Abstract
The tumor suppressor gene 101 (tsg101) regulates vesicular trafficking processes in yeast and mammals. We report a novel protein, Tal (Tsg101-associated ligase), whose RING finger is necessary for multiple monoubiquitylation of Tsg101. Bivalent binding of Tsg101 to a tandem tetrapeptide motif (PTAP) and to a central region of Tal is essential for Tal-mediated ubiquitylation of Tsg101. By studying endocytosis of the epidermal growth factor receptor and egress of the human immunodeficiency virus, we conclude that Tal regulates a Tsg101-associated complex responsible for the sorting of cargo into cytoplasm-containing vesicles that bud at the multivesicular body and at the plasma membrane.
Keywords: Endocytosis, growth factor, HIV, ubiquitin, signal transduction
Conjugation of monomeric ubiquitins to receptors and adaptor proteins plays an essential role in sorting cargo for vesicular trafficking along both the endocytic and exocytic pathways (for review, see Hicke and Dunn 2003). Genetic defects that disrupt ubiquitin-controlled sorting are often linked to aberrations in cell growth and may result in oncogenic transformation. Examples include c-Cbl, an E3 ubiquitin ligase involved in sorting growth factor receptors to endocytosis (for review, see Waterman and Yarden 2001), and Tsg101, an E2-like molecule originally isolated in a random screen for tumor suppressor genes (Li and Cohen 1996). Cbl proteins attach ubiquitin moieties to tyrosine-phosphorylated receptors for the epidermal growth factor (EGF), thereby accelerating their internalization and delivery to the lysosome for degradation. Oncogenic truncation mutants of Cbl either block receptor internalization or enhance recycling of active receptors. Likewise, although genomic alterations in tsg101 are rare in human cancer, functional inactivation of the gene enhances metastatic growth of murine fibroblasts (Li and Cohen 1996). Potentially, Tsg101 may cause mitotic abnormalities by modulating nuclear receptor-mediated transcription, or by disrupting cell cycle regulation (Hittelman et al. 1999). Studies of the yeast Tsg101 ortholog, namely, Vps23p, implicated yet a third potential mechanism. Class E vacuolar protein sorting (vps) genes such as vps23/tsg101 and vps28 are necessary for formation of the multivesicular body (MVB) in yeast (for review, see Katzmann et al. 2002). Mammalian and yeast Tsg101/Vps23p assemble into a large complex, termed ESCRT-1, which recognizes ubiquitylated cargo originating from either the Golgi apparatus or the plasma membrane, and sorts them into the vacuolar/lysosomal lumen (Katzmann et al. 2001). Accordingly, reduced expression of Tsg101 shunts active EGFRs from the normal degradative pathway to a recycling route (Babst et al. 2000). Hence, like oncogenic forms of Cbl, aberrant expression of Tsg101 may transform cells by enhancing growth factor signaling.
Whereas both Cbl- and Tsg101-mediated processes sort ubiquitylated cargoes into invaginating buds, only the latter protein packages portions of the cytoplasm in budding vesicles. A topologically similar process takes place when viruses bud at the surface of infected host cells, and recent reports implicated Tsg101 in the budding process of certain enveloped viruses like the human immunodeficiency virus 1 (HIV-1; for review, see Pornillos et al. 2002c). Retroviral Gag proteins are necessary and sufficient for assembly and budding of virus-like particles (VLPs) from Gag-expressing cells, but host proteins like Tsg101 and the E3 ligase Nedd4 must be recruited to and function at sites of Gag assembly and budding. Tsg101 binds a tetrapeptide motif, Pro-(Thr/Ser)-Ala-Pro (PTAP), which is located within the p6 domain of HIV-1 Gag. PTAP binding is mediated by the N-terminally located ubiquitin E2 variant (UEV) domain of Tsg101 (Garrus et al. 2001; Martin-Serrano et al. 2001; VerPlank et al. 2001; Demirov et al. 2002; Myers and Allen 2002; Pornillos et al. 2002a). Because of the absence of a critical cysteine, no ubiquitin-conjugating activity is associated with this domain, but it can also bind ubiquitin with low affinity (Bishop and Woodman 2001; Garrus et al. 2001; Katzmann et al. 2001).
The exact mechanism that underlies the ability of Tsg101 to sort cargoes into newly formed membrane buds is incompletely understood, but identification of functional partners of Tsg101 may shed light on the biochemical machinery. In addition to Vps28, a component of yeast and mammalian ESCRT-I (Katzmann et al. 2001; Martin-Serrano et al. 2003), an as-yet-unidentified mammalian ortholog of yeast Vps37 likely complements the action of Tsg101. Recent reports identified two additional partners of Tsg101, namely, AIP1, the ortholog of Bro1 (Strack et al. 2003; von Schwedler et al. 2003), and Hrs (Bache et al. 2003a; Katzmann et al. 2003; Pornillos et al. 2003). In an effort to resolve the sorting action of Tsg101, we screened cDNA libraries for Tsg101-associated proteins by using the yeast two-hybrid system. Here we report on the identification of Tal, a novel RING finger E3 ubiquitin ligase that physically associates with, and selectively ubiquitylates, Tsg101 both in vitro and in living cells. Importantly, ubiquitylation of Tsg101 by Tal inactivates the ability of Tsg101 to sort both endocytic (EGF receptors) and exocytic (viral proteins) cargoes.
Results
Tsg101 interacts with a novel E3 ubiquitin ligase
To identify Tsg101-interacting proteins, we fused full-length Tsg101 to the LexA-binding domain, and used this bait to screen a human brain cDNA library. From 6.5 × 106 transformants screened, several clones of the motor protein KIF5A and of Intersectin, an adaptor of the endocytic machinery, were found to interact with Tsg101. A third interacting clone represents a novel protein, which we named Tal, for Tsg101-associated ligase. The full-length cDNA of human Tal (hTal) predicts an N-terminal leucine-rich repeat (LRR), followed by several recognizable motifs, including an ezrin-radixin-moezin (ERM) domain, a coiled-coil (CC) region, a SAM domain, and a C-terminal C3HC4-type RING finger domain (Fig. 1A). Interestingly, hTal also contains two closely located PTAP motifs in its C terminus. Screening of genomic and EST databases revealed that tal occurs as a single copy gene in all vertebrate genomes, and sequence alignments indicated very high conservation of all structural motifs (see Supplemental Material; Supplementary Fig. 1). To confirm the amino acid sequence, we raised antibodies to synthetic peptides derived from the N and the C terminus of hTal. Both antibodies identified an ectopic hTal transiently overexpressed in HEK-293T cells, as well as a similar 80-kDa species in mouse brain (Fig. 1B). In addition, Northern blot analysis identified a murine Tal mRNA species of 3.5 kb in all tissues examined (data not shown).
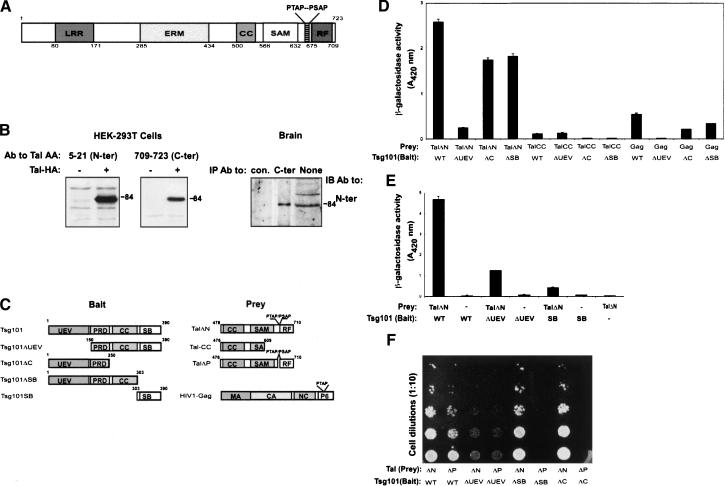
Figure 1.
Domain structure of human Tal and binding with Tsg101 in yeast. (A) Schematic diagram of human Tal, showing the approximate boundaries of the leucine-rich repeats (LRRs), ezrin-radixin-moesin (ERM) domain, coiled-coil (CC) region, α sterile alpha motif (SAM), a RING finger (RF), and a double PTAP motif. (B) HA-tagged hTal was immunoprecipitated (IP) from extracts of HEK-293T cells transfected with either the respective plasmid, or an empty vector (-). The rabbit antibodies used for IP are specific to peptides corresponding to the indicated amino acids (AA) of hTal. An antibody to HA was used for immunoblotting (IB). Alternatively, extracts of mouse brain were subjected to IB, either directly (None) or after IP. (C) Schematic diagrams of Tsg101, Tal, and Gag proteins, along with their mutants. The following domains of Tsg101 are indicated: ubiquitin E2 variant (UEV) region, proline-rich domain (PRD), coiled coil (CC), and steadiness box (SB). The PTAP motif of Gag is indicated inside the p6 domain. Other Gag domains are matrix (MA), capsid (CA), and nucleocapsid (NC). (D,E) The interaction between Tal (or a LexA control protein; -) and Tsg101, either wild-type (WT) or the indicated mutants, was determined in yeast by using reporter gene activation. The histogram represents the average (±S.D.) activity of β-galactosidase measured in three separate assays using O-nitrophenyl-β-D-galactopyranoside as a substrate. (F) Yeast cells transformed by the indicated pairs of plasmids were subjected to serial dilutions and plated on selective (Trp-Leu-His minus) medium.
