Here, Wilk et al. examined ∼8000 mRNA transcripts throughout Drosophila embryogenesis. They found that almost all RNAs, both coding and noncoding RNAs, are subcellularly localized at some stage of development, thus providing an important resource for functional gene analysis.
Keywords: Drosophila, in situ hybridization, lncRNA, localization, mRNA, subcellular
Abstract
In a previous analysis of 2300 mRNAs via whole-mount fluorescent in situ hybridization in cellularizing Drosophila embryos, we found that 70% of the transcripts exhibited some form of subcellular localization. To see whether this prevalence is unique to early Drosophila embryos, we examined ∼8000 transcripts over the full course of embryogenesis and ∼800 transcripts in late third instar larval tissues. The numbers and varieties of new subcellular localization patterns are both striking and revealing. In the much larger cells of the third instar larva, virtually all transcripts observed showed subcellular localization in at least one tissue. We also examined the prevalence and variety of localization mechanisms for >100 long noncoding RNAs. All of these were also found to be expressed and subcellularly localized. Thus, subcellular RNA localization appears to be the norm rather than the exception for both coding and noncoding RNAs. These results, which have been annotated and made available on a recompiled database, provide a rich and unique resource for functional gene analyses, some examples of which are provided.
The first observations of localized transcripts were confined, not surprisingly, to large and/or highly polarized cells that could be easily resolved, microscopically or physically, into distinct regions. Examples included RNAs differentially localized to the animal and vegetal poles of Xenopus oocytes (Rebagliati et al. 1985), bicoid transcripts at the anterior pole of Drosophila embryos (Macdonald and Struhl 1988; St. Johnston et al. 1989), and actin transcripts at the motile leading edges of migrating fibroblasts (Lawrence and Singer 1986). The discovery of MAP II transcript localization in neuronal axons soon followed (Garner et al. 1988), leading the way to the discovery of many more easily resolved dendrite- or axon-localized transcripts. By the end of the century, the total number of identified localized transcripts for all organisms had climbed to ∼100 (Bashirullah et al. 1998; Jansen 2001), the majority of which was still confined to oocytes and neurons.
With the data then available, the general perception was that mRNA localization was a specialized process confined to cells with unusual properties and needs and limited to a critical minority of transcripts. However, subsequent, more global approaches began to hint at the diversity and extent of RNA localization in other cell types. For example, careful observation of the mRNAs encoding proteins localized to the daughter cells of dividing yeast cells or Drosophila neuroblasts showed that some of these were also specifically partitioned into the daughter cells (Li et al. 1997; Gonzalez et al. 1999). A more systematic analysis in yeast then revealed 22 additional transcripts that localized to daughter cells (Ni and Snyder 2001). Similar types of larger-scale biochemical and microarray approaches began to show that numerous transcripts could also be specifically isolated together with subcellular organelles such as endoplasmic reticulum (ER), microtubules, and mitochondria even when translation was inhibited (for review, see Lecuyer et al. 2009; Martin and Ephrussi 2009). These types of studies began to suggest significantly higher rates of subcellular RNA trafficking than those previously suggested.
To gain a better idea of the prevalence and diversity of localization events, we previously developed a high-throughput, tyramide signal amplification (TSA)-based, fluorescent in situ hybridization (FISH) protocol that could be applied to Drosophila embryos in 96-well microtiter plates (Lecuyer et al. 2008) and analyzed the distributions of ∼2300 mRNAs during the first 4 h of embryogenesis (embryogenesis takes ∼24 h). During much of this time interval, nuclei divide in a syncytial environment, and the majority of gene products is provided maternally, which ensured that all nonuniform patterns observed would involve some form of subcellular trafficking. By the end of the third hour, the embryo cellularizes, which also allowed us to assess localization in the relatively large blastoderm cells that first form. This initial survey of early stage embryos (Lecuyer et al. 2007) revealed nonuniform subcellular RNA distributions for ∼70% of the observed transcripts, suggesting that RNA localization plays a far more important role than previously thought. However, the unusual syncytial nature of early Drosophila embryogenesis raised the question of whether these numbers are peculiar to Drosophila, perhaps representing a necessary evolutionary adaptation required for this type of early development. Consistent with this possibility, the majority of the observed localized transcripts (∼70% of the reported localized transcripts) was localized due to some degree of exclusion from the forming blastoderm layer or extruded pole cells.
To address the prevalence and need for RNA localization in other cell and tissue types, we assessed the distributions of ∼8000 transcripts, with approximately half analyzed during all stages of embryonic development. We also initiated analyses of the majority of third instar larval tissues, with ∼800 transcripts analyzed thus far. Due to the diversity of cell types now analyzed and the increased resolution afforded by larger third instar cells, these analyses have provided a better indication of the true number of localized transcripts as well as new insights into the associated mechanisms and uses. We now conclude that subcellular trafficking and compartmentalization of mRNAs is the rule rather than the exception, with virtually all transcripts examined localized in some cell type at some stage of development.
We also analyzed the distributions and subcellular localization of ∼100 long noncoding RNAs (lncRNAs). There is much debate about the functional roles and degree of importance of this large and rapidly evolving class of genes (Eddy 2001; Wilusz et al. 2009; Pauli et al. 2011; Fatica and Bozzoni 2014; Palazzo and Lee 2015). The expression levels and cellular/subcellular distributions of the 103 transcripts examined suggest that all or most may have important functional roles.
Results
RNA expression patterns at the cellular level
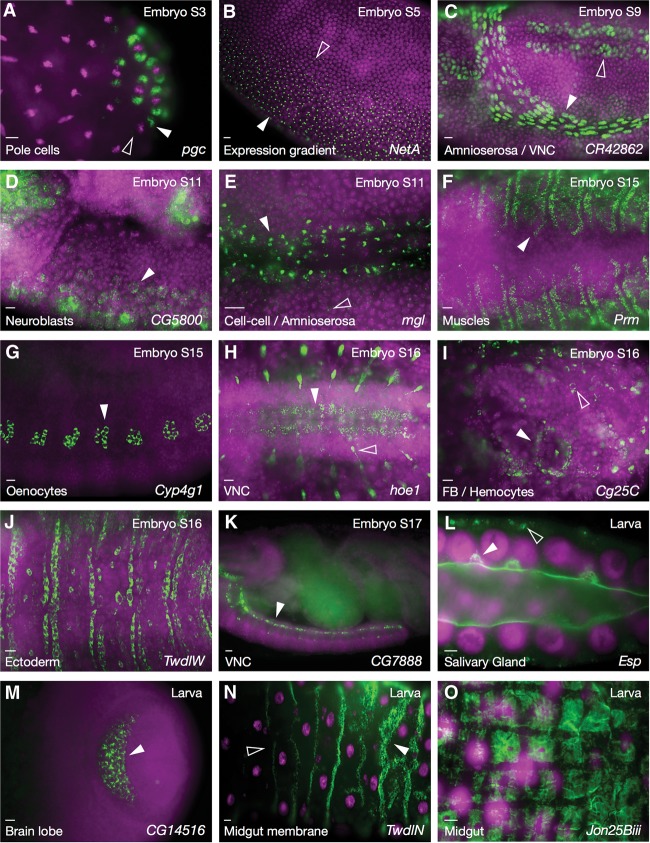
The analysis of later stages of embryogenesis and larval development, with the hundreds of new cellular and subcellular structures and RNA distribution patterns involved, has necessitated a number of advances in our methodology, annotation, and database structure (see the Materials and Methods). While the focus of this project is on subcellular transcript localization, the analysis of later developmental stages is yielding images and information that are equally useful for gene expression analyses at the cell and tissue levels. To help with interesting gene discoveries and analyses, genes that we feel show particularly striking or revealing expression patterns are now highlighted (shaded in green when viewed in alphabetical order listings) in our publicly available online database (http://fly-fish.ccbr.utoronto.ca). A number of examples have been compiled in Figure 1 to illustrate the potential and impact of these patterns. For example, the dorsal–ventral gradient of expression observed for NetrinA (NetA) (Fig. 1B) suggests a significant role in driving the cell intercalation or invagination movements that occur during ventral furrow formation and/or germ band extension. Also consistent with predicted functions and potential mechanisms of action are the expression of Megalin (mgl) in amnioserosa cell–cell junctions (Fig. 1E), paramyosin (Prm) in bands within syncytial musculature (Fig. 1F), and chitin-encoding TweedleW (TwdlW) at the apical surfaces of denticle-forming ectodermal cells (Fig. 1J). Other striking examples include the localization of Epidermal stripes and patches (Esp) in large vesicles as they fuse to the apical lumen of the larval salivary gland (Fig. 1L) and Jonah 25Biii (Jon25Biii), a member of the Jonah peptidase family, within larval midgut muscle fibers (Fig. 1O). The interesting patterns of expression of less-well-characterized genes, such as CR42862 (Fig. 1C), CG5800 (Fig. 1D), CG7888 (Fig. 1K), and CG14516 (Fig. 1M), should also prove highly useful for predicting important cellular and biological functions.
Figure 1.
Interesting and unusual expression patterns. (A) Posterior stage 3 embryo showing polar granule component (pgc) expression around the centrosomes of budding pole cells. When nuclear division is oriented anterior–posterior, pgc is preferentially located around the posterior centrosome (white vs. empty arrowheads). (B) Graded NetA expression in a ventral stage 5 embryo (filled and empty arrowheads indicate higher and lower expression levels). (C) Nuclear localization of CR42862 in the amnioserosa (white arrowhead) and ventral nerve cord (VNC; empty arrowhead). (D) Anterior region stage 11 embryo showing CG5800 expression in delaminating neuroblasts (white arrowhead). (E) mgl transcripts in amnioserosa cell–cell junctions (white arrowhead). Scattered “foci” are also present in the nearby ectoderm (empty arrowhead). (F) Dorsal view showing bands of Prm expression in syncytial somatic musculature. (G) Lateral view showing Cytochrome P450-4g1 (Cyp4g1) in oenocytes. (H) Ventral view of mRNA encoding the transmembrane transporter hoepel1 (hoe1) in the ventral nerve cord (white arrowhead), peripheral nervous system (PNS), and connecting axons (empty arrowhead). (I) Collagen type IV (Cg25C) transcripts in fat body (FB; white arrowhead) and hemocytes (empty arrowhead). (J) Dorsal view showing expression of the chitin-based cuticle component TwdlW in denticle belt-forming ectodermal cells. (K) Lateral view showing CG7888 expression at the periphery of the ventral nerve cord (arrowhead). (L) Third instar larval salivary gland showing Esp (sulphate transporter) localization to membrane and fusing vesicles (white arrowhead) as well as basal clusters (empty arrowhead). (M) Expression of CG14516 in the brain lobe of a third instar larva (arrowhead). (N) Third instar larval midgut showing TwdlN expression in a series of subcellular bands of membrane-associated expression, with increasing anterior-to-posterior levels of expression (arrowheads). (O) Third instar larval midgut showing checkerboard expression of Jon25Biii in midgut musculature. Names of transcripts are indicated at the bottom right of each panel, developmental stage is indicated at the top right, and type of expression or tissue is at the bottom left. RNA is in green, and DAPI-stained nuclei are in magenta. Bars, 10 µm.
