Abstract
Metastatic disease is the spread of malignant tumor cells from the primary cancer site to a distant organ and is the primary cause of cancer associated death 1. Common sites of metastatic spread include lung, lymph node, brain, and bone 2. Mechanisms that drive metastasis are intense areas of cancer research. Consequently, effective assays to measure metastatic burden in distant sites of metastasis are instrumental for cancer research. Evaluation of lung metastases in mammary tumor models is generally performed by gross qualitative observation of lung tissue following dissection. Quantitative methods of evaluating metastasis are currently limited to ex vivo and in vivo imaging based techniques that require user defined parameters. Many of these techniques are at the whole organism level rather than the cellular level 3-6. Although newer imaging methods utilizing multi-photon microscopy are able to evaluate metastasis at the cellular level 7, these highly elegant procedures are more suited to evaluating mechanisms of dissemination rather than quantitative assessment of metastatic burden. Here, a simple in vitro method to quantitatively assess metastasis is presented. Using quantitative Real-time PCR (QRT-PCR), tumor cell specific mRNA can be detected within the mouse lung tissue.
Keywords: Medicine, Issue 107, mammary tumor, lung, metastasis, Real-Time PCR, xenograft, genetically engineered mouse models
Introduction
QRT-PCR analysis is proposed as a method for assessing tumor metastasis. This method is proposed as an alternative for users that are interested in evaluating metastasis but may not have access to specific equipment such as in vivo imaging equipment or a fluorescence capable stereoscope. A discussion of commonly used methods is presented followed by a demonstration for how QRT-PCR analysis can be used either as a separate or as a companion method to evaluate metastasis. This procedure has the potential to provide a quantitative analysis of metastatic burden.
Standard methods of gross analysis, including visualization of lungs under a stereomicroscope as well as serial sectioning followed by hematoxylin and eosin (H&E) staining of lung tissue, are quantifiable but rely heavily on user defined parameters for counting 2-5. When evaluating whole lungs using a stereomicroscope, only large surface metastases are visible and analysis requires the investigator to have reasonable knowledge of lung anatomical structure to determine what constitutes a metastatic lesion. Fluorescent labeling of tumor cells with a marker such as GFP and use of a stereomicroscope that contains a light cube with the appropriate excitation/emission maxima (e.g. near 470/510 nm for GFP) assists in this process, but only surface tumor nodules are detectable. Additionally, fluorescence from blood contamination, which is visible under the same parameters as GFP, may lead to false identification of possible metastatic lesions.
Sectioning of the lung followed by H&E staining to visualize lung metastasis is a useful method to evaluate micrometastases and other microscopic processes including immune cell infiltration but often requires use of the entire lung tissue for paraffin embedding, sectioning, and staining procedures. Therefore, downstream procedures are not ideal following this method. Although quantifiable, this procedure requires the investigator to evaluate a large number of stained lung sections per animal to ensure that the analysis accounts for the entire 3D structure of the lung. Consequently, this type of examination is time consuming, can lead to counting error, and analysis relies heavily on investigator discretion.
Several in vivo imaging techniques (e.g. MRI, PET, SPECT) are currently used to perform or test biological processes in experimental rodent models 8. In vivo bioluminescent imaging is a common method used to acquire a gross view of metastasis 9. This technique is generally applied to evaluate the presence of luciferase reporter activity due to the accumulation of tumor cells, which are engineered to contain a luciferase response element, that reside in specific organs like the mammary gland after tumor cell implantation and the lung upon spontaneous metastasis 10. Visualization of luciferase reporter activity is induced by the presence of luciferin substrate (D-luciferin). Luciferase catalyzes the oxidative decarboxylation of D-luciferin to oxyluciferin generating bioluminescence. While informative, this method is limited by several factors including substrate stability (i.e. short half-life), adequate distribution of substrate which depends on how it is delivered to experimental animals, and low sensitivity of detection 9. A main merit to this technique is that it is non-invasive, can be performed on live animals, and can lead to the detection of tumor cell metastases in multiple organs that may not have been normally harvested at dissection 9,10.
One positive aspect of in vivo imaging techniques is that the lung tissue is undisturbed allowing for secondary procedures like paraffin embedding or as presented here, QRT-PCR analysis. However, because QRT-PCR is theoretically a more sensitive measure of detection, gross evaluation may not reveal low numbers of tumor cells present in the lung. While useful, the imaging techniques described above can be substituted or supplemented with the QRT-PCR method currently described. QRT-PCR has the potential to provide a sensitive measure of tumor-derived mRNA within a lung.