To map their interaction, we mutated Tsg101 and hTal (Fig. 1C) and tested them in yeast. As prey, we used the partial-length hTal (residues 476-710; ΔN), which was identified in the original screen. As a control we used HIV-Gag, because the single PTAP motif of this viral protein strongly binds to the UEV of Tsg101 (Garrus et al. 2001; Martin-Serrano et al. 2001; VerPlank et al. 2001; Demirov et al. 2002; Myers and Allen 2002; Pornillos et al. 2002a). Indeed, using HIV-Gag as prey, we confirmed binding to full-length Tsg101, as well as to two C-terminal truncation mutants (Tsg101-ΔC and ΔSB), but not to a mutant lacking the UEV (Tsg101-ΔUEV; Fig. 1D). In contrast, in the same assay, Tsg101-ΔUEV reproducibly retained reduced, but significant, recognition of Tal-ΔN. This observation suggested that hTal and Tsg101 maintain both PTAP-dependent and PTAP-independent interactions. PTAP-independent recognition was displayed by an hTal mutant lacking the C terminus (hTal-CC; Fig. 1D), and further analysis localized the interaction to the steadiness box (SB) of Tsg101. Tal-CC weakly bound Tsg101-ΔUEV, but not Tsg101-ΔSB, implying involvement of the SB in binding to Tal (Fig. 1D). In line with this model, an isolated SB retained weak, but specific, binding to hTal (Fig. 1E), and a PTAP-defective mutant of hTal (Tal-ΔP) recognized WTTsg101, but not a mutant lacking the SB (Fig. 1F). In conclusion, hTal and Tsg101 maintain bimodal interactions: In addition to the UEV-PTAP recognition, the highly conserved SB mediates a secondary Tal-Tsg101 interaction by binding to a central region of hTal, which includes a CC domain.
To test interactions in mammalian cells, we analyzed coimmunoprecipitates of HA-tagged hTal and a modified glutathione-S-transferase (mGST)-tagged Tsg101. As shown in Figure 2A, hTal underwent specific coprecipitation with mGST-Tsg101. Further, an ectopic hTal underwent coimmunoprecipitation with the endogenous Tsg101 of HEK-293T cells (Fig. 2B), and Tal of A431 human epidermoid carcinoma cells was precipitated by anti-Tsg101 antibodies (Supplementary Fig. 2A). Further, a coimmunoprecipitation assay confirmed the importance of the double PTAP motif of Tal, as well as the SB region of Tsg101 (Fig. 2C). Interestingly, a CC deletion mutant of hTal displayed enhanced binding to Tsg101, whereas deletion of the SB of Tsg101 abolished the interaction in mammalian cells, unlike its mild effect in yeast. On the other hand, deletion of the UEV did not affect binding to hTal in HEK-293T cells, although it severely impaired interactions in yeast (Fig. 1D). Taken together, these observations imply that Tal-Tsg101 interactions in mammalian cells are more complex than what is reflected in yeast, which may relate to interacting factors, or to subcellular localization.
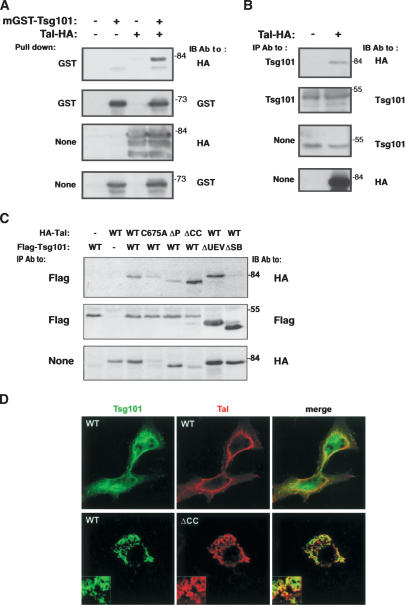
Figure 2.
Bimodal interaction and partial colocalization of Tal and Tsg101. (A) Extracts of HEK-293T cells coexpressing mGST-Tsg101 and HA-hTal were subjected to a pull-down assay using glutathione-agarose beads and IB, as indicated. (B) Extracts of HEK-293T cells transiently expressing HA-hTal were subjected to IP with an antibody directed to the endogenous Tsg101 protein. Cell lysates and IPs were analyzed as indicated. (C) HEK-293T cells coexpressing various forms of HA-hTal and Flag-Tsg101 were analyzed for coprecipitation. (D) HeLa cells expressing HA-hTal (either wild type [WT] or ΔCC) and Flag-Tsg101 were fixed, permeabilized, and stained with primary and fluorescently labeled secondary antibodies, prior to confocal microscopy.
When ectopically expressed in HeLa cells and visualized with antibodies to HA, hTal displayed a punctate distribution and also localized to a submembranal ring (Fig. 2D). In contrast, Tsg101 exhibited broader distribution (Fig. 2D), in line with previous reports (Bishop and Woodman 2001; Goila-Gaur et al. 2003). The relatively peripheral fraction of Tsg101 partly colocalized with hTal, an observation we confirmed by using untransfected HeLa cells and antibodies to the endogenous Tal and Tsg101 proteins (Supplementary Fig. 2B). Interestingly, hTal mutants defective in the RING or CC domains mislocalized, together with WT-Tsg101, to the outer rim of large vesicular structures, which stained negatively for the early endosome antigen (EEA1; Fig. 2D; data not shown). In conclusion, Tal and Tsg101 display partially overlapping patterns of distribution, and mutant Tal proteins comislocalize with Tsg101, which may reflect a concerted action in vesicular trafficking.
Tal ubiquitylates Tsg101 in cells and in vitro
Because Tal contains a RING finger similar to motifs found within E3 ubiquitin ligases (Joazeiro and Weissman 2000), we determined whether it promotes ubiquitylation of Tsg101. Flag-tagged Tsg101 was transiently expressed in HEK-293T cells together with hTal and Myc-tagged ubiquitin, and the presence of ubiquitylated Tsg101 was tested by immunoblotting. As shown in Figure 3A, expression of an ectopic hTal significantly increased the levels of Tsg101 ubiquitylation. In contrast to WT-hTal, two RING mutants (C675A and H695A) could not increase Tsg101 ubiquitylation (Fig. 3A,C). Because the SB of Tsg101 mediates part of the association with hTal, we tested the corresponding deletion mutant. As expected, no hTal-induced ubiquitylation of Tsg101-ΔSB was detectable (Fig. 3A,C). Likewise, deletion of either the PTAP motifs or the CC of hTal abolished ubiquitylation of Tsg101 (Fig. 3C,E). The reciprocal mutant of Tsg101, namely, a protein whose UEV domain was impaired by replacing a TYN motif (residues 67-69) with a triplet of alanines (VerPlank et al. 2001; Pornillos et al. 2002a,b), underwent no ubiquitylation (Fig. 3F), probably because of monovalent binding of the mutant to hTal. Taken together, these results indicate that bimodal binding of hTal to Tsg101 is essential for enhanced Tsg101 ubiquitylation.
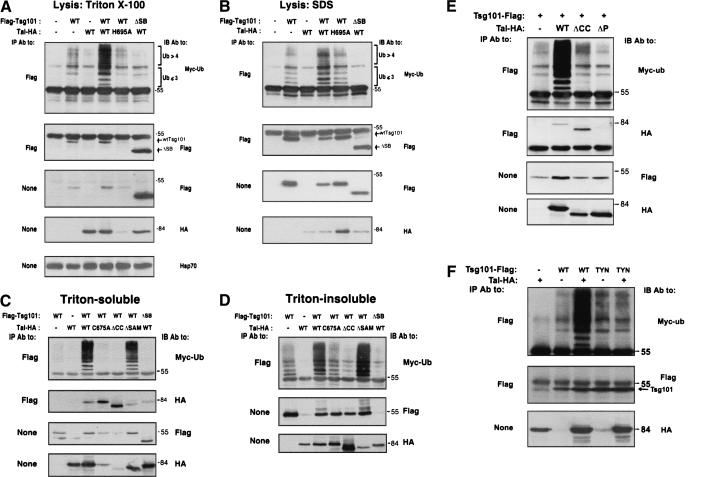
Figure 3.
