Benyoucef et al. reveal the existence of a subtype-specific epigenetic vulnerability in T-cell acute lymphoblastic leukemia (T-ALL) by which a particular subgroup of T-ALL characterized by expression of the oncogenic transcription factor TAL1 is uniquely sensitive to variations in the dosage and activity of the histone 3 Lys27 (H3K27) demethylase UTX/KDM6A.
Keywords: UTX, TAL1, histone demethylation, leukemia, histone demethylase inhibitor, epigenetics
Abstract
T-cell acute lymphoblastic leukemia (T-ALL) is a heterogeneous group of hematological tumors composed of distinct subtypes that vary in their genetic abnormalities, gene expression signatures, and prognoses. However, it remains unclear whether T-ALL subtypes differ at the functional level, and, as such, T-ALL treatments are uniformly applied across subtypes, leading to variable responses between patients. Here we reveal the existence of a subtype-specific epigenetic vulnerability in T-ALL by which a particular subgroup of T-ALL characterized by expression of the oncogenic transcription factor TAL1 is uniquely sensitive to variations in the dosage and activity of the histone 3 Lys27 (H3K27) demethylase UTX/KDM6A. Specifically, we identify UTX as a coactivator of TAL1 and show that it acts as a major regulator of the TAL1 leukemic gene expression program. Furthermore, we demonstrate that UTX, previously described as a tumor suppressor in T-ALL, is in fact a pro-oncogenic cofactor essential for leukemia maintenance in TAL1-positive (but not TAL1-negative) T-ALL. Exploiting this subtype-specific epigenetic vulnerability, we propose a novel therapeutic approach based on UTX inhibition through in vivo administration of an H3K27 demethylase inhibitor that efficiently kills TAL1-positive primary human leukemia. These findings provide the first opportunity to develop personalized epigenetic therapy for T-ALL patients.
T-cell acute lymphoblastic leukemias (T-ALLs) are aggressive hematological malignancies resulting from the oncogenic transformation of thymocytes at distinct stages of differentiation. Different subtypes of T-ALL arise from the aberrant expression of specific transcription factors in T-cell progenitors (Van Vlierberghe and Ferrando 2012). One of the most prevalent oncogenic transcription factors in T-ALL is the class II basic helix–loop–helix (bHLH) protein TAL1 (also called SCL) that is aberrantly expressed in 40%–60% of T-ALL patients (Lecuyer and Hoang 2004; Van Vlierberghe and Ferrando 2012). Clinically, the TAL1-positive T-ALL subtype displays a particularly poor prognosis compared with other subtypes that do not express TAL1 (Ferrando et al. 2002). The oncogenic role of TAL1 has been demonstrated through forced expression of TAL1 in T-cell progenitors that leads to the development of T-cell leukemia (Condorelli et al. 1996; Kelliher et al. 1996). Furthermore, TAL1 is necessary for the maintenance of leukemia, as shown by the dramatic increase in apoptosis observed in T-ALL cells upon TAL1 knockdown (Palii et al. 2011b; Sanda et al. 2012). We (Palii et al. 2011b) and others (Kusy et al. 2010; Thoms et al. 2011; Sanda et al. 2012) have previously identified TAL1 genomic binding sites in T-ALL, revealing that TAL1 controls the expression of specific genes necessary for cell growth and maintenance of T-ALL. However, the molecular mechanism through which TAL1 regulates transcription of these genes remains unclear.
Results
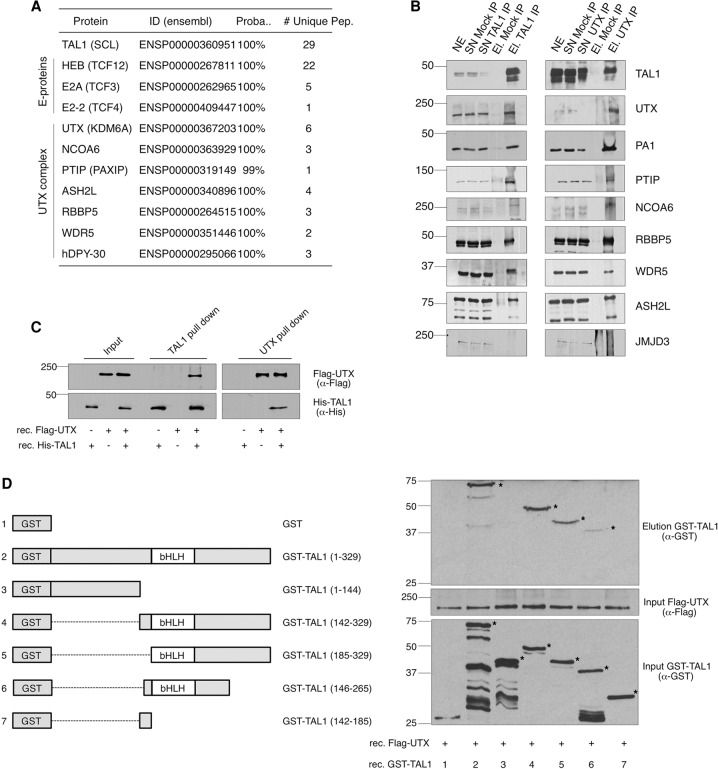
To elucidate the function of TAL1 in leukemia, we immunoprecipitated the endogenous TAL1 protein from T-ALL nuclear extracts (NEs) and identified TAL1-interacting proteins by mass spectrometry (MS) (Supplemental Table 1). This experiment confirmed previously known TAL1-interacting partners (Hu et al. 2009) and revealed components of the UTX complex (Cho et al. 2007; Issaeva et al. 2007) as a novel set of TAL1-interacting proteins (Fig. 1A). Reciprocal immunoprecipitations using antibodies (Abs) against endogenous TAL1 and UTX proteins confirmed the interaction of TAL1 with subunits of the UTX complex (Fig. 1B), and in vitro binding experiments with recombinant proteins revealed a direct interaction between UTX and the bHLH domain of TAL1 (Fig. 1C,D). UTX (also called KDM6A) is a Jumonji C domain-containing demethylase that removes the repressive histone mark trimethylated histone 3 Lys27 (H3K27me3) (Agger et al. 2007; Hong et al. 2007). Previous studies have shown that UTX plays important roles in embryonic development (Lee et al. 2012; Welstead et al. 2012) and adult hematopoiesis (Thieme et al. 2013). Our identification of an interaction between TAL1 and UTX (Fig. 1) suggests that UTX could act as a coactivator for the TAL1-mediated oncogenic program in T-ALL.
Figure 1.
Identification of the UTX complex as a TAL1-interacting partner. (A) TAL1-interacting proteins identified by MS after TAL1 immunoprecipitation in Jurkat NEs. (Proba.) Probability of identification determined by ProteinProphet (Nesvizhskii et al. 2003). (B) Western blot analysis of reciprocal immunoprecipitations performed in Jurkat NEs with Abs against endogenous TAL1 and UTX proteins. (SN) Supernatant; (El.) elution. (C) Western blot analysis of TAL1 and UTX pull-downs performed on recombinant purified Flag-UTX and His-TAL1 fusion proteins. (D) Western blot analysis (right panel) of Flag pull-downs performed on recombinant purified Flag-UTX and the indicated GST-TAL1 fusion proteins (left panel). The asterisk indicates the recombinant protein of expected size. (B–D) Molecular masses are indicated (in kilodaltons). Representative examples of three biological replicates are shown.
