Abstract
In mammalian cells, both microRNAs (miRNAs) and small interfering RNAs (siRNAs) are thought to be loaded into the same RNA-induced silencing complex (RISC), where they guide mRNA degradation or translation silencing depending on the complementarity of the target. In Drosophila, Argonaute2 (AGO2) was identified as part of the RISC complex. Here we show that AGO2 is an essential component for siRNA-directed RNA interference (RNAi) response and is required for the unwinding of siRNA duplex and in consequence assembly of siRNA into RISC in Drosophila embryos. However, Drosophila embryos lacking AGO2, which are siRNA-directed RNAi-defective, are still capable of miRNA-directed target RNA cleavage. In contrast, Argonaute1 (AGO1), another Argonaute protein in fly, which is dispensable for siRNA-directed target RNA cleavage, is required for mature miRNA production that impacts on miRNA-directed RNA cleavage. The association of AGO1 with Dicer-1 and pre-miRNA also suggests that AGO1 is involved in miRNA biogenesis. Our findings show that distinct Argonaute proteins act at different steps of the small RNA silencing mechanism and suggest that there are inherent differences between siRNA-initiated RISCs and miRNA-initiated RISCs in Drosophila.
Keywords: RNAi, siRNA, miRNA, AGO1, AGO2
Double-stranded (ds) RNA induces the sequence-specific posttranscriptional gene silencing of cognate genes in numerous organisms (Cogoni and Macino 2000; Hutvagner and Zamore 2002a; Denli and Hannon 2003). The multidomain ribonuclease III enzyme Dicer excises long dsRNA into duplexes of 21-23 nucleotides (nt) termed short interfering RNAs (siRNAs; Bernstein et al. 2001; Ketting et al. 2001; Knight and Bass 2001), which direct the cleavage of complementary mRNA targets, a process known as RNA interference (RNAi; Fire et al. 1998). Prior to target mRNA recognition, an siRNA duplex goes through an ATP-dependent unwinding process and one strand over the other is often preferentially loaded onto the RNA-induced silencing complex (RISC), the multiple-turnover enzyme complex that mediates endonucleolytic cleavage in the RNAi pathway. The RISC is guided to cleave target mRNAs sharing perfect complementarity across the center of the complementary siRNA strand in the absence of high-energy cofactors (Hammond et al. 2001; Nykanen et al. 2001; Hutvagner and Zamore 2002b; Martinez et al. 2002). siRNAs are not the only products of Dicer. Natural dsRNA-encoding genes, named microRNA (miRNA) genes, encode RNA products of ∼70 nt that are predicted to form imperfect hairpin structures and are processed by Dicer to mature 21-23-nt miRNAs (Grishok et al. 2001; Hutvagner et al. 2001). Only one Dicer enzyme is found in Caenorhabditis elegans and humans, therefore indicating that the same Dicer is required for both RNAi and for the processing of miRNA precursors in these organisms (Grishok et al. 2001; Hutvagner et al. 2001; Knight and Bass 2001). The expression of miRNAs is often developmentally regulated, suggesting an important role for miRNAs in the regulation of endogenous gene expression (Lagos-Quintana et al. 2001; Lau et al. 2001; Lee and Ambros 2001; Reinhart et al. 2002; Brennecke et al. 2003; Xu et al. 2003). Target mRNAs containing sequences imperfectly complementary to the miRNA can be subject to translational repression without altering mRNA stability (Lee et al. 1993; Wightman et al. 1993; Olsen and Ambros 1999; Reinhart et al. 2000; Bartel 2004).
Recent findings point to a tight connection between miRNA and RNAi molecular machineries. Both miRNAs and siRNAs have been shown to be capable of target mRNA degradation or translation silencing in mammalian cells and plants (Hutvagner and Zamore 2002b; Llave et al. 2002; Doench et al. 2003; Zeng et al. 2003); additionally, it has been shown that both siRNAs and miRNAs can be loaded onto the same RISC complex (Hutvagner and Zamore 2002b; Martinez et al. 2002). These findings imply that, regardless of the maturation process, once the small RNA is loaded, the RISC uses it to degrade or inhibit translation depending on the degree of complementarity between the small RNA and its mRNA target.
In C. elegans, genetic analyses suggested that RDE-1, a member of the Argonaute family of proteins (Carmell et al. 2002), is required for the initiation of RNAi with injected dsRNAs (Grishok et al. 2000), whereas Alg-1 and Alg-2, other Argonaute family members, are required for the accumulation of stable mature miRNAs in C. elegans, but not for RNAi driven by dsRNA (Grishok et al. 2001). These results suggest that distinct members of the Argonaute family of proteins may provide specificity to their respective pathways, RNAi and translation inhibition, in C. elegans. However, the underlying molecular mechanisms are not well understood.
In Drosophila, it has been shown that Argonaute2 (AGO2) protein, a member of the Argonaute family of proteins (Carmell et al. 2002), is essential for RNAi driven by exogenously introduced dsRNA and is the first protein component to be identified as part of the RISC complex in cultured Drosophila S2 cells (Hammond et al. 2001). Although AGO2 is known to associate with several proteins such as the Drosophila homolog (dFMR1) of the human fragile X mental retardation protein and VIG (Caudy et al. 2002; Ishizuka et al. 2002), the precise role that AGO2 plays in RNAi is not well understood. In this paper, we have produced AGO2 deletion mutant flies and found that embryos lacking AGO2 are siRNA-directed RNAi defective but are still capable of miRNA-directed target RNA cleavage. We also show that AGO2 mutant embryos are impaired in the assembly of siRNA into RISC. In contrast, Argonaute1 (AGO1), another Argonaute protein in fly (Kataoka et al. 2001), is dispensable for siRNA-directed RNA cleavage but is necessary for the accumulation of stable mature miRNAs, and thus impacts on miRNA-directed target RNA cleavage. Our findings suggest that distinct Argonaute proteins act at different steps of the small RNA silencing mechanism, and provide specificity to their respective pathways in Drosophila.
