Abstract
The liver is an essential organ that plays a pivotal role in metabolism, digestion and nutrient storage. Major efforts have been made to develop zebrafish (Danio rerio) as a model system to study the pathways regulating hepatic growth during liver development and regeneration. Zebrafish offer unique advantages over other vertebrates including in vivo imaging at cellular resolution and the capacity for large-scale chemical and genetic screens. Here, we review the cellular and molecular mechanisms that regulate hepatic growth during liver development in zebrafish. We also highlight emerging evidence that developmental pathways are reactivated following liver injury to facilitate regeneration. Finally, we discuss how zebrafish have transformed drug discovery efforts and enabled the identification of drugs that stimulate hepatic growth and provide hepatoprotection in pre-clinical models of liver injury, with the ultimate goal of identifying novel therapeutic approaches to treat liver disease.
Introduction
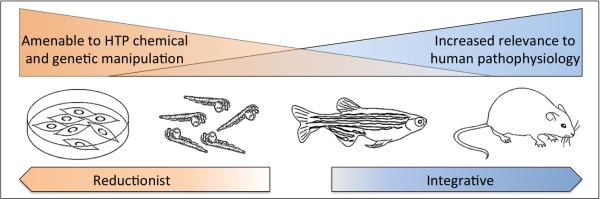
Over the last two decades zebrafish have become a primary model system to study vertebrate liver development. One of the major advantages of zebrafish as a model system is the rapid ex utero development of transparent embryos enabling liver development to be imaged at the cellular level. Furthermore, zebrafish matings produce large clutches of embryos (>200), which facilitate chemical and genetic screens to identify novel genes and small molecules that regulate liver formation. Finally, the zebrafish genome has been sequenced and the annotation has revealed that ~70% of human genes have a zebrafish orthologue[1]. Consequently, the genes and developmental pathways underpinning liver development and disease are highly conserved among vertebrates. Together, these features have placed zebrafish in a unique position filling the void in liver research between reductionist mechanistic studies in cultured cells and integrative murine studies that provide a higher degree of relevance to human pathophysiology (Figure 1).
Figure 1.
The advantages of zebrafish as a model system to study liver physiology.
The human liver is the only solid organ capable of regeneration. The process of liver regeneration is multifaceted, as it requires a complex tissue comprised of multiple cell types to sense the extent of injury and mount an appropriate compensatory regrowth response, while maintaining the tissue architecture required for liver function[2]. In the context of liver disease, acute liver injury necessitates a rapid regenerative response to avoid acute liver failure, whereas chronic liver injury is often associated with maladaptive scarring (fibrosis) that impairs liver regeneration. Despite the clinical significance, surprisingly little is understood regarding the molecular mechanisms regulating liver regeneration. Most of our knowledge regarding liver regeneration has been learned from studies performed in rodents. However, recent insights using zebrafish, which have a greater capacity to regenerate than mammals, have expanded the modern field of liver research. This review will highlight the value of zebrafish as a complementary model organism and outline innovative approaches that could yield novel insights into the molecular and cellular underpinnings of liver regeneration.
Regulation of hepatic growth during development
Introduction to zebrafish liver development
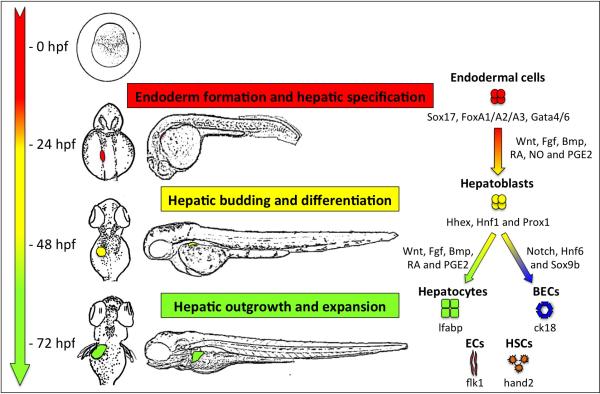
Classic studies by Stainier and colleagues[3,4] using transgenic Tg(XIa.Eef1a1:GFP) (gut:GFP) fish, which express GFP throughout the endoderm, defined the morphological phases of liver development in zebrafish and they include i) hepatic specification, ii) hepatic budding, and iii) hepatic outgrowth. Hepatic specification occurs at 24 hpf as cells on the ventral surface of the anterior section of the endodermal rod begin to express hepatoblast markers such as prox1 and hhex. Specified hepatoblasts subsequently begin budding to the left side of the embryo and at 36 hpf the connection between the primordial liver bud and the intestine becomes restricted as the cells begin to take on a biliary fate as they ultimately form the common bile duct. From 48 hpf onward the outgrowth phase of liver development is accompanied by hepatocyte differentiation, the formation of a biliary ductal network and the vascularization of the liver, which is fully functional by 72 hpf. The zebrafish liver is mainly comprised of hepatocytes (labeled cells in Tg(fabp10a:GFP))[5,6], which carry out most functions of the liver, including bile and serum protein secretion, blood detoxification and metabolic regulation of lipids, glucose and amino acids. Non-parenchymal cells such as endothelial cells (labeled cells in Tg(kdrl:GFP))[7,8] biliary epithelial cells (BECs) (labeled cells in Tg(krt18:GFP)) [9,10] and hepatic stellate cells (HSCs) (labeled cells in Tg(hand2:GFP))[11,12] facilitate liver function by transporting blood and bile whilst transmitting mitogenic cues for hepatic growth. Together, these landmark studies demonstrate the power of fluorescent transgenic lines to image the cells involved in liver function in vivo.
Pathways regulating hepatic specification
The regulation of hepatic specification is highly conserved throughout the vertebrate kingdom. The nuclear receptor hepatocyte nuclear factor (hnf1) and the transcription factor hematopoietically expressed homeobox (hhex) are both required for hepatoblast specification[13,14]. Among the other factors impacting on early liver development are the GATA transcription factors, particularly gata6, which is required for hepatoblast specification[15]. Epigenetic regulation also plays a key role in programming hepatic cell fate as hdac1 mutant embryos fail to specify hepatoblasts[16]. Together, these cell autonomous factors program the fate of cells in the foregut endoderm towards the hepatic lineage.
Emerging evidence suggests that mesodermally-derived Wnt, FGF and Bmp ligands play a fundamental role in hepatoblast specification (Figure 2). Ober, Stainier and colleagues revealed that wnt2bb (prt mutant) is essential for liver specification, showing for the first time that the Wnt pathway plays a key role in regulating liver development[17]. Wnt2bb is expressed in the lateral plate mesoderm adjacent to the developing endoderm and plays an inductive role in hepatic specification. Subsequent studies showed that overexpression of wnt2bb enhanced hepatoblast proliferation and induced hepatomegaly by acting on the receptor frizzled homologue 5 (fzd5)[18]. Wnt2bb enhances hepatoblast bud formation by activating expression of nav3a, which guides endodermal migration away from the gut endoderm[19]. Mechanistic studies have shown that Sox32 is required for eventual liver formation in wnt2bb mutant embryos[20]. Recent insights have revealed that other factors such as EpCam and hnf1ba genetically interact with wnt2bb to specify hepatopancreatic progenitors[21,22]. We recently used apc mutant and wnt8-inducible transgenic zebrafish to discover that liver formation required dynamic regulation of the Wnt pathway, with suppression of Wnt activity in early somitogenesis followed by elevated Wnt signaling during the hepatic specification phase[23]. Induction of wnt8 during mid-somitogenesis negatively regulated endoderm formation, whereas, activation of wnt8 during hepatic specification resulted in hepatomegaly. Recent studies have shown that wnt8 directly converts non-hepatic endodermal cells into hepatoblasts in a cell-autonomous manner[24]. In light of these studies, it is clearly evident that the Wnt pathway is a master regulator of hepatic differentiation and growth during development.
