Young et al. show that AMPK plays critical roles in lineage specification. AMPK−/− embryoid bodies exhibited reduced levels of Tfeb, a master transcriptional regulator of lysosomes, leading to diminished endolysosomal function. The compromised endolysosomal system resulting from AMPK or Tfeb inactivation blunted Wnt signaling, while up-regulating this pathway restored expression of endodermal markers.
Keywords: AMPK, embryonic stem cells, germ layer specification, endoderm, Tfeb, lysosomes, Wnt
Abstract
Faithful execution of developmental programs relies on the acquisition of unique cell identities from pluripotent progenitors, a process governed by combinatorial inputs from numerous signaling cascades that ultimately dictate lineage-specific transcriptional outputs. Despite growing evidence that metabolism is integrated with many molecular networks, how pathways that control energy homeostasis may affect cell fate decisions is largely unknown. Here, we show that AMP-activated protein kinase (AMPK), a central metabolic regulator, plays critical roles in lineage specification. Although AMPK-deficient embryonic stem cells (ESCs) were normal in the pluripotent state, these cells displayed profound defects upon differentiation, failing to generate chimeric embryos and preferentially adopting an ectodermal fate at the expense of the endoderm during embryoid body (EB) formation. AMPK−/− EBs exhibited reduced levels of Tfeb, a master transcriptional regulator of lysosomes, leading to diminished endolysosomal function. Remarkably, genetic loss of Tfeb also yielded endodermal defects, while AMPK-null ESCs overexpressing this transcription factor normalized their differential potential, revealing an intimate connection between Tfeb/lysosomes and germ layer specification. The compromised endolysosomal system resulting from AMPK or Tfeb inactivation blunted Wnt signaling, while up-regulating this pathway restored expression of endodermal markers. Collectively, these results uncover the AMPK pathway as a novel regulator of cell fate determination during differentiation.
During embryogenesis, a series of temporal and spatially regulated cell fate decisions must be precisely executed to ensure proper progression through the developmental program. These changes largely involve pluripotent cells, comprising the bulk of nascent embryos, transitioning to a more restricted, differentiated state. The formation of epiblast and primitive endoderm cells within blastocysts represents an early divergence in cell identity, which is then followed by further specification of the epiblast into three germ layers (the ectoderm, mesoderm, and endoderm), creating the progenitors for all of the diverse lineages found in fully developed organisms (Lanner 2014).
Embryonic stem cells (ESCs) are derived from a very early developmental stage prior to the onset of many of these decision points and therefore possess the capability to differentiate into a multitude of cell types (Evans and Kaufman 1981; Martin 1981; Murry and Keller 2008). This makes them an attractive tool to study the mechanisms underlying the acquisition of distinct cell fates. One particularly useful system that recapitulates several aspects of embryonic development consists of growing ESCs in suspension, which causes them to aggregate into clusters called embryoid bodies (EBs). Cells within EBs spontaneously differentiate, eventually forming separate lineages corresponding to all three germ layers (Leahy et al. 1999). Along with this multilineage specification, dramatic anatomical changes occur, transforming the dense mass of cells into hollow structures surrounded by layers of distinct cell types, highly similar to the process by which the egg cylinder arises from the blastocyst in vivo (Coucouvanis and Martin 1995).
Many studies from both animal models and ESC systems have revealed that cell fate determination, including the formation of the different germ layers from pluripotent cells, is largely orchestrated by the spatial and temporal integration of numerous signaling cascades. Distinct levels and combinations of signals through pathways such as Wnt, Nodal/TGFβ, BMP, and FGF/MAPK direct the exit from pluripotency and govern the induction of lineage-specific transcription factors, which ultimately dictate cell type. For example, high flux through the Wnt and Nodal pathways cooperatively engages master transcriptional regulators of endoderm formation, including Sox17 and GATA4 (Gadue et al. 2006; Grapin-Botton and Constam 2007; Li et al. 2011a). These factors subsequently induce more terminal differentiation makers, such as Hnf4α and albumin, to generate the specialized cells associated with this germ layer (Bossard and Zaret 1998; Kanai-Azuma et al. 2002).
In the past decade, there has been a newfound appreciation for the intricate connections between cellular metabolism and a wide variety of biological processes (Metallo and Vander Heiden 2013). In addition to providing nutrients and raw materials, pathways regulating energy homeostasis also impinge on an array of signaling and transcription factor modules through either direct interactions or the control of metabolite/cofactor levels (Wellen and Thompson 2012). As a result, energy perturbations, either naturally occurring or from environmental insults, can dramatically alter molecular networks and regulate cell proliferation, growth, and survival. Interestingly, early embryonic stages are marked by significant alterations in nutrient status and cellular metabolism, although the importance of these changes is unclear (Chason et al. 2011; Shyh-Chang et al. 2013). Furthermore, while several studies have documented large-scale metabolic reprogramming as cells transition between pluripotent and differentiated states in vitro, a direct role for the pathways mediating these shifts was largely unexplored (Cho et al. 2006; Folmes et al. 2011; Moussaieff et al. 2015).
AMP-activated protein kinase (AMPK) is a heterotrimeric complex consisting of α, β, and γ subunits that detects diminished ATP levels through direct interaction with AMP and ADP, allowing it to acutely sense energy stress (Hardie 2015). Upon nucleotide binding, AMPK is directly phosphorylated by the tumor suppressor LKB1, resulting in AMPK activation and phosphorylation of a number of downstream targets, including important regulators of cell growth, metabolism, and autophagy (Inoki et al. 2003; Gwinn et al. 2008; Egan et al. 2011; Li et al. 2011b). This multifaceted response comprises an important metabolic checkpoint, inhibiting energy-intensive processes while simultaneously promoting those that generate ATP (Mihaylova and Shaw 2011). AMPK is a well-established regulator of metabolic homeostasis in a wide variety of cells and tissues in response to mitochondrial insults, metabolic hormones, and physiological stressors, including exercise. However, the extent to which this pathway functions in other settings marked by metabolic reprogramming, such as during embryonic development, remains unresolved. Although previous studies briefly reported embryonic lethality as a result of combined deletion of either both α or both β subunits, no mechanistic details were provided (Viollet et al. 2009; O'Neill et al. 2011). In addition, whether AMPK directly impacts cellular differentiation and lineage choice is completely unknown.
Here we describe a role for the AMPK pathway in regulating cell fate. Mouse ESCs (mESCs) lacking both AMPK α subunits appeared normal when maintained in the pluripotent state but exhibited severe defects during differentiation—most notably an inability to contribute to chimeras in vivo and a marked germ layer skewing away from endoderm and toward ectoderm during EB formation. AMPK-deficient EBs displayed a dramatic reduction in lysosomal function due to diminished levels of Tfeb, a master transcriptional regulator of lysosomal genes. Genetically inactivating Tfeb also led to endodermal defects, whereas overexpressing Tfeb in AMPK-null ESCs rescued their differentiation potential, highlighting the central role of Tfeb/lysosomes in endodermal specification. Both AMPK and Tfeb mutant EBs showed attenuated canonical Wnt signaling, and augmenting this pathway rescued endodermal gene expression. In summary, these results demonstrate that AMPK, through its regulation of Tfeb and the lysosomal compartment, plays an important role in cell fate determination during differentiation.
Results
Dynamic AMPK signaling during ESC differentiation
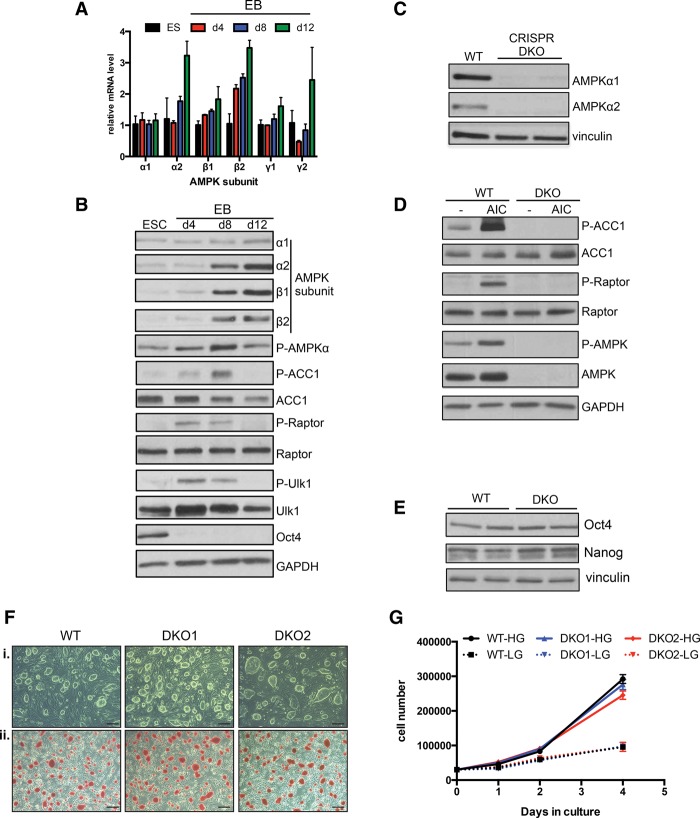
We began by examining the mRNA expression pattern of the subunits that comprise the mature AMPK heterotrimeric complex during an EB differentiation time course. Notably, at least one isoform of each subunit showed significant transcriptional up-regulation as differentiation progressed, and several of these changes were validated at the protein level (Fig. 1A,B). To monitor AMPK pathway activation during this process, we analyzed the phosphorylation status of several direct targets of AMPK, including ACC1, raptor, and ULK1. In order to rule out the possibility that these changes in AMPK signaling were due to limitations in nutrient availability due to medium exhaustion, which can dramatically affect AMPK activity in cell culture, EBs were lysed 1–2 h after a fresh medium change in each condition. Interestingly, each substrate exhibited dynamic phosphorylation, indicative of a wave of AMPK activity that rises from relatively low levels in the pluripotent state and at day 4, peaks at around day 8, and then returns to a basal state by day 12 (Fig. 1B). The decrease in target phosphorylation at late time points occurred despite a steady increase in kinase subunit expression, suggesting a complex interplay between the AMPK pathway and antagonistic signaling events, such as phosphatase up-regulation, as development progresses.
Figure 1.
Dynamic AMPK signaling during EB differentiation and generation and characterization of AMPK double-knockout ESCs. (A) RT-qPCR analysis of AMPK subunits during EB differentiation. γ3 was not detected at any time point. Data are from two independent experiments. Bar graphs depict mean ± SEM. (B) ESCs and differentiating EBs were lysed on the indicated days 1 h after a medium change and subjected to Western blotting with the antibodies listed. (C) Western blot analysis of AMPKα1 and AMPKα2 in wild-type (WT) parental and two independent AMPK double-knockout (DKO) CRISPR clones. (D) Immunoblot on lysates from wild-type and AMPK double-knockout ESCs following vehicle or 1 h of 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR) treatment (0.5 mM). (E) Levels of pluripotency markers Oct4 and Nanog in wild-type and AMPK double-knockout ESCs as determined by Western blot. (F) Bright-field images of wild-type and two AMPK double-knockout ESC lines grown on feeders indicating normal ESC-like morphology (panel i) and equivalent amounts of alkaline phosphatase activity between the different genotypes (panel ii). Bar, 100 µm. (G) Proliferation curves of wild-type and AMPK double-knockout ESCs grown in the absence of feeders in both high glucose (HG; 25 mM) and low glucose (LG; 2.5 mM). n = 2 samples per condition.
