In this study from Arnes et al., the authors provide novel insight into the role of a long noncoding RNA in pancreas development and development of diabetes. They demonstrate that βlinc1 is necessary for the specification and function of insulin-producing cells through the coordinated regulation of a number of islet-specific transcription factors, and deletion of βlinc1 results in defective islet development and disruption of glucose homeostasis in the adult.
Keywords: β cell, long noncoding RNA, pancreas development
Abstract
Pancreatic β cells are responsible for maintaining glucose homeostasis; their absence or malfunction results in diabetes mellitus. Although there is evidence that long noncoding RNAs (lncRNAs) play important roles in development and disease, none have been investigated in vivo in the context of pancreas development. In this study, we demonstrate that βlinc1 (β-cell long intergenic noncoding RNA 1), a conserved lncRNA, is necessary for the specification and function of insulin-producing β cells through the coordinated regulation of a number of islet-specific transcription factors located in the genomic vicinity of βlinc1. Furthermore, deletion of βlinc1 results in defective islet development and disruption of glucose homeostasis in adult mice.
Type 2 diabetes (T2D) arises when pancreatic islets are no longer able to compensate for the increasing demand of insulin to maintain glucose homeostatic conditions. Despite the improvement in current therapies, >300 million people live with T2D, and those numbers are expected to almost double in the next 20 years. Accordingly, it is necessary to increase our understanding of the underlying biology of islet dysfunctions associated with T2D and identify novel druggable targets.
Studies in pancreas development have identified numerous transcription factors and lineage-specific regulatory networks that are required for the specification of the different pancreatic cell types, including acini, ducts, and endocrine cells (Arda et al. 2013). This functionally diverse group of cells arises from a common pool of pancreatic progenitors in two major waves of differentiation termed the primary and secondary transitions. Differentiation of the pancreatic β cells occurs during the secondary transition, and the signaling pathways and regulatory factors involved in this important developmental process have been well characterized (for review, see Pan and Wright 2011). These studies have provided the platform to generate functional β cells in vitro for regenerative medicine and suggested novel therapeutic approaches to prevent and treat β-cell dysfunction.
The study of β-cell biology has been recently complemented by genome-wide transcriptome analysis and characterization of epigenetic modifications, suggesting that non-protein-coding regions of the genome are integral to the transcriptional network regulating development and function of β cells (for review, see Arnes and Sussel 2015). Furthermore, a large number of T2D-associated common variants that were identified in genome-wide association studies map to noncoding genomic regions (Pasquali et al. 2014). Although there is increasing evidence that long noncoding RNAs (lncRNAs) play an important role in development and disease (Batista and Chang 2013), none have been investigated in vivo in the context of pancreas development and β-cell function. In this study, we describe the functional characterization of a newly identified conserved lncRNA termed βlinc1 (β-cell long intergenic noncoding RNA 1). We demonstrated that βlinc1 is required for the proper specification and function of endocrine cells through the coordinated regulation of a number of islet-specific transcription factors located in the genomic vicinity of βlinc1. Furthermore, deletion of βlinc1 results in defective islet development and disrupted glucose homeostasis in the adult. These results have important implications for the identification of novel regulatory mechanisms underlying T2D susceptibility and suggest that ncRNAs could represent novel therapeutic targets for the treatment of diabetes.
Results and Discussion
Characterization of βLINC1
lncRNAs expressed in the pancreatic islet are often highly tissue-specific, associated with clusters of open chromatin, and located in the genomic vicinity of transcription factors involved in β-cell development and/or function (Ku et al. 2012; Moran et al. 2012). We analyzed βLINC1 (formally HI-LNC15), a 6.8-kb post-transcriptionally processed human islet-specific transcript without coding potential (Moran et al. 2012) that is located in a region of open chromatin ∼20 kb upstream of NKX2.2, an essential islet homeobox transcription factor gene (Supplemental Fig. 1; Sussel et al. 1998). βLINC1 resides in a large syntenic block located on chromosome 20 in humans and chromosome 2 in mice (Fig. 1A). It is conserved in mammals, with the largest stretches of homology located at the putative promoter region, similar to many characterized lncRNAs (Carninci et al. 2005). This high level of conservation enabled us to identify an orthologous βlinc1 transcript in mice (Fig. 1B). Mouse βlinc1 is also predicted to have no coding potential (CPC score −0.261; CPAT 0.055) (Supplemental Fig. 2a), and comparative sequence analysis between the mouse and human βlinc1 transcripts did not reveal any conserved small ORFs. In mouse islets, the genomic locus surrounding βlinc1 is enriched in H3K4me1/3 and H3K27ac marks (Supplemental Fig. 2b), and there is evidence for NeuroD1, Pdx1, and Foxa2 binding at the putative βlinc1 promoter region (Khoo et al. 2012; Jia et al. 2015). Despite these features, this 4.2-kb region of DNA within the βlinc1 locus did not confer enhancer activity in luciferase reporter assays in MIN6 cells (Supplemental Fig. 3).
