Abstract
Population-level conservation is required to prevent biodiversity loss within a species, but it first necessitates determining the number and distribution of populations. Many whale populations are still depleted due to 20th century whaling. Whales are one of the most logistically difficult and expensive animals to study because of their mobility, pelagic lifestyle and often remote habitat. We tackle the question of population structure in the Antarctic blue whale (Balaenoptera musculus intermedia) – a critically endangered subspecies and the largest extant animal – by capitalizing on the largest genetic dataset to date for Antarctic blue whales. We found evidence of three populations that are sympatric in the Antarctic feeding grounds and likely occupy separate breeding grounds. Our study adds to knowledge of population structure in the Antarctic blue whale. Future research should invest in locating the breeding grounds and migratory routes of Antarctic blue whales through satellite telemetry to confirm their population structure and allow population-level conservation.
Population-level conservation is increasingly being recognized as a requirement for preventing biodiversity loss1. Ecological and evolutionary processes operate on a population level, therefore populations need to be conserved for ecosystems to continue to function and for organisms to adapt to today’s rapidly changing environment2,3,4. Knowledge of population structure – such as the number of populations, their distribution and their degree of interbreeding – is needed to define population-based units for management and conservation purposes.
Baleen whales were killed in the hundreds of thousands during 20th century whaling. It has long been recognized that their conservation should operate on a population level5. Understanding their population structure is complicated because of their pelagic and highly mobile lifestyle6. They typically feed at higher latitudes during summer and migrate to breed at lower latitudes during winter. The population structure possibilities span from each population having a separate non-breeding ground or grounds, to sharing of a non-breeding ground or grounds between different populations. Populations probably remain distinct in the latter case due to individuals returning to where they were born in order to breed. An additional challenge in unravelling the population structure of baleen whales for those feeding off Antarctica is their circumpolar distribution because it inhibits forming a priori boundaries of populations for analysis. Also, Antarctic baleen whales are highly expensive and logistically difficult to study. This is because of the remote and extreme weather conditions of the Antarctic, the high level of expertise required for expeditions, and the effort it takes to locate rare animals in an area encompassing millions of square kilometres.
Research methods have and continue to be developed to mitigate the intrinsic difficulties in studying the population structure of baleen whales. A method used during whaling was Discovery marking, where a whale was marked with a uniquely numbered Discovery tag and the tag recovered later when the whale was killed7. This provided two locations during the months and from the geographic areas of whaling, which was typically the austral summer and around Antarctica for baleen whales feeding in the Antarctic8. Additional technology emerged that allowed more than two locations of an individual: photo-identification by natural markings9,10 and satellite tagging11,12. Satellite tagging allows remote tracking of movements and continues to be optimized to increase tag longevity12,13. Acoustic recordings of vocalizations from whales have also been used to putatively identify populations based on geographically distinct calls and, when these calls occur repetitively, songs14,15,16 that are only known in males17,18,19,20. However, a lack of acoustic differences does not necessarily equate to a lack of population structure, and therefore requires confirmation using genetic approaches21. More cost-effective, informative, and high-throughput molecular markers continue to be developed, with microsatellites, mtDNA, and SNPs widely used today22. Genetic samples can be collected by remote biopsy sampling of live whales, and populations can be identified based on genetic differentiation between groups of individuals in accordance with population genetic theory23.
The largest animal is a species of baleen whale, the blue whale (Balaenoptera musculus), and the largest subspecies of blue whale is the Antarctic blue whale (B. m. intermedia)24,25. Antarctic blue whales reduced in abundance from 239,000 before hunting commenced in the 1904/05 austral summer season to a low of 360 when they were last hunted in the 1972/73 season26. The most recent abundance estimate was 2,280 from surveys conducted between the 1992/93 and 2003/04 austral summer27. This is only about 1% of pre-exploitation abundance. The subspecies is classified as Critically Endangered in the International Union for Conservation of Nature (IUCN) Red List of Threatened Species.
Under the International Whaling Commission (IWC), Antarctic blue whales are currently protected from commercial whaling and have not been killed under Special Permit (scientific) whaling. However, other anthropogenic activities may impede their recovery in numbers. Climate change may have consequences throughout the distribution of all populations28. At the feeding grounds in the Antarctic, potentially threatening anthropogenic activities include research operations, tourism, and the krill fishery29. At the breeding grounds and during migration, threats include but are not limited to seismic exploration for offshore oil and gas30, shipping noise31, and entanglement in fishing gear32. If Antarctic blue whales comprise of different populations, they would presumably inhabit different lower latitude areas during breeding and migration, so each population would be under a different array of anthropogenic threats outside the feeding season. Differences in threats between populations makes population-level conservation especially important to allow the persistence of each population.
The population structure of Antarctic blue whales can currently only be assessed where they feed, in the Antarctic, due to very limited knowledge of the locations of their breeding grounds. Movement of Antarctic blue whales studied using Discovery marks, photo-identification, and satellite tagging show large-scale movements as well as a degree of site fidelity to areas off Antarctica33,34,35,36,37,38. There are no known differences in song types within Antarctic blue whales39, which suggests that any populations within Antarctic blue whales are not acoustically distinct. Multiple populations of Antarctic blue whales are suspected based on analyses of genetic samples from the Antarctic that showed significant genetic differences at fixation indices between some IWC management Areas40. Defining management Areas was discussed since the 1930s and the Areas finally implemented from the 1974/75 season onwards by the IWC to aid in management of Southern Hemisphere baleen whales41. Each Area encompasses 50° to 70° longitude and stretches in latitude from the South Pole to the equator. Fixation index analyses require a priori putative populations, which are often geographic-based, and those that have been used (the IWC management Areas) as well as any other potential a priori geographic groupings are not necessarily biologically relevant given the continuous distribution of blue whales around the Antarctic. There has been no evidence of multiple populations based on Bayesian clustering assignment analyses40,42,43, which are a standard genetic method to assess population structure that does not require a priori geographic groupings. The lack of evidence could be due to insufficient sample sizes (n = 4742), insufficient number of markers (seven microsatellite loci40,42), or inclusion of samples from the pygmy blue whale subspecies (B. m. brevicauda)42,43.
Genetic samples of Antarctic blue whales have been collected since 1990 through highly costly and logistically difficult boat-based surveys conducted through the IWC. Genetic data collection from these samples should be maximized to provide high quantity and quality data that will inform conservation of this critically endangered animal. Here we examine the poorly defined population structure of Antarctic blue whales at their feeding grounds using the largest genetic dataset to date – 142 individuals of the Antarctic subspecies and information from 20 microsatellite markers and the mtDNA control region. We report novel findings that should be used to direct future research on the population structure of Antarctic blue whales.
Results
The Bayesian analysis of STRUCTURE inferred three genetic clusters that occur in sympatry off Antarctica. First, the likelihood in STRUCTURE across different values of K showed a peak at a K of three (−10406.48 (SD 39.99)) and one (−10385.12 (SD 0.56)) (Fig. 1). Second, the ΔK method estimated three genetic clusters (ΔKMAX = 17.20; Fig. 1). Third, there was unequal (i.e. not approximately one third) estimated membership of each individual to each genetic cluster at a K of three, with some individuals strongly classified into a cluster (Fig. 2). Fourth, there was multiple evidence that clusters were not spurious. There was uneven geographic distribution of clusters; in the Pacific Ocean basin sector of the Antarctic there was evidence of a lower proportion of cluster 3 as only three of the 26 individuals there were identified as belonging to that cluster (Fig. 3). Additionally, these individuals had low maximum memberships of 0.547 to 0.597 inclusive, which means there may be no ‘pure’ individuals from this cluster in the Pacific Ocean basin sector of the Antarctic. All ten simulated panmictic populations had evidence of only one genetic cluster in STRUCTURE based on the likelihood peaking at a K of one (mean −10385.13 to −10385.40) and equal membership of individuals to each genetic cluster when K was set to more than one.