During EB differentiation, aggregates of cells form dense clusters that ultimately undergo cavitation to generate distinct lineages surrounding a hollow interior (Coucouvanis and Martin 1995). We wondered whether the unique pattern of AMPK activity described above was localized to particular anatomical regions of EBs. For example, prior to cavitation, cells in the interior may have limited access to nutrients, resulting in increased AMPK activity. However, phospho-ACC1 immunohistochemistry (IHC) revealed strong signal throughout densely packed EBs (Supplemental Fig. 1A, panels i–iii). In addition, well-differentiated EBs displayed highly variable staining across diverse structures and cell types, suggesting that AMPK signaling is not necessarily limited to specific lineages (Supplemental Fig. 1A, panels iv–vi). Together, these results indicate that the AMPK pathway is dynamically regulated during ESC differentiation irrespective of cell culture nutrients.
Generation and characterization of AMPKα1−/−;AMPKα2−/− double-knockout ESCs
To begin to address whether AMPK plays an important role in development, we set out to generate AMPK-deficient ESCs using the CRISPR/Cas9 system. Separate guide RNAs targeting the two genes encoding the α catalytic subunits of AMPK were introduced into the v26.2 ESC line, and we were able to isolate several independent clones that lacked expression of both AMPK α1 and α2 (Fig. 1C; Supplemental Fig. 1B,C). Treating these clones (hereafter referred to as AMPK double-knockout or double-knockout cells) with the AMP-mimetic 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR) failed to induce phosphorylation of AMPK targets, confirming that they had become functionally deficient with respect to the AMPK pathway (Fig. 1D). Initial characterization of AMPK double-knockout ESCs did not reveal any overt differences from their wild-type counterparts. The cells retained normal ESC-like morphology when passaged with and without feeders and displayed equivalent levels of pluripotency-related alkaline phosphatase staining as well as pluripotency markers Oct4 and Nanog. (Fig. 1E,F; data not shown). Furthermore, cell proliferation was unaffected by AMPK deletion (Fig. 1G). In other contexts, AMPK-dependent phenotypes are often exacerbated when cells are placed into energy stress conditions, such as glucose deprivation (Shaw et al. 2004). However, while lowering the glucose concentration 10-fold led to a reduction in cell division, both wild-type and AMPK double-knockout cells responded similarly (Fig. 1G). Finally, culturing both genotypes of cells in the absence of glucose for 2 d failed to unmask AMPK-dependent effects, as both populations displayed equivalent levels of cell death (Supplemental Fig. 1D). Collectively, these data suggest that the AMPK pathway plays a relatively minor role in the basal ESC state or their proliferative response to glucose deprivation.
Impaired differentiation of AMPK double-knockout ESCs
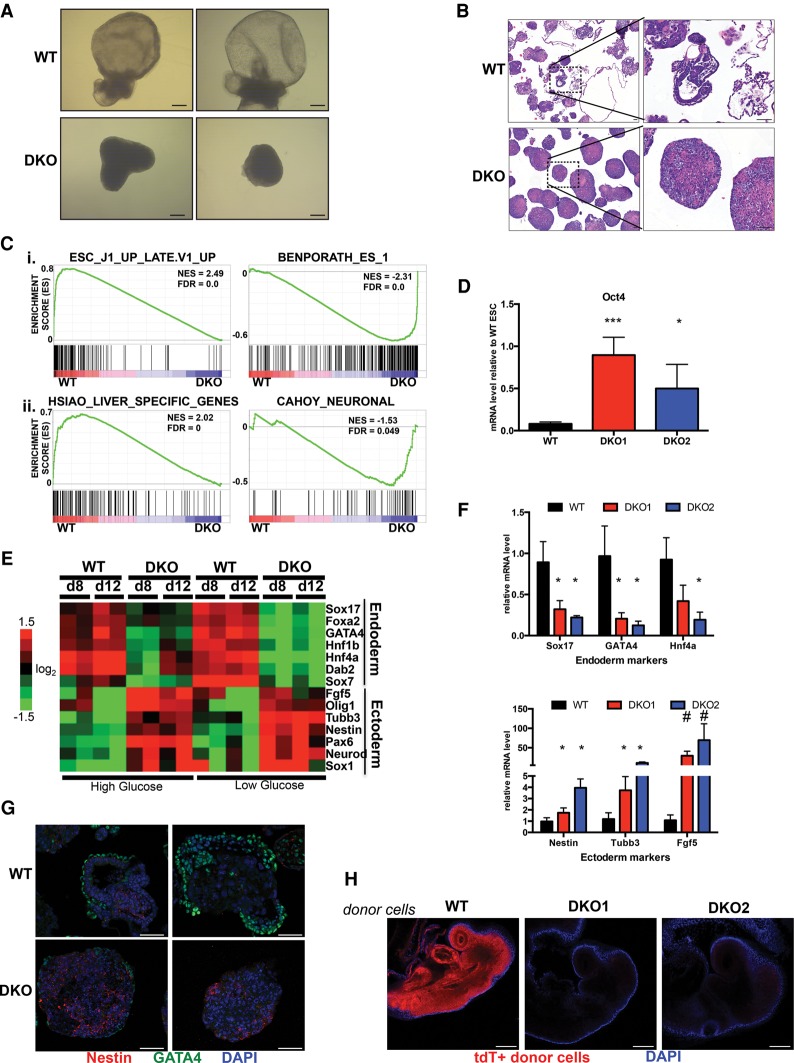
Our results showing increased AMPK signaling during EB formation suggested a potential role for this pathway during cellular differentiation. To test this, we generated EBs from both wild-type and AMPK double-knockout ESCs and began by looking for effects on gross morphology. Cells were grown in both high- and low-glucose conditions to examine how energy stress would affect AMPK-deficient cells. During the first several days, wild-type and double-knockout-derived EBs were indistinguishable from each other (data not shown). However, at mid to late stages of EB differentiation starting at day 8, regardless of glucose concentration, many wild-type structures had formed large internal cavities surrounded by outer layers of cells, a process that corresponds to the creation of the egg cylinder in post-implantation embryos, whereas almost all double-knockout EBs remained as small, dense clusters (Fig. 2A; data not shown). Analyzing fixed sections at both day 8 and day 12 of differentiation revealed an array of structurally diverse wild-type EBs, many of which contained several distinct cell morphologies, suggesting robust multilineage differentiation. In contrast, histological sections of double-knockout-derived EBs predominantly showed tightly packed structures of mostly homogenous cells at both time points and regardless of glucose concentration (Fig. 2B; Supplemental Figs. 2A, 3A).
Figure 2.
Differentiation defects of AMPK double-knockout ESCs. (A) Bright-field images of representative wild-type (WT) and AMPK double-knockout (DKO) EBs differentiated for 12 d in high-glucose conditions. Bar, 100 µm. (B) Hematoxylin and eosin (H&E)-stained sections of wild-type and AMPK double-knockout day 12 EBs in high glucose. Images at the right correspond to boxed sections from the left. Bar, 50 µm. (C) Gene set enrichment analysis (GSEA) plots from high-throughput whole-transcriptome sequencing (mRNA-seq) profiles of wild-type and AMPK double-knockout EBs differentiated for 12 d in high glucose. (Panel i). The top two plots correspond to general differentiation defects of AMPK double-knockout cells. (Panel ii). The bottom plots highlight endoderm (liver) versus ectoderm (neuronal) germ layer skewing in wild-type versus double-knockout EBs, respectively. (NES) Normalized enrichment score; (FDR) false discovery rate. (D) Elevated levels of Oct4 mRNA in day 12 AMPK double-knockout EBs compared with wild type, as determined by RT-qPCR. (***) P < 0.005; (*) P < 0.05, compared with wild type. Relative abundance of Oct4 was normalized to wild-type ESC levels. (E) Heat map depicting relative mRNA expression of several endoderm and ectoderm markers in wild-type and double-knockout EBs at day 8 (d8) and day 12 (d12) of differentiation in high and low glucose. Values were calculated from mRNA-seq data. (F) RT-qPCR analysis of selected endoderm (top) and ectoderm (bottom) markers in high-glucose day 12 EBs derived from either wild-type or AMPK double-knockout cells confirming germ layer skewing between different genotypes. Data are from three independent experiments. Average ± SEM is plotted. (*) P < 0.05; (#) P < 0.005, compared with wild type. (G) Immunofluorescence (IF) on fixed sections of wild-type and AMPK double-knockout samples after 12 d of EB differentiation in high glucose. GATA4 and Nestin served as endoderm and ectoderm markers, respectively. Bar, 50 µm. (H) Representative whole-mount fluorescence images of embryonic day 8.5 (E8.5) embryos that were injected with tdTomato-positive wild-type or AMPK double-knockout ESCs ex vivo at E2.5 prior to blastocyst implantation into recipient mothers. DAPI staining was performed to outline the embryo. Bar, 200 µm. Statistical significance was determined by Student's t-test.
We performed high-throughput whole-transcriptome sequencing (mRNA-seq) on developing wild-type and double-knockout EBs in order to comprehensively characterize their differentiation potential within this assay. Gene set enrichment analysis (GSEA) on expression profiles from both day 8 and day 12 EBs indicated a substantial depletion of general differentiation-associated gene sets in double-knockout samples as well as a strong enrichment of ESC signatures, supporting the notion that AMPK deletion retards the ability of cells to exit the pluripotent state and progress through differentiation (Fig. 2C; Supplemental Fig. 2B). Indeed, Oct4 mRNA levels were elevated in double-knockout versus wild-type EBs (Fig. 2D). Interestingly, other signatures corresponding to the development of specific germ layers appeared uniquely up-regulated in wild-type or double-knockout samples. For example, a gene set specific to the liver, an endoderm-derived tissue, clustered in wild-type EBs, whereas transcripts associated with ectoderm-derived neuronal cell types dominated among double-knockout-specific mRNAs (Fig. 2C; Supplemental Fig. 2B). Plotting the expression of a subset of well-defined endoderm and ectoderm genes further substantiated this germ layer skewing (Fig. 2E). Moreover, by combining data sets from both glucose conditions, we noted even more dramatic changes, especially with regards to endoderm formation, when cells were exposed to glucose deprivation (Fig. 2E). These distinct lineage choices among wild-type and double-knockout cells were validated at day 8 and day 12 in subsequent rounds of EB formation using qPCR of several germ layer-specific genes (Fig. 2F; Supplemental Figs. 2C, 3B). To examine these lineage-specific markers at the protein level, indirect immunofluorescence (IF) analysis of EBs with antibodies against endogenous GATA4 (endoderm) and Nestin (ectoderm) was performed, revealing profound differences in their expression patterns in the wild-type as compared with AMPK double-knockout EBs (Fig. 2G; Supplemental Fig. 3C).
