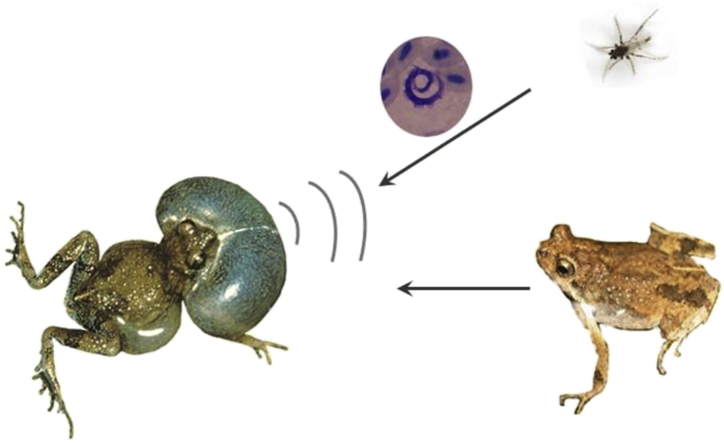
Abstract
Trypanosomes are a diverse group of protozoan parasites of vertebrates transmitted by a variety of hematophagous invertebrate vectors. Anuran trypanosomes and their vectors have received relatively little attention even though these parasites have been reported from frog and toad species worldwide. Blood samples collected from túngara frogs (Engystomops pustulosus), a Neotropical anuran species heavily preyed upon by eavesdropping frog-biting midges (Corethrella spp.), were examined for trypanosomes. Our results revealed sexual differences in trypanosome prevalence with female frogs being rarely infected (<1%). This finding suggests this protozoan parasite may be transmitted by frog-biting midges that find their host using the mating calls produced by male frogs. Following previous anuran trypanosome studies, we examined 18S ribosomal RNA gene to characterize and establish the phylogenetic relationship of the trypanosome species found in túngara frogs. A new species of giant trypanosome, Trypanosoma tungarae n. sp., is described in this study. Overall the morphometric data revealed that the trypomastigotes of T. tungarae n. sp. are similar to other giant trypanosomes such as Trypanosoma rotatorium and Trypanosoma ranarum. Despite its slender and long cell shape, however, 18S rRNA gene sequences revealed that T. tungarae n. sp. is sister to the rounded-bodied giant trypanosome, Trypanosoma chattoni. Therefore, morphological convergence explains similar morphology among members of two non-closely related groups of trypanosomes infecting frogs. The results from this study underscore the value of coupling morphological identification with molecular characterization of anuran trypanosomes.
Keywords: Engystomops pustulosus, Corethrella, Frog-biting midges, Panamá, Physalaemus, Species delimitation, Trypanosome phylogeny
Graphical abstract
Highlights
-
•
There is higher prevalence of trypanosome in male than female túngara frogs.
-
•
Sexual differences in infection suggest potential transmission by frog-biting midges.
-
•
Trypanosoma tungarae n. sp. is a new species infecting túngara frogs.
-
•
This parasite resembles other giant frog trypanosomes from the Aquatic clade.
1. Introduction
Trypanosomes are protozoan parasites that are ubiquitous across invertebrate and vertebrate species. Indeed, trypanosomes infect species across all vertebrate classes. Anuran trypanosomes, however, have received considerably less attention than those in other vertebrates even though they infect frog and toad species worldwide (Bardsley and Harmsen, 1973, Desser and Yekutiel, 1986, Werner, 1993, Desser, 2001, Žičkus, 2002, Lemos et al., 2008). Since many anurans spend at least their early developmental stages in aquatic environments and return to breed as adults, leeches have long been considered the main vectors of trypanosomes in this group (Reilly and Woo, 1982). As adults, however, many species of frogs are preyed upon by a variety of opportunistic and specialized hematophagous insects that may act as possible vectors of blood parasites. Phlebotomine sandflies (Phlebotomus squamirostris), for instance, transmit Trypanosoma bocagei França 1911 to European toads, Bufo bufo (Feng and Chung, 1940). Similarly, trypanosomes may be mosquito-borne parasites for anurans. Mosquitoes, such as Culex territans, that feed mainly on anuran hosts have been implicated in the transmission of Trypanosoma ranarum Lankester 1871 (Desser et al., 1973) but their role as trypanosome vectors is still controversial (Ferguson and Smith, 2012). Other mosquito species such as Aedes aegypti and Culex pipiens can transmit trypanosomes (Trypanosoma rotatorium Mayer 1843 complex) to some frogs even though they do not usually feed on anurans (Ramos and Urdaneta-Morales, 1977). Closely related to mosquitoes, frog-biting midges (Corethrellidae) are small hematophagous flies specialized at feeding on anurans (Borkent, 2008). These midges are thus potentially important vectors of blood parasites of this vertebrate clade (McKeever and French, 2000). In fact, in the Southeastern United States, one species of frog-biting midge (Corethrella wirthi) transmits trypanosomes to green treefrogs, Hyla cinerea (Johnson et al., 1993). The family Corethrellidae contains 107 species of frog-biting midges, in which females are specialized in using the mating call of frogs to localize them and obtain a blood meal (Borkent, 2014). The frog's mating call is the main cue used by the midges for long-distance host detection (Bernal and de Silva, 2015). Further studies that examine the role of other species of frog-biting midges at transmitting trypanosomes are necessary to understand the evolutionary ecology of these interactions. In this study we investigate trypanosome infection in a Neotropical anuran species, the túngara frog (Engystomops pustulosus), which is heavily preyed upon by frog-biting midges.
Túngara frogs are small anurans that occur from southern Mexico to northern South America (Colombia, Venezuela, and Belize) and Trinidad and Tobago. Males aggregate during the rainy season at temporary puddles from where they produce mating calls (Ryan, 1985). While calling to attract a mate, túngara frog males also attract frog-biting midges (Corethrella spp). These eavesdroppers prey upon túngara frogs in great numbers (Fig. 1a). A speaker broadcasting calls equivalent to those produced by a motivate túngara frog male, attracts up to 511 midges in 30 min (average = 142 midges/30 min; Bernal et al., 2006). Túngara frogs represent an ideal opportunity to investigate trypanosome infection potentially transmitted by frog-biting midges.
Fig. 1.
