Abstract
In contrast to commonly reported human glioma xenograft animal models, GL261 murine glioma xenografts recapitulate nearly all relevant clinical and histopathologic features of the human disease. When GL261 cells are implanted intracranially in syngeneic C57BL/6 mice, the model has the added advantage of maintaining an intact immune microenvironment. Stable expression of luciferase in GL261 cells allows non-invasive cost effective bioluminescence monitoring of intracranial tumor growth. We have recently demonstrated that luciferase expression in GL261 cells does not affect the tumor growth properties, tumor cell immunomodulatory cytokine expression, infiltration of immune cells into the tumor, or overall survival of animals bearing the intracranial tumor. Therefore, it appears that the GL261 luciferase glioma model can be useful in the study of novel chemotherapeutic and immunotherapeutic modalities. Here we report the technique for generating stable luciferase expression in GL261 cells and how to study the in vitro and in vivo growth of the tumor cells by bioluminescence imaging.
Keywords: Medicine, Issue 107, immunocompetent animal, mouse, glioblastoma, intracranial xenograft, in vivo bioluminescence imaging, GL261
Introduction
Malignant glioma is the most common and most lethal human brain tumor. Even when treated with maximal surgical resection, radiotherapy, and chemotherapy, median survival remains 15-20 months at major brain tumor referral centers1, 2, 3. The most aggressive form of malignant glioma, glioblastoma, is characterized by mitotic figures, neovascularization, invasion into adjacent brain, and pseudopalisading necrosis. Orthotopic brain tumor xenografts using human brain tumor cells have served an essential function in neuro-oncology research and facilitated pre-clinical translation for testing of new agents, prior to entry into human clinical trials. Whereas most of human xenograft models recapitulate many features of the human disease, a fundamental limitation with all human-based xenograft model systems is the use of immunodeficient animals4.
The absence of an intact host response clearly modifies tumor growth both in terms of tumor morphology, and efficiency of engraftment. While certain assumptions are acceptable in the interpretation of results from xenografted models, achieving relevant conclusions in the area of therapeutic assessment is a challenge. Certain fundamental biologic features are likely to be similar in immunodeficient and immunocompetent animals, but others such as tumor associated inflammation, tissue invasion, response to injury are probably very different. In contrast, GL261 is a chemically induced murine glioma cell line that accurately recapitulates glioma when implanted into the brains of immunocompetent syngeneic C57BL/6 mice5. Similar to the human disease, intracranial tumor growth rapidly and reproducibly leads to neurologic symptoms and animal demise6-10.
Subcutaneous tumor growth can be directly measured. Intracranial tumor growth can only be measured with animal sacrifice or with costly imaging studies. Stable expression of the enzyme luciferase allows non-invasive and cost-effective intracranial bioluminescence imaging11. Bioluminescence of luciferase transfected GL261 cells correlates well with intracranial tumor growth. We have recently directly compared GL261 to luciferase modified GL261 cells and demonstrated no difference in in vitro growth and invasion, immunologic cytokine profile, in vivo survival of intracranial tumor bearing C57BL/6 mice, or immune cell infiltrate. This technique is applicable to glioblastoma preclinical studies which involve non-invasive monitoring of tumor growth.
Protocol
All procedures described below have been reviewed and approved by the Institutional Animal Use and Care Committee at Northwestern University and have been performed under sterilized conditions.
1. Modification of GL261 cells with Firefly Luciferase Expressing Reporter
NOTE: HIV-1-based lentiviral vectors express firefly luciferase (Fluc) under the control of the spleen focus-forming virus (SFFV) promoter. The optical reporter gene has been cloned into the vector plasmid pHRSIN-CSGW-dlNotI.
Maintain 293-T cells in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and GL261 cells in RPMI medium supplemented with 10% FBS and 1% penicillin-streptomycin at 37 °C in a humidified atmosphere containing 95% air and 5% carbon dioxide (CO2).
When the 293-T cell culture reaches 80% confluency in a T-75 flask, wash the cells once with phosphate buffer saline (PBS) without Ca2+ and Mg2+, incubate the cells with 3 ml of trypsin (0.05%) at 37 °C for 5 min, triturate with a 5 ml pipette by holding the flask slightly slanted and collect all the cells from the bottom of flask. Transfer the 3 ml of trypsinized cell suspension into a 15 ml conical tube, and add 7 ml of flesh RPMI medium (a total of 10 ml of the cell suspension).