Tal increases ubiquitylation of Tsg101 and affects its solubility in a RING- and PTAP-dependent manner. (A,B) HEK-293T cells were cotransfected with plasmids encoding Flag-Tsg101 (either wild type [WT] or ΔSB), HA-Tal (either wild type [WT] or H695A), and Myc-ubiquitin. Forty-eight hours after transfection, cells were divided into two portions: The first (75% of cell population) was extracted by using Triton X-100 (A), and the remainder was extracted in SDS (B). IB of immunoprecipitates or whole extracts was performed with the indicated antibodies. Oligo- and multiubiquitylated forms of Tsg101 are labeled. (C,D) HEK-293T cells transiently expressing the indicated proteins, along with Myc-Ub, were sequentially extracted in Triton X-100 (C) and SDS (D) and analyzed as indicated. (E,F) HEK-293T cells transiently expressing the indicated proteins, along with Myc-Ub, were extracted in Triton X-100 and analyzed as indicated. The TYN mutant of Tsg101 represents a protein whose TYN motif of the UEV domain (residues 67-69) has been replaced with a triplet of alanines.
When ectopically expressed, the catalytically inactive H695A mutant of hTal was hardly detectable, and a parallel reduction in extractable Tsg101 was noted in cell extracts (Fig. 3A). Notably, Tsg101 normally associates with detergent-insoluble lipid rafts (Licata et al. 2003), and the steady-state levels of Tsg101 are regulated post-translationally through an unknown mechanism involving the SB (Feng et al. 2000). Hence, we raised the possibility that ubiquitylation modifies the localization of a Tsg101-Tal complex, which normally resides in detergent-insoluble structures. Several observations support this notion. Side-by-side comparison of solubility in ionic and nonionic detergents confirmed reduced solubility of H695A-hTal (cf. anti-HA blots in Fig. 3A,B) and demonstrated that ubiquitylation increases the solubility of Tsg101 (cf. oligo- [<3] and multi- [>3] ubiquitylated species of Tsg101 in Fig. 3A,B). Likewise, sequential extraction with Triton X-100 followed by treatment of the insoluble fraction with sodium dodecylsulfate (SDS) revealed that deletion of the CC, but not the SAM region, of hTal almost completely abolished both ubiquitylation and solubility of Tsg101 (Fig. 3C,D). In this vein, ΔPTAP-hTal was unable to interact with Tsg101 and therefore could not affect its ubiquitylation or solubility (Fig. 3E; data not shown). Interestingly, ΔSB-hTsg101 displayed high solubility but no ubiquitylation (Fig. 3, cf. C and D), and subcellular fractionation, which is not presented, revealed enrichment of Tsg101 in the membrane fraction of Tal-overexpressing HEK-293T cells. Conceivably, on ubiquitylation, Tsg101 translocates from the cytoplasm and from lipid rafts to a detergent-soluble membranal subdomain, and the SB is involved in this process.
Two major partners of Tsg101 are the hepatocyte growth factor-regulated tyrosine kinase substrate (Hgs/Hrs; Bache et al. 2003a,b; Katzmann et al. 2003; Pornillos et al. 2003) and Vps28 (Bishop and Woodman 2001; Martin-Serrano et al. 2003). However, despite their physical associations with Tsg101, ubiquitylation of neither Hrs nor Vps28 was affected by an overexpressed hTal (Fig. 4A). Hence, we conclude that Tsg101 serves as a specific ubiquitylation substrate of Tal. Like other RING-containing E3 ligases, hTal underwent self-ubiquitylation, which was dependent on the integrity of the RING but not the PTAP or the CC domains (Fig. 4B). An in vitro autoubiquitylation assay using an isolated hTal, recombinant E1, E2, and 125I-labeled ubiquitin confirmed self-ubiquitylation of hTal and verified the importance of the RING finger (Fig. 4C). A similar protocol was used to assess the ligase activity of hTal toward Tsg101. Substantial ubiquitylation of an immobilized Tsg101 was observed following preincubation with WT-hTal but not its RING mutants (Fig. 4D). Taken together with experiments performed in living cells, these results identify hTal as an E3 ubiquitin ligase specific for Tsg101.
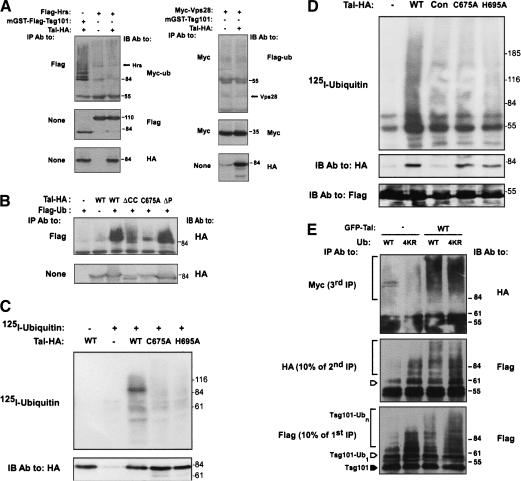
Figure 4.
Tal monoubiquitylates Tsg101 and undergoes self-ubiquitylation. (A) HEK-293T cells transiently expressing combinations of Hrs or Vps28, along with Tal, Tsg101, and either Myc- (left) or Flag- (right) tagged ubiquitin, were extracted in Triton X-100 and analyzed. (B) The state of ubiquitylation of WT-hTal or its indicated mutants was examined by coexpressing a Flag-tagged ubiquitin in HEK-293T cells. (C) The indicated forms of HA-Tal were immunoprecipitated from extracts of HEK-293T cells and incubated with recombinant E1, E2 (Ubc-H5B), and 125I-labeled ubiquitin. Ubiquitylated products were resolved by gel electrophoresis and detected by autoradiography. (D) An immunoprecipitated Flag-Tsg101 was incubated with extracts of HEK-293T cells transfected with either a control plasmid (Con), or vectors encoding the indicated forms of hTal. After extensive washing, the Tsg101-Tal complex was subjected to ubiquitylation in vitro using a radiolabeled ubiquitin. A control reaction was performed in the absence of cell extract (lane labeled -). (E) Extracts of HEK-293T cells expressing Flag-Tsg101, HA-, and Myc-tagged ubiquitin (either wild type [WT] or 4KR), and either GFP-Tal or a control plasmid, were subjected to immunoprecipitation (first IP) with an anti-Flag antibody. A portion (10%) of the beads was directly analyzed and the rest eluted at 95°C and subjected to IP with an anti-HA antibody (second IP). Likewise, a portion (10%) of the beads was directly analyzed and the rest eluted, subjected to IP with an anti-Myc antibody (third IP), and analyzed by electrophoresis. A closed arrowhead indicates the location of nonubiquitylated Tsg101, and an open arrowhead marks the monoubiquitylated form.
To determine whether the smeary appearance of ubiquitylated Tsg101 corresponds to polyubiquitylation or to multiple monomeric ubiquitins (multiubiquitylation), we used a polymerization-defective mutant of ubiquitin (Ub-4KR), which lacks all four branching lysines, in conjunction with a recently described three-step immunoprecipitation protocol (Haglund et al. 2003). Essentially, HEK-293T cells coexpressing Flag-Tsg101 and a fluorescent hTal (GFP-Tal) were transfected with plasmids encoding either wild-type or 4KR ubiquitin. Two forms of exogenous ubiquitins were coexpressed, namely, Myc-Ub and HA-Ub, which enabled tag-specific sequential isolation of ubiquitylated Flag-Tsg101. As expected, the first IP (with anti-Flag antibodies) confirmed higher ubiquitylation of Tsg101 in cells coexpressing hTal (Fig. 4E), and the second IP (with anti-HA antibodies) detected a monoubiquitylated form of Tsg101, along with more richly modified forms. Consistent with specific isolation, the third IP (with anti-Myc antibodies) could not detect the monoubiquitylated form but identified the smeary bands as multiubiquitylated forms of Tsg101 because they could be isolated with anti-Myc antibodies. In other words, the existence in living cells of Tsg101 molecules, which are simultaneously modified by two distinct forms of a polymerization-defective ubiquitin, indicates multiubiquitylation. In conclusion, hTal decorates Tsg101 with multiple monomeric ubiquitins, consistent with a role in vesicular trafficking or protein folding, rather than proteasomal degradation.