Photographs of túngara frogs (Engystomops pustulosus) and frog-biting midges (Corethrella spp). (a) Calling male túngara frog preyed upon by frog-biting midges; (b) female (bottom) in amplexus with a male (top) covered with biting midges; (c) female (bottom) with a biting midge on her nostril that was passed from the male during amplexus. Túngara frogs are about 30 mm long while the frog-biting midges are only about 1.5 mm. Photos taken by Alexander Baugh (a) and Ximena E Bernal (b,c).
The goals of this study were twofold: firstly, to determine the presence of trypanosomes in túngara frogs along with characterizing these parasites, and secondly, to examine whether trypanosome prevalence differs between females and males. Since as in most anuran species, female túngara frogs do not produce mating calls (Ryan, 1985), eavesdropping frog-biting midges most likely only feed on male frogs. We thus expected differences in trypanosome prevalence between male and female túngara frogs reflecting the feeding habits of the frog-biting midges. As predicted, we found trypanosome infected male túngara frogs and thus implemented morphological and molecular methods to characterize and infer the phylogenetic relationship of this Trypanosoma species to other trypanosomes that parasitize other vertebrates that inhabit aquatic and marine environments. The characterization and phylogenetic relationships of this new Trypanosoma species provide new information on anuran trypanosomes, a group with poorly known taxonomic relationships (Martin et al., 2002). In addition, we provide insights about the prevalence of this trypanosome species on its type host.
2. Materials and methods
2.1. Study site and sample collection
Túngara frogs were captured at their breeding areas during the rainy season around the Smithsonian Tropical Research Facilities in Gamboa (9° 79′ N, 79° 42.9′ W), Panama. Individuals were brought to the laboratory where they were measured and blood samples were collected by toe-clipping as well as via the orbital sinus following Lynch et al. (2006). After collecting blood samples, the frogs were placed in individual containers with sufficient amounts of water and released within 24 h at the exact location where they were captured. This procedure was approved by the Smithsonian Tropical Research Institute IACUC (#2009-0616-2012-11). To examine the presence of trypanosomes in túngara frogs and test our prediction of sexual differences in infection, we collected 25 calling males and 15 females approaching the puddle or in amplexus. We performed 2–5 blood smears per individual to include both thin and thick smears for each frog, for a total of 112 blood smears (2.8 blood smears/individual). Given that some trypanosomes in anurans are known to have nocturnal peripheral parasitemia, bleeding of all túngara frogs was performed between 2000 and 0100 h when trypanosome parasitemia is higher in other anuran species (Johnson et al., 1993). Túngara frogs are not preyed upon by other biting insects and liver-baited traps at the small temporary pools in which they breed revealed that leeches are absent (N = 5 nights, two traps a night). In addition, usually when leeches feed on amphibian hosts they leave distinct hematomas on their skin. Careful inspection of túngara frogs did not revealed signs of skin lesions such as those that result from leeches (McCallum et al., 2011, Rhoden and Bolek, 2012).
To characterize the trypanosome species using molecular techniques, additional blood samples were collected from individuals that had been confirmed to be infected with the trypanosome species described here using microscopy. Those samples were stored in lysis buffer and preserved at 4 °C for molecular analysis (Innis et al., 1990, Longmire et al., 1997). Some frogs were kept in captivity for longer periods to conduct behavioral experiments as part of an additional study.
2.2. Morphological characterization
After performing the blood smears, the slides were air dried, fixed with absolute methanol and later stained using Giemsa stain following Mohr (1981). Blood smears were thoroughly screened, covering the entire smear at 400× magnification (1–3 h per slide) using a Nikon Eclipse E 200 (Nikon, Chiyoda, Tokyo, Japan) microscope. Once trypomastigotes were found, they were photographed at 400× and 1000× magnification using a Nikon high-definition color camera head DS-Fi2 and the images were transferred onto a computer screen via a Nikon Camera Control unit DS–L2. We measured trypomastigote morphology (total body length and maximum width, N = 39) with Nikon's NIS-Elements D research application. Given the dark and uniform coloration of the stained trypomastigotes, other morphological characters could not be measured in a reliable way for any of the specimens. Additional blood samples from ten individuals were collected and blood smears prepared and stained using Hemacolor® Giemsa stain kit (Merck KGaA, Darmstadt, Hesse, Germany) in an attempt to obtain images revealing kinetoplastic morphology. Both stain techniques, however, had limited success revealing the kinetoplast and nucleus in the stained trypomastigote. Therefore, we could only make morphological measures of the internal structure in a subset of the specimens (N = 14). Measurements are given as the mean ± standard deviation in micrometers.
All blood smears were labeled and arranged in such a way to prevent biased screening of the slides. Statistical analysis was performed on the proportion of individuals infected across each sex, using a two-tailed Z-test for population proportion implemented through STATA 10 (StataCorp, College Station, TX, USA) (StataCorp, 2007).
2.3. Phylogenetic relationships
We extracted DNA directly from blood samples using DNeasy kits (Qiagen, Valencia, CA, USA) following the manufacturer's recommendations. Following Martin et al. (2002), we examined 18S ribosomal RNA gene (18S rRNA). We amplified by PCR two overlapping fragment of 18S rRNA with newly designed primers from an alignment of frog trypanosomes. The first fragment—955 bp—was amplified with primers SSU1_F (TCTGGTTGATTCTGCCAGTAG) and SSU1_R (AAAACCAACAAAAGCCGAAA); the second fragment—980 bp—was amplified with primers SSU2_F (CCAAAGCAGTCATCCGACTT) and SSU2_R (AGGAGCATCACAGACCTGCT). These primers were designed from a large alignment of trypanosome species (Hamilton et al., 2007); these primer sequences are highly conserved among trypanosomes, likely are able to amplify multiple species of anuran trypanosomes. Both PCR amplifications were conducted with a touchdown PCR profile (Murphy and O'Brien, 2007). After cleaning the PCR product with ExoSAP-IT (Affymetrix, Santa Clara, CA, USA), we completed sequencing reactions in both directions with the ABI BigDye chemistry (Applied Biosystems, Inc., Foster City, CA, USA), and sequenced the fragments on an ABI 3730xl DNA Analyzer automatic sequencer (Applied Biosystems, Inc., Foster City, CA, USA). We assembled contigs with the obtained sequence chromatograms in Geneious 6.1.6 (Biomatters, Auckland, New Zealand), resulting in sequences of 1688 bp for male #165504 (GenBank accession number: KM406915) and 1689 bp for male #165507 (GenBank accession number: KM406916).