Mix the cell suspension well with a 5 ml pipette. Take 100 μl of the cell suspension from the tube and count the number of the cells using hemocytometer. To do so, mix 100 μl of the cell suspension with 300 μl of trypan blue solution and insert 2 μl of the mixture into a hemocytometer. Count the number of the cells without blue staining from four separate views under a microscope. Average the cell number from each view, as this number represent 104 cells/ml.
Meanwhile, centrifuge the tube containing the cell suspension at 300 x g for 7 min. Aspirate the media and resuspend the cell pellets with fresh RPMI medium to make a GL261 cell suspension at density of 1.0 x 106/ml. Seed 2.5 x 106 GL261 cells with 2.5 ml of RPMI media into a T-175 flask with 27.5 ml DMEM (day 1).
After 24 hr, replace the medium with 20 ml fresh DMEM (day 2).
To produce the lentiviral vector, mix 54 μl transfection reagent with 2 ml serum free DMEM for 5 min. Add 3 μg of gag-pol, 3 μg of vesicular stomatitis virus G envelope (VSV-G), and 4.5 μg of Fluc (pSIN-Fluc) into the DMEM with transfection reagent, and incubate for 20 min at RT. Drop the entire DNA mixture into 293-T flask (T-175) and incubate at 37°C in a humidified chamber containing 95% air and 5% CO2 for 48 hr (day 2).
Add 10 ml of fresh DMEM at 24 hr after transfection (total volume of 293-T cell culture medium should be approximately 30 ml) (day 3).
To split GL261 cells into a 24-well plate, wash the cells once with PBS without Ca2+ and Mg2+, incubate the cells with 5 ml of trypsin (0.05%) at 37 °C for 5 min, triturate with a 5 ml pipette by holding the flask slightly slanted, collect all the cells from the bottom of flask, and count cells via hemocytometer as in step 1.2. Meanwhile, centrifuge the tube at 300 x g for 7 min. Aspirate the media and resuspend the cell pellets with fresh RPMI medium to make a GL261 cell suspension at density of 2.5 x 104/ml. Seed 2.5 x 104 GL261 cells with 1 ml of RPMI media in a 24-well plate (day 3).
Collect 30 ml of the lentiviral supernatant with 10 ml pipette from the 293-T flask (T-175) at 48 hr following transfection and transfer the supernatant into 50 ml conical tube. Aspirate the 30 ml lentiviral supernatant via a 50 ml syringe, through a 0.22 μm syringe filter, and aliquot the viral supernatant into 1 ml aliquots (day 4).
Replace the 1 ml of RPMI medium in GL261 cell plate (24-well) with 1 ml of the lentiviral supernatant and incubate at 37 °C in a humidified chamber (day 4).
After 48 hr of lentiviral infection, replace the medium with 1 ml of fresh RPMI (day 6).
After 24 hr, repeat the lentiviral infection of the GL261 cells (day 7).
After the 2nd 48 hr of lentiviral infection, replace the medium with 1 ml of fresh RPMI (day 9).
Continue to grow the luciferase modified GL261 cells (to be referred to as GL261.luc cells) and expand the cell culture to a T-75 flask.
When the GL261.luc cell culture reaches 70% confluency in a T-75 flask, harvest the cells by trypsinization and count by hemocytometer as in step 1.2. Plate the GL261.luc cell into a 24-well plate at graduating densities (1.0 x 106, 5.0 x 105, 2.5 x 105, 1.0 x 105, 5.0 x 104, 2.5 x 104 cells/well) and incubate at 37°C in a humidified chamber containing 95% air and 5% CO2 for 3 hr.
- Measure cellular luciferase activity relative to luciferase modified U-87 MG (U87.luc) cells using bioluminescence imaging (Figure 1).
- Start the image software (click icon on the desktop) and initialize the imaging system (click "Initialize" button). Initialization will begin automatically and may take several min for the system check-up and the camera to cool down to -90 °C.
- Add 25 μl of luciferin (D-Luciferin potassium salt, 150 mg/kg) into each well.
- Incubate the cells with luciferin for 1 min at RT and image the plate for 10 sec. To set up the imaging system, select "Luminescent", "10 sec" exposure time, and "Medium" binning. Place the 24-well plate on the imaging station and click the "Acquire" button to start image.
