Abstract
Seizure models have demonstrated that neuroinflammation and neurodegeneration are preponderant characteristics of epilepsy. Considering the lack of clinical studies, our aim is to investigate the extrinsic pathway of apoptosis in pharmacoresistant temporal lobe epilepsy (TLE) associated with hippocampal sclerosis (HS) patients, TLE(HS). By a specific death receptor-mediated apoptosis array plate, 31 upregulated targets were revealed in the sclerotic hippocampus from TLE(HS) patients. Amongst them are the encoding genes for ligands (FASLG, TNF, and TNFSF10) and death receptors (FAS, TNFRSF1A, TNFRSF10A, and TNFRSF10B). In addition, we evaluated the hippocampal relative mRNA expression of the two TNF receptors, TNFRSF1A and TNFRSF1B, in patients, being both upregulated (n = 14; P < 0.01 and P < 0.04, resp.) when compared to the post mortem control group (n = 4). Our results have clearly suggested that three different death receptor apoptotic systems may be associated with the maintenance and progression of TLE-associated HS: (1) TNF-TNFRSF1A, (2) FASLG-FAS, and (3) TNFSF10-TNFRSF10A/B. Their effects on epilepsy are still scarcely comprehended. Our study points out to TNF and TNF receptor superfamily pathways as important targets for pharmacological studies regarding the benefits of an anti-inflammatory therapy in these patients.
1. Introduction
The connection between neurodegeneration and inflammation in the epileptic brain has emerged as an important axis for the comprehension of the pathomechanisms involved in seizure associated neuronal cell death. Hippocampal sclerosis (HS) is the main pathohistological abnormality found in resected tissue from temporal lobe epilepsy (TLE) patients. It is characterized by astrogliosis: an irregular proliferation of astrocytes due to the apoptosis of nearby neurons [1]. Several studies assert that inflammation has a crucial role in epileptogenesis [2], and there is a high probability that hippocampal chronic inflammation could exacerbate astrogliosis in TLE patients.
Apoptosis is divided into two main specific pathways: intrinsic and extrinsic. In spite of requesting particular triggers to start a cascade of molecular events, both pathways converge in the activation of CASP3, and both are present in TLE-associated HS. The apoptotic intrinsic signaling pathways are initiated by non-receptor-mediated intracellular signals (e.g., DNA damage, radiation) that result in changes in the inner mitochondrial membrane. Seizures by themselves can represent an interesting example of these kinds of signals. One of the primary events in hippocampal seizure-induced apoptosis is the excessive release of glutamate with consequent intracellular calcium overload, culminating in downstream swelling and rupture of intracellular organelles and activated proteolytic enzymes leading to cell death [3].
Unlike the intrinsic signaling pathways that initiate apoptosis, the extrinsic signaling pathways involve transmembrane receptor-mediated interactions. These include death receptors which are members of the TNF receptor gene superfamily [4]. Apoptotic or survival signals are the consequence of the death receptor family activation by death ligands. Various members of this family have been described thus far, including TNFRSF1A, FAS, TNFRSF25, TNFRSF10A, TNFRSF10B, and TNFRSF21 [5]. The execution pathway can be initiated by these receptors through CASP8 or CASP10 whose activation could trigger the proapoptotic proteins BID or BAX, via TP53, culminating in a cross talk with the mitochondrial or intrinsic pathway of apoptosis [4].
Recurrent seizures induce not only neuronal cell loss but also inflammation [6]. However, neuronal cell loss is not a prerequisite for inflammation to happen; rather, the liberation of proinflammatory cytokines can contribute to cell death [7], and dying cells may perpetuate inflammation [6]. Previously, we reported a marked TNF upregulation in TLE(HS) patients [8], which is indicative of chronic hippocampal inflammation.
We believe that the neuronal apoptosis through death receptor pathways highlights the key molecular events involved in triggering astrogliosis and TLE-associated HS. Understanding which are the soluble mediators and the molecular mechanisms crucially involved in the link between inflammation and neuronal cell death is instrumental to shed light on how seizures may contribute to HS in epilepsy and to identify new therapeutic targets for the treatment and cure of pharmacoresistant epilepsy, particularly in TLE(HS) patients.
2. Methods
2.1. Subjects and Tissue Collection
Ethical approval was certified by the “Comitê de Ética da Faculdade de Ciências Médicas da Unicamp” (CEP n 470/2003). Subjects' data was reported in a recent study [8]. The present study was performed with the same patient and post mortem control samples. Briefly, TLE and HS were detected by telemetry/video electroencephalogram and magnetic resonance image, respectively, in 14 patients. Due to the pharmacoresistance of the syndrome, they went through an amygdalohippocampectomy for treatment. All hippocampal tissue samples were immediately collected and divided into two parts. One portion was fixed for histopathological analysis and HS/astrogliosis was confirmed in all of them [H&E staining and the terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay; data not shown]. The second portion was immediately snap-frozen in liquid nitrogen after surgery and stored at −80°C until RNA isolation.
Four post mortem control hippocampal tissue samples (1 female, 3 males; 22.75 ± 5.56 years old, ranging from 19 to 31) were kindly provided by the “Instituto Médico Legal, IML” (Forensic Institute) of Campinas. Post mortem control subjects passed away instantaneously or quickly. Although their deaths were traumatic, which runs against the occurrence and progression of neuroinflammation, neurological findings were not detected and the post mortem delay averaged 7.8 h (range: 6.0–9.0 h).
2.2. Gene Expression
All reagents were purchased from Thermo Fisher Scientific Inc.; Waltham, MA 02451, USA (former Life Technologies; Foster City, CA 94404, USA).
2.2.1. RNA Extraction, Array Gene Expression, and RT-qPCR
Total RNA extraction and RT-qPCR were carried out according to our previous work [8]. TRIzol® Reagent was used for RNA extraction according to the manufacturer's instructions. The RNA integrity number (RIN) average in control and patient groups was 7.525 ± 0.5437 and 6.155 ± 0.2484, respectively. Afterwards, 1 μg of total RNA of each sample was reverse transcribed into cDNA using 200 U of Superscript® III Reverse Transcriptase and 3 μg of Random Primers. Sterilized and filtered DEPC treated water was used in all RNA procedures.
To identify potential targets associated with apoptosis through death receptor signaling, we used a TaqMan® Array Human Apoptosis through death receptors 96-well plate (PN: 4414105). The plate contained 44 assays of genes associated with death receptor-mediated apoptosis and four assays of reference gene (endogenous control) candidates (Table 1). Gene names are in accordance with the approved symbol from the HUGO Gene Nomenclature Committee (HGNC) database. All assays were plated in duplicates: one for cDNA pooled from the TLE(HS) patients (n = 12) versus the other for cDNA pooled from the post mortem controls (n = 4). Among several factors which can influence C T (cycle threshold) and consequently qPCR results credibility, we emphasized instrument calibration and cDNA quality. The template concentration (e.g., concentration of total RNA converted to cDNA) should always be homogeneous among samples, particularly when working with pools. cDNA concentration and purity were assessed by spectrophotometry (NanoDrop ND1000; NanoDrop Technologies, Wilmington, DE). The cDNA concentration means were 858.1 ± 30.07 and 871.9 ± 27.10 for control and patient samples, respectively. cDNA absorbance (A260/A280) ranged from 1.84 to 1.90 considering all 16 samples. Reactions were carried out according to manufacturer's instructions. We used a total volume of 20 μL, comprising 10 μL of TaqMan Gene Expression Master Mix (Life Technologies) and 10 μL of pooled cDNA diluted in DNase-free water. The final concentration of the pooled cDNA samples used was 10 ng in 10 μL for each 20 μL PCR reaction.
