Abstract
Zebrafish are an important model organism with inherent advantages that have the potential to make zebrafish a widely applied model for the study of energy homeostasis and obesity. The small size of zebrafish allows for assays on embryos to be conducted in a 96- or 384-well plate format, Morpholino and CRISPR based technologies promote ease of genetic manipulation, and drug treatment by bath application is viable. Moreover, zebrafish are ideal for forward genetic screens allowing for novel gene discovery. Given the relative novelty of zebrafish as a model for obesity, it is necessary to develop tools that fully exploit these benefits. Herein, we describe a method to measure energy expenditure in thousands of embryonic zebrafish simultaneously. We have developed a whole animal microplate platform in which we use 96-well plates to isolate individual fish and we assess cumulative NADH2 production using the commercially available cell culture viability reagent alamarBlue. In poikilotherms the relationship between NADH2 production and energy expenditure is tightly linked. This energy expenditure assay creates the potential to rapidly screen pharmacological or genetic manipulations that directly alter energy expenditure or alter the response to an applied drug (e.g. insulin sensitizers).
Keywords: Molecular Biology, Issue 107, Energy Expenditure, Oxygen Consumption, Danio rerio, Zebrafish, Drug Discovery, High-throughput, Obesity, Metabolic Syndrome
Introduction
The mouse is currently the predominant model for obesity research. The short generation interval and genetic tools available in the mouse have been unmatched to date. However, the zebrafish also has a short generation interval (3 - 4 months) and surpasses even the mouse in ease of genetic manipulation 1,2. The zebrafish maintains nearly 90% of mammalian genes, while far exceeding the mouse in number of offspring and potential for use in genetic and drug screens 3.
To tap the potential of the zebrafish model for studies in obesity, assays must be developed to investigate factors that influence body weight regulation, including energy expenditure. As metabolites are processed through β-oxidation and the tricarboxylic acid cycle oxygen is consumed and NADH2 is produced. Thus, NADH2 is a direct indicator of the flux of metabolites through metabolic pathways (metabolite oxidation). In poikilotherms, H+ leak through the inner mitochondrial membrane, which uncouples NADH2 oxidation from ATP synthesis, is 4 - 5 fold lower than in homeotherms 4. Accordingly, in zebrafish NADH2 is very tightly linked to ATP production through oxidative phosphorylation. Herein, we describe an assay that measures NADH2 production in larval zebrafish as a proxy for energy expenditure5.
Oxygen consumption is the gold standard for measuring energy expenditure. Yet, to best take advantage of the high throughput potential of the zebrafish, assays of energy expenditure must be amenable to high-throughput. Oxygen consumption systems that depend on a closed chamber circulating system are limited in throughput by the number of chambers available6. Open air O2 consumption/CO2 production assays have also been applied in the zebrafish5,7. These open air 96-well plate based systems are amenable to high throughput. Unfortunately, gas exchange with the environment limits sensitivity of these assays. We recently published the application of an assay that monitors NADH2 using the redox indicator alamarBlue5. This assay overcomes the limitations in throughput and sensitivity common to analyses of oxygen consumption in the zebrafish.
The zebrafish is becoming an increasingly important model for studies of whole body energy homeostasis. In part, because zebrafish are amenable to use in forward genetic screens and drug screens. Moreover, targeted genetic manipulation, including knockdown and knock-in, can be quickly applied. We have previously shown that this assay can be combined with bath drug application and genetic knockdown or knockout to identify compounds and genes that alter metabolic rate5. Moreover, this assay is designed to exploit the high throughput advantages inherent to the zebrafish.
Protocol
Note: This protocol follows the guidelines of the University of Arizona Office for the Responsible Conduct of Research and has been approved by the Institutional Animal Care and Use Committee
1. Zebrafish Breeding and Embryo Maintenance
Prepare E3 embryo medium8. To Make 60x stock solution, dissolve 34.8 g NaCl, 1.6 g KCl, 5.8g CaCl2●2H2O, and 9.78 g MgCl2●6H2O in 1.95 L ddiH2O. Adjust pH to 7.2 using NaOH and HCl Adjust volume to 2 L using ddiH2O.
To prepare 1x E3 embryo medium, dilute 16.5 ml 60x stock in 1 L ddiH20. Add 100 µl 1% methylene blue.