Figure 1. Inference of the number of genetic clusters detected by STRUCTURE for Antarctic blue whales using microsatellites.
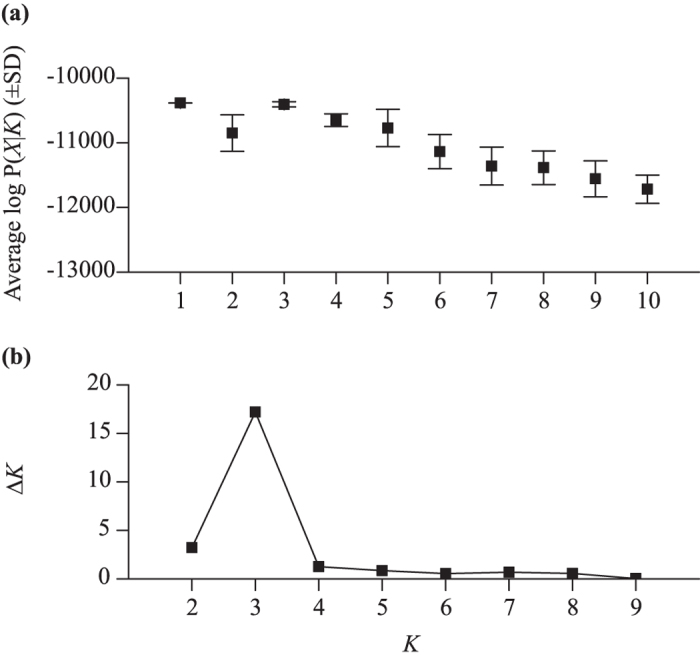
(a) Average estimate (±standard deviation) of the log of the probability of the data for each tested value of K, which was used to calculate (b) ΔK for tested values of K.
Figure 2. Clustering results of the STRUCTURE analysis for Antarctic blue whales using microsatellites when K is set to 3 and as summarized using CLUMPAK.
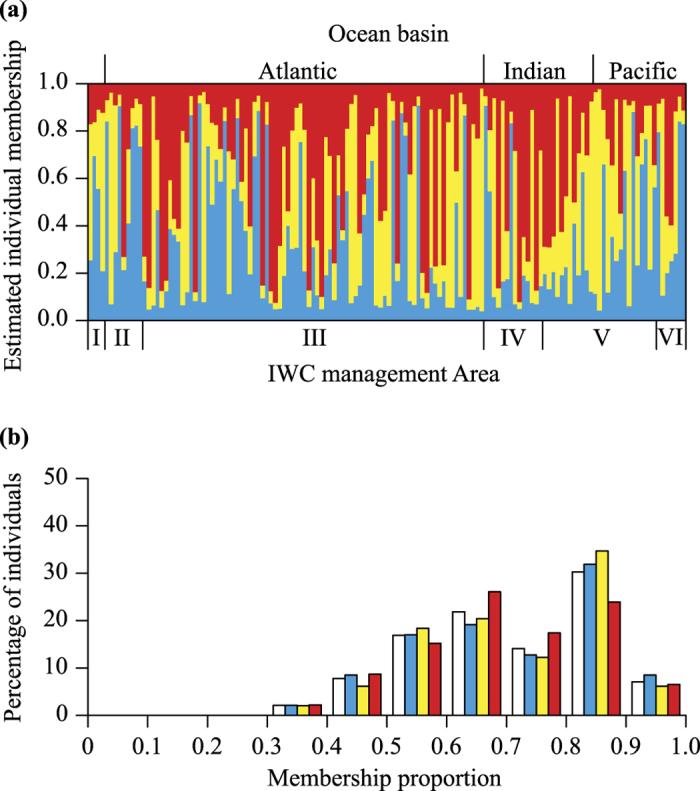
Population 1, blue or medium grey in greyscale; population 2, yellow or light grey in greyscale; population 3, red or dark grey in greyscale. (a) Estimated membership of each individual to each cluster. Each individual is represented by a column. Individuals are ordered according to the longitude at which they were sampled, starting from the western border of IWC management Area I. Individuals sampled at the same longitude were ordered from the oldest to the newest sample. The colouring of each column represents the proportion of estimated membership of each individual to each population. (b) Percentage distribution histogram of the highest membership proportion of Antarctic blue whales (white) and blue whales from each population.
Figure 3. Polar map showing the biopsy sampling locations of individual blue whales off Antarctica.
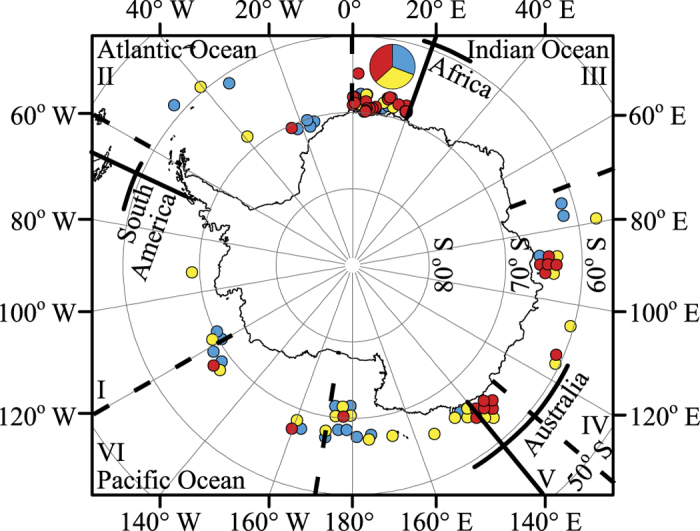
Each individual is represented by a circle, which is shaded according to its highest cluster membership as estimated in STRUCTURE using microsatellites (population 1, blue or medium grey in greyscale; population 2, yellow or light grey in greyscale; population 3, red or dark grey in greyscale). Only the first recorded location is shown for individuals sampled more than once. The map position of individuals sampled from the same or similar locations has been slightly altered so that all individuals are visible, with the exception of individuals located from 0° to 20°E due to extensive sampling from this area (n = 81). A pie chart shows the proportion of individuals that belong to each STRUCTURE cluster in the area from 0° to 20°E. Borders of IWC management Areas I to VI (dashed, black latitudinal lines) and ocean basins (solid, black latitudinal lines; borders according to International Hydrographic Organization definitions110) are shown. The longitudes of the southern coasts of South America, Africa and Australia are indicated (solid, black longitudinal lines). The map was created using ARCGIS 10.0 (Esri).
For the meaningful K value of three, CLUMPAK identified one major clustering solution (21 of 30 replicate runs; Fig. 2) and two minor clustering solutions (each 3 of 30 replicate runs). The relatively low maximum memberships of most individuals for the two minor clustering solutions indicate these clustering solutions are biologically infeasible. They are likely due to difficulty in searching the space of possible membership values.