Results
AGO2 is essential for RNAi in embryos
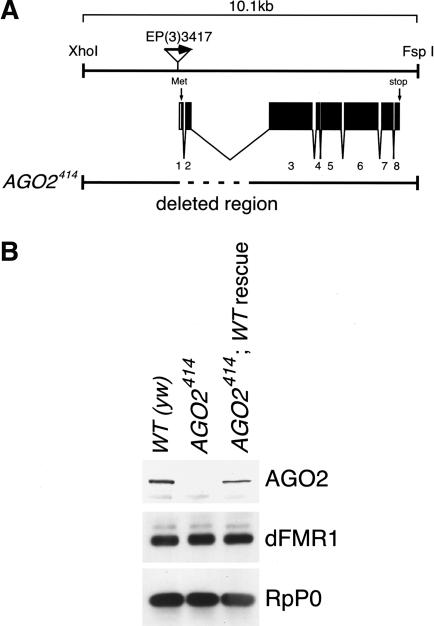
The Drosophila AGO2 protein is an essential factor for RNAi as a component of the RISC complex in cultured S2 cells (Hammond et al. 2001). However, the precise role that AGO2 plays in RNAi is not well understood. To gain an insight into the molecular functions of AGO2 in RNAi, we produced fly strains that lack AGO2. To obtain fly strains bearing deletions in AGO2, we mobilized a P{EP}element (Rorth et al. 1998) inserted in the first exon of the AGO2 gene on the EP(3)3417 chromosome to produce several partial deletions of AGO2. One such mutation named AGO2414 was selected for further characterization (Fig. 1A). Western blot analysis with anti-AGO2 antibody revealed that there is no AGO2 protein in homozygous AGO2414 flies (Fig. 1B). We also confirmed the absence of AGO2 mRNA by Northern blot analysis in homozygous AGO2414 flies (data not shown). These results demonstrated that AGO2414 are AGO2 null flies. We found that homozygous AGO2414 flies proceed into adulthood, are fertile, and appear outwardly normal. Because it is not known whether AGO2 is required for RNAi in embryos, we tested for RNAi in vivo by assaying the ability of long dsRNA corresponding to the fushi tarazu (ftz) gene to produce a ftz phenotype (Kennerdell and Carthew 1998) when injected into wild-type and AGO2 mutant embryos. We used AGO2 mutant adult females to make mutant eggs to remove the maternal contribution of AGO2 (simply called the “AGO2 mutant embryo” or AGO2414 embryos, hereafter). Wild-type embryos injected with ftz dsRNA exhibit segmentation defects in their cuticle (Fig. 2B; Table 1). In contrast, AGO2 mutant embryos showed a complete absence of interference in response to ftz dsRNA, indicating that they are RNAi defective (Fig. 2C). This is consistent with previous findings made with cultured S2 cells (Hammond et al. 2001; Caudy et al. 2002; Ishizuka et al. 2002). To confirm that disruption of the AGO2 gene is directly responsible for the RNAi-defective phenotype, we transformed mutant flies with a P element containing wild-type AGO2 genomic sequences (Fig. 1A,B). The RNAi-defective phenotype was ameliorated by the introduction of this AGO2 minigene (Fig. 2D; Table 1), demonstrating that the RNAi-defective phenotype is caused by the AGO2 mutation rather than a second site mutation elsewhere on the chromosome.
Figure 1.
Characterization of the AGO2 deletion mutant AGO2414. (A) Structure of the AGO2 locus. Exons are indicated with open boxes; the closed portion is the protein-coding region. The translation initiation start site (Met) is located in the first exon. The position of EP(3)3417 is represented as triangle with an arrow pointing in the direction of GAL4-induced transcription. The deleted genomic region of the AGO2414 chromosome is shown as a dotted line. In this mutant, imprecise excision of the EP element generated a 2.3-kb deletion of genomic DNA, which included exons 1 and 2 of the AGO2 gene. Sequence information from the Berkeley Drosophila Genome Project reveals that the AGO2 locus is positioned in a small region on the cytological location 71E1, on the left arm of the third chromosome. (B) Western blotting on protein extracts from adult ovaries. Western blotting was carried out using anti-AGO2 (4D2), anti-dFMR1 (5A11), and antiribosomal protein P0 antibodies. [WT(yw)] Yellow-white wild type; (AGO2414) AGO2414 homozygote; (AGO2414;WTrescue) AGO2414 homozygote with two copies of wild-type rescue. The genomic rescue construct contained a 10.1-kb XhoI-FspI fragment of the AGO2 locus in A in which no other known genes are included.
Figure 2.
RNA interference of ftz activity in embryos. Representative embryonic phenotypes of wild-type and AGO2414 mutant embryos injected with ftz dsRNA are shown. (A) A wild-type embryo injected with water. (B) A wild-type embryo injected with ftz dsRNA has a phenotype similar to that of ftz. (C) An AGO2414 mutant embryo injected with ftz dsRNA has a wild-type phenotype. (D) An AGO2414;WTrescue embryo has a phenotype similar to that of ftz.
Table 1.
Effects of ftz dsRNA and ftz siRNA injected into Drosophila embryos
| Genotype | Injected RNA | Wild type | ftz |
|---|---|---|---|
| WT(yw) | mock (water) | 29 | 0 (0%) |
| WT(yw) | ftz dsRNA | 5 | 30 (86%) |
| AGO2414 | ftz dsRNA | 43 | 0 (0%) |
| P[w+ : AGO2];AGO2414 | ftz dsRNA | 1 | 22 (96%) |
| WT(yw) | ftz siRNA | 2 | 30 (94%) |
| AGO2414 | ftz siRNA | 21 | 0 (0%) |
Numbers indicate embryos with wild-type or ftz phenotype after injection. Percentages are given in parentheses. Embryos were scored as ftz if they had six or fewer ventral denticle belts. P[w+: AGO2];AGO2414 is the genotype of “rescued' flies (AGO2414;WT rescue).
AGO2 is required for the assembly of siRNA into RISC in embryos
To determine whether AGO2 is required for the formation of the siRNA duplex or for interference thereafter, we injected synthetic ftz siRNA into AGO2414 embryos. siRNA produced a ftz phenotype in wild-type embryos (Fig. 3A; Table 1). However, AGO2414 embryos were not responsive to the ftz siRNA (Fig. 3B; Table 1). Similarly, in vitro, AGO2414 embryo lysate processed long ftz dsRNA into short ∼21-nt fragments, similar to lysates from wild-type embryos (Fig. 3C). These findings suggest that AGO2 is necessary for RNAi after the formation of the siRNA duplex.
Figure 3.
AGO2 is required for interference after the formation of the siRNA duplex. (A) Wild-type embryo injected with synthetic ftz siRNA has a phenotype similar to ftz. (B) AGO2414 embryo injected with synthetic ftz siRNA has a wild-type phenotype. (C) AGO2 is not required for the production of the siRNA duplex. Uniformly 32P-labeled dsRNA corresponding to the ftz gene, incubated in extracts from yw and AGO2414 embryos. RNA products of the reaction were analyzed on a polyacrylamide gel. Incubation of lysates with labeled dsRNA generates RNA fragments ∼22 nucleotides (nt) long. [WT(yw)] Yellow-white wild type; (AGO2414) AGO2414 homozygote. (D) Analysis of siRNA unwinding in AGO2414 embryo lysate. The siRNA duplex was incubated in the extracts from wild-type (yw) and AGO2414 embryos. RNA products of the reaction were analyzed on a native acrylamide gel. The migration patterns of the siRNA duplex, the starting material, and the single-stranded siRNA, the expected product after unwinding, are shown on the left. (Lower panel) The amounts of input siRNA duplex and unwound single-stranded (ss) siRNA at 0 and 3 h incubation are shown. The average of five independent experiments is shown here. (E) Native gel analysis of labeled siRNA-protein complexes from wild-type and AGO2414 embryo lysates. B and A complexes are observed in both lanes from wild-type and AGO2414 reactions. These two complexes correspond to intermediate precursors to RISC (Tomari et al. 2004). The complex corresponding to RISC (Tomari et al. 2004) is not detected in the lane from an AGO2414 reaction.