To confirm that these developmental abnormalities resulted from AMPK deletion and were not indirect effects of random selection during clone generation, we took two approaches. First, either wild-type or kinase-dead (KD) versions of AMPK α1 cDNA were reconstituted in our double-knockout lines. Cells expressing wild-type but not KD AMPKa1 cDNA restored AMPK signaling (Supplemental Fig. 4A) and also partially regained the ability to up-regulate endoderm makers during EB formation, whereas effects on ectoderm markers were more variable (Supplemental Fig. 4B). These partial effects suggest that proper differentiation may require physiological regulation of endogenous AMPK.
In addition, we generated a distinct independent line of AMPK double-knockout ESCs by deriving ESCs de novo from intercrosses between UbcCreER; AMPKα1fl/fl ;AMPKα2fl/fl and AMPKα1fl/fl ;AMPKα2fl/fl mice (hereafter UAA). UAA cells were then treated with either ethanol or 4-hydroxytamoxifen (4-OHT) prior to EB differentiation. Only CreER-positive cells incubated with 4-OHT deleted AMPKα (Supplemental Fig. 4C), and, upon EB differentiation, this resulted in the same type of germ layer skewing seen with our CRISPR-derived double knockouts (Supplemental Fig. 4D). Together, these data strongly support that AMPK is genetically required for proper germ layer development during EB differentiation.
ESCs possess the ability to contribute to the development of most embryonic tissues when implanted into early stage blastocysts, resulting in chimeric embryos. To rigorously test the potency of AMPK double-knockout cells, we attempted to generate chimeras using our CRISPR lines. First, both wild-type and double-knockout cells were stably transfected with the mT/mG reporter construct (Muzumdar et al. 2007), creating tdTomato-positive ESCs that allowed us to track their fate in vivo. Remarkably, while wild-type cells widely contributed to developing embryos, we were unable to detect any double-knockout cells in a majority of animals at embryonic day 8.5 (E8.5) (Fig. 2H; Supplemental Fig. 5A). Short-term ex vivo culture after ESC microinjection showed that double-knockout cells successfully implanted into blastocysts, suggesting a defect later in the developmental process (Supplemental Fig. 5B). This failure of AMPK-deficient ESCs to contribute to chimeras further supports the notion that this pathway is required for proper differentiation.
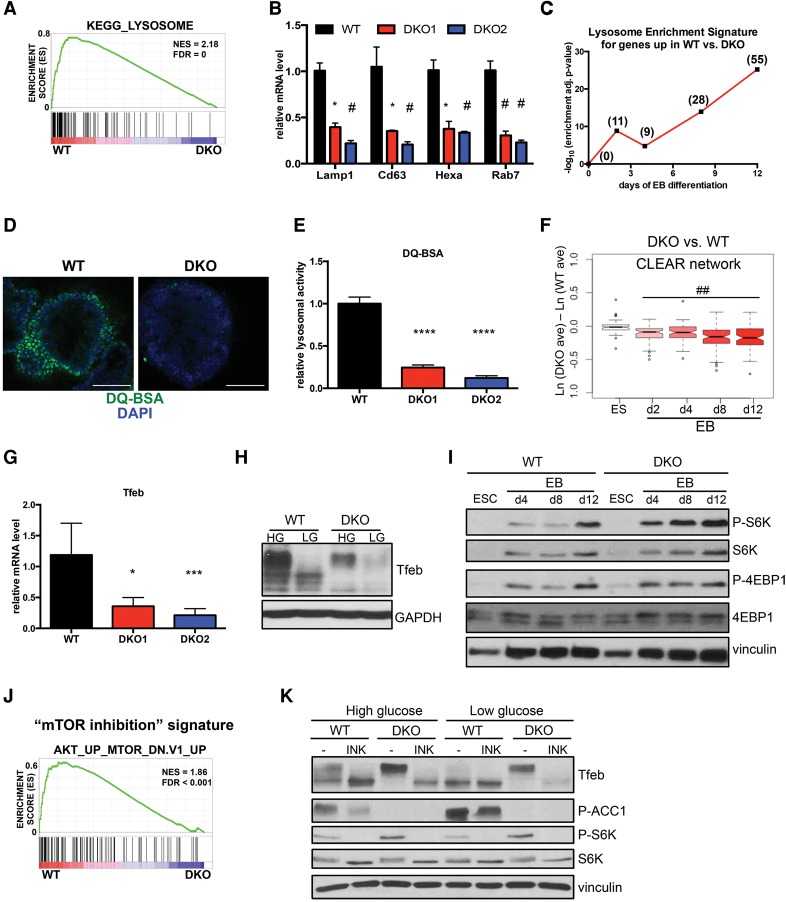
Lysosomes are deregulated in differentiating AMPK double-knockout cells
To search for molecular mechanisms that could explain the defective developmental potential of AMPK double-knockout ESCs, we mined our mRNA-seq data for additional differentially enriched gene sets apart from those relating to the distinct cell types present in late-stage EBs. Surprisingly, this analysis uncovered lysosomal genes as some of the most highly deregulated transcripts in double-knockout cells, being dramatically decreased, which we validated using several markers (Fig. 3A,B). We also performed expression profiling on ESCs and early stages of EB differentiation (days 2 and 4) in order to identify differences that precede the acquisition of unique cell fates observed at later time points, as these changes may represent initiating events that lead to germ layer skewing as opposed to being indirect markers of cell type. Strikingly, at every stage of differentiation, a lysosomal-associated signature was significantly enriched among those genes up-regulated in wild-type EBs, while no such pattern was observed in the ESC state (Fig. 3C). In a separate analysis searching for genes regulated by energy stress, we were surprised to find that although hundreds of transcripts had altered expression in response to differing glucose conditions in wild-type EBs, only a small subset required an intact AMPK pathway. Among this AMPK-dependent, glucose-sensitive gene set, the lysosome scored among the most highly enriched gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) terms, further suggesting that the AMPK pathway is required for proper function of this organelle in developing EBs (Supplemental Fig. 6A).
Figure 3.
AMPK double-knockout EBs exhibit defects in lysosome function and Tfeb regulation. (A) GSEA plot showing enrichment of the gene set associated with the KEGG term “lysosome” in wild-type (WT) versus AMPK double-knockout (DKO) day 12 EBs in high glucose. (B) Relative mRNA levels of selected lysosomal genes in wild-type and AMPK double-knockout day 12 EBs in high glucose as assessed by qRT–PCR. n = 3 independent experiments. (*) P < 0.05; (#) P < 0.005 compared with wild type. (C) Plot of corrected log-transformed P-values of KEGG lysosome pathway enrichment in sets of genes identified to be up-regulated in wild type versus double knockout under high-glucose conditions. At each time point (X-axis), the statistical significance of enrichment of genes associated with the KEGG term “lysosome” in wild-type versus AMPK double-knockout samples was determined as described in the Materials and Methods. The resulting P-values were log-transformed and are plotted on the Y-axis. Numbers in parentheses indicate the number of lysosomal genes significantly up-regulated in wild-type samples at each time point. (D) Direct fluorescence images of day 12 wild-type and AMPK double-knockout EBs after 1 h of incubation with DQ-BSA-Green followed by a 2-h chase prior to fixation. Bar, 100 µm. (E) Relative lysosomal activity, as determined by DQ-BSA assay, in day 12 wild-type and AMPK double-knockout EBs in high glucose. Results are from two independent experiments, with at least 10 EBs analyzed for each sample within an experiment. (****) P < 0.0001 compared with wild type . (F) Box and whisker plot of mRNA expression differences across 81 coordinated lysosomal expression and regulation (CLEAR) network genes in AMPK double-knockout versus wild-type samples at the indicated time points. High-glucose conditions only. See the Materials and Methods for details. (##) P < 10−8, one sample t-test for nonzero mean. (G) RT-qPCR analysis of Tfeb in day 8 wild-type and AMPK double-knockout EBs in high glucose. Three independent experiments were performed. (*) P < 0.05; (***) P < 0.0005 compared with wild type . (H) Immunoblot analysis of Tfeb in wild-type and AMPK double-knockout EBs differentiated in either high (HG) or low (LG) glucose for 8 d. (I) Time-course analysis of selected mTOR signaling components during high-glucose EB differentiation of wild-type and double-knockout ESCs. Lysates were immunoblotted with the indicated antibodies. (J) GSEA plot depicting enrichment of a “mTOR inhibition” gene set (derived from everolimus-treated vs. untreated mouse tissues) in wild-type versus double-knockout EBs at day 12. (K) Western blot analysis of day 6 wild-type and AMPK double-knockout EBs. After overnight incubation in the indicated glucose medium, EBs were treated with either vehicle (DMSO) or 500 nM INK128 for 2 h prior to lysate preparation and blotting with indicated antibodies. For standard bar graphs, average ± SEM is plotted. Statistical significance was determined by Student's t-test unless otherwise noted.
To determine whether these dramatic gene expression differences translated to distinct functional outcomes, we used a well-established lysosomal activity assay involving DQ-BSA. Under basal conditions, this fluorogenic reagent is quenched due to the high degree of dye labeling on protein aggregates. Once digested by intracellular proteases, many of which are present in lysosomes, individual peptides are released, leading to bright fluorescence. In both high- and low-glucose conditions, wild-type EBs produced strong signals following incubation with DQ-BSA. However, EBs derived from two separate AMPK double-knockout clones generated much weaker fluorescence, suggesting diminished lysosome function (Fig. 3D,E; Supplemental Fig. 6B,C). Importantly, wild-type but not KD AMPKα1 reconstitution in double-knockout cells was sufficient to boost lysosome activity (Supplemental Fig. 6D). Together with the gene expression analysis, these results demonstrate that AMPK is a major regulator of lysosomes during EB differentiation.
The coordinated lysosomal expression and regulation (CLEAR) network and Tfeb are regulated by AMPK
Lysosomes are highly dynamic organelles with the ability to acutely respond to a variety of stresses to mediate recycling of damaged macromolecules, nutrient replenishment, and a return to homeostasis (Saftig and Klumperman 2009; Platt et al. 2012). Recent work has uncovered that lysosome biogenesis is predominantly controlled at the level of transcription, with most relevant genes belonging to a network (CLEAR) regulated by the MiT/TFE family of transcription factors, including Tfeb, Mitf, Tfe3, and Tfec (Sardiello et al. 2009; Settembre et al. 2011; Martina et al. 2014; Ploper et al. 2015). Within our mRNA-seq time course, the CLEAR network was significantly down-regulated in AMPK double-knockout cells across all stages of differentiation, starting at day 2 and progressively worsening throughout the process (Fig. 3F; Supplemental Fig. 6E). Additional analysis of these expression profiles indicated that among all of the MiT/TFE members, Tfeb was the most consistently diminished in double-knockout cells, which we confirmed at the mRNA and protein level (Fig. 3G,H data not shown).
