubMed]
- 146.Karin M. How NF-kappaB is activated: the role of the IkappaB kinase (IKK) complex. Oncogene. 1999;18:6867–6874. doi: 10.1038/sj.onc.1203219. [DOI] [PubMed] [Google Scholar]
- 147.Ozes ON, Akca H, Mayo LD, Gustin JA, Maehama T, Dixon JE, Donner DB. A phosphatidylinositol 3-kinase/Akt/mTOR pathway mediates and PTEN antagonizes tumor necrosis factor inhibition of insulin signaling through insulin receptor substrate-1. Proc Natl Acad Sci USA. 2001;98:4640–4645. doi: 10.1073/pnas.051042298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lee DF, Kuo HP, Chen CT, Hsu JM, Chou CK, Wei Y, Sun HL, Li LY, Ping B, Huang WC, He X, Hung JY, Lai CC, Ding Q, Su JL, Yang JY, Sahin AA, Hortobagyi GN, Tsai FJ, Tsai CH, Hung MC. IKK beta suppression of TSC1 links inflammation and tumor angiogenesis via the mTOR pathway. Cell. 2007;130:440–455. doi: 10.1016/j.cell.2007.05.058. [DOI] [PubMed] [Google Scholar]
- 149.Dan HC, Baldwin AS. Differential involvement of IkappaB kinases alpha and beta in cytokine- and insulin-induced mammalian target of rapamycin activation determined by Akt. J Immunol. 2008;180:7582–7589. doi: 10.4049/jimmunol.180.11.7582. [DOI] [PubMed] [Google Scholar]
- 150.Dan HC, Adli M, Baldwin AS. Regulation of mammalian target of rapamycin activity in PTEN-inactive prostate cancer cells by I kappa B kinase alpha. Cancer Res. 2007;67:6263–6269. doi: 10.1158/0008-5472.CAN-07-1232. [DOI] [PubMed] [Google Scholar]
- 151.Singer WD, Brown HA, Sternweis PC. Regulation of eukaryotic phosphatidylinositol-specific phospholipase C and phospholipase D. Annu Rev Biochem. 1997;66:475–509. doi: 10.1146/annurev.biochem.66.1.475. [DOI] [PubMed] [Google Scholar]
- 152.McDermott M, Wakelam MJ, Morris AJ. Phospholipase D. Biochem Cell Biol. 2004;82:225–253. doi: 10.1139/o03-079. [DOI] [PubMed] [Google Scholar]
- 153.Andresen BT, Rizzo MA, Shome K, Romero G. The role of phosphatidic acid in the regulation of the Ras/MEK/Erk signaling cascade. FEBS Lett. 2002;531:65–68. doi: 10.1016/s0014-5793(02)03483-x. [DOI] [PubMed] [Google Scholar]
- 154.Fang Y, Vilella-Bach M, Bachmann R, Flanigan A, Chen J. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science. 2001;294:1942–1945. doi: 10.1126/science.1066015. [DOI] [PubMed] [Google Scholar]
- 155.Sun Y, Chen J. mTOR signaling: PLD takes center stage. Cell Cycle. 2008;7:3118–3123. doi: 10.4161/cc.7.20.6881. [DOI] [PubMed] [Google Scholar]
- 156.Toschi A, Lee E, Xu L, Garcia A, Gadir N, Foster DA. Regulation of mTORC1 and mTORC2 complex assembly by phosphatidic acid: competition with rapamycin. Mol Cell Biol. 2009;29:1411–1420. doi: 10.1128/MCB.00782-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Fang Y, Park IH, Wu AL, Du G, Huang P, Frohman MA, Walker SJ, Brown HA, Chen J. PLD1 regulates mTOR signaling and mediates Cdc42 activation of S6K1. Curr Biol. 2003;13:2037–2044. doi: 10.1016/j.cub.2003.11.021. [DOI] [PubMed] [Google Scholar]
- 158.Ha SH, Kim DH, Kim IS, Kim JH, Lee MN, Lee HJ, Kim JH, Jang SK, Suh PG, Ryu SH. PLD2 forms a functional complex with mTOR/raptor to transduce mitogenic signals. Cell Signal. 2006;18:2283–2291. doi: 10.1016/j.cellsig.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 159.Lehman N, Ledford B, Di Fulvio M, Frondorf K, McPhail LC, Gomez-Cambronero J. Phospholipase D2-derived phosphatidic acid binds to and activates ribosomal p70 S6 kinase independently of mTOR. Faseb J. 2007;21:1075–1087. doi: 10.1096/fj.06-6652com. [DOI] [PubMed] [Google Scholar]
- 160.Sun Y, Fang Y, Yoon MS, Zhang C, Roccio M, Zwartkruis FJ, Armstrong M, Brown HA, Chen J. Phospholipase D1 is an effector of Rheb in the mTOR pathway. Proc Natl Acad Sci USA. 2008;105:8286–8291. doi: 10.1073/pnas.0712268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Exton JH. Regulation of phospholipase D. FEBS Lett. 2002;531:58–61. doi: 10.1016/s0014-5793(02)03405-1. [DOI] [PubMed] [Google Scholar]
- 162.Chou MM, Blenis J. The 70 kDa S6 kinase complexes with and is activated by the Rho family G proteins Cdc42 and Rac1. Cell. 1996;85:573–583. doi: 10.1016/s0092-8674(00)81257-x. [DOI] [PubMed] [Google Scholar]
- 163.Nishimoto T. Upstream and downstream of ran GTPase. Biol Chem. 2000;381:397–405. doi: 10.1515/BC.2000.052. [DOI] [PubMed] [Google Scholar]
- 164.Schurmann A, Brauers A, Massmann S, Becker W, Joost HG. Cloning of a novel family of mammalian GTP-binding proteins (RagA, RagBs, RagB1) with remote similarity to the Ras-related GTPases. J Biol Chem. 1995;270:28982–28988. doi: 10.1074/jbc.270.48.28982. [DOI] [PubMed] [Google Scholar]