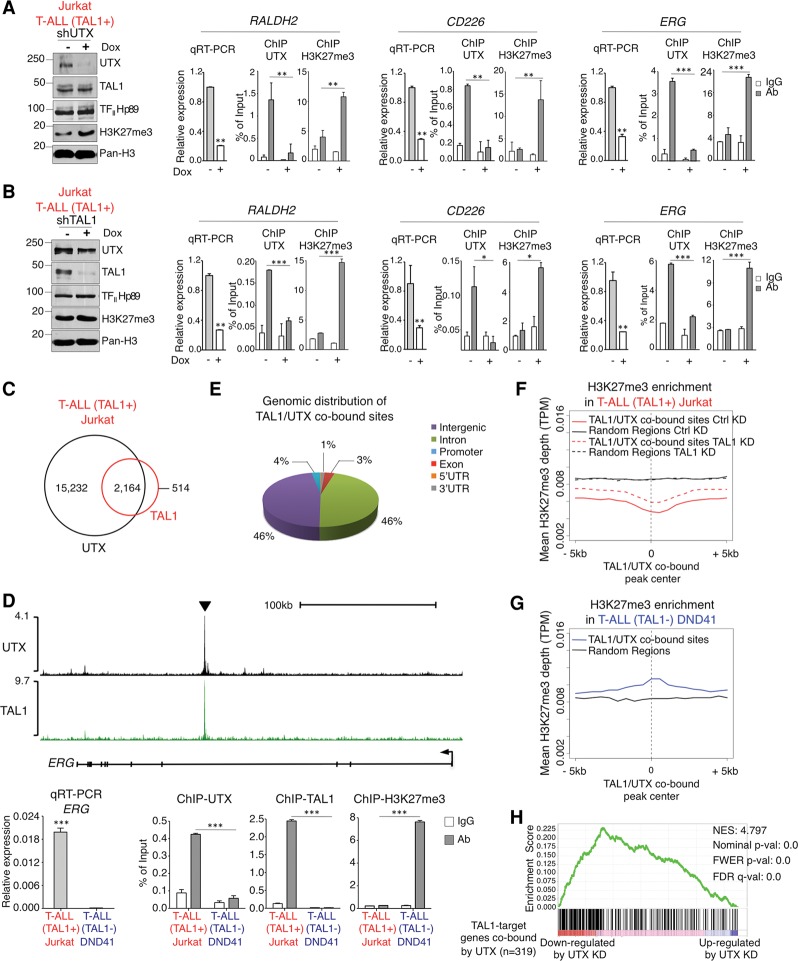
To test this hypothesis, we first examined UTX binding to several previously characterized TAL1-activated genes, including RALDH2 (also called ALDH1A2), the proto-oncogene MYB, and genes involved in cell proliferation (e.g., ERG) and apoptosis inhibition (e.g., CD226) (Palii et al. 2011b; Thoms et al. 2011; Sanda et al. 2012). Chromatin immunoprecipitation (ChIP) experiments indicate that UTX is bound to all TAL1 target genes that were examined (Fig. 2A,B; Supplemental Fig. 1). Furthermore, expression of these genes is significantly reduced upon UTX knockdown, concomitant with an increase in the level of H3K27me3 (Fig. 2A). Finally, the knockdown of TAL1 leads to a significant loss of UTX binding at its target loci accompanied by an increase in H3K27me3 and a decrease in gene expression (Fig. 2B). Collectively, these results indicate that TAL1-mediated recruitment of UTX facilitates the expression of specific genes involved in proliferation and apoptosis inhibition through active removal of the repressive histone mark H3K27me3.
Figure 2.
TAL1 recruits UTX to aberrantly activate transcription of its target genes through H3K27me3 demethylation. (A) UTX binds to and activates TAL1 target genes through active removal of H3K27me3. UTX knockdown (measured by Western blot) induced by doxycycline (Dox)-mediated expression of an anti-UTX shRNA leads to a decrease in transcription of the indicated TAL1 target genes (measured by quantitative RT–PCR [qRT–PCR]) and an increase in H3K27me3 (measured by ChIP-qPCR). (B) TAL1 recruits UTX to activate transcription of its target genes. TAL1 knockdown (measured by Western blot) induced by Dox-mediated expression of an anti-TAL1 shRNA disrupts UTX binding, which leads to an increase in H3K27me3 (measured by ChIP-qPCR) accompanied by decreased transcription of the indicated genes (measured by qRT–PCR). (C) Venn diagram showing the overlap of TAL1 and UTX binding genome-wide. P-value < 0.001. (D, top panel) ChIP-seq (ChIP combined with deep sequencing) density plots (normalized by reads per million mapped reads) for UTX and TAL1 at the ERG gene locus in TAL1-positive T-ALL cells. The arrowhead indicates the region tested by ChIP-qPCR below. (Bottom panel) Validation of the differential expression, H3K27me3 enrichment, and UTX and TAL1 binding at the ERG gene locus in TAL1-positive versus TAL1-negative T-ALL. (E) Pie chart of the genome-wide distribution of TAL1/UTX-cobound sites based on RefSeq in the TAL1-positive T-ALL. (F) TAL1 recruits an active H3K27me3 demethylase to its binding sites genome-wide in TAL1-positive T-ALL. Histone H3K27me3 ChIP-seq profiles are shown in control (Ctrl) and TAL1 knockdown (KD) Jurkat cells around the 2164 TAL1/UTX-cobound sites. Randomly chosen sites not cobound by TAL1 and UTX are shown for comparison. (TPM) Tags per million. (G) Genomic locations depleted of the H3K27me3 histone mark in the TAL1-positive T-ALL cell line Jurkat (shown in F) are enriched for the H3K27me3 mark in the TAL1-negative T-ALL cell line DND41. Histone H3K27me3 ChIP-seq profiles around the same genomic locations as in F are shown in the TAL1-negative T-ALL cell line DND41. Randomly chosen sites (the same as in F) are shown for comparison. (TPM) Tags per million. (H) UTX positively regulates the expression of TAL1 target genes that are cobound by UTX. Gene set enrichment analysis indicates the degree to which genes that are associated with TAL1/UTX-cobound sites and down-regulated upon TAL1 knockdown are overrepresented at the left (down-regulated upon UTX knockdown) or right (up-regulated upon UTX knockdown) of the entire ranked list (defined by RNA sequencing [RNA-seq] upon UTX knockdown). (NES) Normalized enrichment score. (A,B) Molecular masses (in kilodaltons) are indicated on Western blots. A representative example of three biological replicates is shown. (A,B,D), qRT–PCR results are expressed as mean values relative to the internal control ß2microglobulin (ß2M), with error bars corresponding to standard deviations (SDs). Mean ChIP-qPCR values are presented as a fraction of input, with error bars corresponding to SDs. n = 3. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001.
To evaluate the extent to which UTX contributes to the TAL1 oncogenic program genome-wide, UTX genomic binding was analyzed by ChIP combined with deep sequencing (ChIP-seq) and compared with TAL1 binding in TAL1-expressing T-ALL cells. This experiment revealed that the vast majority of TAL1-bound sites (>80%; P-value < 0.001) are co-occupied by UTX (Fig. 2C–E; Supplemental Fig. 1). Furthermore, histone ChIP-seq experiments indicate that genomic regions co-occupied by TAL1 and UTX are depleted of the repressive mark H3K27me3 (Fig. 2F), confirming that UTX is enzymatically active at these sites. Remarkably, the knockdown of TAL1 leads to the re-establishment of H3K27me3 marks at the TAL1/UTX-cobound sites (Fig. 2F, cf. dashed and solid red lines), revealing that TAL1 is involved in actively maintaining low levels of the repressive H3K27me3 mark at its genomic locations. Interestingly, examination of H3K27me3 in a TAL1-negative molecular subtype of T-ALL revealed that this repressive histone mark is enriched at these same genomic locations (Fig. 2G). This suggests that the depletion of H3K27me3 observed at TAL1/UTX-cobound sites (Fig. 2F) is specific to TAL1-positive T-ALL. Consistent with this, ChIP experiments showed that UTX is not bound to TAL1 sites in a TAL1-negative subtype of T-ALL (Fig. 2D, bottom panel for ERG; Supplemental Fig. 1 for other genes).
To determine whether UTX is functionally involved in the regulation of genes cobound by TAL1 and UTX, RNA sequencing (RNA-seq) analyses upon knockdown of each factor were performed in TAL1-positive T-ALL. These experiments reveal that TAL1/UTX-cobound genes (i.e., defined as genes having a UTX/TAL1-cobound peak in a region spanning 50 kb upstream of the transcription start site [TSS] to the 3′ end of the gene) (Supplemental Table 2) that are down-regulated upon TAL1 knockdown also tend to be down-regulated upon UTX knockdown (Fig. 2H), which confirms that UTX is required for the efficient activation of TAL1 target genes. Collectively, the above results show that UTX is a major coactivator of the TAL1 transcriptional regulatory program in T-ALL through removal of the repressive histone mark H3K27me3.