We built an 18S rRNA gene matrix with the newly generated data and previously published sequences of members of the aquatic clade of Trypanosoma, using Trypanosoma avium Danilewsky 1885, Trypanosoma lewisi Kent 1880 and Trypanosoma theileri Laveran 1902 as outgroups (Martin et al., 2002, Ferreira et al., 2007, Ferreira et al., 2008, Hayes et al., 2014). We aligned the sequences using the MUSCLE (Edgar, 2004) plugin within Geneious 6.1.6, and edited manually obvious misplacements and removed suspicious ends of sequences (i.e., ends with abundant substitutions while the remaining of the alignment is conserved). The aligned matrix comprised 67 terminals and a length of 2364 bp. We ran Bayesian and maximum likelihood analyses with a single partition with the model GTR+Γ in MrBayes 3.1.2 (Ronquist and Huelsenbeck, 2003) and RAxML 8.0.12 (Stamatakis, 2014) respectively. For the Bayesian analysis we did two independent runs with 1 cold and 3 heated chains, with sampling the chains every 100 generations. The analysis was allowed to run until reaching estationarity—stopval set at 0.01, and later confirmed by the potential scale reduction factor values close to 1—which occurred at 1,185,000 generations, and 10% of generated trees were discarded as burn in. Nodal support was estimated with posterior probabilities. For the Maximum Likelihood we estimated the nodal support with 1000 bootstrap pseudo replicates.
As an additional confirmation of the species status of this new trypanosome we ran a coalescent-based species delimitation analysis using Poisson tree processes (PTP) model (Zhang et al., 2013). New probabilistic approaches for species delimitation provide alternatives to using arbitrary genetic thresholds and arbitrary monophyletic groupings. In particular, the PTP analysis is a fast species delimitation approach that attempts to identify putative species using a single input phylogenetic tree—usually built with a single locus by modeling speciation rates directly from the number of substitutions. We run the analysis in the bPTP web server with our maximum likelihood tree, using 500,000 Markov chain Monte Carlo generations, a thinning of 100 and a burn-in of 0.25.
3. Results
3.1. Species description
The trypanosomes observed in the blood smears have a unique set of morphological characters that differentiate them from previously described species. Morphology, however, often does not allow researchers to distinguish trypanosomes species and is problematic for determining species relationships. We obtained DNA sequences that revealed this lineage constitute a new species of trypanosome that we describe below.
Taxonomic summary: Phylum Euglenozoa, Cavalier-Smith, 1981; class Kinetoplastea, Honigberg, 1963; order Trypanosomatida, (Kent, 1880) Hollande, 1952; family Trypanosomatidae, Doflein, 1951. Trypanosoma tungarae n. sp. Bernal and Pinto (201×).
Type material: type blood smears of three infected frogs are deposited in the Smithsonian National Museum of Natural History (USNM Numbers TBD). Type Host: Vertebrate host is the túngara frogs E. pustulosus (Amphibia: Leuperidae); putative vectors are Corethrella spp. midges (Diptera: Corethrellidae). Type Locality: Panamá, Colon Province, Gamboa (30 m.a.s.l., 9° 79′ N, 79° 42.9′ W) (Fig. 2). Location on hosts: In the vertebrate hosts peripheral blood. The location in their putative vector frog-biting midges is unknown (possibilities include the digestive tract, the hemocele and the salivary glands). Distribution: Currently known only from the type locality, Gamboa, Panama. Diagnosis: Monomorphic trypanosome with an elongated body (52.13 ± 12.94 μm) and thin soma (5.41 ± 3.62 μm). Free flagellar length (FF), 13.20 ± 5.11 μm; midnucleus to anterior end (MA), 42.71 ± 13.77 μm; midnucleus to posterior end (MP), 29.67 ± 10.59 μm; midnucleus to kinetoplast, 20.31 ± 7.41 μm; posterior end to kinetoplast, 9.71 ± 3.50 μm; relative size of flagellum (FF/MA), 0.34 ± 0.14 μm; length of nucleus, 3.63 ± 1.67 μm; nuclear index (MP/MA), 0.97 ± 0.60 μm (Fig. 3). In general, this species resembles other anuran trypanosomes from Central and South America (Desser, 2001, Ferreira et al., 2007, McKenzie and Starks, 2008). This species is longer and thinner that T. rotatorium – like species found in other leptodactilyd anuran host in South America (Lemos et al., 2008). In particular, this species corresponds to the morphology of anuran trypanosomes with elongated trypomastigotes with pointed ends observed in Bufonidae, Leiuperidae and Leptodactylidae from Brazil (Group I, Ferreira et al., 2007). The morphology of this species, however, is most similar in general to Trypanosoma sp. (e) and Trypanosoma sp. (f) described from Lithobates vaillanti syn. Rana vaillanti by Desser (2001). Although the measurements of the species described here match closely some characteristics of Trypanosoma sp. (e) such as the relative length of the free flagellum, other features, including total body length and the distance from the posterior end to the kinetoplast, are closer to the morphology of Trypanosoma sp. (f). Some other features, however, are distinct from both Trypanosoma sp. (e) and (f) (e.g. distance from the center of the nucleus to the anterior end). A Trypanosoma montrealis-like species was found to be transmitted by North American frog-biting midges (C. wirthi) in Florida (Johnson et al., 1993). Although the body length and width of Trypanosoma montrealis (Fantham et al., 1942) fall within the dimensions of the species described here, that previously described species has a much shorter free flagellum than T. tungarae n. sp (3–5.5 μm vs 13.20 ± 5.11 μm). The validity of T. montrealis, however, has been questioned (Werner et al., 1988). More detailed morphological comparisons with previously described species of anuran trypanosomes from the same geographical area are unfeasible given that detailed morphological measurements are not often reported and recent, updated species descriptions frequently focus on the species genotypes (e.g Ferreira et al., 2007). Intraspecific morphological variation of amphibian trypanosomes, however, is so high that precludes its use for species identification. For example, amphibian trypanosomes can significantly change their morphotype when infecting different hosts (Hysek and Zizka, 1976).
