Abstract
Investigating the cellular processes underlying tendon healing can allow researchers to improve long-term outcomes after injury. However, conducting meaningful studies to uncover the injury healing mechanism at cellular and molecular levels remains challenging. This is due to the inherent difficulty in isolating, culturing, and expanding sufficient primary tenocytes, due to their limited proliferative capacity and short lifespan. In this study, we sought to establish a novel line of immortalized mouse Achilles tenocytes (iMATs) with primary tenocyte properties, but increased proliferative capacity suitable for extensive in vitro experimentation. We show that isolated primary mouse Achilles tenocytes (pMATs) can be effectively immortalized using a piggyBac transposon expressing SV40 large T antigen flanked by FLP recombination target site (FRT). The resulting iMATs exhibit markedly greater proliferation and survival, which can be reversed with FLP recombinase. Furthermore, iMATs express the same set of tendon-specific markers as that of primary cells, although in lower levels, and respond similarly to exogenous stimulation with bone morphogenetic protein 13 (BMP13) as has been previously reported with pMATs. Taken together, our results suggest that iMATs acquire long-term proliferative capacity while maintaining tenogenic properties. We believe that iMATs are a suitable model for studying not only the native cellular processes involved in injury and healing, but also potential therapeutic agents that may augment the stability of tendon repair.
Introduction
Long-term, stable repair of tendon injuries remains a considerable challenge, especially in cases involving reinjury and/or reoperation.1 As such, gaining a better understanding of tendon repair mechanisms can have considerable implications for developing regenerative therapies and improving clinical outcomes. Researchers have established the mouse Achilles tendon as suitable in vivo and ex vivo mechanical model for simulating human injuries, but studying subsequent repair processes at the molecular level can be challenging due to the inherent hypocellularity of the tissue.2–7 Therefore, obtaining precise insight into injury response requires studying tendon cells (tenocytes) in vitro, where extracellular conditions are tightly controlled and gene expression levels can be accurately assessed at specific time points.8,9 In the past, mouse and rat tenocytes have been characterized by expression of type I and type III collagen, fibronectin 1, tenomodulin, scleraxis, and tenascin-C.10–13 A recent study by Lamplot et al. also found that bone morphogenetic protein 13 (BMP13) upregulates some of these genes and subsequently augments early tendon healing in a rat model.14
However, tenocytes are difficult to isolate from tissue and subsequently maintain in culture, proliferating slowly and often failing to survive for more than a few generations. Obtaining and expanding the cells necessary for conducting meaningful studies is often not feasible.11,15 Thus, there is a need to develop a suitable cell line with rapid and long-term proliferative potential while maintaining original tenogenic properties. One approach for creating such a model is by conditional or reversible immortalization of primary cells, often through the expression of an oncogene or inactivation of a tumor suppressor.16 One of the most commonly used oncogenes is the SV40 large T antigen (LTA), stimulating robust proliferation without altering native characteristics.17 In the past, a retroviral vector has been used to express the gene, but with low immortalization efficiency.18–21
The piggyBac transposon system has recently emerged as a promising nonviral alternative. Originally isolated from the cabbage looper moth Trichoplusiani, this transposon is capable of introducing transgenes into a mammalian host cell with great efficiency.21–23 The piggyBac system is comprised of a donor plasmid, carrying the gene of interest flanked by two terminal repeat domains (transposon), and a helper plasmid with the transposase that catalyzes movement of the genetic element.24 Our laboratory engineered the piggyBac-based immortalization vector MPH86 expressing SV40, flanked with a FLP recombinase recognition target (FRT) site for removal, and a hygromycin drug selection marker. We have also previously shown that this system is effective for mouse embryonic fibroblasts.20
In this study, we aim to use this vector to immortalized mouse Achilles tenocytes (iMATs), hoping to create a novel cell line suitable for extensive in vitro experimentation that is comparable to primary mouse Achilles tenocytes (pMATs) in both endogenous and induced gene expression. We also show that the process is reversible, demonstrating the considerable value of using these cells for important basic and translational studies involving tendon healing.
Materials and Methods
Cell culture and chemicals
Human HEK-293 cells and mouse NIH/3T3 cells were obtained from ATCC (Manassas, VA). Both lines were maintained in complete Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS; Invitrogen, Carlsbad, CA), 100 units/mL penicillin, and 100 μg/mL streptomycin at 37°C in 5% CO2. The recently engineered 293pTP line was used for adenovirus amplification.25 Unless indicated otherwise, all chemicals were purchased from either Sigma-Aldrich (St. Louis, MO) or Thermo Fisher Scientific (Pittsburgh, PA).
Isolation of tenocytes
The use and care of animals for this study was approved and overseen by the Institutional Animal Care and Use Committee. Tenocytes were isolated from the Achilles tendon of 4-week-old CD-1 mice. The Achilles tendon was first transected distal to the musculotendinous junction and then transected proximal to its bony insertion at the calcaneus. The tendon sheath and surrounding paratenon were stripped off. The tendons were minced into small pieces and digested in phosphate-buffered saline (PBS) with 1% penicillin–streptomycin solution with 4 mg/mL collagenase type I (Roche, Basel, Switzerland) for 2 h. pMATs were isolated through centrifugation at 1500 g for 5 min and then plated in six-well plates with complete DMEM (Cellgro, Manassas, VA) supplemented with 10% FBS (Sigma-Aldrich). Cells were incubated at 37°C, 95% humidified air, and 5% CO2. Colonies were visible at 3 days, and at 7 days, pMATs were passaged by enzymatic digestion (0.1% trypsin [Sigma-Aldrich]) into 25-cm2 flasks containing DMEM supplemented with 10% FBS. The passage numbers of cell lines used in assays are specified in the following sections of this article as “P#.”