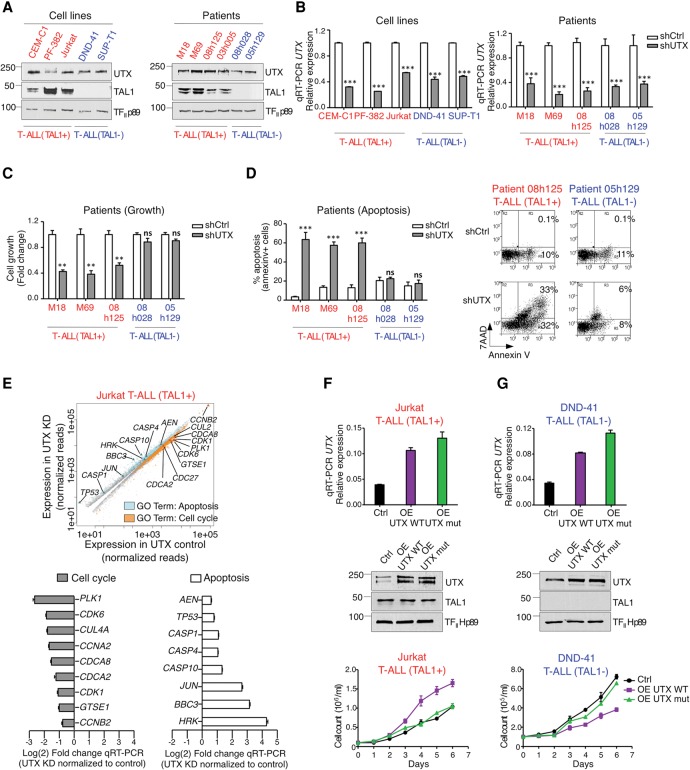
Since TAL1 is a critical oncogene in T-ALL, we hypothesized that its cofactor, UTX, may directly contribute to the maintenance of leukemia. Furthermore, we reasoned that if a potential proleukemic function of UTX in TAL1-positive T-ALL is mediated through its specific recruitment to TAL1 target genes, UTX would not significantly contribute to leukemia maintenance in other molecular subtypes of T-ALL that do not express TAL1. To address these questions, the knockdown of UTX was induced in TAL1-positive T-ALL cells from multiple origins, including three cell lines (CEM-C1, PF-382, and Jurkat ) (see Fig. 3A,B; Supplemental Fig. 2A,C,F) and three primary blast samples from leukemia patients (M18, M69, and 08h125) (see Fig. 3A,B; Supplemental Fig. 2B,I). In all cases of TAL1-positive T-ALL, we observed that the knockdown of UTX leads to a strong increase in apoptosis accompanied by a decrease in cell growth (Fig. 3C,D; Supplemental Fig. 2D,E,G,H,J), a phenotype similar to that previously observed upon the knockdown of TAL1 (Palii et al. 2011b; Sanda et al. 2012). Furthermore, gene ontology (GO) analyses after RNA-seq confirm that proapoptotic genes are up-regulated, while proproliferation genes are down-regulated, upon UTX knockdown (Fig. 3E; Supplemental Fig. 3; Supplemental Table 2). In striking contrast, the knockdown of UTX did not induce apoptosis and had no effect on the growth of T-ALL cells that do not express TAL1, including two primary blast samples from leukemia patients (08h028 and 05h129) (Fig. 3A–D; Supplemental Fig. 2B,I,J) and two cell lines (DND-41 and SUP-T1) (Fig. 3A,B; Supplemental Fig. 2A,F,G,H). Thus, the knockdown of UTX selectively kills TAL1-expressing T-ALL cells.
Figure 3.
UTX is involved in leukemia maintenance of TAL1-positive but not TAL1-negative T-ALL cells. (A) UTX is expressed in TAL1-positive and TAL1-negative T-ALL subtypes. Western blot analysis of UTX and TAL1 expression in NEs prepared from the indicated cell lines and patients’ primary blasts. Representative examples of three biological replicates are shown. (B) Knockdown of UTX by lentiviral delivery of anti-UTX shRNA in T-ALL cell lines and primary blasts from patients. qRT–PCR results are presented as mean values relative to the internal control ß2M ±SD. n = 3. (***) P < 0.001. (C) UTX knockdown decreases the growth of primary blasts from TAL1-positive but not TAL1-negative T-ALL patients. Viable cells after UTX knockdown (shUTX) relative to viable cells without knockdown (shCtrl) are reported as mean ± standard error of the mean (SEM). n = 4. (**) P < 0.01; (ns) not significant. (D) UTX knockdown triggers apoptosis in primary blasts from TAL1-positive but not TAL1-negative T-ALL patients. Apoptosis is reported as the percentage of Annexin V-positive cells. Data are shown as mean ± SEM. n = 4. (***) P < 0.001; (ns) not significant. Representative FACS plots are shown at the right. (E, top panel) Scatter plot summarizing genome-wide changes in the expression of genes (adjusted P-value ≤ 0.05) that belong to the indicated GO categories upon UTX knockdown (KD). RNA-seq was performed in duplicate in control and UTX knockdown Jurkat cells. (Bottom panels) Validation of RNA-seq results by qRT–PCR. Fold changes in expression (knockdown/control) are presented as mean values relative to the internal control ß2M ±SD. n = 3. (F,G) Overexpression (OE) of UTX wild-type but not its enzymatically dead mutant (mut) counterpart increases the growth of TAL1-positive Jurkat T-ALL cells (F) but decreases the growth of TAL1-negative DND41 T-ALL cells (G). (Top panels) qRT–PCR results are presented as mean values relative to the internal control ß2M ±SD. n = 3. (Middle panels) Western blots are shown as representative examples of three biological replicates. (Bottom panels) Concentrations of viable cells are presented as mean ± SEM. n = 4. (Red) TAL1-positive T-ALL; (blue) TAL1-negative T-ALL.
Next, we reasoned that if UTX is truly pro-oncogenic in the TAL1-positive subtype of T-ALL, its overexpression should increase proliferation. Consistent with this, we observed that a twofold overexpression of UTX is sufficient to strongly increase the growth of TAL1-positive T-ALL cells (Fig. 3F). Furthermore, this effect is dependent on the demethylase activity of UTX, since overexpression of an enzymatically dead mutant has no effect on cell growth (Fig. 3F). Interestingly, UTX overexpression in TAL1-negative T-ALL leads to the opposite phenotype (i.e., decreased cell growth), also in a demethylase-dependent manner (Fig. 3G), which reinforces our previous conclusion that UTX is selectively pro-oncogenic in TAL1-positive T-ALL. Furthermore, this last result suggests that UTX could have a tumor suppressor role in TAL1-negative subtypes of T-ALL. Taken together, these results demonstrate for the first time that the mechanism of leukemia maintenance differs between distinct molecular subtypes of T-ALL, with UTX selectively acting as a pro-oncogenic cofactor to maintain leukemia in TAL1-positive cases. This suggests that personalized therapies may be required to efficiently treat the different subtypes of T-ALL.
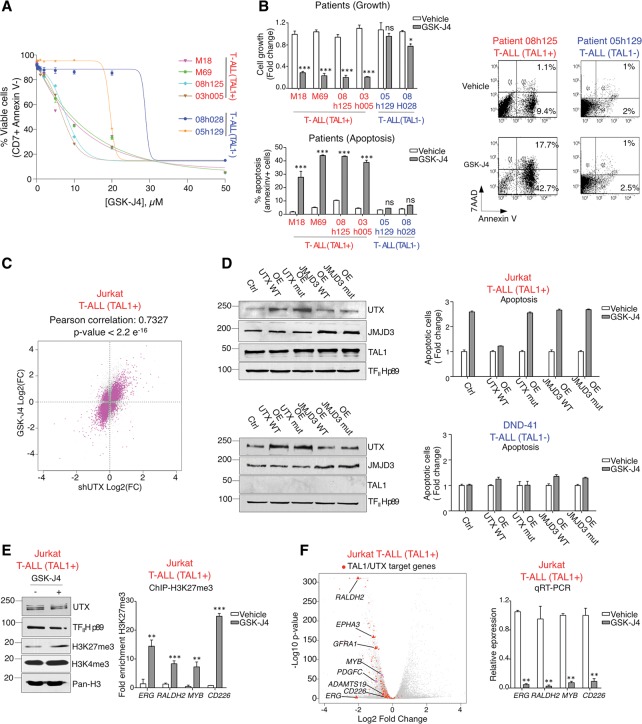
As UTX is important for leukemia maintenance in TAL1-positive T-ALL (Fig. 3; Supplemental Figs. 2, 3), we sought to use pharmacological inhibition of UTX with the H3K27 demethylase inhibitor GSK-J4 (Kruidenier et al. 2012, 2014) as a potential therapeutic strategy to eliminate TAL1-positive leukemic blasts. The dependency of TAL1-positive (but not TAL1-negative) leukemias on UTX level and activity for their survival and growth (Fig. 3; Supplemental Figs. 2, 3) predicts that TAL1-expressing leukemic cells will be more sensitive to UTX inhibition. Indeed, dose response curves measuring cell growth and apoptosis at increasing concentrations of GSK-J4 revealed a disproportionate sensitivity of TAL1-positive T-ALL cells compared with their TAL1-negative counterparts (Fig. 4A,B; Supplemental Fig. 4A–C,E). Specifically, a low dose of GSK-J4 is efficient at killing leukemic blasts from four distinct TAL1-positive T-ALL patients without inducing apoptosis in TAL1-negative blasts (Fig. 4B). The toxic effect of GSK-J4 observed at higher concentrations in blasts from TAL1-negative T-ALL patients (Fig. 4A) may be due to the inhibitory effect of this drug on another H3K27 demethylase (JMJD3, also called KDM6B) or, to a lesser extent, on the H3K4 demethylases KDM5B and KDM5C (Kruidenier et al. 2012, 2014).
Figure 4.
