Abstract
The p38 MAP kinases play a critical role in regulating stress-activated pathways, and serve as molecular targets for controlling inflammatory diseases. Computer-aided efforts for developing p38 inhibitors have been hampered by the necessity to include the enzyme conformational flexibility in ligand docking simulations. A useful strategy in such complicated cases is to perform ensemble-docking provided that a representative set of conformers is available for the target protein either from computations or experiments. We explore here the abilities of two computational approaches, molecular dynamics (MD) simulations and anisotropic network model (ANM) normal mode analysis, for generating potential ligand-bound conformers starting from the apo state of p38, and benchmark them against the space of conformers (or the reference modes of structural changes) inferred from principal component analysis of 134 experimentally resolved p38 kinase structures. ANM-generated conformations are found to provide a significantly better coverage of the inhibitor-bound conformational space observed experimentally, compared to MD simulations performed in explicit water, suggesting that ANM-based sampling of conformations can be advantageously employed as input structural models in docking simulations.
1. Introduction
The p38 mitogen-activated protein (MAP) kinase, referred to as p38, is a key signaling protein activated in response to external stress; it regulates the production of proinflammatory cytokines, and as such serves as an important target for the treatment of inflammatory diseases (1). The structure of p38 in the presence of a variety of inhibitors/ligands has been resolved. However, the intrinsic flexibility of the enzyme has been a major challenge in accurate design and docking of potent inhibitors, and the necessity to gain a better understanding of the conformational variability of p38 has been pointed out (2). Our recent analysis of a set of p38 X-ray structures suggests that the structural changes observed in different ligand-bound forms of the enzyme correlate with its conformational motions intrinsically accessible in the ligand-free form (3). Effective generation of a representative set of conformers that would be utilized for flexible docking appears therefore as a feasible task. The development of such efficient tools for generating representative subsets of potentially bound conformers would greatly facilitate computational efforts for drug discovery, not only for this particular family, but for many target proteins, especially in the absence of sufficient structural data on their alternative conformers (4).
There is a multitude of approaches at different resolutions for generating conformational ensembles. Molecular dynamics (MD) simulations are broadly used in investigating specific interactions and structural changes at atomic scale, but they may be prohibitively time consuming if large-scale changes are explored (5). Elastic network models (ENMs), on the other hand, efficiently explore large-scale changes for large systems, but this comes at the cost of losing atomic precision, and the observed changes are restricted to the neighborhood of the global energy minimum (6, 7). With the ever growing size of Protein Data Bank (PDB) (8), we are able to assess the utility of these methods by benchmarking them against the structural changes detected for well-studied proteins in the presence of different inhibitors.
We use here an ensemble of 134 X-ray structures resolved for p38 in different forms as the reference for the conformational space accessible to p38 upon binding its ligands. p38 has two small-molecule binding sites (Fig. 1A): the ATP-binding site where competitive binding of inhibitors takes place, and the lipid/compound binding site at the MAPK insert, which offers alternative targeting strategies (9). p38 is bound to a structurally diverse set of inhibitors in this dataset. To gain a simplified view of the dataset structural variability, we identified dominant directions of structural changes (modes) via principal component analysis (PCA) (10) (Fig. 1 panels B – E). PCA is a powerful technique for extracting recurrent modes of structural changes from sets of structures (11). Its use in assessing functional dynamics is clearly demonstrated by a recent study of substrate-bound X-ray structures of ubiquitin (12). Hereafter, we will refer to the modes identified by PCA as reference modes.
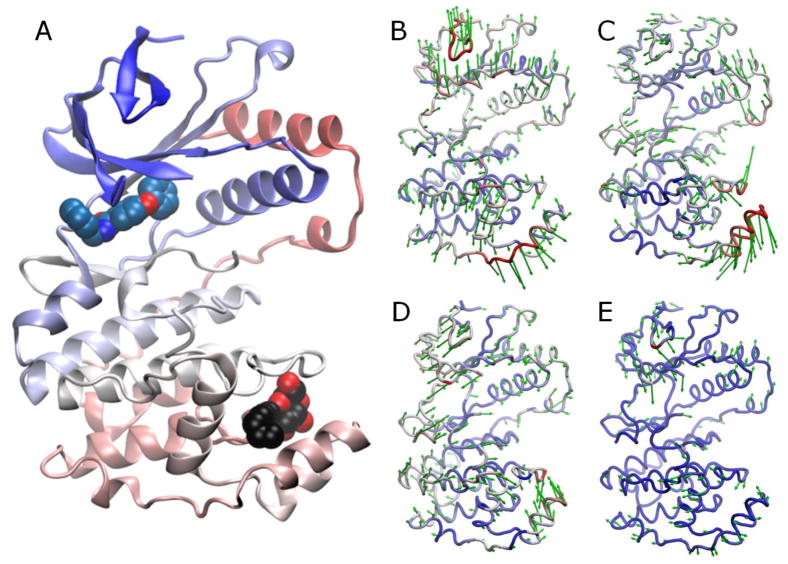
Fig. 1. p38 structure and reference modes.