- After the image is successfully acquired, you will see an image window and tool palette on the screen. Click "ROI (regions of interest) Tools" and select 6 x 4 from the slit icon. Create 6 x 4 slits to cover the area of signal on the image of the 24-well plate and click the measurements tab (pencil icon). Observe a window with a table of ROI measurements as unit of photons per second per steradian per square cm (photons/sec/sr/cm2).
- Use U87.luc cells, previously modified with the same lentivirus used here for GL261 modification11, as a control for assessing luciferase activity in transduced GL261.luc cells.
2. In Vitro Proliferation Assay
When the GL261 and GL261.luc cell cultures reach 70% confluency in a T-75 flask, harvest the both cell lines by trypsinization and count as in step 1.2, and plate cells into a 96-well plate at a density of 1500 cells in 80 μl of RPMI medium per well using a multichannel pipette to minimize time and pipetting error. Seed each cell line into 20 wells total. To limit the evaporation in the wells populated by cells, fill empty wells with 100 μl of fresh RPMI medium.
Assess the cellular proliferation using a colorimetric cell proliferation assay. Here we use the MTS cell proliferation assay. Using a multi-channel pipette, add 20 μl of MTS reagent into each well of the 96-well plate that contains cells.
Incubate the plate at 37°C in a humidified chamber containing 95% air and 5% CO2 for 3 hr. Measure absorbance at 490 nm using a microplate reader. This is repeated at days 1, 2, 4, and 7, after plating. To normalize absorbance values, the values of each day are divided by the corresponding readings obtained at day 1 (Figure 2).
3. Tumor Cell Implantation
Note: Autoclave all equipment. Prepare surgical area by spraying a disinfectant (2% chlorhexidine solution) and cover with absorbent drapes. In order to maintain sterile conditions, wear sterile surgical gloves during surgery, and divide the surgical area into two areas by color tape. Area 1 is labeled "Clean" and contains sterilized supplies; Area 2 is labeled "Dirty" and contains used materials.
Prior to animal surgery, prepare a cell suspension for intracranial injection. Harvest cells from a 70% confluent T-75 flask by trypsinization and count cells via hemocytometer as in step 1.2, and make a cell suspension at a density of 1.0 x 105 cells/µl in Hanks' Balanced Salt Solution without Ca2+ and Mg2+ (HBSS).
Anesthetize mice with intraperitoneal injection of 200 μl mixture containing 100 mg/kg of ketamine and 10 mg/kg of xylazine in 0.9% saline. To confirm proper anesthetization, pinch the toe of the mouse. If the mouse is completely under anesthesia, the mouse should not respond to the stimulus.
Administer 0.1 mg/kg buprenorphine via subcutaneous injection to relieve the post-operative pain.
Shave the top of the head of the animals using clippers and clean with a cotton-tip applicator dipped in 2% chlorhexidine solution. Apply eye ointment to maintain adequate lubrication during the surgery.
Make a sagittal incision approximately 1 cm long over the parieto-occipital bone using a sterile scalpel. To visualize the bregma, wipe the surface of the skull with a cotton-tip applicator soaked in a 3% hydrogen peroxide solution.
Drill the skull with a sterile 25-G needle to make a small hole 3 mm to the right of the bregma and just behind the coronal suture.
To ensure that intracranial injection site is to a depth of 3 mm from the skull, cut 3 mm off the pointed end of a 20 μl pipette tip using scissors and insert a syringe into the pipette tip.
Insert the syringe perpendicularly to the hole previously created, and slowly inject the tumor cell suspension containing 3.0 x 105 cells in 3 µl HBSS, over a 1 min period. Leave the needle in place for another minute, and then slowly pull out the needle.
Swab the skull with cotton-tip applicator dipped in 3% hydrogen peroxide and the 2% chlorhexidine solution. Dry the surface of skull with the cotton-tip applicator. Cut out the sterile bone wax approximately 3mm3 in size and attach the bone wax on the end side of the cotton-tip applicator. Apply the bone wax to cover the hole with the cotton-tip applicator.
Draw the each side of the scalp together over the skull using forceps, and staple to close the wound. Clean the wound with a cotton-tip dipped in 2% chlorhexidine solution.
Monitor all mice post-operatively until they become ambulant and retain normal activity. Typically, recovery time is approximately 30 min. Do not return the mice to the cage with other animals until they have fully recovered from anesthesia.