Table 1.
Array target genes: apoptosis through death receptors in TLE(HS).
| Target genes | |||
|---|---|---|---|
| Symbol, approved name, and Assay ID | Synonyms and aliases | Function | RQ |
| ACTA1 actin, alpha 1, skeletal muscle Hs00559403_m1 |
NEM3, “nemaline myopathy type 3” | Cell motility | 1.12 |
|
| |||
| ACTB actin, beta Hs99999903_m1 |
Cell motility | 1.74 | |
|
| |||
| APAF1 apoptotic peptidase activating factor 1 Hs00559441_m1 |
APAF-1, CED4 | Apoptosis induction by CASP3 activation | 1.59 |
|
| |||
| BAX BCL2-associated X protein Hs00180269_m1 |
BCL2L4 | Apoptosis induction by CASP3 activation | 1.50 |
|
| |||
| BCL2 B-cell CLL/lymphoma 2 Hs99999018_m1 |
Bcl-2, PPP1R50, “protein phosphatase 1, and regulatory subunit 50” | Apoptosis inhibition | 1.87 |
|
| |||
| BID BH3 interacting domain death agonist Hs00609632_m1 |
Apoptosis induction | 1.40 | |
|
| |||
| BIRC2 baculoviral IAP repeat containing 2 Hs01112284_m1 |
“Apoptosis inhibitor 1”, c-IAP1, cIAP1, hiap-2, MIHB, “NFR2-TRAF signalling complex protein”, and RNF48 | Apoptosis inhibition | 1.31 |
|
| |||
| BIRC3 baculoviral IAP repeat containing 3 Hs00154109_m1 |
“Apoptosis inhibitor 2”, c-IAP2, cIAP2, hiap-1, “inhibitor of apoptosis protein 1”, MALT2, “mammalian IAP homolog C”, MIHC, RNF49, and “TNFR2-TRAF signaling complex protein” | Apoptosis inhibition | 1.27 |
|
| |||
| CASP2 caspase 2 Hs00234982_m1 |
ICH1, MGC2181, PPP1R57, and “protein phosphatase 1, regulatory subunit 57” | Involved in the activation cascade of caspases responsible for apoptosis execution | 1.38 |
|
| |||
| CASP3 caspase 3 Hs00234387_m1 |
Apopain, CPP32, CPP32B, and Yama | Involved in the activation cascade of caspases responsible for apoptosis execution | 1.44 |
|
| |||
| CASP7 caspase 7 Hs00169152_m1 |
CMH-1, ICE-LAP3, and MCH3 | Involved in the activation cascade of caspases responsible for apoptosis execution; overexpression promotes programmed cell death | 2.13 |
|
| |||
| CASP8 caspase 8 Hs01018151_m1 |
Casp-8, FLICE, MACH, and MCH5 | Most upstream protease of the activation cascade of caspases responsible for the FAS mediated and TNFRSF1A induced cell death | 2.45 |
|
| |||
| CASP9 caspase 9 Hs00154260_m1 |
APAF-3, ICE-LAP6, MCH6, PPP1R56, and “protein phosphatase 1, regulatory subunit 56” | Involved in the activation cascade of caspases responsible for apoptosis execution | 1.55 |
|
| |||
| CFLAR CASP8 and FADD like apoptosis regulator Hs01116280_m1 |
c-FLIP, CASH, Casper, CLARP, FLAME, FLIP, I-FLICE, and MRIT | Apoptosis regulator protein as FAS mediated apoptosis inhibitor | 1.59 |
|
| |||
| CHUK conserved helix-loop-helix ubiquitous kinase Hs00175141_m1 |
IKBKA, IKK-alpha, IKK1, IKKA, and NFKBIKA | Serine kinase. Essential role in the NFkB signaling pathway | 1.22 |
|
| |||
| DAXX death-domain associated protein Hs00154692_m1 |
DAP6 | JNK pathway and apoptosis mediator via MAP3K5 [FAS and TGFBR2 (transforming growth factor beta receptor II) signaling] | 1.53 |
|
| |||
| DIABLO diablo, IAP-binding mitochondrial protein Hs00219876_m1 |
DFNA64, DIABLO-S, FLJ10537, FLJ25049, “second mitochondria-derived activator of caspase”, and SMAC | Apoptosis promoter by caspase activation in the cytochrome c/APAF1/CASP9 pathway | 1.45 |
|
| |||
| FADD Fas associated via death domain Hs00538709_m1 |
“Fas-associating death domain-containing protein,” “Fas-associating protein with death domain,” GIG3, “growth-inhibiting gene 3 protein,” “mediator of receptor-induced toxicity,” and MORT1 | CASP8 and CASP10 apoptotic adaptor recruiter to activated FAS and TNFRSF1A | 1.13 |
|
| |||
| FAS Fas cell surface death receptor Hs00531110_m1 |
APO-1, CD95, and “TNF receptor superfamily member 6” | Receptor with death domain for FASLG | 1.65 |
|
| |||
| FASLG Fas ligand Hs00181225_m1 |
CD178, FasL | Cytokine ligand for FAS | 5.46 |
|
| |||
| IKBKB inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta Hs00233287_m1 |
IKK-beta, IKK2, IKKB, and NFKBIKB | NFkB activator | 1.96 |
|
| |||
| IKBKG inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma Hs00415849_m1 |
FIP-3, FIP3, Fip3p, IKK-gamma, NEMO, and ZC2HC9 | NFkB activator | 1.65 |
|
| |||
| MAP3K5 mitogen-activated protein kinase kinase kinase 5 Hs00178726_m1 |
“Apoptosis signal regulating kinase 1,” ASK1, and MAPKKK5 | Signal transduction mediator by oxidative stress and receptor-mediated inflammatory signals (TNF) | 1.55 |
|
| |||
| MAPK8 mitogen-activated protein kinase 8 Hs01548508_m1 |
JNK, JNK1, “JUN N-terminal kinase,” and SAPK1 | Stressed cell apoptosis promoter through TP53 and YAP1 | 1.05 |
|
| |||
| MAPK9 mitogen-activated protein kinase 9 Hs00177102_m1 |
JNK2, “Jun kinase,” p54a, and SAPK | Stressed cell apoptosis promoter through TP53 and YAP1 | 0.86 |
|
| |||
| NFKB1 nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 Hs00765730_m1 |
KBF1, NF-kappaB, NF-kB1, NFkappaB, NFKB-p50, p105, and p50 | Rel protein-specific transcription inhibitor (105 kD) and DNA binding subunit of the transcription factor NFkB (50 kD) | 1.50 |
|
| |||
| NFKB2 nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) Hs00174517_m1 |
LYT-10, NF-kB2, p105, and p52 | Subunit of the transcription factor NFkB | 3.21 |
|
| |||
| NFKBIA nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha Hs00153283_m1 |
IkappaBalpha, IKBA, and MAD-3 | NFkB inhibitor | 2.36 |
|
| |||
| NGF nerve growth factor (beta polypeptide) Hs01113193_m1 |
Ligand for NTRK1 and NGFR; neuronal proliferation, differentiation and survival regulator | 0.90 | |
|
| |||
| NGFR nerve growth factor receptor Hs00609976_m1 |
CD271, “low affinity nerve growth factor receptor,” p75NTR, “TNFR superfamily, member 16,” and TNFRSF16 | Neural cell death or survival mediator | 3.53 |
|
| |||