- Breeding
- House Male and Female brood stock (7 - 18 months of age) separately to maximize fecundity.
- Turn the lights off 1 - 2 hr before on the day before eggs are desired. Set up mating (1 male and 1 - 2 females) in crossing cages as previously described9. Crossing cages allow for the eggs to escape predation as they pass through the perforated base of the tank insert in which the adult fish are held.
- Optional: For timed breeding, (as would be needed for studies involving morpholino injection), separate the male and female fish by a clear plastic divider., Remove this in the morning to result in timed laying of the eggs and fertilization. For morpholino studies, perform power calculations to assess the number of embryonic fish required per treatment. Without an estimate of the magnitude of effect or variation, a preliminary trial can be run with an n = 8.
Pour the eggs from the crossing cage into a petri dish (100 - 150 embryos/10 cm petri dish). Allow the eggs to settle to the bottom of the dish and pour off the water. Remove any remaining water using a fine tip disposable transfer pipette. Add back E3 embryo medium made in step 1.1.
Make a flame polished disposable pipette for future egg and fish transfer. Using scissors, cut a graduated disposable transfer pipette at the 0.25 ml graduation. To remove rough edges, flame polish the cut tip over a Bunsen burner.
Maintain embryonic fish to 4 days post-fertilization (dpf) in 10 cm petri dishes in zebrafish E3 embryo medium at 28.5 ○C. Twice each day, use a flame polished graduated disposable transfer pipette to remove any unfertilized eggs or dead embryos (opaque white appearance). Once each day, replace E3 embryo medium. Pour off E3 embryo medium, remove remaining liquid using a fine tip disposable transfer pipette, and add back pre-warmed (28.5 ○C) E3 embryo medium.
2. Prepare Assay Solution
Prepare the assay solution according to Table 1. Sterile filter assay solution using a 0.22 µM filter. Prior to assay initiation, warm the sterile assay solution in a 28.5 ○C water bath.
| Ingredient | % of Total | Volume (µl/10 ml) |
| 60x E3 Embryo Medium | 1.667 | 166.7 |
| Double deionized (ddi) H20 | 96.233 | 9623.3 |
| Alamar Blue | 1 | 100 |
| 400 mM NaOH | 1 | 100 |
| DMSO | 0.1 | 10 |
Table 1. Assay Medium
3. Perform Assay
Rinse zebrafish embryos 3 times with sterile filtered E3 embryo medium. Combine all viable zebrafish embryos from the clutch into a beaker. Sterile filter 75 ml E3 embryo medium. Using a fine tip disposable transfer pipette, remove as much of the E3 embryo medium from the zebrafish embryos as possible. Add back 20 ml sterile filtered E3 embryo medium. Remove and replace E3 embryo medium 3 times.
Plate 1 fish into each of 93 wells of a 96-well black sided clear bottom plate. Transfer the embryonic fish to the plate by individually capturing an embryo within a flame polished graduated plastic disposable transfer pipette (step 1.5) and gently ejecting into the well. Transfer E3 embryo medium taken from the beaker containing rinsed fish into the 3 blank wells. This ensures that blank wells have been exposed to the exact same water as the wells that contain fish. Both wells that contain fish and blank wells should be at least ½ full (200 µl) at the conclusion of this step.
Replace E3 embryo medium with assay solution in all 96 wells, including blank wells. This step should be performed 1 column at a time for each of the 12 columns in a 96 well plate. Using a fine tip disposable transfer pipette, remove E3 embryo medium from every well in one column of the plate (8 wells). Take care to not touch the zebrafish embryo. Add 300 µl of assay solution to each well from which water was removed. Repeat the removal of E3 embryo medium and replacement with assay solution for all 12 columns of the plate.
- Optional: To test the metabolic rate change in response to a compound, make a solution that is 100 times (100X) the desired treatment concentration. Of note, ensure that the 100x solution does not exceed 10% DMSO to limit the final DMSO concentration to no more than 0.1%. Add 3 µl of the 100x solution to each of the treatment wells. NOTE: Perform power calculations to assess the number of treatment wells. However, without an estimate of the magnitude of response or variation, a preliminary trial can be run in 8 compound treated and 8 vehicle treated control wells that contain fish.