4. Bioluminescence Imaging of GL261 Tumor Growth
Spray a generous amount of disinfectant (0.05% dimethyl ammonium chloride solution) on a clean paper towel. Use the disinfectant-soaked paper towel to thoroughly wipe the black sheet, nose cones and divider inside the imaging chamber where the animals will be in contact with the device. Thoroughly wipe all surfaces with a clean, dry paper towel. Repeat this once more and wait 5 min.
Initialize the imaging station as in step 1.14.1.
Anesthetize the mouse with a 300 μl mixture containing 200 μl of Ketamine/Xylazine and 100 μl of luciferin (D-Luciferin potassium salt, 150 mg/kg) by intraperitoneal injection.
Wait 10 min after injection and confirm if the mice are completely under anesthesia by pinching the tail. To set up the imaging system, select "Luminescent", "Auto" exposure time, and "Medium" binning. Place the mice on the imaging station and click the "Acquire" button to start image. Ensure that the time after injection is consistent with each imaging session.
After the image is successfully acquired, an image window and tool palette should appear. Click "ROI Tools" and select "auto" from the circle icon. To define ROIs, encircle the area of signal on the image, and then take measurements of the ROI as unit of photons/sec/sr/cm2.
Take the mice out of the imaging chamber and monitor the mice until they become ambulant and retain normal activity. Typically recovery time is approximately 15 min. Do not return the mice to the cage with other animals until they have fully recovered from anesthesia.
- Monitor tumor growth by bioluminescence imaging twice a week until animals present the following symptoms: weight loss (loss of more than 20% relative to the weight prior to implantation), inappetance (complete anorexia for 24 hr), and lack of sustained purposeful response to gentle stimuli (weak attempt to get up), at which time they should be euthanized. Euthanize animals with these symptoms using a CO2 chamber followed by cervical dislocation.
- Place mice into the euthanasia chamber (do not overcrowded the chamber) and inflow compressed CO2 for 5-6 min. Observe that all animals are unconsciousness and not breathing after approximately 5 min.
- Take the mice out of the chamber. Ensure that the heart is not beating by feeling the chest between your thumb and forefinger and check that there is no blink reflex.
- Restrain the mouse in a prone position on a flat surface and grasp the back of the neck at the base of skull with one hand and base of the tail with your thumb and first finger of the other hand.
- Restrain the head and quickly pull the tail backward. Ensure that skull is separated from the first cervical vertebra using your thumb and first finger.
Normalize luminescence values for each day against corresponding readings obtained at the first day of bioluminescence imaging as in step 2.3 (Figure 3A).
For statistical analysis, generate survival curves using the Kaplan-Meier estimator12 and compare the differences between survival curves using a log-rank test13.
Representative Results
GL261 cells were infected with luciferase containing lentivirus as described above in steps 1. In vitro bioluminescence imaging demonstrates robust luciferase expression similar to levels in the positive control U87.luc human glioblastoma cell line (Figure 1). As expected, uninfected GL261 cells demonstrate no background luciferase expression. This step is simple yet critical to perform prior to further studies using the infected cell line to confirm stable luciferase expression.
Luciferase expression after intracranial implantation.
Before intracranial implantation, we demonstrated no difference in vitro growth rate of GL261.luc compared to GL261 cells (Figure 2). For intracranial growth and survival analysis, cells were injected into the brains of C57BL/6 mice as per protocol step 3. Tumor growth in mice bearing GL261.luc tumors was serially analyzed for bioluminescence imaging using protocol step 4 and demonstrated detectable tumor growth (Figure 3A). Mice bearing GL261.luc tumors rapidly and consistently became moribund and were euthanized. Importantly, there is no difference in overall survival between GL261.luc and GL261 tumor bearing animals (Figure 3B). In vivo bioluminescence imaging can therefore be used to monitor response to experimental glioblastoma treatments. Rapid reliable tumor growth and short overall survival can yield large amounts of data in a relatively short amount of time.