Immortalization of tenocytes
Cells were immortalized as previously described.20,21,23 Briefly, early passage pMATs (P1) were seeded in 25-cm2 flasks and cotransfected with piggyBac vector MPH8620 and the piggyBac transposase expression vector, Super piggyBac (SBI, Mountain View, CA), as previously described.20,21,23,26 Stable iMAT cell pools were established by selecting the transfected cells with hygromycin B (Life Technologies, Grand Island, NY) for 1 week. Aliquots of the iMATs were kept in liquid nitrogen tanks.
Recombinant adenovirus construction and cell treatment
Recombinant adenoviruses were generated using the AdEasy technology as previously described.27 Briefly, the coding regions of human BMP13 and FLP recombinase were PCR amplified and subcloned into adenoviral shuttle vectors and subsequently used to generate recombinant adenoviruses in HEK293 cells, resulting in adenoviruses AdBMP13 and AdFLP, both of which also express green fluorescence protein (GFP) as a marker for monitoring infection efficiency. An analogous control adenovirus expressing GFP and RFP (AdGRFP) was previously constructed and used as a negative control.28–30 As necessary, recombinant adenoviruses were amplified using HEK-293pTP cells as described.25,27
Subconfluent cells (usually at 30% density, e.g., 8 × 104 cells/well for 24-well plates, or 1 × 106 cells per 25-cm2 flask) were treated with adenovirus and 4 μg/mL polybrene to improve infection efficiency.30 Equal infection efficiency of cells across viruses was achieved by dose titration and comparing the fluorescence signal at 24 h.
Cell proliferation WST-1 assay
Subconfluent iMATs (P3) and pMATs (P2) were plated at equal density (at 1.5 × 104 cells/well) into 96-well plates. Wells without cells were used as background controls. At the indicated time points, 4 μL of premixed WST-1 (BD Clontech, Mountain View, CA) was added to each well and incubated at 37°C for 1 h. After 2 min of vigorous mixing, an absorbance reading of each well was taken at 440 nm using a plate reader. The obtained A440 nm values were subjected to background reading subtractions. Each assay condition was done in triplicate.
Crystal violet cell proliferation assay
Subconfluent cells (iMAT P3, and pMAT P2) were seeded at equal density (1.5 × 105 cells/well) in 12-well plates and simultaneously infected with adenovirus (if applicable). For wells with adenovirus, media were changed at 24 h to prevent viral lysate toxicity. Cells were subjected to crystal violet staining for 30 min at the indicated time points. Each assay condition was done in triplicate. Pictures of representative wells were taken.
Trypan blue cell counting assay
Subconfluent iMATs (P3) and pMATs (P2) were seeded at equal density (1.5 × 105 cells/well) in 12-well plates, with three wells for each time point. Cells were visualized using light microscopy, trypsinized, and stained with Trypan Blue (Sigma-Aldrich). Unstained, viable cells were counted using a hemocytometer. Counts were averaged at each time point.
FACS analysis
After being seeded and maintained in 1% FBS serum for 24 h, subconfluent cells (iMAT P3, pMAT P2, at 3 × 105 cells/well in six-well plates) were harvested by forceful pipetting, washed with PBS, and stained with Hoechst 33342 (Life Technologies). Cell cycles were analyzed using the BD LSR II Flow Cytometer and the FlowJo software (FlowJo, Ashland, OR).
RNA isolation and quantitative polymerase chain reaction
After seeding (at 1 × 106 cells per 25-cm2 flask) and maintaining cells in 1% FBS serum for 24 h, total RNA was isolated using the TRIzol Reagent (Invitrogen; Life Technologies) and 5–10 μg total RNA for each sample were subjected to reverse transcription reaction with hexamer and M-MuLV Reverse Transcriptase (New England Biolabs, Ipswich, MA) as described.31 The cDNA products were diluted 10- to 50-fold and used as PCR templates. The integrity of the diluted cDNA products was confirmed using Bio-Rad RNA Quality Assays (RQ1 and RQ2) (Hercules, CA), as per assay manufacturer's guidelines, a cycle threshold difference of less than three between forward and reverse primer reactions was indicative of high-integrity RNA.31 Touchdown quantitative polymerase chain reaction (TqPCR) was carried out using mouse gene-specific primers (Table 1). Transcripts were designed by using Primer3 Plus program (www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi) to amplify the genes of interest. Reactions were carried out using the Bio-Rad CFX-Connect machine, with 2× SsoFast qPCR Supermix with EvaGreen (Bio-Rad) or iTaq Universal SYBR Green Supermix (Bio-Rad), in triplicate using the following conditions: 95°C × 3" for one cycle; 95°C × 20", 66°C × 10", for four cycles by decreasing 3°C per cycle; 95°C × 20", 55°C × 10", 70°C × 1", followed by a plate read, for 40 cycles. All samples were normalized to the expression level of GAPDH.
Table 1.
Primers Used for Touchdown qPCR (Mouse)
| Name | Sequence |
|---|---|
| GAPDH | CTACACTGAGGACCAGGTTGTCT |
| TTGTCATACCAGGAAATGAGCTT | |
| Col1a1 | GAGCGGAGAGTACTGGATCG |
| GCTTCTTTTCCTTGGGGTTC | |
| Col3a1 | GCACAGCAGTCCAACGTAGA |
| TCTCCAAATGGGATCTCTGG | |
| Fibronectin 1 | AATGGAAAAGGGGAATGGAC |
| CTCGGTTGTCCTTCTTGCTC | |
| Scleraxis | CTGGCCTCCAGCTACATTTCT |
| GTCACGGTCTTTGCTCAACTT | |
| Tenascin-C | CCATGAAGGGATTCGAAGAA |
| TCATGCAGCTCGTACTCCAC | |
| Tenomodulin | GCCACACCAGACAAGCAA |
| GGTTGCCTCGACGACAGT |
qPCR, quantitative polymerase chain reaction.
Statistical analysis
Excel (Microsoft, Redmond, WA) was used to calculate mean values and statistically significant differences between samples using the Student's t-test. A p-value of less than 0.05 was considered statistically significant. To test for normal distribution of our input data, we performed D'Agostino–Pearson omnibus normality tests. All quantitative data sets passed the normality tests.