Figure 1.

βlinc1 is a conserved endocrine-specific lncRNA. (A) βlinc1 is located in a large syntenic block on human chromosome 20 and mouse chromosome 2 (purple lines). The position and direction of βlinc1 and the nearest adjacent genes are indicated. (B) Snapshot depicting βlinc1 transcript structure generated by de novo assembly of RNA sequencing (RNA-seq) data from mouse embryonic day 14.5 (E14.5) pancreas and islet samples and 30-way Multiz Alignment and Conservation. The mouse βlinc1 locus spans 8 kb, located in a gene desert between Nkx2.2 and Pax1 on the long (q) arm of chromosome 2 (chr2: 147,030,314–147,038,352, mm9), with a 73.6% sequence conservation with the human βLINC1 locus as determined by LiftOver. (C) βlinc1 RNA expression was determined by quantitative RT–PCR (qRT–PCR) in a tissue panel isolated from E15.5 embryos and adult islets. (D) RNA in situ hybridization of βlinc1 in pancreatic sections of E18.5 embryos and adult pancreata showing enrichment of the βlinc1 transcript in the trunk endocrine compartment and adult islets. White dotted lines depict the endocrine area and islets. The image is representative of at least three experiments. (E) Cellular fractionation of MIN6 cells showing that βlinc1 is highly enriched in nuclear versus cytosolic fractions. Gapdh and Malat1 were included as negative and positive controls of nuclear transcript retention, respectively. Samples without the addition of reverse transcriptase (noRT) were included to control for genomic contamination. n = 4. (F) βlinc1 and Nkx2.2 expression in MIN6 cells treated with two different siRNAs against βlinc1. n = 4. Error bars represent ±SEM. (*) P < 0.05, Student's t-test.
Expression studies revealed that βlinc1 is enriched in embryonic pancreata and adult islets (Fig. 1C). RNA in situ analysis confirmed that βlinc1 expression is restricted to adult islets and the trunk region of the developing pancreas (Fig. 1D). Furthermore, assessment of βlinc1 expression in several islet cell lines demonstrated that βlinc1 is enriched in insulin-producing cells (Supplemental Fig. 4a), corresponding to its expression in FACS-purified human β cells (Supplemental Fig. 1).
βlinc1 RNA is retained in the nuclear fraction of β cells, further suggesting a role for βlinc1 in transcriptional regulation (Fig. 1E). Although βlinc1 is expressed at relatively low levels, the half-life of the βlinc1 transcript is equivalent to that of Tbp (Supplemental Fig. 4b,c). This suggests that low transcript levels are not due to the degradation of aberrant transcripts, similar to what has been documented for other low-expressing lncRNAs (Clark et al. 2012).
Although there are limited tools available to predict the function of lncRNAs based on nucleotide sequence or genomic location, there is growing evidence that a subset of nuclear lncRNAs functions locally to regulate neighboring genes (Sauvageau et al. 2013; Vance et al. 2014). Consistently, siRNA-mediated knockdown of βlinc1 RNA in MIN6 cells resulted in the down-regulation of the adjacent coding gene Nkx2.2 (Fig. 1F), suggesting that the βlinc1 transcript positively regulates the expression of Nkx2.2.
βlinc1 knockout mice are glucose-intolerant
To determine the in vivo function of βlinc1, we generated βlinc1-null mice (Supplemental Fig. 5). βlinc1+/− and βlinc1−/− mice were viable and fertile. Consistent with the expression of βlinc1 being restricted to the developing endocrine pancreas and adult islet, βlinc1−/− mice were mildly glucose-intolerant compared with βlinc1+/− or βlinc1+/+ littermate controls (Fig. 2A). Furthermore, βlinc1−/− mice displayed abnormal fasting plasma insulin levels and increased insulin secretion under low-glucose conditions and failed to elevate circulating insulin in response to glucose stimulation (Fig. 2B; Supplemental Fig. 6a). Interestingly, βlinc1−/− mice were also mildly insulin-resistant despite the absence of βlinc1 expression in muscle and subcutaneous white adipose tissue (Supplemental Fig. 6b,c). We did not detect additional phenotypes in βlinc1−/− mice, particularly in other tissues where Nkx2.2 is expressed, such as the brain and intestine, which is also consistent with the restricted expression and function of the βlinc1 transcript to the endocrine pancreas.