- 165.Sekiguchi T, Hirose E, Nakashima N, Ii M, Nishimoto T. Novel G proteins, Rag C and Rag D, interact with GTP-binding proteins, Rag A and Rag B. J Biol Chem. 2001;276:7246–7257. doi: 10.1074/jbc.M004389200. [DOI] [PubMed] [Google Scholar]
- 166.Nakashima N, Noguchi E, Nishimoto T. Saccharomyces cerevisiae putative G protein, Gtr1p, which forms complexes with itself and a novel protein designated as Gtr2p, negatively regulates the Ran/Gsp1p G protein cycle through Gtr2p. Genetics. 1999;152:853–867. doi: 10.1093/genetics/152.3.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wang Y, Nakashima N, Sekiguchi T, Nishimoto T. Saccharomyces cerevisiae GTPase complex: Gtr1p-Gtr2p regulates cell-proliferation through Saccharomyces cerevisiae Ran-binding protein, Yrb2p. Biochem Biophys Res Commun. 2005;336:639–645. doi: 10.1016/j.bbrc.2005.08.108. [DOI] [PubMed] [Google Scholar]
- 168.Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Gao M, Kaiser CA. A conserved GTPase-containing complex is required for intracellular sorting of the general amino-acid permease in yeast. Nat Cell Biol. 2006;8:657–667. doi: 10.1038/ncb1419. [DOI] [PubMed] [Google Scholar]
- 170.Buerger C, DeVries B, Stambolic V. Localization of Rheb to the endomembrane is critical for its signaling function. Biochem Biophys Res Commun. 2006;344:869–880. doi: 10.1016/j.bbrc.2006.03.220. [DOI] [PubMed] [Google Scholar]
- 171.Takahashi K, Nakagawa M, Young SG, Yamanaka S. Differential membrane localization of ERas and Rheb, two Ras-related proteins involved in the phosphatidylinositol 3-kinase/mTOR pathway. J Biol Chem. 2005;280:32768–32774. doi: 10.1074/jbc.M506280200. [DOI] [PubMed] [Google Scholar]
- 172.Schu PV, Takegawa K, Fry MJ, Stack JH, Waterfield MD, Emr SD. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science. 1993;260:88–91. doi: 10.1126/science.8385367. [DOI] [PubMed] [Google Scholar]
- 173.Kapeller R, Cantley LC. Phosphatidylinositol 3-kinase. Bioessays. 1994;16:565–576. doi: 10.1002/bies.950160810. [DOI] [PubMed] [Google Scholar]
- 174.Stack JH, Herman PK, Schu PV, Emr SD. A membrane-associated complex containing the Vps15 protein kinase and the Vps34 PI 3-kinase is essential for protein sorting to the yeast lysosome-like vacuole. EMBO J. 1993;12:2195–2204. doi: 10.1002/j.1460-2075.1993.tb05867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Byfield MP, Murray JT, Backer JM. hVps34 is a nutrient-regulated lipid kinase required for activation of p70 S6 kinase. J Biol Chem. 2005;280:33076–33082. doi: 10.1074/jbc.M507201200. [DOI] [PubMed] [Google Scholar]
- 176.Gulati P, Gaspers LD, Dann SG, Joaquin M, Nobukuni T, Natt F, Kozma SC, Thomas AP, Thomas G. Amino acids activate mTOR complex 1 via Ca2+/CaM signaling to hVps34. Cell Metab. 2008;7:456–465. doi: 10.1016/j.cmet.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Stein MP, Feng Y, Cooper KL, Welford AM, Wandinger-Ness A. Human VPS34 and p150 are Rab7 interacting partners. Traffic. 2003;4:754–771. doi: 10.1034/j.1600-0854.2003.00133.x. [DOI] [PubMed] [Google Scholar]
- 178.Drenan RM, Liu X, Bertram PG, Zheng XF. FKBP12-rapamycin-associated protein or mammalian target of rapamycin (FRAP/mTOR) localization in the endoplasmic reticulum and the Golgi apparatus. J Biol Chem. 2004;279:772–778. doi: 10.1074/jbc.M305912200. [DOI] [PubMed] [Google Scholar]
- 179.Zhang X, Shu L, Hosoi H, Murti KG, Houghton PJ. Predominant nuclear localization of mammalian target of rapamycin in normal and malignant cells in culture. J Biol Chem. 2002;277:28127–28134. doi: 10.1074/jbc.M202625200. [DOI] [PubMed] [Google Scholar]
- 180.Desai BN, Myers BR, Schreiber SL. FKBP12-rapamycin-associated protein associates with mitochondria and senses osmotic stress via mitochondrial dysfunction. Proc Natl Acad Sci USA. 2002;99:4319–4324. doi: 10.1073/pnas.261702698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Liu X, Zheng XF. Endoplasmic reticulum and Golgi localization sequences for mammalian target of rapamycin. Mol Biol Cell. 2007;18:1073–1082. doi: 10.1091/mbc.E06-05-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Park IH, Bachmann R, Shirazi H, Chen J. Regulation of ribosomal S6 kinase 2 by mammalian target of rapamycin. J Biol Chem. 2002;277:31423–31429. doi: 10.1074/jbc.M204080200. [DOI] [PubMed] [Google Scholar]