Abstract
gmhA encodes a phosphoheptose isomerase that catalyzes the biosynthesis of heptose, a conserved component of lipopolysaccharide (LPS). GmhA plays an important role in Yersinia pestis biofilm blockage in the flea gut. waaA, waaE, and coaD constitute a three-gene operon waaAE-coaD in Y. pestis. waaA encodes a transferase that is responsible for binding lipid-A to the core oligosaccharide of LPS. WaaA is a key determinant in Y. pestis biofilm formation, and the waaA expression is positively regulated by the two-component regulatory system PhoP/PhoQ. WaaE is involved in LPS modification and is necessary for Y. pestis biofilm production. In this study, the biofilm-related phenotypic assays indicate that the global regulator CRP stimulates Y. pestis biofilm formation in vitro and on nematodes, while it has no regulatory effect on the biosynthesis of the biofilm-signaling molecular 3′,5′-cyclic diguanosine monophosphate. Further gene regulation experiments disclose that CRP does not regulate the hms genes at the transcriptional level but directly promotes the gmhA transcription and indirectly activates the waaAE-coaD transcription through directly acting on phoPQ-YPO1632. Thus, it is speculated that CRP-mediated carbon catabolite regulation of Y. pestis biofilm formation depends on the CRP-dependent carbon source metabolic pathways of the biosynthesis, modification, and transportation of biofilm exopolysaccharide.
Keywords: Yersinia pestis, CRP, gmhA, waaAE-coaD, phoPQ-YPO1632, biofilm
Introduction
Yersinia pestis, the causative agent of plague, is transmitted primarily by fleas and has been responsible for three plague pandemics in human history (Perry and Fetherston, 1997). Biofilms formed by Y. pestis can attach to and block the proventriculus of a flea, which promotes the flea-borne transmission of this pathogen (Zhou and Yang, 2011; Chouikha and Hinnebusch, 2012). HmsHFRS is responsible for the biosynthesis and translocation of poly-β-1,6-N-acetylglucosamine exopolysaccharide (EPS), a primary dry component of the Y. pestis biofilm matrix (Bobrov et al., 2008). In Y. pestis, HmsT and HmsD (hmsD is located in the hmsCDE operon) are the two sole diguanylate cyclases that catalyze the biosynthesis of 3′,5′-cyclic diguanosine monophosphate (c-di-GMP), a second messenger stimulating biofilm EPS production (Bobrov et al., 2011; Sun et al., 2011). Conversely, c-di-GMP is degraded by the sole phosphodiesterase HmsP in Y. pestis (Kirillina et al., 2004; Bobrov et al., 2005).
Lipopolysaccharide (LPS) is an integral component of the outer membrane of Gram-negative bacteria. Generally, it is composed of three domains: lipid-A, core oligosaccharide, and O-specific antigen or O side chain. However, the LPS from Y. pestis contains only lipid-A bound to the core oligosaccharide by 3-deoxy-D-manno-octulosonic acid (Kdo) (Prior et al., 2001). The gmhA gene encodes a phosphoheptose isomerase that is required for heptose biosynthesis, a conserved component of the core oligosaccharide, and the deletion of gmhA in Y. pestis leads to inadequate biofilm production for flea blockage (Darby et al., 2005). In Y. pestis, waaA, waaE, and coaD constitute a three-gene operon waaAE-coaD. The waaA gene encodes a Kdo transferase involved in the attachment of lipid-A to the core oligosaccharide, and waaA mutants show a severely biofilm-defective phenotype of Y. pestis (Tan and Darby, 2005). The waaE gene encodes a protein that is required for adding a substitution on the inner core heptose, and the waaE deletion leads to 40% attenuation of biofilm production of Y. pestis (Izquierdo et al., 2002).
PhoP and PhoQ constitute a typical two-component regulatory system (Groisman, 2001). The PhoPQ system is necessary for the intracellular survival of Y. pestis in phagocytes during the early stages of infection because the PhoP-regulated genes promote the resistance to antimicrobial peptides and the adaptation to low-Mg2+ conditions (Oyston et al., 2000; Grabenstein et al., 2006). However, the deletion of phoP in Y. pestis contributes little to plague pathogenesis in mice (Bozue et al., 2011). In addition, PhoPQ is induced by low pH in the flea gut, and controls physiological adaptation to acid and other stresses encountered during infection of the fleas; it simultaneously stimulates the formation of flea-borne infectious Y. pestis biofilms (Rebeil et al., 2013; Vadyvaloo et al., 2015). Nevertheless, PhoP-mediated regulation of Y. pestis biofilm formation relies on its modulation of the waaAE-coaD operon rather than the hms genes (Liu et al., 2014).
Cyclic AMP receptor protein (CRP) has been characterized as a virulence-associated regulator in many pathogens (Skorupski and Taylor, 1997; Petersen and Young, 2002; Rickman et al., 2005; Zhan et al., 2008; Oh et al., 2009). CRP controls the expression of multiple bactieral virulence genes and responds to environmental changes by sensing the availability of carbons (Busby and Ebright, 1999). CRP can only be activated by binding a small-molecule inducer cyclic AMP (cAMP), and the cAMP-CRP complex usually serves as a DNA-binding transcription factor by directly binding to the promoter regions of its target genes to activate or repress their transcription (Busby and Ebright, 1999). The cAMP-CRP complex modulates more than 6% of Y. pestis genes that are involved in a large variety of functions (Zhan et al., 2008). CRP is required for the development of bubonic and pneumonic plague, which may rely on CRP-dependent expression of pla and that of the sycO-ypkA-yopJ operon of the Yop-Ysc type III secretion system in Y. pestis (Kim et al., 2007; Liu et al., 2009; Zhan et al., 2009). Meanwhile, crp expression is positively regulated by PhoP at the transcriptional level and by Hfq (a major posttranscriptional regulator that controls biofilm formation and virulence in Y. pestis) at the posttranscriptional level (Zhang et al., 2013a; Lathem et al., 2014), indicating the significant role of CRP in the virulence gene regulation of Y. pestis. CRP or its homologous regulators control biofilm production in various pathogens such as Listeria monocytogenes (Salazar et al., 2013), Xanthomonas campestris pathovar campestris (Lu et al., 2012), Aggregatibacter actinomycetemcomitans (Torres-Escobar et al., 2013), Vibrio cholera (Fong and Yildiz, 2008), and Serratia marcescens (Kalivoda et al., 2008). The loss of CRP in Y. pestis results in a significant reduction of biofilm production, and the carbon storage regulator CsrA can enhance CRP-mediated regulation of Y. pestis biofilm formation (Willias et al., 2015). However, the crucial CRP-dependent factors that contribute to this alteration of biofilm formation are still unknown.
