Abstract
Signaling molecules such as Cdc42 and mitogen-activated protein kinases (MAPKs) can function in multiple pathways in the same cell. Here, we propose one mechanism by which such factors may be directed to function in a particular pathway such that a specific response is elicited. Using genomic approaches, we identify a new component of the Cdc42- and MAPK-dependent signaling pathway that regulates filamentous growth (FG) in yeast. This factor, called Msb2, is a FG-pathway-specific factor that promotes differential activation of the MAPK for the FG pathway, Kss1. Msb2 is localized to polarized sites on the cell surface and interacts with Cdc42 and with the osmosensor for the high osmolarity glycerol response (HOG) pathway, Sho1. Msb2 is glycosylated and is a member of the mucin family, proteins that in mammalian cells promote disease resistance and contribute to metastasis in cancer cells. Remarkably, loss of the mucin domain of Msb2 causes hyperactivity of the FG pathway, demonstrating an inhibitory role for mucin domains in MAPK pathway activation. Taken together, our data suggest that Msb2 is a signaling mucin that interacts with general components, such as Cdc42 and Sho1, to promote their function in the FG pathway.
Keywords: Morphogenesis, cell polarity, signal transduction, pseudohyphal growth, specificity
How specificity is achieved by signaling pathways that contain common factors is a question that grows in complexity with the discovery of each new common factor. For example, in the budding yeast Saccharomyces cerevisiae, the filamentous growth MAPK (mitogen-activated protein kinase) pathway (FG pathway) is composed chiefly of general components (Fig. 1A; Kron 1997; Madhani and Fink 1998; Madhani 2000; Pan et al. 2000), including the ubiquitous polarity establishment Rholike GTPase Cdc42, its PAK Ste20, the MAPK components Ste11/Ste50 and Ste7, and transcription factor Ste12 (Liu et al. 1993; Roberts and Fink 1994; Peter et al. 1996; Leberer et al. 1997; Jansen et al. 2001; Lamson et al. 2002). These core components are also required in the mating pathway (Elion 2000; Dohlman and Thorner 2001), and at least four of them—Cdc42, Ste20, Ste50, and Ste11—are components of the high osmolarity glycerol response (HOG) pathway (O'Rourke and Herskowitz 1998; Posas et al. 1998; Raitt et al. 2000; Reiser et al. 2000). Cdc42, which is associated with the plasma membrane by isoprenylation at its C terminus, functions not only in these signal transduction pathways but also as a general trigger for polarized growth (Wedlich-Soldner et al. 2003) and the cell cycle. Thus, regulating the activity of factors that function in multiple pathways, such as Cdc42, is crucial for signal propagation and polarity reorganization that leads to correct cell morphogenesis and cell fate.
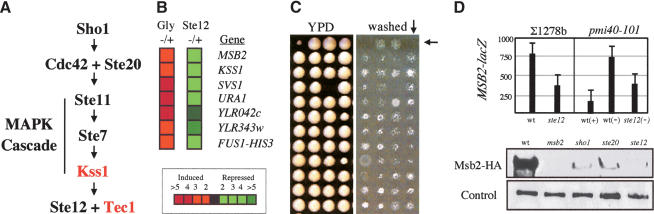
Figure 1.
MSB2 is an FG-pathway target. (A) The FG pathway with general factors (black) and FG-pathway-specific factors (red). (B) DNA microarray analysis. Targets of a glycosylation defect (left column, Gly-/+, pmi40-101 induced in YPD-/+ Man) that were also Ste12-dependent (right column, Ste12-/+, pmi40-101 ± ste12 induced in YPD-Man) are shown. (Red) Induced expression; (green) repressed expression (inset shows fold change). FUS1-HIS3 is a predicted target used as a control (Cullen et al. 2000). (C) Part of the genomic screen to isolate agar-invasion defective mutants. (Left panel) Colonies pinned to YPD for 4 d; (right panel) washed plate. Arrows point to the msb2 mutant. (D) MSB2 expression. (Upper panel) MSB2-lacZ expression. Cells of the indicated genotype and background containing pMSB2-lacZ were assayed for β-galactosidase activity. Wild-type (wt) or ste12 mutant ∑1278b cells were assayed over time for MSB2 expression; the 16-h time point is shown. Glycosylation mutant pmi40-101 containing or lacking the ste12 mutation (wild type [wt], pmi40-101, and ste12, pmi40-101 ste12) was incubated for 8 h ±Man as indicated. As shown, MSB2 expression in glycosylation mutants is partially induced by another factor. Numbers are in Miller Units, and error bars show standard deviation. (Lower two panels) Western blots. ∑1278b cells of the indicated genotypes expressing Msb2-HA were grown to mid-log phase in YPD, and extracts were assayed using antibodies against HA (upper panel) or a control protein (Dpm1, lower panel).
Among the proteins that influence specificity in signaling pathways are scaffolding proteins. Ste5 is a scaffold for the mating pathway (Whitmarsh and Davis 1998; Elion 2001) and Pbs2 for the HOG pathway (Posas and Saito 1997). Such scaffolds tether general MAPK cascade components to specific MAPKs to promote differential pathway activation (Harris et al. 2001; van Drogen and Peter 2001). Indeed, each MAPK pathway in yeast has its own MAPK: Kss1 for the FG pathway, Fus3 for the mating pathway, and Hog1 for the HOG pathway (Cook et al. 1997; Madhani et al. 1997; Bardwell et al. 1998a; Breitkreutz et al. 2001; Sabbagh et al. 2001; for reviews, see Madhani and Fink 1998; Sprague 1998; Breitkreutz and Tyers 2002). Factors that insulate signaling pathways have also been identified at the transcription factor level. Ste12, for example, has a distinct binding partner, Tec1, that specifies it to function in the FG pathway (Madhani and Fink 1997; Bardwell et al. 1998a). Ste12 is also influenced by negative regulatory proteins, Dig1/Rst1 and Dig2/Rst2 (Cook et al. 1996; Tedford et al. 1997; Bardwell et al. 1998b) and by a specific cyclin-dependent kinase, Srb10 (Nelson et al. 2003).
Remarkably little is known about factors that contribute to specificity and activation at the head of the FG pathway. A receptor for the FG pathway has yet to be identified. The presumptive osmosensor for the HOG pathway, Sho1 (Maeda et al. 1995; Raitt et al. 2000; Reiser et al. 2000), is required for diploid pseudohyphal growth and for activation of an Ste12-dependent pathway in response to a glycosylation defect (Fig. 1A; Lee and Elion 1999; O'Rourke and Herskowitz 1998; Cullen et al. 2000). Thus, Sho1 may be a general component of both the HOG and FG pathways. And although Cdc42 and Ste20 function in the FG pathway, how they are directed to function specifically in that pathway has yet to be determined. It is known that 14-3-3 proteins interact with Ste20 and may influence Ste20 function in the FG pathway (Roberts et al. 1997).
We undertook complementary genomics approaches to better characterize the FG pathway and discovered a cell-surface protein, Msb2, that functions at the pathway's head. Msb2 interacts with both Sho1 and Cdc42 and appears to be a FG-pathway-specific factor. Thus, Msb2 may specify Sho1 and Cdc42 to function in the FG pathway. Moreover, Msb2 is a member of the signaling mucin class of proteins, and we discovered that deletions within the mucin domain of Msb2 induced hyperactivity. Therefore, our results point to new functions for signaling mucins in controlling cell polarity and signaling in Cdc42- and MAPK-dependent pathways. Finally, because signaling mucins play an important role in metastasis in human cancer cells, our results suggest a simple mechanism whereby mutation of signaling mucins can cause MAPK pathway hyperactivation and possibly cancer in human cells.
Results
Independent genomic approaches identify MSB2 as an FG pathway target
DNA microarray analysis was used to identify targets of the FG pathway. Two microarray comparisons were performed: one to uncover targets of a glycosylation defect (pmi40-101 ± Man; mannose), and a second to identify those targets that were Ste12-dependent (pmi40-101 ± Ste12). This exploited the fact that glycosylation defects activate the FG pathway and that in a particular glycosylation mutant (pmi40-101) such defects are suppressed in mannose-supplemented medium (Cullen et al. 2000). A small number of potential targets of the FG pathway were identified after a rigorous statistical analysis (Fig. 1B; Supplementary Table S5). Most of the induced genes were known FG pathway targets, including KSS1, YLR042c, SVS1, and Ty1 elements (Madhani et al. 1999; Morillon et al. 2000; Roberts et al. 2000). However, three potentially new FG pathway targets were identified: MSB2, URA1, and YLR343w (Fig. 1B).