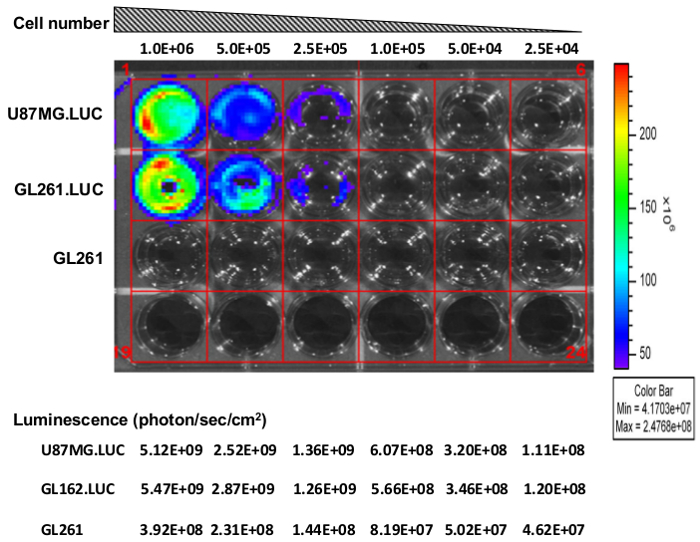 Figure 1. Luciferase activity in GL261.luc cells. U87.luc, GL261.luc and GL261 (negative control) cells were plated at densities of 1.0 x 106, 5.0 x 105, 2.5 x 105, 1.0 x 105, 5.0 x 104, and 2.5 x 104 cells/well from left to right. Luminescence (photon/sec/sr/cm2) was measured by the lumina imaging station. Please click here to view a larger version of this figure.
Figure 1. Luciferase activity in GL261.luc cells. U87.luc, GL261.luc and GL261 (negative control) cells were plated at densities of 1.0 x 106, 5.0 x 105, 2.5 x 105, 1.0 x 105, 5.0 x 104, and 2.5 x 104 cells/well from left to right. Luminescence (photon/sec/sr/cm2) was measured by the lumina imaging station. Please click here to view a larger version of this figure.
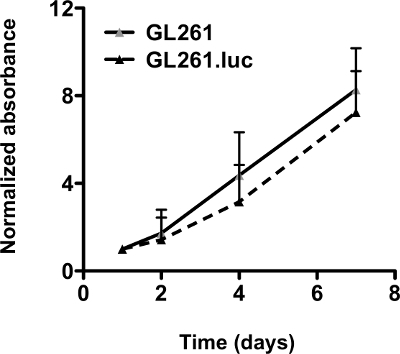 Figure 2. Luciferase expression does not affect GL261 cell proliferation. GL261.luc cells do not cause adifference in proliferation as demonstrated by MTS cell proliferation assay. The graph shows fold increase, relative to day 1, as determined by comparing the average absorbance value (mean ± SEM) at thespecified time point to the average value at day 1 (unpaired t-test values for comparisons between each cell line: P = 0.7796)14. Error bars represent the average values (mean± SEM) from quadruplicate samples on each day. Please click here to view a larger version of this figure.
Figure 2. Luciferase expression does not affect GL261 cell proliferation. GL261.luc cells do not cause adifference in proliferation as demonstrated by MTS cell proliferation assay. The graph shows fold increase, relative to day 1, as determined by comparing the average absorbance value (mean ± SEM) at thespecified time point to the average value at day 1 (unpaired t-test values for comparisons between each cell line: P = 0.7796)14. Error bars represent the average values (mean± SEM) from quadruplicate samples on each day. Please click here to view a larger version of this figure.
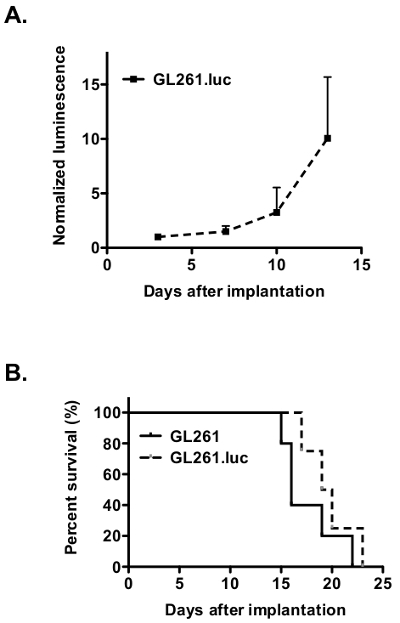 Figure 3. Luciferase expression dose not affect on in vivo tumor growth and animal survival. (A) Bioluminescence monitoring demonstrates progressive growth of intracranial GL261.luc tumor in C57BL/6 mice. (B) Kaplan-Meier survival analysis demonstrates no difference in overall survival for mice implanted intracranially with either GL261 (solid line) or GL261.luc cells (dashed line). Please click here to view a larger version of this figure.