Tal-mediated ubiquitylation of Tsg101 controls a late sorting event in EGFR endocytosis
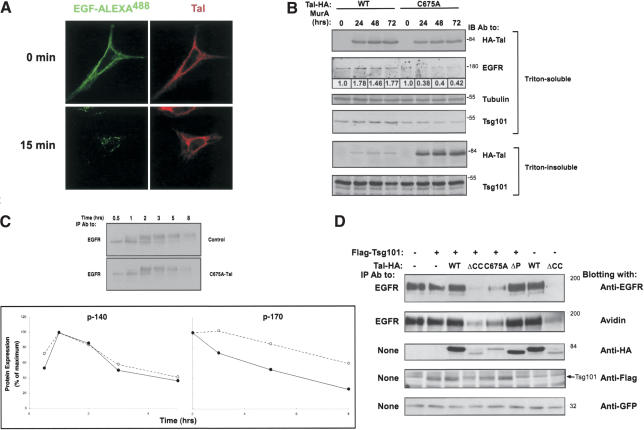
Normal internalization of EGFRs was observed in fibroblasts depleted of Tsg101, but instead of trafficking to lysosomes, EGFRs were shunted in a late endocytic compartment to a recycling pathway (Babst et al. 2000). In line with lack of involvement in early steps of endocytosis, overexpression of hTal did not alter the rapid translocation of a fluorescent derivative of EGF to endocytic vesicles (Fig. 5A). Similarly, ligand-induced ubiquitylation of EGFR was not significantly changed when hTal was overexpressed (data not shown). To link the endocytic fate of EGFR to Tal, we established an HEK-293T cell system that expresses WT-hTal, or a catalytically inactive mutant, from an inducible promoter. On induction with Muristerone A, an overexpressed WT-hTal led to increased expression of endogenous EGFR, whereas induction of C675A-hTal significantly reduced EGFR expression (Fig. 5B). As expected, the relatively insoluble C675A-hTal reduced the solubility of the endogenous Tsg101 in Triton X-100. To test the prediction that hTal regulates endocytic degradation of EGFR, rather than maturation and delivery to the plasma membrane, we subjected EGFR to a short pulse of metabolic labeling and followed subsequent receptor maturation and degradation (Fig. 5C). Ectopic C675A-hTal did not affect the rate of synthesis of the precursor p140EGFR, which gradually matured to p170EGFR. The latter, however, disappeared more rapidly in the presence of C675A-hTal. Because the primary site of interactions between Tsg101 and EGFR is the MVB (Babst et al. 2000), the results presented in Figure 5A-C imply that Tal normally ubiquitylates, and thereby inactivates, or recycles, a late endosomal sorting complex that includes Tsg101.
Figure 5.
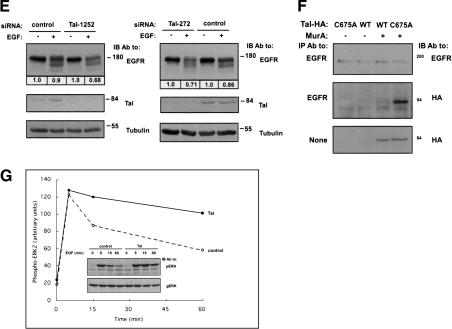
A catalytically inactive mutant of hTal stably associates with EGFRs and accelerates their endocytic degradation. (A) HeLa cells expressing HA-hTal were preincubated at 4°C with EGF conjugated to AlexaFluor 488. Thereafter, cells were incubated for 15 min at 37°C, permeabilized, and stained with anti-HA antibodies, followed by a fluorescent secondary antibody. (B) HEK-293 cells stably expressing the ecdysone receptor were transfected with plasmids that express HA-hTal (wild type [WT] or C675A) under the control of an ecdysone-inducible promoter. The indicated stable clones were incubated without or with Muristerone A (2 μM) for the indicated time intervals and thereafter cells were extracted in Triton X-100. The insoluble material was extracted in SDS, and both fractions were directly analyzed. Numbers below lanes indicate quantification of signals normalized to the respective tubulin signal. (C) Chinese hamster ovary (CHO) cells transiently transfected with an EGFR plasmid, along with either a C675A-Tal (solid line) or an empty vector (Control; broken line), were preincubated in cysteine- and methionine-free medium prior to a 20-min-long pulse of [35S]-labeled amino acids. Thereafter, cells were chased at 37°C in fresh medium for the indicated time intervals. An autoradiogram of the immunoprecipitated EGFR is shown, along with the respective quantification of the precursor (p140) and mature (p170) forms of EGFR. (D) CHO cells transiently coexpressing EGFR, Flag-Tsg101, and the indicated forms of HA-hTal were surface biotinylated 48 h after transfection. A GFP expression vector was used to monitor transfection efficiency. Cell extracts were analyzed as indicated. (E) HeLa-SS6 cells were transfected with the indicated siRNA oligonucleotides (50 nM each). Forty-eight hours posttransfection, cells were starved for 6 h in the absence of serum, and then stimulated with EGF (20 ng/mL) for 1 h. Whole-cell extracts were immunoblotted with the indicated antibodies. The EGFR signals were quantified and normalized to the respective levels of tubulin. (F) HEK-293 cells expressing HA-hTal (wild type [WT] or C675A) from a Muristerone-inducible promoter were incubated without or with Muristerone A (2 μM) for 48 h. Cell extracts were tested for coimmunoprecipitation of hTal and EGFR. (G) HEK-293T cells were cotransfected with a GFP-ERK2 plasmid and either a vector encoding for HA-hTal or a control empty plasmid. Thirty-six hours posttransfection, cells were starved for 8 h in the absence of serum, and then stimulated with EGF (100 ng/mL) for the indicated time intervals. Whole-cell extracts were immunoblotted with antibodies to the doubly phosphorylated ERK (pERK) or a general ERK antibody (gERK). Shown are the resulting immunoblots (inset) and quantification of the active ERK signal.
To firmly establish this conclusion, we undertook two approaches. The gain-of-function approach entailed transient overexpression of wild-type or mutant hTal proteins in CHO cells whose surface EGFRs were labeled with biotin. Although WT-hTal moderately increased receptor stability, and the inactive ΔP-hTal exerted no effect, two dominant negative mutants of hTal (C675A and ΔCC) remarkably accelerated degradation of surface biotinylated EGFRs (Fig. 5D). The loss-of-function approach used Tal-specific small interference RNA (siRNA). Initial screens enabled us to identify two siRNA oligonucleotides that effectively reduced (>80%) expression of the endogenous hTal in human cells, as revealed by using anti-Tal antibodies (Fig. 5E). When transfected into HeLa-SS6 cells, the oligonucleotides significantly and comparably accelerated EGF-induced degradation of endogenous EGFR molecules, consistent with a role for hTal in restraining late sorting of EGFR to degradation.
Next, we addressed the mechanism underlying the ability of catalytically inactive mutants of hTal to accelerate endocytic degradation of EGFR. According to one model, Tsg101 directly binds monoubiquitylated cargoes like EGFR and sorts them to the lumen of MVBs (Katzmann et al. 2002). Presumably, the sorting function of Tsg101 is inactivated on ubiquitylation by Tal, but catalytically defective forms of Tal can maintain Tsg101 in its active, deubiquitylated state. This model predicts formation of a stable sorting complex containing both EGFR and C675A-hTal. Indeed, by using our inducible hTal, we were able to coimmunoprecipitate EGFR with C675A-hTal (Fig. 5F). No complex was detectable in cells expressing WT-hTal or in uninduced cells, in line with the proposed model of Tal's action. In light of the ability of WT-hTal to stabilize EGFR at the cell surface (Fig. 5C,E), we envisioned that Tal will alter the kinetics of EGF-induced signaling, a process regulated primarily through receptor endocytosis (for review, see Waterman and Yarden 2001). Indeed, the onset phase of MAPK activation by EGF was similar in control and in hTal-expressing cells, but the inactivation phase almost disappeared when hTal was overexpressed (Fig. 5G). Notably, a similar behavior of MAPK signaling was observed in cells defective in Tsg101 (Babst et al. 2000), in line with the proposition that Tal inactivates Tsg101 by means of multiubiquitylation.
Tal and Tsg101 cooperatively regulate release of HIV-1
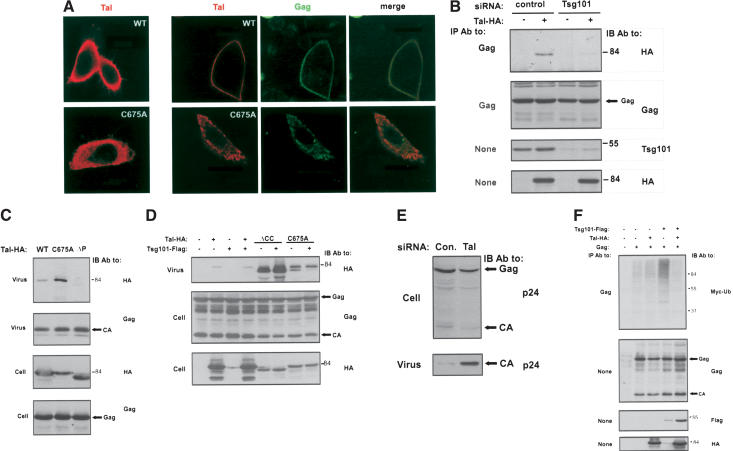
Because Tsg101 is crucial for budding of HIV-1 VLPs (Garrus et al. 2001; Martin-Serrano et al. 2001; Demirov et al. 2002), we postulated that hTal is involved in viral budding. Hence, we expressed in HeLa-SS6 cells a Gag protein fused to the green fluorescence protein (GFPGag), which recapitulates many aspects of viral assembly and budding (Hermida-Matsumoto and Resh 2000). When coexpressed with WT-hTal, Gag significantly narrowed the submembranal distribution of hTal to a fine peripheral layer containing both proteins (Fig. 6A). Notably, another partner of Tsg101, namely, Vps28, is similarly recruited by Gag to the plasma membrane (Martin-Serrano et al. 2003). In contrast with WT-hTal, the mislocalized C675A-hTal altered the distribution of Gag, and both colocalized to circular structures similar to those observed with other dominant negative mutants of Tal (Fig. 2D).