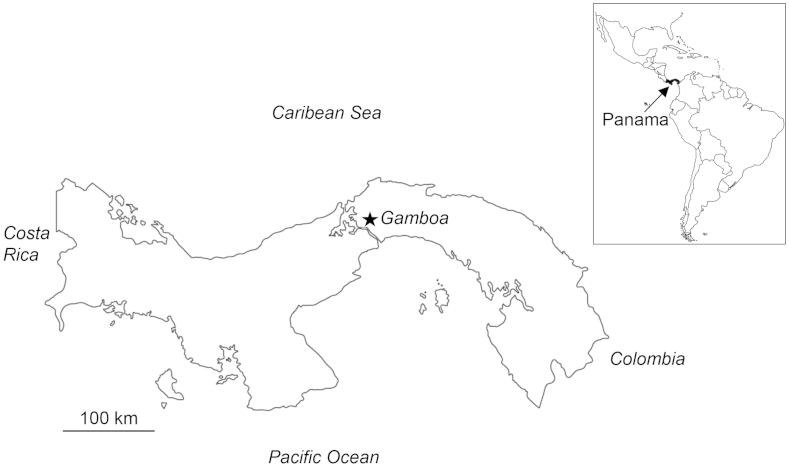
Fig. 2.
Map of the Republic of Panamá indicating with a star the location of Gamboa, the type locality of Trypanosoma tungarae n. sp. Insert shows the location of Panamá in the New World.
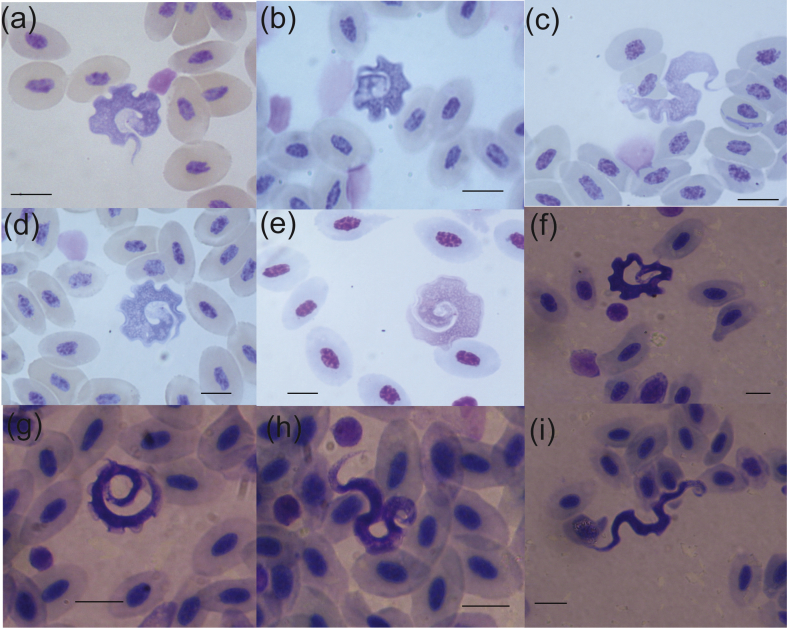
Fig. 3.
Light microscopy of Trypanosoma tungarae n. sp. (Giemasa-staining). (a–e) Trypomastigotes stained using Hemacolor® Giemsa stain kit (Voigt Global Distribution Inc, USA); (f–i) Trypomastigotes stained using Giemsa stain following Mohr (1981). Scale bars: 10 μm.
This species does not resemble in morphology Trypanosoma chattoni, the closest related species known to date (see under Phylogenetic relationships below), that has a characteristic round to oval body (Lemos et al., 2008). Trypomastigotes of both species, however, have large size and this new species thus becomes a new member of the giant trypanosomes that includes species such as Trypanosoma mega, T. ranarum and T. rotatorium (Martin et al., 2002). Despite the widespread distribution of T. chattoni including Asia (China, Werner, 1993; Kyushu and Ryukyu Islands, Miyata, 1978; Thailand, Sailasuta et al., 2011), North America (United Sates, Diamond, 1965; Canada, Jones and Woo, 1986) and South America (Brazil, Lemos et al., 2008), this species is monomorphic with little geographic variation. Both T. chattoni and T. tungarae n. sp. have heavily stained cytoplasms that often obscure the nucleus and kinetoplast. When visible, the kinetoplast lays towards the anterior end at about a fourth of the total length of the cell. Glass slides of Giemsa-stained smears from túngara frog blood samples and DNA samples are kept at the Smithsonian National Museum of Natural History, Washington, DC. To comply with the regulations of the International Code of Zoological Nomenclature (ICZN), details of this species have been submitted to ZooBank with the Life Science Identifier (LSID) zoobank.org:pub:TBD.
Etymology: Túngara (English pronunciation: toon-gah-rra) is the common name of the frog E. pustulosus, the vertebrate host of this new species of trypanosome. Túngara is a feminine Spanish onomatopoeic word resembling part of the singing repertoire of the E. pustulosus males. We treat tungarae as a feminine noun in the genitive case.
3.2. Host prevalence
Consistent with our prediction, we found sexual differences in trypanosomes infection in túngara frogs (Z-test, Z = 2.28, p = 0.022).While 40% of male túngara frogs sampled were infected with this blood parasite, only 6.6% of the females were infected (males: 10/25; females: 1/15). We were, however, expecting that no females would be infected since female túngara frogs do not vocalize. Frog-biting midges are attracted to the mating calls produced by males (Bernal et al., 2006, Borkent, 2008, McKeever and Hartberg, 1980), so our results beg the question, if frog-biting midges are the vectors, how did a female become infected with this new species of trypanosomes? Careful inspections of our records confirmed this result and field observations revealed a potential path of transmission for female frogs to be infected. When túngara frog are in amplexus, frog-biting midges attempting to feed on the calling male have an opportunity to move directly from their original victim, the male, to the female and obtain a blood meal (Fig. 1b,c).
3.3. Phylogenetic relationships
The maximum likelihood and the Bayesian inference phylogenies of the 18S rRNA gene are highly concordant, and show strong support for the placement of the new species, Trypanosoma tungarae, in the clade with aquatic trypanosomes; however, several internal branches are poorly supported for both methods. Trypanosoma tungarae n. sp. is sister to T. chattoni, and both form a highly supported clade sister to other trypanosomes of South American frogs (Fig. 4).