Protocol
The protocol follows the guidelines and animal care standards of the Medical University of South Carolina (MUSC) and its Institutional Animal Care and Use Committee (IACUC).
1. Lung Dissection
- Mammary tumor dissection (optional depending on study goals and whether primary tumor analysis or tail vein injection was performed).
- Euthanize mouse using a lethal dose of isoflurane anesthesia according to IACUC and university regulation. Confirm euthanization by surgical dislocation.
- On foam board, pin with dissection pins by placing mouse on its back with limbs spread.
- Spray mouse with 70% ethanol.
- Using forceps, grasp the lower portion of the animal at the center.
- Using scissor, cut upward toward the neck through the skin, being careful not to pierce the peritoneal cavity of the mouse.
- Cut back skin at the limbs to reveal tumor tissue.
- Dissect out tumor tissue and preserve by preferred method, such as freezing or fixation, for further analysis (not discussed further in this protocol).
- Lung tissue dissection.
- If tumor tissue was harvested as described above, pin down any excess skin.
- Using forceps, grasp peritoneum at lower end of the animal and begin cutting upwards toward the base of the neck.
- Carefully cut along the left and right sides of the rib cage being careful to avoid severing any blood vessels that may bleed into the thoracic cavity. Remove the rib bones by cutting to the left and right through the diaphragm and upper portions of the rib cage.
- Using forceps grasp the trachea of the mouse and pull forward.
- Sever the trachea with dissection scissors and remove lungs from mouse. Note: The heart may remain attached to the lung. If this occurs, carefully dissect heart away from lungs being careful to collect all five lobes (see Figure 1).
- Wash gently with PBS to remove excess blood.
- Gross evaluation of lung tissue.
- Option 1: In vivo imaging.
- For luciferase imaging, ~10 min prior to imaging, give animals a 150 mg/kg dose of luciferin by intraperitoneal (i.p.) injection. Image lungs in vivo in anesthetized animals or upon removal after dissection (Figure 2) 5.
- Option 2: Gross evaluation.
- Examine lungs under stereo microscope to visualize tumor nodules. Note: If cells are GFP labeled, a fluorescence capable stereomicroscope containing a light cube with the appropriate excitation/emission maxima (e.g. near 470/510 nm) to detect GFP may be used to examine lungs stereoscopically for fluorescence prior to processing.
- If lungs are to be analyzed immediately, proceed to 'RNA Isolation' section. Otherwise, snap freeze tissue using liquid nitrogen or dry ice. Store at -80 °C.
2. RNA Isolation
Note: For the representative analysis, RNA was isolated using a RNA isolation kit (see Materials List). While the current analysis uses one specific manufacturer's product, a variety of isolation kits are available from several reputable vendors. Additionally, non-kit based centrifugation methods are also an option. cDNA should also be prepared from a positive control RNA sample, such as a cell line or plasmid, for standard curve analysis. Alternatively, a positive control specific for the primer probe set may be purchased (see Materials List).
If using previously frozen tissue, gather frozen tissue on dry ice.
- Homogenize tissue using one of the following methods such as: mortar and pestle grinding performed by hand over dry ice after freezing tissue in liquid nitrogen; tissue homogenizer or mechanical dissociation device per manufacturer's suggestion; sonication, or other preferred method. Note: For the representative analysis, the preferred method of homogenization is sonication.
- For the analysis presented in Figure 3, prepare lysis buffer (provided in RNA isolation kit) and 2-mercaptoethanol at a ratio of 20 µl 2-mercaptoethanol for every 1 ml of lysis buffer. Resuspend tissue in 300 µl lysis buffer supplemented with 2-mercaptoethanol for every 30 g of tissue and sonicate for 10 sec with sonicator set at 30% amplitude.
- If using mortar and pestle, resuspend ground tissue in 300 µl lysis buffer after grinding.
Add 10 µl proteinase K to 590 µl TE buffer (10 mM Tris-HCl, pH 8.0; 1 mM EDTA). Add 600 µl proteinase K solution to every 300 µl (i.e. for every 30 mg) of tissue. Incubate for 10 min at RT. Note: Digestion time may vary.
Centrifuge samples at ≥13,000 g for 10 min to remove debris.
Transfer supernatant to new tubes.