Results
pMATs are effectively immortalized using SV40 LTA and demonstrate increased proliferative capacity
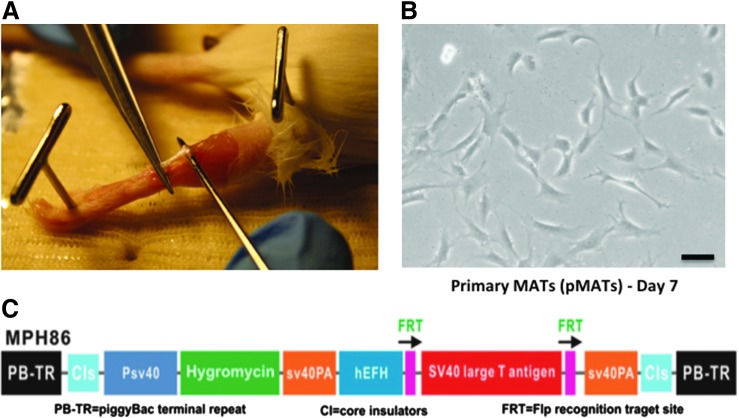
In this study, we isolated tenocytes from the Achilles tendons of 4-week-old CD1 mice (Fig. 1A). The isolated pMATs were able to proliferate in culture, although slowly, and the morphology of cells is shown at day 7 (Fig. 1B). To obtain an immortalized cell line, we utilized the piggyBac vector (pMPH86) that expresses SV40 LTA flanked with FLP recombinase (FLP) recognition sites (Fig. 1C). We previously used this system to successfully obtain reversibly immortalized MEFs (piMEFs).20 Even after multiple passages, iMATs maintained a similar morphological phenotype to the freshly isolated pMATs, indicating that the immortalization process did not significantly change the morphological phenotypes (Fig. 2A). However, after just a few passages, the pMATs were noted to have significant changes in morphology, including a flattened appearance and increased cytoplasmic-to-nuclear ratio, indicating poor health (Fig. 2A). In fact, pMATs did not survive in culture beyond three passages (roughly 2 weeks between passages). In contrast, iMATs have been passaged for more than 30 generations and retain original morphology, indicating that we successfully immortalized MATs.
FIG. 1.
Primary mouse Achilles tenocytes (pMATs) can be isolated from tissue and cultured in vitro. (A) Picture showing harvest of an Achilles tendon from a 4-week-old CD1 mouse. (B) Light microscopy image of pMATs in culture 7 days after isolation from tendons. Black bar denotes 100 μM (C) schematic representation of piggyBac immortalization vector MPH86 showing the SV40 large T antigen (LTA) expression cassette flanked by FLP recombination target (FRT) sites and hygromycin resistance gene. Color images available online at www.liebertpub.com/tec
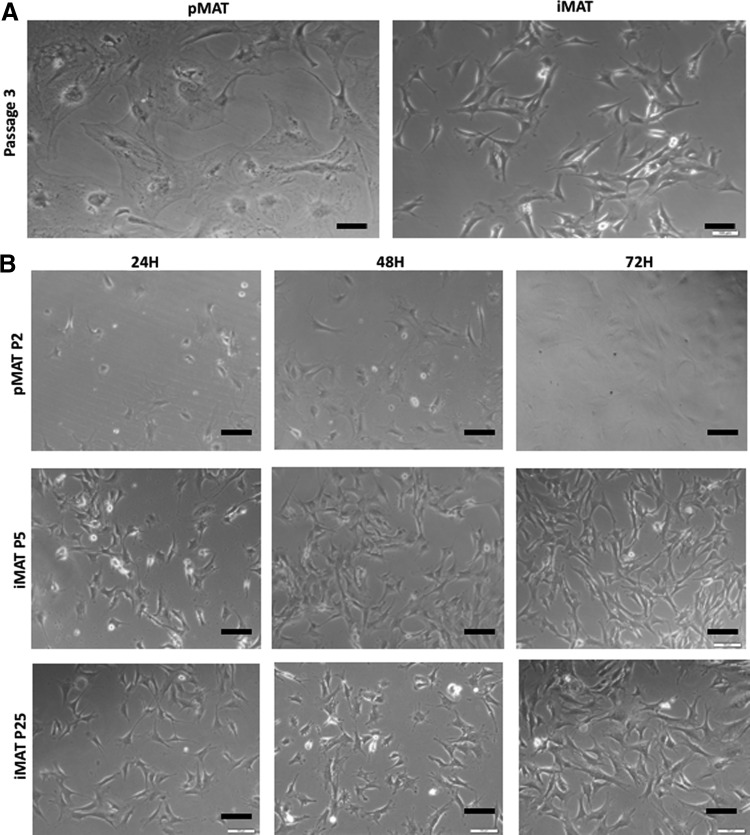
FIG. 2.
Light microscopy shows differences in morphology and proliferation between primary (pMAT) and immortalized (immortalized mouse Achilles tenocyte [iMAT]) cells [Black bars denote 100 μM]. (A) pMATs and iMATs demonstrating differences in morphology over several passages. pMATs appear unhealthy with increased cytoplasmic-to-nuclear ratio, whereas iMATs retain native morphology. (B) Increased proliferation of iMATs (early and late passages) compared to pMATs at 24-h intervals.
We next compared the proliferative activity of pMATs and iMATs. Using light microscopy, it was clear that iMATs proliferated much faster than pMATs, with differences in cell density apparent as early as 24 h (Fig. 2B). Furthermore, this increased proliferative capacity was preserved over numerous iMAT passages (Fig. 2B). Direct cell counting using Trypan Blue staining further showed that iMATs grew faster than pMATs (Fig. 3A). Likewise, iMATs exhibited a higher proliferation rate than pMATs in the quantitative WST-1 assay after pMATs and iMATs were seeded at the same low density (Fig. 3B). Crystal violet staining indicated that iMATs reached confluence by day 3 while pMATs failed to reach confluence over 7 days, when both started with a similar cell density (Fig. 3C). Finally, flow cytometry analysis indicated that the iMAT population had a greater percentage of cells in the S or G2 phases of the cell cycle (p = 0.0053), explaining the increased proliferation rates seen in the above assays (Fig. 3D). Taken together, these results demonstrate that immortalized MATs proliferate considerably faster than pMATs.