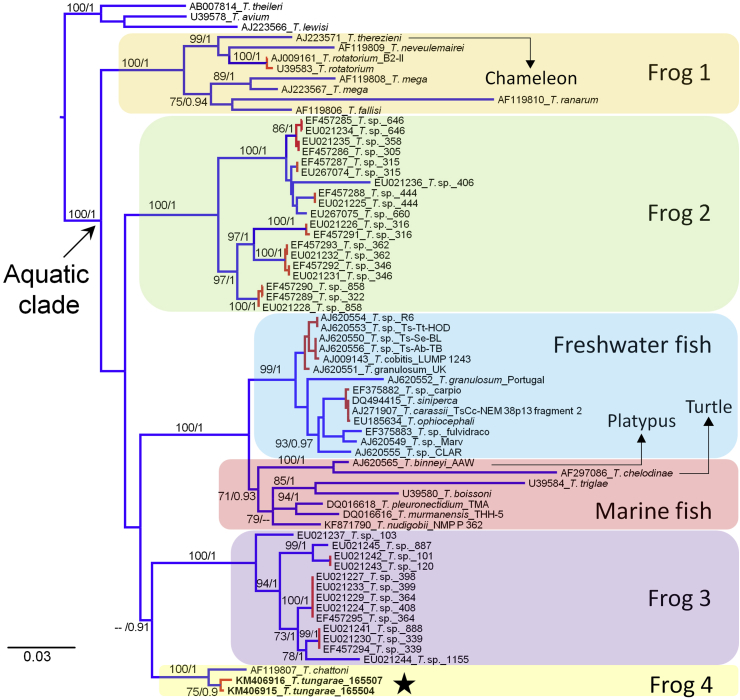
Fig. 4.
Phylogeny of the aquatic clade, and PTP species delimitation results. Best maximum likelihood tree of the18S rRNA gene of member of the aquatic clade and selected outgroups. Numbers on the branches represent support values corresponding to ≥70% bootstrap replicates (left) and ≥0.9 Bayesian posterior probabilities (right). Subclades are highlighted with colored boxes to indicate host associations. Color of the branches indicate the PTP species delimitation results; monophyletic groups in red indicate members of a single species, blue terminal branches indicate that only one sample is included in such species. Names of the terminals indicate the GenBank accession numbers, scientific name, and sample or isolate code. Star indicates the position of T. tungarae n. sp. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Both, the maximum likelihood and Bayesian solutions of the putative species delimitation analysis in PTP indicate that T. tungarae n. sp. is a different species from other trypanosomes for which molecular data is available. Also, the PTP analyses indicate that it might be some over splitting of species in fish trypanosomes, and several unrecognized species of frog trypanosomes (Fig. 4). The two sequences of T. tungarae n. sp. diverge in eight nucleotides, and it is likely that additional genetic variation can be found within the study area. Despite that the 18S rRNA gene is a slowly evolving marker, the variation that we found is not surprising given the complex patterns of intra and inter specific trypanosome diversity found in this geographic region (Pinto et al., 2012, Cottontail et al., 2014).
4. Discussion
Our results revealed that while male túngara frogs are frequently infected with trypanosomes, females rarely carry these parasites. Since females do not vocalize, they do not attract frog-biting midges (Bernal et al., 2006) and are thus rarely in contact with this putative vector. Similarly, sexual differences in prevalence of trypanosomes in green treefrogs, H. cinerea, were reported in the Southeastern United States where frog-biting midges (C. wirthi) were implicated as vectors of this parasite (Johnson et al., 1993). Transmission of T. tungarae n. sp. by vectors other than frog-biting midges seems unlikely. Leeches, common vectors of trypanosomes of the aquatic clade (Hamilton et al., 2007), are absent from the breeding puddles of túngara frogs in the study population. Although we collected leeches at our study site in larger ponds where other anurans breed, no leeches were found using the same traps in the puddles where túngara frogs breed. During the time we have spent observing túngara frogs in the field and collecting insects biting them (>100 h), no other blood-sucking insects or lesions potentially inflicted by leeches have been detected. The high numbers of frog-biting midges that bite túngara frogs (Bernal et al., 2006), combined with their ability to transmit this parasite to other frogs (Johnson et al., 1993), strongly suggest frog-biting midges may be the main vectors of T. tungarae n. sp. The advertisement call of túngara frogs attracts at least seven species of frog-biting midges (Bernal et al., 2006) and it is unclear if all, or only some, of those species may act as vectors of T. tungarae n. sp. Further studies that confirm the presence of T. tungarae n. sp. in the midgut or salivary glands of frog-biting midges and examine species differences in transmission of trypanosomes among frog-biting midges are necessary to confirm that the midges are indeed the vectors of T. tungarae n. sp. These studies would also provide valuable insights by clarify the degree of species specificity of trypanosomes and the midges.
In addition to frog-biting midges, there are other dipterans that are potential vectors of blood parasites that in general should be considered when investigating the transmission patterns of amphibian trypanosomes. There are, for instance, at least two species of mosquitos that use the mating calls of frogs to find their victim and feed exclusively on anurans (Uranotaenia lowii, Borkent and Belton, 2006; C. territans, Bartlett-Healy et al., 2008). Other frog-biting insects such as Forcipomyia species specialize on amphibians (Thompson, 1969) and could also act as vectors of anuran trypanosomes. Although at our study site túngara frogs are only preyed upon by frog-biting midges, frogs and toads are often bitten by a wide range of insects. Considering all potential vectors of anuran trypanosomes is essential to understand the dynamics of these protozoan parasites.