Figure 2.
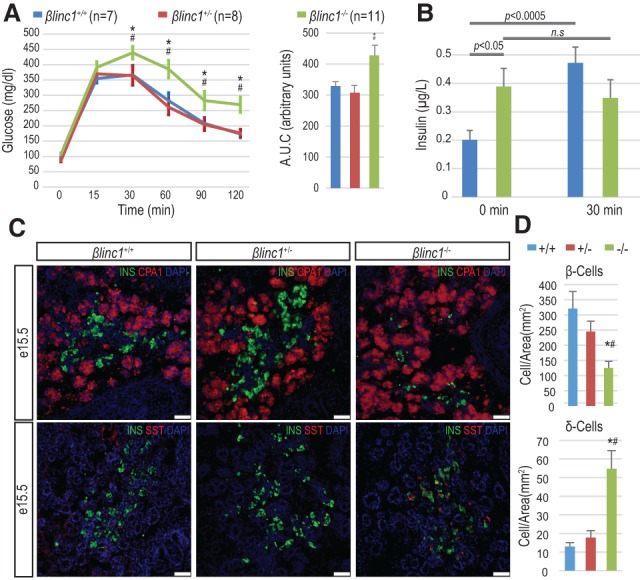
βlinc1 knockout mice are glucose-intolerant and have defects in endocrine specification. (A) βlinc1−/− 16-wk-old mice are mildly glucose-intolerant. (AUC) Area under the curve. (B) Plasma insulin levels in 16-wk-old βlinc1−/− mice (green) (n = 10) and littermate βlinc1+/+ control mice (blue) (n = 7) at 0 and 30 min after glucose injection. (C) Immunofluorescence analysis of pancreas development at E15.5 showing a reduction of β cells and an increase of somatostatin-producing δ cells in βlinc1−/− mice compared with βlinc1+/+ and βlinc1+/− mice. There are no apparent morphological defects in the exocrine compartment, which was visualized by immunostaining against CPA1. Images are representative of n > 3 mice. Bar, 50 µm. (D) Quantification of hormone-producing cells in E15.5 pancreata. n = 4. Error bars represent ±SEM. (*) P < 0.05 βlinc1−/− versus βlinc1+/+; (#) P < 0.05 βlinc1−/− versus βlinc1+/−, Student's t-test.
Endocrine development is affected in βlinc1−/− mice
Given the expression of βlinc1 in the embryonic pancreas, it was possible that aberrant glucose homeostasis was caused by defects in pancreas development. Perinatal βlinc1−/− mice did not exhibit overt defects in pancreas morphology but displayed a 50% reduction in β cells and an increase in the number of somatostatin-expressing cells (Supplemental Fig. 7a,b; data not shown). We did not detect alterations in islet cell ratios in βlinc1+/− neonates (data not shown). The changes in islet cell ratios in the βlinc1−/− animals appeared to be initiated during development at the secondary transition. At embryonic day 15.5 (E15.5), the number of insulin-producing β cells was already significantly reduced, and there was an approximately threefold increase in the number of somatostatin-producing cells (Fig. 2C,D; Supplemental Fig. 7c). There was also a smaller but significant reduction in the number of glucagon- and ghrelin-producing cells (Supplemental Fig. 7d–g). Changes in the expression of the corresponding hormone genes and β-cell-specific transcription factors were consistent with the changes in cell numbers (Fig. 3A,B). Interestingly, we did not observe up-regulation of δ-cell markers, such as Hhex and Crhr2, suggesting that deletion of βlinc1 may not cause the formation of excess bona fide δ cells. However, we also did not detect coexpression of SST with the other endocrine hormones (Supplemental Fig. 8), indicating that the increased numbers of SST-expressing cells may arise from precocious or de novo differentiation of immature and/or noncanonical δ-cell populations.
Figure 3.