Expression at the subcellular level
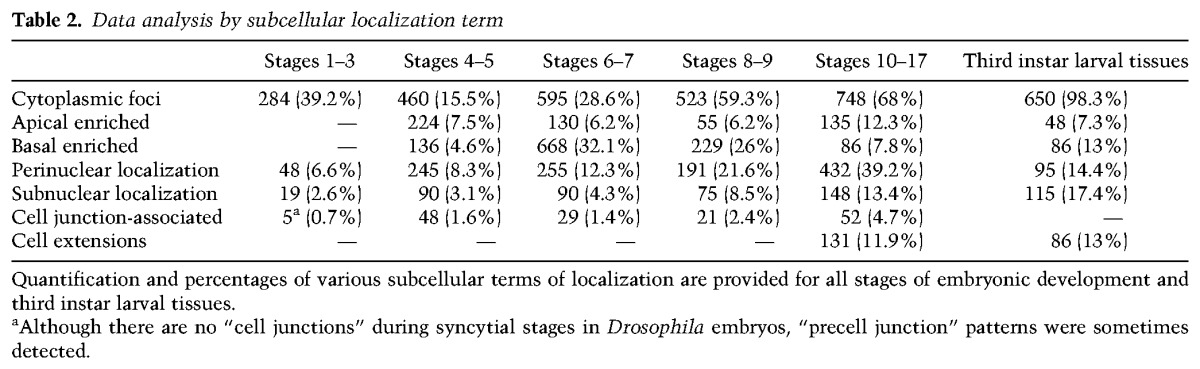
As of this writing, we have completed the analysis and annotation of ∼8000 different genes, approximately half of which have been examined over the full course of embryogenesis. Prevalences of subcellular localization by developmental stage are provided in Table 1. Most notably, the percentage of transcripts that are detectably subcellularly localized after gastrulation is significantly lower than before gastrulation (41% during stages 10–17 vs. 64% during stages 1–8). This lower number may be ascribed to several factors. The most obvious of these is that late stage embryonic cells are much smaller than a syncytial embryo or the blastoderm cells that first form, which makes the detection of less obvious subcellular distributions more difficult to detect. This relative ease of earlier detection may be particularly true for the apical and pole cell exclusion mechanisms and patterns that were the predominant category of subcellular distributions observed in the early embryo. Interestingly, though, all subcellular distribution categories common to both early and late embryonic stages showed a twofold to fourfold increase in prevalence during the later stages (Table 2). This could reflect a greater requirement for these localization mechanisms later in development or simply an increased likelihood of detection due to the increased diversity or polarity of later cell types and tissues. The exception to this increase was the basal enrichment category, which was extremely high in the early embryo (Table 2) and, as with apical exclusion, may be particularly needed by, or a consequence of, the early cellularization process, which progresses from apical to basal. Interestingly, 22% of transcripts found to be localized during stages 10–17 were not found to be localized during earlier stages.
Table 1.
Fly-FISH screen summary
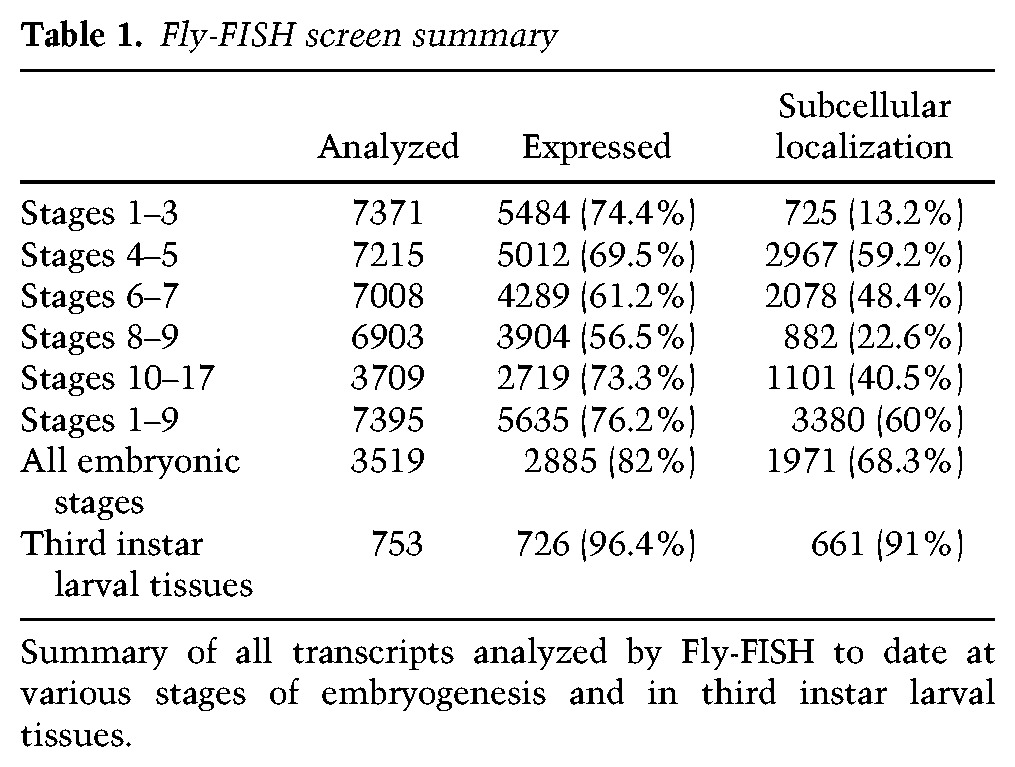
Table 2.
Data analysis by subcellular localization term
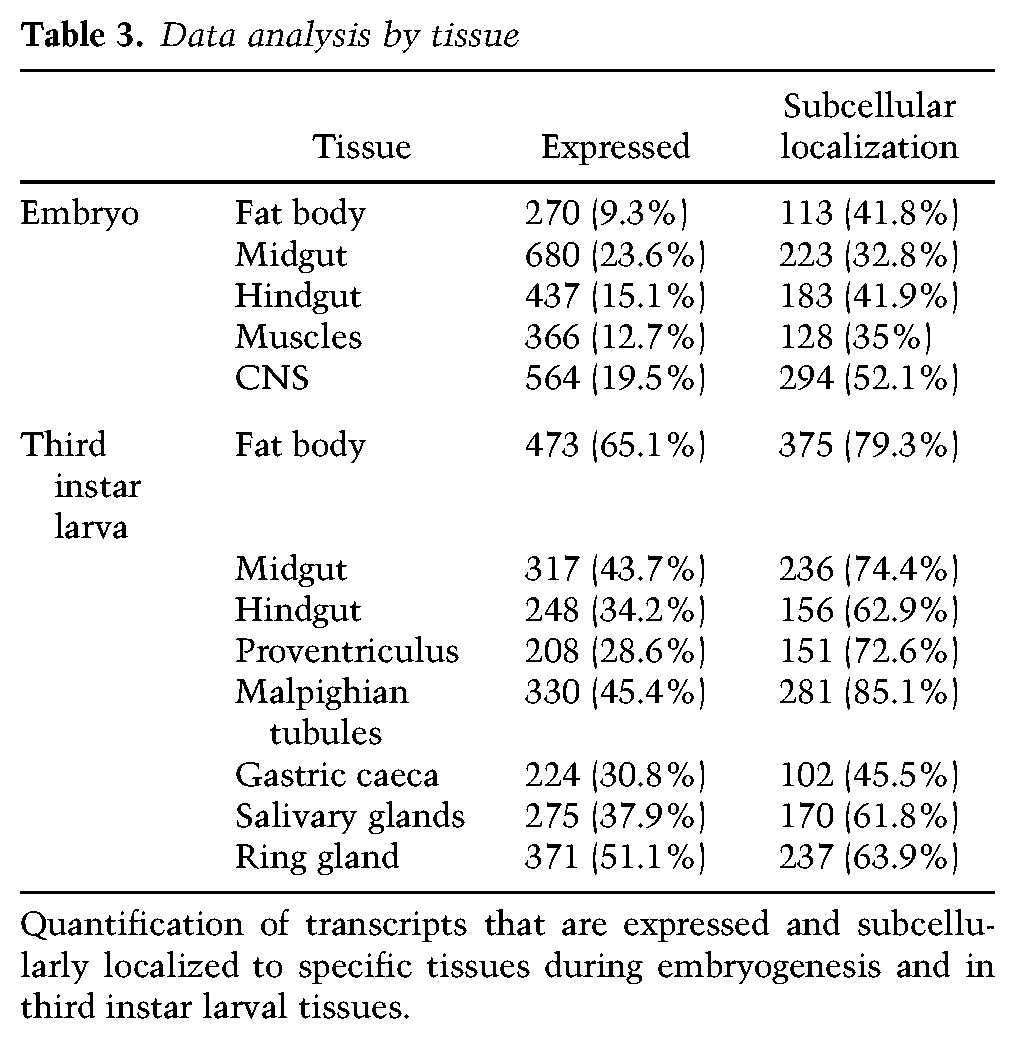
Embryogenesis is followed by three consecutive sets of larval instar stages. At the end of the third larval instar, tissues and cells prepare for metamorphosis. It is during this stage that many interesting hormonal and metabolic changes occur, which has made it a particularly important and prevalent developmental stage for Drosophila research (Ashburner 1973; Muskavitch and Hogness 1980; Basler and Struhl 1994; Mirth et al. 2005; Baker and Thummel 2007; Gerber and Stocker 2007; Gomez-Marin and Louis 2012; Makhijani and Bruckner 2012; Tennessen et al. 2014). To gain some insight into transcript expression and localization during this stage, we developed a method for semi-high-throughput TSA-FISH that includes the majority of larval tissues (Materials and Methods; Wilk et al. 2010, 2013). To date, we have analyzed ∼800 genes and defined >150 new terms to describe these new expression and localization patterns. Ninety-six percent of all successfully generated probes provided detectable expression patterns in these tissues, and, somewhat surprisingly, 91% of these exhibited some type of subcellular distribution (Tables 1, 2). In terms of tissues, the subcellular localization rates range from a high of 79% and 85% for larval fat body and malpighian tubules, respectively, to a low of 46% in the gastric caeca (Table 3). In tissues that were present and examined in both embryos and larvae, the prevalence of observed subcellular localization was always much higher in larvae, with 42% of transcripts that are subcellularly localized in larval tissues not noted to be localized in embryos (see Tables 1, 3). This suggests that localization may also occur earlier but is not sufficiently obvious due to the smaller cell sizes. An alternative possibility is that the localization mechanisms or requirements observed in larval cells may not exist or be needed at earlier stages (see the Discussion).