- Add 3 µl of vehicle control to control (no drug treatment) wells that contain fish. To ensure that the fluorescent response to compound is a result of actions within the fish, apply drug and vehicle treatments to 3 blank wells in a 96 well plate.
Read fish filled plate on fluorescent plate reader with excitation wavelength at 530 nM and emission wavelength at 590 nM.
Place plate into humidified incubator set to 28.5 ○C (maintain plate in darkness throughout assay to avoid photobleaching of assay solution). The assay can be run for durations of time that range from 1 hr to 24 hr. The duration of assay depends on the objective of the study. For testing the response to a short acting non-stable compound a short duration may be most suitable. Yet, for stable compound tests or tests of genetic effects, maximizing assay duration is preferable to exploit the advantages associated with the cumulative signal.
At a pre-determined time point (see step 3.6 to help determine timing) read fluorescence of fish-filled plate again with the same settings as step 3.5.
4. Assess the Relative Change in Fluorescence Associated with Treatment
Calculate change in fluorescence of each well by subtracting the fluorescence at time 0 from the fluorescence at the conclusion of the study. Wells without fish (blanks) should have values near 0, far below values from wells that contain fish. Change in Fluorescence = Fluorescence at conclusion - Fluorescence at time 0
To ascertain the relative change in fluorescence, calculate the average change in fluorescence for control wells then divide the change in fluorescence of each well by the average change in fluorescence of the control wells. Of note, controls could be either vehicle controls or genetic controls.
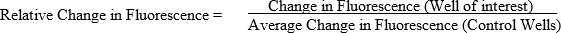
Representative Results
Assay solution does not increase fluorescence in the absence of embryonic fish (Figure 1). However, the assay is highly sensitive to small changes in metabolic rate within the well. A single fish within a well creates a signal significantly different from blank wells within 1 hr (P<0.0001). Figure 2 provides a visual representation of signals generated by embryonic fish after 24 hr of incubation. For the purpose of this picture clear sided wells were used. Yet, black sided wells are preferred for assay performance to prevent leaching of the fluorescent signal from nearby wells.
Because the NADH2 induced reduction of alamarBlue is non-reversible, the signal accumulates with time. This allows for small changes to be amplified. To show that differences accumulate and thus increase with time, we monitored the relative change in fluorescence induced by 1 to 5 fish at 1, 2, and 4 hr (Figure 3). At each number of fish/well the relative change in fluorescence significantly increased with time (P<0.001). More importantly, because signal accumulates with time, the magnitude of differences in fluorescence change that result from manipulating the number of fish/well is more robust at 4 hr than at 1 hr of incubation. Thus, by increasing the duration of exposure we can enhance the ability to detect differences associated with a given treatment.
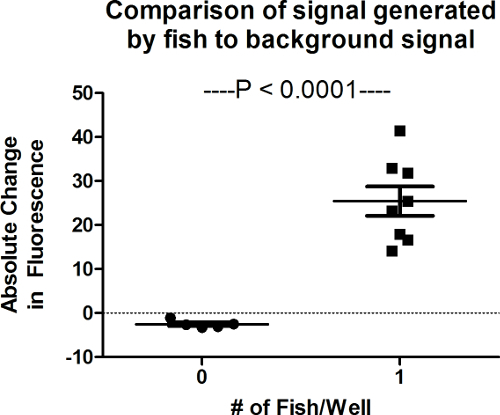 Figure 1. Comparison of Noise to Signal. A detectable change in fluorescence is generated by 1 zebrafish in a 96 well plate at 1 hr. Wells that do not contain fish (blanks) generate little signal. Error bars represent SEM (n = 5 - 8). Please click here to view a larger version of this figure.
Figure 1. Comparison of Noise to Signal. A detectable change in fluorescence is generated by 1 zebrafish in a 96 well plate at 1 hr. Wells that do not contain fish (blanks) generate little signal. Error bars represent SEM (n = 5 - 8). Please click here to view a larger version of this figure.
 Figure 2. Visual Representation of Assay Plate at 24 hr. Representative picture of results obtained using this assay. Pink wells are indicative of fish with a high metabolic rate, purple wells a moderate metabolic rate, and blue wells a low metabolic rate. Please click here to view a larger version of this figure.