Figure 6.
Tal interacts with Tsg101 and Gag along the exocytic pathway of HIV-1. (A) Cultures of HeLa-SS6 cells expressing either wild type (WT) or C675A-hTal were treated with either a control vector (left column), or a Gag-GFP-encoding plasmid. Fixation, staining, and confocal visualization were performed 6 h later. (B) HeLa-SS6 cells were transfected with the indicated siRNA oligonucleotides, and 24 h later a second transfection was performed with vectors encoding HIV-1 Gag (pNLenv-1; Schubert et al. 1995) and HA-hTal (or a control plasmid). Cells were extracted 24 h later, and coimmunoprecipitation of Tal and Gag was tested by using the respective antibodies. (C,D) HEK-293T cells were cotransfected with the pNLenv-1 vector encoding HIV-1 Gag, along with the indicated Tal and Tsg101 plasmids. Cells and VLPs were analyzed 48 h posttransfection (CA; capsid). (E) HeLa-SS6 cells were transfected with a Tal-specific siRNA, which starts at nucleotide 1252, and a control inverted sequence (50 nM each). Forty-eight hours later, cells were cotransfected with the indicated oligonucleotides (25 nM) along with pNLenv-1 (1 μg). The presence of Gag in VLPs or in cytoplasmic extracts was analyzed 24 h later. (F) HEK-293T cells were cotransfected with pNLenv-1, along with a Myc-ubiquitin vector and the indicated Flag-Tsg101 and HA-hTal plasmids. Forty-eight hours posttransfection, cells were extracted and analyses performed either directly or after IP with anti-Gag antibodies.
Unlike EGFR, Gag contains a PTAP motif, which recruits Tsg101 to virus budding sites (Garrus et al. 2001; Martin-Serrano et al. 2001; VerPlank et al. 2001; Demirov et al. 2002; Myers and Allen 2002; Pornillos et al. 2002a). In addition, it has been shown that Tsg101 multimerizes via its CC domain (Martin-Serrano et al. 2003). Hence, the observed codistribution of hTal and GFP-Gag might be mediated by a Gag-(Tsg101)2-Tal complex. Indeed, the Gag polyprotein (Pr55Gag) and Tal underwent coimmunoprecipitation when coexpressed, but blocking expression of Tsg101 by using a specific siRNA practically abolished complex formation (Fig. 6B). Hence, it is likely that a Tsg101-Tal complex is recruited by Gag to sites of virus egress. This conclusion predicts insertion of Tal into VLPs, as has been reported for Tsg101 (Myers and Allen 2002). To test their cobudding, we coexpressed WT-hTal and Pr55Gag in HEK-293T cells and harvested VLPs from the medium. Unlike a PTAP deletion mutant of hTal, which cannot bind Tsg101, WT-hTal was detectable in VLPs (Fig. 6C). Moreover, more effective insertion into VLPs was observed with C675A-hTal (Fig. 6C), a ubiquitylation-defective mutant that strongly binds to and decreases solubility of Tsg101 (Fig. 3C).
These observations suggested that a stable, perhaps unubiquitylated, Tal-Tsg101 complex is essential for insertion of Tal into VLPs. Hence, we tested ΔCC-hTal, a ubiquitylation-defective mutant that binds Tsg101 even more strongly than C675A-hTal. We observed extremely effective insertion of ΔCC-hTal into VLPs (Fig. 6D), which reinforces the interpretation that Tsg101 drives Tal into budding HIV-1 particles. Because Tsg101 plays an essential role in budding of HIV-1 (Garrus et al. 2001; Martin-Serrano et al. 2001; Demirov et al. 2002) and Tal profoundly alters Tsg101 partitioning by means of multiubiquitylation (Figs. 3, 4), we addressed Tal's effect on VLP release. A Tal-specific siRNA (denoted Tal-1252) dramatically increased budding (Fig. 6E), consistent with ubiquitylation-mediated inactivation of Tsg101. Interestingly, only limited inhibition of VLP release was observed in cells overexpressing hTal, but coexpression of Tal and Tsg101 almost blocked egress (data not shown). In conclusion, Tal-mediated ubiquitylation of Tsg101 likely inactivates its virus release function, in analogy with the inhibitory action of Tal in late-stage endocytosis (Fig. 5).
Ubiquitin plays an important, albeit poorly understood, role in viral budding (Ott et al. 2003). Consistent with the essential role for Tsg101 in HIV budding, overexpression of Tsg101 increases ubiquitylation of Gag of HIV-2 (Myers and Allen 2002). By coexpressing Pr55Gag and Tsg101 in HeLa cells, we extended this observation to the Gag protein of HIV-1 (Fig. 6F). Because Tal appears to inhibit other activities of Tsg101, we addressed the possibility that Tal can reduce the effect of Tsg101 on Gag ubiquitylation. As shown in Figure 6F, coexpression of hTal with Tsg101 decreased Gag ubiquitylation to the basal level. These observations are consistent with the possibility that Tsg101 recruits to Gag an E3 ubiquitin ligase distinct from Tal. However, by enhancing ubiquitylation of Tsg101, hTal inactivates this function of Tsg101, in analogy with Tal's ability to inactivate Tsg101-mediated cargo sorting in the endocytic and exocytic pathways.
Discussion
Receptor endocytosis and subsequent degradation in lysosomes underlie the major mechanism of desensitization following cellular stimulation via receptor tyrosine kinases or G protein-coupled receptors (Katzmann et al. 2002). In the case of receptors for growth factors, two distinct sorting events are critical for the determination of the receptor's fate. The first occurs at the plasma membrane and entails recruitment of active receptors to clathrin-coated regions of the plasma membrane. The postinternalization sorting event takes place at a late endosomal compartment, the MVB, and it involves budding of vesicles into the lumen of endosomes (Futter et al. 1996). Importantly, these two sorting events differ in their topology: whereas the initial event represents an inward budding and fission process, the late outward budding engulfs portions of the cytoplasm. A topologically similar process might drive sequestration of viral particles within secretory vesicles of macrophages (Raposo et al. 2002) and egress of enveloped RNA viruses from lymphocytes (Pornillos et al. 2002c). The identification of Tal and characterization of its interaction with Tsg101, a major regulator of outward vesicular budding, shed light on the underlying mechanism of cargo sorting and underscore the critical regulatory role played by monoubiquitylation of the sorting machinery.
Tal is an evolutionary conserved ubiquitin ligase that bimodally recognizes Tsg101
Tal orthologs are very distant from any other known gene family, and the respective gene appears to occur as a single copy gene in all vertebrate genomes, including a single ortholog in the draft Fugu rubripes genome. In contrast, invertebrate species outside of the chordate phylum do not contain a Tal homolog. Hence, Tal clearly represents a vertebrate lineage development. Interestingly, the draft genome assembly of the nonvertebrate chordate Ciona intestinalis suggests that Tal appeared at a relatively early stage of prevertebrate evolution of the chordate phylum. Thus, although the function of Tsg101 is conserved from yeast to mammals, the regulation by Tal evolved more recently.