Tfeb is tightly controlled by nutrient status, with energy-rich conditions leading to hyperphosphorylation and cytoplasmic retention and low-energy stores promoting hypophosphorylation and nuclear localization (Roczniak-Ferguson et al. 2012; Settembre et al. 2012). Wild-type EBs displayed a mixture of hyperphosphorylated and hypophosphorylated Tfeb species in high glucose, and lowering this energy source produced only the hypophosphorylated form. In contrast, in high glucose, the double-knockout cells exclusively expressed the hyperphosphorylated form, while glucose deprivation failed to produce the lower species and instead reduced overall Tfeb levels (Fig. 3H). This decrease in protein levels most likely results from an uncoupling of the autoregulatory loop recently described for this transcription factor (Settembre et al. 2013). As expected, alterations in Tfeb phosphorylation status led to pronounced differences in its nuclear localization, as shown by the marked reduction in nuclear Tfeb in double-knockout cells (Supplemental Fig. 6F). Importantly, AMPK deletion in our UAA-derived EBs also led to the same deregulation of Tfeb phosphorylation (Supplemental Fig. 6G), whereas re-expression of wild-type AMPKα1 in the CRISPR-derived double-knockout cells restored normal Tfeb regulation (Supplemental Fig. 6H). Collectively, these data demonstrate significant Tfeb deregulation upon AMPK loss in developing EBs.
We next sought to elucidate the molecular mechanism underlying this AMPK–Tfeb connection. Several signaling pathways have been implicated in controlling Tfeb phosphorylation, including MAPK, mTOR, and calcineurin (Settembre et al. 2011, 2012; Roczniak-Ferguson et al. 2012; Medina et al. 2015). Because of the well-established cross-talk between AMPK and mTOR, including AMPK-mediated phosphorylation of core mTOR components (Mihaylova and Shaw 2011), we focused on this relationship first. Indeed, AMPK-deficient EBs exhibited elevated mTOR signaling, as determined by higher levels of mTOR substrates P-S6K and P-4EBP1 (Fig. 3I). Furthermore, a gene set comprised of transcripts up-regulated by mTOR inhibition was depleted in double-knockout EBs, providing additional evidence of higher flux through the mTOR pathway in these samples (Fig. 3J). To examine whether this hyperactive signaling contributed to Tfeb deregulation, we treated developing EBs with the mTOR inhibitor INK128. Incubating double-knockout EBs with INK128 completely collapsed Tfeb from the hyperphosphorylated to the hypophosphorylated state, resulting in a banding pattern similar to treated wild-type cells (Fig. 3K). Interestingly, in low-glucose conditions, although acute mTOR inhibition caused this band shift, overall Tfeb levels were much lower. This suggests additional layers of complexity with respect to Tfeb regulation, most likely including the autoregulation mentioned above, especially when nutrients are limited. Nonetheless, these results demonstrate AMPK's inhibition of mTOR as one mechanism dictating proper Tfeb levels during EB differentiation.
Tfeb and proper lysosomal function are required for endoderm differentiation
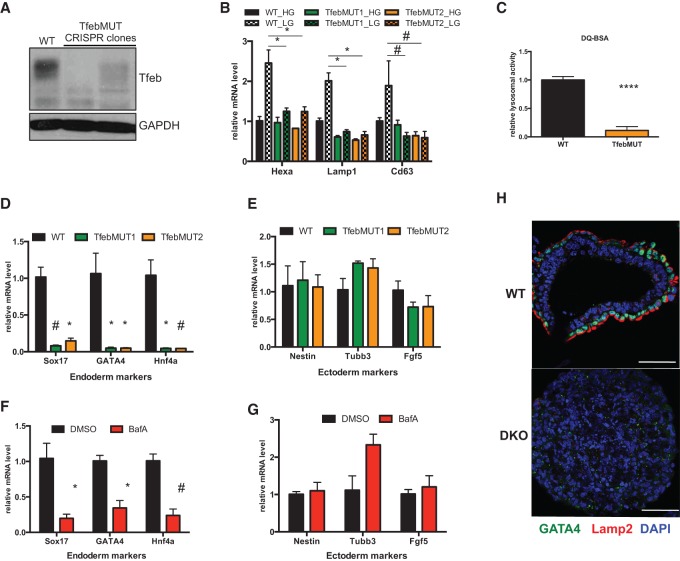
Genetic disruption of Tfeb as well as many lysosomal components is known to result in embryonic lethality, although precise details regarding the underlying developmental defects are unknown (Steingrimsson et al. 1998; Sun-Wada et al. 2000; Eskelinen et al. 2004). We were intrigued by the possibility that the profound Tfeb/lysosomal abnormalities present in our AMPK double-knockout cells could directly affect differentiation potential. To assess the role of Tfeb in lineage specification, we first used the CRISPR/Cas9 system to generate two independent mutant ESC lines (referred to here as TfebMUT) that showed loss of Tfeb mRNA and protein (Fig. 4A; Supplemental Fig. 7A,B). EBs derived from both TfebMUT lines were unable to induce several CLEAR target genes upon nutrient deprivation (Fig. 4B), indicating that no other MiT/TFE family members can compensate for Tfeb deficiency to control these targets. In addition, DQ-BSA assays revealed minimal lysosomal activity in mutant EBs compared with their wild-type counterparts (Fig. 4C). Together, these results confirmed a defective lysosomal compartment in TfebMUT cells.
Figure 4.
Proper lysosomal function is required for endoderm differentiation. (A) Immunoblot analysis of Tfeb in EBs derived from wild-type (WT) parental and two independent TfebMUT CRISPR clones. (B) Wild-type and TfebMUT ESCs underwent EB differentiation for 7 d in high (HG) or low (LG) glucose followed by RT-qPCR of selected CLEAR network genes. (*) P < 0.05; (#) P < 0.005 compared with wild-type LG. (C) Relative lysosomal activity, as determined by DQ-BSA assay, in wild-type and TfebMUT2 EBs. Data are from three independent experiments, with 10 EBs analyzed in each experiment. (****) P < 0.0001 compared with wild type . (D,E) RT-qPCR analysis of endoderm (D) and ectoderm (E) markers in day 7 EBs from wild-type and two TfebMUT clones. (*) P < 0.05; (#) P < 0.005 compared with wild type . (F,G) Wild-type ESCs were differentiated into EBs for 7 d in the presence or absence of 2.5 nM bafilomycin A (BafA). The compound was added daily. mRNA levels of selected endoderm (F) and ectoderm (G) genes were assessed by RT-qPCR. (*) P < 0.05; (#) P < 0.005 compared with DMSO. (H) IF analysis depicting wild-type EB-specific colocalization of GATA4, an endoderm marker, with the highly polarized staining of lysosomal marker Lamp2 in the outer layer of cells. Most EBs derived from AMPK double-knockout (DKO) ESCs lack appreciable staining throughout the structure, as shown in the bottom image. Bar, 50 µm. For B and D–G, data are from three independent experiments. For all bar graphs, average ± SEM is plotted. Statistical significance was determined by Student's t-test.
To examine the developmental capabilities of TfebMUT ESCs, we measured the expression of specific germ layer markers in developing EBs. Strikingly, both TfebMUT lines failed to up-regulate several endoderm markers during the differentiation process, similar to what was observed in AMPK double-knockout cells (cf. Figs. 4D and 2F). However, in contrast to AMPK-deficient cells, this defective endoderm formation did not coincide with an induction of ectoderm markers, which remained at wild-type levels, suggesting that some developmental defects in AMPK double-knockout cells are Tfeb-independent (Fig. 4E). In agreement with this notion, TfebMUT cells possessed an intermediate ability to contribute to embryonic tissues during chimera formation (Supplemental Fig. 7C,D). Therefore, although not as severe as AMPK deletion, Tfeb disruption in ESCs does lead to differentiation defects, most notably phenocopying an inability to properly induce the endodermal gene expression program during EB formation.
While the most established function of Tfeb relates to up-regulating lysosome-associated functions through control of the CLEAR network, it is possible that, within the context of EB differentiation, either newly ascribed or unknown functions of Tfeb could be responsible for its ability to regulate endoderm formation (Settembre et al. 2013). To further investigate the link between endodermal differentiation and lysosomes, we sought an independent method to disrupt this organelle. To this end, developing wild-type EBs were treated with bafilomycin A1 (BafA), a compound that targets vacuolar H+ ATPases and inhibits lysosomal function. Strikingly, cells treated with a chronic, low dose of BafA exhibited considerable defects in endodermal gene expression, while ectoderm markers were largely unaffected (Fig. 4F,G). This unique relationship between endoderm and lysosomes was further substantiated when examining the localization of Lamp2, a well-established lysosomal marker, in developing EBs. As expected, differentiated wild-type EBs contained an outer layer of GATA4-positive endoderm cells surrounding inner layers comprised of other cell types that border the hollow interior formed by cavitation. Remarkably, robust Lamp2 staining was observed only in the GATA4+ endoderm compartment and was mostly localized to the apical side of these cells. In contrast, most double-knockout EBs lacked both markers (Fig. 4H). These results are consistent with the model that the role of Tfeb in lysosomal maturation is critical for its ability to regulate endoderm formation and further suggest that specification of this germ layer is especially reliant on this organelle.
Rescue of lysosomal and endodermal defects in AMPK double-knockout cells by Tfeb overexpression
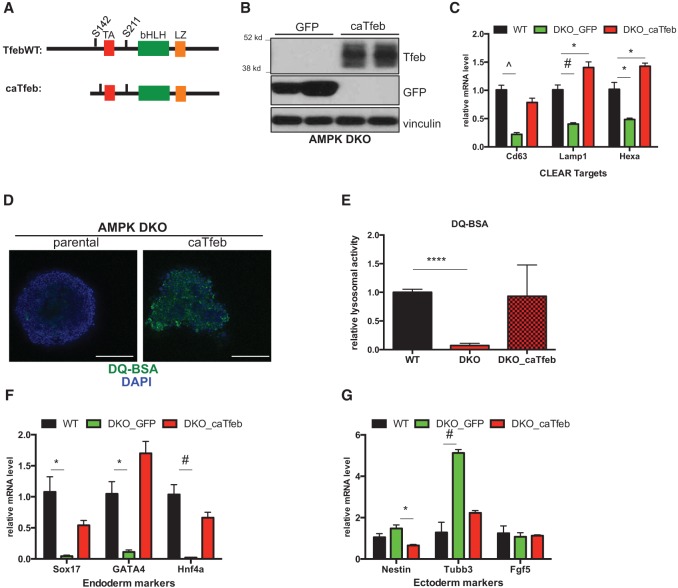
Combining our biochemical data showing that Tfeb is downstream from AMPK together with the fact that endoderm differentiation is highly sensitive to lysosomal function, we next wondered whether correcting the defects within this cellular compartment in AMPK double-knockout cells would rescue their aberrant differentiation potential. In several experimental systems, overexpression of Tfeb by itself is sufficient to up-regulate a majority of the CLEAR network and significantly increase lysosomal activity in target cells (Sardiello et al. 2009; Settembre et al. 2011). Therefore, we set out to generate AMPK double-knockout ESCs stably overproducing Tfeb. Our initial efforts to create cell lines containing exogenous full-length Tfeb were unsuccessful. Based on previous experience, we speculated that reducing the size of the cDNA would increase our efficiency. We deleted the first 110 amino acids on the N terminus, leaving intact the domains responsible for DNA binding and transactivation (Fig. 5A). Importantly, this truncated version of Tfeb (“caTfeb”) was expressed and able to induce CLEAR target genes to levels higher than full-length constructs when transfected into 293T cells (Supplemental Fig. 8A–C). This increased expression and activity is most likely due to the fact that the N-terminal region of the protein targets Tfeb to lysosomes, where it is phosphorylated and inhibited by mTOR (Roczniak-Ferguson et al. 2012). Using this construct, we were able to generate several independent clones of AMPK double-knockout ESCs expressing caTfeb along with GFP targeted cells as controls (Fig. 5B).