Figure 2. Visual Representation of Assay Plate at 24 hr. Representative picture of results obtained using this assay. Pink wells are indicative of fish with a high metabolic rate, purple wells a moderate metabolic rate, and blue wells a low metabolic rate. Please click here to view a larger version of this figure.
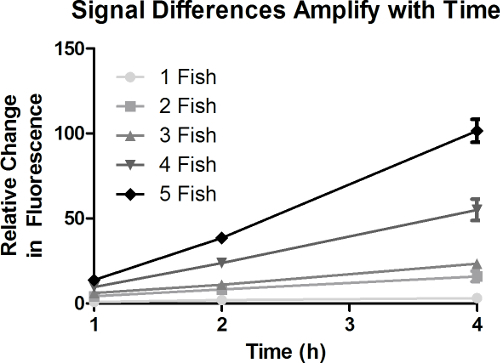 Figure 3. Signal Differences Amplify With Time. The difference in fluorescent signal generated by varying the number of fish/well increases with time. Because signal accumulates with time, the magnitude of fluorescence change diverges with time betweensamples that differ in rate of signal generation. Error bars represent SEM (n = 7).
Figure 3. Signal Differences Amplify With Time. The difference in fluorescent signal generated by varying the number of fish/well increases with time. Because signal accumulates with time, the magnitude of fluorescence change diverges with time betweensamples that differ in rate of signal generation. Error bars represent SEM (n = 7).
Discussion
Contamination with bacteria or fungi will greatly limit application of this assay. The methylene blue in the E3 embryo medium limits the possibility of fungal contamination. Throughout embryo rearing it is critical that care is taken to remove dead embryos twice each day. Additionally, it is essential to thoroughly rinse the embryos with sterile E3 embryo medium on the day of assay initiation. These 2 steps limit the potential for bacterial contamination of the embryos. The inclusion of blank wells described in step 3.2 allows for detection of any bacterial contamination. Bacterial contamination that yields a measurable signal is rare. However, if the blank wells do show a robust signal, the assay should be repeated with care taken to maintain embryo cleanliness. If the user expects that contamination is arising prior to embryo collection, the embryos may be bleached as previously described (p. 22) 10. Alternatively, if the experimenter suspects that contamination is occurring after collection the user may use E2 with penicillin and streptomycin to rear the embryos to 4 dpf (p. 23)10.
Although this assay is valuable for application in embryos, we have not tested for validity in adult zebrafish. We suspect that functionality of this assay would be limited in adult fish by poor tissue penetration of AlamarBlue. Moreover, in feeding fish the gut microbiota may produce a profound signal, not indicative of the fish metabolism. Thus, this assay is restricted to use in yolk feeding embryonic fish. The relative inactivity of embryonic fish provides a unique advantage, as this limits the variability that would be associated with activity induced energy expenditure 11. Thus this assay is a unique tool to screen the metabolic rate response to genetic or pharmacological manipulation in a whole animal system.
The zebrafish is rapidly becoming a leading model for the metabolic perturbations that accompany obesity. In fact, there are published models of both transgenic and diet-induced zebrafish obesity 12,13. Similar to mouse models, diet induced obesity in the zebrafish increases serum triglycerides and hepatic lipid accumulation12. Furthermore, overfed zebrafish have elevated fasting glucose, suggesting glucose intolerance 14. Using the assay described here we have shown that 24 hr of pretreatment with Metformin, an insulin sensitizer, increases the metabolic rate response to insulin treatment 5. Thus, the zebrafish may prove to be an ideal whole animal model to discover drugs that enhance insulin sensitivity (embryo) and to establish their effectiveness in alleviating obesity related insulin resistance (adult).
The development of genetic and diet induced obese zebrafish models that mimic the phenotypes observed in obese mammals creates the impetus for initiating zebrafish studies focused on metabolic disease. High throughput assays are essential for fully appreciating the value of the zebrafish model to studies of energy homeostasis. Innovative tools designed to assess phagic drive will allow for screens focused on energy intake, while high throughput lipid storage assays will lead to studies on whole body lipid homeostasis 15-17. The assay we describe here provides a high throughput tool to assess the role of genes, compounds or their interactions on whole body energy expenditure.