The H3K27 demethylase inhibitor GSK-J4 represses TAL1/UTX target genes and selectively kills TAL1-positive T-ALL cells through UTX inhibition. (A,B) TAL1-positive T-ALL cells are more sensitive to GSK-J4 treatment. (A) Dose-dependent effect of GSK-J4 on the viability of primary blasts from T-ALL patients. Data are presented as mean values of the percentage of CD7-positive Annexin V-negative cells ±SEM. n = 3. The reduction of apoptosis in TAL1-positive T-ALL is statistically significant compared with TAL1-negative T-ALL (e.g., at 5 µM GSK-J4, P-value = 0.016; at 10 µM GSK-J4, P-value = 8.8 × 10−3 by t-test [unpaired, two-tail, nonequal variance]). (B) GSK-J4 (5 µM) decreases growth (top panel) and increases apoptosis (bottom panel) of primary blasts from TAL1-positive T-ALL patients. (Top panel) Viable cell numbers after GSK-J4 treatment relative to viable cell numbers after vehicle control treatment are reported as mean values ± SEM. n = 4. (Bottom panel) Apoptosis is reported as the percentage of Annexin V-positive cells. Data are shown as mean ± SEM. n = 4. (*) P < 0.05; (***) P < 0.001; (ns) not significant. Representative FACS plots are shown at the right. (C,D) In TAL1-positive T-ALL, GSK-J4 acts predominantly through UTX inhibition. (C) Correlation of changes in gene expression between UTX knockdown (shUTX) and GSK-J4 treatment. RNA-seq was performed with pairs of duplicates for (1) control and UTX knockdown Jurkat cells and (2) vehicle-treated and GSK-J4-treated Jurkat cells. (D) Overexpression (OE) of UTX, but not JMJD3, rescues GSK-J4-mediated apoptosis in TAL1-positive T-ALL. Overexpression of UTX (wild type or enzymatically dead mutant) or JMJD3 (wild type or enzymatically dead mutant) was induced in TAL1-positive Jurkat cells (top) or TAL1-negative DND41 cells (bottom) followed by treatment with GSK-J4 or a vehicle control. (Left panels) Western blot analyses of UTX and JMJD3 overexpression. Representative examples of three biological replicates are shown. (Right panels) The percentage of apoptotic cells (Annexin V-positive) after GSK-J4 treatment is reported relative to the percentage of apoptotic cells after treatment with vehicle control. Data are shown as mean ± SEM. n = 4. (E) GSK-J4 treatment increases the repressive histone mark H3K27me3 globally (as measured by Western blot; representative example of three biological replicates) (left panel) and on specific TAL1/UTX target genes (as measured by ChIP-qPCR) (right panel). Mean values are presented as a fraction of input ± SD. n = 3. (**) P < 0.01; (***) P < 0.001). (F) GSK-J4 treatment leads to down-regulation of the TAL1–UTX transcriptional regulatory network. (Left panel) Volcano plot showing genome-wide changes in gene expression upon GSK-J4 treatment in Jurkat cells as measured by RNA-seq. Adjusted P-value (−log10) versus fold change (log2). The entire set of TAL1/UTX target genes (defined as TAL1/UTX-cobound genes that are significantly down-regulated upon TAL1 knockdown and UTX knockdown) are highlighted in red, with gene names shown for representative examples. (Right panel) Validation of RNA-seq by qRT–PCR for a subset of genes. Results are reported as mean values relative to the internal control ß2M ±SD. n = 3. (**) P < 0.01.
To determine the extent to which the effect of GSK-J4 treatment in TAL1-positive cells is mediated through UTX inhibition, changes in gene expression profile upon GSK-J4 treatment were measured by RNA-seq and compared with changes in gene expression after UTX knockdown. This experiment revealed a strong correlation between the two gene expression signatures (Fig. 4C), suggesting that, in TAL1-positive cells, GSK-J4 acts mainly through the inhibition of UTX. Consistent with this, overexpression of wild-type UTX (but not its enzymatically dead mutant) is sufficient to prevent GSK-J4-mediated induction of apoptosis in TAL1-positive T-ALL cells, whereas overexpression of JMJD3 is unable to rescue GSK-J4-induced apoptosis (Fig. 4D). Therefore, the induction of apoptosis by GSK-J4 in the TAL1-positive T-ALL subtype is mediated through inhibition of UTX demethylase activity, a result that is consistent with our finding that TAL1 interacts with UTX but not JMJD3 (Fig. 1B). Importantly, GSK-J4 treatment also leads to an increase in the repressive H3K27me3 mark (Fig. 4E) and results in global repression of the TAL1–UTX gene expression program, with 87% of the TAL1/UTX target genes being down-regulated upon GSK-J4 treatment (Fig. 4F), which confirms that GSK-J4 acts at least partially through reversing TAL1-mediated aberrant gene activation. Collectively, these results indicate that, in the TAL1-positive subtype of T-ALL, leukemia is maintained through the TAL1–UTX axis, which confers on these leukemic cells a disproportionate sensitivity to the demethylase inhibitor GSK-J4. In this regard, it is interesting to note that the leukemic blasts that we used are resistant to dexamethasone (Supplemental Fig. 4F), a glucocorticoid that is currently used as a treatment for T-ALL (Piovan et al. 2013). Thus, the disproportionate sensitivity to GSK-J4 may provide a therapeutic window to eliminate TAL1-positive leukemic cells, including dexamethasone-resistant blasts.
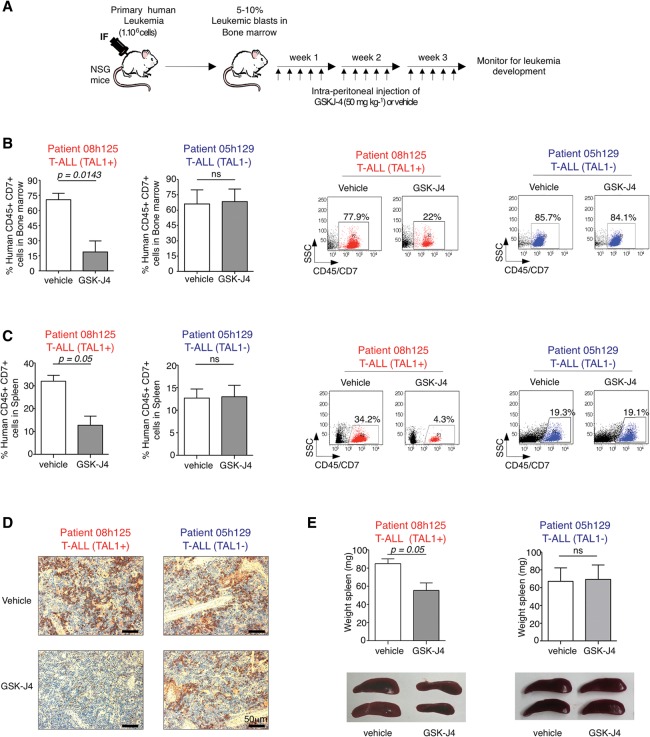
Since GSK-J4 has not yet been tested in vivo as a leukemia treatment, we designed a novel therapeutic approach in which the drug is administered to patient-derived xenograft models of T-ALL. In this approach, immunodeficient NOD-scid IL2Rgnull (NSG) mice are first engrafted with primary blasts from leukemic patients (TAL1-positive and TAL1-negative) and then treated by intraperitoneal injection of 50 mg kg−1 GSK-J4 for 3 consecutive weeks (Fig. 5A). GSK-J4 treatment of TAL1-positive T-ALL-derived xenografted mice results in a dramatic decrease in the percentage of human leukemic blasts (CD45+ CD7+) in the bone marrow (BM) (Fig. 5B) while strongly reducing infiltration of human leukemic cells in the spleen (Fig. 5C,D). Furthermore, GSK-J4 significantly attenuates leukemia-induced splenomegaly of TAL1-positive T-ALL xenografted mice (Fig. 5E). In contrast, GSK-J4 has no effect on TAL1-negative patient-derived xenografts (Fig. 5B–E), providing in vivo validation of the drug selectivity for TAL1-positive T-ALL. Our finding in cell culture that the effect of GSK-J4 is mediated through UTX inhibition in TAL1-positive T-ALL (Fig. 4C,D) strongly suggests that GSK-J4 also works through UTX inhibition in vivo. However, we cannot exclude that, in the context of systemic intraperitoneal injection, GSK-J4 may also inhibit other demethylases, including JMJD3/KDM6B or, to a lesser extent, KDM5B and KDM5C (Kruidenier et al. 2012, 2014). Importantly, GSK-J4 treatment is well tolerated in mice, with no weight loss and no adverse effect detected in the intestine, spleen, liver, kidney or the hematopoietic system (Supplemental Fig. 5). Therefore, GSK-J4 is a novel, highly promising epigenetic therapy against TAL1-positive T-ALL.
Figure 5.
The H3K27 demethylase inhibitor GSK-J4 is efficient in vivo against patient-derived xenotransplant models of TAL1-positive T-ALL. (A) Experimental strategy. NSG mice were transplanted by intrafemoral (IF) injection of primary human T-ALL cells. When the engraftment level reached 5%–10% of human leukemic blasts in the BM, mice were injected intraperitoneally with GSK-J4 or a vehicle control. (B–D) In vivo administration of GSK-J4 strongly reduces the percentage of TAL1-positive human leukemic blasts in the BM (B) and spleen (C,D) but has no visible effect on TAL1-negative T-ALL transplanted mice. (B,C) The percentages of human CD45+ CD7+ leukemic blasts are presented as mean values ± SEM. (ns) Not significant (P ≥ 0.5); four mice per group. Representative FACS plots are shown at the right. (D) Immunohistochemistry using a human CD45 Ab on spleen histological sections. The results are representative of three biological replicates. (E) In vivo administration of GSK-J4 significantly reduces leukemia-induced splenomegaly in TAL1-positive but not TAL1-negative T-ALL transplanted mice. Spleen weights are presented as mean values ± SEM. (ns) Not significant (P ≥ 0.5); four mice per group.