In the present work, the biofilm-related phenotypic assays showed that deletion of crp in a Y. pestis Microtus strain led to a dramatic reduction in biofilm production, but had no effect on the c-di-GMP biosynthesis. The subsequent gene regulation experiments indicated that CRP directly activated the gmhA transcription and meanwhile indirectly stimulated the waaA transcription through directly acting on phoPQ, while CRP had no regulatory effect on the hms genes at the transcriptional level.
Materials and Methods
Ethics Statement
All the animal experiments were carried out in accordance with the protocols approved by the Ethics Committee in the Beijing Institute of Microbiology and Epidemiology.
Bacterial Strains
The wild-type Yersinia pestis Microtus strain 201 (WT) is avirulent to humans but highly virulent to mice (Zhou et al., 2004). The non-polar crp mutant (Δcrp) and its complemented mutant strain (C-crp) have been described previously (Zhan et al., 2008). Given the previous observation that deletion of hmsS led to a biofilm-defective phenotype in Y. pestis (Forman et al., 2006), the hmsS mutant ΔhmsS was used as a reference biofilm-defective strain in this work. All of the primers designed in this study are listed in Supplementary Table S1.
Bacterial Growth and RNA Isolation
Overnight cell cultures in Luria-Bertani (LB) broth with an optical density (OD620) of about 1.0 were diluted 1:20 into 18 ml of fresh LB broth for further cultivation at 26°C with shaking at 230 rpm to reach middle stationary phase (an OD620 of 1.0 to 1.2), then cells were harvested for gene regulation or phenotypic assays. Immediately before harvesting of bacterial cells for RNA isolation, a double-volume of RNAprotect reagent (Qiagen) was added to one-volume of cell culture, and total RNA was extracted using TRIzol Reagent (Invitrogen). RNA quality was monitored by agarose gel electrophoresis, and RNA quantity was determined by spectrophotometry.
Primer Extension Assay
As described in our previous studies (Sun et al., 2012; Zhang et al., 2013a,b) a 5′-32P-labeled oligonucleotide primer complementary to a portion of the RNA transcript of each indicated gene was employed to synthesize cDNAs from total RNA templates using the Promega Primer Extension System. If different Y. pestis strains were involved in a single experiment, equal amounts of total RNA were used as starting materials. Sequence ladders were prepared with the same 5′-32P-labeled primers using the AccuPower and Top DNA Sequencing Kit (Bioneer). Radioactive species were detected by autoradiography. The 5′-terminus of the RNA transcript (i.e., transcription start) of each target gene was mapped according to the size of primer extension products, while the relative mRNA levels were determined by the intensities of the primer extension products.
Quantitative RT-PCR
Gene-specific primers were designed to produce amplicons for target genes. Contaminating DNA in the RNA samples was removed using the Ambion DNA-freeTM Kit (Applied Biosystems). cDNAs were generated using 5 μg of RNA and 3 μg of random hexamer primers. Real-time PCR was performed using the LightCycler system (Roche) and the SYBR Green master mix (Zhan et al., 2008; Sun et al., 2014). Based on the standard curves of 16S rRNA expression, the relative mRNA level was determined by calculating the threshold cycle (ΔCt) of target genes via the classic ΔCt method. Negative controls used cDNA generated without reverse transcriptase as a template. Reactions containing primer pairs without template were also included as blank controls. The 16S rRNA gene was used as an internal control for normalization.
LacZ Fusion and β-Galactosidase Assay
A promoter-proximal DNA region of each indicated gene was cloned into the low-copy-number transcriptional fusion vector pRW50 (Lodge et al., 1992) that harbors a promoterless lacZ reporter gene. Y. pestis strains transformed with the recombinant plasmid or the empty pRW50 (negative control) were cultivated to measure β-galactosidase activity in the cellular extract using the β-galactosidase Enzyme Assay System (Promega) (Sun et al., 2012; Zhang et al., 2013a,b).
Purification of 6x His-Tagged CRP (His-CRP) Protein
The entire coding region of crp of strain 201 was directionally cloned into the BamHI and HindIII sites of plasmid pET28a (Novagen) as described previously (Zhan et al., 2008). The recombinant plasmid encoding the His-CRP protein was transformed into Escherichia coli BL21 (DE3) cells (Novagen). Expression of His-CRP was induced by the addition of 1 mM isopropyl-beta-D-thiogalactoside. The overproduced proteins were purified under native conditions using a Ni-NTA agarose column (Qiagen). The purified, eluted protein was concentrated with the Amicon Ultra-15 (Millipore) to a final concentration of 0.8–1.0 mg/ml in storage buffer containing phosphate buffered saline (PBS, pH 9.5) plus 20% glycerol. The protein purity was verified by sodium dodecyl sulfate polyacrylamide gel electrophoresis with silver staining.
Electrophoretic Mobility Shift Assay (EMSA)
For EMSA (Sun et al., 2012; Zhang et al., 2013a), promoter-proximal DNA regions were prepared by PCR amplification. EMSA was performed using the Gel Shift Assay Systems (Promega). The 5′ ends of DNA were labeled using [γ-32P] ATP and T4 polynucleotide kinase. DNA binding was performed in a 10 μl reaction volume containing binding buffer [100 μM MnCl2, 1 mM MgCl2, 0.5 mM DTT, 50 mM KCl, 10 mM Tris-HCl (pH 7.5), 0.05 mg/ml sheared salmon sperm DNA, 0.05 mg/ml BSA, and 4% glycerol], labeled DNA (1000 to 2000 c.p.m/μl), 1 mM cAMP, and increasing amounts of His-CRP. We included two control reactions: one contained the specific DNA competitor (unlabeled promoter DNA regions; cold probe), while the other was the non-specific protein competitor (rabbit anti-F1-protein polyclonal IgG antibody). After incubation at room temperature for 30 min, the products were loaded onto a native 4% (w/v) polyacrylamide gel and electrophoresed in 0.5x Tris-borate buffer containing 100 μM MnCl2 for 60 min at 150 V. Radioactive species were detected by autoradiography.