Disclosures
The authors have no competing financial interests to disclose.
Acknowledgments
The authors would like to thank Drs. Roger Cone and Chao Zhang for their contributions to validating the application of this assay as previously published. This work was supported by NIH 1F32DK082167-01 (BJR).
References
- Jao LE, Wente SR, Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. PNAS. 2013;110:13904–13909. doi: 10.1073/pnas.1308335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y, Hisano Y, Kawahara A, Higashijima S. Efficient generation of knock-in transgenic zebrafish carrying reporter/driver genes by CRISPR/Cas9-mediated genome engineering. Sci. Rep. 2014;4:6545. doi: 10.1038/srep06545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieschke GJ, Currie PD. Animal models of human disease: zebrafish swim into view. Nat. Rev. Genet. 2007;8:353–367. doi: 10.1038/nrg2091. [DOI] [PubMed] [Google Scholar]
- Brand MD, Couture P, Else PL, Withers KW, Hulbert AJ. Evolution of energy metabolism. Proton permeability of the inner membrane of liver mitochondria is greater in a mammal than in a reptile. Biochem. J. 1991;275(Pt 1):81–86. doi: 10.1042/bj2750081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renquist BJ, Zhang C, Williams SY, Cone RD. Development of an assay for high-throughput energy expenditure monitoring in the zebrafish. Zebrafish. 2013;10:343–352. doi: 10.1089/zeb.2012.0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingston RJ. A volumetric respirometer for long-term studies of small aquatic animals. J.Mar. Biol. Assoc. U.K. 1968;48:485–497. [Google Scholar]
- Makky K, Duvnjak P, Pramanik K, Ramchandran R, Mayer AN. A whole-animal microplate assay for metabolic rate using zebrafish. J. Biomol. Screen. 2008;13:960–967. doi: 10.1177/1087057108326080. [DOI] [PubMed] [Google Scholar]
- E3 medium (for zebrafish embryos) Cold Spring Harb. Protoc. 2011. http://cshprotocols.cshlp.org/citmgr?gca=protocols%3B2011%2F10%2Fpdb.rec66449.
- Nasiadka A, Clark MD. Zebrafish breeding in the laboratory environment. ILAR J. 2012;53:161–168. doi: 10.1093/ilar.53.2.161. [DOI] [PubMed] [Google Scholar]
- Brand M, Granato M, Nusslein-Volhard C. In: Zebrafish, A practical approach. Nusslein-Volhard C, Dahm R, editors. Oxford University Press; 2002. [Google Scholar]
- Fry FEJ, Hart JS. Cruising speed of goldfish in relation to water temperature. J. Fish. Res. Board Can. 1948;7:169–175. [Google Scholar]
- Oka T, et al. Diet-induced obesity in zebrafish shares common pathophysiological pathways with mammalian obesity. BMC physiol. 2010;10(21) doi: 10.1186/1472-6793-10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Cone RD. Creation of a genetic model of obesity in a teleost. FASEB J. 2007;21:2042–2049. doi: 10.1096/fj.06-7503com. [DOI] [PubMed] [Google Scholar]
- Maddison LA, Joest KE, Kammeyer RM, Chen W. Skeletal muscle insulin resistance in zebrafish induces alterations in beta-cell number and glucose tolerance in an age and diet dependent manner. Am. J. physiol. Endocrinol. 2015. [DOI] [PMC free article] [PubMed]
- Flynn EJ, 3rd, Trent CM, Rawls JF. Ontogeny and nutritional control of adipogenesis in zebrafish (Danio rerio) J. lipid res. 2009;50:1641–1652. doi: 10.1194/jlr.M800590-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada Y, Hirano M, Nishimura Y, Tanaka T. A high-throughput fluorescence-based assay system for appetite-regulating gene and drug screening. PloS one. 2012;7:e52549. doi: 10.1371/journal.pone.0052549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tingaud-Sequeira A, Ouadah N, Babin PJ. Zebrafish obesogenic test: a tool for screening molecules that target adiposity. J. lipid res. 2011;52:1765–1772. doi: 10.1194/jlr.D017012. [DOI] [PMC free article] [PubMed] [Google Scholar]