In a related approach, genomic screens of the yeast ordered deletion collection were performed to identify mutants that displayed differential FG from wild type in a plate-washing assay. One of the genes identified by this mutant screen, MSB2, was also identified by the microarray analysis. In particular, msb2 mutants were agarinvasion defective (Fig. 1C). Several genes identified in the genomic screens that were initially thought promising were disrupted, and the resulting mutants were characterized for phenotypes in FG. The msb2 mutant stood out among these as having the strongest phenotype, equivalent to ablation of known FG pathway functions. Therefore, MSB2 was extensively characterized.
To confirm that MSB2 is an FG pathway target, expression of the MSB2 gene was monitored using an MSB2-lacZ transcriptional fusion. In strains of the filamentous background (∑1278b), full MSB2 expression was dependent on Ste12 (Fig. 1D). Glycosylation defects also induced Ste12-dependent MSB2 expression (Fig. 1D). We examined the abundance of the Msb2 protein by measuring the levels of a functional Msb2-HA fusion under the control of its native promoter. Msb2-HA levels were significantly reduced in ∑1278b strains lacking FG pathway components Sho1, Ste20, and Ste12 (Fig. 1D). We examined the upstream sequence of the MSB2 gene and identified two consensus Ste12-binding sites ([A]TGAAACA) at 474-481 and 522-530 bp upstream of the start site, suggesting that Ste12 has the potential to bind directly to the MSB2 promoter to induce its expression.
Msb2 is a component of the FG pathway
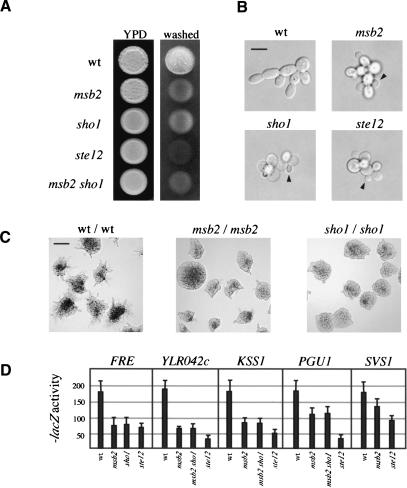
That MSB2 is a target of the FG pathway suggests that Msb2 may have a role in FG. We disrupted MSB2 in haploid ∑1278b cells, which caused a defect in agar invasion by the plate-washing assay (Fig. 2A). The defect was equivalent to FG pathway null mutations such as those in presumptive upstream (Sho1) and downstream (Ste12) components (Fig. 2A), providing the first suggestion that Msb2 is not only a target but also a component of the FG pathway. Disruption of MSB2 and SHO1 together conferred a phenotype equivalent to either single mutation (Fig. 2A). The single-cell invasive-growth assay (Cullen and Sprague 2000) demonstrated that Msb2 was required for unipolar budding and polarized growth (Supplementary Table S3) and had phenotypes that were equivalent to FG pathway mutants (Fig. 2B). Diploid cells undergo pseudohyphal growth in response to limiting nitrogen (Gimeno et al. 1992), and homozygous msb2/msb2 mutant diploids were defective for pseudohyphal growth to the same degree as sho1/sho1 mutants (Fig. 2C) and other homozygous FG-pathway mutants (data not shown).
Figure 2.
Msb2 is required for filamentous growth. (A) Plate-washing assay. Equal concentrations of cells of the indicated genotypes and ste4 were spotted onto YPD, incubated for 4 d (left), and washed (right). (B) Single-cell invasive-growth assay. Cells as in A were spread onto SC medium (lacking glucose), incubated for 16 h, and photographed at 100×. Arrows point to buds emerging from the proximal pole. Bar, 10 μm. (C) Pseudohyphal growth assay. Cells were spread onto low nitrogen (SLAHD) medium for 48 h and photographed at 20×. Bar, 50 μm. (D) Expression of FG-pathway reporters as indicated in given strains. Extracts prepared from cells in midlog phase SCD-LEU (SCD, synthetic complete dextrose) to select for reporter plasmids. All wild-type values were set at 180, and mutant values were adjusted by ratiometric division so that all values could be compared on the same scale. Numbers are in Miller Units.
We verified the possibility that Msb2 is an FG-pathway component by assaying expression of FG-pathway-dependent reporters. Expression of FRE-lacZ (FRE, filamentous response element; Laloux et al. 1994) and presumptive FG-pathway targets YLR042c, KSS1, PGU1, and SVS1 (Madhani et al. 1999; Roberts et al. 2000) were Msb2-, Sho1-, and Ste12-dependent (Fig. 2D). Agar invasion and expression of the reporters was not reduced in sho1 msb2 mutants to the same level as in ste12 mutants, indicating that a third factor may contribute to full Ste12-dependent expression (Fig. 2A,D). DNA microarray analysis confirmed that the expression of known FG pathway targets is Msb2-dependent (data not shown).
As previously described, the FG pathway is activated in glycosylation mutants and is required for their viability (Cullen et al. 2000). We found that Msb2 was required for FG-pathway activation in such mutants and likewise was required for their viability (Supplementary Fig. S1), indicating that Msb2 is an FG-pathway component by these criteria as well.
Msb2 is localized to polarized sites at the cell surface and is an integral-membrane protein
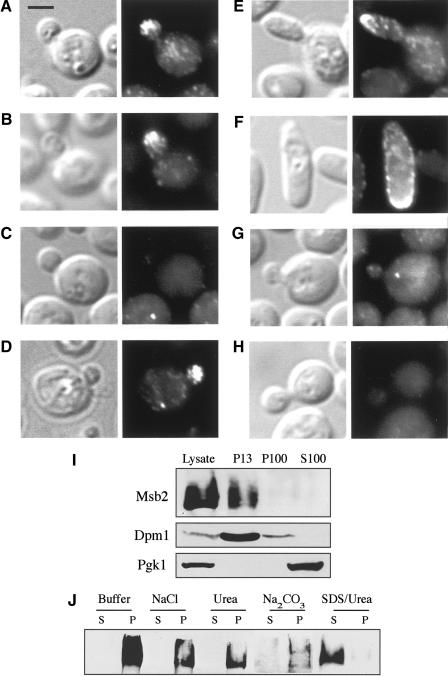
We characterized Msb2 to define its function in the FG pathway. Msb2 is a predicted cell-surface protein containing a single transmembrane domain near the C terminus of the protein (Bender and Pringle 1992; O'Rourke and Herskowitz 2002), resulting in a large extracellular domain (1190 amino acids) and a small cytoplasmic domain (98 amino acids). Immunofluorescence was performed to determine the cellular location of Msb2. Msb2-HA was detected in small and medium-sized buds at the distal pole (Fig. 3A). Msb2-HA was also observed within cells as cables extending to the distal pole, presumably reflecting its delivery to the cell surface via the actin cytoskeleton (Fig. 3B). Indeed, treatment of cells with the actin-depolymerizing agent Latrunculin A (Lat A) prevented distal-pole localization of Msb2 (Fig. 3C), whereas treatment of cells with DMSO (dimethyl sulfoxide) alone, in which Lat A is solubilized, did not impair Msb2-HA localization (Fig. 3D). In cells overproducing Msb2-HA (by the GAL1 promoter), Msb2 was observed at the distal pole, at the cell periphery, and at sites of presumptive bud emergence (Fig. 3E,F). Msb2 was not detected in FG-pathway mutants such as ste12 (Fig. 3G), consistent with the fact that MSB2 gene expression and hence abundance is controlled by the FG pathway. The fluorescent staining pattern was not observed in cells lacking the HA epitope (Fig. 3H).
Figure 3.
Msb2 localization. (A-H) Immunolocalization of Msb2-HA. Treatments or genotypes are as follows: wild type (A,B); Latrunculin A (Lat A; C); DMSO (D); GAL-MSB2-HA (E,F); ste12 (G); no tag (H). (Left panels) DIC; (right panels) FITC. Bar, 5 μm. (I,J) Subcellular localization of Msb2. (I) Cells expressing Msb2-HA were solubilized and cell extracts were separated by centrifugation. Lysate, supernatnat (S), and pellet (P) fractions are shown ([13] 13,000XG; [100] 100,000XG). Western blotting was performed using antibodies against HA, Dpm1 (an ER integral-membrane protein), and Pgk1 (a soluble cytoplasmic protein) as indicated. (J) P13 fraction analysis. Treatments were lysis buffer alone (Buffer) or with NaCl (0.5 M), urea (5 M), sodium bicarbonate (100 mM Na2CO3 at pH 11), or SDS (5%) and urea (8 M).
Subcellular fractionation confirmed the topological prediction that Msb2 is an integral-membrane protein. Supernatant and pellet fractions were prepared from cell lysates containing Msb2-HA, which was detected in the pellet, P13, fraction (Fig. 3I), a location consistent with the plasma membrane and associated proteins. Treatments that solubilize peripheral membrane proteins, such as salt, sodium bicarbonate, and urea failed to liberate Msb2 into supernatant fractions, whereas treatments that disrupt membranes solubilized Msb2 (Fig. 3J), confirming that Msb2 is an integral-membrane protein.