FIG. 3.
Assays show that iMATs demonstrate higher proliferative activity when compared to pMATs. (A) Trypan Blue cell counting performed in triplicate at each time point after seeding cells at equal density on day 0. (B) WST-1 cell proliferation assay performed in triplicate at each time point after seeding cells at equal density on day 0. Line of best fit overlaid for each cell type. (C) Crystal violet staining at multiple time points after seeding cells at equal density on day 0. Assay performed in triplicate and representative wells shown. (D) Flow cytometry analysis shows that iMATs have a significantly greater proportion of cells in the S and G2 phases of the cell cycle (n = 3, *p < 0.05). Color images available online at www.liebertpub.com/tec
FLP removal of large T antigen from iMATs significantly reduces cell survival and proliferation
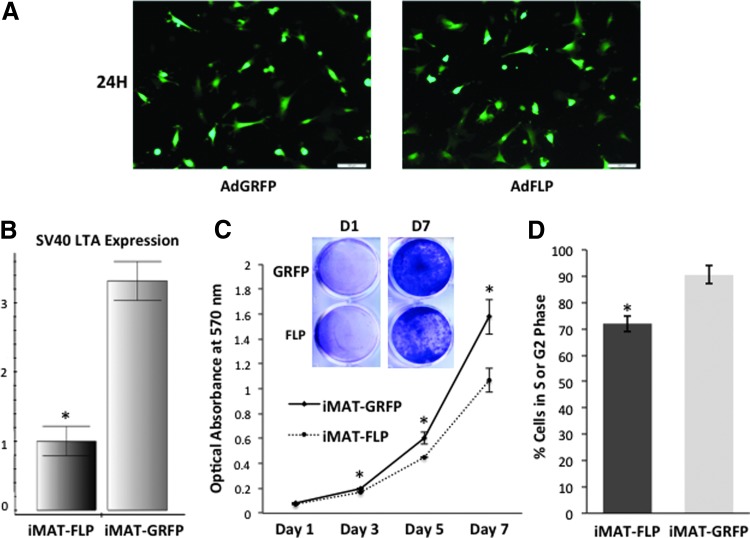
We tested if the immortalized phenotype of iMATs couldbe reversed by introducing FLP recombinase. To efficiently deliver FLP, we used a recombinant adenovirus that expresses the recombinase.20 We demonstrated effective and equal infection of iMATs using the adenoviral vectors AdFLP and AdGRFP (negative control) (Fig. 4A). When the RNA from the FLP-transduced iMAT cells was subjected to reverse transcription PCR and subsequent TqPCR analysis, the expression of SV40 T antigen was significantly reduced compared to control cells (p < 0.05) (Fig. 4B). Furthermore, we found that FLP-mediated removal of the SV40 LTA significantly reduced the survival and proliferation as assessed by crystal violet staining qualitatively and quantitatively (p < 0.05 at all time points after day 1) (Fig. 4C). Flow cytometry analysis of cells expressing GFP revealed that at 72 h, cells infected with FLP had a greater proportion of cells in the G1 phase (20.09% compared to 5.66% of GRFP) and fewer cells in the S/G2 phases (76.11% compared to 85.04% of GRFP), explaining the striking differences seen in cell proliferation activity (Fig. 4D). Thus, our results strongly suggest that the immortalization phenotype of the iMATs may be reversed by the introduction of FLP recombinase with notable effects on cell cycling.
FIG. 4.
Removal of SV40 T antigen reduces the proliferative activity of iMTAs. (A) Fluorescence microscopy images showing equal transfection of iMATs with AdFLP and AdGRFP (negative control) after 24 h. (B) Quantitative polymerase chain reaction (qPCR) analysis demonstrates significantly lower expression of SV40 LTA in iMAT population infected with AdFLP compared to that of AdGRFP (n = 3, *p < 0.05). (C) Crystal violet staining at multiple time points after seeding cells at equal density on day 0. The assay was performed in triplicate and representative wells from day 1 to 7 are shown. The stain was dissolved and quantitatively measured for absorbance at 570 nm. Cell viability difference in iMATs infected with AdFLP versus AdGRFP was statistically significant at day 3, 5, and 7 (*p < 0.05). (D) Green fluorescence protein (GFP)-gated cell cycle analysis shows a greater proportion of iMAT-GRFP cells in the S or G2 phases compared to iMAT-FLP cells, indicating successful removal of large T-antigen in the latter group. Color images available online at www.liebertpub.com/tec
It is noteworthy that FLP recombinase exhibited a varied efficiency in removing the SV40 LTA from iMATs. This was evidenced by the presence of quantifiable FLP cDNA in transduced cells, while little to no change in SV40 T antigen expression (although we did not know the stability of the SV40 T transcript [data not shown]). Nonetheless, we observed that removal of SV40 T antigen in the FLP well-expressed subpopulation resulted in rapid cell death. This was seen with a sharp decline in the number of green fluorescent cells in the FLP group compared to the GRFP group over the first few days after infection (data not shown). We addressed this issue by isolating RNA promptly after FLP transfection, at around 24 h, when SV40 T antigen expression would have been decreased, but cell death was yet to occur.