Table 3.
Data analysis by tissue
New and prevalent subcellular distribution categories
One of the most common localization categories observed during later developmental stages is “cytoplasmic foci” (refer to Table 2). Supplemental Figure S1 shows examples of transcripts with and without foci accumulation. Patterns without foci are expected to have relatively evenly dispersed transcripts, while foci are expected to contain multiple transcripts, with size and intensity proportional to transcript numbers and additional components. The numbers of foci per cell and their subcellular locations also varied significantly between different transcripts (note that most RNAs localized in foci also shared other subcellular terms). Although there was considerable variability in foci characteristics, each type observed was highly reproducible in that all embryos or larval tissues showed the same distributions for a given probe, and experimental repeats also produced the same results.
Although there appear to be many types of foci, the limited resolution of our fluorescent microscopy analyses has made the definitive separation of the foci category into clear subcategories difficult. That said, many of these focus transcripts encode proteins found in different subcellular organelles or structures to which RNAs are now known to localize (see the Discussion). Together with the use of specific organelle markers, these functional associations may allow future stratification of the foci category.
Figure 2 shows examples of transcript foci whose products are localized within a variety of organelles to which mRNAs may also be localized. The foci in Figure 2, A–F, all encode proteins that are localized within or transported by various components of the cellular secretion machinery. For example, the cytochrome P450-encoding transcripts phantom (phm) and Cyp6a21 (Fig. 2A,B) both encode ER proteins. ruby (rb) (Fig. 2C) encodes a coated pit-localized protein, and Figure 2D shows enrichment of fat transcripts along membranes that face parasegmental grooves. fat encodes a transmembrane protein that controls cell planar polarity and growth. In contrast, Neuroligan1 (Nlg1) foci are polarized away from invaginating cell surfaces (Fig. 2E). Finally, Tequila (Fig. 2F) encodes a secreted peptidase.
Figure 2.
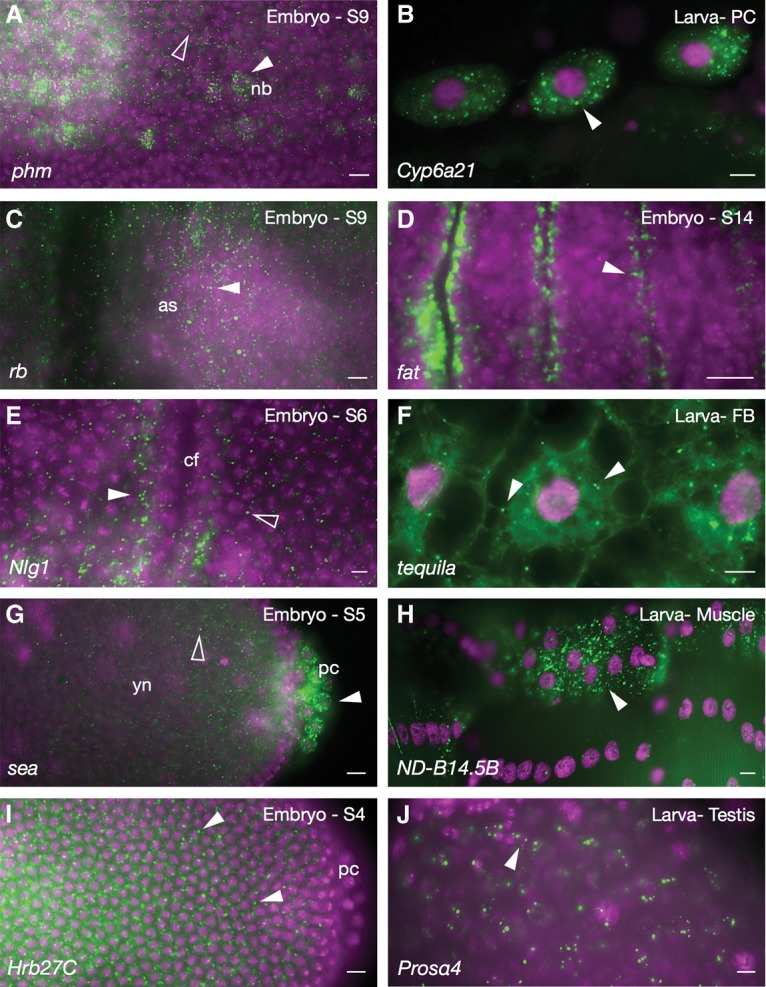
Cytoplasmic foci. (A) phm mRNA transcripts in foci surrounding neuroblast nuclei (nb; white arrowhead). Occasional foci are also detected in nondelaminating cells (empty arrowhead). (B) Larval pericardial cells (PC) showing Cyp6a21 cytoplasmic foci (arrowhead). (C) rb transcript foci in the amnioserosa (as). (D) fat mRNA in small foci enriched at membranes facing parasegmental grooves (arrowhead). (E) Nlg1 foci (white arrowhead) enriched at membranes facing away from the invaginating cephalic furrow (cf); foci are less abundant in neighboring cells (empty arrowhead). (F) tequila cytoplasmic foci in third instar larval fat body (FB). (G) scheggia (sea) transcripts around pole cell nuclei (white arrowhead) and at lower levels in the yolk (empty arrowhead). (yn) Yolk nuclei. (H) Larval muscle showing the mitochondrial NADH dehydrogenase (ubiquinone) B14.5 B subunit (ND-B14.5B) transcript in cytoplasm. (I) Heterogeneous nuclear ribonucleoprotein at 27C (Hrb27C) foci in an early embryo (arrowheads). (J) proteasome α subunit 4 (prosα4) foci (arrowhead) in larval testis. Names of transcripts are indicated at the bottom left of each panel, and developmental stage is indicated at the top right. Colors and labels are as described in Figure 1.
Foci encoding proteins associated with other types of subcellular structures include scheggia (sea) (Fig. 2G) and NADH dehydrogenase B subunit (ND-B14.5B) (Fig. 2H), which encode mitochondrial proteins; Heterogeneous nuclear ribonucleoprotein at 27C (Hrb27C) (Fig. 2I), which encodes an RNA-binding protein enriched in P bodies (Thomson et al. 2008); and the unusual foci of proteasome α4 (prosα4) mRNA (Fig. 2J), which encodes a major component of the proteasome complex.
Another frequent localization category during later developmental stages was “perinuclear” (Table 2). In this case, distinctive subcategories were more readily defined (Fig. 3). These subcategories varied from limited numbers of perinuclear foci or clusters to larger numbers of foci or clusters that either completely encircle the nucleus or show consistently polarized distributions. The potential impact for many of these patterns on related functions was often clear. For example, dilute class unconventional myosin (didum) (Fig. 3B) encodes a myosin protein involved in the movement of transcripts and mitochondria along microtubules in a minus-to-plus direction (Toth et al. 2005; Yang et al. 2006; Krauss et al. 2009). Since microtubules are attached to the nucleus at their minus ends, translation of didum transcripts at the nuclear surface would position the protein perfectly for use. In another example, the polarized distribution of Hsp83 transcripts on one side of the nucleus (Fig. 3E) looks remarkably similar to a number of nuclear receptor transcripts, which we previously colocalized together with subregions of ER (Wilk et al. 2013). This colocalization could be associated with the known roles of Hsp83 as a chaperone and regulator of nuclear receptor folding and function, with perhaps an additional level of regulation at the level of translation. Catalase (Cat) (Fig. 3F) encodes an enzyme that has been localized to peroxisomes, and Mothers against dpp (Mad) encodes a transcription factor that translocates to the nucleus in response to Dpp signaling.
Figure 3.
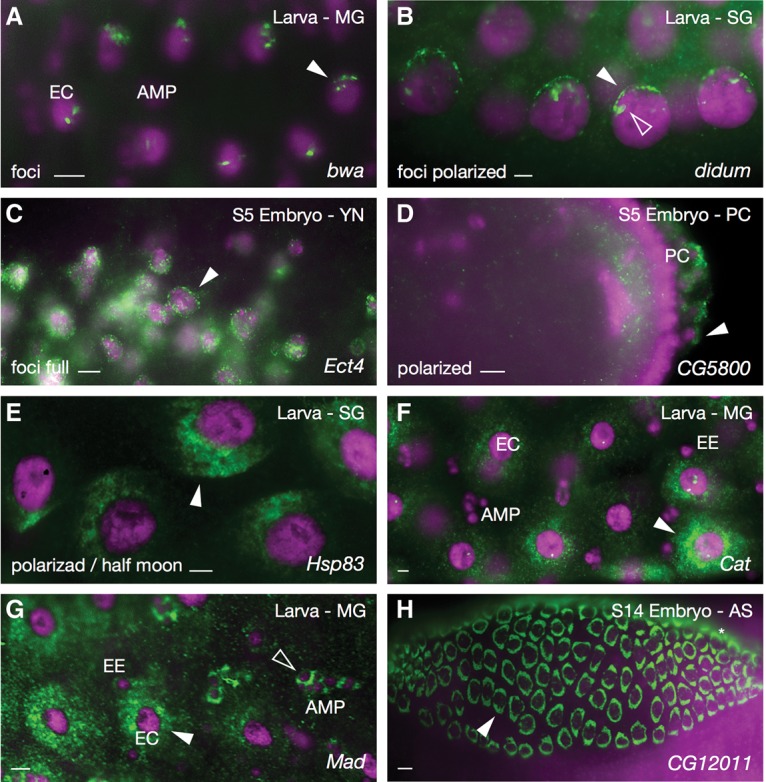
Perinuclear patterns. (A) brain washing (bwa) foci at the edges of enterocyte (EC) nuclei in third instar larval midgut (MG). (B) Foci of didum enriched at the apical surfaces (arrowheads) of larval salivary gland (SG) nuclei. (C) Ectoderm-expressed 4 (Ect4) foci surrounding yolk nuclei (arrowhead). (D) Polarized localization of CG5800 transcripts around embryonic pole cells (PC; arrowhead). (E) Polarized “half moon” pattern of Hsp83 transcripts around larval salivary gland nuclei (arrowhead). (F) Catalase (Cat) transcripts surrounding enterocyte nuclei (arrowhead) and partially surrounding adult midgut cell precursor (AMP) and endocrine (EE) nuclei of larval midgut cells. (G) Mothers against dpp (Mad) transcripts surrounding nuclei of large enterocyte (white arrowhead), small endocrine, and adult midgut cell precursor (empty arrowhead) cell nuclei in third instar larval midgut. (H) Solid perinuclear rings of CG12011 transcripts around amnioserosa (AS) cell nuclei (arrowhead). Colors and labels are as described in Figure 1.