Before target mRNA recognition, the siRNA duplex is unwound (Nykanen et al. 2001), and each RISC contains only one of the two strands of the siRNA duplex (Martinez et al. 2002). We tested whether AGO2 activity is necessary for this unwinding process. Unwound siRNA was detected in wild-type embryo lysate, whereas no such activity was detected in AGO2414 embryo lysate (Fig. 3D), suggesting that AGO2 is required for the unwinding of the siRNA duplex. An alternative explanation of the results could be that the absence of AGO2 leaves the unwound single-stranded RNA exposed to nucleolytic degradation and therefore the single-stranded RNA could not be detected. However, this is unlikely because the amount of input siRNA duplex left in AGO2414 embryo lysate was not significantly reduced even after 3 h incubation as opposed to that seen in wild-type embryo lysate (Fig. 3D, lower panel), indicating that input siRNA duplex is indeed not unwound in AGO2 mutant embryo lysate. Using a recently developed RISC assembly assay (Tomari et al. 2004), we next analyzed RISC formation in AGO2 mutant embryo lysates. ds-siRNA was incubated with embryo lysates in a standard RNAi reaction, and then RISC complexes were resolved by electrophoresis through a native gel. As shown in Figure 3E, three major complexes (B, A, and RISC) were detected in wild-type embryo lysates as reported previously (Tomari et al. 2004). Complexes B and A are thought to be intermediate precursors to active RISC and contain ds-siRNA (Tomari et al. 2004). Both complexes B and A were readily detected in AGO2 mutant embryo lysates. However, RISC formation was impaired in AGO2 mutant embryo lysates (Fig. 3E). Together, these results suggest that AGO2 is required in a step(s) in RISC assembly after binding of the siRNA duplex to RISC precursors (complexes B and A).
miRNA-initiated RNA cleavage does not need AGO2 but siRNA-initiated RNA cleavage does
Recently, it has been demonstrated that both siRNAs and miRNAs are associated with the same RISC and that miRNAs can even direct cleavage of perfectly base-paired substrates in human cells (Hutvagner and Zamore 2002b; Martinez et al. 2002). To see if this is also the case in Drosophila, we investigated whether the lack of AGO2 affects the miRNA-mediated cleavage of target RNAs. 32P-radiolabeled target mRNA, containing a sequence fully complementary to an endogenous miRNA, miR-2b, as well as a sequence exactly complementary to one strand of a ftz siRNA duplex (Fig. 4A), was incubated with AGO2414 embryo lysate in an in vitro RNAi reaction. Both siRNA-directed and miRNA-directed cleavage products are seen in the wild-type lysate (Fig. 4B) as shown previously (Schwarz et al. 2002). However, ftz siRNA did not direct cleavage in AGO2414 embryo lysate (Fig. 4B); in contrast, miR-2b still directed cleavage of the target RNA in AGO2414 embryo lysates (Fig. 4B). These results demonstrate that although AGO2 is an essential factor in siRNA-directed RNAi, AGO2 is dispensable for miRNA-directed RNA cleavage in RNAi.
Figure 4.
AGO2 is not required for miRNA-initiated RNA cleavage. (A) Schematic drawing of the target RNA and its pairing with ftz siRNA and miRNA (miR-2b or let-7). (Magenta) miR-2b, or let-7 and their complementary sequences (below); (green) ftz siRNA and its complementary sequence. An asterisk indicates the 5′-32P radiolabel. (B) miRNA-directed target RNA cleavage is catalyzed by embryo lysates with and without AGO2. In vitro RNAi assays were carried out with 14-16-h embryo lysates of yw and AGO2414. Target RNA contains the ftz siRNA and the miR-2b target sequences as shown in A. (C) In vitro processing of pre-let-7 RNAs internally labeled with α-32P-GTP. Mature let-7-generating activities were compared between lysates from 0- to 2-h yw and AGO2414 embryos. Both synthetic 21-nt let-7 RNA (not shown) and in vitro processed let-7 migrate at ∼25 nt relative to the markers. The discrepancy between molecular mass and mobility in polyacrylamide gel electrophoresis is frequently observed for small RNAs (Hutvagner et al. 2001; Lee et al. 2003). (D) In vitro RNAi assays with lysates prepared from 0- to 2-h yw and AGO2414 embryos. The RNA target contains let-7 and ftz siRNA target sequences as shown in A.
In this experiment, however, the timings of miRNA loading and siRNA loading into RISC are different. miR-2b is already incorporated into the RISC by the embryo before the lysate is made, whereas siRNA is added to the reaction mixture. We therefore added synthetic Drosophila melanogaster let-7 precursor RNA to an in vitro RNAi reaction mixture containing the let-7 complementary target RNA. It should be noted that the sequence of the synthetic let-7 precursor used here (Fig. 4C, bottom panel) is slightly longer than that of naturally processed pre-let-7, which contains short 3′ overhangs (Basyuk et al. 2003; Lee et al. 2003). However, the synthetic let-7 precursor has been previously shown to be processed and produce mature let-7 with precisely the same 5′ end as authentic let-7 in the Drosophila embryo lysates (Hutvagner et al. 2001). In this experiment, the synthetic let-7 precursor RNA was also converted to mature let-7 in vitro, not only in the wild-type embryo lysate, but also in AGO2414 embryo lysate (Fig. 4C; data not shown). In the in vitro RNAi assay, target RNA was cleaved within the let-7 complementary sequence in AGO2414 embryo lysate as efficiently as in wild type (Fig. 4D). Thus, miRNA that is not preloaded by the embryo is still capable of entering into the RNAi pathway without AGO2 activity. These results demonstrate that AGO2 is not required for miRNA production or the loading of miRNAs into the RISC and suggest that AGO2 has a specific role for siRNA activity, likely as a specific unwinding and loading factor of siRNA into RISC as shown earlier, and possibly as a necessary component of an siRNA-initiated RISC.