iMATs express tenocyte markers
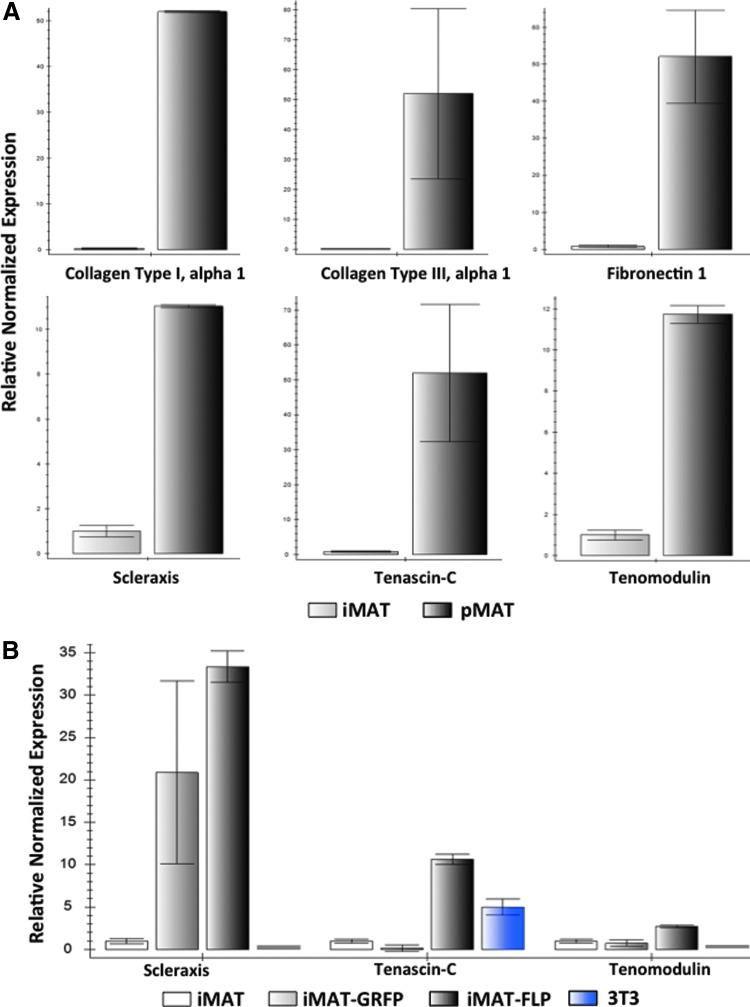
We compared the expression of tenogenic markers in pMATs (P2) and iMATs (P5). While no single marker can be used to identify tenocytes, some of the reported genes include collagen type I, collagen type III, fibronectin 1, scleraxis, tenascin-C, and tenomodulin. We found that all of these markers were readily detectable in iMATs by TqPCR analysis, but expression levels were lower than those observed in pMATs (Fig. 5A). Furthermore, we compared the expression levels of these same markers between early and late iMAT passages, with no significant differences seen (data not shown). Compared to control 3T3 fibroblast cells, iMATs showed higher expression of collagen type III, scleraxis, and tenomodulin, all of which are highly specific to tendon cells (Fig. 5B). Thus, these results demonstrate that iMATs express all of the common tenocyte markers, although at lower levels, suggesting that they may retain tenogenic properties.
FIG. 5.
qPCR analysis demonstrates that iMAT cells express the same tendon-specific markers as pMATs. (A) Endogenous gene expression in iMATs and pMATs. RNA was isolated from primary and immortalized MAT cells and subjected to reverse transcription. Touchdown qPCR (TqPCR) shows that iMATs express the same tendon-specific genes as pMATs, although in lower levels due to the less differentiated nature of immortalized cells. These genes include collagen type I alpha 1, collagen type III alpha 1, fibronectin 1, scleraxis, tenascin-C, and tenomodulin. iMAT expression level is set to 1 in all plots, and error bars do not cross zero. (B) Changes in iMAT endogenous gene expression after infection with FLP recombinase and GRFP. Twenty-four hours after infection with adenovirus, RNA was isolated from iMATs, subjected to reverse transcription, and analyzed using TqPCR. When immortalization is reversed using FLP, expression of tendon-specific markers, scleraxis, tenascin-C, and tenomodulin, is partially reverted to higher levels when compared to AdGRFP infection. NIH/3T3 cells were used as a control group, demonstrating that iMAT cells are closer to pMAT cells in identity based on expression levels of these tendon-specific genes. Color images available online at www.liebertpub.com/tec
To demonstrate that these relative differences in endogenous gene expression were transient, we performed TqPCR analysis on cells before and after FLP recombinase transduction. As expected, we found that iMATs (P7) treated with AdFLP demonstrated increased expression levels of tenascin-C and tenomodulin compared to both untreated and AdGRFP-treated iMATs (Fig. 5B). A similar trend was also observed for both collagen type I and collagen type III, although the differences between FLP- and GRFP-infected cells were less apparent (data not shown). Overall, it appears that changes in native gene expression levels caused by the immortalization process can be partially reversed.