| PARP1 poly(ADP-ribose) polymerase 1 Hs00242302_m1 |
PARP | DNA repair pathway initiation; apoptosis promoter in response to genotoxic stress | 1.51 |
|
| |||
| RELA v-rel avian reticuloendotheliosis viral oncogene homolog A Hs00153294_m1 |
p65 | Subunit of the transcription factor NFkB | 2.02 |
|
| |||
| TNF tumor necrosis factor Hs00174128_m1 |
DIF, “TNF superfamily, member 2,” TNF-alpha, and TNFSF2 | Multifunctional proinflammatory cytokine ligand for TNFRSF1A and TNFRSF1B | 39.96 |
|
| |||
| TNFRSF10A tumor necrosis factor receptor superfamily member 10a Hs00269492_m1 |
Apo2, CD261, DR4, and TRAILR-1 | Receptor with death domain for TNFSF10; NFkB activator | 2.39 |
|
| |||
| TNFRSF10B tumor necrosis factor receptor superfamily member 10b Hs00366278_m1 |
CD262, DR5, KILLER, TRAIL-R2, TRICK2A, and TRICKB | Receptor with death domain for TNFSF10; ER stress-induced apoptosis promoter; NFkB activator | 1.55 |
|
| |||
| TNFRSF10C tumor necrosis factor receptor superfamily member 10c Hs00182570_m1 |
CD263, DcR1, LIT, TRAILR3, and TRID | Decoy receptor for TNFSF10; apoptosis inhibitor | 4.82 |
|
| |||
| TNFRSF10D tumor necrosis factor receptor superfamily member 10d Hs00533560_m1 |
CD264, DcR2, TRAILR4, and TRUNDD | Decoy receptor for TNFSF10; apoptosis inhibitor | 3.74 |
|
| |||
| TNFRSF1A tumor necrosis factor receptor superfamily member 1A Hs00205419_m1 |
CD120a, TNF-R, TNF-R-I, TNF-R55, TNFAR, and TNFR60 | Receptor with death domain for TNF and LTA (lymphotoxin alpha); NFkB activator, apoptosis mediator, and inflammation regulator | 3.07 |
|
| |||
| TNFRSF21 tumor necrosis factor receptor superfamily member 21 Hs00237054_m1 |
CD358, “death receptor 6,” and DR6 | Receptor with death domain; apoptosis promoter | 1.09 |
|
| |||
| TNFRSF25 tumor necrosis factor receptor superfamily member 25 Hs00187070_m1 |
APO-3, DDR3, DR3, LARD, TR3, TRAMP, WSL-1, and WSL-LR | Receptor with death domain for TNFSF12; mediator of NFkB activation and apoptosis promoter | 1.72 |
|
| |||
| TNFRSF6B tumor necrosis factor receptor superfamily member 6b Hs00234356_m1 |
DcR3, DCR3, M68, and TR6 | Decoy receptor for FASLG; apoptosis inhibitor | 2.54 |
|
| |||
| TNFSF10 tumor necrosis factor superfamily member 10 Hs00174664_m1 |
Apo-2L, CD253, TL2, and TRAIL | Cytokine ligand for TNFRSF10A, TNFRSF10B, TNFRSF10C, and TNFRSF10D | 2.50 |
|
| |||
| TP53 tumor protein p53 Hs01034249_m1 |
LFS1, “Li-Fraumeni syndrome,” and p53 | Tumor suppressor, growth arrest, or apoptosis promoter depending on the physiological circumstances and cell type | 2.77 |
|
| |||
| TRAF2 TNF receptor associated factor 2 Hs00184192_m1 |
TRAP3 | Mediator of the antiapoptotic signals from TNF receptors; NFkB and JNK activation regulator | 1.54 |
List of all target genes investigated in the array, involved in the extrinsic apoptotic pathway. The gene expression data [relative quantification (RQ) column], also shown in Figure 1, corresponds to the cDNA pooled from TLE(HS) patients (n = 12) versus the cDNA pooled from post mortem controls (n = 4). The latter was used as a calibrator with gene expression equal to 1.0. The values in the table can be converted to fold change values, where the negative inverse (−1/x) is taken for values between 0 and 1 (e.g., 0.5 is converted to −2). Values greater than 1 will not be affected. A −2 value indicates that the molecule is 2-fold downregulated. Only MAPK9 and NGF were downregulated. The reference gene candidates in the plate were 18S (eukaryotic 18S rRNA; Hs99999901_s1), GAPDH (glyceraldehyde-3-phosphate dehydrogenase; Hs99999905_m1), GUSB (glucuronidase, beta; Hs99999909_m1), and HPRT1 (hypoxanthine phosphoribosyltransferase 1; Hs99999908_m1) although only GAPDH and HPRT1 were effectively used as reference genes (in combination).
Two target genes were chosen to be tested separately by subject: TNFRSF1A and TNFRSF1B (Assay ID: Hs00153550_m1). The latter was not included in the array plate. The PCR efficiency validation was also performed according to our previous work [8]. The amplification efficiencies were close to 1.0 (100%). cDNA samples derived from the investigated genes were detected using an ABI PRISM® 7500 Sequence Detection system and TaqMan Gene Expression Assays. For RT-qPCRs, reference genes were selected according to Wierschke et al. [9]. Therefore, HPRT1 and the geometric mean of ENO2 and TBP were used as our normalization factors. Both were indicated as being the most stable reference genes in epileptogenic tissue [9]. Each qPCR was run as triplicates with 10 ng cDNA sample in 6.25 μL TaqMan Gene Expression Master Mix, 0.625 μL of the respective probe/primer mix and 0.625 μL purified and deionized H2O. All reactions were run as triplicates, and measurements with a difference of more than 0.3 C T-values were excluded from analysis.
2.3. Data Analysis
Relative gene expression data was generated and analyzed by the 7500 Software version 2.0.5 (Life Technologies). The software GraphPad Prism 5 was used for the statistical analysis (GraphPad Prism version 5.04 for Windows, GraphPad Software, San Diego California USA, 〈 http://www.graphpad.com 〉). The Mann-Whitney U test was used for comparison between data from the control group (n = 4) versus the patient group (n = 14). For all analyses, differences of P < 0.05 were considered significant.
A molecular pathway related to differentially expressed genes was created by the IPA-Ingenuity Pathway Analysis (Ingenuity Systems, 〈 http://www.ingenuity.com 〉) application via the “Core Analysis” function. The array data sheet generated by the 7500 Software was uploaded into the IPA application. The differentially expressed genes were connected based on the reported association among genes or proteins and their functional roles.