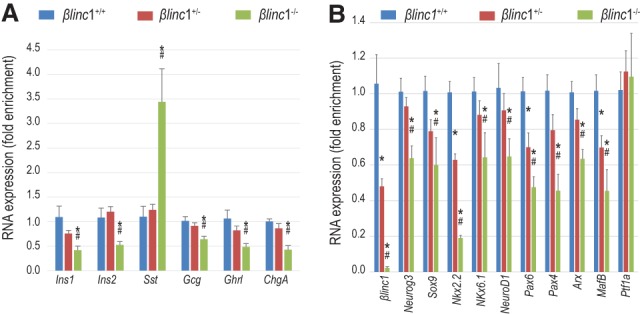
βlinc1 regulates the expression of endocrine-specific genes. (A) qPCR analysis of hormones in E15.5 pancreata. General reduction of hormone expression with the up-regulation of somatostatin. n ≥ 5. (B) qPCR analysis of βlinc1 and several transcription factors involved in pancreas development in E15.5 pancreata. n ≥ 5. Error bars represent ±SEM. (*) P < 0.05 βlinc1−/− versus βlinc1+/+; (#) P < 0.05 βlinc1+/− versus βlinc1+/+, Student's t-test.
The altered changes in islet cell ratios in βlinc1−/− embryos also did not appear to be caused by aberrant proliferation or apoptosis, since there were no detectable differences in pHistone H3 or cleaved caspase 3 staining between the mutant and wild-type embryos (data not shown). In support of a role for βlinc1 in the proper specification of some endocrine cell lineages, Neurog3 is also decreased (Fig. 3B). Importantly, there was no effect on expression of the exocrine marker Ptf1a (Fig. 3B), confirming the endocrine specificity of βlinc1 function.
βlinc1 deficiency affects the expression of several genes related to endocrine cell differentiation and β-cell function
To identify the pathways regulated by βlinc1, we performed global transcriptome analysis on E15.5 βlinc1−/− versus βlinc1+/+ pancreata (Gene Expression Omnibus [GEO] accession no. GSE73711) (Supplemental Table 1). Consistent with the changes in endocrine cell ratios, these data indicated a general dysregulation of endocrine system development in the absence of βlinc1 (Supplemental Fig. 9a,b). Notably, we did not detect changes in genes coding for transcription factors involved in the specification and maintenance of pancreas progenitors; however, in the absence of βlinc1, a large number of genes involved in the specification of endocrine progenitors and in the maturation and function of β cells were dysregulated (Supplemental Fig. 9c). This supports a role for βlinc1 in the proper specification of endocrine progenitors during the secondary transition of pancreas development, when the majority of β-cell differentiation occurs.
Although we did not detect altered islet cell ratios or glucose metabolism defects in the heterozygous βlinc1+/− pancreata, (Fig. 2a; Supplemental Fig. 7), βlinc1+/− embryos had reduced βlinc1 transcript and displayed defects in the expression of several islet transcription factors, including Nkx2.2, Pax6, and Mafb (Fig. 3B). To identify additional gene expression changes that were not confounded by a change in islet cell type ratios, we performed transcriptome analysis on E15.5 βlinc1+/− pancreata. Strikingly, there were many genes dysregulated in pancreata lacking one copy of βlinc1 (Fig. 4A; Supplemental Table 1). This analysis also revealed that, in addition to Nkx2.2, Pax6, and Mafb, five of the top 10 most significantly dysregulated genes in the βlinc1+/− mice were also located within an ∼55-Mb region on chromosome 2 (Fig. 4B,C). qPCR analysis verified that these five down-regulated genes on chromosome 2 were significantly decreased in both βlinc1+/− and βlinc1−/− pancreata (Fig. 4D).
Figure 4.
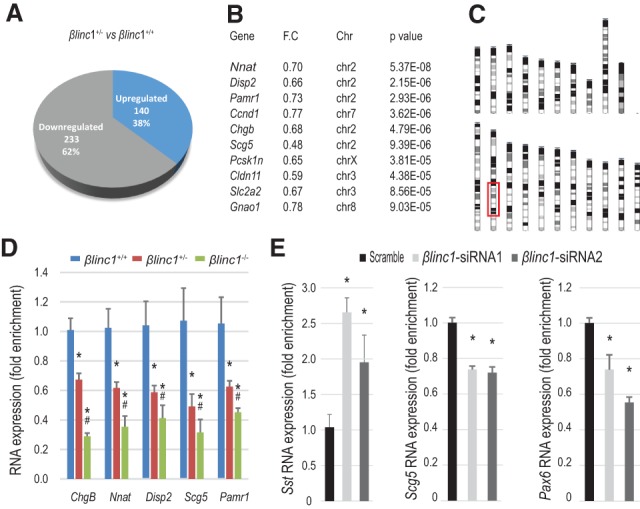
βlinc1 predominantly regulates the expression of β-cell-enriched genes that map to the same chromosomal region as βlinc1. (A) Pie chart showing the total numbers and relative percentages of up-regulated and down-regulated genes (P < 0.05) in βlinc1+/− versus βlinc1+/+. (B) Fold change and chromosomal location of the 10 most significantly down-regulated genes in βlinc1+/− compared with βlinc1+/+. A large proportion of these genes is in chromosome 2 (Supplemental Fig. 10a). (C) Schematic representation of the mouse karyotype. The red box denotes the location of the dysregulated genes on chromosome 2 in βlinc1+/− compared with βlinc1+/+ mice. (D) qRT–PCR validation of RNA-seq results for the genes located in chromosome 2. (*) P < 0.05 versus βlinc1+/+; (#) P < 0.05 versus βlinc1+/−. (E) Down-regulation of βlinc1 with two different siRNAs in MIN6 cells recapitulates the increase in somatostatin expression and down-regulation of Scg5 and Pax6 seen in vivo. n = 4. Error bars represent ±SEM. (*) P < 0.05 versus scrambled. Student's t-test.