DNase I Footprinting
For DNase I footprinting (Zhang et al., 2013a,b), the target promoter-proximal DNA regions with a single 32P-labeled end were incubated with increasing amounts of purified His-CRP, which was followed by partial digestion with RQ1 RNase-Free DNase I (Promega). The digested DNA samples were extracted with phenol/chloroform, precipitated with ethanol, and analyzed in a 8 M urea-6% polyacrylamide gel. Protected regions were identified by comparison with the sequence ladders. For sequencing, we used the Top DNA Sequencing Kit (BIONEER). The templates for sequencing were the same as the DNA fragments in the DNase I footprinting assay.
Biofilm-Related Assays
Four different methods (Fang et al., 2013, 2015) were employed for biofilm-related assays: (i) crystal violet staining of the in vitro biofilm masses attached to the well walls when bacteria were grown in polystyrene microtiter plates; (ii) determination of the percentages of fourth-stage larvae and adults (L4/adult) of Caenorhabditis elegans (Darby et al., 2002; Fang et al., 2013) after the incubation of nematode eggs on Y. pestis lawns, which negatively reflected the bacterial ability to produce biofilms; (iii) observation of the rugose colony morphology of bacteria grown on LB agar plates, which positively reflected the bacterial ability to synthesize biofilm matrix biofilm EPS; and (iv) determination of intracellular c-di-GMP levels by a chromatography-coupled tandem mass spectrometry (HPLC-MS/MS) method.
Experimental Replicates and Statistical Methods
For real-time RT-PCR, LacZ fusion, crystal violet staining of biofilms, and the determination of L4/adult nematodes or c-di-GMP, experiments were performed with at least three independent bacterial cultures/lawns, and values were expressed as the mean ± SD. The paired Student’s t-test was performed to determine significant differences; P < 0.01 was considered to indicate statistical significance. For primer extension, EMSA, DNase I footprinting, and colony morphology observation, representative data from at least two independent biological replicates are shown.
Results
CRP Enhances Biofilm Formation, But Does Not Influence c-di-GMP Production
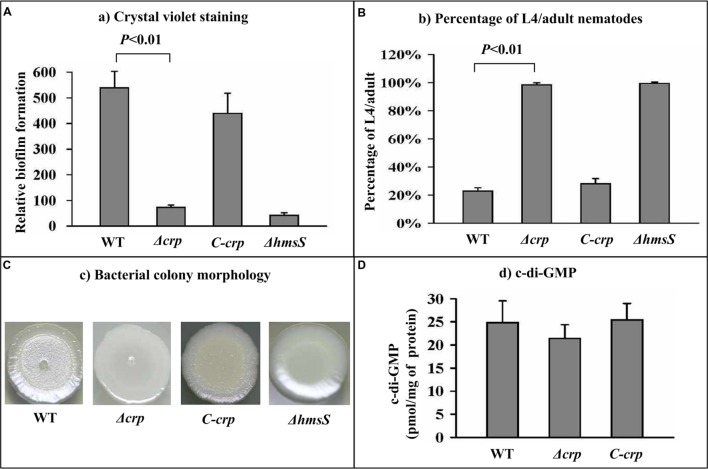
Both the Δcrp and ΔhmsS mutant strains showed a dramatic reduction in biofilm crystal violet staining compared with the WT and the complemented mutant strain C-crp, indicating the significantly decreased production of in vitro biofilms due to the deletion of crp (Figure 1A). After incubation of nematode eggs on bacterial lawns of WT or C-crp, only a small portion (about 20%) of larvae grew and developed into L4/adult nematodes owing to abundant attachment of Y. pestis biofilms on nematode heads; while the value for the Δcrp and ΔhmsS mutants was nearly 100% (Figure 1B). When grown on LB agar plates, Δcrp developed much smoother colony morphology than WT that was comparable to C-crp (Figure 1C). This observation suggested that Δcrp produced much less biofilm EPS relative to WT. The intracellular levels of c-di-GMP were determined in the WT, Δcrp, and C-crp strains by a HPLC-MS/MS method. However, Δcrp showed no difference in the c-di-GMP production compared with WT and C-crp (Figure 1D). In summary, CRP was an activator of Y. pestis biofilm development, but had no effect on c-di-GMP production.
FIGURE 1.
Involvement of CRP in biofilm formation and c-di-GMP biosynthesis. (A) Crystal violet staining. Yersinia pestis was grown in 24-well polystyrene dishes, and the bacterial biomass (in vitro biofilms) attached to well walls were stained with crystal violet to determine OD570 values. The planktonic cells were used to determine OD620 values. The relative capacity for biofilm formation of each strain was determined as follows: 100 × OD570/OD620. (B) C. elegans biofilms. After incubation of nematode eggs on lawns of the indicated Y. pestis strains, the developmental stages of nematodes on each lawn were scored to calculate the percentage of L4/adults. (C) Bacterial colony morphology. Aliquots of bacterial glycerol stocks were spotted onto LB plates, followed by incubation for one week. (D) Intracellular c-di-GMP concentration. The intracellular c-di-GMP concentrations were determined by a HPLC-MS/MS method, and the determining values were expressed as pmol/mg of bacterial protein.
The first genes of the multi-gene operons hmsHFRS, hmsCDE, phoPQ-YPO1632, and waaAE-coaD as well as the individual genes hmsT, hmsP, and gmhA, which encode the major biofilm determinants of Y. pestis, were subjected to the following gene regulation experiments to analyze the detailed regulatory action of CRP on these target genes.