3. Results
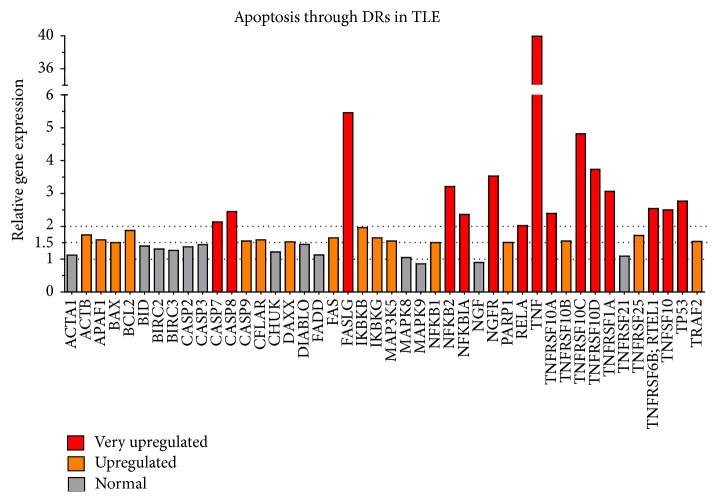
Array gene expression quantifications are shown in Table 1 and Figure 1. From 44 target genes associated with apoptosis through death receptors, we found 31 upregulated genes (70.45%). Our criterion to define an upregulated gene was based on an increase of a minimum of 50% on mRNA levels in comparison to controls, which corresponds to a cutoff of 1.5-fold change. Choosing a precise cutoff is always a challenge in array studies and it is crucial to analyze results and select the most important targets. Considering our previous study on NFKB1 expression in the same group of TLE(HS) patients (n = 14) [8], a 1.5 cutoff is statistically acceptable since the NFKB1 expression in the array was exactly 1.5-fold.
Figure 1.
Hippocampal expression of genes associated with apoptosis through death receptors in TLE(HS) patients versus post mortem controls. The combination of GAPDH and HPRT1 was used as the reference gene. The pool of post mortem control samples (n = 4) was the reference sample (calibrator) and its relative quantification (RQ) was always 1.0. Different colors represent gene expression in RQ values (or fold-change values, when RQ is a positive number) and correspond to the following parameters: red = very upregulated (RQ ≥ 2.0); orange = upregulated (1.99 ≥ RQ ≥ 1.5); gray = normal (1.49 ≥ RQ ≥ 0.67). None of the 44 target genes analyzed was downregulated (0.66 ≥ RQ ≥ 0.51) or very downregulated (RQ ≤ 0.5).
The number of upregulated genes is narrowed to 15 if a cutoff of 2.0 is considered (at least an augmentation of 100% on mRNA levels). This more conservative cutoff reveals critical target genes that can be divided into 3 categories: (1) ligands and receptors, (2) mediators, and (3) molecules related to the transcription factor nuclear factor kappa B (NFkB). Discussion will be guided by these categories and focused on the controversies and novelties related to these most upregulated targets.
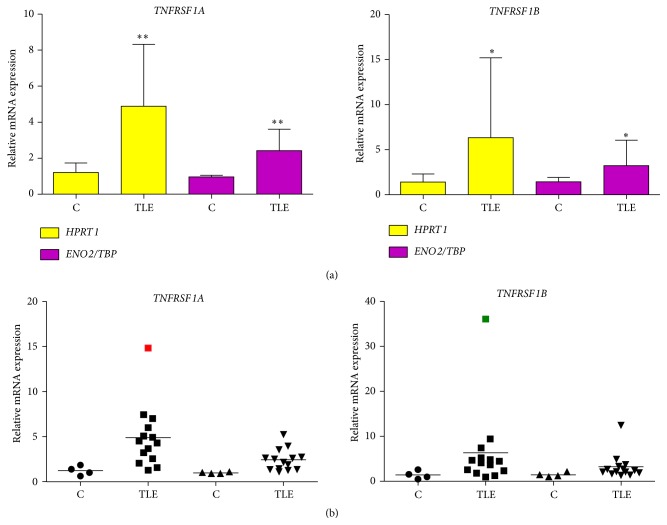
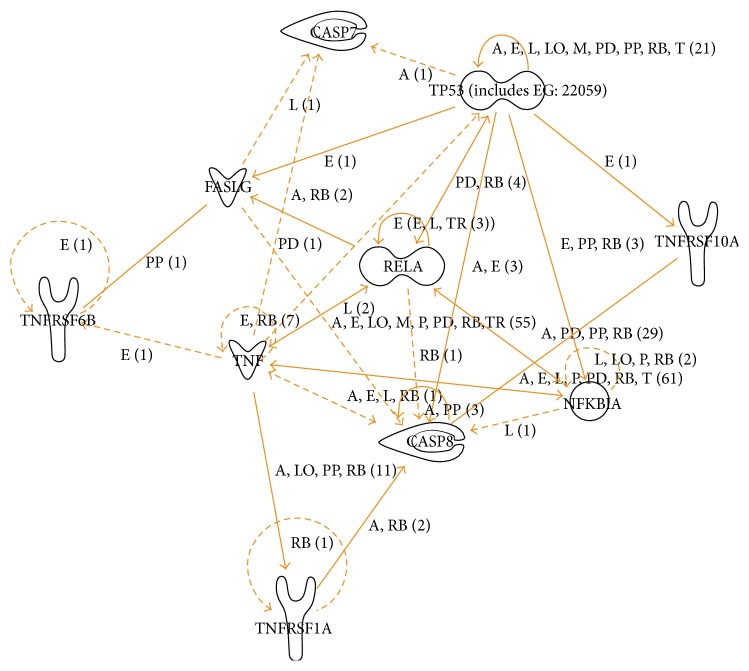
Regardless of the cutoff or even the reference genes, array TNF expression (39.96-fold) was very meaningful, particularly in comparison to the expression of the other genes. This significant overregulation led us to quantify separately the expression of its receptors, TNFRSF1A and TNFRSF1B, being both statistically upregulated (P < 0.01 and P < 0.04, resp.) (Figure 2). In Figure 3, we proposed a schematic pathway which involves TNF, TNFRSF1A, and eight of the other 13 very upregulated genes.
Figure 2.
Hippocampal gene expression of TNFRSF1A and TNFRSF1B in TLE(HS) patients versus post mortem controls. HPRT1 and ENO2/TBP were used as reference genes. One of the post mortem control samples was randomly chosen as the reference and its relative quantification was always 1.0. All quantification data for the remaining samples, including controls and patients, was benchmarked to the reference sample. Samples were separated in two groups: TLE(HS) patients (n = 14) and post mortem controls (n = 4). (a) Columns are means with SD. Mann-Whitney U tests were used for the comparison between groups. ∗ P < 0.05; ∗∗ P < 0.01. (b) Marks are different geometric figures, which represent the gene expression mean of the samples. The horizontal line is the mean of the group analyzed. Circles and squares correspond to gene expression with HPRT1 as the reference. The two kinds of triangles correspond to gene expression with ENO2/TBP as the reference. The red and green squares (outliers) correspond to TLE 09 and TLE 03 patients, respectively (see Table 1 in Teocchi et al., 2013 [8]).
Figure 3.
Apoptosis through death receptors: a signaling pathway in TLE(HS). This pathway was generated by the IPA software, based on 10 of the 15 upregulated genes (cutoff 2.0-fold change; see Table 1) in hippocampal tissues from TLE(HS) patients (n = 12) compared to post mortem controls (n = 4). The biological relationship between two genes, represented as nodes, is shown as a line. Nodes with different shapes indicate different functional class. The pathway created did not include the following genes: NFKB2, NGFR, TNFSF10, TNFRSF10C, and TNFSF10D. The function “nervous relaxed” was used. A, activation; E, expression regulation; I, inhibition; L, proteolysis; LO, localization; M, biochemical modification; P, phosphorylation; PD, protein-DNA interaction; PP, protein-protein interaction; RB, regulation of binding; T, transcription; and TR, translocation.