Discussion
T-ALL is a heterogeneous disease comprising several molecular subgroups characterized by the aberrant expression of distinct oncogenic transcription factors, unique gene expression signatures, and different prognoses (Van Vlierberghe and Ferrando 2012). While the existence of specific molecular subtypes of T-ALL has long been established (Ferrando et al. 2002), to date, there has been no description of functional differences between them, and therefore therapeutic strategies (both clinical [van Grotel et al. 2006] and preclinical [Palomero and Ferrando 2009]) are applied uniformly across subtypes, leading to variable responses between patients coupled with high toxicity (Pui et al. 2008; Liu et al. 2011).
Here we describe for the first time a functional difference between the TAL1-positive and TAL1-negative subtypes of T-ALL. Specifically, we found that, in the TAL1-positive T-ALL subtype, UTX promotes cell growth and is necessary for leukemia maintenance (both in vitro and in vivo) through its ability to coactivate genes targeted by the oncogenic transcription factor TAL1. Conversely, in the TAL1-negative subgroups of T-ALL, even though UTX is expressed, it does not participate in leukemia maintenance (Fig. 6). Furthermore, we showed that the demethylase activity of UTX is essential for leukemia maintenance in TAL1-positive but not TAL1-negative T-ALL. Taken together, these results have important therapeutic implications, as they point toward the need for personalized therapies to treat distinct subtypes of T-ALL. Consistent with this, we showed that pharmacological inhibition of UTX with the H3K27 demethylase inhibitor GSK-J4 selectively ablates TAL1-expressing human leukemic blasts in both cell culture and xenograft models of patient-derived leukemia, providing proof of principle that subtype-specific therapy in T-ALL is not only desirable but also feasible (Fig. 6). This is all the more important, as TAL1-positive T-ALL patients generally have a poor prognosis with current treatments (Ferrando et al. 2002), and TAL1-positive leukemic cells appear to resist treatment with other recently developed anti-leukemia drugs, including the Brd4 inhibitor JQ1 (Zuber et al. 2011), the DOT1L inhibitor EPZ004777 (Daigle et al. 2011), and the BCL2 inhibitor ABT199 (Peirs et al. 2014). Importantly, GSK-J4 is well tolerated in vivo at doses that are therapeutically relevant, further supporting the clinical potential of this class of inhibitory molecules for selective epigenetic therapies against TAL1-positive T-ALL. Along those lines, a number of studies have noted that, even though UTX and JMJD3 are required for development and differentiation, these functions are largely independent of their demethylase activity (Miller et al. 2010; Shpargel et al. 2012; Wang et al. 2012; Thieme et al. 2013). Furthermore, transgenic mice with a homozygous knock-in of an enzymatically dead UTX mutant are viable and fertile (C Wang and K Ge, unpubl.), confirming that UTX enzymatic activity is dispensable for mouse development. Thus, transient inhibition of H3K27 demethylase activity in vivo would be expected to present limited long-term secondary effects.
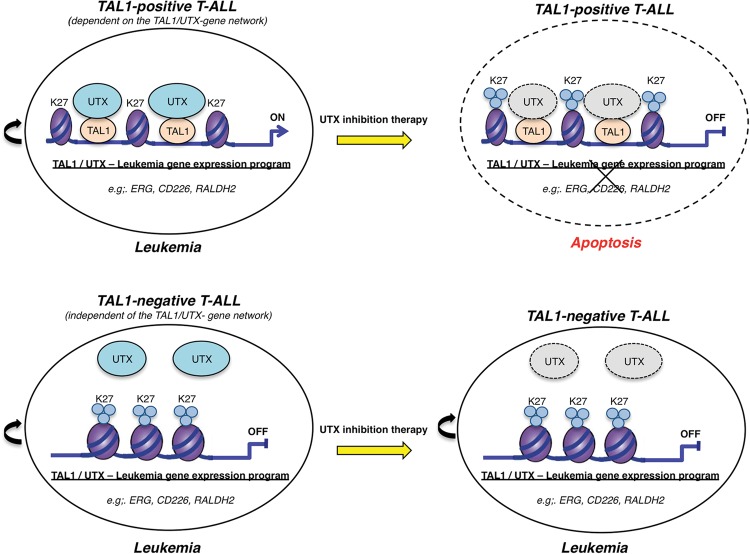
Figure 6.
Model of UTX inhibition as a selective therapy in T-ALL. In TAL1-positive T-ALL, leukemia is maintained through a failure to down-regulate the oncogenic transcription factor TAL1 that in turn tethers the H3K27me3 demethylase UTX to genomic sites that should normally be silenced. Aberrant maintenance of an open chromatin configuration at these sites through active removal of the repressive histone mark H3K27me3 permits the establishment of a TAL1-mediated leukemic gene expression program. As such, in the TAL1-positive molecular subtype of T-ALL, UTX is a major pro-oncogenic cofactor, and a therapy based on UTX inhibition is efficient at eliminating leukemic blasts through down-regulation of the TAL1 leukemic gene expression program. On the other hand, in TAL1-negative T-ALL, leukemia is maintained independently of UTX, which explains that, even though UTX is expressed in these cells, inhibition of its enzymatic activity is not efficient as a therapy to eliminate TAL1-negative leukemic blasts. Thus, UTX inhibition may provide a useful therapeutic strategy for TAL1-positive (but not TAL1-negative) T-ALL patients.
The disproportionate sensitivity of TAL1-expressing T-ALL cells to variations in UTX activity, together with the selectivity and efficacy of GSK-J4 to eliminate TAL1-positive blasts while preserving nonleukemic cells in vivo, supports the concept of “epigenetic vulnerability,” according to which cancer cells are highly dependent on a specific molecular axis for their survival (e.g., the TAL1–UTX axis for TAL1-positive T-ALL), while normal cells have several redundant mechanisms to attenuate or bypass external perturbations of their regulatory pathways (Dawson and Kouzarides 2012). While we identified the TAL1–UTX axis as the Achilles’ heel of the TAL1-positive T-ALL subtype, it remains to be determined whether alternate axes of epigenetic vulnerability exist in other T-ALL subtypes. Furthermore, since UTX is part of a COMPASS-like protein complex that also contains the H3K4 methyltransferases MLL3/KMT2C and MLL4/KMT2D (Shilatifard 2012), one could envisage that these enzymes may also be involved in TAL1 leukemic function, a possibility that remains to be investigated.
Histone-modifying enzymes play complex roles in cancer as either oncogenes or tumor suppressors, depending on the cellular context (Dawson and Kouzarides 2012; Ezponda and Licht 2014). For instance, the H3K27 methyltransferase EZH2 is oncogenic in a variety of cancers, including prostate and lung, but acts as a tumor suppressor in others, such as myeloid malignancies and T-ALL (Simon et al. 2012; Ezponda and Licht 2014). In the same way, UTX has been shown to act as an oncogene in breast cancer (Kim et al. 2014) while playing a tumor suppressor role in solid cancers (such as bladder) and hematological malignancies (including T-ALL) (Van der Meulen et al. 2014). Surprisingly, our data have revealed yet another level of complexity, as we show for the first time that the same cofactor can play opposing roles as either a pro-oncogene or a protumor suppressor within the same cancer type. Indeed, our results indicate that UTX is required for the expansion of blasts in TAL1-positive T-ALL, whereas, in TAL1-negative T-ALL, UTX overexpression leads to a decrease in cell growth that is consistent with its previously described role as a tumor suppressor (Ntziachristos et al. 2014; Van der Meulen et al. 2015). While we showed that the oncogenic function of UTX in TAL1-positive T-ALL is mediated by TAL1-directed activation of a leukemic program, the mechanism of UTX function as a tumor suppressor in TAL1-negative subtypes of T-ALL remains to be elucidated.
Recently, UTX mutations have been identified in a small subset of T-ALL patients (De Keersmaecker et al. 2013; Ntziachristos et al. 2014; Van der Meulen et al. 2015). Interestingly, the described mutations are limited to TAL1-negative T-ALL patients (De Keersmaecker et al. 2013; Van der Meulen et al. 2015). Furthermore, we note that the presence of UTX mutations is not necessarily mutually exclusive with a pro-oncogenic function for UTX in TAL1-positive T-ALL. Indeed, our results show that the presence of a heterozygous inactivating mutation of UTX in the TAL1-positive T-ALL cell line PF-382 (van Haaften et al. 2009; Van der Meulen et al. 2015) does not eliminate the pro-oncogenic, TAL1 coactivator function of UTX expressed from the wild-type allele (Supplemental Fig. 6). Therefore, the presence of UTX mutations does not preclude a pro-oncogenic activity for this cofactor in TAL1-expressing T-ALL as long as the mutations do not result in the complete absence or complete inactivation of UTX demethylase activity.