Figure 2.
Pathways regulating liver development.
Groundbreaking studies by Dong, Stainier and colleagues revealed that Fgf10 is expressed in adjacent mesenchyme and regulates hepatopancreatic biliary morphogenesis and hepatocyte differentiation[25]. In this study, the authors found that Fgf10a-deficient embryos displayed expanded biliary formation and enhanced hepatic differentiation. Follow-up work has shown that Fgf10a is expressed in the pancreatic bud where it negatively regulates hepatic competence[26]. In line with this notion, compound fgf10−/−;fgf24−/− mutant embryos develop ectopic hepatocytes that form near the pancreas[27]. Fgf10b is expressed in the liver and its depletion inhibits liver formation suggesting that the two paralogues have spatial subfunctionalization[28]. Stimulation of FGF signaling during gastrulation phosphorylates and inactivates sox32, thereby impairing endoderm specification[29]. However, inhibition of FGF signaling, by inducible expression of a dominant negative FGF receptor 1 (dnFGFR1), has demonstrated that FGF is essential for hepatoblast specification[15]. Interestingly, FGF signaling is also required cell-autonomously for hepatic growth during the outgrowth phase of liver development[30]. Overall, these studies suggest that FGF signaling plays a complex role regulating zones of hepatic competence whilst contributing to the milieu of mitogenic factors essential for liver formation.
Several studies have shown that Bmp overexpression during mid-somitogenesis compromises endoderm specification and disrupts anterior-posterior patterning of endoderm, which can lead to organ laterality defects[29,31,32]. Bmp2b is expressed in the lateral plate mesoderm and signals through the receptor alk8 to specify hepatoblasts[33]. Decreased Bmp signaling in alk8 mutants inhibits liver development, whereas bmp2b overexpression causes pancreatic-fated cells to transdifferentiate towards the hepatic fate[33]. Studies using dominant negative bmp receptor 1 (dnBmpr1a) inducible transgenics revealed that Bmp signaling is essential for hepatoblast development, but not maintenance[15]. Bmp2b overexpression can partially compensate for the loss of FGF signaling[15]. Peng and colleagues have shown that ectopic expression of bmp2b rescued liver development in protein phosphatase 1, regulatory subunit 12A (ppp1r12a or mypt1) mypt1 mutant embryos that exhibit defects in lateral plate mesoderm formation[34]. These studies highlight the importance of Bmp signaling in organ laterality and liver specification.
Pathways regulating biliary specification and expansion
During liver formation, transcriptional programs instruct bipotential hepatoblasts to differentiate into hepatocytes or biliary epithelial cells (BECs) (Figure 2). Founding studies by Pack and colleagues characterized biliary development in zebrafish and found that Notch signaling plays a key role in regulating biliary fate[35]. In addition to Notch, loss of function studies have demonstrated that the Onecut family of transcription factors, which includes hnf6, play an essential role in biliary differentiation and morphogenesis[36,37]. Elegant studies have recently established that sox9b is a master regulator of hepatopancreatic ductal system[38,39]. These studies reveal that sox9b mutant zebrafish exhibit defects in biliary morphogenesis and bile duct canaliculi during embryogenesis and develop cholestasis and fibrosis in adulthood. These studies define the core transcriptional networks programming BECs (Notch, Onecut and sox9b) during hepatobiliary development.
Deregulated cell proliferation and survival impacts upon liver outgrowth
Genetic defects in fundamental aspects of cell biology tend to manifest during zebrafish development as a failure to thrive with reduced survival or proliferation in specific cell types. For example, sorting nexon 7 (snx7) and annexin A4 (anxa4) are both required for hepatoblast survival[40,41]. Similarly, the small nuclear RNA (snRNA) transcription complex component snap4c is required for BEC survival[42]. Mitochondrial homeostasis plays an important role in the maintenance of hepatocyte survival as illustrated by the rapid liver degeneration observed in zebrafish embryos with defects in mitochondrial transport (tom22)[43], mitochondrial RNA helicase activity (supv3L1)[44] and mitochondrial antioxidant activity (trx2)[45]. Some zebrafish mutants such as def[46,47], elys[48,49], ssrp1a[50] and bms1l[51] exhibit liver hypoplasia due to nucleolar defects that culminate in replication stress or a DNA damage response. In similar work, Sadler and colleagues have determined that uhrf1 mutants developed a small-for-size liver during development due to DNA hypomethylation, which leads to cell cycle arrest and the induction of apoptosis[52-55]. Several genes that are highly enriched in the developing liver such as matrix metalloproteinase mmp23b[56], as well as the secretory pathway components leg1[57,58] and sec13[59] are required for hepatocyte proliferation. Other genes including augmenter of liver regeneration (alr)[60], zfyve9a[61] and klf6[62] seem to be specifically requires for proliferation during the outgrowth phase of liver development. Together, these studies begin to identify factors that are required for the survival and proliferation during liver development.
Small molecules that modulate liver growth during liver development
One of the key advantages of studying liver formation in zebrafish is that the embryos develop ex utero, which facilitates chemical screening. We conducted a chemical screen in zebrafish embryos and identified numerous small molecules that modulate hepatic growth during development (Figure 2). Among the hits, we observed that small molecules impacting on retinoic acid (RA), prostaglandin (PGE2) and nitric oxide (NO) signaling regulated liver formation. We found that RA receptor (RAR) agonists such as ATRA enhanced liver size during development, whereas the RA pathway inhibiter DEAB impaired liver development[32]. Related work by Negishi et al. has shown that RA signaling positively regulates liver development by inducing wnt2bb[63]. Our lab has published a series of studies that have uncovered a previously unappreciated role for PGE2 signaling in regulating cell fate and hepatic growth[64-67]. These studies have shown that PGE2 exposure stimulates Wnt signaling to enhance hepatic growth, whereas cyclooxygenase inhibition impairs liver development. These findings build on previous investigations by Jones and colleagues that showed that PGE2 regulates β-catenin stability in Apc-deficient zebrafish[68,69]. We recently identified that NO signaling is required for optimal liver development[70]. At the mechanistic level, NO did not regulate hepatic growth by classical cGMP-mediated vasodilation, but rather via S-nitrosoglutathione reductase (GSNOR)-regulated S-nitrosothiol (SNO) signaling. An innovative chemical screen was recently performed by Yin and colleagues to identify small molecules that regulate the number of hepatic stellate cells[11]. This study found that RA signaling or the inhibition of the VEGF pathway reduced the number of HSCs, whereas the RXR agonist methoprene acid increased HSC formation. Early studies by Farooq et al. found that the HDAC inhibitor valproic acid impaired liver development by inhibiting hepatoblast specification and differentiation[71]. Embryonic exposure to 5-azacytidine has a deleterious effect on biliary development due to DNA hypomethylation and stimulation of an interferon-γ mediated inflammatory response[72,73]. Recent work using the mTORC inhibitors Torin1 or rapamycin have revealed a critical role for mTOR signaling in Wnt-mediated liver hyperplasia[74]. Together, these studies show how small molecules can be used in conjunction with genetic models to gain new insights into aspects of liver development.