CRP Has No Regulatory Effect on hms Genes at the Transcriptional Level
As revealed by the primer extension and real-time RT-PCR assays, the mRNA levels of each of hmsH (Supplementary Figures S1a,b), hmsT (Supplementary Figures S2a,b), hmsC (Supplementary Figures S3a,b), and hmsP (Supplementary Figures S4a,b) were unaltered in Δcrp compared with WT. The LacZ fusion assay also revealed that the deletion of crp had no effect on the promoter activity of each of hmsH (Supplementary Figure S1c), hmsT (Supplementary Figure S2c), hmsC (Supplementary Figure S3c), and hmsP (Supplementary Figure S4c). Moreover, the EMSA assay indicated that His-CRP could not bind to the promoter-proximal region of each of hmsH (Supplementary Figure S1d), hmsT (Supplementary Figure S2d), hmsC (Supplementary Figure S3d), and hmsP (Supplementary Figure S4d). Taken together, CRP had no regulatory effect on hmsHFRS, hmsT, hmsCDE, and hmsP at the transcriptional level.
CRP Directly Induces gmhA Transcription
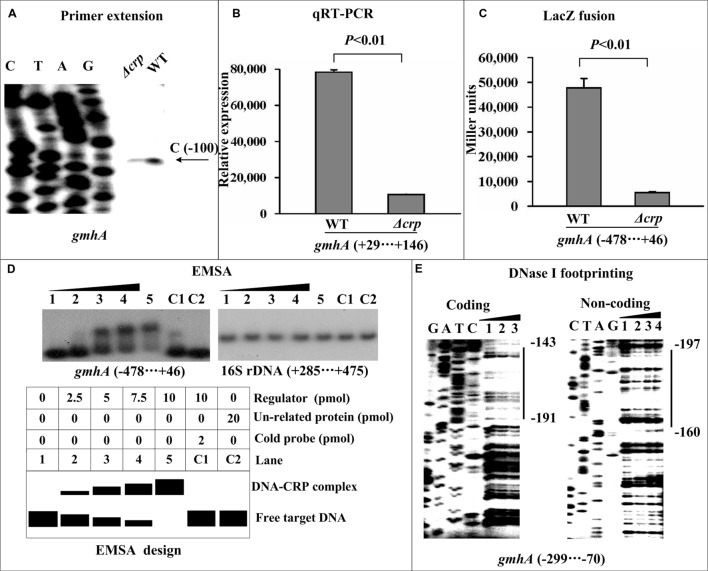
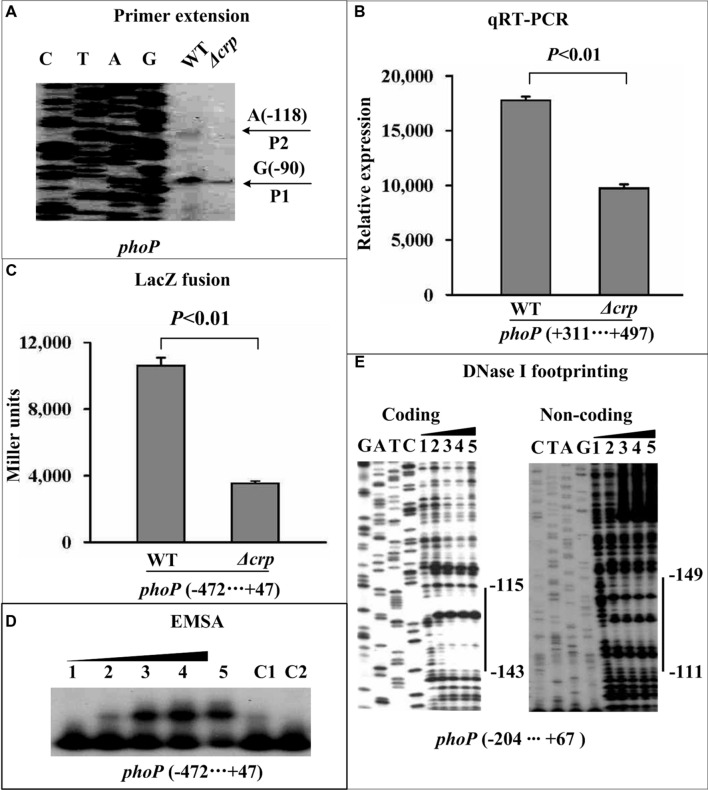
The primer extension assay detected a single transcriptional start site for gmhA, located 100 bp upstream of gmhA, and the mRNA levels of gmhA were found to be reduced in Δcrp compared with WT (Figure 2A), which was confirmed by real-time RT-PCR (Figure 2B). Similarly, the LacZ fusion assay showed that deletion of crp induced markedly attenuated promoter activity of gmhA (Figure 2C). The EMSA assay indicated that a purified His-CRP bound to the labeled gmhA promoter DNA in a dose-dependent manner. To confirm the specificity of the CRP–DNA association, the EMSA experiments included a partial coding region of the 16S rRNA gene and negative results were obtained (Figure 2D). To further locate the precise CRP sites, DNase I footprinting experiments were performed with both coding and non-coding strands of target DNA fragments. The results showed that His-CRP protected a single region within each of the two target DNA fragments tested against DNase I digestion in a dose-dependent manner. The footprint was located from 197 to 143 bp upstream of gmhA (Figure 2E). Thus, CRP directly promotes the transcription of the gmhA gene.
FIGURE 2.