Msb2 and Sho1 function together in the FG pathway
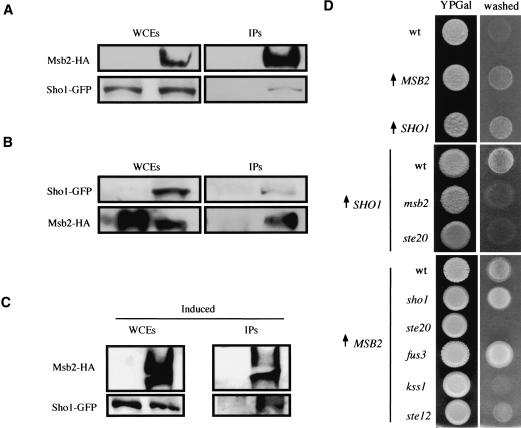
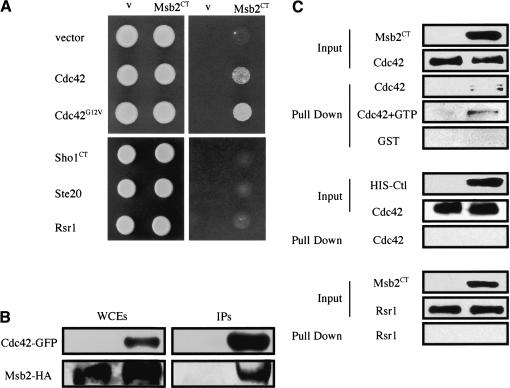
The localization pattern of Msb2 suggests that it functions as a cell-surface component of the FG pathway. Because Sho1 is also a presumptive cell-surface protein that functions in the FG pathway, we tested the possibility that Msb2 and Sho1 interact. Protein interaction was tested by coimmunoprecipitation (coIP) of permeabilized cell extracts containing epitope-tagged and functional Msb2-HA and Sho1-GFP proteins in 1% NP-40. In exponentially growing cells, we observed an interaction between Msb2 and Sho1. That is, precipitation of Msb2-HA using antibodies against the HA epitope coprecipitated Sho1-GFP (Fig. 4A). We substantiated this finding by the reverse coIP: precipitation of Sho1-GFP using anti-GFP antibodies coprecipitated Msb2-HA (Fig. 4B). We also examined the interaction between Msb2 and Sho1 under conditions in which the FG pathway is active, hypothesizing that the interaction may be dynamic. Indeed, precipitation of Msb2 in extracts prepared from cells undergoing filamentous growth coprecipitated Sho1-GFP in greater abundance than from cells in vegetative growth (Fig. 4, cf. C and A). One contributing factor to the more robust interaction may be that more Msb2 is present under this condition because of induction of MSB2 gene expression during FG.
Figure 4.
Msb2 and Sho1 interact and function together in the FG pathway. (A) Immunoprecipitation of Msb2-HA also precipitates Sho1. Western blots of whole-cell extracts (WCEs) and immunoprecipitations (IPs) are shown, probed using antibodies specific for HA (Msb2-HA) and GFP (Sho1-GFP). (B) Immunoprecipitation of Sho1-GFP also precipitates Msb2-HA. Labels are as in A. (C) Sho1-GFP is coprecipitated at higher concentrations by Msb2-HA in cells undergoing FG. Induced refers to cells undergoing FG from which extracts for coIPs were prepared; see Materials and Methods for details. (Msb2 is more abundant under this condition, presumably because of to FG-pathway induction of MSB2 expression. However, the amount of IPed Msb2 is similar under induced and uninduced conditions; under inducing conditions, more Sho1 is precipitated. (D) Cells overproducing Msb2 or Sho1 in FG-pathway mutant backgrounds. Cells were spotted onto YP + 2% galactose (YPGal; left panels; [↑] GAL1 promoter) and washed after 48 h (right panels).
Placement of Msb2 and Sho1 together at the head of the FG pathway is consistent with genetic evidence obtained using FG-pathway reporters and by examining agar-invasion phenotypes. First, as already shown, the mutant phenotypes of msb2 and sho1 disruptions in FG assays are equivalent (see Fig. 2A-C). Second, overexpession of Sho1 caused hyperinvasive growth that was Msb2- and Ste20-dependent (Fig. 4D). Likewise, overexpression of Msb2 also caused hyperinvasive growth (Fig. 4D); however, such overexpression restored agar invasion to the sho1 mutant (Fig. 4D), suggesting that Msb2 can bypass the requirement for Sho1 in FG when overexpressed. Agar invasion caused by overexpression of Msb2 was dependent on Ste20 (Fig. 4D), as was FG-pathway activity (Supplementary Fig. S2). Third, a hyperactive allele of Msb2 induced FG-pathway-reporter activation and morphological abnormalities that were Sho1- and Ste20-dependent (see below). Therefore, Msb2 and Sho1 function together upstream of the PAK Ste20 in the FG pathway.
Msb2 interacts with Cdc42
We addressed the possibility that Msb2 interacts with known downstream components of the FG pathway. Because Msb2 is an integral-membrane protein containing a presumptive cytoplasmic domain, directed two-hybrid analysis was performed using its cytoplasmic tail (CT, amino acids 1209-1306), called Msb2CT. Msb2CT was tested for interaction with FG-pathway components, and a specific interaction was found between Msb2CT and Cdc42 (Fig. 5A). Msb2CT interacted better with an activated allele of Cdc42, Cdc42G12V (Fig. 5A). Msb2CT did not interact with another GTPase, Rsr1, the PAK Ste20, or with the cytoplasmic tail of Sho1 (Fig. 5A). Western blot confirmed equivalent levels of protein in all cells (data not shown).
Figure 5.
Msb2 interacts with Cdc42. (A) Two-hybrid analysis. GBD-Msb2CT in strains containing GAD fusions to GTPases Cdc42 and Rsr1 and FG-pathway components as shown. Growth was scored on SCD-URA-LEU-HIS + 4 mM aminotriazole (AT). (v) Vector. (B) Msb2-HA and Cdc42-GFP interact by coIP analysis. IP of Cdc42-GFP in IP buffer with 1% NP-40 using antibodies specific for GFP (upper panels) coprecipitates Msb2-HA (lower panels). The WCE and IPs are shown. (C) Cdc42 and Msb2CT interact in vitro. Fusion proteins were expressed and purified from E. coli and incubated together in binding buffer containing BSA (Input) and GTP as indicated. After a 15-min incubation, Co+ beads that recognize the HIS epitope on the Msb2CT and Ctl [Urm1] fusions were added to the reaction for 15 min, and HIS-tagged proteins were isolated by low-speed centrifugation (Pull Down). Western blotting was used to detect the GST epitopes on the Cdc42, GST, and Rsr1 fusions, and the HIS epitopes on the Msb2CT and Ctl [Urm1] fusions. The inputs for Cdc42 + GTP and GST reactions had equivalent input protein amounts to that shown for the inputs in the top two panels (data not shown).
CoIP analysis confirmed the interaction between Msb2 and Cdc42. Precipitation of GFP-Cdc42 in IP buffer with 1% NP-40 using antibodies that recognize the GFP epitope coprecipitated Msb2-HA (Fig. 5B). In contrast to the Msb2-Sho1 interaction, the Msb2-Cdc42 interaction was not influenced by conditions that activate the FG pathway (data not shown). We also confirmed the interaction between Msb2 and Cdc42 by pulldowns using epitope-tagged proteins expressed and purified from bacterial cells. Specifically, isolation of a purified HIS-Msb2CT fusion protein using beads that recognize the HIS epitope also pulled down GST-Cdc42 (Fig. 5C), but at substoichiometric levels. Addition of GTP did not significantly influence the interaction (Fig. 5C). HIS-Msb2 did not differentially pull down GST or another Rho-like GTPase, GST-Rsr1, under these conditions, and a control HIS-tagged protein (HIS-Urm1) did not pull down Cdc42 (Fig. 5C). Because the coIP experiment showed a more substantial interaction between Msb2 and Cdc42 than the two-hybrid or in vitro experiments, the in vivo context of these membrane-associated proteins might contribute to their interaction.
Interaction between Msb2 and Cdc42 is also supported by genetic evidence. First, MSB2 is a high-copy suppressor of alleles of CDC24, which encodes the guanine nucleotide exchange factor (GEF) for Cdc42 (Bender and Pringle 1992). (We did not observe an interaction between Msb2 and Cdc24 by coIP; data not shown.) Second, overexpression of Cdc42 exacerbated the morphological abnormalities present in cells containing a hyperactive allele of Msb2 (data not shown). Third, an allele of STE20 lacking the Cdc42-interaction domain, ste20Δ335-370 (Peter et al. 1996; Leberer et al. 1997), which is specifically defective for FG, blocked the elevated signaling in cells overproducing Msb2 (Supplementary Fig. S2) and was required for Msb2-dependent FUS1 expression in glycosylation mutants (Supplementary Table S4). Because this allele does not abrogate mating pathway activity, its ability to block Msb2 is an indication of a specific role for Msb2 in influencing Cdc42 function in the FG pathway. By these results, we surmise that Msb2 requires Cdc42 to activate the FG pathway, and presumably it does so directly.