Liver injury models to study hepatic regeneration in zebrafish
Introduction to adult liver physiology
The anatomy of the adult zebrafish liver has been the focus of several recent reviews[75,76]. The zebrafish liver is a trilobar structure that lies at the anterior end of the intestinal tract. Similar to other vertebrates, zebrafish hepatocytes are arranged as bi-layered hepatic cords separated by sinusoids distributed radially around a central vein. Consistent with their directivity of absorption and secretion, zebrafish hepatocytes are polarized with the basal membrane adjacent to endothelial cells that encompass the sinusoids and the apical membrane forming the canaliculi that drain into the bile preductular cells. Unlike mammals, the zebrafish liver is not organized into zonated liver lobules with a central vein and portal triads. Instead, the intercellular biliary canaliculi anastomose into the biliary tree, which transports bile through the common hepatic duct into the gallbladder and ultimately the intestine.
Liver regeneration following partial hepatectomy
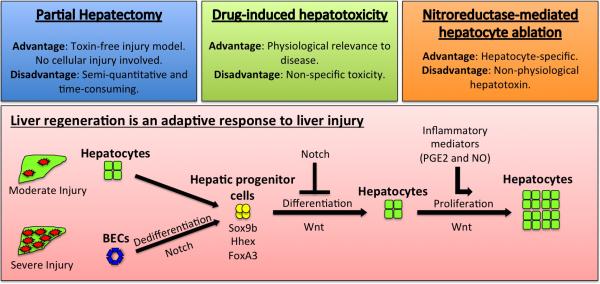
By far the most well established model of liver regeneration is the partial hepatectomy model[2]. Innovative studies by Sadler and colleagues characterized liver regeneration following one-third partial hepatectomy in adult zebrafish[53,54]. In this model, the inferior lobe running along the ventral side of the fish is surgically removed. In contrast to the compensatory hyperplasia observed in mammals, liver regeneration in zebrafish involves the recovery of the original lobular structure (Figure 3). By measuring inferior lobe regrowth the authors showed that liver regeneration was impaired in uhrf1-deficient (uhrf1+/−) zebrafish. In parallel studies, we used the one-third partial hepatectomy model to reveal that the Wnt pathway plays a key role in liver regeneration[23]. In these studies, inferior lobe regrowth was accelerated in apc+/− mutants and heat-shocked Tg(hsp70:wnt8) transgenics, whereas liver regeneration was stunted in heat-shocked Tg(hsp70:dnTCF) transgenics. Corresponding partial hepatectomy experiments revealed an elevated regenerative capacity in Apc+/min mice compared to wild-types, illustrating that the Wnt pathway plays an evolutionarily conserved role as a master regulator of liver regeneration. In addition, we recently revealed that PGE2 pathway impacts upon Wnt activity in the context of liver regeneration[64,65]. Building on developmental studies implicating S-nitrosothiol signaling in hepatic growth, we recently examined the impact of GSNOR inhibition on inferior lobe regeneration following partial hepatectomy and found that it accelerated hepatic regrowth, whereas inhibition of NO signaling stunted regeneration[70]. Studies by Izpisua Belmonte and colleagues examined the dynamics of liver regeneration following partial hepatectomy and found that BMP and FGF signaling are required for optimal liver regeneration[77]. In another report, Dovey et al. found that top2a is haploinsufficient for liver regrowth following partial hepatectomy[78]. Recent efforts have enriched our understanding of maladaptive regeneration following partial hepatectomy by revealing that def is required for the resolution of the fibrotic scar forms at the amputation site[79]. Together, these studies have revealed new insights into the regulation of hepatic growth following partial hepatectomy and laid the platform for future studies.
Figure 3.
Models of liver injury that can be used to study liver regeneration.
Adaptive and maladaptive regenerative responses to drug-induced hepatotoxicity
In the Goessling laboratory, we have developed a clinically relevant zebrafish model of acetaminophen (APAP) induced liver injury[66,70,80] (Figure 3). In the clinic, APAP (Tylenol) is the most common cause of acute liver failure and the only FDA-approved therapy, N-acetylcysteine (NAC), works as an antidote to limit oxidative damage[81]. We reasoned that drug screening in a zebrafish model of APAP toxicity could lead to the identification of small molecules that promote regeneration, which could be used in conjunction with NAC. A chemical modifier screen in APAP-exposed larvae identified prostaglandin (PGE2) as a novel compound that acts synergistically with NAC to inhibit liver damage and enhance survival[66]. At the mechanistic level, we found that PGE2 activates the Wnt pathway to stimulate regeneration in the context of APAP-induced liver injury. More recently, we determined that S-nitrosothiol signaling enhances survival after APAP-induced liver injury[70]. GSNOR inhibition provided hepatoprotection by causing sustained activation of the Nrf2 oxidative stress response pathway. GSNOR inhibition synergized with NAC and enhanced survival even after delayed treatment. GSNOR-deficient mice were resistant to APAP-induced liver injury, confirming the conservation of hepatoprotective properties provided by S-nitrosothiol signaling across vertebrates. These thought-provoking studies lend support to the hypothesis that mediators of inflammation (PGE2 and NO), which are generated upon liver injury, orchestrate regeneration (Figure 3).
Perhaps the best studied hepatotoxin in zebrafish is ethanol[82], which induces hepatic steatosis[83,84]. At the molecular level, ethanol metabolism generates ROS and induces the unfolded protein response (UPR) in the endoplasmic reticulum (ER), which activates the lipogenic transcription factors SREBP and ATF6[83,85-88]. Ethanol promotes maladaptive regeneration by activating HSCs, which proliferate and increase the deposition of extracellular matrix components leading to scar formation[11]. These studies illustrate how amenable the zebrafish system is to study the pathways regulating maladaptive regeneration in the context of ethanol-induced hepatotoxicity.