This description of a new species of Trypanosoma here highlights an interesting pattern of convergence in morphology among members of two non-closely related groups of trypanosomes infecting frogs. The morphometric data revealed that the trypomastigotes of T. tungarae n. sp. have overall similarity to other giant trypanosomes such as T. rotatorium and T. ranarum. Despite its slender and long cell shape, however, T. tungarae n. sp. is sister to T. chattoni—a highly derived trypanosome with a large rounded body, lacks a free flagellum, and lack of undulating membrane (Martin et al., 2002, Lemos et al., 2008). This convergence in morphology, however, could be explained by functionality; the sizes of the host's erythrocytes are correlated with the morphology of trypanosomes suggesting adaptations of the trypanosomes to the host environment (Wheeler et al., 2013). We limit our discussion to comparisons between T. tungarae n. sp. and species of trypanosomes from the phylogenetic tree used here because (i) we are confident they represent separate lineages, and (ii) it is difficult to rely on morphology to discern between blood trypanosomes (Lima et al., 2012, Fermino et al., 2015). Sequences, however, are not available for all anuran trypanosomes described to date. Therefore, it is possible that T. tungarae n. sp. may be equivalent to a previously described, unnamed trypanosome for which no molecular data is yet available. Further studies of trypanosome diversity in anurans that include a combination of morphological and molecular work would provide an opportunity to identify further cases of morphological convergence and overall patterns of evolution within members of the aquatic clade.
Despite significant efforts to revise the phylogenetic relations and taxonomic status of anuran trypanosomes (Diamond, 1965, Ayala, 1970, Ayala, 1971, Desser and Yekutiel, 1986, Desser, 2001, Martin et al., 2002, Ferreira et al., 2007, Ferreira et al., 2008, Lemos et al., 2008), there is still an urgent need for an extensive revision of this group of parasites. The phylogeny of anuran trypanosomes needs in particular the advancement of the development of tools to include additional genes. Traditionally only the 18S rRNA and gGAPDH genes have been used for trypanosome phylogenetics (e.g. Hamilton et al., 2007), and most of the work conducted on the aquatic clade has relied only on data from one gene (this study, Martin et al., 2002, Ferreira et al., 2007, Ferreira et al., 2008, Hayes et al., 2014). Perhaps the difficulties associated with amplifying markers different to the 18S rRNA directly from DNA extracted from blood and tissues have hampered the efforts to build stronger phylogenies of this clade. In this study, we failed to amplify the gGAPDH gene using published and new primers. As a consequence, our analysis only includes one gene and several relationships are thus not well supported in our phylogeny. For example, there is little support for the relationships among the subclades that we identified. Despite this challenge, however, fragments of the 18S rRNA gene have been successfully used to characterize trypanosome species in other systems (e.g., Hayes et al., 2014) and the DNA sequences found in this study indicate the trypanosome examined here represents a single, new lineage.
The PTP species delimitation approach we used here is a reliable method to tentatively identify trypanosome species using phylogenetic data. Another study explored the usefulness of this method in uncovering several species of trypanosomes in a single location providing convincing evidence of its reliability (Cottontail et al., 2014). Multiple loci and multiple delimitation approaches, however, are necessary to confirm these inferences (Carstens et al., 2013). Nonetheless, for organisms as poorly studied as the trypanosomes of wildlife, the PTP method is promising—at least until generating data from multiple genes is a common practice.
Trypanosoma tungarae n. sp. is currently only known from its type host, túngara frogs. Although a trypanosome was previously reported to be transmitted by frog-biting midges to another anuran in the Southeastern US (Johnson et al., 1993), its relationship to T. tungarae n. sp. is unknown because it was not characterized. Few studies have investigated host specificity of anuran trypanosomes. Studies have described the presence of the same trypanosome species across a broad range of frogs and toads (Ray and Choudhury, 1983) and, given that vectors are often associated with several species of vertebrate hosts (Ferreira et al., 2008), their association to only one or few anuran species seems unlikely. The diversity of hosts used by T. tungarae n. sp. requires further examination. Other potential anuran hosts include, for instance, include the hourglass frog (Dendrosophus ebbracatus) and yellow cricket treefrog (D. microcephalus) that are also victims of frog-biting midges (de Silva et al., 2014), the putative vector of T. tungarae n. sp. in this area. Investigating the diversity of hosts of T. tungarae in further studies will be important to understand the patterns of this blood parasite's dynamics in this anuran community.
Conflicts of interest
None declared.
Acknowledgments
We thank the Smithsonian Tropical Research Institute (STRI) for logistical support. Mabelle Chong, Johanna Goyes, Simone Loss, Rachel Page and Miryam Venegas provided invaluable help during the field and laboratory work. Daniel Castillo from the Universidad de Panama provided valuable advice about staining techniques. We are also grateful to Cameron Smith and Erik Galindo who helped inspect blood smears. Susan Perkins, Matthew Bolek, and Agustín Jiménez read different versions of this manuscript and provided insightful comments. Two anonymous reviewers provided valuable comments that helped improve this study. This research was approved by the Panamanian Authorities (Autoridad Nacional del Medio Ambiente, ANAM permit #SE/A-29-09, SEX/A-60-09). XEB was supported by a grant from the National Science Foundation (IOS#1433990).
Contributor Information
Ximena E. Bernal, Email: xbernal@purdue.edu.