iMATs respond to exogenous stimulation as expected
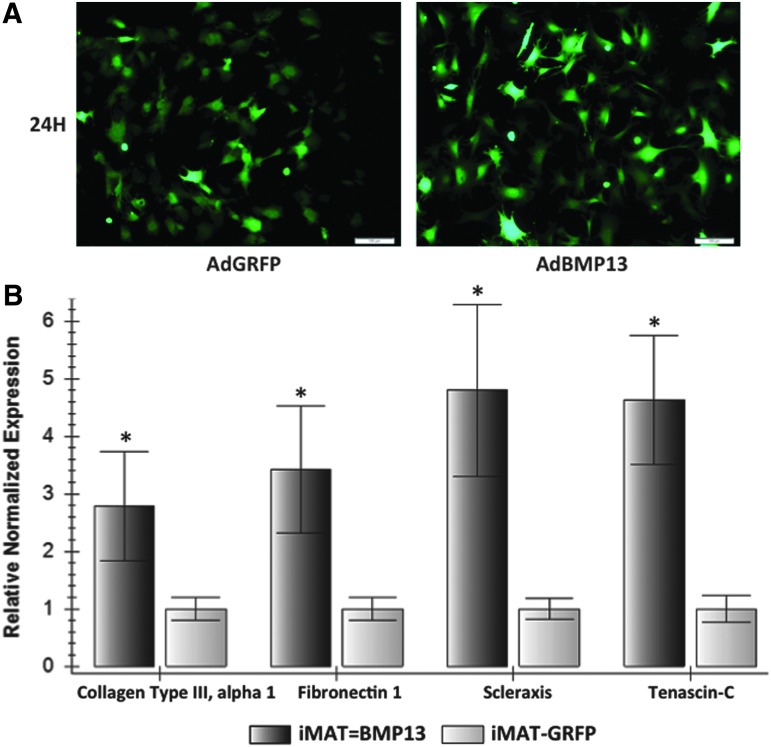
To further characterize the tenogenic properties of the immortalized cells, we assessed whether iMATs respond to exogenous stimulation as has been previously reported using pMATs.14 We infected iMATs (P8) with AdBMP13 and analyzed the resulting changes in gene expression for collagen type III, fibronectin 1, scleraxis, and tenascin-C. When compared to a GRFP control with equal infection efficiency, AdBMP13 upregulated the expression of collagen type III (p = 0.0004), fibronectin 1 (p = 0.0003), scleraxis (p = 0.0006), and tenascin-C (p = 0.0001) (Fig. 6). In contrast, 3T3 fibroblast cells stimulated with BMP13 did not result in a similar upregulation of all four aforementioned genes (data not shown). This indicates that iMATs respond in a similar manner to exogenous stimulation as pMATs, reinforcing that the immortalized cells retain native tendon cell characteristics.
FIG. 6.
Bone morphogenetic protein 13 (BMP13) stimulation of iMATs upregulates the expression of genes critical to early tendon healing as previously described with pMATs. (A) iMATs showing equal AdBMP13 and AdGRFP infection efficiency after 24 h. (B) Changes in gene expression after BMP13 stimulation. At 72 h after infection with AdBMP13 or AdGRFP, RNA was isolated from iMATs, subjected to reverse transcription, and analyzed using TqPCR. Collagen type III alpha 1, fibronectin 1, scleraxis, and tenascin-C were all upregulated in the BMP13 group compared to GRFP control (n = 3, *p < 0.001), as has been previously observed with primary MATs.14 Color images available online at www.liebertpub.com/tec
Discussion
Tenocytes have been valuable to study in culture, but with significant experimental limitations due to their slow growth, short lifespan, and difficulty being isolated from tissue.11 Developing a novel tenocyte model with increased proliferative capacity can help researchers gain more insight into the molecular underpinnings of tendon healing. Such a cell line would not only have vast implications for investigating native cellular processes occurring in response to injury, but also for testing novel therapeutics with clinical applications. Therefore, in this study, we sought to iMATs using the SV40 LTA and piggyBac expression system. We succeeded in not only creating a cell line with robust growth in culture over numerous generations, but also demonstrated that this phenotype is reversible through the use of FLP recombinase. Finally, gene expression analysis has shown that immortalized tenocytes express the same tenogenic markers as primary cells, although in lower levels, and demonstrate a similar response to exogenous stimulation.
We have previously reported that the piggyBac system is an attractive option for carrying out immortalization when compared to other protocols, such as retroviral vectors.20 To summarize, this transposon system allows for packaging of larger cargo up to 100 kb, higher efficiency using liposome-based transfection, delivery of multiple copies of the vector resulting in high transgene expression levels, and greater integration specificity for regions surrounding transcriptional start sites (i.e., integration is less random).21,22,32 Finally, the immortalization process has the potential to be completely reversible and leave behind no footprints.33,34
Nonetheless, we experienced certain technical dilemmas using FLP recombinase (FLP) to effectively reverse the immortalization phenotype. On one hand, we found that FLP appears to have low efficiency in removing the SV40 LTA from the overall populations of iMATs, as demonstrated by qPCR analysis confirming the presence of FLP cDNA, but a negligible change in SV40 expression. On the other hand, the immortalized cells appear to be so dependent on SV40 T antigen expression that removal of T antigen resulted in rather rapid cell death, which may also account for our inability to demonstrate the reversibility of the immortalized cells. It has been well documented that either FLP and/or Cre recombinase exhibit significantly variable excision efficiency, which may be largely dependent on their genomic integration sites and accessibility of the recombinases. We are currently developing a comprehensive strategy to accomplish more effective reversibility by using adenoviral vectors to express siRNAs that silence SV40 T antigen expression26 and/or an integration-defective mutant piggyBac transposase,34 in addition to the use of FLP recombinase. It is conceivable that this combinatorial approach would enable us to reverse the immortalization phenotype with high efficiency.
Despite the current technical limitation, our novel cell line remains a promising model for studying tenocytes in culture. We were able to definitively show that, even in the presence of SV40 T antigen expression, iMATs express the same set of tendon-specific markers as primary cells, although at lower levels. This may be expected since immortalized cells exhibit high proliferative activity and are less differentiated when compared to the slow-growing, terminally differentiated primary cells.35 Nonetheless, we demonstrated that FLP recombinase-based removal of SV40 did partially revert gene expression levels to those observed in pMATs. Furthermore, we have shown that BMP13 stimulation of iMATs results in upregulation of the same genes involved in early tendon healing as reported in another work that studied primary tenocytes.14 This is critically important, since the appeal of working with such a model is being able to equivocate any results to those expected of native cells.
In conclusion, we have demonstrated that reversibly iMATs show long-term proliferative capacity while retaining native tenogenic properties. Additionally, FLP recombinase may potentially serve as a tool to reverse the immortalization phenotype. iMATs express tenocyte-specific markers and further show upregulation of tendon-healing makers when stimulated with BMP13, as has been previously shown using primary tenocytes. Taken together, these findings support the use of this novel cell line for studying the normal function and characteristics of tenocytes, molecular mechanisms underlying tendon injury and healing, and the use of biofactors or synthetic agents to augment tendon repair.