AGO1 is necessary for efficient miRNA-directed RNA cleavage
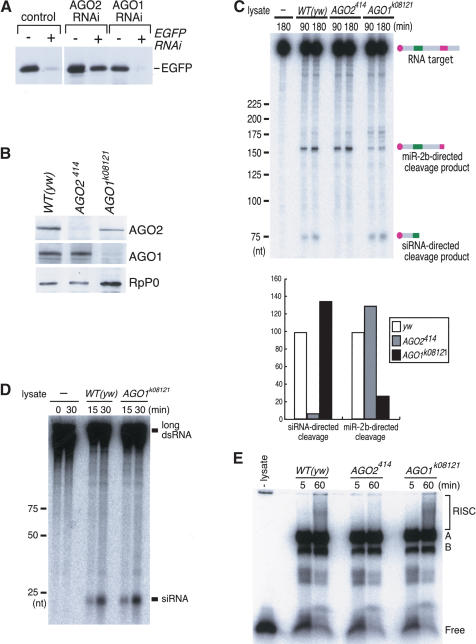
We next tested whether siRNA- and miRNA-directed RNA cleavage pathways have differential requirements for AGO1. Mutations in AGO1 result in late embryonic/early larval lethality with developmental defects (Kataoka et al. 2001). Although AGO1 has been shown necessary for efficient RNAi in embryos (Williams and Rubin 2002), AGO1 appears to have little or no effect on the efficiency of RNAi in cultured S2 cells (Fig. 5A; see also Caudy et al. 2002). AGO1k08121 is a strong allele (Kataoka et al. 2001). AGO1k08121 was balanced over a CyO chromosome carrying a kruppel (Kr)-Gal4 and a UAS-GFP transgene. Homozygous embryos were separated from heterozygous and CyO-Kr-Gal4-UAS-GFP homozygous embryos on the basis of GFP expression. Because there is strong reduction of AGO1 transcripts in AGO1k08121 embryos (Kataoka et al. 2001; data not shown) and AGO1 protein was not detected in lysate from 14- to 16-h staged AGO1k08121 embryos (Fig. 5B), we tested 14-16-h staged AGO1k08121 embryo lysate for its ability to direct cleavage of siRNA or miRNA complementary RNA targets. ftz siRNA directed its cleavage in AGO1k08121 embryo lysate as effectively as in wild type (Fig. 5C). AGO1k08121 embryo lysate also processed long ftz dsRNA into short ∼21-nt fragments (Fig. 5D). Furthermore, RISC formation directed by siRNA duplex was not impaired in AGO1k08121 embryo lysate as judged by the native gel shift assay (Fig. 5E). These findings suggest that AGO1 is not necessary for the production of the siRNA duplex, siRNA-initiated RISC formation, or siRNA-directed cleavage. However, cleavage of the target RNA directed by miR-2b was suppressed in AGO1k08121 embryo lysate by a factor of fivefold compared with that in wild type (Fig. 5C), suggesting that AGO1 is necessary for efficient target RNA cleavage mediated by miRNAs. Residual miRNA activity seen in AGO1k08121 could be due to some degree of functional complementation among Argonaute family proteins expressed in embryos (Williams and Rubin 2002). Our in vitro experiments with AGO1k08121 mutant embryo lysates argue against AGO1 being required for siRNA-mediated RNAi in embryos when injected with either long dsRNA or siRNA (Williams and Rubin 2002). Although we currently do not understand the discrepancy, our findings are at least consistent with the findings using cultured S2 cells (Fig. 5A; Caudy et al. 2002).
Figure 5.
AGO1 is required for efficient miRNA-initiated RNA cleavage. (A) When AGO2 is suppressed by introducing specific dsRNA in S2 cells expressing EGFP, the ability of the cells to silence EGFP by RNAi is severely reduced. In contrast, when AGO1 is suppressed, the EGFP silencing effect is unaffected, indicating that AGO1 is not essential for the RNAi pathway in S2 cells. (B) Lysates were prepared from 14- to 16-h yw, AGO2414 mutant, and AGO1k08121 mutant embryos. Western blots were performed using the antibodies against AGO2 (upper), AGO1 (middle), and ribosomal protein P0 (lower) as a loading control. (C) In vitro RNAi assays with lysates prepared from 14- to 16-h yw, AGO2414, and AGO1k08121 embryos. The RNA target was the same as that used in Figure 4B. The amount of specifically cleaved bands was quantified and normalized to that of bands in wild-type embryos. (D) AGO1 is not required for the production of the siRNA duplex. Uniformly 32P-labeled dsRNA corresponding to the ftz gene, incubated in extracts from yw and AGO1k08121 embryos. RNA products of the reaction were analyzed on a polyacrylamide gel. Incubation of lysates with labeled dsRNA generates RNA fragments ∼22 nt long. [WT(yw)] Yellow-white wild type; (AGO1k08121) AGO1k08121 embryos. (E) RISC formation is not impaired in AGO1k08121 embryo lysates.
AGO1 is involved in the stable maturation of miRNAs
We hypothesized that AGO1 might be involved in the maturation and/or function of miRNAs. To directly determine the contribution of AGO1 and AGO2 to miRNA production in Drosophila cells, we depleted each from S2 cells by dsRNA soaking (Fig. 6A) and performed Northern blot analyses to monitor the abundance of bantam miRNA (miR-ban; Brennecke et al. 2003). A marked reduction of mature miR-ban was observed in AGO1-depleted S2 cells but not in AGO2-depleted S2 cells (Fig. 6A). We also confirmed that the miRNA level was significantly reduced in the AGO1k08121 embryos (Fig. 6B). The accumulation of pre-miR-ban was not observed in both cases (data not shown; see also Fig. 6C). Similar results were obtained when the same RNA preparations were probed for the expression of another miRNA, miR-2b, which is also known to be expressed both in S2 cells and embryos (data not shown). These results suggest that AGO1 is involved in the maturation and/or stability of miRNAs.
Figure 6.
AGO1 is involved in miRNA biogenesis. (A) AGO1 is required for stable, mature miRNA production. Expression of AGO1, AGO2, or EGFP (control) was suppressed by RNAi in S2 cells. Western blots (lower panels) show that the Argonaute proteins are indeed down-regulated. RNAs were obtained from each cell and subjected to Northern blots to analyze the amounts of miR-ban using a specific probe. (B) The miRNA level in the wild-type, AGO1, and AGO2 mutant embryos. Northern blot analysis on the AGO1 mutant (AGO1k08121) shows a marked reduction of mature miR-ban. (C) Dicer-1 and AGO1 are related to miRNA processing and stabilization, respectively. Expression of either Dicer-1 (Dcr-1), Dicer-2 (Dcr-2), AGO1, AGO2, or EGFP (control) was reduced by RNAi in S2 cells for indicated days, then total RNAs were obtained from each cell and subjected to Northern blots to visualize mature and pre-miR-ban. (D) AGO1 associates with Dicer-1. TEV extract prepared from S2 cells expressing AGO1-TAP, AGO2-TAP, or dFMR1-TAP, or the parental S2 cells (control), were subjected to Western blots using antibodies against Dicer-1, AGO1, dFMR1, and AGO2. Protein bands with asterisks indicate AGO1, AGO2, and dFMR1 with a CBP tag, a converted form of a TAP tag after TEV cleavage. (E) AGO1 associates with both pre-miRNA and mature miRNA. Northern blots show that all AGO1-TAP, AGO2-TAP, and dFMR1-TAP complexes contain mature miR-ban, but the precursor is associated only with the AGO1-TAP complex.