Figure 5.
Tfeb overexpression corrects lysosomal defects and increases endodermal gene expression in AMPK double-knockout cells. (A) Schematic of wild-type (WT) Tfeb and caTfeb cDNA constructs. (TA) Transactivation domain; (bHLH) basic helix–loop–helix domain; (LZ) leucine zipper. S142 and S211 denote phosphorylation sites that control nuclear–cytoplasmic shuttling. (B) Western blots of representative GFP and caTfeb-expressing clones following stable transfection of cDNAs into an AMPK double-knockout (DKO) ESC CRISPR line. (C) RT-qPCR analysis of selected CLEAR target genes in wild-type parental and AMPK double-knockout clones expressing either GFP or caTfeb following 7 d of EB differentiation. (D) Direct fluorescence images of day 12 EBs derived from parental or caTfeb-expressing AMPK double-knockout ESCs after 1 h of incubation with DQ-BSA-Green followed by a 2-h chase prior to fixation. Bar, 100 µm. (E) Relative lysosomal activity, as determined by DQ-BSA assay, in day 12 wild-type and AMPK double-knockout (parental or caTfeb-expressing) EBs in high glucose. See the Materials and Methods for details. Results are from two independent experiments, with at least 10 EBs analyzed for each sample within an experiment. (****) P < 0.0001. (F,G) Wild-type parental and AMPK double-knockout ESCs expressing either GFP or caTfeb were differentiated for 7 d followed by RT-qPCR analysis of selected endoderm (F) and ectoderm (G) markers. C, F, and G show data from one representative experiment in which multiple GFP and caTfeb clones (n = 3 per cDNA) were analyzed and compared with wild-type parental controls. Similar results were observed in four independent experiments with additional independently derived cDNA clones. (*) P < 0.05; (#) P < 0.005; (^) P < 0.0005. For all bar graphs, average ± SEM is plotted. Statistical significance was determined by Student's t-test.
Initially, we confirmed the up-regulation of several CLEAR network members in caTfeb-expressing clones upon differentiation into EBs (Fig. 5C). In addition to the expected gene expression restoration, we also observed significant rescue of lysosomal activity as measured by the DQ-BSA assay (Fig. 5D,E). Most importantly, with respect to germ layer differentiation, caTfeb overexpression reproducibly led to elevated levels of endodermal markers while simultaneously reducing several ectodermal genes in developing EBs, a reversal of the skewing initially documented in AMPK double-knockout cells (Fig. 5F,G). Another hallmark of differentiation, EB cavitation, was only partially restored in caTfeb clones, suggesting that either Tfeb-independent processes or proper physiological regulation of Tfeb prevented by our constitutively activated allele is required for some aspects of EB development (Supplemental Fig. 8D). To further corroborate our results linking Tfeb overexpression and endoderm induction, additional cell lines were created expressing a full-length Tfeb cDNA, referred to as TfebAA, which contained serine-to-alanine mutations at two residues whose phosphorylation was previously shown to be responsible for the mobility shifts described earlier (Supplemental Fig. 8A–C). Similarly to caTfeb clones, TfebAA-expressing double-knockout ESCs also exhibited strong induction of endodermal gene expression relative to GFP controls (Supplemental Fig. 8E,F). Collectively, these rescue experiments demonstrate that endodermal differentiation defects of AMPK double-knockout EBs result from diminished Tfeb levels and associated reductions in lysosomal activity.
Impaired canonical Wnt signaling in AMPK and Tfeb mutant cells contributes to endodermal defects
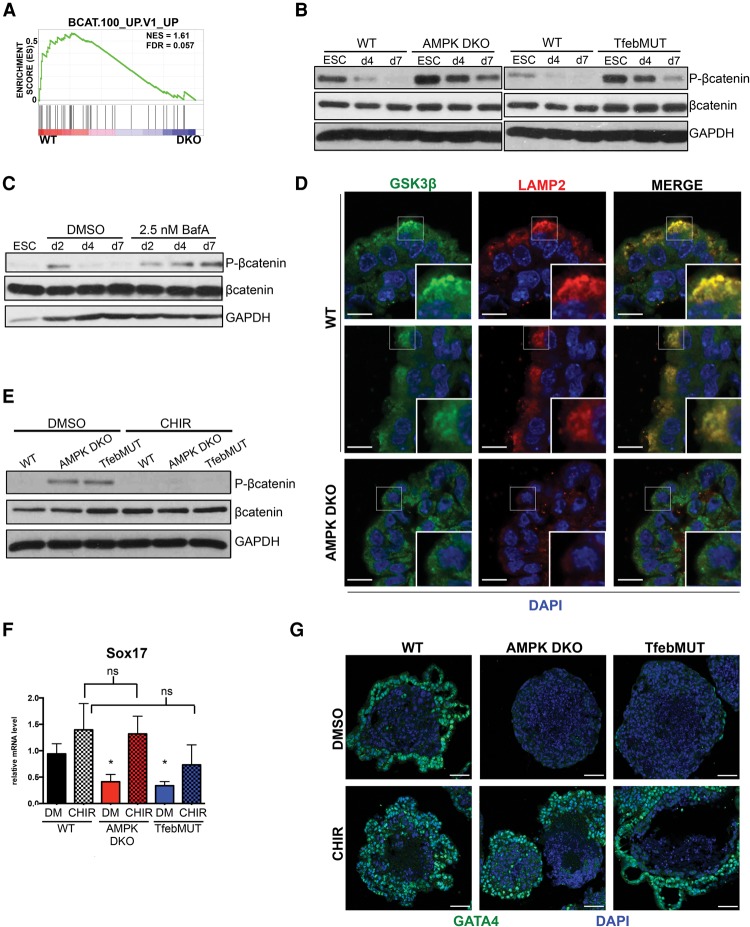
Specification of the different germ layers from multipotent progenitors is dictated by the integration of distinct signaling pathways that in turn drive induction of lineage-restricted transcription factors. For acquisition of an endodermal fate, Wnt, Nodal (Tgfβ), and Fgf pathways are known to be particularly important (Grapin-Botton and Constam 2007). To more fully characterize the molecular mechanisms underlying the altered developmental potential of AMPK-deficient ESCs, we examined whether any of these signaling cascades were aberrantly regulated during EB differentiation. Initial analysis of the Fgf pathway indicated no significant changes in our mutant lines, whereas Nodal signaling dynamics were highly variable (Supplemental Fig. 9A; data not shown). Intriguingly, GSEA of early time-point expression profiles revealed substantial enrichment of a β-catenin-associated gene set in wild-type samples, suggesting defects in canonical Wnt signaling in AMPK double-knockout cells (Fig. 6A). To validate these findings, we monitored the status of the Wnt pathway throughout EB differentiation by assessing levels of the inhibitory marker phospho-β-catenin Ser33/37/41 in whole-cell lysates. Developing wild-type EBs exhibited a progressive reduction in phospho-β-catenin, indicative of increasing Wnt activation, whereas, in AMPK double-knockout cells, this pattern was greatly attenuated (Fig. 6B). Importantly, we observed similar results in differentiating TfebMUT cells (Fig. 6B). Therefore, diminished canonical Wnt signaling is a hallmark of both AMPK and Tfeb mutant EBs.
Figure 6.
Attenuated canonical Wnt signaling resulting from lysosomal deregulation contributes to endodermal differentiation defects in AMPK double-knockout and TfebMUT cells. (A) GSEA plot depicting a β-catenin-associated gene set up-regulated in wild-type (WT) versus AMPK double-knockout (DKO) EBs following 4 d of differentiation. (B) Western blots showing dynamics of phospho-β-catenin during the EB differentiation time course of wild-type, AMPK double-knockout, and TfebMUT ESCs. ESCs, day 4 (d4) EBs, and day 7 (d7) EBs were harvested and immunoblotted with the indicated antibodies. (C) Wild-type ESCs were differentiated into EBs in the presence of vehicle (DMSO) or 2.5 nM BafA for the indicated times before being processed and analyzed as in B. (D) IF of Wnt signaling component GSK3β and lysosomal marker Lamp2 in day 8 EBs of the indicated genotypes. Note the subcellular colocalization of GSK3β/ Lamp2 in several cells on the outer layer of wild-type EBs. Insets in the bottom right of each panel depict zoomed-in images from the boxed-in sections. Bar, 10 µm. (E) Immunoblot analysis of day 7 wild-type , AMPK double-knockout, and TfebMUT EBs treated daily with either vehicle (DMSO) or 3 µM CHIR99021 (CHIR). One hour after the last treatment, lysates were harvested and blotted with the indicated antibodies. (F) RT-qPCR analysis of the endoderm marker Sox17 in EBs from E (DM-DMSO). (*) P < 0.05 compared with wild-type DM; (ns) not significant. n = 4 independent experiments. (G) IF analysis of GATA4 in day 8 wild-type, AMPK double-knockout, or TfebMUT EBs either vehicle- or CHIR-treated throughout the differentiation protocol. Bar, 50 µm. The bar graph represents average ± SEM. Statistical significance was determined by Student's t-test.
The endolysosomal system, through its ability to recycle and/or degrade pathway components, critically controls flux through numerous signaling cascades (Wada and Sun-Wada 2013). Interestingly, recent evidence has highlighted MiT/TFE-dependent regulation of late endosomes and lysosomes as particularly important for optimal canonical Wnt signaling (Ploper et al. 2015). Indeed, treating wild-type EBs with BafA, the lysosomal inhibitor, significantly blunted β-catenin activation, demonstrating a connection between lysosomes and Wnt in this context (Fig. 6C). Furthermore, caTfeb expression partially corrected the abnormal pattern of Wnt activation present in AMPK double-knockout cells, suggesting that Tfeb plays a direct role in regulating this signaling pathway during EB differentiation (Supplemental Fig. 9B).
Previous studies indicate that binding of Wnt ligands to their receptors mediates the targeting of the destruction complex (DC), a multiprotein unit containing GSK3β, the kinase responsible for phosphorylating and inhibiting β-catenin, into endolysosomal compartments known as multivesicular bodies (MVBs), allowing for newly synthesized β-catenin to enter the nucleus (Taelman et al. 2010; Dobrowolski and De Robertis 2012). To determine whether this process occurred during EB differentiation, we examined GSK3β and Lamp2 localization by indirect IF. Notably, many cells in the outer layer of wild-type EBs displayed areas of punctate GSK3β, and these regions almost completely overlapped with robust Lamp2 staining. In contrast, double-knockout EBs lacked this pattern of GSK3β, while Lamp2 was also much weaker and failed to show colocalization (Fig. 6D). These results are consistent with the idea that GSK3β sequestration into Lamp2-positive structures is a part of normal EB development and that this process is compromised in the absence of AMPK.