Msb2 lacking its mucin domain is hyperactive and dominant
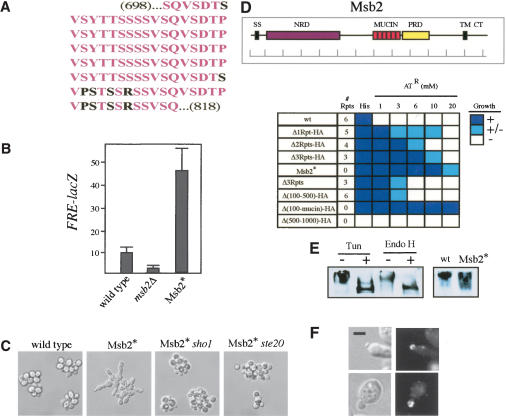
Msb2 contains six tandem Ser/Thr/Pro-rich repeats (amino acids 698-818) in its presumed extracellular domain (Fig. 6A) that are characteristic of domains found in glycosylated cell-surface adhesion molecules called mucins (Hollingworth and Swanson 2004). We asked if the mucin-homology (mucin) domain of Msb2 is required for its function in the FG pathway. We deleted the mucin domain of Msb2 and replaced it with the HA epitope (called Msb2*). Surprisingly, Msb2* caused elevated FRE-lacZ expression (Fig. 6B) and morphological phenotypes such as hyperelongation and unipolar budding (Fig. 6C). These phenotypes were dependent on Sho1, Ste20, and Ste12 (Fig. 6C; Supplementary Fig. S3). Wild-type MSB2, introduced on a plasmid, did not suppress Msb2*, indicating the mutation is dominant (Supplementary Fig. S3). Nutrient-rich conditions did, however, suppress the morphological phenotypes, demonstrating that the mutation was hyperactive (as opposed to constitutive; Supplementary Fig. S3). These results suggest that Msb2 is a positive factor in the FG pathway, and the mucin domain of Msb2 inhibits or regulates the active form of the protein.
Figure 6.
Properties of the Msb2 mucin. (A) The mucin domain of Msb2. Identical amino acids (pink) and nonidentical amino acids (black) are shown. (B) FRE-lacZ expression in strains lacking Msb2 or containing Msb2*. Numbers are in Miller Units. (C) Morphological phenotypes associated with Msb2*. Cells were grown to saturation in YEPD medium and examined at 100×. (D) Hyperactive variants of the Msb2 protein. (Inset) The Msb2 protein. Shown are the N-terminal signal sequence (SS), the N-terminal negative regulatory domain (NRD, purple), the mucin repeats (MUCIN, pink), the positive regulatory domain (PRD, yellow), the transmembrane domain (TM), and the cytoplasmic tail (CT). Bar in 100-amino acid increments. Strains with indicated alleles of Msb2 were tested for expression of the FUS1-HIS3 reporter by growth on SCD-His + aminotriazole (AT) at the concentrations shown. Growth was scored over separate trials: (+) growth, dark blue; (+/-) spotty growth, light blue; (-) no growth, white. (E) Msb2 is glycosylated. (Left panel) Cells containing Msb2-HA were grown to mid-log phase in YPD and treated with DMSO (lane 1) or tunicamycin in DMSO (lane 2) for 2 h, and extracts were prepared for SDS-PAGE and Western analysis using anti-HA antibodies. (Lanes 3,4) Endo H treatment (right two lanes) was performed on cell extracts derived from mid-log-phase cells. (Right panel) Msb2-HA (wt) or Msb2* cells were grown to mid-log phase, and extracts were prepared and analyzed as above. (F) Localization of Msb2-HA in cells treated with tunicamycin (upper two panels). Localization of Msb2* (lower two panels). Bar, 5 μm. (Left panels) DIC; (right panels) FITC. For wild-type reference, see Figure 3A.
Additional mutations were created to study the role of the mucin domain in regulating Msb2 function. A variant of Msb2 lacking only one of the six tandem repeats was sufficient to induce hyperactivity (Fig. 6D; Δ1Rpt-HA). Variants lacking progressively more repeats caused a progressive increase in hyperactivity, with the original variant, Msb2* (lacking all 6 repeats), being the most hyperactive (Fig. 6D). One hyperactive variant was identified that lacked several repeats and the HA epitope (Fig. 6D; Δ3Rpts). This deletion was presumably created by recombination between mucin repeat sequences that exhibit between 84% and 100% identity at the nucleotide sequence level. This result suggests that hyperactive variants of Msb2 may arise by spontaneous deletion of the mucin domain. Changes in the length of the mucin domain were not the cause of the hyperactivity, because hyperactive variants were identified that were the same size as wild-type Msb2 (e.g., Δ3Rpts-HA; Fig. 6D), nor was the HA epitope required for hyperactivity (Fig. 6D; Δ3Rpts). Thus, both the number and particular amino acid sequence of the repeats contribute to their regulatory function.
Additional deletions were made in the MSB2 gene. Msb2Δ(100-500)-HA, which deletes an N-terminal region separate from the mucin domain, also caused detectable hyperactivity. Thus, a distinct negative regulatory domain (NRD; Fig. 6D and inset), like the mucin domain, has a function in inhibiting Msb2 function. A deletion encompassing both inhibitory domains, Msb2Δ(100-mucin)-HA, was the most hyperactive allele isolated (Fig. 6D). In contrast, a variant lacking an additional region of Msb2, Msb2Δ(500-1000)-HA, resulted in a nonfunctional Msb2 protein (although it was expressed at wild-type levels; Fig. 6D). We presume this domain plays a positive regulatory role in Msb2 function (PRD in Fig. 6D, inset). Mutants of Msb2 lacking its N-terminal signal sequence (SS), presumptive transmembrane domain (TM), or cytoplasmic tail (CT) were defective for FG-pathway activity (data not shown). These data demonstrate that the presumptive extracellular region of Msb2 contains positive (PRD) and negative (NRD and MUCIN) regulatory domains.
Mucins are heavily modified by glycosyl side-chain addition. Msb2 has nine predicted sites for N-linked glycosylation, seven of which reside in its presumed extracellular domain, and immunoblot analysis showed that Msb2-HA migrates higher (>250 kDa) than its predicted molecular mass (140 kDa). Treatment of cells with the N-linked glycosylation inhibitor, tunicamycin, or treatment of cell extracts with an enzyme that cleaves N-linked glycosyl side chains, Endo H, caused an increase in the mobility of Msb2 (Fig. 6E), confirming that Msb2 is glycosylated.
Two pieces of evidence suggest that glycosylation of Msb2 may inhibit its function. First, a glycosylation defect activates the FG pathway in an Msb2-dependent fashion (Supplementary Fig. S1); Msb2 itself is presumably underglycosylated under this condition. Second, mucin domains themselves are glycosylated (Silverman et al. 2001), and the Msb2* protein, which lacks its mucin domain, migrates more rapidly than wild type (Fig. 6E), indicating that Msb2* is underglycosylated. Perhaps lack of glycosylation is one reason that Msb2* is hyperactive. Both wild-type Msb2 following tunicamycin treatment and Msb2* have normal localization (Fig. 6F, upper and lower panels, respectively); thus changes in subcellular locale are not responsible for Msb2 activation.
Specificity of Msb2 for activation of the FG-pathway MAPK
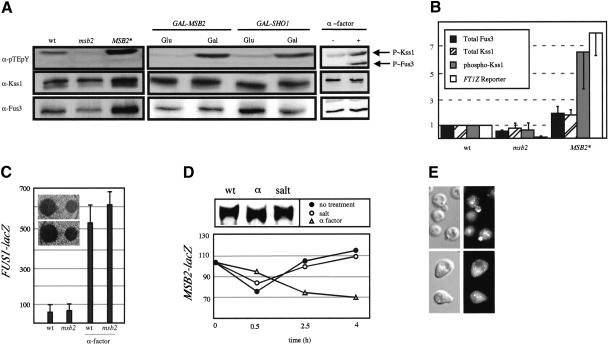
That Msb2 is at the head of the FG pathway suggests that it is required for the phosphorylation and activation of the MAPK Kss1. We examined Kss1∼P levels in strains containing alleles of MSB2. Kss1 was underphosphorylated in msb2 mutants, particularly in those lacking an intact mating pathway (Fig. 7A). Msb2* caused hyperphosphorylation of Kss1 (threefold; Fig. 7A,B) and higher abundance of the Kss1 protein (twofold), resulting in a sixfold net increase in Kss1∼P. Msb2-dependent induction of Kss1∼P correlated with induced FRE-lacZ expression (Fig. 7B). Likewise, overproduction of Msb2 caused hyperphosphorylation of Kss1 (Fig. 7A). As for Msb2, overproduction of Sho1 induced Kss1∼P (Fig. 7A).