Liver regeneration following nitroreductase-mediated hepatocyte ablation
The most recent methodology adopted to induce liver injury in zebrafish is nitroreductase (NTR)-mediated hepatocyte ablation. This approach emerged from cancer gene therapy studies that used NTR as a prodrug gene suicide system to eliminate cancer cells exposed to the substrate CB1954[89]. Pioneering studies by the Stainier[90,91] and Parsons[92,93] laboratories provided the proof-of-principle that the NTR system could be adapted to ablate specific cell types in zebrafish (Figure 3). These studies took advantage of an alternate substrate metronidazole (MTZ), which was well tolerated and had greatly reduced off-target and bystander effects. Transgenic fish were developed for use in liver regeneration studies that expressed a Cyan Fluorescent Protein (CFP)-NTR fusion driven by the hepatocyte promoter fabp10a[90,91]. Upon exposure of the Tg(fabp10a:CFP-NTR) fish to MTZ the hepatocytes were rapidly ablated, while leaving the other cell types unaffected[90,91]. Recent studies have taken advantage of Tg(fabp10a:CFP-NTR) fish to show that the biliary epithelial cells (BECs) transdifferentiate to mature hepatocytes during liver regeneration following hepatocyte ablation[94,95]. These elegant studies used a Cre/lox approach to indelibly label BECs and follow their fate during liver regeneration after hepatocyte ablation. BECs dedifferentiated into hepatic progenitors and then differentiated to highly proliferative hepatocytes. Mechanistic studies with mutant NTR lines demonstrated that sox9b was necessary for BEC transdifferention[95], whereas wnt2bb was required to stimulate hepatocyte proliferation during regeneration[94]. Recent work by Huang et al. has further developed the MTZ-NTR-mediated hepatocyte ablation model to encompass maladaptive regeneration and fibrosis[96]. In this model, sustained fibrosis is achieved by pretreating larvae with ethanol, which activates hepatic stellate cells and leads to the deposition of laminin and collagen. The authors used the MTZ-NTR fibrosis model to demonstrate an antagonistic interactionn between Notch and Wnt in which stimulation of the Wnt pathway is required to promote liver regeneration. Together, these studies highlight the benefits of using the MTZ-NTR-mediated hepatocyte ablation model to enhance our understanding of the cellular and molecular basis of liver regeneration.
Conclusion
Liver research in the zebrafish field has expanded from its origins in embryonic development to a growing cadre of clinically relevant liver injury models that can be used to study regeneration. Fueled by the development of innovative chemical genetic approaches and in vivo imaging, zebrafish have greatly contributed to our understanding of the molecular underpinnings of liver development and regeneration. Ultimately, it is hoped that such studies will provide us with novel therapeutic approaches to treat liver disease.
Papers of Outstanding interest
-
94Choi, T.Y., Ninov, N., Stainier, D.Y. & Shin, D. Extensive conversion of hepatic biliary epithelial cells to hepatocytes after near total loss of hepatocytes in zebrafish. Gastroenterology 146, 776-788 (2014).
- -Together with ref 95, this manuscript describes the contribution of BECs to liver regeneration following hepatocyte ablation. This paper also demonstrates that Wnt2bb is required for hepatocyte regeneration.
-
95He, J., Lu, H., Zou, Q. & Luo, L. Regeneration of liver after extreme hepatocyte loss occurs mainly via biliary transdifferentiation in zebrafish. Gastroenterology 146, 789-800 e788 (2014).
- -Together with ref 94, this manuscript demonstrates that BECs transdifferentiate into hepatocytes during liver regeneration following severe hepatocyte ablation. This paper also shows that Sox9b is required for BEC transdifferentiaion.
-
96Huang, M. et al. Antagonistic interaction between Wnt and Notch activity modulates the regenerative capacity of a zebrafish fibrotic liver model. Hepatology 60, 1753-1766 (2014).
- -In this paper the authors further develop the NTR/MTZ hepatocyte ablation model to encompass fibrosis. The authors identify an antagonistic interplay between Wnt and Notch during liver regeneration.
Papers of special interest
-
1Howe, K. et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 496, 498-503 (2013).
- -This manuscript describes a high-quality sequence of the zebrafish genome and reveals similarities with the human genome.
-
10Wilkins, B.J., Gong, W. & Pack, M. A novel keratin18 promoter that drives reporter gene expression in the intrahepatic and extrahepatic biliary system allows isolation of cell-type specific transcripts from zebrafish liver. Gene Expr Patterns 14, 62-68 (2014).
- -In this paper, the authors explore biliary development using transgenic Tg(krt18:GFP) zebrafish and illustrate the utility of translating ribosome affinity purification (TRAP) transgenic lines to enrich hepatocyte or BEC mRNA.
-
12Yin, C., Evason, K.J., Asahina, K. & Stainier, D.Y. Hepatic stellate cells in liver development, regeneration, and cancer. J Clin Invest 123, 1902-1910 (2013).
- -This review focuses on recent insights into hepatic stellate cells using zebrafish as a model system.
-
21Lu, H., Ma, J., Yang, Y., Shi, W. & Luo, L. EpCAM is an endoderm-specific Wnt derepressor that licenses hepatic development. Dev Cell 24, 543-553 (2013).
- -In this paper, the authors show that EpCAM cell-autonomously cooperates with Wnt2bb signaling to dictate hepatic competence.
-
22Lancman, J.J. et al. Specification of hepatopancreas progenitors in zebrafish by hnf1ba and wnt2bb. Development 140, 2669-2679 (2013).
- -This manuscript demonstrates that Hnf1ba enables Wnt signaling in the foregut endoderm.
-
24So, J., Martin, B.L., Kimelman, D. & Shin, D. Wnt/beta-catenin signaling cell-autonomously converts non-hepatic endodermal cells to a liver fate. Biol Open 2, 30-36 (2013).
- -This paper demonstrates that endodermal cells are directly converted into hepatoblasts upon induction of Wnt8a.
-
64Nissim, S. et al. Prostaglandin E2 regulates liver versus pancreas cell-fate decisions and endodermal outgrowth. Dev Cell 28, 423-437 (2014).
- -This manuscript provides evidence that PGE2 may act as a morphogen regulating endodermal cell fate decisions.
-
70Cox, A.G. et al. S-nitrosothiol signaling regulates liver development and improves outcome following toxic liver injury. Cell Rep 6, 56-69 (2014).
- -This paper identifies that S-nitrosothiol signaling regulates liver growth during development and regeneration. The authors provide evidence that S-nitrosothiol signaling activates the Nrf2 pathway.
-
85Tsedensodnom, O., Vacaru, A.M., Howarth, D.L., Yin, C. & Sadler, K.C. Ethanol metabolism and oxidative stress are required for unfolded protein response activation and steatosis in zebrafish with alcoholic liver disease. Dis Model Mech 6, 1213-1226 (2013).
- -In this paper, the authors demonstrate the ethanol metabolism and oxidative stress play a key role in alcoholic liver disease.
-
87Howarth, D.L. et al. Activating transcription factor 6 is necessary and sufficient for alcoholic fatty liver disease in zebrafish. PLoS Genet 10, e1004335 (2014).
- -This manuscript finds that the unfolded protein response (UPR) sensor ATF6 is both necessary and sufficient to induce fatty liver disease.