Dysregulated genes in βlinc1+/− localize to the genomic vicinity of βlinc1
To determine whether the genes regulated by βlinc1were biased based on their genomic location, we positionally mapped all genes that were significantly dysregulated (P < 0.05) in βlinc1+/− pancreata versus the wild-type controls. Remarkably, a disproportionate number of dysregulated genes mapped to a discrete region of chromosome 2, within 40 Mb of the βlinc1 locus. Importantly, this is not due to an increased number of genes that are expressed on chromosome 2 during pancreas development (Supplemental Fig. 10a,b). Moreover, gene ontology analysis showed that the dysregulated genes on chromosome 2 are associated with endocrine development and islet morphology. Ingenuity Pathway Analysis also indicated that the predominant upstream regulators affected by loss of one allele of βlinc1 are the known islet-specific transcription factors Pdx1 and NeuroD1. These pancreas-related categories are lost if the same analysis is performed with all of the genes that are located in this region of chromosome 2 (Supplemental Fig. 10a,b).
The remarkable observation that many βlinc1-regulated genes were located on the same chromosome as βlinc1 prompted us to investigate whether the observed gene expression changes were due to loss of βlinc1 transcript rather than the deletion of a DNA enhancer element, similar to what was observed with Nkx2.2 regulation (Fig. 1F). Assessment of the expression of additional candidate βlinc1target genes located on chromosome 2 in response to siRNA-mediated knockdown of βlinc1 in MIN6 cells revealed the up-regulation of Sst expression and down-regulation of Scg5 and Pax6, confirming the biological role of the βlinc1 transcript in gene regulation (Fig. 4E). Moreover, shRNA-mediated knockdown of the orthologous human βLINC1 transcript in the human insulin-producing EndoC-βH1 cell line also causes altered expression of genes dysregulated in βlinc1−/− mice (I Akerman and J Ferrer, in prep.). Therefore, it appears that the βlinc1 transcript regulates the expression of a set of functionally related genes that are important for the development and/or function of β cells.
In this study, we describe the first in vivo characterization of a conserved islet-specific lncRNA. Homozygous deletion of βlinc1 in mice causes the down-regulation of important islet transcription factors and impaired β-cell specification during embryonic development. Furthermore, adult mice lacking βlinc1 are glucose-intolerant and display defective insulin secretion. Although it is possible that a subset of the observed βlinc1−/− phenotypes could be due to the deletion of a regulatory element within the genomic locus, siRNA- and shRNA-mediated knockdown experiments of the βlinc1RNA in mouse MIN6 cells (Figs. 1F, 4E) and human EndoC-βH1 cells recapitulated some of the gene changes observed in vivo. Furthermore, the βlinc1 genomic locus does not appear to have enhancer activities in MIN6 cells (Supplemental Fig. 3); however, limitations of these in vitro studies prevented us from ascribing all of the βlinc1 phenotypes to loss of the βlinc1 transcript. Interestingly, although we demonstrated that the βlinc1 transcript regulates several important islet transcription factors, only a subset of the βlinc1−/− phenotypes could be attributed to reduction of Nkx2.2, Pax6, and/or MafB expression (Supplemental Table 2; Pan and Wright 2011), suggesting that βlinc1 is involved in novel lineage regulatory pathways.