Genetic variation was similar between each inferred cluster detected by STRUCTURE. This was regardless of whether variation was based on microsatellites or the mtDNA control region (Table 1). There was significant and similar genetic differentiation based on fixation indices between the clusters (microsatellite FST = 0.020–0.024; mtDNA FST = 0.016–0.029) (Table 2). There was also significant genetic differentiation between some IWC management Area pairwise comparisons (Table 2).
Table 1. Genetic variation at 20 microsatellites and the mtDNA control region of Antarctic blue whales and the populations identified using STRUCTURE.
| Microsatellites | mtDNA control region | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| n | NA | HO | HE | AR | n | NH | h | HR | |
| AllAntarctica | 142 | 11.65(5.46) | 0.758(0.130) | 0.763(0.133) | 9.99 | 140 | 46 | 0.968(0.005) | 26.58 |
| Population 1 | 47 | 9.85(4.04) | 0.764(0.148) | 0.761(0.148) | 9.82 | 46 | 23 | 0.958(0.013) | 22.76 |
| Population 2 | 49 | 9.05(3.72) | 0.751(0.144) | 0.748(0.130) | 8.96 | 49 | 25 | 0.958(0.014) | 23.97 |
| Population 3 | 46 | 9.50(4.65) | 0.760(0.122) | 0.747(0.126) | 9.50 | 45 | 20 | 0.938(0.017) | 20.00 |
n, number of samples; NA, mean number of alleles; HO, mean observed heterozygosity; HE, mean unbiased expected heterozygosity; AR, mean allelic richness (based on the minimum sample size of 46 across populations and loci); NH, number of haplotypes; h, haplotype diversity; HR, haplotype richness (based on the minimum sample size across populations). Standard deviations are in parentheses.
Table 2. Genetic differentiation (FST) of Antarctic blue whales between (a) the populations identified using STRUCTURE, and (b) the IWC management Areas.
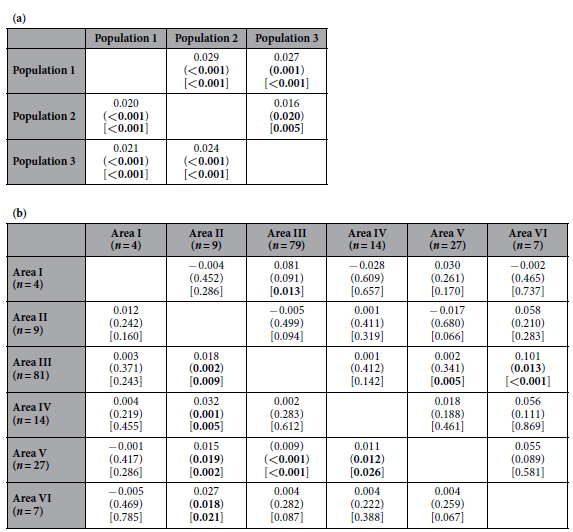
Analyses were conducted based on microsatellites (below diagonal) and the mtDNA control region (above diagonal).
P values from permutation are in parentheses and from exact tests are in square brackets. Significant P values are in bold.
Discussion
Our study helps refine knowledge of population structure in the Antarctic blue whale, B. m. intermedia. A Bayesian clustering assignment method based on microsatellite DNA provided evidence that three genetically differentiated populations occur sympatrically off Antarctica during the austral summer feeding season. For simplicity, these are henceforth referred to as populations. The levels of differentiation were low and similar between the populations, and consistent between analyses of microsatellites and the mtDNA control region. Genetic variation based on microsatellites and the mtDNA control region was similar in the three populations.
Previous studies were unable to detect the multiple, sympatric populations of Antarctic blue whales, even though they also used the same standard Bayesian clustering method40,42,43. This is because of either smaller sample sizes, smaller number of markers, not specifically investigating population structure within the Antarctic subspecies, or a combination of these. Fixation indices, which require a priori putative populations, have previously40 and in the current study detected genetic differentiation between some IWC management Areas. Given the results of the current study for analyses that do not require a priori groupings, this genetic differentiation between Areas could be due to different proportions of the populations in different management Areas.
The only longitudinal range where the proportion of different populations can be accurately assessed is between 0° and 20°E since 57% of samples were from this area. Here, there is an almost equal ratio of individuals from each population (number of samples from population 1 = 25, population 2 = 26, population 3 = 30). In contrast, in the Pacific Ocean basin sector there is perhaps greater proportions of populations 1 and 2 compared with population 3 as only three of the 26 individuals there were identified as belonging to population 3, and these had low memberships (estimated in STRUCTURE) to population 3 of 0.547 to 0.597 inclusive. These individuals and others with relatively low membership to their assigned population may have admixed ancestry to multiple populations43; in other words, they may have immediate ancestors (e.g. parents, grandparents) from different populations. An alternative explanation for low membership is insufficient power for accurate assignment given the low levels of population differentiation.
The movement data available for Antarctic blue whales corroborate the current study’s findings. They show large-scale movements off Antarctica – including movements that cross IWC management Areas and ocean basins – as well as small-scale movements. The data are from a wide range of methodologies and are across a wide time span: Discovery marks deployed from the 1934/35 to 1962/63 season and recaptured until the 1966/67 season33, photo-identifications from the 1987/88 to 2014/15 season34,35,36,37, Olson pers. comm. and satellite tagging and tracking within the 2012/13 season38 (Table 3). Longitudinal movements around the Antarctic require less travelling due to the high latitudes than the same amount of longitudinal movement at low latitudes, allowing the blue whales to move relatively easily between longitudes when in the Antarctic. The geographic distance was typically closer for intra-seasonal recaptures compared with inter-seasonal recaptures, though inter-seasonal recaptures still included small-scale movements33,35. The small-scale movements indicate site fidelity and the potential for different proportions of populations in different areas, and the large-scale movements indicate the potential for sympatry of populations.
Table 3. Summary of movement data for Antarctic blue whales generated by previous studies through Discovery marking, photo-identification and satellite tagging.
| Discovery marks33 | Photo-identifications34,35,36,37, Olson pers. comm. | Satellite tagging33 | |
|---|---|---|---|
| Austral summer seasons | 1934/35–1966/67 | 1987/88–2014/15 | 2012/13 |
| Number of individuals | 2295 | 399 | 2 |
| Number recaptured within a season | 49 | 36 | NA |
| Minimum movement within a season(degrees longitude; km) | 0.10; 32 | 0.04; 3 | 16.12; 1433 |
| Maximum movement within a season(degrees longitude; km) | 76.45; 3516 | 24.69; 1172 | 75.06; 5300 |
| Number recaptured between seasons | 46 | 14 | NA |
| Minimum movement between seasons(degrees longitude; km) | 1.77; 122 | 0.47; 19 | NA |
| Maximum movement between seasons(degrees longitude; km) | 172.58; 6250 | 141.81; 6550 | NA |
The distances provided as km include latitudinal movements, and the movements within a season exclude those recorded on the same day. The “number of individuals” for Discovery marking is the number of mark deployments, however note that four individuals recaptured within a season, four recaptured between seasons, and one recaptured on the same day had two marks, and that one recaptured between seasons was morphologically identified as a pygmy blue whale33. The whale with the minimum movement detected by satellite tagging was also the whale with the maximum intra-seasonal photo-identification, with a photo-identification taken prior to tagging and on the day of tagging36. Two of the inter-seasonal photo-identification recaptures35 were also identified as recaptures using genetic methods40,43. One was a female identified in the current study as belonging to population 3 (estimated membership 0.474) and moved 131.70° longitude, the other was a male identified as belonging to population 1 (estimated membership 0.846) and moved 6.69° longitude. NA, not applicable.