Production of both siRNAs and miRNAs require Dicers, which interact with RISC components (Hammond et al. 2001; Tabara et al. 2002), suggesting that Dicer action is coupled to loading small RNAs onto the RISC. Two Dicers, Dicer-1 and Dicer-2, have been identified in Drosophila. siRNA production is associated with Dicer-2, but not Dicer-1 (Liu et al. 2003). Dicer-2, together with the dsRNA-binding protein R2D2, facilitates siRNA loading onto RISC (Liu et al. 2003). We found that the targeted destruction of only one of the two Dicers, Dicer-1, leads to an accumulation of the miR-ban precursor in S2 cells (Fig. 6C), consistent with the recent genetic studies showing that Dicer-1 is required for miRNA processing (Lee et al. 2004). The amount of pre-miR-ban was further increased after prolonged Dicer-1 knockdown for 8 d. In contrast, prolonged suppression of AGO1 expression did not result in the accumulation of pre-miR-ban (Fig. 6C). These findings suggest that mature miRNAs are processed from pre-miRNAs by Dicer-1 and are stabilized by AGO1. We next asked whether AGO1 is physically associated with Dicer-1 by purifying AGO1-associated complexes from S2 cells using a TAP method (Rigaut et al. 1999). It was found that AGO1 was associated with Dicer-1 (Fig. 6D). This interaction of AGO1 with Dicer-1 seems to be quite specific for AGO1 because Dicer-1 was not detected in AGO2- and in dFMR1-associated complexes under the conditions used for the purification (Fig. 6D). RNA preparations from AGO1-, AGO2-, and dFMR1-associated complexes were then probed for the presence of miR-ban. AGO2- and dFMR1-associated complexes contained the mature form of miR-ban (Fig. 6E). Only the AGO1-associated complex contained the pre-miR-ban and mature forms (Fig. 6E). The interaction of AGO1 with Dicer-1 and pre-miRNA further suggests that AGO1 is involved in miRNA biogenesis.
Discussion
In mammals, there appear to be no real distinctions between the siRNA and miRNA pathways downstream of Dicer (Hutvagner and Zamore 2002b). In contrast, genetic studies in C. elegans have shown that distinct Argonaute homologs appear to be dedicated to distinct RNAi and translation repression pathways (Grishok et al. 2001). Using target RNA cleavage assays, we show that siRNA-directed RNA cleavage depends on AGO2 but does not require AGO1, whereas miRNA-directed RNA cleavage depends on AGO1 but does not need AGO2 in Drosophila embryos. Our results suggest that maturation and the function of siRNAs and miRNAs have differential requirements for Argonaute proteins in Drosophila. These findings also suggest that Argonaute proteins regulate entry points of small RNAs to RISC but may not act as determinants for target RNA cleavage or translation repression. AGO2 is an essential component of RNAi and part of the RISC complex (Hammond et al. 2001). We show that the particular function of AGO2 lies downstream of siRNA duplex production in the RNAi pathway (Fig. 3). It has been shown that duplex siRNAs are incorporated in RISC precursors and ATP-dependent unwinding of siRNAs converts RISC precursors into active RISC that then degrades the specific target mRNAs (Hammond et al. 2000; Nykanen et al. 2001; Tomari et al. 2004). Thus, AGO2 is thought to function at some or all of these steps in RNAi. In this study, we demonstrate that both the unwinding of siRNA duplex and the assembly of siRNA into functional RISC are impaired in AGO2 mutant embryos (Fig. 3). Therefore, these results indicate that AGO2 functions at a step(s) in RISC assembly after binding of the siRNA duplex to RISC precursors. Recently, Liu et al. (2003) demonstrated that Dicer-2 not only associates with siRNA production, but also, together with R2D2, facilitates siRNA loading onto RISC in Drosophila. These data suggest that AGO2, together with Dicer-2 and R2D2, assembles siRNA into functional RISC in Drosophila embryos.
AGO2414 flies that we produced are developmentally normal, which provides circumstantial evidence that AGO2 is not important for development and by inference not essential for utilization and function of miRNA that often regulates the developmental pathways (Bartel 2004). In contrast, our findings that AGO1 is required for the stable production of mature miRNAs might explain the fact that AGO1 is essential for normal development, particularly in the nervous system (Kataoka et al. 2001). The physical association of AGO1 with Dicer-1 and pre-miRNA suggests that AGO1 is involved in miRNA biogenesis. In fact, recent genetic studies of Dicers in Drosophila have shown that mutations in Dicer-1 block processing of miRNA precursors (Lee et al. 2004). Together, these results suggest that in Drosophila, distinct pathways exist for siRNA and miRNA production and their concomitant assembly into RISC complexes. Do AGO1 and AGO2 functional specificities reflect physically distinct miRNA-associated RISC and siRNA-associated RISC, or differential processing and/or loading of small RNAs into generic RISC? Although there is no definitive evidence that there is a generic RISC for both small RNAs, recent studies have shown that the RISC containing active miRNAs and the RISC involved in siRNA-directed RNAi are very similar, if not identical, as endogenous miRNAs can cleave mRNAs with perfect complementarity (Hutvagner and Zamore 2002b; Llave et al. 2002), and exogenously introduced siRNAs can translationally repress mRNAs bearing imperfectly complementary binding sites (Doench et al. 2003; Zeng et al. 2003). Our findings show that the miRNA-associated RISC that cleaves RNA does not need AGO2, whereas siRNA-associated RISC does. This argues that there are inherent differences between siRNA-initiated RISC and miRNA-initiated RISC in Drosophila. Physically, they might be the same generic complex (for instance, both containing AGO2) but one does not need AGO2 activity. It is also possible that they might be physically distinct with respect to Argonaute proteins. AGO1 is not present in AGO2-associated complexes (Fig. 6D) and can be biochemically separated from siRNA-loaded RISCs (Caudy et al. 2002). Therefore, AGO1 and AGO2, conceivably, are not incorporated into the same RISC. However, miRNAs and siRNAs are found, to some extent, in both AGO1 and AGO2 complexes (Fig. 6E; see also Caudy et al. 2002), suggesting that association of Argonaute proteins with small RNAs is preferential rather than absolutely specific. Alternatively, a fraction of small RNAs might be exchangeable between the two complexes, probably during recycling of their silencing function.