C. Miguel Pinto, Email: pintom@si.edu.
References
- Ayala S.C. Two new trypanosomes from California toads and lizards. J. Protozool. 1970;17:370–373. [Google Scholar]
- Ayala S.C. Trypanosomes in wild California sandflies, and extrinsic stages of Trypanosoma bufophlebotomi. J. Protozool. 1971;18:433–438. doi: 10.1111/j.1550-7408.1971.tb03349.x. [DOI] [PubMed] [Google Scholar]
- Bardsley J.E., Harmsen R. The Trypanosomes of Anura. Adv. Parasitol. 1973;11:1–73. doi: 10.1016/s0065-308x(08)60184-0. [DOI] [PubMed] [Google Scholar]
- Bartlett-Healy K., Crans W., Gauger E. Phonotaxis to amphibian vocalizations in Culex territans (Diptera: Culicidae) Ann. Entomol. Soc. Am. 2008;101:95–103. [Google Scholar]
- Bernal X.E., Rand A.S., Ryan M.J. Acoustic preferences and localization performance of blood-sucking flies (Corethrella Coquillett) to túngara frog calls. Behav. Ecol. 2006;17:709–715. [Google Scholar]
- Bernal X.E., de Silva P. Cues used in host-seeking behavior by frog-biting midges (Corethrella spp Coquillet) J. Vector Ecol. 2015;40:122–128. doi: 10.1111/jvec.12140. [DOI] [PubMed] [Google Scholar]
- Borkent A. The frog-biting midges of the world (Corethrellidae: Diptera) Zootaxa. 2008;1804 1–456. [Google Scholar]
- Borkent A. World catalog of extant and fossil Corethrellidae (Diptera) Zootaxa. 2014;3796:453–468. doi: 10.11646/zootaxa.3796.3.3. [DOI] [PubMed] [Google Scholar]
- Borkent A., Belton P. Attraction of female Uranotaenia lowii (Diptera: Culicidae) to frogs in Costa Rica. Can. Entomol. 2006;138:91–94. [Google Scholar]
- Carstens B.C., Pelletier T.A., Reid N.M., Satler J.D. How to fail at species delimitation. Mol. Ecol. 2013;22:4369–4383. doi: 10.1111/mec.12413. [DOI] [PubMed] [Google Scholar]
- Cottontail V.M., Kalko E.V., Cottontail I., Wellinghausen N., Tschapka M., Perkins S., Pinto C.M. High local diversity of Trypanosoma in a common bat species, and implications for the biogeography and taxonomy of the T. cruzi clade. PLoS One. 2014;9:e108603. doi: 10.1371/journal.pone.0108603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Silva P., Jaramillo C.A., Bernal X.E. Selection of biting sites on anuran hosts by Corethrella Coquillett species. J. Insect Behav. 2014;27:302–316. [Google Scholar]
- Desser S.S., Yekutiel D. Blood parasites of amphibians and reptiles in Israel. Isr. J. Zool. 1986;34:77–90. [Google Scholar]
- Desser S.S. The blood parasites of anurans from Costa Rica with reflections on the taxonomy of their trypanosomes. J. Parasitol. 2001;87:152–160. doi: 10.1645/0022-3395(2001)087[0152:TBPOAF]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Desser S.S., Mciver S.B., Ryckman A. Culex territans as a potential vector of Trypanosoma rotatorium. I. Development of the flagellate in the mosquito. J. Parasitol. 1973;59:353–358. [PubMed] [Google Scholar]
- Diamond L.S. Studies on the morphology, biology and taxonomy of the trypanosomes of Anura. Wildl. Dis. 1965;44:1–77. [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantham H.B., Porter A., Richardson L.R. Some Haematozoa observed in vertebrates in eastern Canada. Parasitology. 1942;34:199–226. [Google Scholar]
- Feng L.C., Chung H.L. Phlebotomus squamirostris Newstead, transmitter of Trypanosoma bocagei França in the toad, Bufo bufo gargarizans (Cantor) China Med. J. 1940;3:198–211. [Google Scholar]
- Ferguson L.V., Smith T.G. Reciprocal trophic interactions and transmission of blood parasites between mosquitoes and frogs. Insects. 2012;3:410–423. doi: 10.3390/insects3020410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fermino B.R., Paiva F., Soares P., Tavares L.E.R., Viola L.B., Ferreira R.C., Botero-Arias R., de-Paula C.D., Campaner M., Takata C.S., Teixeira M.M. Field and experimental evidence of a new caiman trypanosome species closely phylogenetically related to fish trypanosomes and transmitted by leeches. Int. J. Parasitol. Parasites Wildl. 2015;4:368–378. doi: 10.1016/j.ijppaw.2015.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira R.C., Campaner M., Viola L.B., Takata C.S.A., Takeda G.F., Teixeira M.M.G. Morphological and molecular diversity and phylogenetic relationships among anuran trypanosomes from the Amazonia, Atlantic Forest and Pantanal biomes in Brazil. Parasitology. 2007;134:1623–1638. doi: 10.1017/S0031182007003058. [DOI] [PubMed] [Google Scholar]
- Ferreira R.C., De Souza A.A., Freitas R.A., Campaner M., Takata C.S.A., Barrett T.V., Shaw J.J., Teixeira M.M.G. A phylogenetic lineage of closely related Trypanosomes (Trypanosomatidae, Kinetoplastida) of Anurans and sand flies (Psychodidae, Diptera) sharing the same Ecotopes in Brazilian Amazonia. J. Eukaryot. Microbiol. 2008;55:427–435. doi: 10.1111/j.1550-7408.2008.00342.x. [DOI] [PubMed] [Google Scholar]
- Hamilton P.B., Gibson W.C., Stevens J.R. Patterns of co-evolution between trypanosomes and their hosts deduced from ribosomal RNA and protein-coding gene phylogenies. Mol. Phylogenet. Evol. 2007;44:15–25. doi: 10.1016/j.ympev.2007.03.023. [DOI] [PubMed] [Google Scholar]
- Hayes P.M., Lawton S.P., Smith N.J., Gibson W.C., Davies A.J. Morphological and molecular characterization of a marine fish trypanosome from South Africa, including its development in a leech vector. Parasit. Vectors. 2014;7:1–11. doi: 10.1186/1756-3305-7-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hysek J., Zizka Z. Transmission of Trypanosoma rotatorium from frogs to white mice. Nature. 1976;260:608–609. doi: 10.1038/260608a0. [DOI] [PubMed] [Google Scholar]
- Innis M.A., Gelfand D.H., Sninsky J.J., White T.J. Academic Press; London: 1990. PCR Protocols: a Guide to Methods and Applications. [Google Scholar]
- Johnson R.N., Young D.G., Butler J.F. Trypanosome transmission by Corethrella wirthi (Diptera, Chaoboridae) to the green treefrog, Hyla cinerea (Anura, Hylidae) J. Med. Entomol. 1993;30:918–921. doi: 10.1093/jmedent/30.5.918. [DOI] [PubMed] [Google Scholar]