Another frequently observed mode of transcript localization in later developmental stages, particularly in tissues such as yolk plasm, neurons, trachea, and muscle, was within or along cellular processes (Fig. 4). Examples shown include transcripts encoding the anion transporter Organic anion transporting polypeptide 58Dc (Oatp58Dc) and the cytochrome P450 Cyp6d5 in yolk processes (Fig. 4A,B), Ankyrin 2 (Ank2) and GABA transporter (Gat) in axons (Fig. 4C,D), Osiris 19 (osi19) and CG13627 in processes extending between tracheal precursor cells (Fig. 4E,F), and Wings up (WupA) and bent (bt) within embryonic muscles (Fig. 4G,H). Once again, the molecular roles of the encoded proteins, taken together with transcript localization, have major implications for the use and importance of these various subcellular localization patterns.
Figure 4.
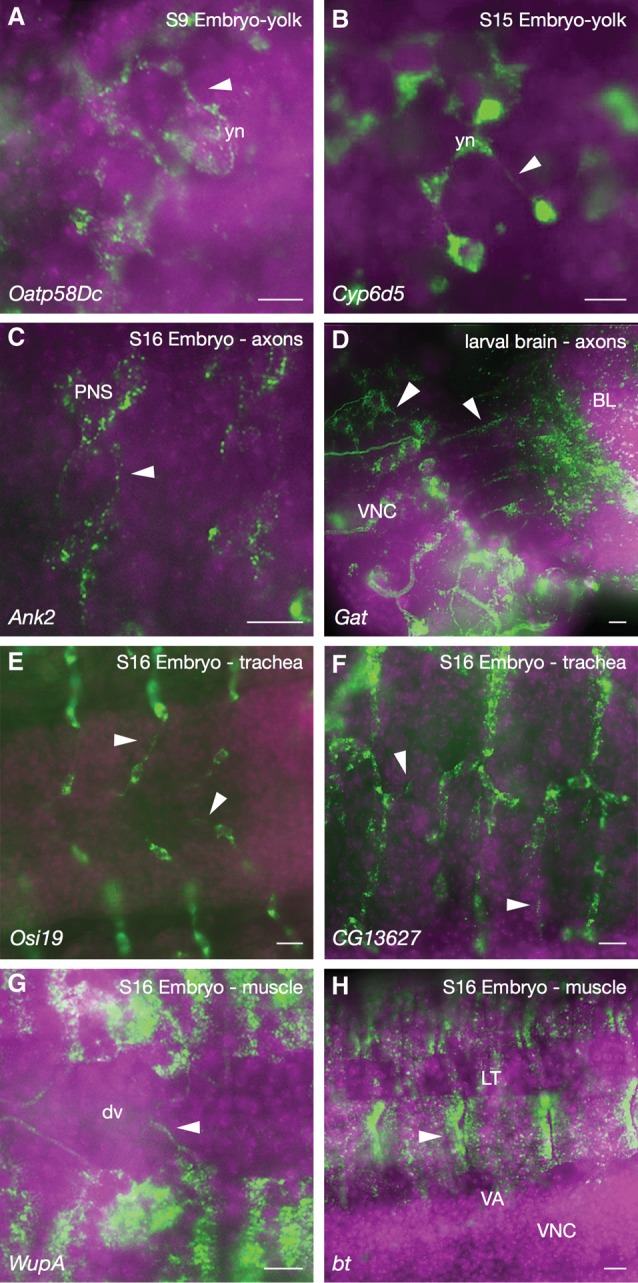
Cell extensions and processes. (A,B) Oatp58Dc (A) and Cyp6d5 (B) localized along filamentous yolk processes in embryonic yolk tissue. (yn) Yolk nuclei. (C) Ank2 transcripts in axons emanating from the peripheral nervous system (PNS) neurons (arrowheads). (VNC) Ventral nerve cord. (D) Transcripts encoding the neurotransmitter Gat in axons (white arrowheads) in third instar larval brain lobes (BL). (E,F) Osi19 (E) and CG13627 (F) transcripts within tracheal cell extensions (arrowheads). (G,H) WupA (G) and bt (H) transcripts in muscle cell structures and extensions (arrowhead). (dv) Dorsal vessel; (LT) lateral transverse muscle; (VA) ventral acute muscle. Colors and labels are as described in Figure 1.
In the case of muscles, which, in their mature form, are syncytial in nature, we expect to see a great deal more detail and examples of localization in the larger larval muscle tissues. Until recently, however, the majority of this larval tissue was lost upon removal of the cuticle/carcass during sample preparation.
Expression and subcellular localization of lncRNAs
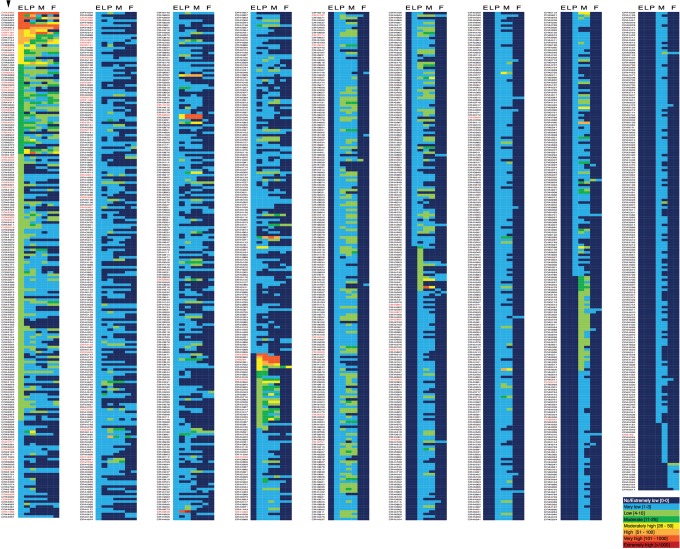
A query of the FlyBase database for lncRNAs identified 2419 CR (noncoding) genes that are >200 nucleotides (nt) in length. These produce 2858 different RNA products, with an average size of 1008 nt (Supplemental Fig. S2). Of these 2419 annotated lncRNA genes, expression data for 1823 were found in the recent developmental expression profiling project carried out by modENCODE (Brown et al. 2014). These data were consolidated to generate Figure 5, which is a compilation of expression data for embryo, larva, pupa, and adult stages of development. A couple of the immediately noticeable features of this data set are the number of lncRNAs that are undetectable or expressed at low levels of expression and the number that are expressed in males and not females (Fig. 5, cf. columns M and F). Many of the latter are likely to be due to expression in testes, as modENCODE has noted that ∼30% of identified lncRNAs are expressed there, with ∼6% expressed exclusively in testes (Brown et al. 2014; Brown and Celniker 2015).
Figure 5.
List of Drosophila lncRNAs and associated expression profiles. List of 1834 lncRNAs noted in the FlyBase database for which RNA sequencing (RNA-seq) expression data (obtained by modENCODE) are available. CR gene numbers are listed at the left in each of the nine columns (first column indicated with an arrowhead), with expression data heat maps at the right. Expression levels for embryonic (E), larval (L), pupal (P), adult male (M; 1 + 5 d old and 30 d old), and adult female (F; 1 + 5 d old and 30 d old) are color-coded, ranging from undetected (dark blue) to very high (red), as described in the bottom right inset. The list is ordered from highest (top left) to lowest (right) expression. Gene names in red text indicate genes examined in this study. Twenty-seven additional lncRNAs not examined by modENCODE were also examined in this study.
For our initial localization analyses, probes were generated for all lncRNAs for which templates were found within the DGC-1 or DGC-2 cDNA libraries that we have been using to generate probes. Along with these, we also generated probes for the nine annotated lncRNAs located within the Antennapedia (AntP-C) or Bithorax (Bx-C) complexes (see the Materials and Methods). Not unexpectedly, the majority of the 103 transcripts examined showed interesting and complex subcellular distribution patterns, suggesting a corresponding diversity in subcellular functions and localization mechanisms. Approximately 65% of the lncRNAs tested were provided maternally, 34% were exclusively expressed zygotically, and 75% were expressed during some stage of embryonic development. All but one were detected in at least one third instar larval tissue.
In terms of subcellular localization, ∼90% were clearly subcellularly localized during embryogenesis, and 98% were subcellularly localized in larvae. While it has been contended that the majority of lncRNAs is nuclearly localized (Palazzo and Lee 2015), we observed only 4% localized exclusively within the nucleus. Forty percent were exclusively cytoplasmic, and 56% were found in both. Examples of these cellular and subcellular distributions are shown in Figures 6 and 7.
Figure 6.
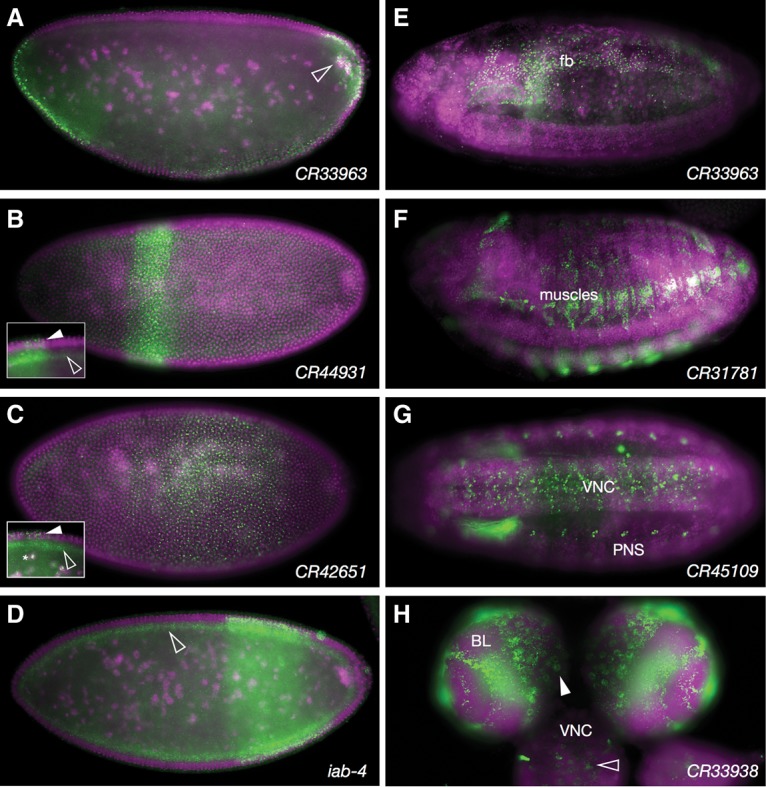
lncRNA expression patterns. (A–D) Lateral views of stage 5 embryos showing nascent transcript expression from lncRNAs expressed in stripes. The empty arrowhead in A points to cytoplasmic transcripts enriched around yolk nuclei. Solid and empty arrowheads in B (inset), C (inset), and D highlight nuclear localization in stripes and ubiquitous cytoplasmic localization both in and outside stripes. (E–H) Representative lncRNA expression patterns in common tissues of expression. (E) CR33963 expression in fat body (fb). (F) CR31781 expression in somatic muscles. (G) CR45109 expression in the ventral nerve cord (VNC) and peripheral nervous system (PNS). (H) CR33938 expression in brain lobes (BL) and ventral nerve cord of a third instar larva. Colors and labels are as described in Figure 1.