Figure 3. Luciferase expression dose not affect on in vivo tumor growth and animal survival. (A) Bioluminescence monitoring demonstrates progressive growth of intracranial GL261.luc tumor in C57BL/6 mice. (B) Kaplan-Meier survival analysis demonstrates no difference in overall survival for mice implanted intracranially with either GL261 (solid line) or GL261.luc cells (dashed line). Please click here to view a larger version of this figure.
Discussion
GL261 murine glioma cells, when implanted intracranially into syngeneic immunocompetent C57BL/6 mice, offer several advantages compared to human glioma xenograft animal models. Many of xenografted tumors grow as encapsulated lesions that do not accurately recapitulate the invasive human disease. In contrast, GL261 tumor not only demonstrates invasion into adjacent brain, but also neovascularization, mitotic figures, and profound necrosis7. Most importantly when studying tumor immunology or immunotherapeutic strategies15, 16, the C57BL/6 mouse retains an intact immune system8. Previous studies have shown robust expression of the immunosuppressive cytokine TGF-β and intracranial tumors contain immunosuppressive T-regulatory cells similar to human glioblastoma9, 10.
The simple technique described here allows for stable expression of luciferase by GL261 cells. We have recently reported similar in vitro and in vivo growth of GL261.luc cells compared to GL261 cells, in addition to similar tumor histologic characteristics and immune cell infiltrate17. GL261.luc cells stably express luciferase which catalyzes the oxidation of the substrate luciferin converting chemical energy to photons and therefore detectable light. Luciferin can be safely administered to animals and crosses the blood brain barrier after intraperitoneal or intravenous injection. In small research animals such as mice, the bioluminescence can be detected externally in a non-invasive manner11. Therefore, tumor growth can be serially assessed without the need for animal sacrifice or costly MRI or CT imaging.
The critical steps in the protocol involve, not only the lentiviral transduction, but also verification of stable expression of luciferase in vitro before beginning any in vivo experiments. Loss of transduction may require repeat lentivirus infection. Introduction of an antibiotic resistance gene into the expression vector could also be used to select for luciferase expression. Meticulous technique is required for intracranial implantation to reproducibly place a consistent number of cells in a precise anatomic location to allow comparison of different treatment groups. A major difference between the current study and previous studies of luciferase expressing GL261 cells is the use of a free hand technique for implanting cells compared to the use of a stereotactic frame18. We have previously demonstrated consistent implantation results with the free hand technique19. The advantage of the free hand technique is the ability to perform high throughput analyses of different treatment agents. In our hands, we can implant approximately 60 animals in 1 hr.
After mastering the technique, the future applications of the technique are limitless. As GL261 demonstrates elevated mitoses and rapid tumor growth similar to human glioblastoma, anti-proliferative treatments can be evaluated. Similarly, anti-invasive and anti-angiogenic therapies can be used as the GL261 tumors are invasive and angiogenic. Caution should be used when assessing the effect of chemotherapeutic agents aimed at human targets. Compared to previous studies utilizing human glioblastoma xenografts expressing luciferase20, this would be considered a limitation of our model. Also, the effect of immunotherapeutic strategies of tumor growth can be studied in the GL261 xenografted model.
Disclosures
There are no conflicts of interest to disclose.
Acknowledgments
We would like to thank Rajwant Kaur for technical assistance. We would like to thank Maxwell Tom for assistance with lentivirus preparation. This work was supported by the Reza and Georgianna Khatib Endowed Chair in Skull Base Tumor Surgery at UCSF, and the Michael J. Marchese Professor and Chair at Northwestern University.