In summary, using a mechanistic-based approach, we identified UTX as a novel cofactor that mediates the leukemic activity of TAL1 and showed that, by targeting the oncogenic transcription factor TAL1 through its cofactor UTX, it is possible to selectively kill leukemic cells of the particular subtype of T-ALL that expresses TAL1. Importantly, this finding was validated in vivo, showing for the first time that a UTX inhibitor can be used systemically for the treatment of leukemia. Thus, our study has given rise to the development of a promising therapeutic approach to selectively treat a specific molecular subtype of T-ALL, paving the way for personalized medicine in T-cell acute leukemia.
Materials and methods
Cell culture, UTX knockdown, and in vitro drug treatment
Human T-ALL cell lines were purchased from American Type Culture Collection, Deutsche Sammlung von Mikroorganismen und Zellkulturen, or Invitrogen (Supplemental Fig. 2A) and cultured in RPMI 1640 supplemented with 10% FBS and 100 U/mL penicillin and 100 mg/mL streptomycin. All cell lines tested negative for mycoplasma. Primary human leukemic T-ALL blasts (Supplemental Fig. 2B) were obtained from the Quebec Leukemia Cell Bank (Simon et al. 2012) and the Pflumio laboratory (Armstrong et al. 2009; Gerby et al. 2011) with informed consent from patients or their legal guardians and according to protocols approved by the Research Ethics Board of all participating institutions. All analyses were approved by The Ottawa Hospital Research Ethics Board. Primary T-ALL cells were cultured on MS5 stromal cells expressing the NOTCH ligand Delta-like-1 (DL1) as previously described (Armstrong et al. 2009). Briefly, T-ALL cells were plated in reconstituted α minimum essential medium with 10% fetal calf serum (StemCell Technologies, 06450) and 10% human AB serum (Sigma-Aldrich, H4522) supplemented with 50 ng/mL SCF (Prospecbio, CYT-255), 20 ng/mL Flt3-L (Miltenyibiotec, 130-096-474), 20 nM insulin (Sigma-Aldrich, I9278), 10 ng/mL IL-7 (Miltenyibiotec, 130-095-367), and 100 U/mL penicillin and 100 mg/mL streptomycin. Lentiviruses expressing an anti-UTX shRNA (5′-GCCTCAGGATGCCATTAAA-3′) or a control shRNA (5′-GCCAGCCAATCCGTTGAGT-3′) were prepared as previously described (Palii et al. 2011a) and used to infect T-ALL cell lines and primary cells. T-ALL cell lines were infected by spinoculation (800 g for 30 min) at a multiplicity of infection (MOI) of 50 in the presence of 8 μg/mL hexadimethrine bromide (Polybrene, Sigma). The infection was performed twice at 24-h intervals, and the cells were harvested 72 h after the first infection or at the indicated time points for RNA extraction and phenotypic analyses. A similar protocol was used to infect primary T-ALL cells (removed from their MS5-DL1 layer) except that the cells were infected three times at an MOI of 100.
GSK-J4 was obtained initially from the Structural Genomics Consortium and then from Cayman Chemicals (no. 12073) and used at the concentrations indicated. T-ALL cell lines were treated with GSK-J4 for 48 h. Primary T-ALL cells were removed from the MS5-DL1 layer and plated on Matrigel (Corning) followed by 72 h of treatment with GSK-J4 or a vehicle control (DMSO).
UTX and JMJD3 overexpression and drug rescue experiment
Lentiviral particles expressing UTX or an enzymatically dead UTX mutant (H1146A and E1148) (Wang et al. 2012) were prepared as previously described (Palii et al. 2011a). Retroviral particles expressing JMJD3 (Addgene plasmid no. 21212) or an enzymatically dead JMJD3 mutant (H1390A) (Addgene plasmid no. 21214) (Sen et al. 2008) were generated in 293-GP cells using the Retro-X universal packaging system with the Ampho envelope protein vector (Clontech). For the drug rescue experiment, cells were infected with lentiviruses or retroviruses expressing UTX or JMJD3 by spinoculation twice at 24-h intervals prior to GSK-J4 treatment for 48 h (as described above).
Inducible knockdown of UTX in Jurkat cells
Two Jurkat cell lines stably expressing doxycycline (Dox)-dependent shRNA sequences targeting UTX (5′-GCCTCAGGATGCCATTAAA-3′ for shUTX#3 and 5′-GCCAAAGGACAAGTTGAAT-3′ for shUTX#14) were established from TetR-expressing Jurkat cells as previously described (Demers et al. 2007) and maintained in RPMI 1640 medium supplemented with 10% (v/v) Tet-free FBS, 100 U/mL penicillin and 100 mg/mL streptomycin, 20 µg/mL blasticidin, and 1 mg/mL G418. The knockdown of UTX was induced by incubation with Dox at 5 μg/mL for 72 h.
Drug treatment of patient-derived xenograft models of T-ALL
NSG female mice (8–10 wk of age) were purchased from The Jackson Laboratory and maintained under sterile conditions at the University of Ottawa Animal Care Facility. Mice were irradiated at 3 Gy and anesthetized with a solution of 10% ketamine and 5% xylasine in PBS. Leukemic blasts from patients (Supplemental Fig. 2B) were expanded in NSG mice by transplanting 0.5 × 106 to 2 × 106 cells via intrafemoral injection as previously described (Armstrong et al. 2009). Engraftment of human leukemic blasts in the BM was assessed by FACS staining for human CD45 and human CD7 (Gerby et al. 2011) using phycoerythrin (PE)-conjugated CD45 Ab (eBioscience, 12-0459-42) and allophycocyanin (APC)-conjugated CD7 Ab (eBioscience, 8017-0078-12) on a Becton Dickinson (BD) LSRFortessa system. Results were analyzed using BD FACSDiva. Mice were euthanized when the engraftment of leukemic blasts was >90%, and BM and spleen cells were harvested.
For drug treatment experiments, NSG mice were injected with 1 × 106 leukemic blasts, and the BM was analyzed weekly. When the percentage of human CD45+ CD7+ blasts reached 5%–10% in the BM, mice were randomized and injected intraperitoneally with either vehicle or GSK-J4 (50 mg kg−1 diluted in 10:90 DMSO:20% captisol) 5 d a week for 3 wk. Mice were euthanized, the spleen and BM were harvested, spleens were weighed, and the percentage of human CD45+ CD7+ leukemic blasts in the BM and spleen was determined by FACS. Statistical analyses were performed using GraphPad Prism software (version 4.0). A nonparametric, one-sided Mann-Whitney test was used to assess significance. To assess drug toxicity, nonengrafted NSG mice were injected with GSK-J4 following the same protocol. The University of Ottawa Animal Care and Veterinary Service Committee approved all mouse procedures.
Nuclear extraction, immunoprecipitation, and protein identification by MS
Nuclear extraction, immunoprecipitation, and MS were performed essentially as described (Chaturvedi et al. 2012). Briefly, Ab cross-linked beads were incubated with a precleared NE overnight at 4°C with rotation followed by two washes with 1 M immunoprecipitation buffer (25 mM Tris at pH 7.9, 5 mM MgCl2, 10% [v/v] glycerol, 0.1 % [v/v] NP40, 1 M KCl, 0.3 mM DTT, protease inhibitors), and two washes with 150 mM immunoprecipitation buffer (25 mM Tris at pH 7.9, 5 mM MgCl2, 10% [v/v] glycerol, 0.1 % [v/v] NP40, 150 mM KCl, 0.3 mM DTT, protease inhibitors). Bound proteins were eluted with 5% acetic acid, dried by speedvac, and resolved by SDS-PAGE. Proteins were then identified by Western blot or MS. For MS identification, silver-stained proteins were excised from the gel, digested with trypsin, and analyzed by liquid chromatography (LC)-tandem MS (MS/MS) on a linear trap quadrupole (LTQ)-Orbitrap XL mass spectrometer with a nanospray source and Surveyor high-performance liquid chromatography (HPLC). Data analysis was performed using the Trans-Proteomics Pipeline (TPP) (Deutsch et al. 2010) with SEQUEST and the International Protein Index (IPI) human database version 3.72. Data were also converted to the Ensembl protein human database. A probability of identification for each protein was determined using the ProteinProphet algorithm (Nesvizhskii et al. 2003) integrated in the TPP. We only report proteins with a probability of identification >0.9. Proteins identified in a mock immunoprecipitation performed in parallel were subtracted from the list. The TAL1 immunoprecipitation MS experiment was performed twice, and we report results of both experiments in Supplemental Table S1. For immunoprecipitation experiments followed by Western blot analysis, we added 100 µg/mL ethidium bromide to the NE to prevent nucleic acid-mediated protein interactions.
Histone extraction was performed as previously described (Shechter et al. 2007).
UTX and TAL1 ChIP and high-throughput sequencing
UTX ChIP was performed from Jurkat cells as previously described (Palii et al. 2011b) using the UTX antibodies described below. For quantitative PCR (qPCR), chromatin-immunoprecipitated DNA was quantified by real-time qPCR with SYBR Green using a standard curve generated with genomic DNA and was normalized by dividing the amount of the corresponding target in the input fraction. For high-throughput sequencing, UTX chromatin-immunoprecipitated DNA was amplified using the TrueSeq DNA kit (Illumina), and paired-end sequencing was performed on an Illumina HiSeq 2000. Reads of 50 base pairs (bp) were mapped to the NCBI build 37.1 (hg19) human reference genome using BWA (version 0.6.2), allowing for a maximum of two mismatches. Using this method, we obtained 122.5 million unique reads. To detect regions of enrichment, we used model-based analysis of ChIP combined with massively parallel sequencing (MACS) version 2.0.10 using default values with a mock ChIP as a background and a P-value threshold for peak significance of <1 × 10−10. At this P-value cutoff, 17,396 UTX peaks were identified genome-wide.