A. p38 structure (PDB id: 1ZYJ) is shown as a ribbon diagram colored by residue index from blue to red. The upper and lower lobes are referred to as the N- and C-terminal lobes. Two ligand-binding sites are distinguished: ATP-binding site, with the bound inhibitor shown in blue spheres; and the site at the MAPK insert, marked by the bound lipid (n-octyl-β-D-glucoside) in black/red space filling representation. B–E. Directions of PCA modes 1-4 (green arrows) retrieved from the analysis of 134 X-ray crystallographically resolved p38 structures in different forms. Coloring is based on mobility along the mode directions, red being most mobile.
We generated alternative conformations by two approaches: MD simulations, subjected to essential dynamics analysis (EDA), (13) and anisotropic network model (ANM) (14–16) analysis. MD simulations were repeated in the presence of explicit water and in solutions of water and probe molecules (small organic molecules to mimic the effects of drugs/inhibitors, as recently performed to investigate target protein druggability (17, 18)). We examined (i) the coverage of reference conformational space by MD and ANM, and (ii) the correspondence of the modes observed in MD-EDA and predicted by ANM to those (Figure 1B–E) inferred from experiments.
2. Materials and Methods
2.1. Structural data
Using the human p38α sequence (GenBank id: CAG38743.1) in Biopython (http://biopython.org) protein Blast module, we retrieved from the PDB a set of 134 p38 MAP kinase isoform α structures, with 95% or more sequence identity (human or mouse proteins). Most structures contained a ligand bound to ATP binding site and/or MAPK insert (Table 1).
Table 1.
Summary of p38 structural ensemble(*).
| MAPK insert | Total | |||||
|---|---|---|---|---|---|---|
| Unbound | Lipid | Compound | ||||
| ATP site | Unbound | 4

|
10

|
14 | ||
| Inhibitor | 87

|
20

|
8

|
115 | ||
| Peptide/protein bound | 5

|
5 | ||||
| Total | 91 | 30 | 8 | 5 | 134 | |
2.2. MD simulations
We performed two types of 20 ns simulations, each repeated twice. The 1st type contained water and counter ions, in addition to p38. The 2nd was performed in a solution of water and small-organic molecules at a fixed ratio of 20:1, summarized in Table 2. The ligand-free p38 structure resolved by Wang et al.(19) (PDB id: 1P38) was used. Missing atoms were modeled using PSFGEN (20). Solvent box padding distance was at least 6 Å. Solvated system coordinates were prepared using VMD (20). Prior to the productive runs, probe-free systems were equilibrated for 30 ps. For probe-containing systems were subjected to 450 ps simulated annealing to achieve uniform spatial distributions of probes, followed by 300 ps equilibration. All simulations were performed using NAMD (21) software with CHARMM (22) force field.
Table 2.
Description of MD simulations performed for p38 in different solvent environments.
| MD Sim | Atoms(*) | Water | Isopropanol | Isopropylamine | Acetate | Acetamide |
|---|---|---|---|---|---|---|
| 1 | 26454 | 6929 | - | - | - | - |
| 2 | 26454 | 6929 | - | - | - | - |
| 3 | 28515 | 6360 | 318 | - | - | - |
| 4 | 28563 | 6400 | 224 | 32 | 32 | 32 |
protein and non-protein molecules. All simulations contained 9 sodium ions to balance the charge.
2.3. PCA/EDA of data from experiments/simulations
The PCA of ensembles of structures is an orthogonal linear transformation that projects data from Cartesian coordinate space onto a space of collective coordinates uniquely defined by the examined ensemble (10). The new coordinate system is such that the greatest variance in the dataset lies along the first principal component (PC) axis, shortly referred to as PC1, followed by PC2, PC3 and so on. The method of approach is identical in the EDA of MD trajectories (13), with the only exception that the analyzed set of conformers consists of MD snapshots, rather than the experimentally resolved structures collected from the PDB.