Acknowledgements
AGC was the recipient of an Irwin Arias Postdoctoral Fellowship by the American Liver Foundation and a Harvard Digestive Diseases Center Pilot Feasibility Grant. .He is currently an American Liver Foundation Liver Scholar. WG is supported by NIH/NIDDK grants R01DK090311 and R01DK095721. WG is a Pew Scholar in the Biomedical Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, Collins JE, Humphray S, McLaren K, Matthews L, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496:498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Michalopoulos GK. Principles of liver regeneration and growth homeostasis. Compr Physiol. 2013;3:485–513. doi: 10.1002/cphy.c120014. [DOI] [PubMed] [Google Scholar]
- 3.Ober EA, Field HA, Stainier DY. From endoderm formation to liver and pancreas development in zebrafish. Mech Dev. 2003;120:5–18. doi: 10.1016/s0925-4773(02)00327-1. [DOI] [PubMed] [Google Scholar]
- 4.Field HA, Ober EA, Roeser T, Stainier DY. Formation of the digestive system in zebrafish. I. Liver morphogenesis. Dev Biol. 2003;253:279–290. doi: 10.1016/s0012-1606(02)00017-9. [DOI] [PubMed] [Google Scholar]
- 5.Her GM, Chiang CC, Chen WY, Wu JL. In vivo studies of liver-type fatty acid binding protein (L-FABP) gene expression in liver of transgenic zebrafish (Danio rerio). FEBS Lett. 2003;538:125–133. doi: 10.1016/s0014-5793(03)00157-1. [DOI] [PubMed] [Google Scholar]
- 6.Her GM, Yeh YH, Wu JL. 435-bp liver regulatory sequence in the liver fatty acid binding protein (L-FABP) gene is sufficient to modulate liver regional expression in transgenic zebrafish. Dev Dyn. 2003;227:347–356. doi: 10.1002/dvdy.10324. [DOI] [PubMed] [Google Scholar]
- 7.Sakaguchi TF, Sadler KC, Crosnier C, Stainier DY. Endothelial signals modulate hepatocyte apicobasal polarization in zebrafish. Curr Biol. 2008;18:1565–1571. doi: 10.1016/j.cub.2008.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korzh S, Pan X, Garcia-Lecea M, Winata CL, Wohland T, Korzh V, Gong Z. Requirement of vasculogenesis and blood circulation in late stages of liver growth in zebrafish. BMC Dev Biol. 2008;8:84. doi: 10.1186/1471-213X-8-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lorent K, Moore JC, Siekmann AF, Lawson N, Pack M. Reiterative use of the notch signal during zebrafish intrahepatic biliary development. Dev Dyn. 2010;239:855–864. doi: 10.1002/dvdy.22220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilkins BJ, Gong W, Pack M. A novel keratin 18 promoter that drives reporter gene expression in the intrahepatic and extrahepatic biliary system allows isolation of cell-type specific transcripts from zebrafish liver. Gene Expr Patterns. 2014;14:62–68. doi: 10.1016/j.gep.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yin C, Evason KJ, Maher JJ, Stainier DY. The basic helix-loop-helix transcription factor, heart and neural crest derivatives expressed transcript 2, marks hepatic stellate cells in zebrafish: analysis of stellate cell entry into the developing liver. Hepatology. 2012;56:1958–1970. doi: 10.1002/hep.25757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin C, Evason KJ, Asahina K, Stainier DY. Hepatic stellate cells in liver development, regeneration, and cancer. J Clin Invest. 2013;123:1902–1910. doi: 10.1172/JCI66369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wallace KN, Yusuff S, Sonntag JM, Chin AJ, Pack M. Zebrafish hhex regulates liver development and digestive organ chirality. Genesis. 2001;30:141–143. doi: 10.1002/gene.1050. [DOI] [PubMed] [Google Scholar]
- 14.Sun Z, Hopkins N. vhnf1, the MODY5 and familial GCKD-associated gene, regulates regional specification of the zebrafish gut, pronephros, and hindbrain. Genes Dev. 2001;15:3217–3229. doi: 10.1101/gad946701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shin D, Shin CH, Tucker J, Ober EA, Rentzsch F, Poss KD, Hammerschmidt M, Mullins MC, Stainier DY. Bmp and Fgf signaling are essential for liver specification in zebrafish. Development. 2007;134:2041–2050. doi: 10.1242/dev.000281. [DOI] [PubMed] [Google Scholar]
- 16.Noel ES, Casal-Sueiro A, Busch-Nentwich E, Verkade H, Dong PD, Stemple DL, Ober EA. Organ-specific requirements for Hdac1 in liver and pancreas formation. Dev Biol. 2008;322:237–250. doi: 10.1016/j.ydbio.2008.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ober EA, Verkade H, Field HA, Stainier DY. Mesodermal Wnt2b signalling positively regulates liver specification. Nature. 2006;442:688–691. doi: 10.1038/nature04888. [DOI] [PubMed] [Google Scholar]
- 18.Poulain M, Ober EA. Interplay between Wnt2 and Wnt2bb controls multiple steps of early foregut-derived organ development. Development. 2011;138:3557–3568. doi: 10.1242/dev.055921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klein C, Mikutta J, Krueger J, Scholz K, Brinkmann J, Liu D, Veerkamp J, Siegel D, Abdelilah-Seyfried S, le Noble F. Neuron navigator 3a regulates liver organogenesis during zebrafish embryogenesis. Development. 2011;138:1935–1945. doi: 10.1242/dev.056861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shin D, Weidinger G, Moon RT, Stainier DY. Intrinsic and extrinsic modifiers of the regulative capacity of the developing liver. Mech Dev. 2012;128:525–535. doi: 10.1016/j.mod.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu H, Ma J, Yang Y, Shi W, Luo L. EpCAM is an endoderm-specific Wnt derepressor that licenses hepatic development. Dev Cell. 2013;24:543–553. doi: 10.1016/j.devcel.2013.01.021. [DOI] [PubMed] [Google Scholar]
- 22.Lancman JJ, Zvenigorodsky N, Gates KP, Zhang D, Solomon K, Humphrey RK, Kuo T, Setiawan L, Verkade H, Chi YI, et al. Specification of hepatopancreas progenitors in zebrafish by hnf1ba and wnt2bb. Development. 2013;140:2669–2679. doi: 10.1242/dev.090993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goessling W, North TE, Lord AM, Ceol C, Lee S, Weidinger G, Bourque C, Strijbosch R, Haramis AP, Puder M, et al. APC mutant zebrafish uncover a changing temporal requirement for wnt signaling in liver development. Dev Biol. 2008;320:161–174. doi: 10.1016/j.ydbio.2008.05.526. [DOI] [PubMed] [Google Scholar]
- 24.So J, Martin BL, Kimelman D, Shin D. Wnt/beta-catenin signaling cell-autonomously converts non-hepatic endodermal cells to a liver fate. Biol Open. 2013;2:30–36. doi: 10.1242/bio.20122857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong PD, Munson CA, Norton W, Crosnier C, Pan X, Gong Z, Neumann CJ, Stainier DY. Fgf10 regulates hepatopancreatic ductal system patterning and differentiation. Nat Genet. 2007;39:397–402. doi: 10.1038/ng1961. [DOI] [PubMed] [Google Scholar]