The recent study has shown that miRNA-associated RISCs and siRNA-associated RISC are different with respect to Dicers in Drosophila (Lee et al. 2004). Our results clearly suggest that in conjunction with Dicers, Argonaute proteins regulate the formation of siRNA-associated RISC or miRNA-associated RISC, as their particular functions lie downstream of Dicers in a step(s) in the assembly of small RNAs into RISC.
Materials and methods
Drosophila strains
Flies were maintained under standard procedures. P element insertion in AGO2 EP(3)3417 was out-crossed to yw to clean up the genetic background. AGO1k08121 (Kataoka et al. 2001) was balanced over a CyO chromosome carrying a Kruppel-Gal4 and a UAS-GFP transgene (CyO-Kr-Gal4-UAS-GFP chromosome).
Generation of AGO2 mutants
EP(3)3417 contains the EP transposon (Rorth et al. 1998) in the 5′ UTR region of the AGO2 locus. To isolate new AGO2 alleles, we generated 400 independent lines from the cross of virgin EP(3)3417 females with males possessing Δ2-3 transposase. Genomic DNA from these lines were prepared and used as PCR templates to screen lines that carried a deletion within the AGO2 locus. The breakpoints of the deletion were determined by sequencing the PCR products. AGO1k08121 was maintained as a heterozygous stock balanced over a CyO-Kr-Gal4-UAS-GFP chromosome. One-quarter of the embryos produced in this stock were AGO1k08121 homozygous mutants, as identified by the absence of zygotic GFP expression. The remaining three-quarters expressed GFP and represented both heterozygous and CyO-Kr-Gal4-UAS-GFP homozygous embryos.
RNAi in embryos
For dsRNA production, a region of the fushi tarazu (ftz) gene was amplified with oligonucleotides 5′-ATCCGATATGGCCACCAC-3′ and 5′-CGCTCTGGGGAAGAGAGTAA-3′ by using an embryonic cDNA pool as a template. The PCR product was cloned into the pGEM-T vector (pGEM-T-ftz) and the insert was amplified with T7 and SP6 primers. The resulting PCR product was used as a template for in vitro transcription reactions using MEGAScript T7 and SP6 kits (Ambion) according to the manufacturer's instructions. Equimolar concentrations of sense and antisense RNAs were boiled and allowed to anneal >12 h at room temperature. siRNA (21 bp) with 2 base 3′ overhangs corresponding to the ftz gene (siFTZ 5′-UGCCUACUAUCAGAACACC-3′) was purchased from Dharmacon. The siRNA was 5′-phosphorylated with T4 polynucleotide kinase (PNK; TaKaRa). dsRNA or siRNA (1 mg/mL in H2O) was injected into syncytial blastoderm embryos (Kennerdell and Carthew 1998). After 20-24 h at room temperature, the surviving embryos were scored for the number of ventral denticle belts. For consistency, any embryos with four, five, or six belts were scored as ftz. For cuticle analysis, embryos were incubated at room temperature under oil for 20-24 h. After dissection from their vitelline membranes, embryos were washed and fixed, and their cuticles were prepared as described (Kennerdell and Carthew 1998).
RNAi in S2 cells
dsRNAs (∼700 bp long) of EGFP, AGO2 and AGO1, Dicer-1, and Dicer-2 were produced by in vitro T7 transcription, denatured by heating, and reannealed in water. The dsRNA of AGO1 or AGO2 was introduced into S2 cells expressing EGFP by soaking (Clemens et al. 2000). After 3 d, cells were harvested and divided into half. To one half, AGO1 or AGO2 dsRNA was again introduced. To the other half, AGO1 or AGO2 dsRNA plus EGFP dsRNA were introduced into the cells. All cell pools were incubated for another 4 d and subjected to Western blot analyses using anti-EGFP antibody. In Figure 6, to start RNAi for AGO1, AGO2, or EGFP in S2 cells expressing EGFP, dsRNA for each gene was introduced into cells by soaking. After 4 d incubation or after double transfection of siRNA or dsRNA and prolonged incubation for 8 d, total RNAs were prepared from each sample and Northern blotting was performed using a bantam antisense oligo labeled with gamma-32P-ATP.
In vitro RNAi assays
To prepare ftz RNA with miR-2b or let-7 target site, we amplified a region of the ftz gene from pGEM-T-ftz with the following primer pairs: 5′-ACAACATGCATCACCCCCACAGCCT-3′ and 5′-AGAGCTCGAGTATCACAGCCAGCTTTGAGGAGCGTTGTAGTAGTAGCAGCTCTCCGAG-3′ (for the miR-2b target) or 5′-AGAGCTCGAGTGAGGTAGTAGGTTGTATAGTTGTTGTAGTAGTAGCAGCTCTCCGAG-3′ (for the let-7 target). The resulting PCR products were cloned into the NsiI-SalI site of pGEM-T-ftz as an NsiI-XhoI fragment (pGEM-T-ftz-2b or pGEM-T-ftz-let-7). The target RNAs were transcribed with a MEGAScript SP6 kit from PCR templates prepared with SP6 and 3′ (5′-CGCCGGGTGATGTATCTATT-3′) primers and pGEM-T-ftz-2b or pGEM-T-ftz-let-7. RNA was radiolabeled at the 5′-G cap by guanylyltransferase (Ambion). In a 20-μL reaction, 500 ng RNA, 40 U RNasin (Promega), 0.2 U inorganic pyrophosphatase (Sigma), and 5 μL of α-32P GTP (800 mCi/mmole) were incubated in 50 mM Tris-HCl (pH 7.9), 12.5 mM MgCl2, 2 mM EDTA, 60 mM KCl, 1 mg/mL BSA, and 25 mM DTT. The reaction mixture was incubated for 2 h at 37°C, and the radiolabeled RNA was then PAGE-gel purified. Pre-let-7 RNA was transcribed from a synthetic deoxynucleotide template with T7 RNA polymerase in the presence or absence of α-32P-GTP (4). Extract used for in vitro RNAi assays were prepared as described in Tuschl et al. (1999) and Hutvagner and Zamore (2002b) with some modifications. In short, embryos were collected for 2 h and immediately homogenized (0-2-h embryo extract), or then aged for 14 h at room temperature and homogenized (14-16-h embryo extract). Homozygous AGO1k08121 embryos were sorted from their AGO1k08121/CyOKr-Gal4-UAS-GFP and CyO-Kr-Gal4-UAS-GFP/CyO-Kr-Gal4-UAS-GFP siblings under fluorescent microscopy. Approximately 100 mg embryos were homogenized in 100 μL of lysis buffer (100 mM potassium acetate, 30 mM HEPES at pH 7.4, 2 mM magnesium acetate, 5 mM DTT, and 0.5 mg/mL Pefabloc SC), centrifuged for 25 min at 14,600 × g at 4°C, and flash-frozen in 10-μL aliquots. Total protein concentration in the extracts was determined with a colorimetric Bio-Rad assay. Cleavage of dsRNA or processing of pre-let-7 was assayed by incubating 1000 cpm of labeled dsRNA or pre-let-7 in a 10-μL reaction containing 15 μg of extract proteins for increasing times at room temperature. Reactions were stopped by the addition of stop buffer (100 mM Tris-HCl at pH 7.9, 50 mM NaCl, 50 mM EDTA, 1% SDS, and 100 μg/mL proteinase K). For miR-2b target RNA cleavage reactions, 100 nM ftz siRNA duplex was incubated with 2000 cpm of cap-labeled miR-2b target RNA in 14-16-h (Figs. 4, 5) embryo lysate under standard conditions for 90 or 180 min at room temperature. For let-7 target RNA cleavage reactions, 100nM ftz siRNA duplex and 100 nM in vitro transcribed pre-let-7 RNA were incubated with 2000 cpm of cap-labeled let-7 target RNA in 0-2-h embryo lysate under standard conditions for 90 or 180 min at room temperature.