Both analyses are based on the cross-correlations (or covariance) observed (in experiments or simulations) between the fluctuations of Cα-atoms. Here, we used 324 Cα-atoms (residues 5-31, 36-116, 121-168,185-352) that were structurally resolved in at least 90% of the examined dataset. The approach in either case is to diagonalize the covariance matrix and examine the dominant modes of structural changes (eigenvectors) which are associated with the largest eigenvalues. Prior to PCA/EDA, structures/snapshots are superposed using the Kabsch algorithm (23) in an iterative procedure (3). Mean positions <Ri> = [<xi> <yi> <zi>]T are determined for each α-carbon i. The departures of α-carbons from their mean positions, ΔRis = [Δxis Δyis Δzis]T (where Δxis = xis –<xi>) are organized in a 3N-dimensional deformation vector ΔRs where (ΔRs)T = [(ΔR1s)T (ΔR2s)T …. (ΔRNs)T]), for each structure, s, in the dataset; and their cross-correlations, averaged over the entire set are organized in a 3N × 3N covariance matrix C. C may be written in terms of N × N submatrices C(ij) (1≤i, j ≤ N), each of size 3 × 3, given by
| (1) |
Here 〈ΔxiΔxj〉 represents the cross-correlation between the x-component of ΔRis and the y-component of ΔRjs averaged over all structures (1 ≤ s ≤ Stot) in the dataset. The trace of C(ij) gives the cross-correlations between the fluctuations of residues i and j as tr{C(ij)} = <ΔRi • ΔRj>, and that of the ith diagonal block C(ii) gives the mean-square fluctuations <(ΔRi)2> of α-carbon i.
Principal/essential modes are obtained by decomposing C for the dataset of conformers (PDB/MD) as where p(i) and σi, are the ith eigenvector and eigenvalue of C, respectively, and m is the total number of nonzero eigenvalues (m = 3N-6 if Stot > 3N-6, and m = Stot otherwise). σ1 corresponds to the largest variance component (i.e. σ1 ≥ σ2 ≥ … ≥ σm). The fractional contribution of fluctuations along p(i) to the overall structural variance in the dataset is given by fi = σi/Σjσj where the summation is performed over all m components.
2.4. ANM analysis and sampling of conformers using ANM modes
In contrast to PCA and EDA, the ANM analysis is performed for a single structure (e.g., the apo structure), not an ensemble. In the ANM, the second-order partial derivatives of the potential energy function (a sum over uniform pairwise harmonic potentials of force constant γ between all ‘connected’ residues in the network) are organized in the Hessian matrix H, which, in turn, is decomposed into 3N-6 nonzero eigenvalues λi and corresponding eigenvectors ui, i.e., (14, 16). H is written in terms of N × N submatrices each of size 3 × 3. The ijth submatrix is given by
| (2) |
and . Here is the equilibrium distance between the α-carbons i and j, and Xij, Yij and Zij are its components; Γij is the ijth element of the Kirchhoff matrix Γ, equal to 1 if i and j are connected (or within a distance rcut), zero otherwise. The ANM covariance matrix is CANM = H−1 such that 1/λ1 is the counterpart of the PCA σ1, and u(i) is the counterpart of p(i).
ANM conformations along mode i are generated using the relation , where s is a scaling parameter proportional to (kBT/γ)½ (24). Thus, the structural changes along the slowest/softest mode (u1) are the largest in size (λ1 ≤ λ2 ≤ … ≤ λ3N-6). We generated ensembles around the initial conformation R0, using the pseudocode given in Textbox 1, with M = 3 modes and s = K(kBT/γ)½ where K varies as 1 ≤ K ≤ 7 and kBT/γ = 2Å2, to obtain 153 = 3375 conformers, the spread of which matches that of the reference space. The root-mean-square deviations (RMSDs) between nearest conformers along modes 1, 2 and 3, were 0.25, 0.21, and 0.14 Å, approximately. Calculations were repeated with different structures to confirm the robustness of the predicted ANM modes. Results obtained with unliganded (PDB id: 1P38 (19)) and inhibitor-bound p38 (PDB id: 2BAJ (25)) yielded practically indistinguishable results.