- 26.Shin D, Lee Y, Poss KD, Stainier DY. Restriction of hepatic competence by Fgf signaling. Development. 2011;138:1339–1348. doi: 10.1242/dev.054395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naye F, Voz ML, Detry N, Hammerschmidt M, Peers B, Manfroid I. Essential roles of zebrafish bmp2a, fgf10, and fgf24 in the specification of the ventral pancreas. Mol Biol Cell. 2012;23:945–954. doi: 10.1091/mbc.E11-08-0664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan C, Zheng W, Gong Z. Zebrafish fgf10b has a Complementary Function to fgf10a in Liver and Pancreas Development. Mar Biotechnol (NY) 2014 doi: 10.1007/s10126-014-9604-x. [DOI] [PubMed] [Google Scholar]
- 29.Poulain M, Furthauer M, Thisse B, Thisse C, Lepage T. Zebrafish endoderm formation is regulated by combinatorial Nodal, FGF and BMP signalling. Development. 2006;133:2189–2200. doi: 10.1242/dev.02387. [DOI] [PubMed] [Google Scholar]
- 30.Tsai SM, Liu DW, Wang WP. Fibroblast growth factor (Fgf) signaling pathway regulates liver homeostasis in zebrafish. Transgenic Res. 2013;22:301–314. doi: 10.1007/s11248-012-9636-9. [DOI] [PubMed] [Google Scholar]
- 31.Tiso N, Filippi A, Pauls S, Bortolussi M, Argenton F. BMP signalling regulates anteroposterior endoderm patterning in zebrafish. Mech Dev. 2002;118:29–37. doi: 10.1016/s0925-4773(02)00252-6. [DOI] [PubMed] [Google Scholar]
- 32.Garnaas MK, Cutting CC, Meyers A, Kelsey PB, Jr., Harris JM, North TE, Goessling W. Rargb regulates organ laterality in a zebrafish model of right atrial isomerism. Dev Biol. 2012;372:178–189. doi: 10.1016/j.ydbio.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chung WS, Shin CH, Stainier DY. Bmp2 signaling regulates the hepatic versus pancreatic fate decision. Dev Cell. 2008;15:738–748. doi: 10.1016/j.devcel.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang H, Ruan H, Aw MY, Hussain A, Guo L, Gao C, Qian F, Leung T, Song H, Kimelman D, et al. Mypt1-mediated spatial positioning of Bmp2-producing cells is essential for liver organogenesis. Development. 2008;135:3209–3218. doi: 10.1242/dev.024406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lorent K, Yeo SY, Oda T, Chandrasekharappa S, Chitnis A, Matthews RP, Pack M. Inhibition of Jagged-mediated Notch signaling disrupts zebrafish biliary development and generates multi-organ defects compatible with an Alagille syndrome phenocopy. Development. 2004;131:5753–5766. doi: 10.1242/dev.01411. [DOI] [PubMed] [Google Scholar]
- 36.Matthews RP, Lorent K, Russo P, Pack M. The zebrafish onecut gene hnf-6 functions in an evolutionarily conserved genetic pathway that regulates vertebrate biliary development. Dev Biol. 2004;274:245–259. doi: 10.1016/j.ydbio.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 37.Matthews RP, Lorent K, Pack M. Transcription factor onecut3 regulates intrahepatic biliary development in zebrafish. Dev Dyn. 2008;237:124–131. doi: 10.1002/dvdy.21407. [DOI] [PubMed] [Google Scholar]
- 38.Delous M, Yin C, Shin D, Ninov N, Debrito Carten J, Pan L, Ma TP, Farber SA, Moens CB, Stainier DY. Sox9b is a key regulator of pancreaticobiliary ductal system development. PLoS Genet. 2012;8:e1002754. doi: 10.1371/journal.pgen.1002754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manfroid I, Ghaye A, Naye F, Detry N, Palm S, Pan L, Ma TP, Huang W, Rovira M, Martial JA, et al. Zebrafish sox9b is crucial for hepatopancreatic duct development and pancreatic endocrine cell regeneration. Dev Biol. 2012;366:268–278. doi: 10.1016/j.ydbio.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu L, Yin W, Xia J, Peng M, Li S, Lin S, Pei D, Shu X. An antiapoptotic role of sorting nexin 7 is required for liver development in zebrafish. Hepatology. 2012;55:1985–1993. doi: 10.1002/hep.25560. [DOI] [PubMed] [Google Scholar]
- 41.Zhang D, Golubkov VS, Han W, Correa RG, Zhou Y, Lee S, Strongin AY, Dong PD. Identification of Annexin A4 as a hepatopancreas factor involved in liver cell survival. Dev Biol. 2014;395:96–110. doi: 10.1016/j.ydbio.2014.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schaub M, Nussbaum J, Verkade H, Ober EA, Stainier DY, Sakaguchi TF. Mutation of zebrafish Snapc4 is associated with loss of the intrahepatic biliary network. Dev Biol. 2012;363:128–137. doi: 10.1016/j.ydbio.2011.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Curado S, Ober EA, Walsh S, Cortes-Hernandez P, Verkade H, Koehler CM, Stainier DY. The mitochondrial import gene tomm22 is specifically required for hepatocyte survival and provides a liver regeneration model. Dis Model Mech. 2010;3:486–495. doi: 10.1242/dmm.004390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ni TT, Lu J, Zhu M, Maddison LA, Boyd KL, Huskey L, Ju B, Hesselson D, Zhong TP, Page-McCaw PS, et al. Conditional control of gene function by an invertible gene trap in zebrafish. Proc Natl Acad Sci U S A. 2012;109:15389–15394. doi: 10.1073/pnas.1206131109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang J, Cui X, Wang L, Liu F, Jiang T, Li C, Li D, Huang M, Liao S, Wang J, et al. The mitochondrial thioredoxin is required for liver development in zebrafish. Curr Mol Med. 2014;14:772–782. doi: 10.2174/1566524014666140724103927. [DOI] [PubMed] [Google Scholar]
- 46.Chen J, Ruan H, Ng SM, Gao C, Soo HM, Wu W, Zhang Z, Wen Z, Lane DP, Peng J. Loss of function of def selectively up-regulates Delta113p53 expression to arrest expansion growth of digestive organs in zebrafish. Genes Dev. 2005;19:2900–2911. doi: 10.1101/gad.1366405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tao T, Shi H, Guan Y, Huang D, Chen Y, Lane DP, Chen J, Peng J. Def defines a conserved nucleolar pathway that leads p53 to proteasome-independent degradation. Cell Res. 2013;23:620–634. doi: 10.1038/cr.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davuluri G, Gong W, Yusuff S, Lorent K, Muthumani M, Dolan AC, Pack M. Mutation of the zebrafish nucleoporin elys sensitizes tissue progenitors to replication stress. PLoS Genet. 2008;4:e1000240. doi: 10.1371/journal.pgen.1000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Jong-Curtain TA, Parslow AC, Trotter AJ, Hall NE, Verkade H, Tabone T, Christie EL, Crowhurst MO, Layton JE, Shepherd IT, et al. Abnormal nuclear pore formation triggers apoptosis in the intestinal epithelium of elys-deficient zebrafish. Gastroenterology. 2009;136:902–911. doi: 10.1053/j.gastro.