Figure 7.
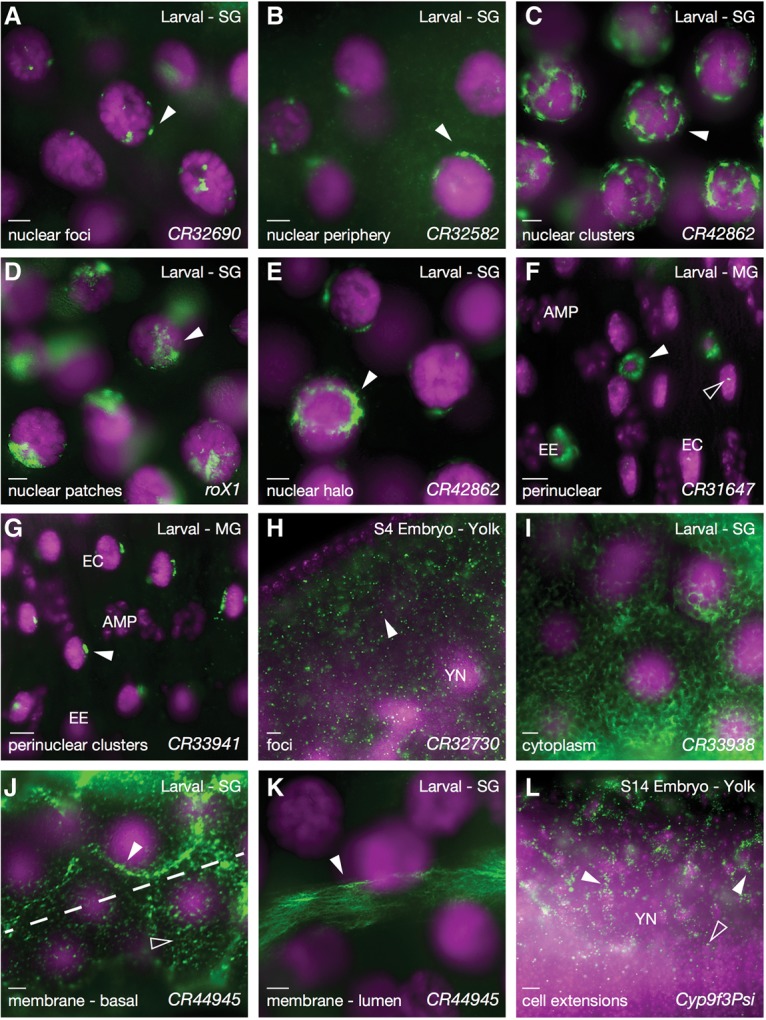
Subcellular lncRNA patterns. (A–E,) Third instar salivary gland cells showing various types of nuclear and perinuclear lncRNA distributions. Gene names are indicated at the bottom right of each panel. (F,G) Third instar larval midgut showing nuclear/perinuclear localization in different cell types. (EE) Endocrine cells; (EC) enterocytes; (AMP) adult midgut progenitors. (H) Stage 4 embryo showing cytoplasmic foci for CR32730 (arrowhead). (I–K) Larva salivary gland cells. (I) Cytoplasmic distribution of CR33938. (J,K) Membrane association for the same RNA, CR44945, at two different stages of differentiation in the same salivary gland, showing an earlier stage in proximal cells (J) and a later stage in distal cells (K). J is also divided into two focal planes (peripheral cell surface in the bottom pane and just under the cell surface in the top pane). The focus in K is on the opposite side of the cell (luminal). (L) Stage 14 embryo showing localization along yolk plasm extensions. Colors and labels are as described in Figure 1.
Figure 6, A–D, reflects the diversity of embryonic anterior–posterior striped expression patterns of the examined lncRNAs, three of which are located in the AntP-C or Bx-C Hox clusters. The exception, CR33963 (Fig. 6A,E), lies within the first intron of the gene Tis11, which encodes an RNA-binding protein that regulates mRNA stability (Choi et al. 2014). Although the Hox complex lncRNA transcripts are generally expected to have nuclear roles in transcriptional regulation, a significant proportion of these RNAs was found in the cytoplasm. Interestingly, while cytoplasmic distribution was generally seen across the embryo, nuclear localization was seen only in the striped expression domains associated with adjacent Hox gene expression. This differential distribution of nuclear and cytoplasmic localization was true for all of the Hox lncRNAs tested (see Fig. 6B,C, insets). All were also seen to be redeployed in mesoderm and gut tissues during mid-embryogenesis, but localization at these stages was exclusively cytoplasmic (see Fly-FISH). The presence of related lncRNAs in the Hox complexes of other metazoa as well as recent demonstrations of function (Rinn et al. 2007; Brock et al. 2009; Gupta et al. 2010) suggest that these varying expression patterns and subcellular distributions may also be evolutionarily conserved aspects of Hox lncRNA functions.
Figure 6, E–H, shows other tissues with relatively frequent and intricate lncRNA expression patterns. These include the nervous system, muscles, and fat body. For the majority of these RNAs, expression was tightly limited to one specific tissue and/or developmental stage.
In terms of subcellular distributions, as with coding RNAs, the lncRNAs examined were detected in virtually all parts of the cell. Figure 7 illustrates this diversity, starting with limited distributions in the nucleus and working out toward the cellular extremities. Figure 7, A–G, shows nuclear localization patterns ranging from very limited foci to more extensive foci, clusters, patches, and distributions that completely encircle the nucleus. The majority of these nucleus-associated patterns likely represents RNAs bound to chromatin or associated structures. Figure 7, H and I, shows several cytoplasmic distributions, and Figure 7, J–L, shows membrane-associated and cellular extension patterns. Notably, CR44945 (Fig. 7J,K) is first seen in the salivary gland at peripheral/basal membranes and later moves to the opposite luminal/apical membrane surface.
In many cases, lncRNA genes overlap with other coding and noncoding genes in either sense or antisense orientation. These overlaps with coding and regulatory regions suggest potential modes of action. In some of these cases, though, this also complicates the interpretation of expression data, as probes may not distinguish between CR expression and overlapping coding gene expression. This is the case for CR45109 (Fig. 6G), which overlaps with the 5′ untranslated region (UTR) of isoforms A and B of the 2mit gene. Interestingly, both CR45109 and 2mit are contained within an intron of the circadian rhythm gene Timeout, which is transcribed in the opposite direction. The expression patterns observed here for CR45109/2mit in the nervous system, taken together with previous evidence of 2mit in the control of behavior and the overlaps between CR45109, 2mit, and timeout, are all consistent with potential cross-regulatory interactions made possible by overlapping sequences and cellular/subcellular expression patterns. In total, 10 of the 108 probes used in this study had the potential to detect both coding and noncoding RNAs.
The majority of lncRNAs shown in Figure 6 was found by modENCODE RNA sequencing (RNA-seq) analyses to be expressed at low, very low, or undetectable levels during the majority of development. This was the case for the majority of lncRNAs that we examined (indicated with red text in Fig. 5), yet we clearly observed robust patterns of expression in each case. For example, the expression patterns shown in Figure 6, A, C, D, and E, are from stages at which modENCODE lists expression as very weak. For the patterns shown in Figure 7, F, H, J, and K, expression is listed as nondetected in those stages and tissues. We attribute these discrepancies to the ability of FISH to preferentially detect expression that is temporally or spatially focused. In addition, the concentration of transcripts within specific subcellular regions can also enhance signal detection over those of more diffusely localized products.
Bioinformatics
Large-scale gene expression studies have provided the basis of many functional analyses that have been able to link related gene functions and pathways. Examples of such studies include genome-wide analyses of particular cell types, tissues, developmental stages, or diseases. The data being compiled in Fly-FISH provide advantages over other gene expression databases in that Fly-FISH includes high-resolution spatial data at both the cellular and subcellular levels. As such, it provides an additional level of association with greater potential to identify genes and products that share common functions and may interact directly. As a quick test of this potential, we performed gene ontology (GO) and association studies for several classes of colocalized transcripts using the publicly available program GeneMANIA (http://www.genemania.org), which also incorporates known physical, genetic, and coexpression data (Warde-Farley et al. 2010). In each case, related and logical GO term functions were clearly enriched as well as numerous, previously identified and unidentified genetic and physical interactions. Figure 8 and Supplemental Figure S3 show several example analyses of RNAs from various subcellular localization categories.
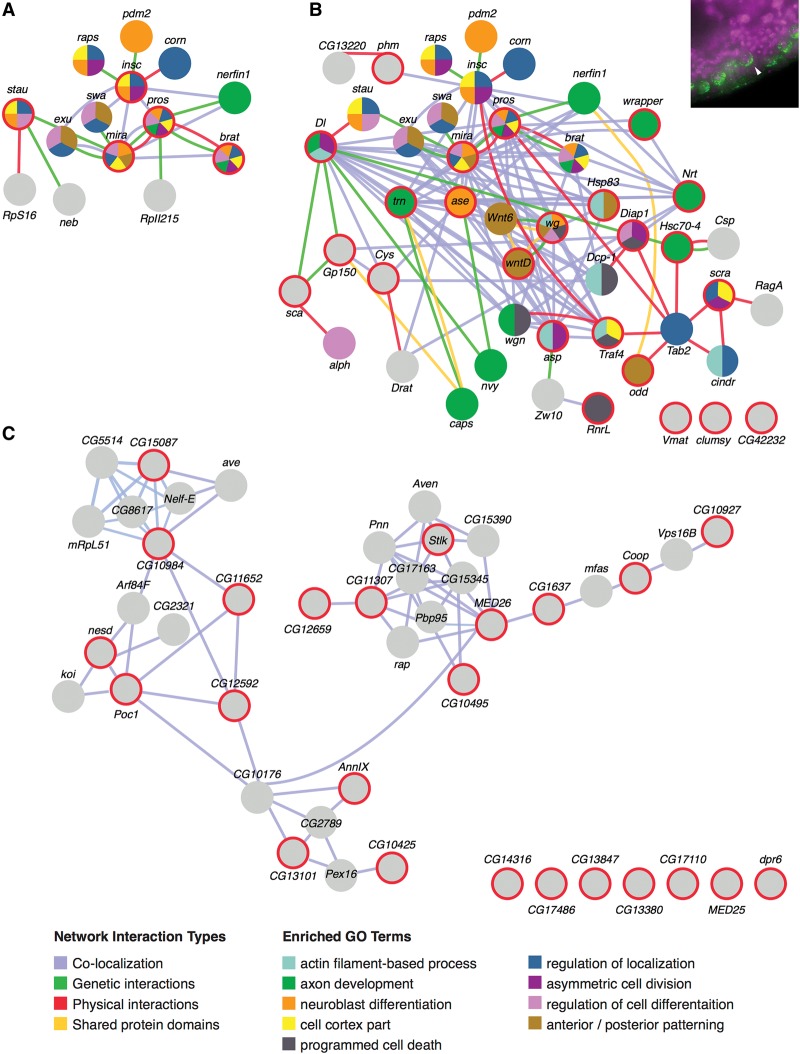
Figure 8.