Direct activation of gmhA transcription by CRP. Lanes C, T, A, and G represent Sanger sequencing reactions. The minus and positive numbers indicated nucleotide positions upstream and downstream of the indicated genes. (A) Primer extension. An oligonucleotide primer was designed to be complementary to the RNA transcript of each indicated gene. The primer extension products were analyzed on an 8 M urea-6% acrylamide sequencing gel. The transcriptional start sites are indicated by arrows, showing the nucleotides. (B) Quantitative RT-PCR. The mRNA levels of gmhA were compared between Δcrp and WT. A standard curve was generated for each RNA preparation with the 16S rRNA gene; the relative mRNA level was determined by calculating the threshold cycle (ΔCt) of target genes via the classic ΔCt method. (C) LacZ fusion. The target promoter-proximal DNA region was cloned into the lacZ transcriptional fusion vector pRW50 and then transformed into the WT or Δcrp strain to determine the promoter activity, i.e., the β-galactosidase activity (miller units) in the cellular extracts. (D) EMSA. The radioactively labeled promoter-proximal DNA fragments were incubated with increasing amounts of a purified His-CRP protein and then subjected to 4% (w/v) polyacrylamide gel electrophoresis. The band of free DNA disappeared with increasing amounts of His-CRP, resulting in a DNA band with decreased mobility, which presumably represented the DNA-CRP complex. Shown also is a schematic representation of the EMSA design. A DNA fragment from the coding region of the 16S rRNA gene served as a negative control. (E) DNase I footprinting. Labeled coding or non-coding DNA probes were incubated with increasing amounts of purified His-CRP and then subjected to a DNase I footprinting assay. The footprint regions are indicated with vertical bars.
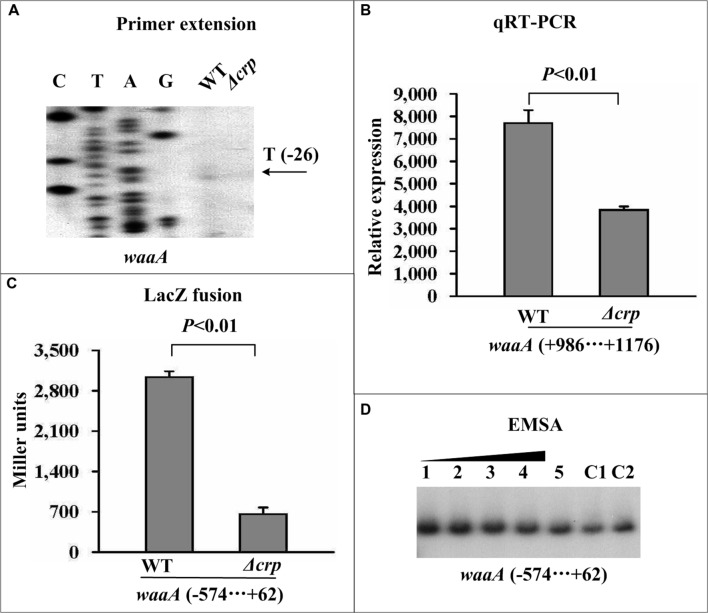
CRP Indirectly Promotes waaA Transcription Through Directly Acting on phoP
The primer extension and real-time RT-PCR assays revealed that the mRNA levels of phoP (Figures 3A,B) and waaA (Figures 4A,B) were dramatically reduced in Δcrp compared with WT. Additionally, two distinct transcriptional start sites for phoP and a single transcriptional start site for waaA were detected by the primer extension experiment. The LacZ fusion assay indicated that the expression of crp was essential for the promoter activity of the phoP (Figure 3C) and waaA (Figure 4C) genes. The EMSA assay indicated that His-CRP was able to bind in a dose-dependent manner to the promoter-proximal region of the phoP gene (Figure 3D), but not to that of the waaA (Figure 4D) gene. The precise CRP-sites for its direct target gene phoP were determined by DNase I footprinting experiments, and the results showed that the footprint was located from 149 to 111 bp upstream of phoP (Figure 3E). Therefore, CRP-dependent activation of phoPQ-YPO1632 occurs in a direct manner, while CRP-dependent activation of waaAE-coaD occurs in an indirect manner. In addition, our previous study showed that PhoP directly stimulates the transcription of waaAE-coaD (Liu et al., 2014), and thereby CRP-mediated activation of waaAE-coaD is most likely to function by directly acting on phoPQ-YPO1632.
FIGURE 3.
Direct activation of phoP transcription by CRP. See Figure 2 for the annotations of primer extension (A), quantitative RT-PCR (B), LacZ fusion (C), EMSA (D), and DNase I footprinting (E).
FIGURE 4.
Indirect activation of waaA transcription by CRP. See Figure 2 for the annotations of primer extension (A), quantitative RT-PCR (B), LacZ fusion (C), and EMSA (D).
Promoter Structure of gmhA, phoP, and waaA
The primer extension experiment was used to depict the 5′ termini (transcriptional starts) of RNA transcripts of gmhA, phoP, and waaA, and the –10 and –35 core promoter elements for RNA polymerase recognition were accordingly predicted. The DNase I footprinting experiments were carried out to determine the precise CRP sites for the direct CRP target genes. Collection of data from the translational/transcriptional start sites, the core promoter –10 and –35 elements, the Shine–Dalgarno sequences for ribosomal binding, and the CRP sites (in this study) and PhoP sites (Zhang et al., 2013a; Liu et al., 2014) enabled us to characterize the organization of the promoters of gmhA (Figure 5A), phoP (Figure 5B), and waaA (Figure 5C).
FIGURE 5.
Organization of promoter-proximal DNA regions. The promoter-proximal DNA regions of the CRP target genes gmhA (A), phoP (B), and waaA (C) are derived from Y. pestis CO92. Shown are translation/transcription starts, predicted core promoter –10 and –35 elements, predicted Shine–Dalgarno sequences, CRP and PhoP sites.
Discussion
In this study, CRP-dependent activation of biofilm production was observed in Y. pestis Microtus strain 201, which confirms the results obtained in Y. pestis KIM6+ (a pCD1- derivative of KIM) and CO92 (Willias et al., 2015). Our findings also indicated that CRP-mediated regulation of Y. pestis biofilm production has no regulatory relationship with the Hms factors, which is in agreement with the results showing that the deletion of crp had no effect on the biosynthesis of c-di-GMP. However, CRP is able to stimulate biofilm development by directly activating the transcription of gmhA and meanwhile indirectly promoting the transcription of waaAE-coaD. CRP stimulates the transcription of waaAE-coaD through acting on PhoPQ, a direct activator of waaAE-coaD (Liu et al., 2014).