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koltowska K, Apitz H, Stamataki D, Hirst EM, Verkade H, Salecker I, Ober EA. Ssrp1a controls organogenesis by promoting cell cycle progression and RNA synthesis. Development. 2013;140:2013–140. doi: 10.1242/dev.093583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Y, Luo Y, Hong Y, Peng J, Lo L. Ribosome biogenesis factor Bms1-like is essential for liver development in zebrafish. J Genet Genomics. 2012;39:451–462. doi: 10.1016/j.jgg.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 52.Tien AL, Senbanerjee S, Kulkarni A, Mudbhary R, Goudreau B, Ganesan S, Sadler KC, Ukomadu C. UHRF1 depletion causes a G2/M arrest, activation of DNA damage response and apoptosis. Biochem J. 2011;435:175–185. doi: 10.1042/BJ20100840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sadler KC, Krahn KN, Gaur NA, Ukomadu C. Liver growth in the embryo and during liver regeneration in zebrafish requires the cell cycle regulator, uhrf1. Proc Natl Acad Sci U S A. 2007;104:1570–1575. doi: 10.1073/pnas.0610774104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chu J, Loughlin EA, Gaur NA, SenBanerjee S, Jacob V, Monson C, Kent B, Oranu A, Ding Y, Ukomadu C, et al. UHRF1 phosphorylation by cyclin A2/cyclin-dependent kinase 2 is required for zebrafish embryogenesis. Mol Biol Cell. 2012;23:59–70. doi: 10.1091/mbc.E11-06-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jacob V, Chernyavskaya Y, Chen X, Tan PS, Kent B, Hoshida Y, Sadler KC. DNA hypomethylation induces a DNA replication-associated cell cycle arrest to block hepatic outgrowth in uhrf1 mutant zebrafish embryos. Development. 2015 doi: 10.1242/dev.115980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qi F, Song J, Yang H, Gao W, Liu NA, Zhang B, Lin S. Mmp23b promotes liver development and hepatocyte proliferation through the tumor necrosis factor pathway in zebrafish. Hepatology. 2010;52:2158–2166. doi: 10.1002/hep.23945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cheng W, Guo L, Zhang Z, Soo HM, Wen C, Wu W, Peng J. HNF factors form a network to regulate liver-enriched genes in zebrafish. Dev Biol. 2006;294:482–496. doi: 10.1016/j.ydbio.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 58.Chang C, Hu M, Zhu Z, Lo LJ, Chen J, Peng J. liver-enriched gene 1a and 1b encode novel secretory proteins essential for normal liver development in zebrafish. PLoS One. 2011;6:e22910. doi: 10.1371/journal.pone.0022910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Niu X, Gao C, Jan Lo L, Luo Y, Meng C, Hong J, Hong W, Peng J. Sec13 safeguards the integrity of the endoplasmic reticulum and organogenesis of the digestive system in zebrafish. Dev Biol. 2012;367:197–207. doi: 10.1016/j.ydbio.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 60.Li Y, Farooq M, Sheng D, Chandramouli C, Lan T, Mahajan NK, Kini RM, Hong Y, Lisowsky T, Ge R. Augmenter of liver regeneration (alr) promotes liver outgrowth during zebrafish hepatogenesis. PLoS One. 2012;7:e30835. doi: 10.1371/journal.pone.0030835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu N, Li Z, Pei D, Shu X. Zfyve9a regulates the proliferation of hepatic cells during zebrafish embryogenesis. Int J Dev Biol. 2013;57:773–778. doi: 10.1387/ijdb.130065xs. [DOI] [PubMed] [Google Scholar]
- 62.Zhao X, Monson C, Gao C, Gouon-Evans V, Matsumoto N, Sadler KC, Friedman SL. Klf6/copeb is required for hepatic outgrowth in zebrafish and for hepatocyte specification in mouse ES cells. Dev Biol. 2010;344:79–93. doi: 10.1016/j.ydbio.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Negishi T, Nagai Y, Asaoka Y, Ohno M, Namae M, Mitani H, Sasaki T, Shimizu N, Terai S, Sakaida I, et al. Retinoic acid signaling positively regulates liver specification by inducing wnt2bb gene expression in medaka. Hepatology. 2010;51:1037–1045. doi: 10.1002/hep.23387. [DOI] [PubMed] [Google Scholar]
- 64.Nissim S, Sherwood RI, Wucherpfennig J, Saunders D, Harris JM, Esain V, Carroll KJ, Frechette GM, Kim AJ, Hwang KL, et al. Prostaglandin E2 regulates liver versus pancreas cell-fate decisions and endodermal outgrowth. Dev Cell. 2014;28:423–437. doi: 10.1016/j.devcel.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goessling W, North TE, Loewer S, Lord AM, Lee S, Stoick-Cooper CL, Weidinger G, Puder M, Daley GQ, Moon RT, et al. Genetic interaction of PGE2 and Wnt signaling regulates developmental specification of stem cells and regeneration. Cell. 2009;136:1136–1147. doi: 10.1016/j.cell.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.North TE, Babu IR, Vedder LM, Lord AM, Wishnok JS, Tannenbaum SR, Zon LI, Goessling W. PGE2-regulated wnt signaling and N-acetylcysteine are synergistically hepatoprotective in zebrafish acetaminophen injury. Proc Natl Acad Sci U S A. 2010;107:17315–17320. doi: 10.1073/pnas.1008209107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shan J, Schwartz RE, Ross NT, Logan DJ, Thomas D, Duncan SA, North TE, Goessling W, Carpenter AE, Bhatia SN. Identification of small molecules for human hepatocyte expansion and iPS differentiation. Nat Chem Biol. 2013;9:514–520. doi: 10.1038/nchembio.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Eisinger AL, Nadauld LD, Shelton DN, Peterson PW, Phelps RA, Chidester S, Stafforini DM, Prescott SM, Jones DA. The adenomatous polyposis coli tumor suppressor gene regulates expression of cyclooxygenase-2 by a mechanism that involves retinoic acid. J Biol Chem. 2006;281:20474–20482. doi: 10.1074/jbc.M602859200. [DOI] [PubMed] [Google Scholar]
- 69.Eisinger AL, Nadauld LD, Shelton DN, Prescott SM, Stafforini DM, Jones DA. Retinoic acid inhibits beta-catenin through suppression of Cox-2: a role for truncated adenomatous polyposis coli. J Biol Chem. 2007;282:29394–29400. doi: 10.1074/jbc.M609768200. [DOI] [PubMed] [Google Scholar]
- 70.Cox AG, Saunders DC, Kelsey PB, Jr., Conway AA, Tesmenitsky Y, Marchini JF, Brown KK, Stamler JS, Colagiovanni DB, Rosenthal GJ, et al. S-nitrosothiol signaling regulates liver development and improves outcome following toxic liver injury. Cell Rep. 2014;6:56–69. doi: 10.1016/j.celrep.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Farooq M, Sulochana KN, Pan X, To J, Sheng D, Gong Z, Ge R. Histone deacetylase 3 (hdac3) is specifically required for liver development in zebrafish. Dev Biol. 2008;317:336–353. doi: 10.1016/j.ydbio.2008.02.034. [DOI] [PubMed] [Google Scholar]