Interaction networks and functional enrichments for RNAs in the “polarized neuroblast” category. GeneMANIA (http://www.genemania.org) was used to generate interaction networks and GO term enrichments for query genes and other genes previously known to associate at various levels. (A) Network generated from a query of genes (spheres circled in red) previously known to be important for differential RNA and protein localization to neuroblast daughter cells. Additional spheres (no red rings) indicate genes found by GeneMANIA that are known to interact physically (red lines) and genetically (green lines), colocalize (blue lines), or share protein domains (yellow lines). Colors within the circles show the most enriched GO terms for each gene (listed in the functions legend). (B) Network generated from a query of 24 transcripts scored with the subcellular localization term “polarized in delaminating neuroblasts” (an example localization is shown in the top right inset). Red circles indicate query genes. Query genes that have no known associations within the network are shown at the bottom right. (C) Negative control network generated from 24 randomly selected genes.
Figure 8 focuses on a localization category with a convenient number of genes that make this type of analysis relatively easy to illustrate. Figure 8A is a GeneMANIA-organized analysis of genes previously known (spheres with red rings) to be involved in the asymmetric localization of RNAs in neuroblasts that are moving out of the blastoderm layer to populate the newly forming ventral nervous system (an example is shown in the inset in Fig. 8B). Note that GeneMANIA also populates the interaction map with additional genes that have previously been shown to encode gene products that interact at some level (Fig. 8, spheres without red rings).
Figure 8B shows that the geneMANIA output generated from the 24 transcripts identified thus far that localize asymmetrically in the same delaminating neuroblasts (spheres with red rings). The first point to be made from this network is the greatly expanded number of nodes and interactions. Not unexpectedly, there is a continued strong enrichment for cell polarity and neurogenic GO term functions. In comparison, Figure 8C shows a typical interaction map generated by the same number of randomly selected query genes. Note that there is no enrichment of GO terms (Fig. 8C, gray spheres) and many fewer interactions, and the majority of interactions noted is limited to coexpression (Fig. 8C, blue lines only). Thus, the many new interactions introduced in Figure 8B are likely to be highly informative and serve major new roles in asymmetric RNA trafficking and function.
One of the more striking new sets of interactions observed in Figure 8B is the additional direct physical interactions (red lines) and, in particular, those that connect the Prospero interaction hub with that of TAK1-associated binding protein 2 (Tab2; Pros-Tab2, and Insc-Traf4). The many new interaction partners linked by these interactions are enriched for asymmetric localization, actin/microtubule regulation, and membrane receptor functions. The presence of Zn fingers in Tab2 and Traf4 also suggests possible additional RNA interaction motifs. Thus, Tab2 may serve as a major scaffold or node for pairing neuron-specific and nonspecific cellular asymmetry structures and processes. Curiously, many of the components in the Tab2 subnetwork are also associated with cell death and ubiquitination functions.
While the number of direct interactions shown in Figure 8B is already far higher than random, we expect that there will be many more direct interactions than those currently indicated. For example, the many genetic and colocalization lines connecting transcripts such as Delta, asymmetric cell processes (asp), asense (ase), abnormal spindle (asp), Cystatin (Cys), Neurotactin (Nrt), and tartan (trn) suggest that some of these are particularly likely to also interact directly.
As noted earlier, GeneMANIA populates clusters with genes (Fig. 8B, spheres without red rings) that have previously been shown to associate with the query genes. When examined in Fly-FISH, some of these, such as staufen, corn, and nerfin-1, were clearly expressed in neuroblasts but not in a notably polarized distribution. The proteins encoded by these mRNAs may achieve localized distributions or activities via interactions with one or more of the other localized transcripts or proteins. In one case, however, we did observe similar subcellular localization for a “GeneMANIA-recruited” transcript that had not initially been noted as localized in this fashion (Laminin B2 in the perinuclear group). For some of the other GeneMANIA-recruited genes, expression in neuroblasts was not observed, suggesting that these interactions may be restricted to other cell types or stages. For example, exu and swa are known to play roles in setting up posterior identity in the early embryo. Additional examples of GeneMANIA-generated clusters are shown in Supplemental Figure S3.
Discussion
Until now, studies on Drosophila RNA localization have focused primarily on transcripts that are localized during the first few hours of embryogenesis and a handful of localization mechanisms. In this study, we opened this area of study to include the most studied stages of Drosophila development, which include all of embryogenesis and the majority of late third instar larval tissues. In doing so, we discovered dozens of new subcellular distribution patterns and implied mechanisms. The enormous amount of new information now documented in our recompiled Fly-FISH database will be useful for the majority of current and future Drosophila research projects.
Subcellular localization prevalence
Many thought that the high rates of RNA localization that we observed previously in early Drosophila embryos (Lecuyer et al, 2009) might be a Drosophila anomaly that arose due to the unusual requirements for defining anterior–posterior and dorsal–ventral coordinates in a syncytial environment. Instead, our results indicate that these mechanisms were already in existence and in use in other developmental stages and tissues. In fact, it was probably the prior existence of these pervasive processes that facilitated the evolutionary advance of syncytial embryonic patterning, which in turn increased the speed and efficiency of axial patterning by one or more orders of magnitude.
While the later stages of embryogenesis examined here have revealed many new types of localization, it was the third instar larval tissues that provided the most new insights and reliable estimates in terms of subcellular localization frequencies and details. Most notably, some form of subcellular localization was observed for essentially all of the RNAs annotated thus far. Although this extremely high frequency of localization may be attributed in part to the relatively large sizes of these cells and the consequential spatial resolution and ease of detection, it is also possible that this prevalence is due to the unusual and perhaps nonconserved properties of some of these specialized cell types. However, a number of observations argue to the contrary. First, the fact that these RNAs can all be differentially localized indicates that the necessary cis elements and cellular machineries are available for potential use in other cell types. Second, polyploid cells are not unique to larval stages of development or Drosophila. Third, when examined in other highly polarized cell types, such as neurons, other studies have also shown extremely high rates of RNA subcellular localization (Bruckenstein et al. 1990; Olink-Coux and Hollenbeck 1996; Mercer et al. 2008). Finally, recent studies have shown that mRNAs encoding common organelle-specific gene products, such as those that function in the ER, mitochondria, and P bodies, can also be copurified bound to these organelles (Decker and Parker 2012; Reid and Nicchitta 2012; Kraut-Cohen et al. 2013; Lesnik et al. 2015). Indeed, these biochemical approaches have suggested that as much as 50% of cytosolic protein-encoding transcripts are translated on ER (Reid and Nicchitta 2012; Jagannathan et al. 2014). We also showed previously, using double labeling, that transcripts encoding transcription factors can also be enriched and translated on ER (Wilk et al. 2013), suggesting that ER localization may be a common means of localizing transcripts that encode numerous types of proteins. In the case of the nuclear receptor transcripts that we found to be associated with ER, this may provide a number of advantages, including proximity to the nucleus, proximity to translational regulators and chaperone cofactors (e.g., Hsp83 RNA also appears to be similarly localized), or proximity to ER-derived ligands (e.g., the E75 ligand heme is enriched in ER) (Reinking et al. 2005). Future studies using suitable markers and higher-resolution detection methods should soon provide more accurate numbers and clearer explanations for these observations and questions.
Subcellular localization relevance
Numerous studies have documented the functional importance of various subcellular RNA localization processes, including roles in various diseases (Augood et al. 1999; Tanimukai et al. 1999; Belanger et al. 2003; Bassell and Kelic 2004; Koensgen et al. 2007; An et al. 2009; Martin and Ephrussi 2009). This importance is also emphasized by studies showing that localized RNAs tend to be translationally repressed and less stable when not at their proper sites of localization (Lipshitz and Smibert 2000; Lecuyer et al. 2009; Martin and Ephrussi 2009). However, the prevalence of localization noted here might suggest that even higher rates of cellular malfunctions and disease should result from disrupted localization events. The fact that this is yet to be shown has several possible explanations. First, the study of RNA localization is still a relatively young field, and more extensive and directed studies will be required to make such assessments. Second, many of these events may make relatively modest contributions to protein localization and function due to redundancy in RNA and protein localization mechanisms. Third, the effects of mislocalization may be manifested only in specific cell types or conditions that have yet to be addressed. Finally, the impacts of mislocalization for many transcripts may only become obvious when multiple transcripts are affected. For example, the cumulative benefits of having most RNAs localized and translated in specific cell regions may be critical for ensuring overall cellular efficiency and organization, but affecting these events in small numbers may have relatively little impact.
Subcellular localization in other organisms
This study clearly shows that subcellular RNA localization is a pervasive and crucial level of gene regulation at all stages of Drosophila development. Other recent studies are also beginning to imply similar levels of localization in other organisms (Taliaferro et al. 2014; Weil 2014; Jambor et al. 2015; Liao et al. 2015). However, functional studies that validate the relative importance and underlying mechanisms behind the majority of these localization events are lagging behind. This is particularly true for lncRNAs.
lncRNAs in development
Although ncRNAs have been receiving a great deal of attention of late, the subclass of lncRNAs has seen considerably less attention, due largely to the relative lack of previous information and interest as well as their heterogeneity in size and function. It has also been suggested that the majority of lncRNAs is genomic “junk.” However, the striking expansion in size of this class of transcripts in higher eukaryotes and the tight correlation of lncRNA numbers with organism complexity have suggested a major role in driving animal evolution and complexity (Mercer et al. 2008; Wilusz et al. 2009). An appreciation of their overall importance and range of functions will require the mapping of both cellular and subcellular distribution patterns.