Textbox 1. Pseudocode for ANM sampling.
| Initialize a list to store conformations, and append the initial conformation: |
| L = [R0] |
| Do for ANM modes 1 ≤ i ≤ M |
| Do for each conformation RC in L |
| Initialize a list to store conformations generated at this step, L_temp = [ ] |
| Do for k in [−K, −(K−1), …, −1, 1, …, K−1, K] |
| Append Rk to L_temp |
| Append conformations in L_temp to L |
2.6. Comparison of dominant modes from PCA, EDA and ANM
In this section, we define the metrics for comparing the modes from ANM (predicted) and PCA (experiments); similar expressions hold for the comparison of EDA (simulations) and PCA modes, as well as EDA and ANM modes. The overlap between ANM and PCA modes is given by the correlation cosine Oij = p(i) · u(j) (26). The cumulative overlap, measures how well a subset of J low frequency ANM modes reproduces the PCA mode i(27). Note that for J = 3N -6, by definition, i.e., the complete set of 3N-6 ANM eigenvectors form an orthonormal basis set. Finally, the essential subspace overlap between the PCA and ANM subspaces spanned by top K modes is evaluated using (13). Finally, the degrees of collectivity of the principal modes derived from either computations (ANM or EDA) or experiments (PCA) were calculated using the definition proposed by Brüschweiler (28).
2.7. Projection of conformations onto a reference subspace and normality test
The projection of a given conformational change ΔRs onto p(i) is found from . The points in Figs 2 and 3 represent the projections onto the subspaces spanned by PC1, PC2, PC3, and/or PC4. In the extreme case of (ΔRs)T perfectly aligned along p(i), , where the double bars designate the magnitude of the enclosed vector. The normality of projections of PDB structures onto the principal modes were tested using A’Agostino and Pearson’s test (29, 30) where skewness and kurtosis are combined into an omnibus test (using SciPy, http://scipy.org/).
Fig. 2. Distribution of the PDB ensemble of structures on the subspaces spanned by reference modes.
134 p38 structures are projected onto PC1-PC2 (A) and PC3-PC4 (B) subspaces. Markers are described in Table 1. The distributions of structures along the individual modes are shown by the histograms.. A conformation on the positive portion of these projections corresponds to a deformation along the direction indicated by the arrows in Fig. 1B–E.
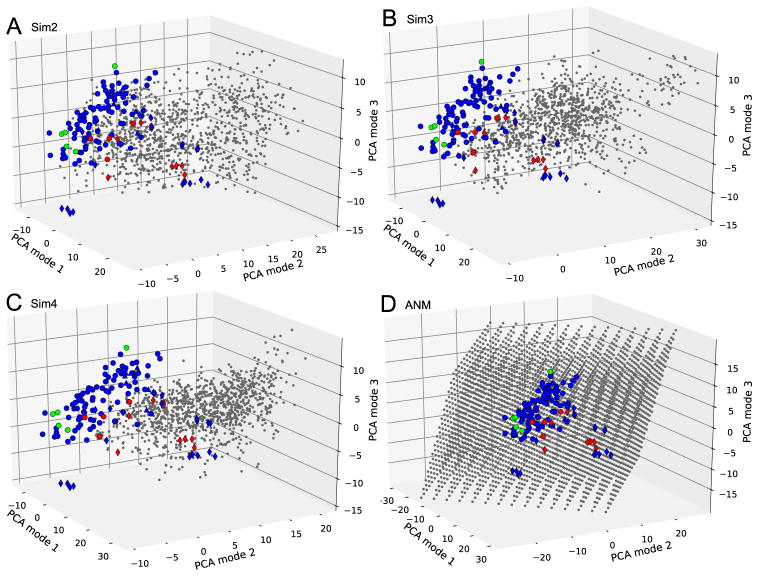
Fig. 3. Projections of MD and ANM ensembles onto the subspace spanned by the reference modes PC1-PC3.
Ensembles from Sim2, Sim3, Sim4, and ANM are shown in panels A, B, C, and D, respectively. PDB structures are marked as in Fig. 2. Conformations generated by computations are shown by gray points. The perspective is the same in all panels for ease of comparison.