Add 450 µl ethanol (96%-100%) for every 900 µl of supernatant to precipitate RNA.
Transfer 700 µl of the lysate/ethanol mix to the column.
Centrifuge samples at ≥13,000 g for 1 min to bind RNA to column.
Discard liquid waste and add remaining supernatant to column and spin again.
Once all the lysate is spun through the column, add 350 µl of Wash Buffer 1 to the upper portion of the column and centrifuge samples at ≥13,000 g for 30 sec.
Discard liquid from column and replace upper portion.
Prepare DNase by mixing 5 units of DNase I enzyme with 5 µl of 10x DNase and 40 µl of DNase/RNase free water per sample (total volume of 50 µl/sample).
Add 50 µl of DNase mix to each column and incubate for 15 min at RT.
Add 350 µl of Wash Buffer 1 to column and centrifuge samples at ≥13,000 g for 30 sec.
Discard liquid from column and replace upper portion.
Add 600 µl of Wash Buffer 2 to the column and spin 30 sec at ≥13,000 g.
Discard liquid from column and replace upper portion.
Add 250 µl of Wash Buffer 2 and centrifuge for 2 min at ≥13,000 g.
Put part of column into new collection tube and discard old collection tube.
Add 50-100 µl of RNase/DNase free water to column and incubate for 1 min.
Spin for 1 min at ≥13,000 g.
Re-run collected eluate through column to increase RNA yield.
For immediate use, keep samples on ice and proceed to quantitation. For use later, snap freeze on dry ice or liquid nitrogen. Store at -80 °C freezer until needed.
3. First Strand Synthesis
Note: For the representative analysis, the reverse transcriptase (RT) reaction was performed a First Strand cDNA synthesis kit designed for QRT-PCR (see Materials List).
If samples were previously collected, thaw tubes on ice.
Measure RNA quantity using a spectrophotometer.
- Using 0.5-2 µg of total RNA, perform first strand synthesis using reverse transcriptase to generate cDNA stock.
- In sterile, RNase/DNase-free PCR tubes/plates, prepare the reaction mix (Table 1). Mix gently and centrifuge. Program a standard PCR machine (Table 2).
Dilute finished reaction to desired volume (generally 50-100 µl) using sterile nuclease-free water.
4. Real-time PCR
Note: For the representative analysis, SYBR green was used. However, any preferred method can be substituted at the user's discretion.
- Using a positive control cDNA, set up a standard curve for each primer set.
- For the analysis shown in Figure 3, prepare the standard curve for human HER2 using the manufacturer recommended positive control cDNA (see Materials List). For mouse gapdh, use the negative control lung cDNA to generate a standard curve. For each probe, serially dilute the cDNA 1:4 to generate 5 standards.
Calculate the number of total samples, which should include the standard curve and experimental samples. Note: For the analysis described here, 2-3 experimental replicates per sample were analyzed. Standard curve analysis was performed to generate a simple linear regression model that can be applied to determine the relative amount of HER2 and gapdh per sample to calculate final normalized quantity. This analysis will also ensure that the primer efficiency is adequate for the analysis at hand.
- In a sterile, RNase/DNase-free tube, prepare master mix (Table 3).
- Calculate master mix amount per sample. Scale up for more than one sample. Use a master mix for each experimental gene of interest, as well as an internal control such as gapdh. Use primers for gapdh at a final concentration of 200 nM. Note: The internal control is particularly useful when trying to distinguish between human and mouse cell origin. Note: Primer concentration for HER2 was optimized and determined by the manufacturer.
Prepare test plate containing 1 µl cDNAs corresponding to each standard or experimental sample. Allocate at least 1 cDNA sample/well for each primer probe. Add 19 µl of master mix per well for each primer probe. Note: For the representative data in Figure 3, the cDNA samples were analyzed in duplicate for each primer probe and graphed as the average of the two samples.
Run reaction in QRT-PCR machine using recommended or previously tested PCR protocol. See Table 4 for the PCR protocol used for the representative data in Figure 3 and Figure 4.
Representative Results
Beyond the time it takes to perform the initial inoculation of tumor cells into the experimental animal and if performing, primary in vivo tumor analysis, the tissue harvest, RNA isolation, and QRT-PCR analysis is a 1-2 day procedure (Figure 1).