Transcriptome analysis of pancreata lacking one copy of βlinc1 also revealed the dysregulation of a set of functionally related β−cell genes located in discrete genomic regions. It is particularly intriguing that βlinc1 specifically regulates three essential islet transcription factors, ChgB, and additional β-cell genes on chromosome 2. Long-range interchromosomal and intrachromosomal interactions through chromosomal looping and transcription factories are known to be involved in the transcriptional coregulation of functionally related genes in a cell type-specific manner (Schoenfelder et al. 2010; Papantonis et al. 2012). For example, insulin expression and calcium signaling have been shown to be coupled through a long-range chromosomal interaction that spans >65 Mb (Xu et al. 2014). lncRNAs have also been implicated in higher-order gene regulation through chromatin organization, and there is evidence of lncRNAs being highly enriched at transcription “factories” (Caudron-Herger et al. 2015). Therefore, it is possible that βlinc1 coordinately regulates β-cell gene expression through the structural organization of the chromatin in β cells. Analysis of the known chromatin features and topologically associated domains (TADs) in and around βlinc1 failed to reveal any obvious domains or features that could explain the regulation of the genes on chromosome 2. HiC analyses suggested that βlinc1 may be located at the edge of a TAD (Supplemental Fig. 11; Shen et al. 2012); however, the presence of this domain does not correlate with the observed in vivo gene expression changes. For example, the TAD data include all of the genes located between Insm1 and Foxa2, but only two of the 13 genes within this domain (Nkx2-2 and Insm1) are down-regulated in βlinc1−/− mice.
Although we are just beginning to understand a role for lncRNAs in the cell, many findings have suggested that lncRNAs provide a layer of cell-specific gene regulation that contributes to cellular diversity (Mattick 2001). It is therefore intriguing that βlinc1 has such a limited expression domain, and the phenotype of the global βlinc1−/− mice is primarily restricted to β-cell formation and function. Furthermore, our discovery of βlinc1 as a novel islet-specific transcriptional regulator has important implications for understanding β-cell biology and suggests that ncRNAs could represent novel therapeutic targets for the treatment of diabetes.
Materials and methods
Generation of the βlinc1 knockout allele
The βlinc1 knockout allele was generated using the Recombinase Mediate Cassette Exchange protocol as previously described (Arnes et al. 2012) with some modifications. Short arms (≈500 base pairs) homologous to the flanking region of βlinc1 and to a downstream region (2.9 kb) were generated by PCR and cloned into pLCA.71/2272 and pMCS-DTA. The BAC clone (RP23-236P19) was modified in two consecutive recombineering steps in SW106 cells: The βlinc1 sequence was replaced with the puΔtk-EM7-kan cassette, and the DTA-Ampr cassette was inserted 2.9 kb downstream from βlinc1, resulting in the replacement of 1 kb of genomic DNA (Supplemental Fig. 5b). Positive clones were validated by PCR and DNA sequencing, and a correctly modified BAC was electroporated into mouse embryonic stem cells at Columbia University (Herbert Irving Comprehensive Cancer Center Transgenic Shared Resource). Potentially recombined clones were screened by PCR with primers P1 and P2 (Supplemental Fig. 5d,e). Two positive clones were used to generate chimeric mice that resulted in germline transmission.
Mice
All mice were maintained on a mixed C57BL6/129SV genetic background. All animals were maintained according to Columbia University Institutional Animal Care and Use Committee approval protocol AAAG3206.
Physiological assays
Glucose tolerance tests, measurement of plasma insulin, and insulin secretion assays were performed as previously described (Gu et al. 2010).
Cell lines and transfection
MIN6, αTC1 clone 6 (American Type Culture Collection), and PG1 cells (provided by Dr. Jeffrey Zigman, University of Texas Southwestern Medical Center) were passaged and maintained following standard techniques in 5% CO2 and 95% air. MIN6 cells were transfected with 10 nM siRNA targeting βlinc1 and a scrambled control (Silencer Select, Ambion) using Lipofectamine 2000 following the manufacturer's instructions (Life Technologies). siRNAs targeting βlinc1 were designed using the algorithm provided by the manufacturer, and sequences are listed in Supplemental Table 3.
Enhancer activity
A 4.2-kb fragment of the βlinc1 genomic locus (chr2: 147,030,443–147,034,638, mm9) was cloned into the PGL4.27 luciferase vector. One microgram of the experimental vector PGL4.27-βlinc1 region and the positive controls (CDKN2BAS enhancer, Ins2 promoter, and NeuroD promoter/enhancer) were individually cotransfected with 0.1 µg of pRL into MIN6 cells in triplicate. Luciferase activity was measured after 48 h. PGL4.23-CDKN2BAS was a gift from Jorge Ferrer (Addgene plasmid no. 60296). PGL3-Ins2 and PGL3-NeuroD have been previously described (Raum et al. 2006; Anderson et al. 2009). Luciferase values were normalized to Renilla activity to account for transfection efficiencies and were expressed as fold increase over the empty vector.