3. Results and Discussion
3.1. p38 reference modes derived from X-ray crystallographic data
The ensemble of 134 p38 structures provides a rich representation of the conformational space accessible to this enzyme under a wide variety of conditions, e.g., differences in ligands, crystal conditions, or mutations. The dominant changes observed in this dataset were extracted by PCA as described above, and displayed in Fig. 1B–E. Table 3 (columns 1–5) provides more information on these modes, including the size of the associated conformational variance, σ2, their contribution to structural variation, and their degree of collectivity. The first mode, PC1 (Fig. 1B) for example, accounts for 24.5% of the structural variability in the dataset (Table 3). The fluctuations along this mode correspond to the anti-correlated movements of the N- and C-terminal lobes of p38. Motions along the PC1 axis in the positive direction (indicated by the arrows in Fig. 1B) favor ‘open’ conformers, and those in the negative direction favor ‘closed’ forms. Movements along PC1 thus directly affect the size of the ATP/inhibitor-binding pocket in the N-terminal lobe.
Table 3.
Properties of the reference (PCA) modes from experiments, and projection of MD snapshots onto them.
| PCA Mode | PCA of PDB ensemble | Sim1 | Sim2 | Sim3 | Sim4 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| σ2(a) | %(b) | p-value(c) | Collectivity(d) | μ(e) | σ2(f) | μ | σ2 | μ | σ2 | μ | μ | |
| 1 | 44.5 | 24.5 | 0.26 | 0.49 | 6.9 | 40.1 | 6.4 | 74.8 | 9.9 | 59.7 | 15.4 | 43.6 |
| 2 | 37.6 | 20.7 | 0.00 | 0.36 | 6.3 | 30.6 | 9.9 | 46.9 | 11.0 | 43.9 | 8.3 | 31.1 |
| 3 | 25.6 | 14.1 | 0.00 | 0.58 | −8.2 | 25.0 | −0.6 | 20.5 | 0.5 | 20.2 | 0.9 | 14.1 |
| 4 | 11.1 | 6.1 | 0.63 | 0.52 | 9.4 | 20.8 | 3.2 | 27.1 | 0.8 | 19.9 | 2.7 | 12.9 |
Variance along the reference mode in the PDB dataset.
Percent of total structural heterogeneity accounted for by the reference mode.
The probability that the projection of structures along the reference mode obeys a normal distribution.
Degree of collectivity.
Mean position of MD snapshots along the reference mode.
Variance of MD snapshots along the reference mode.
Figure 2 displays the projections of the 134 structures, each indicated by a color/shape coded symbol (described in Table 1), on the subspace spanned by PC1 and PC2. The unliganded structures (red dots in Fig. 2A) occupy the region PC1 > 0 of the subspace, consistent with their tendency to assume a relatively open form. Upon inhibitor binding, p38 tends to close down. Normality test of the projection onto PC1 (upper bars plot in Fig. 2A) shows that an approximately Gaussian distribution is obtained. The ensemble is not separated into distinct clusters in this case, suggesting that a continuous spectrum of conformers is visited rather than two distinctive ‘open’ and ‘closed’ states, with the unbound structures exhibiting a tendency to be open.
The second reference mode (PC2), on the other hand, describes the structural changes in the secondary lipid/compound binding pocket at the MAPK insert. This mode explains 20.7% of the structural variability in the dataset. As shown in Fig. 2A, this mode divides the ensemble into two groups: (i) structures with a bound ligand (lipid) molecule at the MAPK insert (red and blue diamonds, mostly clustered in the positive PC2 region), and (ii) structures with empty MAPK inserts (red and blue circles). Normality test confirms that the PDB structures exhibit a bimodal distribution along this mode. Compared to PC1, changes are slightly more localized and pronounced near the lipid-binding site at the C-terminal lobe. The collectivity of this mode is lower (0.36) compared to that of the first mode (0.49).
The structural changes along the 3rd and 4th reference modes (Fig. 1D and E) account for the respective 14.1% and 6.1% of the total variance. Both of these modes are highly collective (Table 2) as may also be seen from the uniform distribution of movements across the enzyme. Lipid-bound structures (diamonds) tend to move toward the negative direction along PC3. This behavior is particularly distinctive in inhibitor-bound structures, which results in a skewed, non-Gaussian distribution. The movements along PC4, on the other hand, exhibit a normal distribution.
In summary, the first four modes provide a description of structural changes associated with binding of ligands (ATP and/or inhibitors) at the ATP-binding sites (PC1), binding of lipids to the MAPK insert (PC2), and the collective rearrangements of the entire enzyme to accommodate different bound forms (PC3 and PC4). Notably, local changes at the binding sites are coupled to global changes in the enzyme structure (Fig. 1 panels C and D), pointing to the functional significance of the global modes favored by the p38 architecture.