For TAL1 genomic binding, we used our previously published TAL1 ChIP-seq raw data from Jurkat cells (Gene Expression Omnibus accession no. GSE25000) (Palii et al. 2011b) that were reanalyzed using the same parameters as the UTX ChIP-seq. At a P-value cutoff of <1 × 10−10, we identified 2678 TAL1 peaks genome-wide from 7.6 million unique reads. Notably, we previously published an analysis of this TAL1 ChIP-seq data set based on the rate at which background signal was converted to foreground signal, demonstrating that this number of reads provides sufficient sequencing depth for genome coverage (Palii et al. 2011b). Furthermore, this number of TAL1 peaks in Jurkat cells is in line with TAL1 ChIP-seq data published by others (Sanda et al. 2012).
H3K27me3 ChIP and high-throughput sequencing
Histone H3K27me3 ChIP was performed in Jurkat cells with and without TAL1 knockdown using a native ChIP protocol (Brand et al. 2008) with the H3K27me3 Ab from Millipore (catalog no. 07-449). For qPCR, chromatin-immunoprecipitated DNA was quantified by real-time qPCR with SYBR Green using a standard curve generated with genomic DNA and was normalized by dividing the amount of the corresponding target in the input fraction. For high-throughput sequencing, chromatin-immunoprecipitated DNA was amplified using the KAPA ChIP library kit (KAPA Biosystems), and paired-end sequencing was performed on an Illumina HiSeq 2000. Paired 50-bp reads were mapped to the NCBI build 37.1 (hg19) human reference genome using Bowtie2 (version 2.2.4), resulting in 81,706,875 and 72,791,410 unique mapped fragments from wild-type and TAL1-knockdown Jurkat cells, respectively.
For H3K27me3 ChIP-seq in DND41 cells, we used the publically available data set from ENCODE (Gene Expression Omnibus accession no. GSM1003521) (The ENCODE Project Consortium 2012) generated using the same H3K27me3 antibodies that we used in Jurkat cells (Millipore, catalog no. 07-449). DND41 H3K27me3 ChIP-seq reads were downloaded from the University of California at Santa Cruz Genome Browser and mapped to NCBI build 37.1 using the same parameters as were used for the H3K27me3 reads from Jurkat, resulting in 56,918,481 unique mapped reads.
Global analysis of H3K27me3 at TAL1/UTX-cobound sites in Jurkat and DND41 cells
H3K27me3 profiles were generated using the BEDTools suite (version 2.17.0) (Quinlan and Hall 2010) to count mapped ChIP-seq reads in extended windows around peaks. The center points of TAL1 ChIP-seq peaks overlapping with UTX ChIP-seq peaks were calculated, and BEDTools slopbed, makewindows, and multicov tools were used, respectively, to calculate extended regions around each peak, divide the extended regions into windows, and count H3K27me3 ChIP-seq reads mapping to each window. To obtain H3K27me3 profiles for non-TAL1/UTX regions, the BEDTools shuffle tool was used to move the TAL1–UTX peaks to random genomic locations, from which H3K27me3 reads were counted in the same way. The H3K27me3 read counts were loaded into R (version 3.1.2) and scaled to account for differences in library size. The mean read depth was calculated across corresponding windows around each peak and background region and used to plot profiles as illustrated. This analysis is shown in Figure 2, F and G.
RNA-seq analyses
RNA was isolated from Jurkat cells using the RNeasy mini extraction kit (Qiagen), including a DNase I digestion step (1) before and after TAL1 knockdown (72-h time point), (2) before and after UTX knockdown (72-h time point), and (3) before and after GSK-J4 treatment (72-h time point), with two biological replicates per library. Libraries were constructed using the TruSeq mRNA enrich stranded RNA library kit (Illumina), and paired-end sequencing was performed on an Illumina HiSeq 2000. Reads were aligned to the NCBI build 37.1 (hg19) genome reference sequence using TopHat2 (version 2.0.12) (Kim et al. 2013), guided by the Ensembl release 75 transcript model, and reads were assigned to transcripts using HTseq-count (version 0.6.1p1). Differential gene expression analyses were performed using DESeq2 (Love et al. 2014), calculating size factors and dispersion estimates separately for each set of control/treatment replicates. Differentially expressed genes were identified using an adjusted P-value (Benjamini-Hochberg) of 0.05.
Data deposition
Data are deposited in Gene Expression Omnibus under the following accessions: GSE64832 (UTX ChIP-seq), GSE25000 (TAL1 ChIP-seq), and GSE72300 (H3K27me3 ChIP-seq and RNA-seq for TAL1 knockdown, UTX knockdown, and GSK-J4).
Supplementary Material
Acknowledgments
We thank Achim Leutz (Max-Delbruck-Center for Molecular Medicine, Germany) for the pcDNA3 plasmid expressing human TAL1, Paul A. Khavari (Stanford University, USA) for JMJD3 plasmids, and Cheryl Arrowsmith (Structural Genomics Consortium; http://www.thesgc.org) for initially providing GSK-J4. We also thank Lawrence Puente (Ottawa Hospital Research Institute Proteomics Facility; http://www.ohri.ca/proteomics) and Gareth Palidwor (Ottawa Bioinformatics Core Facility; http://www.ohri.ca/bioinformatics) for MS and bioinformatic analyses. High-throughput DNA sequencing was performed at the Molecular Biology and Functional Genomics Core Facility of the Institut de Recherche Clinique de Montreal (IRCM, Canada). This project was funded with grants from the Canadian Institutes of Health Research (MOP-82813 to M.B., and MOP-77778 to F.J.D.). C.W. and K.G. were supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health (USA). The Quebec Leukemia Cell Bank (http://www.bclq.org) is supported by a grant from the Cancer Research Network of Fonds de la Recherche du Quebec en Sante (FRSQ). The Ottawa Bioinformatics Core Facility is supported in part by a Government of Ontario Ministry of Research and Innovation (MRI) grant.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.276790.115.