siRNA unwinding assay
siRNA unwinding assay was performed as described in Nykanen et al. (2001) with some modifications. The 5′ end of the antisense strand siRNA was 32P-labeled with PNK. RNAi reactions (10 μL) were quenched with 40 μL of stop buffer and a 100 molar excess of Pp-luc-antisense RNA; incubated for 15 min at 25°C; and immediately ethanol precipitated, redissolved in 4 μL of 1× loading buffer (Takara), and immediately analyzed by electrophoresis at 4°C through 15% native polyacrylamide gel. siRNA sequences were 5′-UCGAAGUAUUCCGCGUACGUG-3′ (Pp-luc-antisense) and 5′-CGUACGCGGAAUACUUC GAAA-3′ (Pp-luc-sense).
RISC assembly assay
RISC assembly assay was performed as described in Tomari et al. (2004). 32P-radiolabeled siRNA duplex was incubated with embryo lysates at room temperature. After incubation, the samples were adjusted to 6% (w/v) glycerol and resolved by submarine native agarose gel electrophoresis. Gels were dried under vacuum onto Hybond-N+ nylon membrane (Amersham Bioscience).
Northern blot analysis
RNA was isolated from S2 cells or the IgG bound fractions with ISOGEN (Nippon gene). Northern blot was performed as described (Ishizuka et al. 2002), except that the RNA was run on 12% acrylamide-denaturing gels and transferred onto Hybond-N+ membrane. The hybridizations were performed at 42°C in 0.2 M sodium phosphate (pH 7.2) and 7% SDS and washed at 42°C in 2× saline sodium citrate and 0.1% SDS.
TAP purification
The expression of AGO1-TAP, AGO2-TAP, or dFMR1-TAP in S2 cells was induced by adding copper ion into the medium. After overnight incubation, the cytoplasmic lysate was prepared in a buffer containing 10 mM HEPES (pH 7.0), 200 mM KOAc, 2 mM MgAc, 0.1% NP40, 2 μg/mL leupeptin, 2 μg/mL pepstatin, and 0.5% aprotinin. AGO1-TAP, AGO2-TAP or dFMR1-TAP, and associated materials to the TAP-tagged fusion protein were bound to IgG beads. After extensive washing, the bound fraction was eluted by treatment with TEV protease. The polyclonal antibodies against Dicer-1 and AGO1 were kind gifts from G. Hannon and T. Uemura, respectively. The AGO2 and dFMR1 monoclonal antibodies (4D2 and 5A11, respectively) were produced by injecting AGO2 or dFMR1 peptides into mice, followed by fusing the mouse spleen cells to myeloma cells to obtain hybridomas producing the specific antibodies.
Acknowledgments
We thank the Bloomington Stock Center for fly stocks, T. Uemura for gifts of AGO1 antisera and AGO1 mutant fly stocks, G. Hannon for anti-Dicer antibodies, S.B. Inoue for help producing AGO2 mutant fly strains, K. Mizumoto for purified capping enzyme, J. Wilusz for an expression vector for capping enzyme, Y. Kurihara for helping produce monoclonal antibodies, and T. Uchiumi for anti-P0 antibody. Some EP fly strains came from Exelixis, Inc. We thank S. Nakielny, A. Nakamura, and members of the Siomi laboratory for discussions and comments on the manuscript. K.O. and A.I. are predoctoral fellows of the 21st century COE Program from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT). Supported in part by grants to M.C.S. and H.S. from MEXT.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1210204.