Figure 7.
Msb2 is an FG-pathway-specific factor. (A) Western blot of P∼Kss1 (upper band, upper panel), P∼Fus3 (lower band, upper panel), total Kss1 (middle panel), and total Fus3 (bottom panel) from strains containing the indicated genotypes and ste4. (B) Total Fus3 protein, total and dually phosphorylated Kss1 protein, and FRE-lacZ from YEpU-FT1Z were quantified in two independent experiments and normalized to wild type. (C) FUS1-lacZ expression and halo assays. Wild-type (wt) or msb2 cells were incubated in ±30 μM α-factor for 4 h and tested for FUS1-lacZ expression. Numbers are in Miller Units. (Inset) Halo assay. Cells were spread onto YPD plates to which 2 or 10 μL of 1 μg/μL α-factor was applied. (D) Wild-type cells containing MSB2-lacZ in mid-log phase were incubated ±0.9 M NaCl or ±30 μM α-factor. Numbers are in Miller Units. (Upper panel) Western blot of Msb2-HA from same cells treated with salt or α-factor: (α) α-factor. (E) Msb2-HA localization in cells treated with 30 μM α-factor for 3 h (lower panels) or untreated (upper panels).
We also examined the effect of Msb2 and Sho1 on phosphorylation of Fus3, the MAPK for the mating pathway. Fus3∼P was not influenced by Msb2 or Sho1 (Fig. 7A), although its abundance was Msb2-dependent. The observation that Msb2 and Sho1 are both required for phosphorylation of Kss1, but not Fus3, is brought into sharp focus by examination of the phosphorylation of the kinases upon mating pathway induction. In this situation, both MAPKs were equivalently phosphorylated (Fig. 7A). This result shows that Msb2 and Sho1 influence phosphorylation of Kss1 to an equivalent degree as does pheromone-stimulated receptor during mating. Consistent with these results, Kss1 was required for the hyperinvasive growth phenotypes of cells overproducing Msb2 (Fig. 4D), whereas Fus3 caused significant inhibition (Fig. 4D).
That Msb2 is not required for inducing Fus3∼P suggests that Msb2 may not play a role in mating. Consistent with this hypothesis, we found that Msb2 was not required for basal or pheromone-induced FUS1-lacZ expression in wild-type cells (Fig. 7C); nor was it required for other pheromone-dependent processes such as cell cycle arrest (Fig. 7C, inset), shmoo formation or morphology, secretion of α-factor, or mating (data not shown). Addition of α-factor to wild-type MATa cells containing the MSB2-lacZ fusion did not induce its expression or the abundance of the Msb2 protein (Fig. 7D). We also found that Msb2-HA was absent from shmoo tips (Fig. 7E), an unusual result given that many membrane proteins become concentrated at the end of the shmoo.
We also tested Msb2 for a function in the HOG pathway. Msb2 was not required for osmotolerance (data not shown). Moreover, expression of the MSB2 gene and abundance of the Msb2 protein were unaffected by osmotic shock (Fig. 7D). These observations are consistent with evidence that Msb2 is not required for phosphorylation or localization of the MAPK for the HOG pathway or for induction of HOG pathway targets (O'Rourke and Herskowitz 2002). Indeed, HOG pathway components Pbs2 (MAPKK) and Hog1 (MAPK) negatively regulate the FG pathway, possibly by interaction with Sho1 (O'Rourke and Herskowitz 1998; Davenport et al. 1999). We found that disruption of PBS2 in ∑1278b cells stimulated Msb2- and Sho1-dependent FG-pathway activity (Supplementary Fig. S2). Moreover, FUS1 reporter induction in rga1 pbs2 mutants (Stevenson et al. 1995), Rga1 being one of three GTPase activating proteins for Cdc42 (Gladfelter et al. 2002; Smith et al. 2002), was Msb2- and Sho1-dependent (Supplementary Fig. S2). Thus, Msb2 functions antagonistically to the HOG pathway to promote Sho1-, Cdc42-, and Kss1-dependent FG-pathway activation.
Discussion
Upstream factors of the FG pathway
Genome-wide investigations aimed at identifying new factors of the FG pathway uncovered Msb2 as a FG-pathway component. Genetic, localization, and biochemical evidence indicate that Msb2 functions at the cell surface as an upstream component of the FG pathway. The osmosensor for the HOG pathway, Sho1, has also been implicated as a component of the FG pathway (O'Rourke and Herskowitz 1998). In this study, we substantiate the requirement for Sho1 as an FG-pathway component that also functions at the head of the pathway. Our results provide the first definitive evidence for cell-surface proteins at the top of the FG pathway. The identification of such proteins is an important step in defining the receptor for the FG pathway and elucidating those stimuli that trigger it.
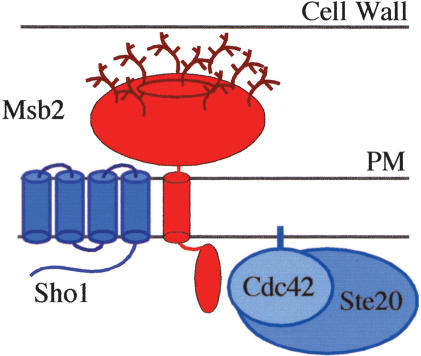
Based on topological predictions and biochemical evidence, a model emerges for FG-pathway proteins at the plasma membrane (Fig. 8). Msb2 interacts with both Cdc42 and Sho1. Because Msb2 is the only FG-pathway-specific component of the three, perhaps it functions to recruit Sho1 and Cdc42 to the pathway. Evidence from hyperactive alleles of Msb2 (which may mimic the active form of the protein) indicates that the extracellular mucin domain of Msb2 communicates to its cytoplasmic domain to inhibit Msb2's function. Changes to the extracellular domain may directly alter the conformation of the cytoplasmic tail. Alternatively, changes to the extracellular domain may induce Msb2 to interact with Sho1 or Cdc42, thus promoting the function of these proteins in the FG pathway.
Figure 8.
Model for the protein complex at the head of the FG pathway. Msb2 (in red) is composed of a large extracellular domain, which contains the mucin repeats (bright red oval) and is glycosylated (as shown), and integral-membrane and cytoplasmic domains. General factors Sho1, Cdc42 (isoprenylated and attached to the plasma membrane; PM), and Ste20 all shown in blue are recruited to the FG pathway by Msb2. The interaction between Cdc42 and the cytoplasmic domain of Msb2 is indicated, as is the interaction between Msb2 and Sho1, which is stimulated by FG-pathway activation. An interaction between the cytoplasmic domains of the Msb2 and Sho1 proteins was not detected; thus, they may interact in their membrane-spanning or extracellular domains.
We suspect that Msb2 and Sho1 function in a sensory capacity for the FG pathway. The N terminus of Msb2, which is glycosylated and contains the mucin domain, comprises a substantial portion of the extracellular component of such a sensor (Fig. 8). What might this protein complex sense at the cell surface? Possibly, the extracellular domain of Msb2 interacts with components of the cell wall to detect stress at this site. Filamentation is thought to occur on solid surfaces; perhaps detection of this environment is one function of the Msb2/Sho1 protein complex.
Msb2 and Sho1 have a dramatic effect on Kss1 phosphorylation but not on Fus3 phosphorylation. This is the first direct evidence that a signal emanating from upstream components of the FG pathway flows through Kss1. Moreover, the differential specificity of Msb2/Sho1 for Kss1 confirms a specific role for these cell-surface factors in FG-pathway activation. Selective activation of Kss1 presumably promotes efficient FG, because active Fus3 antagonizes FG (Sabbagh et al. 2001). However, because the Msb2/Sho1-dependent signal presumably goes through Ste7, the activating kinase for both Kss1 and Fus3, how discrimination between these kinases is achieved remains an open question.