The individual whales likely move depending on the densities of their prey due to their high energetic requirements as the largest extant animal44. Blue whales are specialist predators that feed on krill (order Euphausiacea). The Antarctic has high biological productivity, including Antarctic krill (Euphausia superba)45, due to the Antarctic Circumpolar Current generating upwelling and circumpolar fronts46. In the summer feeding season the Antarctic blue whales are generally south of the Antarctic Polar Front (also known as the Antarctic Convergence)33, a region associated with particularly high biological productivity47 and where blue whales feed on Antarctic krill48. The distribution and density of Antarctic krill changes within and between seasons depending on environmental conditions49, requiring marine predators, such as blue whales, to move to find sufficient amounts of their prey e.g.50,51,52. Such dependence is particularly important to consider given evidence of changes in Antarctic krill demographics due to recent climate change53,54. The dependence of blue whales on high concentrations of krill may also occur outside their feeding grounds33,55,56, unlike traditional thinking that baleen whales fast during migration and at breeding grounds. This means the specific locations of Antarctic blue whale breeding grounds may be influenced by the location and abundance of krill during the austral winter.
The exact breeding ground locations of Antarctic blue whales are unknown. One possibility is that populations breed at different ocean basins. This means that populations would be physically separated by the continental land masses of South America, Africa and Australia during the austral winter breeding season. It has long been suggested that at least one Antarctic population breeds in each of these physically separated regions57. Acoustic recordings of Antarctic blue whale calls in the austral winter indicate their breeding grounds include low latitudes of the Indian Ocean and eastern Pacific Ocean58,59. Their breeding grounds may also include low latitudes of the South Atlantic Ocean, with perhaps no recordings of their calls in that basin due to insufficient effort39. For example, the Atlantic Ocean region off south-west Africa has been suggested as a breeding location based on seasonality of Antarctic blue whale historical catches in south-west Africa24,33. However, since whaling there have been only two recorded blue whale sightings off that coast33. There is also a paucity of data on the population structure of other Antarctic baleen whales. There is evidence that populations of humpback whales (Megaptera novaeangliae) – arguably the most well-understood Antarctic baleen whale – have discrete distributions in the Antarctic, with overlap in the distribution of some populations60. They are thought to breed at lower latitudes segregated across the three ocean basins, but also with multiple populations breeding discretely in each ocean basin60. The Antarctic minke whale (B. bonaerensis) may also have a similar pattern of discrete population distributions in the Antarctic with some overlap61. Outside the Antarctic there is evidence of populations in sympatry at feeding grounds. The common minke whale (B. acutorostrata) appears to have complete sympatry on feeding grounds in the North Atlantic62.
It is also possible that at least one population of Antarctic blue whales is resident in the Antarctic throughout the year. A resident population may follow the ice edge as it expands northwards in the austral winter and recedes in the austral summer. This is suggested by year-round acoustic detections of the Antarctic subspecies off the western coast of the Western Antarctic Peninsula63,64 and off eastern Antarctica at 67°S, 70°E64. Antarctic blue whales have also been recorded year-round off the Crozet Islands59,65 based on acoustics, and off the sub-Antarctic island of South Georgia based on whaling catch data33. Blue whales of other subspecies have also been recorded at some localities year-round59,65,66,67,68, and other baleen whale species have been recorded in the Antarctic69,70,71 and other locations72,73,74,75 year-round. Some of these are likely to be resident populations72,73.
Another explanation for year-round presence at a feeding ground is overlapping timing of departures and arrivals. This overlap could be mediated by temporal segregation of migration based on age, sex, reproductive state, or migratory destination, as is thought for humpback whales76,77. Alternatively, a proportion of a population or populations may remain at the feeding grounds year-round. These could be individuals that are sexually or physically immature, or as also suggested for humpback whales78, mature females that are currently not breeding. Sexual maturity in blue whales is only reached at 10 years, and physical maturity afterwards79,80,81. Female blue whales are not thought to breed each season as they have a two to three year inter-calf interval, gestation lasting at least 10 months, weaning lasting seven months, and simultaneous pregnancy and lactation is rare80,81. However, the vocalizations that are geographically distinct in blue whales and appear as songs are likely only produced by males17,18, and the distinct Antarctic blue whale vocalization was used to report the year-round presence of blue whales in the Antarctic63,64. This means males are present year-round, with or without females.
It is possible that whaling has influenced the population structure detected here. Exploitation can lead to the formation or loss of populations, or for the level of genetic differentiation between populations to increase or decrease82. This can be due to increased genetic drift and associated loss of variation and increased genetic differentiation, and changes in the degree of gene flow between populations. Ideally, a sufficient number of pre-whaling samples from the same locations as the current samples should be used to determine if the population structure of blue whales has changed due to human impacts83, but such samples are not available in the present study. However, the most likely pre-whaling scenarios of blue whale populations can be inferred based on non-genetic data and the biology of this species. It is likely that there were multiple populations of Antarctic blue whales before whaling. While there are different possibilities regarding the breeding ground locations of Antarctic blue whales, acoustic and whaling catch data indicate that they include different ocean basins24,33,58,59. This would promote the formation of different populations, as seen in humpback whales60, because land barriers between ocean basins would hinder movement between breeding grounds. Philopatry to basins may be mediated by maternally-directed cultural learning of migratory routes and destinations, which makes population-level conservation particularly imperative as extirpation of a population could result in no re-colonization of the associated breeding ground84. In addition, it is likely that Antarctic blue whale populations were sympatric in the Antarctic before whaling. This is because there are no obvious differences in the pattern of blue whale movements in the Antarctic according to Discovery mark33 and photo-identification34,35,36,37 datasets, which together span from the 1934/35 to 2014/15 season. The non-genetic data therefore indicate that different populations likely existed within Antarctic blue whales before whaling, and these were sympatric in the Antarctic.
The current level of gene flow between populations may be greater than that prior to whaling, which could account for the low level of genetic differentiation and evidence of admixture found in the current study. Increased gene flow may occur because individuals need to travel further afield to find mates given that the number of Antarctic blue whales reduced from 239,000 to 360 individuals due to whaling26. Indeed, hybridization between the Antarctic blue whale and pygmy blue whale subspecies may have begun occurring or increased in occurrence within the last four decades due to whaling or climate change43. Although, there may have always been high levels of gene flow between the Antarctic populations. The low differentiation can otherwise be explained by the Antarctic populations being founded relatively recently (but still prior to whaling) on an evolutionary timescale, which is unlikely given evidence that Antarctic blue whales are the ancestral subspecies of blue whales in the Southern Hemisphere85. It is also unlikely that whaling has caused an increase in population differentiation through genetic drift because increased genetic drift has not yet existed for long enough to have a major effect; blue whales are long-lived with a generation time of 31 years86, and Antarctic blue whales were hunted from the 1904/05 to 1972/73 season, so the populations have only had a reduced size for about three to four generations.