4. Discussion
Inflammation, degeneration of neurons and HS are closely related [10]. Recently, we reported a marked TNF upregulation in TLE(HS) patients [8], which is indicative of chronic hippocampal inflammation. The importance of this cytokine as one of the main apoptosis “propellers” in TLE-associated HS is reinforced by our Human Apoptosis through death receptors array results (Table 1 and Figure 1) which show the upregulation of 31 related genes. They are associated with the extrinsic apoptosis signaling pathway, among them members of the TNF and TNF receptor gene superfamilies and several factors which play a role in the cell death cascade. The vast majority of them have never been quantified in patient's samples. Below, we present and discuss our finding which suggests the involvement of these genes with neurodegeneration and the consequent astrogliosis in TLE(HS).
4.1. Apoptotic Systems: Ligands and Receptors
4.1.1. The TNF-TNFRSF1A Axis
There are few clinical studies on the TNF system and its effects on epilepsy in spite of intense investigation in animals. Several models indicate that seizures induce the TNF expression in the brain [2, 11–14] although some have shown no important TNF alterations in plasma or CSF after different kinds of seizures [15–18]. In patients, Sinha et al. detected increased serum levels of TNF and other cytokines in several epilepsy syndromes [19]. Our results for TLE(HS) patients differ from those on seizure models only by the fact that the TNF mRNA increase does not seem to be transiently enhanced. Some patients had their last seizure several days before the surgery, implying that the high TNF expression levels were happening often, suggesting that chronic hippocampal inflammation could be intrinsic to refractory TLE(HS).
The most controversial evidence on the TNF role in epilepsy was raised when Balosso et al. reported that intrahippocampal injection of murine-recombinant TNF in mice potently prevented seizures [20]. Moreover, transgenic mice overexpressing TNF by astrocytes showed shorter seizures, whereas deficient mice for TNF receptors had prolonged seizures [20]. The dichotomic effect of TNF on seizures may be related to the tissue microenvironment, including the cellular source (neuronal versus glial) of TNF release, the extent of TNF increase, its concentration and persistence in tissue, and action through its two receptors expressed by the targeted cells [14, 20, 21].
In the hippocampus, TNF is able to activate its two receptors (TNFRSF1A and TNFRSF1B) to regulate cell-signaling pathways [22, 23]. TNFRSF1A is ubiquitously expressed in human tissues and is the principal signaling receptor for TNF. A cytoplasmic death domain, required for apoptotic signaling pathways and NFkB activation, is present in this major receptor. TNFRSF1B does not contain a death domain and actuates in restricted biological processes, being mainly expressed in immune cells. Through intracellular adaptors called TRAFs (TNF Receptor-Associated Factors) associated with this receptor, JNK (c-Jun N-terminal kinase) is activated during the cell survival induction through activation of NIK (NFkB-inducing kinase), a downstream target of TRAF2. Thus, TNFRSF1A has been involved in apoptosis activation, whereas TNFRSF1B is implicated with activation of the NFkB system [24–26]. Likewise, seizure model studies imply that the TNFRSF1A pathway is associated with deleterious effects and that the TNFRSF1B pathway is related to anticonvulsive outcomes [14, 27].
Our hippocampal RT-qPCR results showed the upregulation of both TNF receptors in TLE(HS) patients (Figure 2). The deleterious consequences of TNFRSF1A action are well-known; however, for TNFRSF1B, consequences are still unclear. TNFRSF1B could be triggered as a survival mechanism to compensate for the extensive neuronal cell death found in epilepsy-associated HS or its expression could enable the harmful consequences of TNFRSF1A activation [28]. These authors demonstrated that TNFRSF1B significantly decreased the TNF concentration required for cell death without the generation of an intracellular signal. Instead, TNFRSF1B modulated the rate of TNF association with TNFRSF1A, possibly by increasing the local concentration of TNF at the cell surface through rapid ligand association and dissociation. Functional experiments are necessary to verify this finding in brain cells and clarify the role of TNFRSF1B in TLE(HS). Furthermore, these authors proposed that other cell-surface receptors, such as NGFR, may utilize an analogous “ligand passing” mechanism. Our array results showed the upregulation of NGFR (3.53-fold). Increased expression of NGFR in hippocampal neurons of TLE patients has already been described [29].
4.1.2. The FAS-FASLG Axis
FASLG triggers apoptosis by binding to FAS [30]. Our finding showed the upregulation of both FAS (1.65-fold) and FASLG (5.46-fold), which represents an important apoptotic system scarcely investigated in clinical cases. Only one study reported an augmentation of FAS in the sclerotic hippocampus from TLE patients [31]. In animals, seizures induced a significant augmentation in FAS expression within the ipsilateral hippocampus from 4 to 24 h after levels returned to baseline [32]. Similarly, increased expression of both Fas mRNA and protein were evident in the adult rat brain from 4 h to 5 days after the onset of kainic acid-induced seizures and neurons with increased FAS expression were also immunoreactive for TP53 [33]. Induced by kainate, FASLG expression increased rapidly at 6 h and returned to the basal level at 3 days in CA1 and CA3/DG (dentate gyrus) hippocampal regions [30, 34].
FASLG was the second most upregulated gene according to our results (Figure 1). Considering the chronic status of our patients, this cytokine does not appear to have a transient expression in pharmacoresistant TLE(HS) and might contribute to neuronal loss, being the FAS-FASLG pathway an interesting target for pharmacological studies. In addition, we found that TNFRSF6B was also overregulated in TLE(HS) patients (2.54-fold). The encoded protein by TNFRSF6B is assumed to play a regulatory role in suppressing FASLG-mediated cell death. It acts as a decoy receptor that competes with death receptors for ligand binding [35] (Figure 3). We have not found any studies regarding the association between TNFRSF6B expression and epilepsy.
4.1.3. The TNFSF10-TNFRSF10A/B Axis
Besides apoptosis mediated by TNF and FASLG and its receptors, TNFSF10 is another potent inducer of apoptosis [36, 37]. Both apoptosis-inducing and nonapoptosis-inducing membrane-bound receptors have been described for TNFSF10. Only TNFRSF10A and TNFRSF10B are able to generate a death signal [38, 39], while TNFRSF10C and TNFRSF10D are truncated and have been proposed to function as decoy receptors by binding TNFSF10 without producing a death signal, thereby inhibiting apoptosis [39, 40]. Our results on gene expression accentuate the importance of TNFSF10 (2.50-fold) and its upregulated receptors (Figure 1). They have never been quantified by RT-qPCR in human TLE hippocampal samples. In brain tissues from TLE patients, Dörr and colleagues did not detect the expression of TNFSF10, but both apoptosis-inducing and non-apoptosis-inducing receptors were expressed on neurons, astrocytes, and oligodendrocytes, which indicate their possible susceptibility to TNFSF10-mediated apoptosis [41]. On the other hand, a significant increase on TNFSF10 expression in both patients and animals (seizure model) has been reported [42]. In accord with our results, it is conceivable that there is an important action of TNFSF10 and its receptors in TLE(HS)-associated neuronal apoptosis. The expression and regulation of these receptors might be crucial for death or survival of an individual cell, since both apoptosis-mediating and apoptosis-blocking receptors were present on the different brain parenchymal cells [37, 41]. Our data per se shows an overview on this apoptotic system and is complemented by the two previously discussed studies on proteins.
4.2. Apoptotic Mediators: Caspases and TP53
We found elevated mRNA levels for CASP7, CASP8, and CASP9 in our TLE(HS) samples (Table 1 and Figure 1). This finding extends and reinforces protein data from seizure models and human epileptogenic tissue studies, which have reported a deregulated expression of caspases [32, 43–46]. It is well-known that sequential activation of caspases plays a central role in the execution phase of cell apoptosis.