Msb2- and FG-pathway specificity
The expression profile of the MSB2 gene indicates that it is a specific FG-pathway target. Expression of MSB2 is induced by FG-pathway activation during a glycosylation defect and during FG. Other genome-wide expression profiles have uncovered MSB2 as a target induced by a glycoslyation block (Travers et al. 2000), as a crosstalk-pathway target (O'Rourke and Herskowitz 2002), and as an Ste12-dependent target of the FG pathway (Madhani et al. 1999). Indeed, Ste12 and Tec1 have been shown to bind to the MSB2 promoter in cells exhibiting butanol-induced FG (Zeitlinger et al. 2003). In contrast, MSB2 expression is influenced neither by changes in osmolarity (Posas et al. 2000; O'Rourke and Herskowitz 2002; this paper) nor by mating pheromone (Roberts et al. 2000; this paper). That MSB2 expression is not induced by pheromone is significant: the other genes whose action is specific for the FG pathway, TEC1 and KSS1, exhibit some pheromone-dependent transcriptional induction (Oehlen and Cross 1998; Roberts et al. 2000). Ste12 may bind to the Msb2 promoter in conjunction with Tec1 to promote FG-pathway-dependent and pheromone-independent expression. Although Msb2 appears to be an FG-pathway-specific factor, it may not be a specificity factor per se, because overexpression of Msb2, Msb2*, or loss of Msb2 does not appear to alter the specificity of FG-pathway components, although we have not explored this possibility fully.
The phenotype of MSB2 alleles is consistent with a specific role for this protein in the FG MAPK pathway. Msb2 is required for FG and viability of glycosylation mutants but not for mating or osmotolerance (O'Rourke and Herskowitz 2002; this paper). The morphological phenotypes conferred by hyperactive Msb2 are reminiscent of hyperfilamentation rather than an activated mating pathway or HOG pathway. For example, cells containing the hyperactive Msb2 are not hypersensitive to pheromone and do not show nuclear Hog1 localization in low salt conditions (an indicator of HOG pathway activation; P.J. Cullen and G.F. Sprague, unpubl.). Msb2 may have other cellular functions: Msb2 is expressed and localized to polar sites during vegetative growth and is required for osmotolerance in some strains (O'Rourke and Herskowitz 2002). However, we have not detected a role for Msb2 in osmotolerance or during vegetative growth.
Mucins as signaling pathway regulators and polarity control proteins
Msb2 is a member of the mucin family of proteins, which are glycosylated cell-surface adhesion proteins. In mammalian cells, mucins act as barriers to pathogen infection (Carson et al. 1998) and are key factors in metastasis in a variety of human cancers (Carraway et al. 2001; Corfield et al. 2001). In addition, two membrane-spanning mucins in humans, MUC1 and MUC4, function as signaling molecules (Wreschner et al. 1994; Carraway et al. 2003). Like Msb2, MUC1 and MUC4 are cell-surface integral-membrane proteins whose cytoplasmic tails interact with signaling molecules at the head of a cascade. MUC1 is a docking protein for β-catenin, and tyrosine phosphorylation of the cytoplasmic domain of MUC1 activates an MAPK pathway, the Grb2-Sos-Ras-MEK-ERK2 pathway (Meerzaman et al. 2001). MUC4 binds to the tyrosine kinase ErbB2/HER2/Neu, to trigger phosphorylation of ErbB2 and potentiate signaling through the ErbB2/ErbB3 heterodimeric receptor complex (Komatsu et al. 2001).
Several findings from this study may be extrapolated to signaling mucins in general. First, as for Msb2, the mucin domains of MUC1 and MUC4 (and others) may have inhibitory roles. Hence, mutation of mucin domains may cause pathway activation and contribute to cancer progression in mammalian cells. The sequence similarity of mucin tandem repeats makes them highly susceptible to recombination-mediated deletion, as can occur for Msb2. Moreover, if, as for Msb2, the hyperactivity is dominant, then inappropriate pathway activation in mucin-deleted receptors may be prevalent among human cancers. The duality of mucin function in signaling pathway function should be a consideration in studies of adhesion-dependent developmental responses in normal mammalian cells (Verfaillie 1998) and for appropriate drug design in human tumors (Agrawal et al. 1998b). Indeed, MUC1 has been reported to have positive and negative roles in signaling (Agrawal et al. 1998a; Chang et al. 2000). A second consequence of this study comes from the finding that Msb2 interacts with Cdc42 to redirect cell polarity. This finding suggests a role for signaling mucins in regulating polarized growth. Intriguingly, both MUC1 and MUC4 are localized to the apical surfaces of epithelial cells (Carraway et al. 2003). Elucidation of the roles of signaling mucins in signaling pathway activation and polarized growth will help define the mechanisms by which these molecules induce metastasis in human cancers.
Materials and methods
Strains, plasmids, and microbiological techniques
Yeast strains are listed in Supplementary Table S1 and plasmids used in this study in Supplementary Table S2. Complete microarray data sets are listed in Supplementary Table S5. Plasmid constructions, additional methodology, and standard genetic and biochemical techniques including references can be found in the Supplemental Material.
Genomic screens
The ordered deletion collection containing 4806 haploid MATα strains (Research Genetics) was pinned to YPD medium in a 384-grid format using a Biomek 2000 robot (Beckman). To restore agar invasion to this background (Liu et al. 1996), the pFLO8 plasmid was introduced into the collection by standard mating techniques using strains and techniques provided by C. Boone (University of Toronto, Canada; Tong et al. 2001). Strains containing pFLO8 were pinned onto YPD medium and incubated for 4 d. Plates were washed and scored visually for agar invasion.
DNA microarray analysis
Cells pmi40-101 and pmi40-101 ste12 were induced in YPD ± 50 mM Man for 4 h. RNA was prepared by hot acid phenol and passage over an RNeasy column (QIAGEN). DNA microarray analysis was performed as described (DeRisi et al. 1997; Lashkari et al. 1997). Microarray construction, target labeling, and hybridization protocols were as described (Fazzio et al. 2001). Sample comparisons were independently replicated six times, each of which was derived from a separate induction. Fluoro-reverse experiments were used to identify sequence-specific dye biases. Arrays were scanned using a GenePix 4000 scanner (Axon Instruments). Image analysis was performed using GenePix Pro 3.0. A Bayesian t-statistic derived for microarray analysis (Baldi and Long 2001) and a false discovery rate methodology (FDR) were used to account for multiple testing (Benjamini and Hochberg 1995).
CoIP analysis
CoIP of epitope-tagged proteins was performed based on published procedures (Kemp and Sprague 2003). Epitope-tagged proteins were IPed from isogenic strains grown to mid-log phase, and cells were harvested and stored at -20°C. Cell pellets were thawed, resuspended in IP buffer containing 1% N P-40, lysed, and incubated with primary antibody and protein A-Sepharose beads. Induction of the FG pathway was performed by shifting cells in mid-log phase from YPD to YPGal for 4 h.
Protein purification and analysis
Interactions between HIS-Msb2CT and GST-Cdc42 and controls were tested using proteins that were expressed and purified from Escherichia coli. Purified HIS-Msb2CT and HIS-Urm1 were concentrated by ammonium sulfate precipitation. Purified proteins were incubated together in binding buffer (50 mM Tris at pH 8, 1 mM DTT, 1 mg/mL BSA, and a protease inhibitor cocktail) for 15 min at 25°C. Beads that recognize the HIS epitope were preincubated in binding buffer and added to the reaction for 15 min. Beads were precipitated by centrifugation at 1000 rpm and washed once in binding buffer. Input and precipitated proteins were analyzed using antibodies specific for GST (Novagen) and HIS (Immunology Consultants Laboratory) epitopes. Glycosylation was examined in cells containing Msb2-HA using 25 μg/mL tunicamycin in DMSO for 2 h, or treating cell extracts with EndoHf (New England Biolabs). Subcellular fractionation experiments were performed as described (Horazdovsky and Emr 1993).
Microscopy
Differential-interference-contrast (DIC) and fluorescence microscopy using UV and FITC (fluorescein) filter sets was performed using an Axioplan 2 microscope (Zeiss), a black and white Orca II digital camera (Hamamatsu), and the Openlab software program (Improvision). Only brightness and contrast digital adjustments were performed on photographs.
Acknowledgments
This manuscript is dedicated to Ira Herskowitz. We thank B. Errede, J. Pringle, G. Fink, C. Boone, H. Oak-Park, T. Stevens, E. Leberer, D. Mitchell, and S. O'Rourke for reagents and advice. We thank lab members for suggestions. We thank M. Dorfman, M. Rendell, and D. Rivers for work on the genomic screen and for helpful discussions. This work was supported by research grants (GM-30027 for G.F.S. and GM-60366 for L.B.) from the U.S. Public Health Service and by a fellowship (AHA120635Z for P.C.) from the American Heart Association.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1178604.