Locating the breeding grounds of blue whales feeding off Antarctica is needed to confirm their current population structure. The biopsy samples used in the current study and the raw data for much of the discussed post-whaling Antarctic blue whale research, including photo-identifications34,35,36,37, abundance estimates26,27, past genetic research40,42,43, and satellite tagging38, were collected through international, collaborative vessel surveys in the Antarctic. The breeding grounds of Antarctic blue whales would ideally be located through continuing collaborative efforts to satellite tag Antarctic blue whales while they are feeding and tracking their subsequent movements38. Despite current longevity issues12, there has been much success in using tags to determine movements and migratory destinations87,88, and to assess inter-year and individual variation when enough tags are deployed55. In addition, tags continue to be improved to increase their longevity12,13. Subsequent genetic samples from breeding grounds would allow confirmatory genetic structure analyses with biologically reasonable a priori groupings and a baseline for comparative genetic analyses of samples collected off Antarctica. Monitoring each population’s abundance would then need to be performed at their breeding grounds. Monitoring is needed because the populations may differ in pre- and post-whaling abundance and recovery trends, especially because they occupy different migratory routes and breeding grounds with likely different carrying capacities and anthropogenic threats.
Methods
Data collection
Genetic samples from the 142 Antarctic blue whale individuals used in this study were collected off Antarctica using non-lethal biopsy darts. These samples exclude resamples, migrant pygmy blue whales, and subspecies hybrids found off Antarctica by Attard et al.43. The samples were collected from 1990 to 2009 in December to February during the International Decade of Cetacean Research (IDCR) and Southern Ocean Whale and Ecosystem Research (SOWER) cruises. This was approved by and conducted in accordance with the regulations of the IWC. Samples were preserved in either 20% DMSO saturated with NaCl or 70–100% ethanol, and archived at Southwest Fisheries Science Center (SWFSC) in the USA. DNA was extracted at SWFSC using multiple methods, including a modified salting-out protocol89 and DNeasy® Blood and Tissue Kit (Qiagen).
Genetic data were collected from the samples at 20 microsatellites and a 414 bp fragment of the mtDNA control region. The data used in the current study from the 20 microsatellites were obtained by Attard et al.43 (BM03290 was discarded in the previous study and current study due to evidence of null alleles). Data from the 414 bp fragment of the mtDNA control region was obtained by LeDuc et al.42 and in the current study. The current study obtained the mtDNA data using the methods of LeDuc et al.42 or Attard et al.90, with the alteration of using an exo-SAP (Fermentas) protocol to purify mtDNA PCR products and an ABI 3730xl Genetic Analyzer (Applied Biosystems) for sequencing.
Analyses confirming microsatellite data adhered to the assumptions of subsequent analyses were conducted for the Antarctic subspecies and each genetic cluster identified by Bayesian analyses. This included checking for homozygote excess, and genotyping errors from stuttering and short allele dominance, using MICROCHECKER 2.2.391 (95% confidence intervals, 10,000 runs). Deviations from Hardy-Weinberg equilibrium were tested using ARLEQUIN 3.5.1.292 (10,000 dememorizations, 100,000 Markov chain steps) and linkage disequilibrium between pairs of loci were tested using FSTAT 2.9.3.293, with sequential Bonferroni corrections applied to significance values94. The only potential evidence of unsuitability of a microsatellite locus was evidence of homozygote excess for locus Dde0995 for the whole dataset, but this was likely due to a lack of panmixia.
Population structure
Population structure was assessed using a standard strategy that is suitable for continuously distributed and potentially sympatric populations. The Bayesian clustering assignment method of STRUCTURE 2.3.496 was implemented for microsatellite data with the admixture model of ancestry, the correlated allele frequency model97, and without using sampling locations as priors98 (burn-in of 100,000 iterations then runs of 106). Thirty independent runs were conducted for each value of K from one to 10, where K is the number of inferred genetic clusters. Runs were summarized using CLUMPAK99 (Main Pipeline, default parameters) to obtain the estimated membership of each individual to each cluster. CLUMPAK accounts for differences in replicate runs caused by label switching (from the arbitrary labelling of clusters) and multimodality (from either difficulty in searching the space of possible membership values or real biological factors).
Multiple measures were used to infer the most meaningful number of populations (K) detected by STRUCTURE. First, the probability of the data for each tested value of K was taken into account, with the highest probabilities expected at meaningful values of K. Second, the ΔK method100 was implemented in STRUCTURE HARVESTER 0.6.93101. As this method is based on changes in the probability of successive values of K, it can only detect a true K that is greater than one. Third, the distribution of estimated fraction of membership to each cluster for each individual at a given K was taken into account. Approximately 1/K fractions are expected for each individual at K > 1 when there is no genetic structure. Fourth, spurious genetic clustering was considered a possibility if the geographic distribution of inferred genetic clusters was even or if STRUCTURE detected two or more clusters for any of ten simulated panmictic populations.
The panmictic populations were simulated and analyzed following Quintela et al.102. Ten panmictic populations were simulated by permuting alleles within each locus using package poppr103 in R 3.0.3104 (function shufflepop, permute alleles method). These were analyzed in STRUCTURE using the models, burn-in and run length implemented for the empirical dataset. Ten independent runs were conducted for each value of K from one to five. The most meaningful K was determined based on the probability of the data for each tested value of K and the estimated fraction of membership to each cluster for each individual at a given K. The ΔK method was not implemented as it can only detect a true K that is greater than one.
Pairwise genetic differentiation (FST) was calculated based on microsatellites and the mtDNA control region using ARLEQUIN (significance assessed by 10,000 permutations) between the clusters, as well as between a priori geographic groupings of IWC management Areas. Genetic differentiation was calculated to allow comparisons with previous a priori genetic differentiation analyses of Antarctic blue whales40 rather than to elucidate the genetic structure of Antarctic blue whales. Exact tests of population differentiation were conducted for microsatellites using GENEPOP 4.3105,106 (10,000 dememorizations, 100 batches, 10,000 iterations per batch) and for the mtDNA control region using ARLEQUIN (100,000 Markov chain steps, 10,000 dememorizations).
Genetic variation
Genetic variation was determined for the Antarctic subspecies and each genetic cluster identified by Bayesian analyses. Variation based on microsatellites was determined by calculating the number of alleles, observed heterozygosity (HO) and unbiased expected heterozygosity (HE) using GENALEX 6.501107,108, and allelic richness using FSTAT93. Genetic variation based on the mtDNA control region was determined by calculating the haplotype diversity (h) using ARLEQUIN, and haplotype richness using CONTRIB 1.02109.
Additional Information
How to cite this article: Attard, C. R. M. et al. Towards population-level conservation in the critically endangered Antarctic blue whale: the number and distribution of their populations. Sci. Rep. 6, 22291; doi: 10.1038/srep22291 (2016).
Acknowledgments
Genetic data analyses of samples were funded by the Australian Marine Mammal Centre within the Australian Antarctic Division, and Flinders University and Macquarie University in Australia. C.R.M.A. was supported for part of the study by an Australian Postgraduate Award (APA). Sampling was conducted under non-lethal research programs of the International Whaling Commission (IWC) (see Attard et al.43 for details, including Southwest Fisheries Science Center (SWFSC) sample identification numbers). We thank Paula A. Olson for her input regarding Antarctic blue whale photo-identification, Kelly M. Robertson for her input as sample curator at SWFSC, Jonathan K. Pritchard for assisting with STRUCTURE interpretation, Brian S. Miller for input regarding blue whale acoustics and Elizabeth Eyre for input regarding whale acoustics, Virginia Andrews-Goff for input regarding movement data, and Eduardo R. Secchi for his comments on the manuscript. We thank attendees that provided suggestions at the 20th Biennial Conference on Marine Mammals in Dunedin, New Zealand. We thank Robert L. Brownell Jr. and one anonymous reviewer for their suggestions during the review process.