RNA in situ hybridization
The βlinc1 probe was generated by PCR from E15.5 pancreas cDNA and cloned into pCRII-TOPO (Life Technologies). Sense and antisense probes were labeled with the DIG RNA-labeling mix (Roche Applied Science). RNA in situ hybridization was performed as previously described (Mastracci et al. 2013), including a short treatment of proteinase K digestion.
Immunofluorescence
Tissue processing and immunofluorescence analysis were performed as previously described (Arnes et al. 2012). Primary antibodies are listed in Supplemental Table 3. DAPI (1:1000; Invitrogen) was applied for 30 min following secondary antibody incubation: DyLight-488, DyLight-549, DyLight-649, Alexa-488, and Alexa-647 (Jackson Immunoresearch). Images were acquired with either an epifluorescence (Leica DM5500) or a confocal (Zeiss LSM 710) microscope.
Morphometric analysis and cell counting
Morphometric analysis was performed in E15.5 and postnatal day 2 pancreata. The entire organ was sectioned, and at least six evenly distributed sections were analyzed. The number of endocrine cells was determined relative to the total pancreatic area. Pancreas area was quantified using Image Pro Plus 5.0.1 software (Media Cybernetics).
Cellular fractionation
MIN6 cells were grown to confluency, detached by trypsinization, and pelleted. Half of the pellet was used for total RNA isolation, and the other half was used for nuclear and cytoplasmic isolation using the PARIS kit (Ambion) following the manufacturer's instructions.
RNA extraction and quantitative RT–PCR analysis
Total RNA was isolated and analyzed as previously described (Arnes et al. 2012). Primers and probes are listed in Supplemental Table 3.
RNA sequencing (RNA-seq) analysis
Total RNA from βlinc1+/+, βlinc1+/−, and βlinc1−/− mice was converted into cDNA libraries (TruSeq RNA sample preparation kit version 2, Illumina) using poly-A pull-down for mRNA enrichment. Sequencing was performed to a depth of 30 million pairs in three biological replicates per condition. Differential expression between replicates was assessed using DESeq (R package). All samples had RNA integrity (RIN) values >9.0 as determined with Agilent Bioanalyzer 2100. Complete RNA-seq data are available through GEO accession number GSE73711.
TADs
TADs in the genomic vicinity of βlinc1 were identified from the mouse encode project at the Ren laboratory (http://chromosome.sdsc.edu/mouse).
Statistics
All data were expressed as mean ± SEM. Mean and SEM values were calculated from at least three biological replicates. The statistical significance was measured by two-tailed Student's t-test.
Supplementary Material
Acknowledgments
We thank the Sussel laboratory for critical discussions throughout the project and reading of the manuscript. We are especially grateful to Chyuan-Sheng (Victor) Lin for assistance with the generation of the βlinc1 knockout mice, Jiguang Wang for computational support, and Jeffrey Zigman for providing the PG1 cells. The project was initiated with the National Institutes of Health B-Cell Biology Consortium collaborative project grant (U19 DK072473) to L.S. and J.F. Additional support was provided by Juvenile Diabetes Research Foundation grant CU13-252 (L.A.), the Russell Berrie Foundation (L.A.), the Foundation for Diabetes Research (L.S.), and Juvenile Diabetes Research Foundation grant 117-2012054 (L.S.). L.S. and J.F. conceived the study. I.A. and J.F. collected and analyzed the data on human βLINC1. L.A. generated and processed the data on mouse βlinc1 with assistance from D.A.B. L.S. and L.A. designed the study, analyzed the data, and wrote the manuscript. All authors discussed the results and edited the manuscript.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.273821.115.