The strongest arguments previously made against the functional importance of the majority of lncRNAs are (1) their relatively low levels of expression, (2) their low levels of sequence conservation, and (3) their retention in the nucleus (Palazzo and Lee 2015). As elaborated below, our results counter the assumptions of arguments 1 and 3, and, in terms of their low levels of conservation, this can be explained by their relatively recent and ongoing introduction into the genome and their lack of protein encoding restrictions.
As a beginning, we analyzed the embryonic and larval third instar expression patterns for >100 of the relatively manageable ∼2400 currently predicted Drosophila lncRNAs. For those that we analyzed, all were found to be expressed at some stage of development, with many showing relatively high degrees of tissue selectivity, implying more highly specialized functions than those of most protein-coding genes.
In terms of expression levels and distribution, the modENCODE project provided initial evidence of expression for 1823 (75%) of the 2419 currently annotated Drosophila lncRNAs over the course of development. Many of those missing from this analysis may have been missed due to ongoing annotation changes and refinements. Consistent with this likelihood, 27 of the 103 lncRNAs that we examined were not among those listed in Figure 5. Approximately half of these show recent category changes into CR status or continued listing as pseudogenes. The other half of the 27 not listed in Figure 5 are listed as nonexpressed by modENCODE, yet we observed clear and interesting patterns of expression for all of them. Similarly, most of the other lncRNAs listed as very low or nonexpressed that we examined showed easily detected and often robust patterns of expression when analyzed by FISH. This may reflect the ability of FISH to focus in on specific stages, tissues, cells, and/or subcellular regions to detect relatively high levels of expression that might otherwise be averaged out in time and space. The results obtained here suggest that the majority of existing lncRNAs will likewise be found to be expressed in interesting patterns and tissues during some stage of development.
Another argument that has been made in favor of the majority of lncRNAs being nonfunctional is that most are believed to be restricted subcellularly to the nucleus, where they would not cause additional problems due to the spurious translation of junk polypeptides or participation in deleterious interactions. However, of the 103 lncRNAs that we examined, the majority showed cytoplasmic localization at some time or place, with most being predominantly cytoplasmic. These diverse and dynamic cytoplasmic and membrane-associated distributions suggest not only specific functions in these regions but also a greater diversity of functions and mechanisms of action than has been previously imagined. Even in the nucleus, the diversity of subnuclear distribution patterns observed here suggests functions in addition to the previously studied roles in epigenetic chromatin regulation (Umlauf et al. 2008).
Another observation that does not appear to have been made before is the striking correlation between the increasing numbers of lncRNAs and C2H2 Zn finger proteins over the course of evolution. Current research is focused on the assumption that these Zn finger proteins are DNA-binding transcription factors (Emerson and Thomas 2009; Najafabadi et al. 2015). However, it is perhaps more likely that they function as RNA-binding proteins (or both), with lncRNAs serving as an important set of coevolving targets.
At present, preliminary data suggest that Drosophila is at a relatively early stage in the burst of lncRNA amplification and functionality relative to vertebrates (e.g., ∼2300 lncRNAs in Drosophila vs. ∼59,000 in humans) (Iyer et al. 2015). As such, Drosophila should provide a more tractable system than vertebrate models for the further validation and study of this transcript class as well as its evolutionary role in driving eukaryote complexity and diversity.
Bioinformatic opportunities
As the quantity and quality of data in the Fly-FISH database continue to grow, opportunities to draw out patterns of functional correlation will also grow. Here, we provided a glimpse of the potential information that can be gleaned from these data and such analyses (Fig. 8; Supplemental Fig. S3). These show how the inherent implications of subcellular colocalization can be used to flesh out new or greatly expanded gene, protein, and RNA interaction networks. More extensive and sophisticated analyses will also identify numerous new functions for previously unstudied genes, new subcellular localization elements and pathways, new subcellular organelles and structures, and new cellular processes and pathway intersections.
Materials and methods
Preparation and in situ hybridization of embryos
To generate a relatively homogeneous distribution of embryo stages, equivalent numbers were collected, aged in three groups (0–2.5 h, 0–4.5 h, and 0–20 h), and then mixed together after fixation. Subsequent steps were carried out in batch form until the completion of prehybridization, and then embryos were aliquoted into 96-well plates. Details of embryo, probe, and signal preparation/production are described in Wilk et al. (2010). The following nine oligos (5′ to 3′) were designed for specific lncRNA probe template production via PCR of genomic DNA: CR44945 (Forw, CACCCCTTTTCGATCTGAGG; Rev, GTAATACGACTCACTATAGGGAGACCACAATTCCACTGAACGAGCTGG), CR45750 (Forw, AACAAGAGAAACACGGACGC; Rev, GTAATACGACTCACTATAGGGAGACCACGGTCTCCCAGTTTTTCCAGG), CR45751 (Forw, GCA TGTGGTCTGAGTCTACG; Rev, GTAATACGACTCACTATAGGGAGACCACCCCAGTTTCTATGAGCCGAT), CR31271 (iab-4) (Forw, TAACGTGGAAAATCGGCCCA; Rev, GTAATACGACTCACTATAGGGAGACCACTACCCCAGTGAGTGGCGATA), CR43261 (iab-8) (Forw-GTAATACGACTCACTATAGGGAGACCACATGGGTCATACAGCGAGTGC; Rev-CAGCGCAACTTGACTCAACC), CR31273 (bxd) (Forw, GTAATACGACTCACTATAGGGAGACCACACGCACTGCCAGAAGTTCAT; Rev, TGTACTTGTGGTTCCACCCG), CR43617 (Forw, GAGGAGGGGAAGTGGAGGAT; Rev, GTAATACGACTCACTATAGGGAGACCACAGGGGCATTATGGCAACGAA), CR42651 (Forw, GTAATACGACTCACTATAGGGAGACCACTACAGGGTAGACGTGGAGCA; Rev, TTAAAGAAGTGGCGCCGAGT), and CR44931 (Forw, GTAATACGACTCACTATAGGGAGACCACCAGTAGAGCCAACTCGACCC; Rev, AGTGTCTGAATCACTGGGCG).
Dissection, fixation, and in situ hybridization of third instar larval tissues
Climbing third instar larval tissue preparation and hybridization were performed as described (Wilk et al. 2010, 2013) with the following modifications: 40% paraformaldehyde (PFA) stock that was made fresh prior to use (3.68 g of PFA, 10 mL of DEPC H2O, 70 µL of 2N KOH), 1× PBT (PBS with 0.1% Tween-20), 4% PFA (1 mL of 40% PFA stock diluted in 9 mL of 1× PBT and then filtered through a 0.45-µm filter unit), PBTT fix solution (1 mL of 40% PFA stock diluted in 9 mL of 1× PBT, 30 µL of 0.3% Triton-X100, and 10 µL of 0.1% picric acid solution and filtered through a 0.45-µm filter unit), 0.3% H2O2 in 1× PBS (100 µL of 30% H2O2 stock [Sigma, catalog no. 216763] in 9.9 mL of 1× PBS), and 80% acetone (40 mL of acetone with 10 mL of DEPC-ddH2O chilled at −20°C). Quenching of endogenous HRP was performed for 30 min in 0.3% H2O2 in PBS, with fresh solution added at 15 min.
Antibody use and signal production were as published for embryos instead of larva (including an additional avidin–biotin step) with the exception that TSA staining solution concentration was reduced by 50% (diluted 1:100 instead of 1:50).
Bioinformatics
GeneMANIA (http://www.genemania.org) was used to find functional relationships and networks between genes that exhibit similar subcellular distributions. The Drosophila option was selected, and searches were done without the “coexpression” option. GeneMANIA output data were modified in Adobe Illustrator for presentation clarity.
Microscopy and imaging
All images were acquired with a Leica DMRA2 fluorescence microscope using a Q-Imaging Retiga EX camera and Openlab 3.1.7 software. All images were “pseudocolored” in Adobe Photoshop with optimal colors for general viewing, with DAPI in magenta and RNA in green. All data are publicly available in Fly-FISH (http://fly-fish.ccbr.utoronto.ca).
Fly-FISH database changes
The Fly-FISH database has been completely rewritten using the Python code Django, which improves maintainability, extendibility, and portability and supports dynamic gene/term searches, automatic statistics, and powerful data export features. At the same time, numerous bugs and inaccurate curations have been removed or fixed. Search options have been expanded, with “and/or/not” combinations now enabled, and the choice of stage, tissue, or subcellular localization terms selectable. Links in and out now include FlyBase, Berkeley Drosophila Genome Project, and the new Dresden ovarian table (DOT; http://tomancak-srv1.mpi-cbg.de/DOT/main) databases.
Supplementary Material
Acknowledgments
We thank Craig Smibert, Howard Lipshitz, and Alex Palazzo for comments on the manuscript. This work was funded by an operating grant (MOP-133473) provided to H.M.K. by the Canadian Institutes of Health Research.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.276931.115.