An example of gross analysis is bioluminescent imaging to evaluate tumor cells within the lung tissue. Here, a mammary tumorigenesis experiment was performed to evaluate whether an investigational agent that targets the gap junction protein connexin43, called ACT1, would impair spontaneous metastasis of 4T1-luc mammary tumor cells to the lung. After several weeks of treatment, animals that received placebo or the test agent were injected with luciferin and sacrificed. Upon dissection, lungs were visualized for luciferase bioluminescence. From the representative images it appears that lungs from the drug treated animals are negative for luciferase activity which indicates that no tumor cells are present in the lung, suggesting an inhibition of metastasis after ACT1 treatment (Figure 2A). The main purpose of these representative images is to demonstrate that bioluminescent imaging is an appropriate method of detection for metastasis. However, upon detailed investigation of H&E sections of lungs from animals treated with ACT1, micrometastases were identified that were not illuminated by the luciferase imaging (Figure 2B), perhaps suggesting that this type of imaging may not be sensitive enough to detect low numbers of tumor cells embedded within the lung. Ideally, a companion analysis for QRT-PCR would be performed to confirm these results.
The QRT-PCR analysis presented here uses a probe specific for human HER2 to detect human tumor cells in the lung from HER2+ JIMT-1 cells that were originally transplanted into the mammary fat pad of host animals (Figure 3). Metastasis occurred spontaneously after tumor development. Upon dissection, when viewed under a stereomicroscope, no grossly visible metastases were observed on any of the lungs. However, based on the subsequent QRT-PCR analysis it appears that two of the eleven lungs harvested from tumor bearing animals harbored metastases, denoted by the arrows in Figure 3, suggesting that tumor cells that were undetected by visualization are present. cDNA derived from a piece of JIMT-1-derived tumor was used as a positive control for HER2 expression (Figure 3, (+)) and a lung from a mouse that was not injected with tumor cells was used as a negative control (Figure 3, (-)). A mouse specific probe for gapdh was used as an internal control to determine the proportion of human tumor cells to total mouse lung tissue. The positive control sample was normalized to the gapdh levels from the control lung as a reference. An alternate normalization strategy would be to average the amount of gapdh across all lung samples to acquire more consistent normalization levels for control samples. Alternative probes for non-mouse and non-human genes, such as GFP could be substituted to detect tumor cells that have been labeled accordingly, prior to introduction into the mouse.
A secondary analysis was also performed to provide a relative quantitation of total tumor cell number present in the lung. These results are shown in Figure 4. Here, a standard curve analysis was prepared to determine the relative HER2 quantity normalized to mouse lung gapdh in a cell number series. In theory, this allows the researcher to determine the relative amount of HER2 in 1x106, 1x105, 1x104, 1x103, 1x102, and 10 cells. The results show that the QRT-PCR probe and analysis performed was sensitive enough to detect HER2 levels in as low as 10 cells (Figure 4A). The resulting calculated HER2 values were plotted on a log-scale and a simple linear regression analysis was used to determine the relative cell number in each lung sample, as well as the positive and negative controls. As shown in Figure 4B, cell number specific values were generated for the experimental samples.
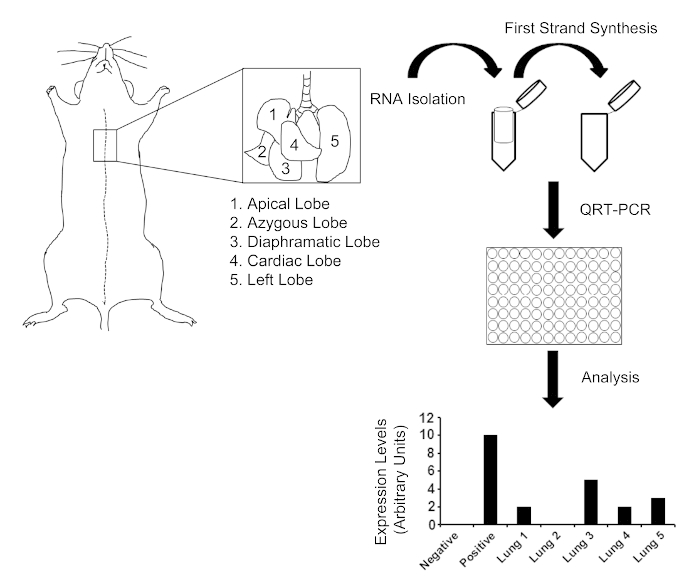 Figure 1. Schematic representation of lung dissection and analysis. The mouse lung contains 5 lobes, as indicated by the inset enlarged drawing. From dissection to analysis the QRT-PCR procedure generally takes 1-2 days to complete. Please click here to view a larger version of this figure.