Each of GmhA, WaaA, and WaaE plays a key role in biofilm EPS biosynthesis or modification and is required for biofilm production in Y. pestis, but the exact mechanisms of action remain to be determined. Biofilm EPS synthesized in Y. pestis cells must be exported through the outer membrane that is predominantly composed of LPS. GmhA, WaaA, and WaaE are all crucial determinants for the biosynthesis of integral LPS. Deletion of each of gmhA, waaA, and waaE results in the formation of an incomplete LPS and, thereby, hinders the export of biofilm EPS, which ultimately leads to a dramatic reduction in biofilm EPS. Given the fact that CRP is a master regulator of the metabolism of alternate carbon sources, it is supposed that CRP-dependent activation of gmhA and waaAE-coaD enhances Y. pestis biofilm production by influencing the carbon source metabolic pathways of the biosynthesis, modification and transportation of biofilm EPS.
The two-component regulatory system PhoP/PhoQ senses and responds to the host environment stimuli of low Mg2+, low pH, and antimicrobial peptides, and plays an important role in the flea gut by stimulating the production of flea-borne infectious Y. pestis biofilms. CRP senses and responds to the switch in carbon sources, and acts as an activator of Y. pestis biofilm formation in vitro and on nematodes. Nevertheless, both CRP (this study) and PhoP (Rebeil et al., 2013) have no regulatory effect on the hms gene transcription, while the expression of gmhA and waaAE-coaD are positively regulated by CRP, and concurrently waaAE-coaD is directly activated by PhoP. Therefore, CRP- and PhoP-mediated regulation of Y. pestis biofilm production may be dependent on a similar mechanism. According to the conclusions obtained in this study and previous studies (Rebeil et al., 2013; Zhang et al., 2013a, 2015; Liu et al., 2014), the complex regulatory interactions of cAMP-CRP and PhoP/PhoQ facilitate the cellular pathways governed by these two regulatory systems to merge into a single global regulatory circuit in Y. pestis, which responds to environmental stimuli and controls the expression of genes involved in various functions in flea vectors, thereby enabling better survival and transmission of this pathogen.
The CRP site for gmhA is upstream of the promoter –35 element, and thereby CRP-mediated regulation of the gmhApromoter may be a class I transcriptional stimulation that depends on the RNA polymerase α subunit C-terminal domain for function (Ishihama, 2000), and PhoP-mediated regulation of waaA may operate via a similar regulatory mechanism. However, PhoP recognizes two distinct sites (site 1 and 2) within the phoP upstream region, and site 1 overlaps the –10 and –35 regions for P2phoP and the –35 region for P1phoP. This may induce the formation of a loop in the phoP upstream DNA region that blocks the entry of the RNA polymerase and thereby leads to the autorepression of phoP. However, the CRP site for phoP partially overlaps with the PhoP site 1, and the interaction of these two regulators may lead to loop relief that facilitates the entry of the RNA polymerase, and thereby stimulates the transcription of phoP.
Author Contributions
DZ and RY conceived the study and designed experimental procedures; LL, HF, HY, YZ, YH performed the experiments and carried out data analysis; DZ, LL, and RY wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31430006, 31471184, and 81302608), the National Basic Research Program of China (2015CB554202), and the Foundation of State Key Laboratory of Pathogen and Biosecurity of China (SKLPBS1402). The English language used in this manuscript was polished by Huaju Culture and Media Co., Ltd (Beijing, China).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00295
CRP had no regulatory effect on hmsH at the transcriptional level. See Figure 2 for the annotations of primer extension (a), quantitative RT-PCR (b), LacZ fusion (c), and EMSA (d).
CRP had no regulatory effect on hmsT at the transcriptional level. See Figure 2 for the annotations of primer extension (a), quantitative RT-PCR (b), LacZ fusion (c), and EMSA (d).
CRP had no regulatory effect on hmsC at the transcriptional level. See Figure 2 for the annotations of primer extension (a), quantitative RT-PCR (b), LacZ fusion (c), and EMSA (d).
CRP had no regulatory effect on hmsP at the transcriptional level. See Figure 2 for the annotations of primer extension (a), quantitative RT-PCR (b), LacZ fusion (c), and EMSA (d).
References
- Bobrov A. G., Kirillina O., Forman S., Mack D., Perry R. D. (2008). Insights into Yersinia pestis biofilm development: topology and co-interaction of Hms inner membrane proteins involved in exopolysaccharide production. Environ. Microbiol. 10 1419–1432. 10.1111/j.1462-2920.2007.01554.x [DOI] [PubMed] [Google Scholar]
- Bobrov A. G., Kirillina O., Perry R. D. (2005). The phosphodiesterase activity of the HmsP EAL domain is required for negative regulation of biofilm formation in Yersinia pestis. FEMS Microbiol. Lett. 247 123–130. 10.1016/j.femsle.2005.04.036 [DOI] [PubMed] [Google Scholar]
- Bobrov A. G., Kirillina O., Ryjenkov D. A., Waters C. M., Price P. A., Fetherston J. D., et al. (2011). Systematic analysis of cyclic di-GMP signalling enzymes and their role in biofilm formation and virulence in Yersinia pestis. Mol. Microbiol. 79 533–551. 10.1111/j.1365-2958.2010.07470.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozue J., Mou S., Moody K. L., Cote C. K., Trevino S., Fritz D., et al. (2011). The role of the phoPQ operon in the pathogenesis of the fully virulent CO92 strain of Yersinia pestis and the IP32953 strain of Yersinia pseudotuberculosis. Microb. Pathog. 50 314–321. 10.1016/j.micpath.2011.02.005 [DOI] [PubMed] [Google Scholar]
- Busby S., Ebright R. H. (1999). Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293 199–213. 10.1006/jmbi.1999.3161 [DOI] [PubMed] [Google Scholar]