- 72.Matthews RP, Eauclaire SF, Mugnier M, Lorent K, Cui S, Ross MM, Zhang Z, Russo P, Pack M. DNA hypomethylation causes bile duct defects in zebrafish and is a distinguishing feature of infantile biliary atresia. Hepatology. 2011;53:905–914. doi: 10.1002/hep.24106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cui S, Eauclaire SF, Matthews RP. Interferon-gamma directly mediates developmental biliary defects. Zebrafish. 2013;10:177–183. doi: 10.1089/zeb.2012.0815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Valvezan AJ, Huang J, Lengner CJ, Pack M, Klein PS. Oncogenic mutations in adenomatous polyposis coli (Apc) activate mechanistic target of rapamycin complex 1 (mTORC1) in mice and zebrafish. Dis Model Mech. 2014;7:63–71. doi: 10.1242/dmm.012625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yao Y, Lin J, Yang P, Chen Q, Chu X, Gao C, Hu J. Fine structure, enzyme histochemistry, and immunohistochemistry of liver in zebrafish. Anat Rec (Hoboken) 2012;295:567–576. doi: 10.1002/ar.22416. [DOI] [PubMed] [Google Scholar]
- 76.Menke AL, Spitsbergen JM, Wolterbeek AP, Woutersen RA. Normal anatomy and histology of the adult zebrafish. Toxicol Pathol. 2011;39:759–775. doi: 10.1177/0192623311409597. [DOI] [PubMed] [Google Scholar]
- 77.Kan NG, Junghans D, Izpisua Belmonte JC. Compensatory growth mechanisms regulated by BMP and FGF signaling mediate liver regeneration in zebrafish after partial hepatectomy. FASEB J. 2009;23:3516–3525. doi: 10.1096/fj.09-131730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dovey M, Patton EE, Bowman T, North T, Goessling W, Zhou Y, Zon LI. Topoisomerase II alpha is required for embryonic development and liver regeneration in zebrafish. Mol Cell Biol. 2009;29:3746–3753. doi: 10.1128/MCB.01684-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhu Z, Chen J, Xiong JW, Peng J. Haploinsufficiency of Def activates p53-dependent TGFbeta signalling and causes scar formation after partial hepatectomy. PLoS One. 2014;9:e96576. doi: 10.1371/journal.pone.0096576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu LY, Fox CS, North TE, Goessling W. Functional validation of GWAS gene candidates for abnormal liver function during zebrafish liver development. Dis Model Mech. 2013;6:1271–1278. doi: 10.1242/dmm.011726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bunchorntavakul C, Reddy KR. Acetaminophen-related hepatotoxicity. Clin Liver Dis. 2013;17:587–607. viii. doi: 10.1016/j.cld.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 82.Howarth DL, Passeri M, Sadler KC. Drinks like a fish: using zebrafish to understand alcoholic liver disease. Alcohol Clin Exp Res. 2011;35:826–829. doi: 10.1111/j.1530-0277.2010.01407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Passeri MJ, Cinaroglu A, Gao C, Sadler KC. Hepatic steatosis in response to acute alcohol exposure in zebrafish requires sterol regulatory element binding protein activation. Hepatology. 2009;49:443–452. doi: 10.1002/hep.22667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Howarth DL, Yin C, Yeh K, Sadler KC. Defining hepatic dysfunction parameters in two models of fatty liver disease in zebrafish larvae. Zebrafish. 2013;10:199–210. doi: 10.1089/zeb.2012.0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tsedensodnom O, Vacaru AM, Howarth DL, Yin C, Sadler KC. Ethanol metabolism and oxidative stress are required for unfolded protein response activation and steatosis in zebrafish with alcoholic liver disease. Dis Model Mech. 2013;6:1213–1226. doi: 10.1242/dmm.012195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cinaroglu A, Gao C, Imrie D, Sadler KC. Activating transcription factor 6 plays protective and pathological roles in steatosis due to endoplasmic reticulum stress in zebrafish. Hepatology. 2011;54:495–508. doi: 10.1002/hep.24396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Howarth DL, Lindtner C, Vacaru AM, Sachidanandam R, Tsedensodnom O, Vasilkova T, Buettner C, Sadler KC. Activating transcription factor 6 is necessary and sufficient for alcoholic fatty liver disease in zebrafish. PLoS Genet. 2014;10:e1004335. doi: 10.1371/journal.pgen.1004335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vacaru AM, Di Narzo AF, Howarth DL, Tsedensodnom O, Imrie D, Cinaroglu A, Amin S, Hao K, Sadler KC. Molecularly defined unfolded protein response subclasses have distinct correlations with fatty liver disease in zebrafish. Dis Model Mech. 2014;7:823–835. doi: 10.1242/dmm.014472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bridgewater JA, Springer CJ, Knox RJ, Minton NP, Michael NP, Collins MK. Expression of the bacterial nitroreductase enzyme in mammalian cells renders them selectively sensitive to killing by the prodrug CB1954. Eur J Cancer. 1995;31A:2362–2370. doi: 10.1016/0959-8049(95)00436-x. [DOI] [PubMed] [Google Scholar]
- 90.Curado S, Anderson RM, Jungblut B, Mumm J, Schroeter E, Stainier DY. Conditional targeted cell ablation in zebrafish: a new tool for regeneration studies. Dev Dyn. 2007;236:1025–1035. doi: 10.1002/dvdy.21100. [DOI] [PubMed] [Google Scholar]
- 91.Curado S, Stainier DY, Anderson RM. Nitroreductase-mediated cell/tissue ablation in zebrafish: a spatially and temporally controlled ablation method with applications in developmental and regeneration studies. Nat Protoc. 2008;3:948–954. doi: 10.1038/nprot.2008.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pisharath H, Rhee JM, Swanson MA, Leach SD, Parsons MJ. Targeted ablation of beta cells in the embryonic zebrafish pancreas using E. coli nitroreductase. Mech Dev. 2007;124:218–229. doi: 10.1016/j.mod.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pisharath H, Parsons MJ. Nitroreductase-mediated cell ablation in transgenic zebrafish embryos. Methods Mol Biol. 2009;546:133–143. doi: 10.1007/978-1-60327-977-2_9. [DOI] [PubMed] [Google Scholar]
- 94.Choi TY, Ninov N, Stainier DY, Shin D. Extensive conversion of hepatic biliary epithelial cells to hepatocytes after near total loss of hepatocytes in zebrafish. Gastroenterology. 2014;146:776–788. doi: 10.1053/j.gastro.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.He J, Lu H, Zou Q, Luo L. Regeneration of liver after extreme hepatocyte loss occurs mainly via biliary transdifferentiation in zebrafish. Gastroenterology. 2014;146:789–800. e788. doi: 10.1053/j.gastro.2013.11.045. [DOI] [PubMed] [Google Scholar]
- 96.Huang M, Chang A, Choi M, Zhou D, Anania FA, Shin CH. Antagonistic interaction between Wnt and Notch activity modulates the regenerative capacity of a zebrafish fibrotic liver model. Hepatology. 2014;60:1753–1766. doi: 10.1002/hep.27285. [DOI] [PMC free article] [PubMed] [Google Scholar]