References
- Bartel D.P. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281-297. [DOI] [PubMed] [Google Scholar]
- Basyuk E., Suavet, F., Doglio, A., Bordonne, R., and Bertrand, E. 2003. Human let-7 stem-loop precursors harbor features of RNase III cleavage products. Nucleic Acids Res. 31: 6593-6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E., Caudy, A.A., Hammond, S.M. and Hannon, G.J. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363-366. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Hipfner, D.R, Stark, A, Russell, R.B., and Cohen, S.M. 2003. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113: 25-36. [DOI] [PubMed] [Google Scholar]
- Caudy A.A., Myers, M., Hannon, G.J., and Hammond, S.M. 2002. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes & Dev. 16: 2491-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell M.A., Xuan, Z., Zhang, M.Q., and Hannon, G.J. 2002. Argonaute family: Tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes & Dev. 16: 2733-2742. [DOI] [PubMed] [Google Scholar]
- Clemens J.C., Worby, C.A., Simonson-Leff, N., Muda, M., Maehama, T., Hemmings, B.A., and Dixon, J.E. 2000. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc. Natl. Acad. Sci. 97: 6499-6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoni C. and Macino, G. 2000. Post-transcriptional gene silencing across kingdoms. Curr. Opin. Genet. Dev. 10: 638-643. [DOI] [PubMed] [Google Scholar]
- Denli A.M. and Hannon, G.J. 2003. RNAi: An ever-growing puzzle. Trends Biochem. Sci. 28: 196-201. [DOI] [PubMed] [Google Scholar]
- Doench J.G., Petersen, C.P., and Sharp, P.A. 2003. siRNAs can function as miRNAs. Genes & Dev. 17: 438-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E., and Mello, C.C. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806-811. [DOI] [PubMed] [Google Scholar]
- Grishok A., Tabara, H., and Mello, C.C. 2000. Genetic requirements for inheritance of RNAi in C. elegans. Science 287: 2494-2497. [DOI] [PubMed] [Google Scholar]
- Grishok A., Pasquinelli, A.E., Conte, D., Li, N., Parrish, S., Ha, I., Baillie, D.L., Fire, A., Ruvkun, G., and Mello, C.C. 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106: 23-34. [DOI] [PubMed] [Google Scholar]
- Hammond S.M., Bernstein, E., Beach, D., and Hannon, G.J. 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404: 293-296. [DOI] [PubMed] [Google Scholar]
- Hammond S.M., Boettcher, S., Caudy, A.A., Kobayashi, R., and Hannon, G.J. 2001. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293: 1146-1150. [DOI] [PubMed] [Google Scholar]
- Hutvagner G. and Zamore, P.D. 2002a. RNAi: Nature abhors a double-strand. Curr. Opin. Genet. Dev. 12: 225-232. [DOI] [PubMed] [Google Scholar]
- ____. 2002b. A microRNA in a multiple-turnover RNAi enzyme complex. Science 297: 2056-2060. [DOI] [PubMed] [Google Scholar]
- Hutvagner G., McLachlan, J., Pasquinelli, A.E., Balint, E., Tuschl, T., and Zamore, P.D. 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293: 834-838. [DOI] [PubMed] [Google Scholar]
- Ishizuka A., Siomi, M.C., and Siomi, H. 2002. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes & Dev. 16: 2497-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka Y., Takeichi, M., and Uemura, T. 2001. Developmental roles and molecular characterization of a Drosophila homologue of Arabidopsis Argonaute1, the founder of a novel gene superfamily. Genes Cells 6: 313-325. [DOI] [PubMed] [Google Scholar]
- Kennerdell J.R. and Carthew, R.W. 1998. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell 95: 1017-1026. [DOI] [PubMed] [Google Scholar]
- Ketting R.F., Fischer, S.E., Bernstein, E., Sijen, T., Hannon, G.J., and Plasterk, R.H. 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes & Dev. 15: 2654-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight S.W. and Bass, B.L. 2001. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 293: 2269-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut, R., Lendeckel, W., and Tuschl, T. 2001. Identification of novel genes coding for small expressed RNAs. Science 294: 853-858. [DOI] [PubMed] [Google Scholar]
- Lau N.C., Lim, L.P., Weinstein, E.G., and Bartel, D.P. 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858-862. [DOI] [PubMed] [Google Scholar]
- Lee R.C. and Ambros, V. 2001. An extensive class of small RNAs in Caenorhabditis elegans. Science 294: 862-864. [DOI] [PubMed] [Google Scholar]
- Lee R.C., Feinbaum, R.L., and Ambros, V. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843-854. [DOI] [PubMed] [Google Scholar]
- Lee Y., Ahn, C., Han, J., Choi, H., Kim, J., Yim, J., Lee, J., Provost, P., Radmark, O., Kim, S., et al. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415-419. [DOI] [PubMed] [Google Scholar]
- Lee Y.S., Nakahara, K., Pham, J.W., Kim, K., He, Z., Sontheimer, E.J., and Carthew, R.W. 2004. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117: 69-81. [DOI] [PubMed] [Google Scholar]
- Liu Q., Rand, T.A., Kalidas, S., Du, F., Kim, H.E., Smith, D.P., and Wang, X. 2003. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 301: 1921-1925. [DOI] [PubMed] [Google Scholar]
- Llave C., Xie, Z., Kasschau, K.D., and Carrington, J.C. 2002. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297: 2053-2056. [DOI] [PubMed] [Google Scholar]
- Martinez J., Patkaniowska, A., Urlaub, H., Luhrmann, R., and Tuschl, T. 2002. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110: 563-574. [DOI] [PubMed] [Google Scholar]
- Nykanen A., Haley, B., and Zamore, P.D. 2001. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell 107: 309-321. [DOI] [PubMed] [Google Scholar]
- Olsen P.H. and Ambros, V. 1999. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 216: 671-680. [DOI] [PubMed] [Google Scholar]
- Reinhart B.J., Slack, F.J., Basson, M., Pasquinelli, A.E., Bettinger, J.C., Rougvie, A.E., Horvitz, H.R., and Ruvkun, G. 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901-906. [DOI] [PubMed] [Google Scholar]
- Reinhart B.J., Weinstein, E.G., Rhoades, M.W., Bartel, B., and Bartel, D.P. 2002. MicroRNAs in plants. Genes & Dev. 16: 1616-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M., and Seraphin, B. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17: 1030-1032. [DOI] [PubMed] [Google Scholar]
- Rorth P., Szabo, K., Bailey, A., Laverty, T., Rehm, J., Rubin, G.M., Weigmann, K., Milan, M., Benes, V., Ansorge, W., et al. 1998. Systematic gain-of-function genetics in Drosophila. Development 125: 1049-1057. [DOI] [PubMed] [Google Scholar]
- Schwarz D.S., Hutvagner, G., Haley, B., and Zamore, P.D. 2002. Evidence that siRNAs function as guides, not primers, in the Drosophila and human RNAi pathways. Mol. Cell 10: 537-548. [DOI] [PubMed] [Google Scholar]
- Tabara H., Yigit, E., Siomi, H., and Mello, C.C. 2002. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell 109: 861-871. [DOI] [PubMed] [Google Scholar]
- Tomari Y., Du, T., Haley, B., Schwarz, D.S., Bennett, R., Cook, H.A., Koppetsch, B.S., Theurkauf, W.E., and Zamore, P.D. 2004. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell 116: 831-841. [DOI] [PubMed] [Google Scholar]
- Tuschl T., Zamore, P.D., Lehmann, R., Bartel, D.P., and Sharp, P.A. 1999. Targeted mRNA degradation by double-stranded RNA in vitro. Genes & Dev. 13: 3191-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B., Ha, I., and Ruvkun, G. 1993. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75: 855-862. [DOI] [PubMed] [Google Scholar]
- Williams R.W. and Rubin, G.M. 2002. ARGONAUTE1 is required for efficient RNA interference in Drosophila embryos. Proc. Natl. Acad. Sci. 99: 6889-6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Vernooy, S.Y., Guo, M., and Hay, B.A. 2003. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr. Biol. 13: 790-795. [DOI] [PubMed] [Google Scholar]
- Zeng Y., Yi, R., and Cullen, B.R. 2003. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc. Natl. Acad. Sci. 100: 9779-9784. [DOI] [PMC free article] [PubMed] [Google Scholar]