Footnotes
Author Contributions C.R.M.A., L.B.B. and L.M.M. conceived, designed and performed the research, analyzed the data, and wrote the paper.
References
- Purvis A. & Hector A. Getting the measure of biodiversity. Nature 405, 212–219 (2000). [DOI] [PubMed] [Google Scholar]
- Stockwell C. A., Hendry A. P. & Kinnison M. T. Contemporary evolution meets conservation biology. Trends Ecol. Evol. 18, 94–101 (2003). [Google Scholar]
- Pressey R. L., Cabeza M., Watts M. E., Cowling R. M. & Wilson K. A. Conservation planning in a changing world. Trends Ecol. Evol. 22, 583–592 (2007). [DOI] [PubMed] [Google Scholar]
- Garner A., Rachlow J. L. & Hicks J. F. Patterns of genetic diversity and its loss in mammalian populations. Conserv. Biol. 19, 1215–1221 (2005). [Google Scholar]
- Clapham P. J., Young S. B. & Brownell R. L. Jr. Baleen whales: conservation issues and the status of the most endangered populations. Mamm. Rev. 29, 35–60 (1999). [Google Scholar]
- Hoelzel A. R. Genetic structure of cetacean populations in sympatry, parapatry, and mixed assemblages: implications for conservation policy. J. Hered. 89, 451–458 (1998). [Google Scholar]
- Brown S. G. Report of the Scientific Committee, Annex L. Consideration of the present technique of whale marking and future marking programmes. Rep. Int. Whal. Comm. 21, 100–105 (1971). [Google Scholar]
- Clapham P. J. & Baker C. S. Whaling, modern in Encyclopedia of Marine Mammals (eds Perrin W. F., Würsig B. & Thewissen J. G. M.) 1328–1332 (Academic Press, 2002). [Google Scholar]
- Hammond P. S., Mizroch S. A. & Donovan G. P. (eds) Individual recognition of cetaceans: use of photo-identification and other techniques to estimate population parameters. Rep. Int. Whal. Comm. Spec. Issue 12 (1990). [Google Scholar]
- Markowitz T. M., Harlin A. D. & Würsig B. Digital photography improves efficiency of individual dolphin identification. Mar. Mamm. Sci. 19, 217–223 (2003). [Google Scholar]
- Montgomery S. (ed.) Report on the 24–26 February workshop to assess possible systems for tracking large cetaceans. Final Report to Minerals Management Service, National Technical Information Service (NTIS) No. PB 87-182135. (Marine Mammal Commission, 1987). [Google Scholar]
- Mate B., Mesecar R. & Lagerquist B. The evolution of satellite-monitored radio tags for large whales: one laboratory’s experience. Deep-Sea Res. II: Top. Stud. Oceanogr. 54, 224–247 (2007). [Google Scholar]
- Robbins J. et al. Satellite tag effectiveness and impacts on large whales: preliminary results of a case study with Gulf of Maine humpback whales. Paper SC/65a/SH05 presented to the IWC Scientific Committee (2013). [Google Scholar]
- Winn H. et al. Song of the humpback whale - population comparisons. Behav. Ecol. Sociobiol. 8, 41–46 (1981). [Google Scholar]
- Payne R. & Guinee L. N. Humpback whale (Megaptera novaeangliae) songs as an indicator of “stocks” in Communication and Behavior of Whales. AAAS Selected Symposium 76. (ed. Payne R.) 333–358 (Westview Press, 1983). [Google Scholar]
- Zimmer W. M. X. Passive acoustic monitoring of cetaceans. (Cambridge University Press, 2011). [Google Scholar]
- McDonald M. A., Calambokidis J., Teranishi A. M. & Hildebrand J. A. The acoustic calls of blue whales off California with gender data. J. Acoust. Soc. Am. 109, 1728–1735 (2001). [DOI] [PubMed] [Google Scholar]
- Oleson E. M. et al. Behavioral context of call production by eastern North Pacific blue whales. Mar. Ecol. Prog. Ser. 330, 269–284 (2007). [Google Scholar]
- Croll D. A. et al. Only male fin whales sing loud songs. Nature 417, 809 (2002). [DOI] [PubMed] [Google Scholar]
- Clapham P. J. The social and reproductive biology of humpback whales: an ecological perspective. Mamm. Rev. 26, 27–49 (1996). [Google Scholar]
- Mellinger D. & Barlow J. Future directions for acoustic marine mammal surveys: stock assessment and habitat use. Report of a workshop held in La Jolla, CA, 20-22 November 2002. NOAA OAR Special Report, NOAA/PMEL Contribution 2557 (NOAA/Pacific Marine Environmental Laboratory, 2003).
- Schlötterer C. The evolution of molecular markers - just a matter of fashion? Nat. Rev. Genet. 5, 63–69 (2004). [DOI] [PubMed] [Google Scholar]
- Hoelzel A. R. Conservation genetics of whales and dolphins. Mol. Ecol. 1, 119–125 (1992). [DOI] [PubMed] [Google Scholar]
- Branch T. A., Abubaker E. M. N., Mkango S. & Butterworth D. S. Separating southern blue whale subspecies based on length frequencies of sexually mature females. Mar. Mamm. Sci. 23, 803–833 (2007). [Google Scholar]
- Gilpatrick J. W. Jr. & Perryman W. L. Geographic variation in external morphology of North Pacific and Southern Hemisphere blue whales (Balaenoptera musculus). J. Cetacean Res. Manage. 10, 9–21 (2008). [Google Scholar]
- Branch T. A., Matsuoka K. & Miyashita T. Evidence for increases in Antarctic blue whales based on Bayesian modelling. Mar. Mamm. Sci. 20, 726–754 (2004). [Google Scholar]
- Branch T. A. Abundance of Antarctic blue whales south of 60°S from three complete circumpolar sets of surveys. J. Cetacean Res. Manage. 9, 253–262 (2007). [Google Scholar]
- Simmonds M. P. & Isaac S. J. The impacts of climate change on marine mammals: early signs of significant problems. Oryx 41, 19–26 (2007). [Google Scholar]
- Leaper R. & Miller C. Management of Antarctic baleen whales amid past exploitation, current threats and complex marine ecosystems. Antarct. Sci. 23, 503–529 (2011). [Google Scholar]
- Di lorio L. & Clark C. W. Exposure to seismic survey alters blue whale acoustic communication. Biol. Lett. 6, 51–54 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcón M. L. et al. Blue whales respond to anthropogenic noises. PLoS ONE 7, e32681 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassoff R. M. et al. Lethal entanglement in baleen whales. Dis. Aquat. Org. 96, 175–185 (2011). [DOI] [PubMed] [Google Scholar]
- Branch T. A. et al. Past and present distribution, densities and movements of blue whales Balaenoptera musculus in the Southern Hemisphere and northern Indian Ocean. Mamm. Rev. 37, 116–175 (2007). [Google Scholar]
- Olson P. A. Blue whale photo-identification from IWC IDCR/SOWER cruises 1987/1988 to 2008/2009. Paper SC/62/SH29 presented to the IWC Scientific Committee (2010).