CASP7 is an effector, responsible for cleaving important apoptotic intracellular substrates. Yamamoto et al. found X-linked inhibitor of apoptosis (XIAP) binding CASP7 in TLE brain, suggesting ongoing antiapoptotic responses, which might be impeding caspases from inducing apoptosis [46]. XIAP can only bind activated CASP7 to modulate its activity [47], and XIAP expression is regulated after experimental seizures [48]. Their finding reinforces the evidence for active CASP7 in the TLE hippocampus. CASP8 is a key initiator of apoptosis via death receptor-mediated pathways, capable of inducing apoptosis by directly processing executioner caspases [32]. In a seizure model, its inhibition significantly reduced neuronal apoptosis, accompanied by a decrease of tBID (truncated BID), cleaved CASP9, and cytosol cytochrome c [45]. Our results are in agreement with the studies mentioned and emphasize the importance of CASP7 and CASP8 in TLE(HS) (Figure 3).
A Tp53 overexpression occurred along with excitotoxicity in an adult rat brain within hours after systemic administration of glutamate analogue kainic acid [49]. Years later and after several other reports on altered TP53 expression in seizure models, including the already mentioned study of Tan and colleagues [33], Xu et al. found TP53 positive cells in the sclerotic hippocampus from TLE patients [31]. In the same year, Engel and colleagues detected significantly higher levels of TP53 by Western blotting in hippocampal tissues from TLE patients [50]. Our result on the elevated TP53 mRNA level extends and reinforces this data. It is still unclear if TP53 could function as a potential target for protection in seizure-induced neuronal death since TP53 regulates a large number of genes, which would need to be carefully evaluated [31].
4.3. Subunits and Regulators of the Transcription Factor NFkB
As previously reported, we found that NFKB1 was upregulated in TLE(HS) patients [8]. In the present study, our array results showed that NFKB1 was 1.5-fold more expressed than controls. Several other genes related to the NFkB complex, such as IKBKB (1.96-fold), IKBKG (1.65-fold), NFKB2 (3.21-fold), NFKBIA (2.36-fold), and RELA (2.02-fold) also showed an increased expression (Figure 1). Taken together, our finding clearly emphasizes the NFkB involvement in TLE(HS) physiopathology (Figure 3). In addition, protein or mRNA expression data on those molecules is scarce, even from seizure models.
A number of signal transduction events, initiated by inflammation, immunity, differentiation, cell growth, tumorigenesis, and apoptosis, converge on NFkB activation [51]. This pleiotropic transcription factor, present in practically all cell types, is a homo- or heterodimeric complex formed by the Rel-like domain-containing proteins RELA/p65, RELB, NFKB1/p105, NFKB1/p50, REL, and NFKB2/p52. The heterodimeric p65-p50 complex appears to be the most common. NFKBIA and NFKBIB (I-kappa-B proteins) inhibit the NFkB complex by trapping it in the cytoplasm [51]. The kinases IKBKA and IKBKB mark I-kappa-B proteins for destruction via the ubiquitination pathway, thereby allowing activation of the NFkB complex. IKBKG is a regulatory subunit of the inhibitor of kappa B kinase (IKK) core complex which phosphorylates NFkB inhibitors, leading to the dissociation of the inhibitor/NFkB complex and ultimately the degradation of the inhibitor [51]. Our finding shows that there is a disturbed expression of activators and repressors of the NFkB complex. Further studies are necessary to understand the actual role of NFkB in TLE(HS).
5. Conclusions
We have demonstrated the upregulation of various genes associated with the apoptosis through death receptor signaling in the hippocampus from TLE(HS) patients. The expression of the vast majority of these genes was studied for the first time in hippocampal human samples despite previous protein results, mainly from seizure models. Our data reveals that both TNF receptor genes (TNFRSF1A and TNFRSF1B) are overexpressed in patients. As key factors in the TNF-induced apoptosis, they may play a crucial role in hippocampal neuroinflammation and neurodegeneration.
Additionally, our results clearly suggest that three different death receptor apoptotic systems may be associated with the maintenance and progression of TLE-associated HS: (1) TNF-TNFRSF1A, (2) FASLG-FAS, and (3) TNFSF10-TNFRSF10A/B. Their effects on epilepsy are still scarcely comprehended. Other targets, such as the NFkB subunits and regulators or the decoy receptor of FAS, TNFRSF6B, whose gene upregulation has never been associated with epilepsy, should be investigated. The understanding of these signaling pathways is essential for the development of new antiepileptic drugs and opens several possible avenues of research that will help us to understand the complex pathophysiology in HS.
Acknowledgments
The authors would like to thank all the patients and the neurosurgeons, Dr. Evandro Pinto da Luz de Oliveira and Dr. Helder Tedeschi, who performed the epilepsy surgery. Quantitative PCRs were carried out at the Hematology and Hemotherapy Center at the University of Campinas. RIN detection was performed by the Central Laboratory of High Performance Technologies-LaCTAD (〈 http://www.lactad.unicamp.br/ 〉). This work was supported by the São Paulo Research Foundation (FAPESP) (Grant no. 2005/565778-4) and UNICAMP. Marcelo Ananias Teocchi was supported by a Graduate Scholarship from CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior). Marcelo Ananias Teocchi wishes to thank David Elieff for helping with the English language review.
Abbreviations
- BAX:
BCL2-associated X protein
- BID:
BH3 interacting domain death agonist
- CASP:
Caspase
- ENO2:
Enolase 2 (gamma, neuronal)
- FAS:
Fas cell surface death receptor
- HPRT1:
Hypoxanthine phosphoribosyltransferase 1
- HS:
Hippocampal sclerosis
- JNK:
c-Jun N-terminal kinase
- NIK:
NFkB-inducing kinase
- RT-qPCR:
Reverse Transcription-Quantitative PCR
- tBID:
Truncated BID
- TBP:
TATA-box binding protein
- TLE(HS):
Temporal lobe epilepsy associated with hippocampal sclerosis
- TLE:
Temporal lobe epilepsy
- TNF:
Tumor necrosis factor
- TNFRSF10A:
Tumor necrosis factor receptor superfamily, member 10a
- TNFRSF10B:
Tumor necrosis factor receptor superfamily, member 10b
- TNFRSF1A:
Tumor necrosis factor receptor superfamily, member 1a
- TNFRSF21:
Tumor necrosis factor receptor superfamily, member 21
- TNFRSF25:
Tumor necrosis factor receptor superfamily, member 25
- TP53:
Tumor protein p53
- TRAF2:
TNF Receptor-Associated Factor 2
- TRAFs:
TNF Receptor-Associated Factors
- TRAIL:
TNF-related apoptosis-inducing ligand
- XIAP:
X-linked inhibitor of apoptosis.
Conflict of Interests
The authors declare that they have no conflict of interests.
Authors' Contribution
Marcelo Ananias Teocchi and Lília D'Souza-Li designed the study. Marcelo Ananias Teocchi collected patients' clinical data, performed the experiments, analyzed the data, prepared the figures and tables, performed the statistical analysis, and drafted the paper. Lília D'Souza-Li conceived the study and supervised all the aspects of its development. Both authors read and approved the final paper.