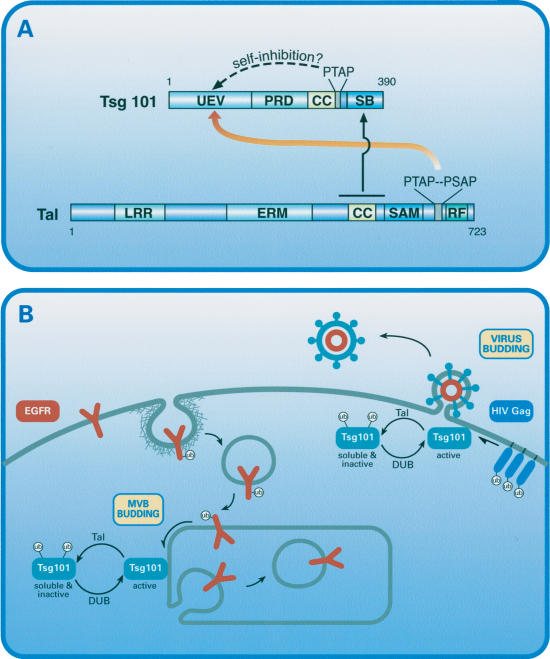
The mode of interaction between hTal and Tsg101 was investigated in both yeast and mammalian cells (Figs. 1, 2). Taken together, our results are consistent with bimodal interactions (Fig. 7A); the UEV domain of Tsg101 interacts with the tandem PTAP motif of hTal, whereas the SB interacts with a region close to the CC domain of hTal. Interestingly, bivalent binding of Tal to Tsg101 is critical for effective transubiquitylation, but monovalent binding involving the CC and SB domains, as well as ubiquitylation-null interactions (e.g., with RING mutants of hTal), are associated with enhancement of Tsg101-Tal interactions (Fig. 3). Hence, it is conceivable that ubiquitylation dissociates the Tal-Tsg101 complex. Remarkably, an analogous bimodal interaction stabilizes a Tsg101-Hrs complex and enables recruitment of Tsg101 to endosomes (Bache et al. 2003a; Katzmann et al. 2003; Pornillos et al. 2003): A PSAP motif of Hrs binds to the UEV of Tsg101, whereas the SB and adjacent regions bind to the C-terminal portion of Hrs. It will be important to test the prediction that Hrs and Tal bind Tsg101 in a mutually exclusive manner. We speculate that the UEV domain is normally autoinhibited by a PTAP motif within Tsg101 (Fig. 7A). Transient release of intramolecular folding allows loading of PTAP-containing cargoes like Gag, or complexes comprising monoubiquitylated EGFRs and adaptors like Hrs. Subsequently, transubiquitylation of Tsg101 by Tal, and binding to Tal's PTAPs may block the nonoverlapping ubiquitin- and PTAP-binding sites of the UEV domain (Pornillos et al. 2002b), thereby instigating cargo unloading.
Figure 7.
Structural and functional interactions between Tal and Tsg101. (A) The domain structures of Tal and Tsg101 are depicted, along with their intermolecular binding specificities (for abbreviations, see Fig. 1). Note that the UEV domain of Tsg101 binds the double PTAP motif of Tal, and a distinct site binds a monomeric ubiquitin (not shown). Secondary interactions between Tal and Tsg101 involve a region encompassing the coiled-coil (CC) domain of Tal and the steadiness box (SB) of Tsg101. Potentially, both binding sites of the UEV domain may be blocked intramolecularly through binding to the C-terminally located PTAP motif and to a monomeric ubiquitin conjugated by Tal. (B) Model illustrating the role of the Tal-Tsg101 complex in budding of vesicles into the lumen of the multivesicular body (MVB) and in virus budding. Accordingly, Tsg101 sorts cargo proteins like the epidermal growth factor receptor (EGFR) and HIV Gag into budding structures. Tal-mediated ubiquitylation of Tsg101 inactivates this sorting function and concomitantly translocates Tsg101 from relatively insoluble membrane subdomains. Presumably, the coordinated action of Tal and a deubiquitylation enzyme (DUB) enables recycling of Tsg101 and reloading of cargo.
Transubiquitylation by Tal enables recycling of the Tsg101-associated sorting complex
The state of ubiquitylation of EGFR is unaffected by Tal (data not shown). Likewise, Tal does not promote ubiquitylation of another cargo, namely, Gag (Fig. 6F). In contrast, Tal robustly ubiquitylates Tsg101 both in vitro (Fig. 4D) and in living cells (Fig. 3), but Tsg101-associated proteins (i.e., Hrs and Vps28) are not modified by Tal (Fig. 4A). Collectively, these observations indicate that the target of Tal is a specific sorting molecule, Tsg101, rather than cargo proteins. Indeed, many other components of sorting complexes are monoubiquitylated (e.g., epsin, Hrs, and Eps15; Polo et al. 2002), similar to Tal-mediated modification of Tsg101, which is limited to monomeric ubiquitins (Fig. 4E). How does the attachment of monomeric ubiquitins modulate the sorting activity of Tsg101? By concentrating on EGFR as an endocytic cargo (Fig. 5) and HIV Gag as a cargo for exocytosis (Fig. 6), we concluded that ubiquitylation restricts the sorting activity of Tsg101. Thus, in line with the ability of Tsg101 to sort EGFR molecules to lysosomal degradation (Babst et al. 2000), catalytically inactive mutants of Tal, as well as two Tal-specific siRNAs (Fig. 5E), accelerated receptor degradation along the endocytic pathway (Fig. 5D). Likewise, the same siRNAs accelerated virus release (Fig. 6E), and overexpression of Tal inhibited two previously described exocytosis activities of Tsg101 (Fig. 6E; data not shown), namely, elevation of Gag ubiquitylation (Myers and Allen 2002) and inhibition of VLP release (Goila-Gaur et al. 2003).
The model for Tal and Tsg101 presented in Figure 7B proposes a mechanism common to endocytosis and exocytosis. Accordingly, ubiquitylation of Tsg101 regulates its shuttling between a membrane-bound active form and an inactive soluble form. Through its UEV domain or associated UIM-containing proteins like Hrs, Tsg101 binds monoubiquitylated cargoes like EGFR and Gag and sorts them into cytoplasm-containing vesicles. Once the associated Tal molecule attaches multiple monomeric ubiquitins to Tsg101, the latter undergoes functional inactivation and partitions into a detergent-soluble membrane subdomain. Thus, transubiquitylation of Tsg101 may enable transient dissociation of the sorting complex from its cargo, ordered assembly of the next ESCRT complex, and reloading of Tsg101 with a new cargo molecule. In line with the proposed model, cargo-containing complexes that include Tal and Tsg101 are detectable, especially when Tal is catalytically defective (Figs. 5F, 6B). Further support for this model is provided by the corelease of catalytically defective forms of Tal in VLPs (Fig. 6C).
Conceivably, the concerted action of deubiquitylating enzymes (DUBs) and membrane anchoring machinery reactivates cargo sorting by Tsg101. A similar mechanism seems to regulate another sorting machinery, whose core is epsin. In insects, the homolog of vertebrate epsin is regulated by a DUB called fat facets, and together they limit to eight the number of photoreceptor cells in each facet of the Drosophila compound eye (Cadavid et al. 2000). Similar to Tsg101, monoubiquitylation of epsin is controlled by several intrinsic ubiquitin-binding domains (Polo et al. 2002), the ubiquitylated molecule cannot mediate endocytosis, and its deubiquitylation is thought to activate sorting of membrane cargoes like Notch (Carthew and Xu 2000). Thus, ubiquitylation/deubiquitylation cycles of the sorting machinery emerges as a general switch that recycles sorting machineries and enables reloading of cargo. In support for this notion, an intact ubiquitylation system is essential for endocytosis of the growth hormone receptor and a Ste2p-ubiquitin fusion protein, receptors that need no direct monoubiquitylation for their endocytosis (Strous et al. 1996; Dunn and Hicke 2001). Similarly, endocytic degradation of EGFR is enhanced when ubiquitylation of Hrs is inhibited (Katz et al. 2002). How monoubiquitylation inhibits the function of sorting molecules like epsin, Hrs, or Tsg101 is currently unknown. However, we note that all three sorting molecules share the presence of ubiquitin-binding domains (either UIM or UEV), which may impose intramolecular folding when ubiquitylated. A similar mechanism of functional inactivation through intramolecular folding may be relevant to MEKK1, a kinase activated in response to extracellular signals. An internal PHD domain mediates self-ubiquitylation of MEKK1 (Lu et al. 2002), thereby inhibiting kinase activity in a mechanism that may involve an intrinsic ubiquitin-interacting motif (Witowsky and Johnson 2003).
In summary, we identified a novel E3 ubiquitin ligase that physically associates with Tsg101. Two domains of Tal independently bind to the UEV and SB domains of Tsg101 in a manner reminiscent of Tsg101 binding to another endocytic regulator, namely, Hrs. Bivalent binding is essential for attachment of multiple monomeric ubiquitins to Tsg101, whose sorting function is consequently inactivated. Analyses of receptor endocytosis and virus budding suggest that Tal enables recycling of Tsg101-containing sorting complexes and cargo reloading. Further, the emerging model predicts that DUBs balance Tal's action and allow ordered assembly of sorting complexes. This possibility, as well as the key role played by Tal-mediated translocation of Tsg101 from detergent-insoluble membrane domains, is a matter for future studies.
Materials and methods
Yeast-two hybrid assays
The coding sequence of Tsg101 was fused to the LexA DNA-binding domain in the pBTM116 bait vector. The L40 yeast strain was first transformed with pBTM116-Tsg101, tested for the absence of autoactivation, and then transformed with a human brain cDNA library cloned into pGAD10. Cotransformants were plated onto Trp-Leu-His selective medium supplemented with 3-aminotriazole (5 mM). His+ colonies were then assayed for β-galactosidase using a filter lift assay. Positive clones were rescued into bacteria and retransformed into the L40 yeast strain to confirm interactions.