References
- Anderson KR, Torres CA, Solomon K, Becker TC, Newgard CB, Wright CV, Hagman J, Sussel L. 2009. Cooperative transcriptional regulation of the essential pancreatic islet gene NeuroD1 (β2) by Nkx2.2 and neurogenin 3. J Biol Chem 284: 31236–31248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arda HE, Benitez CM, Kim SK. 2013. Gene regulatory networks governing pancreas development. Dev Cell 25: 5–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnes L, Sussel L. 2015. Epigenetic modifications and long noncoding RNAs influence pancreas development and function. Trends Genet 31: 290–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnes L, Hill JT, Gross S, Magnuson MA, Sussel L. 2012. Ghrelin expression in the mouse pancreas defines a unique multipotent progenitor population. PLoS One 7: e52026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Chang HY. 2013. Long noncoding RNAs: cellular address codes in development and disease. Cell 152: 1298–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. 2005. The transcriptional landscape of the mammalian genome. Science 309: 1559–1563. [DOI] [PubMed] [Google Scholar]
- Caudron-Herger M, Cook PR, Rippe K, Papantonis A. 2015. Dissecting the nascent human transcriptome by analysing the RNA content of transcription factories. Nucleic Acids Res 43: e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MB, Johnston RL, Inostroza-Ponta M, Fox AH, Fortini E, Moscato P, Dinger ME, Mattick JS. 2012. Genome-wide analysis of long noncoding RNA stability. Genome Res 22: 885–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu C, Stein GH, Pan N, Goebbels S, Hornberg H, Nave KA, Herrera P, White P, Kaestner KH, Sussel L, et al. 2010. Pancreatic β cells require NeuroD to achieve and maintain functional maturity. Cell Metab 11: 298–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia S, Ivanov A, Blasevic D, Muller T, Purfurst B, Sun W, Chen W, Poy MN, Rajewsky N, Birchmeier C. 2015. Insm1 cooperates with Neurod1 and Foxa2 to maintain mature pancreatic β-cell function. EMBO J 34: 1417–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoo C, Yang J, Weinrott SA, Kaestner KH, Naji A, Schug J, Stoffers DA. 2012. Research resource: the pdx1 cistrome of pancreatic islets. Mol Endocrinol 26: 521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku GM, Kim H, Vaughn IW, Hangauer MJ, Myung Oh C, German MS, McManus MT. 2012. Research resource: RNA-seq reveals unique features of the pancreatic β-cell transcriptome. Mol Endocrinol 26: 1783–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastracci TL, Lin CS, Sussel L. 2013. Generation of mice encoding a conditional allele of Nkx2.2. Transgenic Res 22: 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS. 2001. Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep 2: 986–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran I, Akerman I, van de Bunt M, Xie R, Benazra M, Nammo T, Arnes L, Nakic N, Garcia-Hurtado J, Rodriguez-Segui S, et al. 2012. Human β cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab 16: 435–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan FC, Wright C. 2011. Pancreas organogenesis: from bud to plexus to gland. Dev Dyn 240: 530–565. [DOI] [PubMed] [Google Scholar]
- Papantonis A, Kohro T, Baboo S, Larkin JD, Deng B, Short P, Tsutsumi S, Taylor S, Kanki Y, Kobayashi M, et al. 2012. TNFα signals through specialized factories where responsive coding and miRNA genes are transcribed. EMBO J 31: 4404–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquali L, Gaulton KJ, Rodriguez-Segui SA, Mularoni L, Miguel-Escalada I, Akerman I, Tena JJ, Moran I, Gomez-Marin C, van de Bunt M, et al. 2014. Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat Genet 46: 136–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raum JC, Gerrish K, Artner I, Henderson E, Guo M, Sussel L, Schisler JC, Newgard CB, Stein R. 2006. FoxA2, Nkx2.2, and PDX-1 regulate islet β-cell-specific mafA expression through conserved sequences located between base pairs −8118 and −7750 upstream from the transcription start site. Mol Cell Biol 26: 5735–5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvageau M, Goff LA, Lodato S, Bonev B, Groff AF, Gerhardinger C, Sanchez-Gomez DB, Hacisuleyman E, Li E, Spence M, et al. 2013. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife 2: e01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, et al. 2010. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet 42: 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. 2012. A map of the cis-regulatory sequences in the mouse genome. Nature 488: 116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussel L, Kalamaras J, Hartigan-O'Connor DJ, Meneses JJ, Pedersen RA, Rubenstein JL, German MS. 1998. Mice lacking the homeodomain transcription factor Nkx2.2 have diabetes due to arrested differentiation of pancreatic β cells. Development 125: 2213–2221. [DOI] [PubMed] [Google Scholar]
- Vance KW, Sansom SN, Lee S, Chalei V, Kong L, Cooper SE, Oliver PL, Ponting CP. 2014. The long non-coding RNA Paupar regulates the expression of both local and distal genes. EMBO J 33: 296–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Lefevre GM, Gavrilova O, Foster St Claire MB, Riddick G, Felsenfeld G. 2014. Mapping of long-range INS promoter interactions reveals a role for calcium-activated chloride channel ANO1 in insulin secretion. Proc Natl Acad Sci 111: 16760–16765. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