3.2. Do MD snapshots provide good coverage of reference space?
Of interest is to assess how close MD- or ANM-generated conformations are to known PDB structures. In Fig. 3, we show the projection of computationally generated conformations onto the subspace spanned by top three reference modes (PC1-PC3). Panels A–C compare the snapshots from three MD runs (see Table 2), shown by the black dots, to PDB structures (indicated by the symbols in Table 1). In all three cases, we see that the conformations sampled during MD runs drift away from the large majority of the experimentally detected structures. This is also evident from the mean positions of MD snapshots along these three principal axes reported in Table 3. For example, along PC1, the mean position sampled by MD snapshots varies between 6.4 (Sim2) and 15.4 Å (Sim4), which correspond to 0.36 and 0.85 Å RMSD. Likewise, along PC2, the average positions of MD snapshots depart from the experimental dataset by up to 11.0 Å (Sim3). Overall, four independent runs starting from a ‘central’ experimental structure ended up sampling conformational subspaces that do not encompass the majority of experimental structures.
3.3. Do ANM predictions provide good coverage of reference space?
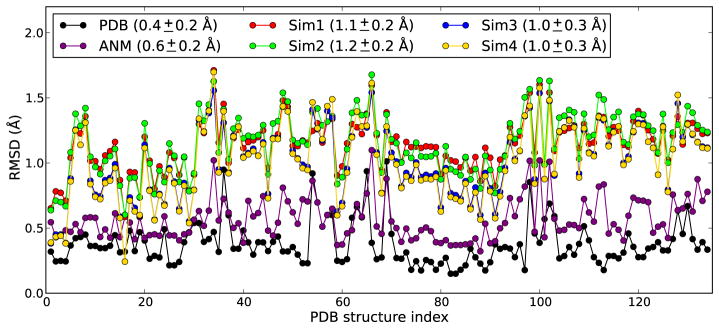
In sharp contrast to results from MD runs, Panel D in Figure 3 shows that conformers generated by deforming the starting structure along ANM modes 1, 2 and 3 are able to cover the reference subspace of conformations comprehensively. In this case ANM sampling is performed using only the slowest three modes. The present comparison clearly shows that the subspace sampled by the three softest ANM modes overlaps with the experimentally accessed subspace of conformations. This remarkable coverage of reference space (from experiments) by ANM predictions also translates into the minimum RMSD plot shown in Fig. 4. In this plot, for each PDB structure, the lowest RMSDs with respect to (i) all other PDB structures (black curve with black dots), (ii) MD snapshots in four different runs (colored as labeled), and (iii) ANM conformations generated along the softest three modes (purple) are shown. The plot for pairs of PDB structures yields an average value of 0.4 Å; ANM sampling yields 0.6 Å. MD runs, on the other hand, yield an average of 1.0Å at least. Simulations performed in the presence of probe molecules (Sim3 and Sim4) yield slightly better results, although the improvement is not significant.
Fig. 4. Minimum RMSD from PDB structures.
Results are shown for all PDB structures (indexed in alphabetical order along the abscissa). The black curve refers to the RMSDs from the PDB structures themselves (experimental data); the purple curve displays the min RMSDs achieved by ANM sampling; and other curves (labeled) refer to MD runs Mean and standard deviations are given in the legend.
3.4. Correspondence of ANM and MD-PCA Modes to Reference Modes
We performed the EDA of the four MD trajectories, and compared the essential modes derived from these runs to the reference modes obtained from the experimental dataset. Fig. 5 panels A and C show for the runs Sim2, Sim3 and Sim4 the overlap (correlation cosine) between each of the essential 10 modes (from EDA of MD) and the top 10-ranking reference modes (from PCA of PDB structures). Panel D displays the correlation between the ANM and PCA modes. If we focus in particular on the top two pairs of modes, the lowest overlaps are observed in Sim2 (53% or less) and similar observations (not shown) were made for Sim1. In both of these two cases, the protein was simulated in water. In Sim3 and Sim4, the correlations with the first two reference modes increase to 63%. This is due to the existence of probe molecules which were able to find either binding pocket on p38, and assisted in the stabilization of bound conformers.
Fig. 5. Correlations between experimentally observed and computationally obtained modes of structural changes.
Panels A–C show the overlap between top-ranking PCA modes (from 134 PDB structures) and the modes yielded by MD runs EDA. Panel D displays the overlaps between ANM and PCA modes.