References
- Clark AJ, et al. Neurosurgical management and prognosis of patients with glioblastoma that progresses during bevacizumab treatment. Neurosurgery. 2012;70(2):361–370. doi: 10.1227/NEU.0b013e3182314f9d. [DOI] [PubMed] [Google Scholar]
- Stupp R, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- Grossman SA, et al. Survival of patients with newly diagnosed glioblastoma treated with radiation and temozolomide in research studies in the United States. Clin Cancer Res. 2010;16(8):2443–2449. doi: 10.1158/1078-0432.CCR-09-3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sughrue ME, et al. Immunological considerations of modern animal models of malignant primary brain tumors. J Transl Med. 2009;7(84) doi: 10.1186/1479-5876-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausman JI, Shapiro WR, Rall DP. Studies on the chemotherapy of experimental brain tumors: development of an experimental model. Cancer Res. 1970;30(9):2394–2400. [PubMed] [Google Scholar]
- Zagzag D, Zhong H, Scalzitti JM, Laughner E, Simons JW, Semenza GL. Expression of hypoxia-inducible factor 1alpha in brain tumors: association with angiogenesis, invasion, and progression. Cancer. 2000;88(11):2606–2618. [PubMed] [Google Scholar]
- Cha S, et al. Dynamic, contrast-enhanced perfusion MRI in mouse gliomas: correlation with histopathology. Magn Reson Med. 2003;49(5):848–855. doi: 10.1002/mrm.10446. [DOI] [PubMed] [Google Scholar]
- Ksendzovsky A, et al. Investigation of immunosuppressive mechanisms in a mouse glioma model. J Neurooncol. 2009;93(1):107–114. doi: 10.1007/s11060-009-9884-6. [DOI] [PubMed] [Google Scholar]
- Biollaz G, et al. Site-specific anti-tumor immunity: differences in DC function, TGF-beta production and numbers of intratumoral Foxp3+ Treg. Eur J Immunol. 2009;39(5):1323–1333. doi: 10.1002/eji.200838921. [DOI] [PubMed] [Google Scholar]
- El Andaloussi A, Han Y, Lesniak MS. Prolongation of survival following depletion of CD4 + CD25+ regulatory T cells in mice with experimental brain tumors. J Neurosurg. 2006;105(3):430–437. doi: 10.3171/jns.2006.105.3.430. [DOI] [PubMed] [Google Scholar]
- Dinca EB, et al. Bioluminescence Monitoring of Intracranial Glioblastoma Xenograft Response to Primary and Salvage Temozolomide Therapy. J. Neurosurg. 2007;107(3):610–616. doi: 10.3171/JNS-07/09/0610. [DOI] [PubMed] [Google Scholar]
- Kaplan EL, Meier P. Non-parametric estimation from incomplete observations. J. Am. Stat. Assoc. 1958;53:457–481. [Google Scholar]
- Peto R, Peto J. Asymptotically efficient rank invariant procedures. J. R. Stat. Soc. Ser. A. Stat. Soc. 1972;135:185207. [Google Scholar]
- Armitage P, Berry G, Matthews JNS. Statistical Methods in Medical Research. 4th ed. Oxford: Blackwell Science; 2001. [Google Scholar]
- Newcomb EW, et al. The combination of ionizing radiation and peripheral vaccination produces long-term survival of mice bearing established invasive GL261 gliomas. Clin Cancer Res. 2006;12(15):4730–4737. doi: 10.1158/1078-0432.CCR-06-0593. [DOI] [PubMed] [Google Scholar]
- Pellegatta S, et al. Neurospheres enriched in cancer stem-like cells are highly effective in eliciting a dendritic cell-mediated immune response against malignant gliomas. Cancer Res. 2006;66(21):10247–10252. doi: 10.1158/0008-5472.CAN-06-2048. [DOI] [PubMed] [Google Scholar]
- Clark AJ, et al. Stable luciferase expression does not alter immunologic or in vivo growth properties of GL261 murine glioma cells. J Transl Med. 2014;12:345–34. doi: 10.1186/s12967-014-0345-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelwahab MG, Sankar T, Preul MC, Scheck AC. Intracranial implantation with subsequent 3D in vivo bioluminescent imaging of murine gliomas. J Vis Exp. 2011. p. e3403. [DOI] [PMC free article] [PubMed]
- Ozawa T, James CD. Establishing intracranial brain tumor xenografts with subsequent analysis of tumor growth and response to therapy using bioluminescence imaging. J Vis Exp. 2010. [DOI] [PMC free article] [PubMed]
- Baumann BC, Dorsey JF, Benci JL, Joh DY, Kao GD. Stereotactic intracranial implantation and in vivo bioluminescent imaging of tumor xenografts in a mouse model system of glioblastoma multiforme. J Vis Exp. 2012. [DOI] [PMC free article] [PubMed]