References
- Agrawal B., Gendler, S.J., and Longenecker, B.M. 1998a. The biological role of mucins in cellular interactions and immune regulation: Prospects for cancer immunotherapy. Mol. Med. Today 4: 397-403. [DOI] [PubMed] [Google Scholar]
- Agrawal B., Krantz, M.J., Parker, J., and Longenecker, B.M. 1998b. Expression of MUC1 mucin on activated human T cells: Implications for a role of MUC1 in normal immune regulation. Cancer Res. 58: 4079-4081. [PubMed] [Google Scholar]
- Baldi P. and Long, A.D. 2001. A Bayesian framework for the analysis of microarray expression data: Regularized t-test and statistical inferences of gene changes. Bioinformatics 17: 509-519. [DOI] [PubMed] [Google Scholar]
- Bardwell L., Cook, J.G., Voora, D., Baggott, D.M., Martinez, A.R., and Thorner, J. 1998a. Repression of yeast Ste12 transcription factor by direct binding of unphosphorylated Kss1 MAPK and its regulation by the Ste7 MEK. Genes & Dev. 12: 2887-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell L., Cook, J.G., Zhu-Shimoni, J.X., Voora, D., and Thorner, J. 1998b. Differential regulation of transcription: Repression by unactivated mitogen-activated protein kinase Kss1 requires the Dig1 and Dig2 proteins. Proc. Natl. Acad. Sci. 95: 15400-15405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender A. and Pringle, J.R. 1992. A Ser/Thr-rich multicopy suppressor of a cdc24 bud emergence defect. Yeast 8: 315-323. [DOI] [PubMed] [Google Scholar]
- Benjamini Y. and Hochberg, Y. 1995. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J.R. Statist. Soc. Ser. B Methodological 57: 289-300. [Google Scholar]
- Breitkreutz A. and Tyers, M. 2002. MAPK signaling specificity: It takes two to tango. Trends Cell Biol. 12: 254-257. [DOI] [PubMed] [Google Scholar]
- Breitkreutz A., Boucher, L., and Tyers, M. 2001. MAPK specificity in the yeast pheromone response independent of transcriptional activation. Curr. Biol. 11: 1266-1271. [DOI] [PubMed] [Google Scholar]
- Carraway K.L., Price-Schiavi, S.A., Komatsu, M., Jepson, S., Perez, A., and Carraway, C.A. 2001. Muc4/sialomucin complex in the mammary gland and breast cancer. J. Mammary Gland Biol. Neoplasia 6: 323-337. [DOI] [PubMed] [Google Scholar]
- Carraway K.L., Ramsauer, V.P., Haq, B., and Carothers Carraway, C.A. 2003. Cell signaling through membrane mucins. Bioessays 25: 66-71. [DOI] [PubMed] [Google Scholar]
- Carson D.D., DeSouza, M.M., Kardon, R., Zhou, X., Lagow, E., and Julian, J. 1998. Mucin expression and function in the female reproductive tract. Hum. Reprod. Update 4: 459-464. [DOI] [PubMed] [Google Scholar]
- Chang J.F., Zhao, H.L., Phillips, J., and Greenburg, G. 2000. The epithelial mucin, MUC1, is expressed on resting T lymphocytes and can function as a negative regulator of T cell activation. Cell Immunol. 201: 83-88. [DOI] [PubMed] [Google Scholar]
- Cook J.G., Bardwell, L., Kron, S.J., and Thorner, J. 1996. Two novel targets of the MAP kinase Kss1 are negative regulators of invasive growth in the yeast Saccharomyces cerevisiae. Genes & Dev. 10: 2831-2848. [DOI] [PubMed] [Google Scholar]
- Cook J.G., Bardwell, L., and Thorner, J. 1997. Inhibitory and activating functions for MAPK Kss1 in the S. cerevisiae filamentous-growth signalling pathway. Nature 390: 85-88. [DOI] [PubMed] [Google Scholar]
- Corfield A.P, Carroll, D., Myerscough, N., and Probert, C.S. 2001. Mucins in the gastrointestinal tract in health and disease. Front. Biosci. 6: 1321-1357. [DOI] [PubMed] [Google Scholar]
- Cullen P.J. and Sprague Jr., G.F. 2000. Glucose depletion causes haploid invasive growth in yeast. Proc. Natl. Acad. Sci. 97: 13619-13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen P.J., Schultz, J., Horecka, J., Stevenson, B.J., Jigami, Y., and Sprague Jr., G.F. 2000. Defects in protein glycosylation cause SHO1-dependent activation of a STE12 signaling pathway in yeast. Genetics 155: 1005-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport K.D., Williams, K.E., Ullmann, B.D., and Gustin, M.C. 1999. Activation of the Saccharomyces cerevisiae filamentation/invasion pathway by osmotic stress in high-osmolarity glycogen pathway mutants. Genetics 153: 1091-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi J.L., Iyer, V.R., and Brown, P.O. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278: 680-686. [DOI] [PubMed] [Google Scholar]
- Dohlman H.G. and Thorner, J.W. 2001. Regulation of G protein-initiated signal transduction in yeast: Paradigms and principles. Annu. Rev. Biochem. 70: 703-754. [DOI] [PubMed] [Google Scholar]
- Elion E.A. 2000. Pheromone response, mating and cell biology. Curr. Opin. Microbiol. 3: 573-581. [DOI] [PubMed] [Google Scholar]
- ____. 2001. The Ste5p scaffold. J. Cell Sci. 114: 3967-3978. [DOI] [PubMed] [Google Scholar]
- Fazzio T.G., Kooperberg, C., Goldmark, J.P., Neal, C., Basom, R., Delrow, J., and Tsukiyama, T. 2001. Widespread collaboration of Isw2 and Sin3-Rpd3 chromatin remodeling complexes in transcriptional repression. Mol. Cell. Biol. 21: 6450-6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno C.J., Ljungdahl, P.O., Styles, C.A., and Fink, G.R. 1992. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: Regulation by starvation and RAS. Cell 68: 1077-1090. [DOI] [PubMed] [Google Scholar]
- Gladfelter A.S., Bose, I., Zyla, T.R., Bardes, E.S., and Lew, D.J. 2002. Septin ring assembly involves cycles of GTP loading and hydrolysis by Cdc42p. J. Cell Biol. 156: 315-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris K., Lamson, R.E., Nelson, B., Hughes, T.R., Marton, M.J., Roberts, C.J., Boone, C., and Pryciak, P.M. 2001. Role of scaffolds in MAPK pathway specificity revealed by custom design of pathway-dedicated signaling proteins. Curr. Biol. 11: 1815-1824. [PubMed] [Google Scholar]
- Hollingworth M.A. and Swanson, B.J. 2004. Mucins in cancer: Protection and control at the cell surface. Nat. Rev. Cancer 4: 45-59. [DOI] [PubMed] [Google Scholar]
- Horazdovsky B.F. and Emr, S.D. 1993. The VPS16 gene product associates with a sedimentable protein complex and is essential for vacuolar protein sorting in yeast. J. Biol. Chem. 268: 4953-4962. [PubMed] [Google Scholar]
- Jansen G., Buhring, F., Hollenberg, C.P., and Ramezani Rad, M. 2001. Mutations in the SAM domain of STE50 differentially influence the MAPK-mediated pathways for mating, filamentous growth and osmotolerance in Saccharomyces cerevisiae. Mol. Genet. Genomics 265: 102-117. [DOI] [PubMed] [Google Scholar]
- Kemp H.A. and Sprague Jr., G.F. 2003. Far3 and five interacting proteins prevent premature recovery from pheromone arrest in the budding yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 23: 1750-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., Jepson, S., Arango, M.E., Carothers Carraway, C.A., and Carraway, K.L. 2001. Muc4/sialomucin complex, an intramembrane modulator of ErbB2/HER2/Neu, potentiates primary tumor growth and suppresses apoptosis in a xenotransplanted tumor. Oncogene 20: 461-470. [DOI] [PubMed] [Google Scholar]
- Kron S.J. 1997. Filamentous growth in budding yeast. Trends Microbiol. 5: 450-454. [DOI] [PubMed] [Google Scholar]
- Laloux I., Jacobs, E., and Dubois, E. 1994. Involvement of SRE element of Ty1 transposon in TEC1-dependent transcriptional activation. Nucleic Acids Res. 22: 999-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamson R.E., Winters, M.J., and Pryciak, P.M. 2002. Cdc42p regulation of kinase activity and signaling by the yeast p21-activated kinase Ste20p. Mol. Cell. Biol. 22: 2939-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lashkari D.A., DeRisi, J.L., McCusker, J.H., Namath, A.F., Gentile, C., Hwang, S.Y., Brown, P.O., and Davis, R.W. 1997. Yeast microarrays for genome wide parallel genetic and gene expression analysis. Proc. Natl. Acad. Sci. 94: 13057-13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leberer E., Wu, C., Leeuw, T., Fourest-Lieuvin, A., Segall, J.E., and Thomas, D.Y. 1997. Functional characterization of the Cdc42p binding domain of yeast Ste20p protein kinase. EMBO J. 16: 83-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B.N. and Elion, E.A. 1999. The MAPKKK Ste11 regulates vegetative growth through a kinase cascade of shared signaling components. Proc. Natl. Acad. Sci. 96: 12679-12684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Styles, C.A., and Fink, G.R. 1993. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science 262: 1741-1744. [DOI] [PubMed] [Google Scholar]