- Olson P. A. Antarctic blue whale photo-idenfication catalogue summary. Paper SC/64/SH8 presented to the IWC Scientific Committee (2012).
- Olson P. A., Ensor P., Schmitt N., Olavarria C. & Double M. C. Photo-identification of Antarctic blue whales during the SORP Antarctic Blue Whale Voyage 2013. Paper SC/65a/SH11 presented to the IWC Scientific Committee (2013).
- Olson P. A. et al. Photo-identification of Antarctic blue whales during the New Zealand-Australia Antarctic Ecosystems Voyage 2015. Paper SC/66a/SH26 presented to the IWC Scientific Committee (2015).
- Andrews-Goff V., Olson P. A., Gales N. J. & Double M. C. Satellite telemetry derived summer movements of Antarctic blue whales. Paper SC/65a/SH03 presented to the IWC Scientific Committee (2013).
- McDonald M. A., Mesnick S. L. & Hildebrand J. A. Biogeographic characterisation of blue whale song worldwide: using song to identify populations. J. Cetacean Res. Manage. 8, 55–65 (2006). [Google Scholar]
- Sremba A. L., Hancock-Hanser B., Branch T. A., LeDuc R. L. & Baker C. S. Circumpolar diversity and geographic differentiation of mtDNA in the critically endangered Antarctic blue whale (Balaenoptera musculus intermedia). PLoS ONE 7, e32579 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan G. P. A review of IWC stock boundaries. Rep. Int. Whal. Comm. Spec. Issue 13, 39–68 (1991). [Google Scholar]
- LeDuc R. G. et al. Patterns of genetic variation in Southern Hemisphere blue whales and the use of assignment test to detect mixing on the feeding grounds. J. Cetacean Res. Manage. 9, 73–80 (2007). [Google Scholar]
- Attard C. R. M. et al. Hybridization of Southern Hemisphere blue whale subspecies and a sympatric area off Antarctica: impacts of whaling or climate change? Mol. Ecol. 21, 5715–5727 (2012). [DOI] [PubMed] [Google Scholar]
- Goldbogen J. A. et al. Mechanics, hydrodynamics and energetics of blue whale lunge feeding: efficiency dependence on krill density. J. Exp. Biol. 214, 131–146 (2011). [DOI] [PubMed] [Google Scholar]
- Laws R. M. The ecology of the Southern Ocean. Am. Sci. 73, 26–40 (1985). [Google Scholar]
- Bost C. A. et al. The importance of oceanographic fronts to marine birds and mammals of the southern oceans. J. Mar. Syst. 78, 363–376 (2009). [Google Scholar]
- Tynan C. T. Ecological importance of the Southern Boundary of the Antarctic Circumpolar Current. Nature 392, 708–710 (1998). [Google Scholar]
- Kawamura A. A review of baleen whale feeding in the Southern Ocean. Rep. Int. Whal. Comm. 44, 261–271 (1994). [Google Scholar]
- Siegel V. Distribution and population dynamics of Euphausia superba: summary of recent findings. Polar Biol. 29, 1–22 (2005). [Google Scholar]
- Santora J. A., Schroeder I. D. & Loeb V. J. Spatial assessment of fin whale hotspots and their association with krill within an important Antarctic feeding and fishing ground. Mar. Biol. 161, 2293–2305 (2014). [Google Scholar]
- Santora J. A., Reiss C. S., Cossio A. M. & Veit R. R. Interannual spatial variability of krill (Euphausia superba) influences seabird foraging behavior near Elephant Island, Antarctica. Fish. Oceanogr. 18, 20–35 (2009). [Google Scholar]
- Nowacek D. P. et al. Super-aggregations of krill and humpback whales in Wilhelmina Bay, Antarctic Peninsula. PLoS ONE 6, e19173 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson A., Siegel V., Pakhomov E. & Rothery P. Long-term decline in krill stock and increase in salps within the Southern Ocean. Nature 432, 100–103 (2004). [DOI] [PubMed] [Google Scholar]
- Kawaguchi S. et al. Will krill fare well under Southern Ocean acidification? Biol. Lett. 7, 288–291 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey H. et al. Behavioural estimation of blue whale movements in the Northeast Pacific from state-space model analysis of satellite tracks. Endanger. Species Res. 10, 93–106 (2009). [Google Scholar]
- Reilly S. B. & Thayer V. G. Blue whale (Balaenoptera musculus) distribution in the eastern tropical Pacific. Mar. Mamm. Sci. 6, 265–277 (1990). [Google Scholar]
- Brown S. G. The movements of fin and blue whales within the Antarctic zone. Discovery Reports 33, 1–54 (1962). [Google Scholar]
- Stafford K. M. et al. Antarctic-type blue whale calls recorded at low latitudes in the Indian and eastern Pacific Oceans. Deep-Sea Res. I: Oceanogr. Res. Pap. 51, 1337–1346 (2004). [Google Scholar]
- Samaran F. et al. Seasonal and geographic variation of southern blue whale subspecies in the Indian Ocean. PLoS ONE 8, e71561 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- IWC. Report of the workshop on the comprehensive assessment of Southern Hemisphere humpback whales. J. Cetacean Res. Manage. Spec. Issue 3, 1–50 (2011). [Google Scholar]
- Pastene L. A., Goto M., Itoh S. & Numachi K. I. Spatial and temporal patterns of mitochondrial DNA variation in minke whales from Antarctic areas IV and V. Rep. Int. Whal. Comm. 46, 305–314 (1996). [Google Scholar]
- Anderwald P. et al. Possible cryptic stock structure for minke whales in the North Atlantic: implications for conservation and management. Biol. Conserv. 144, 2479–2489 (2011). [Google Scholar]
- Širović A. et al. Seasonality of blue and fin whale calls and the influence of sea ice in the Western Antarctic Peninsula. Deep-Sea Res. II: Top. Stud. Oceanogr. 51, 2327–2344 (2004). [Google Scholar]
- Širović A., Hildebrand J. A., Wiggins S. M. & Thiele D. Blue and fin whale acoustic presence around Antarctica during 2003 and 2004. Mar. Mamm. Sci. 25, 125–136 (2009). [Google Scholar]
- Samaran F., Adam O. & Guinet C. Discovery of a mid-latitude sympatric area for two Southern Hemisphere blue whale subspecies. Endanger. Species Res. 12, 157–165 (2010). [Google Scholar]
- Stafford K. M., Nieukirk S. L. & Fox C. G. An acoustic link between blue whales in the eastern tropical Pacific and the northeast Pacific. Mar. Mamm. Sci. 15, 1258–1268 (1999). [Google Scholar]
- Stafford K. M., Chapp E., Bohnenstiel D. R. & Tolstoy M. Seasonal detection of three types of “pygmy” blue whale calls in the Indian Ocean. Mar. Mamm. Sci. 27, 828–840 (2011). [Google Scholar]
- Olson P. A. et al. New Zealand blue whales: residency, morphology, and feeding behavior of a little-known population. Pac. Sci. 69, 477–485 (2015). [Google Scholar]
- Ensor P. H. Minke whales in the pack ice zone, East Antarctica, during the period of maximum ice extent. Rep. Int. Whal. Comm. 39, 219–225 (1989). [Google Scholar]
- Ribic C. A., Ainley D. G. & Fraser W. R. Habitat selection by marine mammals in the marginal ice zone. Antarct. Sci. 3, 181–186 (1991). [Google Scholar]
- Van Opzeeland I., Van Parijs S., Kinderman L., Burkhardt E. & Boebel O. Calling in the cold: pervasive acoustic presence of humpback whales (Megaptera novaeangliae) in Antarctic coastal waters. PLoS ONE 8, e73007 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bérubé M., Urbán R. J., Dizon A. E., Brownell R. L. & Palsbøll P. J. Genetic identification of a small and highly isolated population of fin whales (Balaenoptera physalus) in the Sea of Cortez, Mexico. Conserv. Genet. 3, 183–190 (2002). [Google Scholar]