We speculated that the inability of AMPK double-knockout cells to target GSK3β into endolysosomes would lead to the inappropriate activity of the DC even in the presence of Wnt ligands, resulting in the aberrant Wnt signaling observed in mutant EBs (Fig. 6B). Furthermore, given that key endoderm-specific transcription factors, such as Sox17, are β-catenin target genes (Engert et al. 2013), these signaling defects may be directly responsible for improper germ layer differentiation. To test these possibilities, we treated developing EBs with CHIR99021 (CHIR), a potent GSK3β inhibitor. First, we confirmed that incubation with CHIR normalized Wnt signaling, as seen by the decreased levels of phospho-β-catenin in both mutant lines (Fig. 6E). Strikingly, CHIR treatment restored some cavitation to AMPK double-knockout EBs, indicating a partial normalization of developmental processes in these mutants (Supplemental Fig. 9C). Indeed, exposure to CHIR substantially rescued the ability of both AMPK double-knockout and TfebMUT cells to acquire an endodermal fate, as determined by increased mRNA levels of both Sox17 and GATA4 (Fig. 6F; Supplemental Fig. 9D). Indirect IF experiments confirmed that CHIR-treated mutant EBs displayed a pattern of GATA4 staining highly similar to that seen in wild-type samples (Fig. 6G). Together, these results indicate that attenuated canonical Wnt signaling, most likely due to a down-regulated endosome/lysosomal compartment, contributes to defective endodermal lineage specification in AMPK double-knockout and TfebMUT ESCs.
Discussion
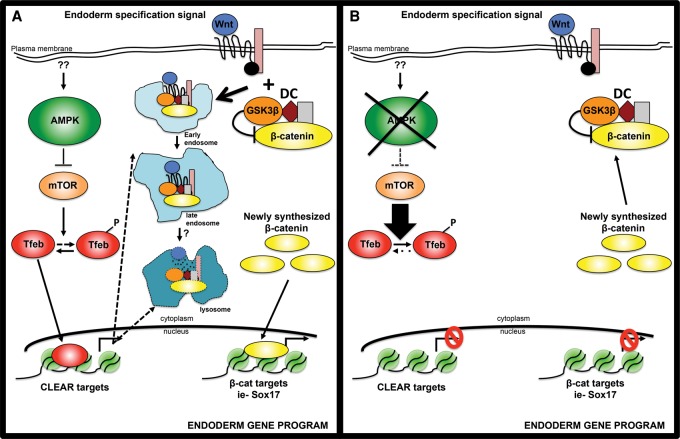
Here we report an important role for AMPK, a central metabolic regulator, in lineage specification of pluripotent cells. Although AMPK-deficient ESCs were largely normal in the pluripotent state, profound defects were noted during EB formation, a process normally marked by differentiation into all three germ layers. Through transcriptional profiling and both loss-of-function and gain-of-function approaches, we identified the endolysosomal compartment as the critical component linking AMPK to these cell fate decisions. Collectively, our data lead to the following model, as depicted in Figure 7. Through its well-established inhibition of the mTORC1 pathway (Mihaylova and Shaw 2011), AMPK counters the negative regulation of Tfeb, a master transcriptional regulator of most lysosomal genes. Optimal Tfeb activation yields a robust endolysosomal system, which in turn is required for proper signaling flux through key developmental pathways that govern germ layer differentiation. Specifically, canonical Wnt signaling, a key orchestrator of endoderm formation, depends on endolysosomal sequestration of the GSK3β-containing DC for maximal activation of β-catenin and downstream engagement of lineage-specific transcription factors.
Figure 7.
Model depicting how AMPK regulates cell fate decisions through lysosome-dependent control of Wnt signaling. (A) In wild-type cells, AMPK inhibition of mTOR allows sufficient levels of Tfeb to enter the nucleus and transcriptionally up-regulate endolysosomes. This organelle system is required for optimal signaling through the canonical Wnt pathway through its ability to sequester the GSK3β-containing DC, freeing β-catenin from degradation. Active β-catenin then translocates into the nucleus to induce target genes, many of which are important mediators of endodermal differentiation, including Sox17. (B) In AMPK double-knockout cells, increased mTOR signaling blunts Tfeb activity, leading to a defective endolysosomal compartment. As a result, DC inhibition of β-catenin remains intact even in the presence of Wnt, preventing robust activation of endodermal genes.
EB differentiation is thought to recapitulate developmental processes that occur early during embryogenesis in vivo. The initial determination of ectodermal and endodermal cells in EBs mirrors one of the first cell fate transitions within the inner cell mass, resulting in embryonic ectoderm surrounded by primitive endoderm, whereas the cavitation of EBs is highly similar to the formation of the hollow egg cylinder shortly after implantation (Coucouvanis and Martin 1999). AMPK double-knockout EBs displayed dramatic defects in both of these processes, skewing toward ectoderm and away from endoderm and also failing to reproducibly cavitate, suggesting that AMPK is required early during development. Indeed, in chimera experiments, AMPK-deficient cells were undetectable in E8.5 embryos, although the extent to which this dramatic loss of development potential relates to EB phenotypes awaits further investigation. Subjecting ESCs to the complex metabolic demands during embryogenesis in vivo may unmask unique liabilities in cells lacking the stress-responsive AMPK pathway.
Previous studies have noted embryonic lethality following deletion of both α or β AMPK subunits, but a comprehensive phenotypic analysis is lacking. Compound mutant AMPKα1−/−;AMPKα2−/− animals have been reported to survive until E9.5–E10.5, which is consistent with, albeit slightly later than, what we would hypothesize given our in vivo results with ESCs (Viollet et al. 2009). Given that our CRISPR-derived AMPK double-knockout cells were generated from C57Bl/6 ESCs, whereas most in vivo studies use mixed background animals, in future studies, it will be important to generate pure C57Bl/6 AMPKα1−/−;AMPKα2−/− embryos to carefully analyze all embryonic stages and compare them with the phenotypes described here.
Our model highlights the nutrient-responsive AMPK–mTOR–Tfeb axis as a critical determinant of germ layer specification, as disruptions in this pathway result in an improper balance of ectodermal versus endodermal cells. Work from human ESCs has revealed that manipulating mTOR either positively by Rheb overexpression or negatively with rapamycin treatment leads to patterns of germ layer skewing similar to what we observed in our system, although it remains to be determined to what extent these effects may be mediated through Tfeb (Zhou et al. 2009). Notably, several members of the MiT/TFE family of transcription factors have previously been implicated in various aspects of pluripotency and differentiation. MITF is a master regulator of the melanocyte lineage, required for the specification, proliferation, and survival of these cells (Steingrimsson et al. 2004). In contrast, TFE3 was recently identified as an important member of the core pluripotency network in mESCs, with its phosphorylation and cytoplasmic retention representing a critical event gating the exit from a stem cell state (Betschinger et al. 2013). In the present study, we ascribe a dominant role for Tfeb in controlling the endodermal differentiation program. Whether these new functions relate to the embryonic lethality of Tfeb−/− animals previously reported remains to be determined (Steingrimsson et al. 1998). Nonetheless, our results further substantiate the claim that MiT/TFE family members are critical determinants of cell fate. Given that nutrient-sensing pathways such as AMPK and mTOR have the capacity to strongly regulate these transcription factors, examining how specific family members may be uniquely controlled under distinct physiological contexts will be an important question to address in the future. This may be a mechanism through which cells couple their metabolic state to particular developmental decisions.
The coordinate regulation of endolysosomes is the most well-established role of Tfeb, but does this explain its function in germ layer differentiation? Our results showing defective endoderm formation in BafA-treated EBs as well as the robust endoderm-specific staining of the lysosomal marker Lamp2 strongly support this notion. Within the endoderm, we noted a strong polarization of Lamp2 toward the apical side of cells, a pattern highly similar to that previously observed in the visceral endoderm (VE) of developing mouse embryos (Kawamura et al. 2012). Furthermore, deletion of several genes involved in endosome–lysosome traffic has been shown to dramatically impair VE formation and function early during embryogenesis (Aoyama et al. 2012; Kawamura et al. 2012). Clearly, our data are consistent with these results from in vivo systems and support a model in which endoderm development critically depends on the endolysosomal compartment.
Maintaining energy homeostasis under conditions of metabolic distress is a hallmark of the AMPK pathway. Similarly, Tfeb, through its ability to up-regulate lysosomes following nutrient starvation, provides important breakdown products that can fuel ATP generation and correct energy imbalances. In addition, these organelles are also integral parts of signaling cascades, effectively recycling or turning over pathway components (Dobrowolski and De Robertis 2012). Indeed, here we show that, under basal conditions, AMPK double-knockout and TfebMUT EBs failed to properly activate Wnt, a master regulator of endoderm, and that simply boosting this pathway rescued germ layer specification in mutant cells. Thus, for cellular differentiation, AMPK and Tfeb appear to be most critical for the generation of a signal transduction node—the endolysosomal system—that fine-tunes signaling strength to generate appropriate transcriptional outputs. These effects were observed in the absence of exogenous energy stress, suggesting that either normal nutrient fluctuations or unrelated inputs direct these developmental functions.
The AMPK pathway has been implicated in several disease states, including diabetes and cancer, and inducing its activity through either compounds or lifestyle changes represents a potential therapeutic option for a number of ailments. However, in many contexts, the relative importance of the multitude of responses controlled by this pathway remains to be determined. Our data directly linking AMPK to lysosomal biogenesis unveil a previously unknown aspect of AMPK biology that may represent a critical mediator of the ability of AMPK to promote cell survival and adapt cellular metabolism under low-nutrient conditions. Furthermore, a deeper understanding of the relationship between AMPK and lysosomes may inform new treatment modalities for the variety of lysosomal inhibitors already in clinical trials (Amaravadi et al. 2011). How critical the fine-tuned control of Tfeb-dependent gene expression is to AMPK function in various cell fate and developmental decisions as well as the metabolic adaptations needed to maintain homeostasis of adult stem cells will be notable areas for future investigation.
Materials and methods
Cell culture
All mESC lines, including the parental C57Bl/6 mESC line v26.2 (a gift from T. Jacks, Massachusetts Institute of Technology) and all derivatives, in addition to those derived from UbcCreER; AMPKα1fl/fl ;AMPKα2fl/fl mice (UAA cells) (see the Supplemental Material) were passaged on gelatinized plates containing irradiated DR4 feeders in standard ES medium (DMEM [Mediatech] supplemented with 15% FBS [Hyclone ES-grade], 1× MEM nonessential amino acids [Mediatech], 100 µM β-mercaptoethanol [Sigma], 100 U/mL penicillin, 100 µg/mL streptomycin [Life Technologies], 1000 U/mL ESGRO-LIF [EMD Millipore]) in 5% CO2 at 37°C. Fresh medium was added daily. Prior to mRNA/protein extractions, proliferation measurements, transfection, and EB formation, mESCs were depleted of feeders for two passages and additionally supplemented with 3 µM CHIR (Stemgent) in order to maintain cells in a pluripotent state, as previously reported (Ye et al. 2012). For high-glucose versus low-glucose experiments, ES medium was made with no-glucose DMEM (Life Technologies), adding D-(+)-glucose (Sigma) to final concentrations of 25 mM (high) or 2.5 mM (low). EBs were generated by trypsinizing feeder-free mESC cultures and plating single-cell suspensions at 5 × 104 to 6 × 104 per square centimeter on low attachment tissue culture plates (Corning) in ES medium lacking ESGRO-LIF. Medium was changed every other day. BafA (Sigma), CHIR, and INK128 (Active Biochem) were dissolved in DMSO; 4-OHT (Sigma) was resuspended in ethanol; and AICAR (Toronto Research Chemicals) was reconstituted in DMEM. To measure cell proliferation, 3 × 104 feeder-free ESCs were seeded in six-well dishes, and trypsinized single-cell suspensions were counted using a Bio-Rad automated cell counter. Alkaline phosphatase staining was performed using the ImmPACT Vector Red alkaline phosphatase substrate kit (Vector Libraries) according to the manufacturer's instructions.