Figure 1. Schematic representation of lung dissection and analysis. The mouse lung contains 5 lobes, as indicated by the inset enlarged drawing. From dissection to analysis the QRT-PCR procedure generally takes 1-2 days to complete. Please click here to view a larger version of this figure.
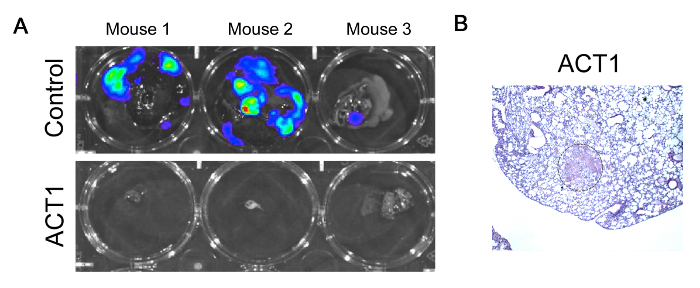 Figure 2. Gross analysis of metastasis. (A) Representative bioluminescent images of lungs from animals that were injected with 4T1-luc cells and treated with either placebo or an investigational drug called ACT1 that targets the gap junction protein, connexin43. (B). Representative H&E section of a micrometastasis in the lung of an animal with a 4T1 tumor that was treated with ACT1. Please click here to view a larger version of this figure.
Figure 2. Gross analysis of metastasis. (A) Representative bioluminescent images of lungs from animals that were injected with 4T1-luc cells and treated with either placebo or an investigational drug called ACT1 that targets the gap junction protein, connexin43. (B). Representative H&E section of a micrometastasis in the lung of an animal with a 4T1 tumor that was treated with ACT1. Please click here to view a larger version of this figure.
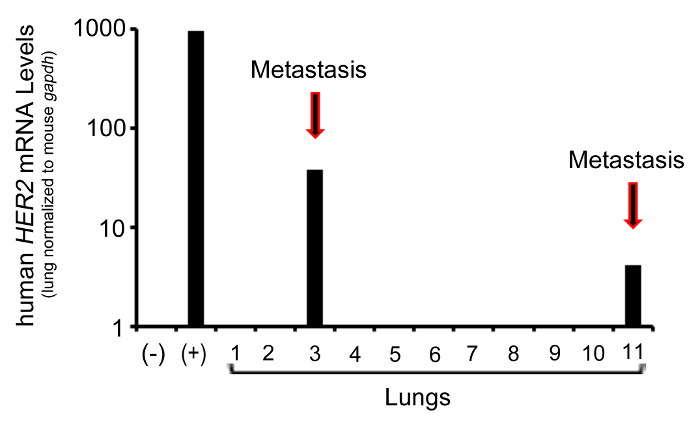 Figure 3. Representative QRT-PCR analysis. Bar graph representing the quantitated data from the QRT-PCR analysis of human HER2, a gene present in the JIMT-1 human breast cancer cell line. Human HER2 levels in the lung are normalized to mouse gapdh. (+) denotes positive control. (-) denotes negative control. Results are plotted on a log-scale. Please click here to view a larger version of this figure.
Figure 3. Representative QRT-PCR analysis. Bar graph representing the quantitated data from the QRT-PCR analysis of human HER2, a gene present in the JIMT-1 human breast cancer cell line. Human HER2 levels in the lung are normalized to mouse gapdh. (+) denotes positive control. (-) denotes negative control. Results are plotted on a log-scale. Please click here to view a larger version of this figure.