References
- An L, Sato H, Konishi Y, Walker DG, Beach TG, Rogers J, Tooyama I. 2009. Expression and localization of lactotransferrin messenger RNA in the cortex of Alzheimer's disease. Neurosci Lett 452: 277–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M. 1973. Sequential gene activation by ecdysone in polytene chromosomes of Drosophila melanogaster. I. Dependence upon ecdysone concentration. Dev Biol 35: 47–61. [DOI] [PubMed] [Google Scholar]
- Augood SJ, Waldvogel HJ, Munkle MC, Faull RL, Emson PC. 1999. Localization of calcium-binding proteins and GABA transporter (GAT-1) messenger RNA in the human subthalamic nucleus. Neuroscience 88: 521–534. [DOI] [PubMed] [Google Scholar]
- Baker KD, Thummel CS. 2007. Diabetic larvae and obese flies—emerging studies of metabolism in Drosophila. Cell Metab 6: 257–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashirullah A, Cooperstock RL, Lipshitz HD. 1998. RNA localization in development. Annu Rev Biochem 67: 335–394. [DOI] [PubMed] [Google Scholar]
- Basler K, Struhl G. 1994. Compartment boundaries and the control of Drosophila limb pattern by hedgehog protein. Nature 368: 208–214. [DOI] [PubMed] [Google Scholar]
- Bassell GJ, Kelic S. 2004. Binding proteins for mRNA localization and local translation, and their dysfunction in genetic neurological disease. Curr Opin Neurobiol 14: 574–581. [DOI] [PubMed] [Google Scholar]
- Belanger G, Stocksley MA, Vandromme M, Schaeffer L, Furic L, DesGroseillers L, Jasmin BJ. 2003. Localization of the RNA-binding proteins Staufen1 and Staufen2 at the mammalian neuromuscular junction. J Neurochem 86: 669–677. [DOI] [PubMed] [Google Scholar]
- Brock HW, Hodgson JW, Petruk S, Mazo A. 2009. Regulatory noncoding RNAs at Hox loci. Biochem Cell Biol 87: 27–34. [DOI] [PubMed] [Google Scholar]
- Brown JB, Celniker SE. 2015. Lessons from modENCODE. Annu Rev Genomics Hum Genet 16: 31–53. [DOI] [PubMed] [Google Scholar]
- Brown JB, Boley N, Eisman R, May GE, Stoiber MH, Duff MO, Booth BW, Wen J, Park S, Suzuki AM, et al. 2014. Diversity and dynamics of the Drosophila transcriptome. Nature 512: 393–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruckenstein DA, Lein PJ, Higgins D, Fremeau RT Jr. 1990. Distinct spatial localization of specific mRNAs in cultured sympathetic neurons. Neuron 5: 809–819. [DOI] [PubMed] [Google Scholar]
- Choi YJ, Lai WS, Fedic R, Stumpo DJ, Huang W, Li L, Perera L, Brewer BY, Wilson GM, Mason JM, et al. 2014. The Drosophila Tis11 protein and its effects on mRNA expression in flies. J Biol Chem 289: 35042–35060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker CJ, Parker R. 2012. P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb Perspect Biol 4: a012286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. 2001. Non-coding RNA genes and the modern RNA world. Nat Rev Genet 2: 919–929. [DOI] [PubMed] [Google Scholar]
- Emerson RO, Thomas JH. 2009. Adaptive evolution in zinc finger transcription factors. PLoS Genet 5: e1000325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A, Bozzoni I. 2014. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet 15: 7–21. [DOI] [PubMed] [Google Scholar]
- Garner CC, Tucker RP, Matus A. 1988. Selective localization of messenger RNA for cytoskeletal protein MAP2 in dendrites. Nature 336: 674–677. [DOI] [PubMed] [Google Scholar]
- Gerber B, Stocker RF. 2007. The Drosophila larva as a model for studying chemosensation and chemosensory learning: a review. Chem Senses 32: 65–89. [DOI] [PubMed] [Google Scholar]
- Gomez-Marin A, Louis M. 2012. Active sensation during orientation behavior in the Drosophila larva: more sense than luck. Curr Opin Neurobiol 22: 208–215. [DOI] [PubMed] [Google Scholar]
- Gonzalez I, Buonomo SB, Nasmyth K, von Ahsen U. 1999. ASH1 mRNA localization in yeast involves multiple secondary structural elements and Ash1 protein translation. Curr Biol 9: 337–340. [DOI] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. 2010. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464: 1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et al. 2015. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet 47: 199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan S, Reid DW, Cox AH, Nicchitta CV. 2014. De novo translation initiation on membrane-bound ribosomes as a mechanism for localization of cytosolic protein mRNAs to the endoplasmic reticulum. RNA 20: 1489–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambor H, Surendranath V, Kalinka AT, Mejstrik P, Saalfeld S, Tomancak P. 2015. Systematic imaging reveals features and changing localization of mRNAs in Drosophila development. Elife 10.7554/eLife.05003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RP. 2001. mRNA localization: message on the move. Nat Rev Mol Cell Biol 2: 247–256. [DOI] [PubMed] [Google Scholar]
- Koensgen D, Mustea A, Klaman I, Sun P, Zafrakas M, Lichtenegger W, Denkert C, Dahl E, Sehouli J. 2007. Expression analysis and RNA localization of PAI-RBP1 (SERBP1) in epithelial ovarian cancer: association with tumor progression. Gynecol Oncol 107: 266–273. [DOI] [PubMed] [Google Scholar]
- Krauss J, Lopez de Quinto S, Nusslein-Volhard C, Ephrussi A. 2009. Myosin-V regulates oskar mRNA localization in the Drosophila oocyte. Curr Biol 19: 1058–1063. [DOI] [PubMed] [Google Scholar]
- Kraut-Cohen J, Afanasieva E, Haim-Vilmovsky L, Slobodin B, Yosef I, Bibi E, Gerst JE. 2013. Translation- and SRP-independent mRNA targeting to the endoplasmic reticulum in the yeast Saccharomyces cerevisiae. Mol Biol Cell 24: 3069–3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JB, Singer RH. 1986. Intracellular localization of messenger RNAs for cytoskeletal proteins. Cell 45: 407–415. [DOI] [PubMed] [Google Scholar]
- Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. 2007. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 131: 174–187. [DOI] [PubMed] [Google Scholar]
- Lecuyer E, Parthasarathy N, Krause HM. 2008. Fluorescent in situ hybridization protocols in Drosophila embryos and tissues. Methods Mol Biol 420: 289–302. [DOI] [PubMed] [Google Scholar]
- Lecuyer E, Yoshida H, Krause HM. 2009. Global implications of mRNA localization pathways in cellular organization. Curr Opin Cell Biol 21: 409–415. [DOI] [PubMed] [Google Scholar]
- Lesnik C, Golani-Armon A, Arava Y. 2015. Localized translation near the mitochondrial outer membrane: an update. RNA Biol 12: 801–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Yang X, Wasser M, Cai Y, Chia W. 1997. Inscuteable and Staufen mediate asymmetric localization and segregation of prospero RNA during Drosophila neuroblast cell divisions. Cell 90: 437–447. [DOI] [PubMed] [Google Scholar]
- Liao G, Mingle L, Van De Water L, Liu G. 2015. Control of cell migration through mRNA localization and local translation. Wiley Interdiscip Rev RNA 6: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipshitz HD, Smibert CA. 2000. Mechanisms of RNA localization and translational regulation. Curr Opin Genet Dev 10: 476–488. [DOI] [PubMed] [Google Scholar]
- Macdonald PM, Struhl G. 1988. cis-acting sequences responsible for anterior localization of bicoid mRNA in Drosophila embryos. Nature 336: 595–598. [DOI] [PubMed] [Google Scholar]
- Makhijani K, Bruckner K. 2012. Of blood cells and the nervous system: hematopoiesis in the Drosophila larva. Fly (Austin) 6: 254–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin KC, Ephrussi A. 2009. mRNA localization: gene expression in the spatial dimension. Cell 136: 719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. 2008. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci 105: 716–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirth C, Truman JW, Riddiford LM. 2005. The role of the prothoracic gland in determining critical weight for metamorphosis in Drosophila melanogaster. Curr Biol 15: 1796–1807. [DOI] [PubMed] [Google Scholar]
- Muskavitch MA, Hogness DS. 1980. Molecular analysis of a gene in a developmentally regulated puff of Drosophila melanogaster. Proc Natl Acad Sci 77: 7362–7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najafabadi HS, Mnaimneh S, Schmitges FW, Garton M, Lam KN, Yang A, Albu M, Weirauch MT, Radovani E, Kim PM, et al. 2015. C2H2 zinc finger proteins greatly expand the human regulatory lexicon. Nature Biotechnol 33: 555–562. [DOI] [PubMed] [Google Scholar]
- Ni L, Snyder M. 2001. A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae. Mol Biol Cell 12: 2147–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olink-Coux M, Hollenbeck PJ. 1996. Localization and active transport of mRNA in axons of sympathetic neurons in culture. J Neurosci 16: 1346–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo AF, Lee ES. 2015. Non-coding RNA: what is functional and what is junk? Front Genet 6: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A, Rinn JL, Schier AF. 2011. Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet 12: 136–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebagliati MR, Weeks DL, Harvey RP, Melton DA. 1985. Identification and cloning of localized maternal RNAs from Xenopus eggs. Cell 42: 769–777. [DOI] [PubMed] [Google Scholar]
- Reid DW, Nicchitta CV. 2012. Primary role for endoplasmic reticulum-bound ribosomes in cellular translation identified by ribosome profiling. J Biol Chem 287: 5518–5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinking J, Lam MM, Pardee K, Sampson HM, Liu S, Yang P, Williams S, White W, Lajoie G, Edwards A, et al. 2005. The Drosophila nuclear receptor e75 contains heme and is gas responsive. Cell 122: 195–207. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D, Driever W, Berleth T, Richstein S, Nusslein-Volhard C. 1989. Multiple steps in the localization of bicoid RNA to the anterior pole of the Drosophila oocyte. Development 107: 13–19. [DOI] [PubMed] [Google Scholar]
- Taliaferro JM, Wang ET, Burge CB. 2014. Genomic analysis of RNA localization. RNA Biol 11: 1040–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimukai H, Sato K, Kudo T, Kashiwagi Y, Tohyama M, Takeda M. 1999. Regional distribution of presenilin-1 messenger RNA in the embryonic rat brain: comparison with β-amyloid precursor protein messenger RNA localization. Neuroscience 90: 27–39. [DOI] [PubMed] [Google Scholar]
- Tennessen JM, Bertagnolli NM, Evans J, Sieber MH, Cox J, Thummel CS. 2014. Coordinated metabolic transitions during Drosophila embryogenesis and the onset of aerobic glycolysis. G3 (Bethesda) 4: 839–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson T, Liu N, Arkov A, Lehmann R, Lasko P. 2008. Isolation of new polar granule components in Drosophila reveals P body and ER associated proteins. Mech Dev 125: 865–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth J, Kovacs M, Wang F, Nyitray L, Sellers JR. 2005. Myosin V from Drosophila reveals diversity of motor mechanisms within the myosin V family. J Biol Chem 280: 30594–30603. [DOI] [PubMed] [Google Scholar]
- Umlauf D, Fraser P, Nagano T. 2008. The role of long non-coding RNAs in chromatin structure and gene regulation: variations on a theme. Biol Chem 389: 323–331. [DOI] [PubMed] [Google Scholar]
- Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, et al. 2010. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 38: W214–W220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil TT. 2014. mRNA localization in the Drosophila germline. RNA Biol 11: 1010–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk R, Sreenivasa UMM, Yan H, Krause HM. 2010. In situ hybridization: fruit fly embryos and tissues. Current Protocols Essential Laboratory Techniques 4: 9.3.1–9.3.24. [Google Scholar]
- Wilk R, Hu J, Krause HM. 2013. Spatial profiling of nuclear receptor transcription patterns over the course of Drosophila development. G3 (Bethesda) 3: 1177–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Sunwoo H, Spector DL. 2009. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev 23: 1494–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Kovacs M, Sakamoto T, Zhang F, Kiehart DP, Sellers JR. 2006. Dimerized Drosophila myosin VIIa: a processive motor. Proc Natl Acad Sci 103: 5746–5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.