Generation of AMPKα1−/−;AMPKα2−/− double-knockout and TfebMUT ESCs using CRISPR
Sequences for Cas9/CRISPR-mediated gene disruption were obtained using the optimized CRISPR design tool (http://crispr.mit.edu). Corresponding oligonucleotides from IDT were phosphorylated, annealed, and ligated into a BbsI-digested pSpCas9(BB)-2A-EGFP vector (Addgene, no. 48138). Sequence-verified clones were transfected into feeder-free ESC cultures using Lipofectamine 2000 (Life Technologies) using standard procedures. Twenty-four hours to 48 h later, GFP-positive cells were collected by FACS, and low-density single-cell suspensions were plated onto feeder-containing plates. Individual clones were picked and screened for disruption of the relevant genes by restriction fragment length polypmorphism of PCR fragments followed by TOPO cloning (Life Technologies) and sequencing of PCR products. Oligonucleotide sequences are listed in Supplemental Table S1.
Tfeb cell line generation
pCIP, an ESC-optimized expression vector containing a CAGGS promoter upstream of a NheI/MfeI cDNA cloning site followed by a IRES-Puro-bpA cassette, was created using standard cloning techniques. Additional details can be provided on request. caTfeb was generated by amplifying a fragment of human Tfeb cDNA (Addgene, no. 44446) corresponding to the C-terminal 341 amino acids, adding a 5′ Kozak sequence and start codon. cDNA-pCIP constructs were linearized by ScaI digestion, repurified, and introduced into feeder-free ESCs using Lipofectamine 2000. One day after transfection, ESCs were plated onto feeders and cultured in ES medium supplemented with 1–2 µg/mL puromycin. After 6–8 d, individual colonies were picked and screened for expression of the relevant cDNAs by Western blot. Primer sequences are listed in Supplemental Table S1.
Western blotting
Cell pellets were resuspended in RIPA buffer (20 mM Tris at pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% Na deoxycholate, 0.1% SDS, 1 mM EDTA, 50 mM NaF, 1 mM Na orthovanadate, 2.5 mM Na pyrophosphate, 2 mM β-glycerophosphate) supplemented with protease inhibitor tables (Roche) followed by two 30-sec rounds of sonication (65% amplitude) using a cup horn (Misonex 3000). After clarification of the lysates by centrifugation, protein concentrations were determined using the BCA assay kit (Pierce). Protein lysates were separated by SDS-PAGE and transferred to PVDF membranes, which were then incubated with the appropriate primary and secondary antibodies. Signal detection was performed with ECL (Genesee Scientific). Antibodies are listed in Supplemental Table S2.
mRNA extraction and qPCR
Snap-frozen ESC and EB pellets were homogenized in Qiazol (Qiagen), and the resulting aqueous phase was further purified by the RNA clean and concentrator kit (Zymo Reasearch). cDNA was synthesized from 1 µg of RNA using SuperScript III (Life Technologies), and qPCR was carried out with diluted cDNA, appropriate primers, and SYBR Green PCR master mix (ThermoFisher Scientific). Relative mRNA levels were calculated using the 2−ΔΔCt method, with TBP serving as the internal control. All primers are listed in Supplemental Table S1.
mRNA-seq
RNA was isolated by Qiazol (Qiagen) and RNA clean and concentrator kit (Zymo Research) and digested with DNase (Qiagen). RNA integrity (RIN) numbers were determined using the Agilent TapeStation prior to library preparation. mRNA-seq libraries were prepared using the TruSeq RNA library preparation kit (version 2) according to the manufacturer's instructions (Illumina). Libraries were then quantified, pooled, and sequenced by single-end 50 base pairs using the Illumina HiSeq 2500 platform at the Salk Next-Generation Sequencing Core. Raw sequencing data were demultiplexed and converted into FASTQ files using CASAVA (version 1.8.2). Libraries were sequenced at an average depth of 15 million reads per sample.
mRNA-seq analysis
Sequenced reads were quality-tested using FASTQC and aligned to the mouse mm10 genome using the STAR aligner version 2.4.0k. Raw gene expression was quantified across all annotated exons, and differential gene expression was carried out using the edgeR package version 3.6.8 using duplicates to compute within-group dispersion. Differentially expressed genes were defined as having a false discovery rate (FDR) <0.05 and a log2 fold change >0.5. GO term and KEGG pathway enrichment analysis was carried out on differentially expressed genes using the HOMER analysis package (Heinz et al. 2010) and the Benjamini and Yekutieli general correction for multiple testing. Alternatively, GSEA (Mootha et al. 2003) was carried out using preranked lists generated from either FDR or log2 fold change values, setting gene set permutations to 1000 and using the c2 and c6 collections in MSigDB version5.0. Differences in natural log expression of CLEAR network genes were calculated using the R 3.1.1 language, and statistical significance for differences between conditions were evaluated using a one-sample t-test for nonzero mean. Clustering was performed with Gene Cluster 3.0 (de Hoon et al. 2004) and visualized by heat maps using Java TreeView (version 1.1.6r4) (Saldanha 2004). DAVID enrichment analysis was performed using the online DAVID tool (https://david.ncifcrf.gov; Huang et al. 2009). Sequencing data have been deposited in the Gene Expression Omnibus with accession number GSE77705.
DQ-BSA lysosomal activity assay
EBs were labeled with 20 µg/mL DQ Green BSA (Thermo Fisher) for 1 h in spent medium at 37°C, washed twice, and then cultured for an additional 2 h in spent medium. Next, EBs were fixed in 4% PFA for 2 h at room temperature and stored in VectaShield with DAPI on chambered cover glass. Images were acquired using a Zeiss LSM 710 confocal microscope and processed with Zen 2011 SP3 software. Matlab was used to calculate the lysosomal activity as the average intensity of DQ Green BSA above threshold per area of DAPI.
Histology, IF, and IHC
EBs were washed once in PBS, fixed in 4% PFA overnight at 4°C, and then embedded in prewarmed HistoGel (Thermo Scientific, specimen processing gel). After cooling, the samples were processed and embedded in paraffin in an Excelsior ES tissue processor (Thermo Scientific). The blocks were sectioned at 5 µm on a rotary microtome (Thermo Scientific) and stained with hematoxylin and eosin if necessary. For IF and IHC, unstained slides underwent standard deparraffinization, rehydration, and antigen retrieval in citrate buffer followed by permeabilization in 0.1% Tx100 and blocking in 5% normal donkey serum (IF) or horse serum (IHC). For IF, after primary antibody incubation overnight at 4°C, sections were stained with appropriate fluorophore-conjugated secondary antibodies (Life Technologies), washed again, DAPI-stained, and mounted in ProLong Gold anti-fade reagent (Life Technologies). For IHC, endogenous peroxidases were quenched with H2O2 followed by overnight primary antibody staining and further processing with ImmPRESS and ImmPact DAB kits from Vector Laboratories. IF images were acquired with a Zeiss LSM700 confocal microscope and processed with Zen 2011 SP2 software, while IHC images were acquired and processed with the Pannoramic MIDI slide scanner from 3DHISTECH. Primary antibody information is in Supplemental Table S2.
Chimera generation and analysis
The Rosa26 mT-mG reporter plasmid (Addgene, no. 17787) was linearized by KpnI digestion, purified, and introduced into feeder-free wild-type , AMPK double-knockout, and TfebMUT cells by transfection with Lipofectamine 2000. Twenty-four hours later, ESCs were plated onto feeders and selected in 250 µg/mL neomycin (Life Technologies) containing ES medium for 1 wk. TdTomato-expressing clones per line were identified by fluorescence microscopy and processed as described below.
Targeted ESCs were cultured for one passage on feeders. On the day of microinjections, the cells were trypsinized and resuspended in M2 medium (Millipore). Three-week-old C57BL/6J females were superovulated with 5 IU of PMS (Millipore) followed by injection of 5 IU of HCG (Sigma) 46 h later and mated with B6D2F1 males from Jackson Laboratory. Blastocyst stage embryos were collected by flashing the uterine horns of 3.5-d post-coitum (dpc) donor females with M2 medium and cultured in the pre-equilibrated drops of KSOM-AA medium (Millipore) covered with mineral oil in a 5% CO2 incubator before and after microinjections. Small and round ESCs were collected with the injection pipette, and 12–15 cells were injected into each blastocyst. Injections were performed at 10°C using cooling stage. Ten to 12 injected embryos were surgically transferred into the uterine horns of 2.5 dpc pseudopregnant CD1 females (Charles River). Embryos were collected at E8.5 under a dissecting microscope, fixed in 4% PFA for 30 min at room temperature, and counterstained for 15 min with DAPI. Whole-mount images were acquired with a Zeiss LSM700 confocal microscope and processed with Zen 2011 SP2 software. All animal procedures were approved by the Salk Animal Care and Use Committee.
Supplementary Material
Acknowledgments
We thank Tsung-Chang Sung in the Salk Transgenic Core for help with ESC targeting and culturing, Mike Adams in the Watt Advanced Biophotonics Center for microscopy assistance, Chris Benner in the Integrative Genomics and Bioinformatics Core for bioinformatics help, and M.C. Ku and the entire staff at the H.A. and Mary K. Chapman Charitable Foundations Genomic Sequencing Core for assistance with mRNA-seq. These cores were supported by the Salk Institute (grant CCSG P30 CA014195). We also thank Kim McIntyre at the University of California at San Diego Histology Core for histology assistance. This work was supported in part by R01 DK080425, R01 CA172229, P01 CA120964, and Leona M. and Harry B. Helmsley Charitable Trust grant 2012-PG-MED002. N.P.Y. and L.J.E were supported by post-doctoral fellowships from the American Cancer Society (123016-PF-PF-12-191-01-TBE to N.P.Y., and PF-15-037-01-DMC to L.J.E.). N.P.Y. and L.J.E. were each previously supported by a training grant (T32 CA009370) to the Salk Institute Center for Cancer Research (P30CA014195). J.V.N is a recipient of a Damon Runyon Post-doctoral Fellowship.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.274142.115.