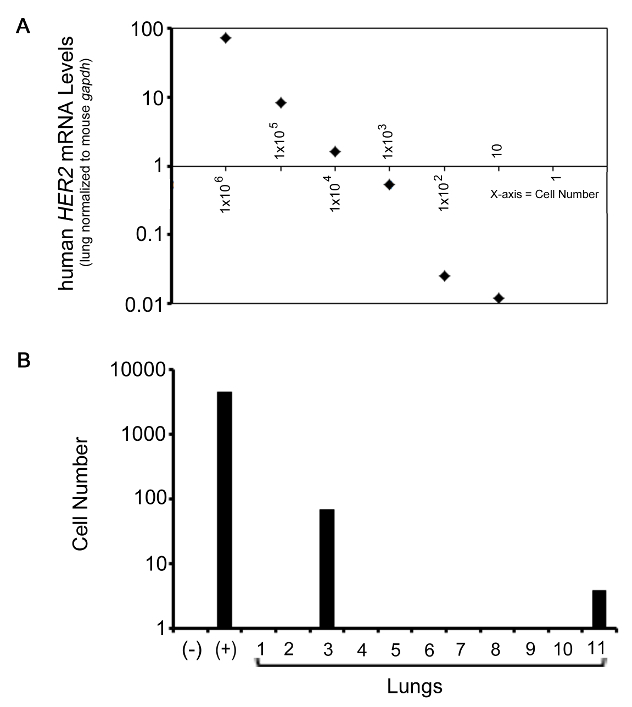 Figure 4. Representative QRT-PCR analysis from cell number analysis. (A) Graph representing HER2 levels in relation to the cell number quantity of JIMT-1 cells from 1x106 to 10 cells. (B) Bar graph representing the quantitated data from the QRT-PCR analysis of human HER2, a gene present in the JIMT-1 human breast cancer cell line. Human HER2 levels in the lung are normalized to mouse gapdh. (+) denotes positive control. (-) denotes negative control. Results are plotted on a log-scale. Please click here to view a larger version of this figure.
Figure 4. Representative QRT-PCR analysis from cell number analysis. (A) Graph representing HER2 levels in relation to the cell number quantity of JIMT-1 cells from 1x106 to 10 cells. (B) Bar graph representing the quantitated data from the QRT-PCR analysis of human HER2, a gene present in the JIMT-1 human breast cancer cell line. Human HER2 levels in the lung are normalized to mouse gapdh. (+) denotes positive control. (-) denotes negative control. Results are plotted on a log-scale. Please click here to view a larger version of this figure.
| Template RNA | 0.5-2 µg |
| iScript Supermix | 4 µl/sample |
| Nuclease-free water | up to 20 µl/sample |
Table 1. First Strand Synthesis reaction mix components (step 3.3.1).
| Step 1: 25 °C | 5 min |
| Step 2: 42 °C | 30 min |
| Step 3: 85 °C (termination) | 5 min |
Table 2. PCR conditions for First Strand Synthesis (step 3.3.3).
| iTAQ Master Mix | 10 µl/sample |
| Primer Mix | 1 µl/sample |
| Nuclease Free water | up to 20 µl/sample |
Table 3. Real-time PCR reaction mix components (step 4.3).
| Step 1 | Activation | 95 °C | 2 min | 1 cycle |
| Step 2 | Denaturation | 95 °C | 5 sec | 40 cycles |
| Annealing/Extension | 60 °C | 30 sec | ||
| Step 3 | Melt Curve (optional) | 65-95 °C (0.5 increments) | 5 sec/step |
Table 4. Real-time PCR conditions (step 4.5).
Discussion
This protocol describes use of QRT-PCR to evaluate mammary tumor cell metastasis to the lung using a xenograft mouse model. It is acknowledged that other techniques, including bioluminescent imaging, are available and are also effective for detecting metastasis despite having their own values and limitations. The data presented suggests that QRT-PCR provides an effective measure of detecting tumor-derived mRNA within lung tissue for quantitation of total metastasis. It is proposed that QRT-PCR analysis provides a secondary method that can complement or be performed in lieu of these imaging techniques. This technique may be best suited for research environments with limited access to certain types of equipment or for research groups with specific interests in metastatic burden but not detailed mechanistic studies.
While informative, the QRT-PCR analysis described here has limitations. It is not possible to perform additional downstream analysis, including immunohistochemistry (IHC) or immunofluorescence (IF) staining because the entire lung tissue is used for RNA isolation. However, if paired with an in vivo imaging technique the lung tissue may be isolated post-imaging to perform this secondary type of analysis. Also, this technique is limited to detecting mRNA and thus protein analysis is not possible unless a multi-prep isolation kit is used to prepare the lung tissue. The analysis of resulting protein is still limited to having to evaluate a total protein mix that includes the tumor and lung tissue all in one, whereas IHC/IF allows the user to delineate proteins stained in tumor tissue from staining found in mouse tissue based on the visualization of the IHC/IF staining. Alternative to using the whole lung, maintaining half of the lung tissue for other analysis such as histology may be beneficial but runs the risk of conflicting findings if metastases are identified in one portion of the lung and not the other.