Acknowledgments
The reported work was supported in part by research grants from the National Institutes of Health (AT004418, AR50142 to T.C.H. and R.C.H.), and the 973 Program of Ministry of Science and Technology (MOST) of China (#2011CB707900 to T.C.H.). S.K.D. was a recipient of the Pritzker Fellowship and Alpha Omega Alpha Carolyn L. Kuckein Fellowship. M.K.M. was a recipient of the Howard Hughes Medical Institute Medical Research Fellowship. This work was also supported in part by The University of Chicago Core Facility Subsidy grant from the National Center for Advancing Translational Sciences (NCATS) of the National Institutes of Health through Grant UL1 TR000430. Funding sources were not involved in the study design; in the collection, analysis, and interpretation of data; in the writing of the report; and in the decision to submit the article for publication.
Disclosure Statement
No competing financial interests exist.
References
- 1.Yang G., Rothrauff B.B., and Tuan R.S. Tendon and ligament regeneration and repair: clinical relevance and developmental paradigm. Birth Defects Res Part C 99, 203, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ansorge H.L., Adams S., Birk D.E., and Soslowsky L.J. Mechanical, compositional, and structural properties of the post-natal mouse Achilles tendon. Ann Biomed Eng 39, 1904, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Riggin C.N., Sarver J.J., Freedman B.R., Thomas S.J., and Soslowsky L.J. Analysis of collagen organization in mouse achilles tendon using high-frequency ultrasound imaging. J Biomech Eng 136, 021029, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gordon J.A., Freedman B.R., Zuskov A., Iozzo R.V., Birk D.E., and Soslowsky L.J. Achilles tendons from decorin- and biglycan-null mouse models have inferior mechanical and structural properties predicted by an image-based empirical damage model. J Biomech 48, 2110, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wood L.K., and Brooks S.V. Ten weeks of treadmill running decreases stiffness and increases collagen turnover in tendons of old mice. J Orthop Res 34, 346, 2016 [DOI] [PubMed] [Google Scholar]
- 6.De Aro A.A., Guerra Fda, R., Esquisatto M.A., Nakagaki W.R., Gomes L., and Pimentel E.R. Biochemical and morphological alterations in the Achilles tendon of mdx mice. Microsc Res Tech 78, 85, 2015 [DOI] [PubMed] [Google Scholar]
- 7.Sharma P., and Maffulli N. Biology of tendon injury: healing, modeling and remodeling. J Musculoskelet Neuronal Interact 6, 181, 2006 [PubMed] [Google Scholar]
- 8.Wang X.T., Liu P.Y., and Tang J.B. Tendon healing in vitro: genetic modification of tenocytes with exogenous PDGF gene and promotion of collagen gene expression. J Hand Surg Am 29, 884, 2004 [DOI] [PubMed] [Google Scholar]
- 9.Zhang J., and Wang J.H. The effects of mechanical loading on tendons—an in vivo and in vitro model study. PLoS One 8, e71740, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Güngörmüş C., and Kolankaya D. Characterization of type I, III and V collagens in high-density cultured tenocytes by triple-immunofluorescence technique. Cytotechnology 58, 145, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shimada A., Wada S., Inoue K., Ideno H., Kamiunten T., Komatsu K., Kudo A., Nakamura Y., Sato T., Nakashima K., and Nifuji A. Efficient expansion of mouse primary tenocytes using a novel collagen gel culture method. Histochem Cell Biol 142, 205, 2014 [DOI] [PubMed] [Google Scholar]
- 12.Shukunami C., Takimoto A., Oro M., and Hiraki Y. Scleraxis positively regulates the expression of tenomodulin, a differentiation marker of tenocytes. Dev Biol 298, 234, 2006 [DOI] [PubMed] [Google Scholar]
- 13.Gungormus C., and Kolankaya D. Gene expression of tendon collagens and tenocyte markers in long-term monolayer and high-density cultures of rat tenocytes. Connect Tissue Res 53, 485, 2012 [DOI] [PubMed] [Google Scholar]
- 14.Lamplot J.D., Angeline M., Angeles J., Beederman M., Wagner E., Rastegar F., Scott B., Skjong C., Mass D., Kang R., Ho S., and Shi L.L. Distinct effects of platelet-rich plasma and BMP13 on rotator cuff tendon injury healing in a rat model. Am J Sports Med 42, 2877, 2014 [DOI] [PubMed] [Google Scholar]
- 15.Maeda T., Sakabe T., Sunaga A., Sakai K., Rivera A.L., Keene D.R., Sasaki T., Stavnezer E., Iannotti J., Schweitzer R., Ilic D., Baskaran H., and Sakai T. Conversion of mechanical force into TGF-beta-mediated biochemical signals. Curr Biol 21, 933, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.May T., Mueller P.P., Weich H., Froese N., Deutsch U., Wirth D., Kröger A., and Hauser H. Establishment of murine cell lines by constitutive and conditional immortalization. J Biotechnol 120, 99, 2005 [DOI] [PubMed] [Google Scholar]
- 17.Ahuja D., Saenz-Robles M.T., and Pipas J.M. SV40 large T antigen targets multiple cellular pathways to elicit cellular transformation. Oncogene 24, 7729, 2005 [DOI] [PubMed] [Google Scholar]
- 18.Lamplot J.D., Liu B., Yin L., Zhang W., Wang Z., Luther G., Wagner E., Li R., Nan G., Shui W., Yan Z., Rames R., Deng F., Zhang H., Liao Z., Liu W., Zhang J., Zhang Z., Zhang Q., Ye J., Deng Y., Qiao M., Haydon R.C., Luu H.H., Angeles J., Shi L.L., He T.C., and Ho S.H. Reversibly immortalized mouse articular chondrocytes acquire long-term proliferative capability while retaining chondrogenic phenotype. Cell Transplant 24, 1053, 2015 [DOI] [PubMed] [Google Scholar]