References
- Amaravadi RK, Lippincott-Schwartz J, Yin XM, Weiss WA, Takebe N, Timmer W, DiPaola RS, Lotze MT, White E. 2011. Principles and current strategies for targeting autophagy for cancer treatment. Clin Cancer Res 17: 654–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama M, Sun-Wada GH, Yamamoto A, Yamamoto M, Hamada H, Wada Y. 2012. Spatial restriction of bone morphogenetic protein signaling in mouse gastrula through the mVam2-dependent endocytic pathway. Dev Cell 22: 1163–1175. [DOI] [PubMed] [Google Scholar]
- Betschinger J, Nichols J, Dietmann S, Corrin PD, Paddison PJ, Smith A. 2013. Exit from pluripotency is gated by intracellular redistribution of the bHLH transcription factor Tfe3. Cell 153: 335–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossard P, Zaret KS. 1998. GATA transcription factors as potentiators of gut endoderm differentiation. Development 125: 4909–4917. [DOI] [PubMed] [Google Scholar]
- Chason RJ, Csokmay J, Segars JH, DeCherney AH, Armant DR. 2011. Environmental and epigenetic effects upon preimplantation embryo metabolism and development. Trends Endocrinol Metab 22: 412–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YM, Kwon S, Pak YK, Seol HW, Choi YM, Park do J, Park KS, Lee HK. 2006. Dynamic changes in mitochondrial biogenesis and antioxidant enzymes during the spontaneous differentiation of human embryonic stem cells. Biochem Biophys Res Commun 348: 1472–1478. [DOI] [PubMed] [Google Scholar]
- Coucouvanis E, Martin GR. 1995. Signals for death and survival: a two-step mechanism for cavitation in the vertebrate embryo. Cell 83: 279–287. [DOI] [PubMed] [Google Scholar]
- Coucouvanis E, Martin GR. 1999. BMP signaling plays a role in visceral endoderm differentiation and cavitation in the early mouse embryo. Development 126: 535–546. [DOI] [PubMed] [Google Scholar]
- de Hoon MJ, Imoto S, Nolan J, Miyano S. 2004. Open source clustering software. Bioinformatics 20: 1453–1454. [DOI] [PubMed] [Google Scholar]
- Dobrowolski R, De Robertis EM. 2012. Endocytic control of growth factor signalling: multivesicular bodies as signalling organelles. Nat Rev Mol Cell Biol 13: 53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, et al. 2011. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 331: 456–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engert S, Burtscher I, Liao WP, Dulev S, Schotta G, Lickert H. 2013. Wnt/β-catenin signalling regulates Sox17 expression and is essential for organizer and endoderm formation in the mouse. Development 140: 3128–3138. [DOI] [PubMed] [Google Scholar]
- Eskelinen EL, Schmidt CK, Neu S, Willenborg M, Fuertes G, Salvador N, Tanaka Y, Lullmann-Rauch R, Hartmann D, Heeren J, et al. 2004. Disturbed cholesterol traffic but normal proteolytic function in LAMP-1/LAMP-2 double-deficient fibroblasts. Mol Biol Cell 15: 3132–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH. 1981. Establishment in culture of pluripotential cells from mouse embryos. Nature 292: 154–156. [DOI] [PubMed] [Google Scholar]
- Folmes CD, Nelson TJ, Martinez-Fernandez A, Arrell DK, Lindor JZ, Dzeja PP, Ikeda Y, Perez-Terzic C, Terzic A. 2011. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab 14: 264–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadue P, Huber TL, Paddison PJ, Keller GM. 2006. Wnt and TGF-β signaling are required for the induction of an in vitro model of primitive streak formation using embryonic stem cells. Proc Natl Acad Sci 103: 16806–16811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grapin-Botton A, Constam D. 2007. Evolution of the mechanisms and molecular control of endoderm formation. Mech Dev 124: 253–278. [DOI] [PubMed] [Google Scholar]
- Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. 2008. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell 30: 214–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG. 2015. AMPK: positive and negative regulation, and its role in whole-body energy homeostasis. Curr Opin Cell Biol 33: 1–7. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Inoki K, Zhu T, Guan KL. 2003. TSC2 mediates cellular energy response to control cell growth and survival. Cell 115: 577–590. [DOI] [PubMed] [Google Scholar]
- Kanai-Azuma M, Kanai Y, Gad JM, Tajima Y, Taya C, Kurohmaru M, Sanai Y, Yonekawa H, Yazaki K, Tam PP, et al. 2002. Depletion of definitive gut endoderm in Sox17-null mutant mice. Development 129: 2367–2379. [DOI] [PubMed] [Google Scholar]
- Kawamura N, Sun-Wada GH, Aoyama M, Harada A, Takasuga S, Sasaki T, Wada Y. 2012. Delivery of endosomes to lysosomes via microautophagy in the visceral endoderm of mouse embryos. Nat Commun 3: 1071. [DOI] [PubMed] [Google Scholar]
- Lanner F. 2014. Lineage specification in the early mouse embryo. Exp Cell Res 321: 32–39. [DOI] [PubMed] [Google Scholar]
- Leahy A, Xiong JW, Kuhnert F, Stuhlmann H. 1999. Use of developmental marker genes to define temporal and spatial patterns of differentiation during embryoid body formation. J Exp Zool 284: 67–81. [DOI] [PubMed] [Google Scholar]
- Li F, He Z, Li Y, Liu P, Chen F, Wang M, Zhu H, Ding X, Wangensteen KJ, Hu Y, et al. 2011a. Combined activin A/LiCl/Noggin treatment improves production of mouse embryonic stem cell-derived definitive endoderm cells. J Cell Biochem 112: 1022–1034. [DOI] [PubMed] [Google Scholar]
- Li Y, Xu S, Mihaylova MM, Zheng B, Hou X, Jiang B, Park O, Luo Z, Lefai E, Shyy JY, et al. 2011b. AMPK phosphorylates and inhibits SREBP activity to attenuate hepatic steatosis and atherosclerosis in diet-induced insulin-resistant mice. Cell Metab 13: 376–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GR. 1981. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci 78: 7634–7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina JA, Diab HI, Lishu L, Jeong AL, Patange S, Raben N, Puertollano R. 2014. The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci Signal 7: ra9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina DL, Di Paola S, Peluso I, Armani A, De Stefani D, Venditti R, Montefusco S, Scotto-Rosato A, Prezioso C, Forrester A, et al. 2015. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat Cell Biol 17: 288–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metallo CM, Vander Heiden MG. 2013. Understanding metabolic regulation and its influence on cell physiology. Mol Cell 49: 388–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaylova MM, Shaw RJ. 2011. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol 13: 1016–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, et al. 2003. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34: 267–273. [DOI] [PubMed] [Google Scholar]
- Moussaieff A, Rouleau M, Kitsberg D, Cohen M, Levy G, Barasch D, Nemirovski A, Shen-Orr S, Laevsky I, Amit M, et al. 2015. Glycolysis-mediated changes in acetyl-CoA and histone acetylation control the early differentiation of embryonic stem cells. Cell Metab 21: 392–402. [DOI] [PubMed] [Google Scholar]
- Murry CE, Keller G. 2008. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell 132: 661–680. [DOI] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. 2007. A global double-fluorescent Cre reporter mouse. Genesis 45: 593–605. [DOI] [PubMed] [Google Scholar]
- O'Neill HM, Maarbjerg SJ, Crane JD, Jeppesen J, Jorgensen SB, Schertzer JD, Shyroka O, Kiens B, van Denderen BJ, Tarnopolsky MA, et al. 2011. AMP-activated protein kinase (AMPK) β1β2 muscle null mice reveal an essential role for AMPK in maintaining mitochondrial content and glucose uptake during exercise. Proc Natl Acad Sci 108: 16092–16097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt FM, Boland B, van der Spoel AC. 2012. The cell biology of disease: lysosomal storage disorders: the cellular impact of lysosomal dysfunction. J Cell Biol 199: 723–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploper D, Taelman VF, Robert L, Perez BS, Titz B, Chen HW, Graeber TG, von Euw E, Ribas A, De Robertis EM. 2015. MITF drives endolysosomal biogenesis and potentiates Wnt signaling in melanoma cells. Proc Natl Acad Sci 112: E420–E429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM. 2012. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal 5: ra42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saftig P, Klumperman J. 2009. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat Rev Mol Cell Biol 10: 623–635. [DOI] [PubMed] [Google Scholar]
- Saldanha AJ. 2004. Java Treeview—extensible visualization of microarray data. Bioinformatics 20: 3246–3248. [DOI] [PubMed] [Google Scholar]
- Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, et al. 2009. A gene network regulating lysosomal biogenesis and function. Science 325: 473–477. [DOI] [PubMed] [Google Scholar]
- Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, et al. 2011. TFEB links autophagy to lysosomal biogenesis. Science 332: 1429–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, et al. 2012. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J 31: 1095–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, De Cegli R, Mansueto G, Saha PK, Vetrini F, Visvikis O, Huynh T, Carissimo A, Palmer D, Klisch TJ, et al. 2013. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol 15: 647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RJ, Bardeesy N, Manning BD, Lopez L, Kosmatka M, DePinho RA, Cantley LC. 2004. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell 6: 91–99. [DOI] [PubMed] [Google Scholar]
- Shyh-Chang N, Daley GQ, Cantley LC. 2013. Stem cell metabolism in tissue development and aging. Development 140: 2535–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steingrimsson E, Tessarollo L, Reid SW, Jenkins NA, Copeland NG. 1998. The bHLH-zip transcription factor Tfeb is essential for placental vascularization. Development 125: 4607–4616. [DOI] [PubMed] [Google Scholar]
- Steingrimsson E, Copeland NG, Jenkins NA. 2004. Melanocytes and the microphthalmia transcription factor network. Annu Rev Genet 38: 365–411. [DOI] [PubMed] [Google Scholar]
- Sun-Wada G, Murata Y, Yamamoto A, Kanazawa H, Wada Y, Futai M. 2000. Acidic endomembrane organelles are required for mouse postimplantation development. Dev Biol 228: 315–325. [DOI] [PubMed] [Google Scholar]
- Taelman VF, Dobrowolski R, Plouhinec JL, Fuentealba LC, Vorwald PP, Gumper I, Sabatini DD, De Robertis EM. 2010. Wnt signaling requires sequestration of glycogen synthase kinase 3 inside multivesicular endosomes. Cell 143: 1136–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viollet B, Athea Y, Mounier R, Guigas B, Zarrinpashneh E, Horman S, Lantier L, Hebrard S, Devin-Leclerc J, Beauloye C, et al. 2009. AMPK: lessons from transgenic and knockout animals. Front Biosci (Landmark Ed) 14: 19–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada Y, Sun-Wada GH. 2013. Positive and negative regulation of developmental signaling by the endocytic pathway. Curr Opin Genet Dev 23: 391–398. [DOI] [PubMed] [Google Scholar]
- Wellen KE, Thompson CB. 2012. A two-way street: reciprocal regulation of metabolism and signalling. Nat Rev Mol Cell Biol 13: 270–276. [DOI] [PubMed] [Google Scholar]
- Ye S, Tan L, Yang R, Fang B, Qu S, Schulze EN, Song H, Ying Q, Li P. 2012. Pleiotropy of glycogen synthase kinase-3 inhibition by CHIR99021 promotes self-renewal of embryonic stem cells from refractory mouse strains. PLoS One 7: e35892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Su P, Wang L, Chen J, Zimmermann M, Genbacev O, Afonja O, Horne MC, Tanaka T, Duan E, et al. 2009. mTOR supports long-term self-renewal and suppresses mesoderm and endoderm activities of human embryonic stem cells. Proc Natl Acad Sci 106: 7840–7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.