Plasmid transfection and surface biotinylation
Transfection (1 μg of DNA) of subconfluent cultures in 100-mm plates was performed using the calcium phosphate method or Lipofectamine 2000 (GIBCO-BRL), and cells were harvested 24-48 h later. The total amount of DNA in each transfection was normalized with the respective empty vectors. EcR-293 (HEK-293 cells constitutively expressing subunits of the ecdysone receptor) stably transfected with pIND-HA-hTal were cultured in medium supplemented with G418 (700 μg/mL) and Zeocin (200 μg/mL). For biotinylation, cells were washed three times with ice-cold phosphate-buffered saline (PBS), and then incubated for 60 min at 4°C with N-hydroxysuccinimide-biotin (biotin-X NHS, 0.5 mg/mL; Calbiochem) dissolved in borate buffer (10 mM boric acid, 150 mM NaCl at pH 8.0). Coupling of biotin was blocked by cell rinsing with a solution of 15 mM glycine in PBS.
Antibodies and analyses of transfected cells
Transfected cells were extracted in lysis buffer and analyzed essentially as described (Levkowitz et al. 1999). Anti-hTal antibodies were raised in rabbits using the synthetic peptides FRKRKPSEEARKRLEYQ-C (N-Ter) and CRQDIAQRLRIYHSS (C-Ter) conjugated via cysteines to a maleimide-activated carrier. The antibodies were affinity purified using the peptide antigens coupled to agarose and they are now available from Sigma. Rabbit anti-HIV-CA antibody was from Seramun Diagnostics.
Construction of mammalian expression vectors
Flag-tagged murine Tsg101 cDNA was cloned into the pEF1α mammalian expression vector. A fusion protein comprising full-length Tsg101 and mGST was generated by PCR amplification and cloning into an mGST expression vector. hTal was cloned from a cDNA library generated from T47D cells and cloned into pcDNA3.1 (Invitrogen). The following deletion mutants were generated by PCR: ΔSAM (deletion of amino acids 566-632), ΔCC (500-547), and ΔPTAP (649-664). Point mutations were introduced by PCR. The pNLenv-1 vector (Schubert et al. 1995) was used in studies of VLP release. pGAG-GFP contains the rev-independent HIV-1HXB2 Gag sequence fused to EGFP (Hermida-Matsumoto and Resh 2000).
Small inhibitory RNAs
The following siRNAs duplexes were synthesized: Tsg101 (sense, CCUCCAGUCUUCUCUCGUCdTdT; and antisense, dTdTGGAGGUCAGAAGAGAGCAG) Control (sense, GUCCAAAGGUUCCGGAGACdTdT; and antisense, dTdTCAGGUUUCCAAGGCCUCUG). Two 21-nucleotide-long RNA duplexes corresponding to hTal (GenBank BC009239) coding nucleotides 272-290 and 1252-1270 relative to the starting codon were designed. Tal siRNA sequences are as follows: 272 sense, UCACCUCACUUCCCUGCUUdTdT; 272 antisense, dTdTAGUGGAGUGAAGGGACGAA; 1252 sense, UGCUGACUGAGAGCUGUAAdTdT; 1252 antisense, UUACAGCUCUCAGUCAGCAdTdT; control Tal siRNA sequences (scrambled 1252) are as follows: sense, AAUGUCGAGAGUCAGUCGUdTdT; antisense, ACGACUGACUCUCGACAUUdTdT. All of the siRNAs were chemically synthesized, purified, and annealed by Dharmacon Research. Usage of the siRNAs was according to the manufacturer's instructions. Cells were transfected with the relevant double-stranded siRNA (50 nM) using Lipofectamine 2000 (Invitrogen).
In vitro ubiquitylation assays
Reactions were performed in ubiquitin wash buffer containing 5 mM MgCl2, 50 mM Tris-HCl (pH 7.5), 2 mM dithiothreitol, 2 mM ATP, and 125I-ubiquitin (0.5 μg per reaction), essentially as described (Levkowitz et al. 1999). For transubiquitylation assays, Flag-Tsg101 immunoprecipitates were tumbled for 1.5 h at 4°C with cleared extracts of HEK-293T cells expressing HA-hTal to allow formation of Tsg101-Tal complexes. Thereafter, the beads and associated proteins were washed and subjected to ubiquitylation assays.
Analysis of the mode of Tsg101 ubiquitylation
To analyze ubiquitylation mode, we constructed Myc- and HA-tagged Ub-4KR, in which Lys 11, Lys 29, Lys 48, and Lys 63 were replaced by arginines. Extracts of cells transiently coexpressing wild-type or mutant ubiquitins, along with Flag-Tsg101 and hTal, were subjected to sequential immunoprecipitation in three steps, essentially as described (Haglund et al. 2003). In brief, the first IP was performed with anti-Flag antibodies and 10% of the immunoprecipitate analyzed by immunoblotting. The rest was eluted at 95°C in 50 mM Tris-HCl buffer (pH 7.5) containing SDS (2%). This sample was diluted 1:10 in solubilization buffer and subjected to a second IP with anti-HA antibodies. Following sampling and elution, a third IP was similarly carried out with anti-Myc antibodies, prior to gel electrophoresis.
Preparation of VLPs
HEK-293T cells were transfected with a vector encoding Gag-GFP (0.5 μg), and the media were collected after 36 h. VLPs were harvested (13,000 rpm for 90 min) by layering filtered culture medium (1.2 mL) onto 0.2 mL of 20% sucrose.
Immunofluorescence
Cells grown on cover slips were fixed with 3% paraformaldehyde, washed, and permeabilized for 10 min at room temperature with PBS containing 1% bovine serum albumin and 0.2% Triton X-100. For staining, cover slips were incubated for 1 h at room temperature with primary antibodies. After extensive washing in PBS, the cover slips were incubated for 40 min with Cy2- or Cy3-conjugated secondary antibodies. For EGF uptake, prior to fixation, transfected cells were washed in PBS and incubated for 3 h in starvation medium supplemented with 0.05% serum. Thereafter, cells were washed once with binding buffer (DME medium supplemented with 1% bovine serum albumin and 20 mM HEPES at pH 7.5) and incubated for 30 min at 4°C with binding buffer containing EGF (2 μg/mL) conjugated to Alexa Fluor 488 (Molecular Probes). After binding, cells were transferred to 37°C for the indicated time intervals, washed in PBS, and fixed. After staining, cover slips were mounted in moviol, and fluorescence was analyzed using a Confocal Zeiss microscope with a 63×/1.4 plane Apochromat objective, attached to the Bio-Rad Radiance 2000 laser scanning system.
Metabolic labeling of cultured cells
Transfected CH0 cells were rinsed twice and preincubated for 3 h in cysteine- and methionine-free medium supplemented with 10% dialyzed serum. Thereafter, cells were labeled for 15 min with a mixture of 35S-labeled amino acids (pulse). Cells were then washed thoroughly and incubated in media containing nonlabeled cysteine and methionine for the indicated time intervals (chase). This was followed by cell lysis, immunoprecipitation, electrophoresis, and autoradiography.
Accession number
The hTal sequence reported in this paper has been submitted to GenBank and may be currently accessed with the number BC009239.
Acknowledgments
We thank Drs. Marilyn Resh, Stanley Cohen, Paul Bieniasz, Wesley Sundquist, and Dirk Bohmann for plasmids and Drs. Daniel Taglicht and Danny Ben-Avraham for siRNA design. This work was supported by research grants provided by the Weizmann Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.294904.