References
- 1.Greenfield J. G., Love S., Louis D. N., Ellison D. Greenfield's Neuropathology. 8th. London, UK: Hodder Arnold; 2008. [Google Scholar]
- 2.Ravizza T., Balosso S., Vezzani A. Inflammation and prevention of epileptogenesis. Neuroscience Letters. 2011;497(3):223–230. doi: 10.1016/j.neulet.2011.02.040. [DOI] [PubMed] [Google Scholar]
- 3.Fujikawa D. G. Prolonged seizures and cellular injury: understanding the connection. Epilepsy and Behavior. 2005;7(supplement 3):S3–S11. doi: 10.1016/j.yebeh.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 4.Elmore S. Apoptosis: a review of programmed cell death. Toxicologic Pathology. 2007;35(4):495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lavrik I., Golks A., Krammer P. H. Death receptor signaling. Journal of Cell Science. 2005;118(2):265–267. doi: 10.1242/jcs.01610. [DOI] [PubMed] [Google Scholar]
- 6.Vezzani A., Aronica E., Mazarati A., Pittman Q. J. Epilepsy and brain inflammation. Experimental Neurology. 2013;244:11–21. doi: 10.1016/j.expneurol.2011.09.033. [DOI] [PubMed] [Google Scholar]
- 7.Allan S. M., Rothwell N. J. Cytokines and acute neurodegeneration. Nature Reviews Neuroscience. 2001;2(10):734–744. doi: 10.1038/35094583. [DOI] [PubMed] [Google Scholar]
- 8.Teocchi M. A., Ferreira A. É. D., da Luz de Oliveira E. P., Tedeschi H., D'Souza-Li L. Hippocampal gene expression dysregulation of Klotho, nuclear factor kappa B and tumor necrosis factor in temporal lobe epilepsy patients. Journal of Neuroinflammation. 2013;10, article 53 doi: 10.1186/1742-2094-10-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wierschke S., Gigout S., Horn P., et al. Evaluating reference genes to normalize gene expression in human epileptogenic brain tissues. Biochemical and Biophysical Research Communications. 2010;403(3-4):385–390. doi: 10.1016/j.bbrc.2010.10.138. [DOI] [PubMed] [Google Scholar]
- 10.Proper E. A., Oestreicher A. B., Jansen G. H., et al. Immunohistochemical characterization of mossy fibre sprouting in the hippocampus of patients with pharmaco-resistant temporal lobe epilepsy. Brain. 2000;123, part 1:19–30. doi: 10.1093/brain/123.1.19. [DOI] [PubMed] [Google Scholar]
- 11.Plata-Salamán C. R., Ilyin S. E., Turrin N. P., et al. Kindling modulates the IL-1β system, TNF-α, TGF-β1, and neuropeptide mRNAs in specific brain regions. Molecular Brain Research. 2000;75(2):248–258. doi: 10.1016/s0169-328x(99)00306-x. [DOI] [PubMed] [Google Scholar]
- 12.Godlevsky L. S., Shandra A. A., Oleinik A. A., Vastyanov R. S., Kostyushov V. V., Timchishin O. L. TNF-α in cerebral cortex and cerebellum is affected by amygdalar kindling but not by stimulation of cerebellum. Polish Journal of Pharmacology. 2002;54(6):655–660. [PubMed] [Google Scholar]
- 13.Vezzani A., Moneta D., Richichi C., et al. Functional role of inflammatory cytokines and antiinflammatory molecules in seizures and epileptogenesis. Epilepsia. 2002;43(supplement 5):30–35. doi: 10.1046/j.1528-1157.43.s.5.14.x. [DOI] [PubMed] [Google Scholar]
- 14.Li G., Bauer S., Nowak M., et al. Cytokines and epilepsy. Seizure. 2011;20(3):249–256. doi: 10.1016/j.seizure.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 15.Peltola J., Palmio J., Korhonen L., et al. Interleukin-6 and interleukin-1 receptor antagonist in cerebrospinal fluid from patients with recent tonic-clonic seizures. Epilepsy Research. 2000;41(3):205–211. doi: 10.1016/s0920-1211(00)00140-6. [DOI] [PubMed] [Google Scholar]
- 16.Haspolat S., Mihci E., Coskun M., Gumuslu S., Ozben T. Interleukin-1β, tumor necrosis factor-α, and nitrite levels in febrile seizures. Journal of Child Neurology. 2002;17(10):749–751. doi: 10.1177/08830738020170101501. [DOI] [PubMed] [Google Scholar]
- 17.Virta M., Hurme M., Helminen M. Increased plasma levels of pro- and anti-inflammatory cytokines in patients with febrile seizures. Epilepsia. 2002;43(8):920–923. doi: 10.1046/j.1528-1157.2002.02002.x. [DOI] [PubMed] [Google Scholar]
- 18.Bauer S., Cepok S., Todorova-Rudolph A., et al. Etiology and site of temporal lobe epilepsy influence postictal cytokine release. Epilepsy Research. 2009;86(1):82–88. doi: 10.1016/j.eplepsyres.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 19.Sinha S., Patil S. A., Jayalekshmy V., Satishchandra P. Do cytokines have any role in epilepsy? Epilepsy Research. 2008;82(2-3):171–176. doi: 10.1016/j.eplepsyres.2008.07.018. [DOI] [PubMed] [Google Scholar]
- 20.Balosso S., Ravizza T., Perego C., et al. Tumor necrosis factor-α inhibits seizures in mice via p75 receptors. Annals of Neurology. 2005;57(6):804–812. doi: 10.1002/ana.20480. [DOI] [PubMed] [Google Scholar]
- 21.Balosso S., Ravizza T., Aronica E., Vezzani A. The dual role of TNF-α and its receptors in seizures. Experimental Neurology. 2013;247:267–271. doi: 10.1016/j.expneurol.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 22.Loetscher H., Steinmetz M., Lesslauer W. Tumor necrosis factor: receptors and inhibitors. Cancer Cells. 1991;3(6):221–226. [PubMed] [Google Scholar]
- 23.Mukai Y., Shibata H., Nakamura T., et al. Structure-function relationship of tumor necrosis factor (TNF) and its receptor interaction based on 3D structural analysis of a fully active TNFR1-selective TNF mutant. Journal of Molecular Biology. 2009;385(4):1221–1229. doi: 10.1016/j.jmb.2008.11.053. [DOI] [PubMed] [Google Scholar]
- 24.Natoli G., Costanzo A., Moretti F., Fulco M., Balsano C., Levrero M. Tumor necrosis factor (TNF) receptor 1 signaling downstream of TNF receptor-associated factor 2. Nuclear factor κB (NFκB)-inducing kinase requirement for activation of activating protein 1 and NFκB but not of c- Jun N-terminal kinase/stress-activated protein kinase. The Journal of Biological Chemistry. 1997;272(42):26079–26082. doi: 10.1074/jbc.272.42.26079. [DOI] [PubMed] [Google Scholar]
- 25.Zhuang L., Wang B., Shinder G. A., Shivji G. M., Mak T. W., Sauder D. N. TNF receptor p55 plays a pivotal role in murine keratinocyte apoptosis induced by ultraviolet B irradiation. The Journal of Immunology. 1999;162(3):1440–1447. [PubMed] [Google Scholar]
- 26.Sheng W. S., Hu S., Ni H. T., Rowen T. N., Lokensgard J. R., Peterson P. K. TNF-α-induced chemokine production and apoptosis in human neural precursor cells. Journal of Leukocyte Biology. 2005;78(6):1233–1241. doi: 10.1189/jlb.0405221. [DOI] [PubMed] [Google Scholar]