- ____. 1996. Saccharomyces cerevisiae S288C has a mutation in FLO8, a gene required for filamentous growth. Genetics 144: 967-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani H.D. 2000. Interplay of intrinsic and extrinsic signals in yeast differentiation. Proc. Natl. Acad. Sci. 97: 13461-13463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani H.D. and Fink, G.R. 1997. Combinatorial control required for the specificity of yeast MAPK signaling. Science 275: 1314-1317. [DOI] [PubMed] [Google Scholar]
- ____. 1998. The riddle of MAP kinase signaling specificity. Trends Genet. 14: 151-155. [DOI] [PubMed] [Google Scholar]
- Madhani H.D., Styles, C.A., and Fink, G.R. 1997. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell 91: 673-684. [DOI] [PubMed] [Google Scholar]
- Madhani H.D., Galitski, T., Lander, E.S., and Fink, G.R. 1999. Effectors of a developmental mitogen-activated protein kinase cascade revealed by expression signatures of signaling mutants. Proc. Natl. Acad. Sci. 96: 12530-12535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T., Takekawa, M., and Saito, H. 1995. Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science 269: 554-558. [DOI] [PubMed] [Google Scholar]
- Meerzaman D., Shapiro, P.S., and Kim, K.C. 2001. Involvement of the MAP kinase ERK2 in MUC1 mucin signaling. Am. J. Physiol. Lung Cell Mol. Physiol. 281: L86-L91. [DOI] [PubMed] [Google Scholar]
- Morillon A., Springer, M., and Lesage, P. 2000. Activation of the Kss1 invasive-filamentous growth pathway induces Ty1 transcription and retrotransposition in Saccharomyces cerevisiae. Mol. Cell. Biol. 20: 5766-5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C., Goto, S., Lund, K., Hung, W., and Sadowski, I. 2003. Srb10/Cdk8 regulates yeast filamentous growth by phosphorylating the transcription factor Ste12. Nature 421: 187-190. [DOI] [PubMed] [Google Scholar]
- Oehlen L. and Cross, F.R. 1998. The mating factor response pathway regulates transcription of TEC1, a gene involved in pseudohyphal differentiation of Saccharomyces cerevisiae. FEBS Lett. 429: 83-88. [DOI] [PubMed] [Google Scholar]
- O'Rourke S.M. and Herskowitz, I. 1998. The Hog1p MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes & Dev. 12: 2874-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2002. A third osmosensing branch in Saccharomyces cerevisiae requires the Msb2p protein and functions in parallel with the Sho1p branch. Mol. Cell. Biol. 22: 4739-4749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Harashima, T., and Heitman, J. 2000. Signal transduction cascades regulating pseudohyphal differentiation of Saccharomyces cerevisiae. Curr. Opin. Microbiol. 3: 567-572. [DOI] [PubMed] [Google Scholar]
- Peter M., Neiman, A.M., Park, H.-O., van Lohuizen, M., and Herskowitz, I. 1996. Functional analysis of the interaction between the small GTP binding protein Cdc42p and the Ste20p protein kinase in yeast. EMBO J. 15: 7046-7059. [PMC free article] [PubMed] [Google Scholar]
- Posas F. and Saito, H. 1997. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: Scaffold role of Pbs2p MAPKK. Science 276: 1702-1705. [DOI] [PubMed] [Google Scholar]
- Posas F., Witten, E.A., and Saito, H. 1998. Requirement of STE50 for osmostress-induced activation of the Ste11p mitogen-activated protein kinase kinase kinase in the highosmolarity glycerol response pathway. Mol. Cell. Biol. 18: 5788-5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posas F., Chambers, J.R., Heyman, J.A., Hoeffler, J.P., de Nadal, E., and Arino, J. 2000. The transcriptional response of yeast to saline stress. J. Biol. Chem. 275: 17249-17255. [DOI] [PubMed] [Google Scholar]
- Raitt D.C., Posas, F., and Saito, H. 2000. Yeast Cdc42p GTPase and Ste20p PAK-like kinase regulate Sho1p-dependent activation of the Hog1p MAPK pathway. EMBO J. 19: 4623-4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser V., Salah, S.M., and Ammerer, G. 2000. Polarized localization of yeast Pbs2p depends on osmostress, the membrane protein Sho1p and Cdc42p. Nat. Cell Biol. 2: 620-627. [DOI] [PubMed] [Google Scholar]
- Roberts R.L. and Fink, G.R. 1994. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: Mating and invasive growth. Genes & Dev. 8: 2974-2985. [DOI] [PubMed] [Google Scholar]
- Roberts R.L., Mosch, H.U., and Fink, G.R. 1997. 14-3-3 proteins are essential for RAS/MAPK cascade signaling during pseudohyphal development in S. cerevisiae. Cell 89: 1055-1065. [DOI] [PubMed] [Google Scholar]
- Roberts C.J., Nelson, B., Marton, M.J., Stoughton, R., Meyer, M.R., Bennett, H.A., He, Y.D., Dai, H., Walker, W.L., Hughes, T.R., et al. 2000. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science 287: 873-880. [DOI] [PubMed] [Google Scholar]
- Sabbagh W., Flatauer, L.J., Bardwell, A.J., and Bardwell, L. 2001. Specificity of MAP kinase signaling in yeast differentiation involves transient versus sustained MAPK activation. Mol. Cell 8: 683-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman H.S., Parry, S., Sutton-Smith, M., Burdick, M.D., McDermott, K., Reid, C.J., Batra, S.K., Morris, H.R., Hollingsworth, M.A., Dell, A., et al. 2001. In vivo glycosylation of mucin tandem repeats. Glycobiology 11: 459-471. [DOI] [PubMed] [Google Scholar]
- Smith G.R., Givan, S.A., Cullen, P.J., and Sprague Jr., G.F. 2002. GTPase-activating proteins for Cdc42. Eukaryot. Cell 1: 469-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague G.F. 1998. Control of MAP kinase signaling specificity or how not to go HOG wild. Genes & Dev. 12: 2817-2820. [DOI] [PubMed] [Google Scholar]
- Stevenson B.J., Ferguson, B., De Virgilio, C., Bi, E., Pringle, J.R., Ammerer, G., and Sprague Jr., G.F. 1995. Mutation of RGA1, which encodes a putative GTPase-activating protein for the polarity-establishment protein Cdc42p, activates the pheromone-response pathway in the yeast Saccharomyces cerevisiae. Genes & Dev. 9: 2949-2963. [DOI] [PubMed] [Google Scholar]
- Tedford K., Kim, S., Sa, D., Stevens, K., and Tyers, M. 1997. Regulation of the mating pheromone and invasive growth responses in yeast by two MAP kinase substrates. Curr Biol. 7: 228-238. [DOI] [PubMed] [Google Scholar]
- Tong A.H., Evangelista, M., Parsons, A.B., Xu, H., Bader, G.D., Page, N., Robinson, M., Raghibizadeh, S., Hogue, C.W., Bussey, H., et al. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364-2368. [DOI] [PubMed] [Google Scholar]
- Travers K.J., Patil, C.K., Wodicka, L., Lockhart, D.J., Weissman, J.S., and Walter, P. 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101: 249-258. [DOI] [PubMed] [Google Scholar]
- van Drogen F. and Peter, M. 2001. MAP kinase dynamics in yeast. Biol. Cell 93: 63-70. [DOI] [PubMed] [Google Scholar]
- Verfaillie C.M. 1998. Adhesion receptors as regulators of the hematopoietic process. Blood 92: 2609-2612. [PubMed] [Google Scholar]
- Wedlich-Soldner R., Altschuler, S., Wu, L., and Li, R. 2003. Spontaneous cell polarization through actomyosin-based delivery of the Cdc42 GTPase. Science 299: 1231-1235. [DOI] [PubMed] [Google Scholar]
- Whitmarsh A.J. and Davis, R.J. 1998. Structural organization of MAP-kinase signaling modules by scaffold proteins in yeast and mammals. Trends Biochem. Sci. 23: 481-485. [DOI] [PubMed] [Google Scholar]
- Wreschner D.H., Zrihan-Licht, S., Baruch, A., Sagiv, D., Hartman, M.L., Smorodinsky, N., and Keydar, I. 1994. Does a novel form of the breast cancer marker protein, MUC1, act as a receptor molecule that modulates signal transduction? Adv. Exp. Med. Biol. 353: 17-26. [DOI] [PubMed] [Google Scholar]
- Zeitlinger J., Simon, I., Harbison, C.T., Hannett, N.M., Volkert, T.L., Fink, G.R., and Young, R.A. 2003. Program-specific distribution of a transcription factor dependent on partner transcription factor and MAPK signaling. Cell 113: 395-404. [DOI] [PubMed] [Google Scholar]