References
- Babst M., Odorizzi, G., Estepa, E.J., and Emr, S.D. 2000. Mammalian tumor susceptibility gene 101 (TSG101) and the yeast homologue, Vps23p, both function in late endosomal trafficking. Traffic 1: 248-258. [DOI] [PubMed] [Google Scholar]
- Bache K.G., Brech, A., Mehlum, A., and Stenmark, H. 2003a. Hrs regulates multivesicular body formation via ESCRT recruitment to endosomes. J. Cell Biol. 162: 435-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bache K.G., Raiborg, C., Mehlum, A., and Stenmark, H. 2003b. STAM and Hrs are subunits of a multivalent ubiquitin-binding complex on early endosomes. J. Biol. Chem. 278: 12513-12521. [DOI] [PubMed] [Google Scholar]
- Bishop N. and Woodman, P. 2001. TSG101/mammalian VPS23 and mammalian VPS28 interact directly and are recruited to VPS4-induced endosomes. J. Biol. Chem. 276: 11735-11742. [DOI] [PubMed] [Google Scholar]
- Cadavid A.L., Ginzel, A., and Fischer, J.A. 2000. The function of the Drosophila fat facets deubiquitinating enzyme in limiting photoreceptor cell number is intimately associated with endocytosis. Development 127: 1727-1736. [DOI] [PubMed] [Google Scholar]
- Carthew R.W. and Xu, C. 2000. Endocytosis: Why not wait to deubiquitinate? Curr. Biol. 10: R532-R534. [DOI] [PubMed] [Google Scholar]
- Demirov D.G., Ono, A., Orenstein, J.M., and Freed, E.O. 2002. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc. Natl. Acad. Sci. 99: 955-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn R. and Hicke, L. 2001. Multiple roles for Rsp5p-dependent ubiquitination at the internalization step of endocytosis. J. Biol. Chem. 276: 25974-25981. [DOI] [PubMed] [Google Scholar]
- Feng G.H., Lih, C.J., and Cohen, S.N. 2000. TSG101 protein steady-state level is regulated posttranslationally by an evolutionarily conserved COOH-terminal sequence. Cancer Res. 60: 1736-1741. [PubMed] [Google Scholar]
- Futter C.E., Pearse, A., Hewlett, L.J., and Hopkins, C.R. 1996. Multivesicular endosomes containing internalized EGF-EGF receptor complexes mature and then fuse directly with lysosomes. J. Cell Biol. 132: 1011-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrus J.E., von Schwedler, U.K., Pornillos, O.W., Morham, S.G., Zavitz, K.H., Wang, H.E., Wettstein, D.A., Stray, K.M., Cote, M., Rich, R.L., et al. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107: 55-65. [DOI] [PubMed] [Google Scholar]
- Goila-Gaur R., Demirov, D.G., Orenstein, J.M., Ono, A., and Freed, E.O. 2003. Defects in human immunodeficiency virus budding and endosomal sorting induced by Tsg101 overexpression. J. Virol. 77: 6507-6519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund K., Sigismund, S., Polo, S., Szymkiewicz, I., Di Fiore, P.P., and Dikic, I. 2003. Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat. Cell Biol. 5: 461-466. [DOI] [PubMed] [Google Scholar]
- Hermida-Matsumoto L. and Resh, M.D. 2000. Localization of human immunodeficiency virus type 1 Gag and Env at the plasma membrane by confocal imaging. J. Virol. 74: 8670-8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke L. and Dunn, R. 2003. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu. Rev. Cell Dev. Biol. 19: 141-172. [DOI] [PubMed] [Google Scholar]
- Hittelman A.B., Burakov, D., Iniguez-Lluhi, J.A., Freedman, L.P., and Garabedian, M.J. 1999. Differential regulation of glucocorticoid receptor transcriptional activation via AF-1-associated proteins. EMBO J. 18: 5380-5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joazeiro C.A. and Weissman, A.M. 2000. RING finger proteins: Mediators of ubiquitin ligase activity. Cell 102: 549-552. [DOI] [PubMed] [Google Scholar]
- Katz M., Shtiegman, K., Tal-Or, P., Yakir, L., Mosesson, Y., Harari, D., Machluf, Y., Asao, H., Jovin, T., Sugamura, K., et al. 2002. Ligand-independent degradation of epidermal growth factor receptor involves receptor ubiquitylation and Hgs, an adaptor whose ubiquitin-interacting motif targets ubiquitylation by Nedd4. Traffic 3: 740-751. [DOI] [PubMed] [Google Scholar]
- Katzmann D.J., Babst, M., and Emr, S.D. 2001. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106: 145-155. [DOI] [PubMed] [Google Scholar]
- Katzmann D.J., Odorizzi, G., and Emr, S.D. 2002. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 3: 893-905. [DOI] [PubMed] [Google Scholar]
- Katzmann D.J., Stefan, C.J., Babst, M., and Emr, S.D. 2003. Vps27 recruits ESCRT machinery to endosomes during MVB sorting. J. Cell Biol. 162: 413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levkowitz G., Waterman, H., Ettenberg, S.A., Katz, M., Tsygankov, A.Y., Alroy, I., Lavi, S., Iwai, K., Reiss, Y., Ciechanover, A., et al. 1999. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol. Cell 4: 1029-1040. [DOI] [PubMed] [Google Scholar]
- Li L. and Cohen, S.N. 1996. Tsg101: A novel tumor susceptibility gene isolated by controlled homozygous functional knockout of allelic loci in mammalian cells. Cell 85: 319-329. [DOI] [PubMed] [Google Scholar]
- Licata J.M., Simpson-Holley, M., Wright, N.T., Han, Z., Paragas, J., and Harty, R.N. 2003. Overlapping Motifs (PTAP and PPEY) within the Ebola Virus VP40 protein function independently as late budding domains: Involvement of host proteins TSG101 and VPS-4. J. Virol. 77: 1812-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z., Xu, S., Joazeiro, C., Cobb, M.H., and Hunter, T. 2002. The PHD domain of MEKK1 acts as an E3 ubiquitin ligase and mediates ubiquitination and degradation of ERK1/2. Mol. Cell 9: 945-956. [DOI] [PubMed] [Google Scholar]
- Martin-Serrano J., Zang, T., and Bieniasz, P.D. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7: 1313-1319. [DOI] [PubMed] [Google Scholar]
- ____. 2003. Role of ESCRT-I in retroviral budding. J. Virol. 77: 4794-4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers E.L. and Allen, J.F. 2002. Tsg101, an inactive homologue of ubiquitin ligase E2, interacts specifically with human immunodeficiency virus type 2 gag polyprotein and results in increased levels of ubiquitinated gag. J. Virol. 76: 11226-11235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott D.E., Coren, L.V., Sowder II, R.C., Adams, J., and Schubert, U. 2003. Retroviruses have differing requirements for proteasome function in the budding process. J. Virol. 77: 3384-3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo S., Sigismund, S., Faretta, M., Guidi, M., Capua, M.R., Bossi, G., Chen, H., De Camilli, P., and Di Fiore, P.P. 2002. A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature 416: 451-455. [DOI] [PubMed] [Google Scholar]
- Pornillos O., Alam, S.L., Davis, D.R., and Sundquist, W.I. 2002a. Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein. Nat. Struct. Biol. 9: 812-817. [DOI] [PubMed] [Google Scholar]
- Pornillos O., Alam, S.L., Rich, R.L., Myszka, D.G., Davis, D.R., and Sundquist, W.I. 2002b. Structure and functional interactions of the Tsg101 UEV domain. EMBO J. 21: 2397-2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pornillos O., Garrus, J.E., and Sundquist, W.I. 2002c. Mechanisms of enveloped RNA virus budding. Trends Cell Biol. 12: 569-579. [DOI] [PubMed] [Google Scholar]
- Pornillos O., Higginson, D.S., Stray, K.M., Fisher, R.D., Garrus, J.E., Payne, M., He, G.P., Wang, H.E., Morham, S.G., and Sundquist, W.I. 2003. HIV Gag mimics the Tsg101-recruiting activity of the human Hrs protein. J. Cell Biol. 162: 425-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo G., Moore, M., Innes, D., Leijendekker, R., Leigh-Brown, A., Benaroch, P., and Geuze, H. 2002. Human macrophages accumulate HIV-1 particles in MHC II compartments. Traffic 3: 718-729. [DOI] [PubMed] [Google Scholar]
- Schubert U., Clouse, K.A., and Strebel, K. 1995. Augmentation of virus secretion by the human immunodeficiency virus type 1 Vpu protein is cell type independent and occurs in cultured human primary macrophages and lymphocytes. J. Virol. 69: 7699-7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack B., Calistri, A., Craig, S., Popova, E., and Gottlinger, H.G. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114: 689-699. [DOI] [PubMed] [Google Scholar]
- Strous G.J., van Kerkhof, P., Govers, R., Ciechanover, A., and Schwartz, A.L. 1996. The ubiquitin conjugation system is required for ligand-induced endocytosis and degradation of the growth hormone receptor. EMBO J. 15: 3806-3812. [PMC free article] [PubMed] [Google Scholar]
- VerPlank L., Bouamr, F., LaGrassa, T.J., Agresta, B., Kikonyogo, A., Leis, J., and Carter, C.A. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag). Proc. Natl. Acad. Sci. 98: 7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schwedler U.K., Stuchell, M., Muller, B., Ward, D.M., Chung, H.Y., Morita, E., Wang, H.E., Davis, T., He, G.P., Cimbora, D.M., et al. 2003. The protein network of HIV budding. Cell 114: 701-713. [DOI] [PubMed] [Google Scholar]
- Waterman H. and Yarden, Y. 2001. Molecular mechanisms underlying endocytosis and sorting of ErbB receptor tyrosine kinases. FEBS Lett. 490: 142-152. [DOI] [PubMed] [Google Scholar]
- Witowsky J.A. and Johnson, G.L. 2003. Ubiquitylation of MEKK1 inhibits its phosphorylation of MKK1 and MKK4 and activation of the ERK1/2 and JNK pathways. J. Biol. Chem. 278: 1403-1406. [DOI] [PubMed] [Google Scholar]