- 27.Weinberg M. S., Blake B. L., McCown T. J. Opposing actions of hippocampus TNFα receptors on limbic seizure susceptibility. Experimental Neurology. 2013;247:429–437. doi: 10.1016/j.expneurol.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tartaglia L. A., Pennica D., Goeddel D. V. Ligand passing: The 75-kDa tumor necrosis factor (TNF) receptor recruits TNF for signaling by the 55-kDa TNF receptor. Journal of Biological Chemistry. 1993;268(25):18542–18548. [PubMed] [Google Scholar]
- 29.Özbas-Gerçeker F., Gorter J. A., Redeker S., et al. Neurotrophin receptor immunoreactivity in the hippocampus of patients with mesial temporal lobe epilepsy. Neuropathology and Applied Neurobiology. 2004;30(6):651–664. doi: 10.1111/j.1365-2990.2004.00582.x. [DOI] [PubMed] [Google Scholar]
- 30.Wei X.-W., Yan H., Xu B., Wu Y.-P., Li C., Zhang G.-Y. Neuroprotection of co-activation of GABA receptors by preventing caspase-3 denitrosylation in KA-induced seizures. Brain Research Bulletin. 2012;88(6):617–623. doi: 10.1016/j.brainresbull.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 31.Xu S., Pang Q., Liu Y., Shang W., Zhai G., Ge M. Neuronal apoptosis in the resected sclerotic hippocampus in patients with mesial temporal lobe epilepsy. Journal of Clinical Neuroscience. 2007;14(9):835–840. doi: 10.1016/j.jocn.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 32.Henshall D. C., Bonislawski D. P., Skradski S. L., Lan J.-Q., Meller R., Simon R. P. Cleavage of bid may amplify caspase-8-induced neuronal death following focally evoked limbic seizures. Neurobiology of Disease. 2001;8(4):568–580. doi: 10.1006/nbdi.2001.0415. [DOI] [PubMed] [Google Scholar]
- 33.Tan Z., Levid J., Schreiber S. S. Increased expression of fas (CD95/APO-1) in adult rat brain after kainate-induced seizures. Neuroreport. 2001;12(9):1979–1982. doi: 10.1097/00001756-200107030-00040. [DOI] [PubMed] [Google Scholar]
- 34.Liu X.-M., Pei D.-S., Guan Q.-H., et al. Neuroprotection of Tat-GluR6-9c against neuronal death induced by kainate in rat hippocampus via nuclear and non-nuclear pathways. The Journal of Biological Chemistry. 2006;281(25):17432–17445. doi: 10.1074/jbc.m513490200. [DOI] [PubMed] [Google Scholar]
- 35.Pitti R. M., Marsters S. A., Lawrence D. A., et al. Genomic amplification of a decoy receptor for Fas ligand in lung and colon cancer. Nature. 1998;396(6712):699–703. doi: 10.1038/25387. [DOI] [PubMed] [Google Scholar]
- 36.Wiley S. R., Schooley K., Smolak P. J., et al. Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity. 1995;3(6):673–682. doi: 10.1016/1074-7613(95)90057-8. [DOI] [PubMed] [Google Scholar]
- 37.Nitsch R., Bechmann I., Deisz R. A., et al. Human brain-cell death induced by tumour-necrosis-factor-related apoptosis-inducing ligand (TRAIL) The Lancet. 2000;356(9232):827–828. doi: 10.1016/s0140-6736(00)02659-3. [DOI] [PubMed] [Google Scholar]
- 38.Pan G., O'Rourke K., Chinnaiyan A. M., et al. The receptor for the cytotoxic ligand TRAIL. Science. 1997;276(5309):111–113. doi: 10.1126/science.276.5309.111. [DOI] [PubMed] [Google Scholar]
- 39.Sheridan J. P., Marsters S. A., Pitti R. M., et al. Control of TRAIL-induced apoptosis by a family of signaling and decoy receptors. Science. 1997;277(5327):818–821. doi: 10.1126/science.277.5327.818. [DOI] [PubMed] [Google Scholar]
- 40.Pan G., Ni J., Wei Y.-F., Yu G.-I., Gentz R., Dixit V. M. An antagonist decoy receptor and a death domain-containing receptor for TRAIL. Science. 1997;277(5327):815–818. doi: 10.1126/science.277.5327.815. [DOI] [PubMed] [Google Scholar]
- 41.Dörr J., Bechmann I., Waiczies S., et al. Lack of tumor necrosis factor-related apoptosis-inducing ligand but presence of its receptors in the human brain. The Journal of Neuroscience. 2002;22(4, article RC209) doi: 10.1523/JNEUROSCI.22-04-j0001.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu Y., Zeng K., Han Y., et al. Altered expression of CX3CL1 in patients with epilepsy and in a rat model. The American Journal of Pathology. 2012;180(5):1950–1962. doi: 10.1016/j.ajpath.2012.01.024. [DOI] [PubMed] [Google Scholar]
- 43.Henshall D. C., Skradski S. L., Bonislawski D. P., Lan J.-Q., Simon R. P. Caspase-2 activation is redundant during seizure-induced neuronal death. Journal of Neurochemistry. 2001;77(3):886–895. doi: 10.1046/j.1471-4159.2001.00291.x. [DOI] [PubMed] [Google Scholar]
- 44.Henshall D. C., Skradski S. L., Meller R., et al. Expression and differential processing of caspases 6 and 7 in relation to specific epileptiform EEG patterns following limbic seizures. Neurobiology of Disease. 2002;10(2):71–87. doi: 10.1006/nbdi.2002.0505. [DOI] [PubMed] [Google Scholar]
- 45.Li T., Lu C., Xia Z., Xiao B., Luo Y. Inhibition of caspase-8 attenuates neuronal death induced by limbic seizures in a cytochrome c-dependent and Smac/DIABLO-independent way. Brain Research. 2006;1098(1):204–211. doi: 10.1016/j.brainres.2006.04.131. [DOI] [PubMed] [Google Scholar]
- 46.Yamamoto A., Murphy N., Schindler C. K., et al. Endoplasmic reticulum stress and apoptosis signaling in human temporal lobe epilepsy. Journal of Neuropathology and Experimental Neurology. 2006;65(3):217–225. doi: 10.1097/01.jnen.0000202886.22082.2a. [DOI] [PubMed] [Google Scholar]
- 47.Deveraux Q. L., Takahashi R., Salvesen G. S., Reed J. C. X-linked IAP is a direct inhibitor of cell-death proteases. Nature. 1997;388(6639):300–304. doi: 10.1038/40901. [DOI] [PubMed] [Google Scholar]
- 48.Korhonen L., Belluardo N., Lindholm D. Regulation of X-chromosome-linked inhibitor of apoptosis protein in kainic acid-induced death in the rat hippocampus. Molecular and Cellular Neuroscience. 2001;17(2):364–372. doi: 10.1006/mcne.2000.0935. [DOI] [PubMed] [Google Scholar]
- 49.Sakhi S., Bruce A., Sun N., Tocco G., Baudry M., Schreiber S. S. p53 Induction is associated with neuronal damage in the central nervous system. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(16):7525–7529. doi: 10.1073/pnas.91.16.7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Engel T., Murphy B. M., Schindler C. K., Henshall D. C. Elevated p53 and lower MDM2 expression in hippocampus from patients with intractable temporal lobe epilepsy. Epilepsy Research. 2007;77(2-3):151–156. doi: 10.1016/j.eplepsyres.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoffmann A., Baltimore D. Circuitry of nuclear factor κB signaling. Immunological Reviews. 2006;210:171–186. doi: 10.1111/j.0105-2896.2006.00375.x. [DOI] [PubMed] [Google Scholar]