Several points of modification and troubleshooting are recommended. The accuracy of the QRT-PCR data is dependent on the primer probe that is chosen for the assay and optimization of probes is recommended including the use of a proper standard curve analysis for transcript quantitation. Additionally, it is important to gauge the quality of the RNA that is isolated. Over-digestion of tissue with proteinase K can reduce the RNA quality and thus optimization is recommended based on the specific tissue of interest and method of RNA preparation. Choosing an appropriate housekeeping reference gene is also important. While gapdh is a commonly used reference gene, every gene serves a normal function in the cell that could be altered due to experimental conditions (i.e. genetic modification of host animals such as gene knockout) and thus alternative choices are useful. Some common reference genes that are reported to have ubiquitous expression and low variation in quantity are the genes encoding TATA-box binding protein (tbp), Retinitis Pigmentosa 2(rp2), Actin-like protein (Act), and Tubby bipartite transcription factor(Tub)11. Furthermore, the critical steps that assure accurate analysis include accurate dissection and washing of the lung (step 1.2.2-1.2.6), correct quantitation of the total RNA isolated (step 3.2), and accurate pipetting during the QRT-PCR setup (step 4.4). Use of a repeat pipettor can aid in this process.
In summary, this protocol aims to demonstrate that QRT-PCR is a quantifiable method to identify lung metastases. Both positive value and limitations are identified with the use of this procedure and it may be best suited to be paired with a secondary method of analysis. However, this method could be a straight forward procedure of quantitation for those research laboratories with specific research needs or lack of extensive equipment to investigate metastasis. . Future applications of this method include modification to look for metastases in target organs beyond the lung, such as lymph node or brain, which are other common sites of metastasis.
Disclosures
ESY and MAA are not employed by FirstString Research Inc. and do not hold any financial interest in the company. FirstString Research Inc. provided the luciferase imaging data that was presented in this manuscript. GSG is president and CEO of FirstString Research. CLG is an employee of FirstString Research. GSG and CLG have stock options issued by FirstString Research.
Acknowledgments
The Yeh lab is supported by research funding from an American Cancer Society Institutional Research Grant (IRG-97-219-14) awarded to the Hollings Cancer Center at MUSC, by research funding from a Department of Defense grant (W81XWH-11-2-0229)at MUSC, and by an award from the Concern Foundation (to ESY).
References
- Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127(4):679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- Bos PD, Nguyen DX, Massague J. Modeling metastasis in the mouse. Curr Opin Pharmacol. 2010;10(5):571–577. doi: 10.1016/j.coph.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim IS, Baek SH. Mouse models for breast cancer metastasis. Biochem Biophys Res Commun. 2010;394(3):443–447. doi: 10.1016/j.bbrc.2010.03.070. [DOI] [PubMed] [Google Scholar]
- Saxena M, Christofori G. Rebuilding cancer metastasis in the mouse. Mol Oncol. 2013;7(2):283–296. doi: 10.1016/j.molonc.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Zhang JJ, Huang XY. Mouse models for tumor metastasis. Methods Mol Biol. 2012;928:221–228. doi: 10.1007/978-1-62703-008-3_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampetsreiter P, Casanova E, Eferl R. Genetically modified mouse models of cancer invasion and metastasis. Drug Discov Today Dis Models. 2011;8(2-3):67–74. doi: 10.1016/j.ddmod.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condeelis J, Segall JE. Intravital imaging of cell movement in tumours. Nat Rev Cancer. 2003;3(12):921–930. doi: 10.1038/nrc1231. [DOI] [PubMed] [Google Scholar]
- Massoud TF, Gambhir SS. Molecular imaging in living subjects: seeing fundamental biological processes in a new light. Genes Dev. 2003;17(5):545–580. doi: 10.1101/gad.1047403. [DOI] [PubMed] [Google Scholar]
- Zinn KR, et al. Noninvasive bioluminescence imaging in small animals. ILAR J. 2008;49(1):103–115. doi: 10.1093/ilar.49.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng JC, Kung AL. Quantitative bioluminescence imaging of mouse tumor models. Cold Spring Harb Protoc. 2015;2015(1) doi: 10.1101/pdb.prot078261. [DOI] [PubMed] [Google Scholar]
- Radonic A, et al. Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun. 2004;313(4):856–862. doi: 10.1016/j.bbrc.2003.11.177. [DOI] [PubMed] [Google Scholar]