- Chouikha I., Hinnebusch B. J. (2012). Yersinia–flea interactions and the evolution of the arthropod-borne transmission route of plague. Curr. Opin. Microbiol. 15 239–246. 10.1016/j.mib.2012.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby C., Ananth S. L., Tan L., Hinnebusch B. J. (2005). Identification of gmhA, a Yersinia pestis gene required for flea blockage, by using a Caenorhabditis elegans biofilm system. Infect. Immun. 73 7236–7242. 10.1128/IAI.73.11.7236-7242.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby C., Hsu J. W., Ghori N., Falkow S. (2002). Caenorhabditis elegans: plague bacteria biofilm blocks food intake. Nature 417 243–244. 10.1038/417243a [DOI] [PubMed] [Google Scholar]
- Fang N., Gao H., Wang L., Qu S., Zhang Y. Q., Yang R. F., et al. (2013). Optimized methods for biofilm analysis in Yersinia pestis. Biomed. Environ. Sci. 26 408–411. 10.3967/0895-3988.2013.05.012 [DOI] [PubMed] [Google Scholar]
- Fang N., Yang H., Fang H., Liu L., Zhang Y., Wang L., et al. (2015). RcsAB is a major repressor of Yersinia biofilm development through directly acting on hmsCDE, hmsT, and hmsHFRS. Sci. Rep. 5 9566 10.1038/srep09566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong J. C., Yildiz F. H. (2008). Interplay between cyclic AMP-cyclic AMP receptor protein and cyclic di-GMP signaling in Vibrio cholerae biofilm formation. J. Bacteriol. 190 6646–6659. 10.1128/JB.00466-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman S., Bobrov A. G., Kirillina O., Craig S. K., Abney J., Fetherston J. D., et al. (2006). Identification of critical amino acid residues in the plague biofilm Hms proteins. Microbiology 152 3399–3410. 10.1099/mic.0.29224-0 [DOI] [PubMed] [Google Scholar]
- Grabenstein J. P., Fukuto H. S., Palmer L. E., Bliska J. B. (2006). Characterization of phagosome trafficking and identification of PhoP-regulated genes important for survival of Yersinia pestis in macrophages. Infect. Immun. 74 3727–3741. 10.1128/IAI.00255-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman E. A. (2001). The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 183 1835–1842. 10.1128/JB.183.6.1835-1842.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama A. (2000). Functional modulation of Escherichia coli RNA polymerase. Annu. Rev. Microbiol. 54 499–518. 10.1146/annurev.micro.54.1.499 [DOI] [PubMed] [Google Scholar]
- Izquierdo L., Abitiu N., Coderch N., Hita B., Merino S., Gavin R., et al. (2002). The inner-core lipopolysaccharide biosynthetic waaE gene: function and genetic distribution among some Enterobacteriaceae. Microbiology 148 3485–3496. 10.1099/00221287-148-11-3485 [DOI] [PubMed] [Google Scholar]
- Kalivoda E. J., Stella N. A., O’Dee D. M., Nau G. J., Shanks R. M. (2008). The cyclic AMP-dependent catabolite repression system of Serratia marcescens mediates biofilm formation through regulation of type 1 fimbriae. Appl. Environ. Microbiol. 74 3461–3470. 10.1128/AEM.02733-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T. J., Chauhan S., Motin V. L., Goh E. B., Igo M. M., Young G. M. (2007). Direct transcriptional control of the plasminogen activator gene of Yersinia pestis by the cyclic AMP receptor protein. J. Bacteriol. 189 8890–8900. 10.1128/JB.00972-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirillina O., Fetherston J. D., Bobrov A. G., Abney J., Perry R. D. (2004). HmsP, a putative phosphodiesterase, and HmsT, a putative diguanylate cyclase, control Hms-dependent biofilm formation in Yersinia pestis. Mol. Microbiol. 54 75–88. 10.1111/j.1365-2958.2004.04253.x [DOI] [PubMed] [Google Scholar]
- Lathem W. W., Schroeder J. A., Bellows L. E., Ritzert J. T., Koo J. T., Price P. A., et al. (2014). Posttranscriptional regulation of the Yersinia pestis cyclic AMP receptor protein Crp and impact on virulence. MBio 5 e01038-13 10.1128/mBio.01038-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Wang H., Qiu J., Wang X., Guo Z., Qiu Y., et al. (2009). Transcriptional profiling of a mice plague model: insights into interaction between Yersinia pestis and its host. J. Basic Microbiol. 49 92–99. 10.1002/jobm.200800027 [DOI] [PubMed] [Google Scholar]
- Liu L., Fang N., Sun Y., Yang H., Zhang Y., Han Y., et al. (2014). Transcriptional regulation of the waaAE-coaD operon by PhoP and RcsAB in Yersinia pestis biovar Microtus. Protein Cell 5 940–944. 10.1007/s13238-014-0110-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodge J., Fear J., Busby S., Gunasekaran P., Kamini N. R. (1992). Broad host range plasmids carrying the Escherichia coli lactose and galactose operons. FEMS Microbiol. Lett. 74 271–276. 10.1111/j.1574-6968.1992.tb05378.x [DOI] [PubMed] [Google Scholar]
- Lu X. H., An S. Q., Tang D. J., McCarthy Y., Tang J. L., Dow J. M., et al. (2012). RsmA regulates biofilm formation in Xanthomonas campestris through a regulatory network involving cyclic di-GMP and the Clp transcription factor. PLoS ONE 7:e52646 10.1371/journal.pone.0052646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh M. H., Lee S. M., Lee D. H., Choi S. H. (2009). Regulation of the Vibrio vulnificus hupA gene by temperature alteration and cyclic AMP receptor protein and evaluation of its role in virulence. Infect. Immun. 77 1208–1215. 10.1128/IAI.01006-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyston P. C., Dorrell N., Williams K., Li S. R., Green M., Titball R. W., et al. (2000). The response regulator PhoP is important for survival under conditions of macrophage-induced stress and virulence in Yersinia pestis. Infect. Immun. 68 3419–3425. 10.1128/IAI.68.6.3419-3425.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry R. D., Fetherston J. D. (1997). Yersinia pestis–etiologic agent of plague. Clin. Microbiol. Rev. 10 35–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen S., Young G. M. (2002). Essential role for cyclic AMP and its receptor protein in Yersinia enterocolitica virulence. Infect. Immun. 70 3665–3672. 10.1128/IAI.70.7.3665-3672.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior J. L., Hitchen P. G., Williamson D. E., Reason A. J., Morris H. R., Dell A., et al. (2001). Characterization of the lipopolysaccharide of Yersinia pestis. Microb. Pathog. 30 49–57. 10.1006/mpat.2000.0411 [DOI] [PubMed] [Google Scholar]