The comparison of ANM modes with the reference modes shows, on the other hand, a much stronger correlation. The slowest mode (ANM1) alone exhibits an overlap of 75% with PC1. Thus, the most prominent conformational variation experimentally observed for p38 upon binding its ligands is in remarkable agreement with the softest mode of motion intrinsically accessible to the enzyme in the unbound state. That is, it is easiest to deform the protein along this ANM mode, or the protein is most likely to sample alternative conformations along this mode under native state conditions (presumably within time scales of the order of μs). In addition, the cumulative overlap between top reference mode and first three ANM modes reaches 87%. We also found significant overlaps between reference modes PC2, PC3, and PC4 and the low frequency ANM modes 1, 2 and 3: PC2 overlaps with ANM1 and ANM3 by 0.49 and 0.55, respectively, yielding a cumulative overlap of 0.74 with these two modes. PC3 correlates with ANM2 by 57%, and PC4 correlates with ANM2 by 52%. It is not thus surprising to see that the alternative conformations generated along ANM1-3 provided a satisfactory coverage of the experimentally observed dataset (Fig. 3D).
Apparently, 65% of the variability in the PDB ensemble is well explained by slowest three ANM modes. A measure of such correspondence is the essential subspace overlap (13). Using three modes, we find that the essential subspace overlap between ANM and reference dataset is 75%. This value is 62% for MD runs Sim1 and Sim2, and increases to 67 and 68%, respectively for Sim3 and Sim4. This suggests that the directions of structural changes observed in MD, represented by EDA modes 1-3, exhibit some correlations with the PCA modes 1-3 extracted from experiments, but there is a drift away from the original subspace during MD runs such that the conformations sampled by MD deviate from the reference state, leading to the relatively high RMSD values shown in Figure 4.
4. Conclusion
We presented a detailed analysis of the structural variations in a large ensemble of p38 MAP kinase X-ray structures, compared to those predicted by snapshots/models generated by MD simulations and by ANM methodology. Our results show that ANM is able to capture the modes of motion that are relevant to ligand/inhibitor/lipid binding much better than MD simulations. The use of probe molecules that mimic the interactions of the protein with inhibitor molecules improves the ability of MD to sample the relevant modes (Fig. 5B–C). Yet, the conformers sampled by MD trajectories of tens of nanoseconds generally fell short of encompassing the space of inhibitor-bound PDB conformations (Fig. 4). MD simulations of 20 ns take about 2 weeks using 12 processors for a typical kinase. Generating a broader MD ensemble would take longer and would demand considerably larger numbers of processors, and this might not prevent the drift away from the experimental structures. The generation of conformers along ANM soft modes, on the other hand, is achieved within minutes, if not seconds, and provides an accurate sampling of experimentally detected subspace. Notably, the latter does not necessitate expensive computations, nor knowledge of multiple structures.
Our work is the most comprehensive comparative analysis of a protein kinase (p38) dynamics using multi-resolution methods. In a previous study, based on small sets of PDB structures for four different kinases, local changes in the glycine rich loop (a β-hairpin that interacts with inhibitors) of the N-terminal lobe were observed (2), and the fast modes were deemed to be used to capture the conformational variability in ligand-bound structures (31). Our current and previous (3) results show that up to 65% of the changes experimentally observed in p38 MAP kinases are actually collective changes explained by 2–3 softest modes. The local motions at ligand-binding site are an integral part of these global movements, and the structural variations observed in different ligand-bound conformers are well represented by the structural rearrangements along the global modes. These observations are in line with work of May and Zacharias (32) in which relaxing a protein kinase along global modes during docking simulations improved the prediction of bound conformers. Their approach circumvents the problem of dealing with conformations potentially irrelevant to ligand binding (decoys). Such decoys are generated by both computational methods, but eliminating ANM conformations with RMSD larger than a threshold to the initial structure can improve the accuracy of sampling. Furthermore, current results are important as they suggest a key coupling between global motions and local binding events, which will need to be systematically examined for a series of proteins. The method and application set forth in the present study may be readily extended to perform such a critical assessment for a large set of proteins.
Acknowledgments
Support from NIH grants 5R01GM086238-02 and 5R01LM007994-06 is gratefully acknowledged by I. Bahar.
Contributor Information
AHMET BAKAN, Email: ahb12@pitt.edu.
IVET BAHAR, Email: bahar@pitt.edu.