- 19.Huang E., Bi Y., Jiang W., Luo X., Yang K., Gao J.-L., Gao Y., Luo Q., Shi Q., Kim S.H., Liu X., Li M., Hu N., Liu H., Cui J., Zhang W., Li R., Chen X., Shen J., Kong Y., Zhang J., Wang J., Luo J., He B.-C., Wang H., Reid R.R., Luu H.H., Haydon R.C., Yang L., and He T.-C. Conditionally immortalized mouse embryonic fibroblasts retain proliferative activity without compromising multipotent differentiation potential. PLoS One 7, e32428, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang N., Zhang W., Cui J., Zhang H., Chen X., Li R., Wu N., Chen X., Wen S., Zhang J., Yin L., Deng F., Liao Z., Zhang Z., Zhang Q., Yan Z., Liu W., Ye J., Deng Y., Wang Z., Qiao M., Luu H.H., Haydon R.C., Shi L.L., Liang H., and He T.C. The piggyBac transposon-mediated expression of SV40 T antigen efficiently immortalizes mouse embryonic fibroblasts (MEFs). PLoS One 9, e97316, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X., Cui J., Yan Z., Zhang H., Chen X., Wang N., Shah P., Deng F., Zhao C., Geng N., Li M., Denduluri S.K., Haydon R.C., Luu H.H., Reid R.R., and He T.-C. Sustained high level transgene expression in mammalian cells mediated by the optimized piggyBac transposon system. Genes Dis 2, 96, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson M.H., Coates C.J., and George A.L. PiggyBac transposon-mediated gene transfer in human cells. Mol Ther 15, 139, 2007 [DOI] [PubMed] [Google Scholar]
- 23.Wen S., Zhang H., Li Y., Wang N., Zhang W., Yang K., Wu N., Chen X., Deng F., Liao Z., Zhang J., Zhang Q., Yan Z., Liu W., Zhang Z., Ye J., Deng Y., Zhou G., Luu H.H., Haydon R.C., Shi L.L., He T.C., and Wei G. Characterization of constitutive promoters for piggyBac transposon-mediated stable transgene expression in mesenchymal stem cells (MSCs). PLoS One 9, e94397, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding S., Wu X., Li G., Han M., Zhuang Y., and Xu T. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell 122, 473, 2005 [DOI] [PubMed] [Google Scholar]
- 25.Wu N., Zhang H., Deng F., Li R., Zhang W., Chen X., Wen S., Wang N., Zhang J., Yin L., Liao Z., Zhang Z., Zhang Q., Yan Z., Liu W., Wu D., Ye J., Deng Y., Yang K., Luu H.H., Haydon R.C., and He T.C. Overexpression of Ad5 precursor terminal protein accelerates recombinant adenovirus packaging and amplification in HEK-293 packaging cells. Gene Ther 21, 629, 2014 [DOI] [PubMed] [Google Scholar]
- 26.Deng F., Chen X., Liao Z., Yan Z., Wang Z., Deng Y., Zhang Q., Zhang Z., Ye J., Qiao M., Li R., Denduluri S., Wang J., Wei Q., Li M., Geng N., Zhao L., Zhou G., Zhang P., Luu H.H., Haydon R.C., Reid R.R., Yang T., and He T.C. A simplified and versatile system for the simultaneous expression of multiple siRNAs in mammalian cells using Gibson DNA assembly. PLoS One 9, e113064, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo J., Deng Z.L., Luo X., Tang N., Song W.X., Chen J., Sharff K.A., Luu H.H., Haydon R.C., Kinzler K.W., Vogelstein B., and He T.C. A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nat Protoc 2, 1236, 2007 [DOI] [PubMed] [Google Scholar]
- 28.Wang N., Zhang H., Zhang B.Q., Liu W., Zhang Z., Qiao M., Zhang H., Deng F., Wu N., Chen X., Wen S., Zhang J., Liao Z., Zhang Q., Yan Z., Yin L., Ye J., Deng Y., Luu H.H., Haydon R.C., Liang H., and He T.C. Adenovirus-mediated efficient gene transfer into cultured three-dimensional organoids. PLoS One 9, e93608, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H., Wang J., Deng F., Huang E., Yan Z., Wang Z., Deng Y., Zhang Q., Zhang Z., Ye J., Qiao M., Li R., Wang J., Wei Q., Zhou G., Luu H.H., Haydon R.C., He T.C., and Deng F. Canonical Wnt signaling acts synergistically on BMP9-induced osteo/odontoblastic differentiation of stem cells of dental apical papilla (SCAPs). Biomaterials 39, 145, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao C., Wu N., Deng F., Zhang H., Wang N., Zhang W., Chen X., Wen S., Zhang J., Yin L., Liao Z., Zhang Z., Zhang Q., Yan Z., Liu W., Wu D., Ye J., Deng Y., Zhou G., Luu H.H., Haydon R.C., Si W., and He T.C. Adenovirus-mediated gene transfer in mesenchymal stem cells can be significantly enhanced by the cationic polymer polybrene. PLoS One 9, e92908, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laboratories B.-R. PrimePCRTM Assays, Panels, and Controls for Real-Time PCR. Instruction Manual 1, 1, 2014 [Google Scholar]
- 32.Kim A., and Pyykko I. Size matters: versatile use of PiggyBac transposons as a genetic manipulation tool. Mol Cell Biochem 354, 301, 2011 [DOI] [PubMed] [Google Scholar]
- 33.Di Matteo, M., Matrai J., Belay E., Firdissa T., Vandendriessche T., and Chuah M.K. PiggyBac toolbox. Methods Mol Biol 859, 241, 2012 [DOI] [PubMed] [Google Scholar]
- 34.Li X., Burnight E.R., Cooney A.L., Malani N., Brady T., Sander J.D., Staber J., Wheelan S.J., Joung J.K., McCray P.B., Bushman F.D., Sinn P.L., and Craig N.L. piggyBac transposase tools for genome engineering. Proc Natl Acad Sci U S A 110, E2279, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hirofumi N., and Naoya K. Controlled Expansion of Mammalian Cell Populations by Reversible Immortalization. J Biotechnol Biomat 3, 1, 2013 [Google Scholar]