- Rebeil R., Jarrett C. O., Driver J. D., Ernst R. K., Oyston P. C., Hinnebusch B. J. (2013). Induction of the Yersinia pestis PhoP-PhoQ regulatory system in the flea and its role in producing a transmissible infection. J. Bacteriol. 195 1920–1930. 10.1128/JB.02000-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickman L., Scott C., Hunt D. M., Hutchinson T., Menendez M. C., Whalan R., et al. (2005). A member of the cAMP receptor protein family of transcription regulators in Mycobacterium tuberculosis is required for virulence in mice and controls transcription of the rpfA gene coding for a resuscitation promoting factor. Mol. Microbiol. 56 1274–1286. 10.1111/j.1365-2958.2005.04609.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar J. K., Wu Z., Yang W., Freitag N. E., Tortorello M. L., Wang H., et al. (2013). Roles of a novel Crp/Fnr family transcription factor Lmo0753 in soil survival, biofilm production and surface attachment to fresh produce of Listeria monocytogenes. PLoS ONE 8:e75736 10.1371/journal.pone.0075736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skorupski K., Taylor R. K. (1997). Cyclic AMP and its receptor protein negatively regulate the coordinate expression of cholera toxin and toxin-coregulated pilus in Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 94 265–270. 10.1073/pnas.94.1.265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F., Gao H., Zhang Y., Wang L., Fang N., Tan Y., et al. (2012). Fur is a repressor of biofilm formation in Yersinia pestis. PLoS ONE 7:e52392 10.1371/journal.pone.0052392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F., Zhang Y., Qiu Y., Yang H., Yang W., Yin Z., et al. (2014). H-NS is a repressor of major virulence gene loci in Vibrio parahaemolyticus. Front. Microbiol. 5:675 10.3389/fmicb.2014.00675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. C., Koumoutsi A., Jarrett C., Lawrence K., Gherardini F. C., Darby C., et al. (2011). Differential control of Yersinia pestis biofilm formation in vitro and in the flea vector by two c-di-GMP diguanylate cyclases. PLoS ONE 6:e19267 10.1371/journal.pone.0019267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L., Darby C. (2005). Yersinia pestis is viable with endotoxin composed of only lipid A. J. Bacteriol. 187 6599–6600. 10.1128/JB.187.18.6599-6600.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Escobar A., Juarez-Rodriguez M. D., Lamont R. J., Demuth D. R. (2013). Transcriptional regulation of Aggregatibacter actinomycetemcomitans lsrACDBFG and lsrRK operons and their role in biofilm formation. J. Bacteriol. 195 56–65. 10.1128/JB.01476-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadyvaloo V., Viall A. K., Jarrett C. O., Hinz A. K., Sturdevant D. E., Joseph Hinnebusch B. (2015). Role of the PhoP-PhoQ gene regulatory system in adaptation of Yersinia pestis to environmental stress in the flea digestive tract. Microbiology 161 1198–1210. 10.1099/mic.0.000082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willias S. P., Chauhan S., Lo C. C., Chain P. S., Motin V. L. (2015). CRP-mediated carbon catabolite regulation of Yersinia pestis biofilm formation is enhanced by the carbon storage regulator protein, CsrA. PLoS ONE 10:e0135481 10.1371/journal.pone.0135481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan L., Han Y., Yang L., Geng J., Li Y., Gao H., et al. (2008). The cyclic AMP receptor protein, CRP, is required for both virulence and expression of the minimal CRP regulon in Yersinia pestis biovar Microtus. Infect. Immun. 76 5028–5037. 10.1128/IAI.00370-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan L., Yang L., Zhou L., Li Y., Gao H., Guo Z., et al. (2009). Direct and negative regulation of the sycO-ypkA-ypoJ operon by cyclic AMP receptor protein (CRP) in Yersinia pestis. BMC Microbiol. 9:178 10.1186/1471-2180-9-178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Sun F., Yang H., Liu L., Ni B., Huang X., et al. (2015). CRP acts as a transcriptional repressor of the YPO1635-phoPQ-YPO1632 operon in Yersinia pestis. Curr. Microbiol. 70 398–403. 10.1007/s00284-014-0736-z [DOI] [PubMed] [Google Scholar]
- Zhang Y., Wang L., Han Y., Yan Y., Tan Y., Zhou L., et al. (2013a). Autoregulation of PhoP/PhoQ and positive regulation of the cyclic AMP receptor protein-cyclic AMP complex by PhoP in Yersinia pestis. J. Bacteriol. 195 1022–1030. 10.1128/JB.01530-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wang L., Fang N., Qu S., Tan Y., Guo Z., et al. (2013b). Reciprocal regulation of pH 6 antigen gene loci by PhoP and RovA in Yersinia pestis biovar Microtus. Future Microbiol. 8 271–280. 10.2217/fmb.12.146 [DOI] [PubMed] [Google Scholar]
- Zhou D., Tong Z., Song Y., Han Y., Pei D., Pang X., et al. (2004). Genetics of metabolic variations between Yersinia pestis biovars and the proposal of a new biovar, Microtus. J. Bacteriol. 186 5147–5152. 10.1128/JB.186.15.5147-5152.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Yang R. (2011). Formation and regulation of Yersinia biofilms. Protein Cell 2 173–179. 10.1007/s13238-011-1024-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
CRP had no regulatory effect on hmsH at the transcriptional level. See Figure 2 for the annotations of primer extension (a), quantitative RT-PCR (b), LacZ fusion (c), and EMSA (d).
CRP had no regulatory effect on hmsT at the transcriptional level. See Figure 2 for the annotations of primer extension (a), quantitative RT-PCR (b), LacZ fusion (c), and EMSA (d).
CRP had no regulatory effect on hmsC at the transcriptional level. See Figure 2 for the annotations of primer extension (a), quantitative RT-PCR (b), LacZ fusion (c), and EMSA (d).
CRP had no regulatory effect on hmsP at the transcriptional level. See Figure 2 for the annotations of primer extension (a), quantitative RT-PCR (b), LacZ fusion (c), and EMSA (d).