- Jones S.R.M., Woo P.T. Trypanosoma chattoni Mathis and Leger, 1911 in Rana pipiens of southern Ontario: morphometrics and a description of the division process. Syst. Parasitol. 1986;9:57–62. [Google Scholar]
- Lemos M., Morais D.H., Carvalho V.T., D'Agosto M. First record of Trypanosoma chattoni in Brazil and occurrence of other Trypanosoma Species in Brazilian frogs (Anura, Leptodactylidae) J. Parasitol. 2008;94:148–151. doi: 10.1645/GE-1095.1. [DOI] [PubMed] [Google Scholar]
- Lima L., Da Silva F.M., Neves L., Attias M., Takata C.S., Campaner M., De Souza W., Hamilton P.B., Teixeira M.M. Evolutionary insights from bat trypanosomes: morphological, developmental and phylogenetic evidence of a new species, Trypanosoma (Schizotrypanum) erneyi sp. nov., in african bats closely related to Trypanosoma (Schizotrypanum) cruzi and allied species. Protist. 2012;163:856–872. doi: 10.1016/j.protis.2011.12.003. [DOI] [PubMed] [Google Scholar]
- Lynch K.S., Crews D., Ryan M.J., Wilczynski W. Hormonal state influences aspects of female mate choice in the túngara frog (Physalaemus pustulosus) Horm. Behav. 2006;49:450–457. doi: 10.1016/j.yhbeh.2005.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longmire J.L., Maltbie M., Baker R.J. Occas. Pap. Museum Texas Tech Univ; 1997. Use of “Lysis Buffer” in DNA Isolation and its Implication for Museum Collections; p. 163. [Google Scholar]
- McCallum M.L., Moser W.E., Wheeler B.A., Trauth S.E. Amphibian infestation and host size preference by the leech Placobdella picta (Verrill, 1872) (Hirudinida: Rhynchobdellida: Glossiphoniidae) from the Eastern Ozarks, USA. Herpetol. Notes. 2011;4:147–151. [Google Scholar]
- McKenzie V.J., Starks H.A. Blood parasites of two Costa Rican amphibians with comments on detection and microfiliaria density associated with adult filarial worm intensity. J. Parasitol. 2008;94:824–829. doi: 10.1645/GE-1433.1. [DOI] [PubMed] [Google Scholar]
- Martin D.S., Wright A.-D.G., Barta J.R., Desser S.S. Phylogenetic position of the giant anuran trypanosomes, Trypanosoma chattoni, Trypanosoma fallisi, Trypanosoma mega, Trypanosoma neveulemairei, and Trypanosoma ranarum inferred from 18S rRNA gene sequences. J. Parasitol. 2002;88:566–571. doi: 10.1645/0022-3395(2002)088[0566:PPOTGA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- McKeever S., Hartberg W.K. An effective method for trapping adult female Corethrella (Diptera, Chaoboridae) Mosq. News. 1980;40:111–112. [Google Scholar]
- McKeever S., French F.E. Int. Virtual Conf. Vet. Med. Proc. Dis. Reptil. Amphib. 2000. Corethrellidae (Diptera), vectors of present and perhaps some of the earliest Anuran trypanosomes. October 1-31, 2000. [Google Scholar]
- Miyata A. Anuran trypanosomes in Kyushu and Ryukyu Islands, with descriptions of six new species. Trop. Med. 1978;20:51–80. [Google Scholar]
- Mohr J.L. Methods for staining protozoans in blood, macrophages or body fluids. In: Baltimore C.G., editor. Staining Procedures. fourth ed. Williams & Wilkins; Philadelphia: 1981. p. 303. [Google Scholar]
- Murphy W.J., O'Brien S.J. Designing and optimizing comparative anchor primers for comparative gene mapping and phylogenetic inference. Nat. Protoc. 2007;2:3022–3030. doi: 10.1038/nprot.2007.429. [DOI] [PubMed] [Google Scholar]
- Pinto C.M., Kalko E.K.V., Cottontail I., Wellinghausen N., Cottontail V.M. TcBat, a bat-exclusive lineage of Trypanosoma cruzi, in the Panama Canal Zone, with comments on its classification and the use of the 18S rRNA gene for lineage identification. Infect. Genet. Evol. 2012;12:1328–1332. doi: 10.1016/j.meegid.2012.04.013. [DOI] [PubMed] [Google Scholar]
- Ray R., Choudhury A. Vol. 8. Zoological Survey of India; Calcutta: 1983. (Trypanosomes of Indian Anurans). [Google Scholar]
- Ramos B., Urdaneta-Morales S. Hernatophagous insects as vectors for frog trypanosomes. Rev. Biol. Trop. 1977;25:209–217. [PubMed] [Google Scholar]
- Reilly B.O., Woo P.T. Susceptibility of the leech Batracobdella picta (Verrill) to Trypanosoma andersoni Reilly and Woo and Trypanosoma gryl. Can. J. Zool. 1982;60:1441–1445. [Google Scholar]
- Rhoden H.R., Bolek M.G. Helminth and leech community structure in tadpoles and caudatan larvae of two amphibian species from western Nebraska. J. Parasitol. 2012;98:236–244. doi: 10.1645/GE-2771.1. [DOI] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Ryan M.J. University of Chicago Press; Chicago: 1985. The Túngara Frog: a Study in Sexual Selection and Communication. [Google Scholar]
- Sailasuta A., Satetasit J., Chutmongkonkul M. Pathological study of blood parasites in rice field frogs, Hoplobatrachus rugulosus (Wiegmann, 1834) Vet. Med. Int. 2011 doi: 10.4061/2011/850568. Article ID 850568 http://dx.doi.org/10.4061/2011/850568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- StataCorp . StataCorp LP; College Station, TX: 2007. Stata Statistical Software: Release 10. [Google Scholar]
- Thompson P.H. Feeding of Forcipomyia fairfaxensis on grass frogs (Rana spp.) in New Jersey. Ann. Entomol. Soc. Am. 1969;62:451–452. [Google Scholar]
- Werner J. Blood parasites of amphibians from Sichuan Province, People's Republic of China. J. Parasitol. 1993;79:356–363. [PubMed] [Google Scholar]
- Werner J.K., Davis J.F., Slaght K.S. Trypanosomes of Bufo americanus from northern Michigan. J. Wild. Dis. 1988;24:647–649. doi: 10.7589/0090-3558-24.4.647. [DOI] [PubMed] [Google Scholar]
- Wheeler R.J., Gluenz E., Gull K. The limits on trypanosomatid morphological diversity. PLoS One. 2013;8:e79581. doi: 10.1371/journal.pone.0079581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Kapli P., Pavlidis P., Stamatakis A. A general species delimitation method with applications to phylogenetic placements. Bioinformatics. 2013;29:2869–2876. doi: 10.1093/bioinformatics/btt499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Žičkus T. The first data on the fauna and distribution of blood parasites of amphibians in Lithuania. Acta Zool. Litu. 2002;12:197–202. [Google Scholar]