References
- 1.Kumar S, Boehm J, Lee JC. Nat Rev Drug Discov. 2003;2:717. doi: 10.1038/nrd1177. [DOI] [PubMed] [Google Scholar]
- 2.Cavasotto CN, Abagyan RA. J Mol Biol. 2004;337:209. doi: 10.1016/j.jmb.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 3.Bakan A, Bahar I. Proc Natl Acad Sci U S A. 2009;106:14349. doi: 10.1073/pnas.0904214106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Totrov M, Abagyan R. Curr Opin Struct Biol. 2008;18:178. doi: 10.1016/j.sbi.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dror RO, Jensen MO, Borhani DW, Shaw DE. J Gen Physiol. 2010;135:555. doi: 10.1085/jgp.200910373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bahar I, Lezon TR, Bakan A, Shrivastava IH. Chem Rev. 2009 doi: 10.1021/cr900095e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bahar I. J Gen Physiol. 2010;135:563. doi: 10.1085/jgp.200910368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berman HM, et al. Nucleic Acids Res. 2000;28:235. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perry JJ, Harris RM, Moiani D, Olson AJ, Tainer JA. J Mol Biol. 2009;391:1. doi: 10.1016/j.jmb.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jolliffe IT. Principal Component Analysis. 2. Springer; New York: 2002. p. 487. [Google Scholar]
- 11.Amadei A, Linssen AB, Berendsen HJ. Proteins. 1993;17:412. doi: 10.1002/prot.340170408. [DOI] [PubMed] [Google Scholar]
- 12.Lange OF, et al. Science. 2008;320:1471. doi: 10.1126/science.1157092. [DOI] [PubMed] [Google Scholar]
- 13.Amadei A, Ceruso MA, Di NA. Proteins. 1999;36:419. [PubMed] [Google Scholar]
- 14.Atilgan AR, et al. Biophys J. 2001;80:505. doi: 10.1016/S0006-3495(01)76033-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doruker P, Atilgan AR, Bahar I. Proteins. 2000;40:512. [PubMed] [Google Scholar]
- 16.Eyal E, Yang LW, Bahar I. Bioinformatics. 2006;22:2619. doi: 10.1093/bioinformatics/btl448. [DOI] [PubMed] [Google Scholar]
- 17.Seco J, Luque FJ, Barril X. J Med Chem. 2009;52:2363. doi: 10.1021/jm801385d. [DOI] [PubMed] [Google Scholar]
- 18.Guvench O, Mackerell AD., Jr PLoS Comput Biol. 2009;5:e1000435. doi: 10.1371/journal.pcbi.1000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z, et al. Proc Natl Acad Sci U S A. 1997;94:2327. doi: 10.1073/pnas.94.6.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Humphrey W, Dalke A, Schulten K. J Mol Graph. 1996;14:33. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 21.Phillips JC, et al. J Comput Chem. 2005;26:1781. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brooks BR, et al. J Comput Chem. 2009;30:1545. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kabsch W. Acta Crystallographica Section A. 1976;32:922. [Google Scholar]
- 24.Xu C, Tobi D, Bahar I. J Mol Biol. 2003;333:153. doi: 10.1016/j.jmb.2003.08.027. [DOI] [PubMed] [Google Scholar]
- 25.Sullivan JE, et al. Biochemistry. 2005;44:16475. doi: 10.1021/bi051714v. [DOI] [PubMed] [Google Scholar]
- 26.Tama F, Sanejouand YH. Protein Eng. 2001;14:1. doi: 10.1093/protein/14.1.1. [DOI] [PubMed] [Google Scholar]
- 27.Yang L, Song G, Carriquiry A, Jernigan RL. Structure. 2008;16:321. doi: 10.1016/j.str.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bruschweiler R. The Journal of Chemical Physics. 1995;102:3396. [Google Scholar]
- 29.D’Agostino R. Biometrika. 1971;58:341. [Google Scholar]
- 30.D’Agostino R, PEARSON ES. Biometrika. 1973;60:613. [Google Scholar]
- 31.Cavasotto CN, Kovacs JA, Abagyan RA. J Am Chem Soc. 2005;127:9632. doi: 10.1021/ja042260c. [DOI] [PubMed] [Google Scholar]
- 32.May A, Zacharias M. J Med Chem. 2008;51:3499. doi: 10.1021/jm800071v. [DOI] [PubMed] [Google Scholar]