- Mikhalev Y. A. Humpback whales Megaptera novaeangliae in the Arabian Sea. Mar. Ecol. Prog. Ser. 149, 13–21 (1997). [Google Scholar]
- Stafford K. M., Mellinger D. K., Moore S. E. & Fox C. G. Seasonal variability and detection range modeling of baleen whale calls in the Gulf of Alaska, 1999–2002. J. Acoust. Soc. Am. 122, 3378–3390 (2007). [DOI] [PubMed] [Google Scholar]
- Straley J. M. Fall and winter occurrence of humpback whales (Megaptera novaeangliae) in southeastern Alaska. Rep. Int. Whal. Comm. Spec. Issue 12, 319–323 (1990). [Google Scholar]
- Craig A. S., Herman L. M., Gabriele C. M. & Pack A. A. Migratory timing of humpback whales (Megaptera novaeangliae) in the central North Pacific varies with age, sex and reproductive status. Behaviour 140, 981–1001 (2003). [Google Scholar]
- Stevick P. T. et al. Segregation of migration by feeding ground origin in North Atlantic humpback whales (Megaptera novaeangliae). J. Zool. 259, 231–237 (2003). [Google Scholar]
- Herman L. M. et al. Resightings of humpback whales in Hawaiian waters over spans of 10–32 years: site fidelity, sex ratios, calving rates, female demographics, and the dynamics of social and behavioral roles of individuals. Mar. Mamm. Sci. 27, 736–768 (2011). [Google Scholar]
- Branch T. A., Mikhalev Y. A. & Kato H. Separating pygmy and Antarctic blue whales using long-forgotten ovarian data. Mar. Mamm. Sci. 25, 833–854 (2009). [Google Scholar]
- Branch T. A. Biologically plausible rates of increase for Antarctic blue whales. Paper SC/60/SH8 presented to the IWC Scientific Committee (2008).
- Branch T. A. Biological parameters for pygmy blue whales. Paper SC/60/SH6 presented to the IWC Scientific Committee (2008).
- Allendorf F. W., England P. R., Luikart G., Ritchie P. A. & Ryman N. Genetic effects of harvest on wild animal populations. Trends Ecol. Evol. 23, 327–337 (2008). [DOI] [PubMed] [Google Scholar]
- Leonard J. A. Ancient DNA applications for wildlife conservation. Mol. Ecol. 17, 4186–4196 (2008). [DOI] [PubMed] [Google Scholar]
- Clapham P. J., Aguilar A. & Hatch L. T. Determining spatial and temporal scales for management: lessons from whaling. Mar. Mamm. Sci. 24, 183–201 (2008). [Google Scholar]
- Attard C. R. M. et al. Low genetic diversity in pygmy blue whales is due to climate-induced diversification rather than anthropogenic impacts. Biol. Lett. 11, 20141037 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor B. L., Chivers S. J., Larese J. & Perrin W. F. Generation length and percent mature estimates for IUCN assessments of cetaceans. Administrative Report LJ-07-01. Southwest Fisheries Science Center (2007).
- Double M. C. et al. Migratory movements of pygmy blue whales (Balaenoptera musculus brevicauda) between Australia and Indonesia as revealed by satellite telemetry. PloS ONE 9, e93578 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block B. A. et al. Tracking apex marine predator movements in a dynamic ocean. Nature 475, 86–90 (2011). [DOI] [PubMed] [Google Scholar]
- Sunnucks P. & Hales D. F. Numerous transposed sequences of mitochondrial cytochrome oxidase I-II in aphids of the genus Sitobion (Hemiptera: Aphididae). Mol. Biol. Evol. 13, 510–524 (1996). [DOI] [PubMed] [Google Scholar]
- Attard C. R. M. et al. Genetic diversity and structure of blue whales (Balaenoptera musculus) in Australian feeding aggregations. Conserv. Genet. 11, 2437–2441 (2010). [Google Scholar]
- Van Oosterhout C., Hutchinson W. F., Wills D. P. M. & Shipley P. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 4, 535–538 (2004). [Google Scholar]
- Excoffier L. & Lischer H. E. L. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567 (2010). [DOI] [PubMed] [Google Scholar]
- Goudet J. FSTAT (version 1.2): a computer program to calculate F-statistics. J. Hered. 86, 485–486 (1995). [Google Scholar]
- Holm S. A simple sequentially rejective multiple test procedure. Scand. J. Stat. 6, 65–70 (1979). [Google Scholar]
- Coughlan J., Mirimin L., Dillane E., Rogan E. & Cross T. F. Isolation and characterization of novel microsatellite loci for the short-beaked common dolphin (Delphinus delphis) and cross-amplification in other cetacean species. Mol. Ecol. Notes 6, 490–492 (2006). [Google Scholar]
- Pritchard J. K., Stephens M. & Donnelly P. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D., Stephens M. & Pritchard J. K. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164, 1567–1587 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubisz M. J., Falush D., Stephens M. & Pritchard J. K. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Resour. 9, 1322–1332 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopelman N. M., Mayzel J., Jakobsson M., Rosenberg N. A. & Mayrose I. CLUMPAK: a program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 15, 1179–1191 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno G., Regnaut S. & Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620 (2005). [DOI] [PubMed] [Google Scholar]
- Earl D. A. & vonHoldt B. M. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4, 359–361 (2012). [Google Scholar]
- Quintela M. et al. Investigating population genetic structure in a highly mobile marine organism: the minke whale Balaenoptera acutorostrata acutorostrata in the North East Atlantic. PLoS ONE 9, e108640 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamvar Z. N., Tabima J. F. & Grünwald N. J. Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2, e281 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core Team. R R: a language and environment for statistical computing. (R Foundation for Statistical Computing, 2014). Available at: www.R-project.org.
- Raymond M. & Rousset F. An exact test for population differentiation. Evolution 49, 1280–1283 (1995). [DOI] [PubMed] [Google Scholar]
- Rousset F. GENEPOP'007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol. Ecol. Resour. 8, 103–106 (2008). [DOI] [PubMed] [Google Scholar]
- Peakall R. & Smouse P. E. GENALEX 6.5: genetic analysis in Excel. Population genetic software for teaching and research - an update. Bioinformatics 28, 2537–2539 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall R. & Smouse P. E. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 6, 288–295 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pétit R. J., El Mousadik A. & Pons O. Identifying populations for conservation on the basis of genetic markers. Conserv. Biol. 12, 844–855 (1998). [Google Scholar]
- International Hydrographic Organization. Limits of Oceans and Seas 3rd edn, Special Publication No. 23. (International Hydrographic Organization, 1953).