References
- Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K. 2007. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449: 731–734. [DOI] [PubMed] [Google Scholar]
- Armstrong F, Brunet de la Grange P, Gerby B, Rouyez MC, Calvo J, Fontenay M, Boissel N, Dombret H, Baruchel A, Landman-Parker J, et al. 2009. NOTCH is a key regulator of human T-cell acute leukemia initiating cell activity. Blood 113: 1730–1740. [DOI] [PubMed] [Google Scholar]
- Brand M, Rampalli S, Chaturvedi CP, Dilworth FJ. 2008. Analysis of epigenetic modifications of chromatin at specific gene loci by native chromatin immunoprecipitation of nucleosomes isolated using hydroxyapatite chromatography. Nat Protoc 3: 398–409. [DOI] [PubMed] [Google Scholar]
- Chaturvedi CP, Somasundaram B, Singh K, Carpenedo RL, Stanford WL, Dilworth FJ, Brand M. 2012. Maintenance of gene silencing by the coordinate action of the H3K9 methyltransferase G9a/KMT1C and the H3K4 demethylase Jarid1a/KDM5A. Proc Natl Acad Sci 109: 18845–18850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YW, Hong T, Hong S, Guo H, Yu H, Kim D, Guszczynski T, Dressler GR, Copeland TD, Kalkum M, et al. 2007. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem 282: 20395–20406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condorelli GL, Facchiano F, Valtieri M, Proietti E, Vitelli L, Lulli V, Huebner K, Peschle C, Croce CM. 1996. T-cell-directed TAL-1 expression induces T-cell malignancies in transgenic mice. Cancer Res 56: 5113–5119. [PubMed] [Google Scholar]
- Daigle SR, Olhava EJ, Therkelsen CA, Majer CR, Sneeringer CJ, Song J, Johnston LD, Scott MP, Smith JJ, Xiao Y, et al. 2011. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell 20: 53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Kouzarides T. 2012. Cancer epigenetics: from mechanism to therapy. Cell 150: 12–27. [DOI] [PubMed] [Google Scholar]
- De Keersmaecker K, Atak ZK, Li N, Vicente C, Patchett S, Girardi T, Gianfelici V, Geerdens E, Clappier E, Porcu M, et al. 2013. Exome sequencing identifies mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T-cell acute lymphoblastic leukemia. Nat Genet 45: 186–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demers C, Chaturvedi CP, Ranish JA, Juban G, Lai P, Morle F, Aebersold R, Dilworth FJ, Groudine M, Brand M. 2007. Activator-mediated recruitment of the MLL2 methyltransferase complex to the β-globin locus. Mol Cell 27: 573–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch EW, Mendoza L, Shteynberg D, Farrah T, Lam H, Tasman N, Sun Z, Nilsson E, Pratt B, Prazen B, et al. 2010. A guided tour of the Trans-Proteomic Pipeline. Proteomics 10: 1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezponda T, Licht JD. 2014. Molecular pathways: deregulation of histone h3 lysine 27 methylation in cancer-different paths, same destination. Clin Cancer Res 20: 5001–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrando AA, Neuberg DS, Staunton J, Loh ML, Huard C, Raimondi SC, Behm FG, Pui CH, Downing JR, Gilliland DG, et al. 2002. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell 1: 75–87. [DOI] [PubMed] [Google Scholar]
- Gerby B, Clappier E, Armstrong F, Deswarte C, Calvo J, Poglio S, Soulier J, Boissel N, Leblanc T, Baruchel A, et al. 2011. Expression of CD34 and CD7 on human T-cell acute lymphoblastic leukemia discriminates functionally heterogeneous cell populations. Leukemia 25: 1249–1258. [DOI] [PubMed] [Google Scholar]
- Hong S, Cho YW, Yu LR, Yu H, Veenstra TD, Ge K. 2007. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci 104: 18439–18444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Ybarra R, Qiu Y, Bungert J, Huang S. 2009. Transcriptional regulation by TAL1: a link between epigenetic modifications and erythropoiesis. Epigenetics 4: 357–361. [DOI] [PubMed] [Google Scholar]
- Issaeva I, Zonis Y, Rozovskaia T, Orlovsky K, Croce CM, Nakamura T, Mazo A, Eisenbach L, Canaani E. 2007. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol Cell Biol 27: 1889–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelliher MA, Seldin DC, Leder P. 1996. Tal-1 induces T cell acute lymphoblastic leukemia accelerated by casein kinase IIα. EMBO J 15: 5160–5166. [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Sharma A, Dhar SS, Lee SH, Gu B, Chan CH, Lin HK, Lee MG. 2014. UTX and MLL4 coordinately regulate transcriptional programs for cell proliferation and invasiveness in breast cancer cells. Cancer Res 74: 1705–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruidenier L, Chung CW, Cheng Z, Liddle J, Che K, Joberty G, Bantscheff M, Bountra C, Bridges A, Diallo H, et al. 2012. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature 488: 404–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruidenier L, Chung CW, Cheng Z, Liddle J, Che K, Joberty G, Bantscheff M, Bountra C, Bridges A, Diallo H, et al. 2014. Kruidenier, et al. reply. Nature 514: E2. [DOI] [PubMed] [Google Scholar]
- Kusy S, Gerby B, Goardon N, Gault N, Ferri F, Gérard D, Armstrong F, Ballerini P, Cayuela JM, Baruchel A, et al. 2010. NKX3.1 is a direct TAL1 target gene that mediates proliferation of TAL1-expressing human T cell acute lymphoblastic leukemia. J Exp Med 207: 2141–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuyer E, Hoang T. 2004. SCL: from the origin of hematopoiesis to stem cells and leukemia. Exp Hematol 32: 11–24. [DOI] [PubMed] [Google Scholar]
- Lee S, Lee JW, Lee SK. 2012. UTX, a histone H3-lysine 27 demethylase, acts as a critical switch to activate the cardiac developmental program. Dev Cell 22: 25–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Chiang MY, Pear WS. 2011. Critical roles of NOTCH1 in acute T-cell lymphoblastic leukemia. Int J Hematol 94: 118–125. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Mohn SE, Weinmann AS. 2010. Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol Cell 40: 594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. 2003. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75: 4646–4658. [DOI] [PubMed] [Google Scholar]
- Ntziachristos P, Tsirigos A, Welstead GG, Trimarchi T, Bakogianni S, Xu L, Loizou E, Holmfeldt L, Strikoudis A, King B, et al. 2014. Contrasting roles of histone 3 lysine 27 demethylases in acute lymphoblastic leukaemia. Nature 514: 513–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palii CG, Pasha R, Brand M. 2011a. Lentiviral-mediated knockdown during ex vivo erythropoiesis of human hematopoietic stem cells. J Vis Exp 10.3791/2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palii CG, Perez-Iratxeta C, Yao Z, Cao Y, Dai F, Davison J, Atkins H, Allan D, Dilworth FJ, Gentleman R, et al. 2011b. Differential genomic targeting of the transcription factor TAL1 in alternate haematopoietic lineages. EMBO J 30: 494–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomero T, Ferrando A. 2009. Therapeutic targeting of NOTCH1 signaling in T-cell acute lymphoblastic leukemia. Clin Lymphoma Myeloma 9: S205–S210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirs S, Matthijssens F, Goossens S, Van de Walle I, Ruggero K, de Bock CE, Degryse S, Canté-Barrett K, Briot D, Clappier E, et al. 2014. ABT-199 mediated inhibition of BCL-2 as a novel therapeutic strategy in T-cell acute lymphoblastic leukemia. Blood 124: 3738–3747. [DOI] [PubMed] [Google Scholar]
- Piovan E, Yu J, Tosello V, Herranz D, Ambesi-Impiombato A, Da Silva AC, Sanchez-Martin M, Perez-Garcia A, Rigo I, Castillo M, et al. 2013. Direct reversal of glucocorticoid resistance by AKT inhibition in acute lymphoblastic leukemia. Cancer Cell 24: 766–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pui CH, Robison LL, Look AT. 2008. Acute lymphoblastic leukaemia. Lancet 371: 1030–1043. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanda T, Lawton LN, Barrasa MI, Fan ZP, Kohlhammer H, Gutierrez A, Ma W, Tatarek J, Ahn Y, Kelliher MA, et al. 2012. Core transcriptional regulatory circuit controlled by the TAL1 complex in human T cell acute lymphoblastic leukemia. Cancer Cell 22: 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen GL, Webster DE, Barragan DI, Chang HY, Khavari PA. 2008. Control of differentiation in a self-renewing mammalian tissue by the histone demethylase JMJD3. Genes Dev 22: 1865–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shechter D, Dormann HL, Allis CD, Hake SB. 2007. Extraction, purification and analysis of histones. Nat Protoc 2: 1445–1457. [DOI] [PubMed] [Google Scholar]
- Shilatifard A. 2012. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem 81: 65–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpargel KB, Sengoku T, Yokoyama S, Magnuson T. 2012. UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLoS Genet 8: e1002964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon C, Chagraoui J, Krosl J, Gendron P, Wilhelm B, Lemieux S, Boucher G, Chagnon P, Drouin S, Lambert R, et al. 2012. A key role for EZH2 and associated genes in mouse and human adult T-cell acute leukemia. Genes Dev 26: 651–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieme S, Gyarfas T, Richter C, Ozhan G, Fu J, Alexopoulou D, Muders MH, Michalk I, Jakob C, Dahl A, et al. 2013. The histone demethylase UTX regulates stem cell migration and hematopoiesis. Blood 121: 2462–2473. [DOI] [PubMed] [Google Scholar]
- Thoms JA, Birger Y, Foster S, Knezevic K, Kirschenbaum Y, Chandrakanthan V, Jonquieres G, Spensberger D, Wong JW, Oram SH, et al. 2011. ERG promotes T-acute lymphoblastic leukemia and is transcriptionally regulated in leukemic cells by a stem cell enhancer. Blood 117: 7079–7089. [DOI] [PubMed] [Google Scholar]
- Van der Meulen J, Speleman F, Van Vlierberghe P. 2014. The H3K27me3 demethylase UTX in normal development and disease. Epigenetics 9: 658–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Meulen J, Sanghvi V, Mavrakis K, Durinck K, Fang F, Matthijssens F, Rondou P, Rosen M, Pieters T, Vandenberghe P, et al. 2015. The H3K27me3 demethylase UTX is a gender-specific tumor suppressor in T-cell acute lymphoblastic leukemia. Blood 125: 13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Grotel M, Meijerink JP, Beverloo HB, Langerak AW, Buys-Gladdines JG, Schneider P, Poulsen TS, den Boer ML, Horstmann M, Kamps WA, et al. 2006. The outcome of molecular-cytogenetic subgroups in pediatric T-cell acute lymphoblastic leukemia: a retrospective study of patients treated according to DCOG or COALL protocols. Haematologica 91: 1212–1221. [PubMed] [Google Scholar]
- van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, Greenman C, Edkins S, Hardy C, O'Meara S, Teague J, et al. 2009. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet 41: 521–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vlierberghe P, Ferrando A. 2012. The molecular basis of T cell acute lymphoblastic leukemia. J Clin Invest 122: 3398–3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Lee JE, Cho YW, Xiao Y, Jin Q, Liu C, Ge K. 2012. UTX regulates mesoderm differentiation of embryonic stem cells independent of H3K27 demethylase activity. Proc Natl Acad Sci 109: 15324–15329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welstead GG, Creyghton MP, Bilodeau S, Cheng AW, Markoulaki S, Young RA, Jaenisch R. 2012. X-linked H3K27me3 demethylase Utx is required for embryonic development in a sex-specific manner. Proc Natl Acad Sci 109: 13004–13009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. 2011. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478: 524–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.