Abstract
Mouse models have become an invaluable tool for understanding human health and disease owing to our ability to exquisitely manipulate the mouse genome. Recent progress in genomic analysis has led to an increase in the number and type of disease-causing mutations detected, and has also highlighted the importance of non-coding regions. As a result there is increasing interest in creating ‘genomically’ humanised mouse models, in which entire human genomic loci are transferred into the mouse genome. The technical challenges to achieving this aim are large but are starting to be tackled with success.
The mouse is the model of choice for recapitulating genetic changes that give rise to human disease. Over the last 30 years mouse molecular genetics has been refined to allow production of an impressive panoply of mutants. These include additive transgenic, knock-out, and knock-in animals (all of which can be conditional or inducible), strains containing chromosomal rearrangements or large megabase-sized deletions and duplications, and even transchromosomic mouse strains1. It is a fast developing area and new technologies are arising all the time, and include methods for modelling ‘sporadic’ disease such as cancer2.
Almost all human disease models have been made to study changes in the coding genome. Typically this has been done by pronuclear injection, to generate transgenics that ectopically express a mutant protein, or by gene targeting in embryonic stem (ES) cells, for example, by creating a gene knock-in. As proteins with a human amino acid sequence can have different biochemical characteristics from their mouse orthologues, transgenics have often been made with human cDNAs, and targeting has involved placing human coding sequences into the orthologous mouse gene. This genetic ‘humanising’ strategy using coding sequences can result in a more accurate mouse model of disease than working with a mutant mouse protein. However, recent progress in genomic analysis has highlighted the importance of the non-coding genome (both transcribed and non-transcribed), making it clear that this category of sequence also needs to be taken into account when modelling disease. In particular, projects such as ENCODE (the ENCylopedia Of DNA Elements3) have discovered extensive transcription of the non-coding genome and human genome-wide association studies (GWASs) demonstrate that variation (including copy number variation) in non-coding regions confers susceptibility and resistance to disease in ways that we do not comprehend. As we learn more of the complexity of the genome it is apparent that understanding human biology, particularly with respect to disease models, will require humanised mouse models that address the potential roles of both coding and non-coding genomic sequence (Box 1).
Box 1. Why humanise mice?
Genetic humanisation
Few proteins are 100% conserved between human and mouse44, and differences in orthologous sequences can have functional consequences. For example, mouse serum amyloid P (SAP) binds to amyloid fibrils with only ~3% of the avidity of the human protein although mouse and human SAP are ~70% conserved45. Similarly, mutant superoxide dismutase 1 (SOD1) is causative for the human neurodegenerative disease amyotrophic lateral sclerosis; the human and mouse proteins share 83% identity, but a tryptophan residue at codon 32 (W32) is found only in humans, where it appears to potentiate SOD1 aggregation and human specific SOD1–SOD1 interaction which may contribute to motor neuron death in humans and in mice with mutant human SOD1 transgenes46.
Similarly, wild-type mice expressing mouse CD81 and occludin (OCLN) are non-permissive to hepatitis C virus (HCV) entry47. However, animals expressing two human orthologues of these two proteins are permissive for HCV infection, while remaining fully immunocompetent. This model greatly eases studies of the immune response to HCV because previously humans and chimpanzees were the only two species known to be permissive for HCV infection47.
Humanisation also gives insight into gene evolution. FOXP2 transcription factor is important for human speech and language. When this protein was humanised in mice it produced abnormal behavioural and other phenotypes in cortico-basal ganglia circuits, suggesting that humanised FOXP2 protein may take on a new function(s) in these regions that is important for the evolution of human language and speech48. These phenotypes were not found in a Foxp2 knock-out, indicating they arose from the function of the wildtype human protein.
For a small number (<200) of human protein-coding genes no mouse orthologue has been found49, and thus one approach to learn more about the biology of these human genes is to introduce them into mice.
Genomic humanisation
Although genetic humanisation has given us great biological insight, genomic humanisation will be necessary to investigate the functional importance of non-coding regions and therefore to fully model aneuploidy, to study disorders in which species-specific splicing patterns play a role, or to determine the functions of untranslated sequences. For example, the different effects of disrupting Hotair orthologues in human and mouse, indicate that this long non-coding RNA has human function(s) that may not easily be determined from non-genomically-humanised mice50,51.
Likewise, genomically humanised mice containing a caspase 12 (CASP12) variant responded in a gender-specific manner when infected with Listeria monocytogenes52, leading to the identification of an oestrogen receptor element (ERE) in intron 7, which appears to be responsible for oestrogen-modulated expression of the CASP12 variant being studied. Treatment of the male humanised CASP12 mice with 17-β-oestradiol (E2) conferred increased resistance to infection, leading to suggestion of the therapeutic use of E2. The oestrogen-response element is not found in mouse intron 7 and wild-type male mice do not respond to oestrogen at the Casp12 locus52. Without the use of genomically humanised mice, this ERE and potential therapy would not have been discovered.
Thus the non-coding genome must be taken into account in studying gene function and genomic humanisation will be essential to create an optimal set of models of human disease.
Laboratories world-wide are developing the technology for creating such ‘genomically’ humanised mice, which are generated by transferring entire human genomic loci (including coding and non-coding regions) into the mouse genome. This is achieved either by the addition of human genomic sequences or by replacing regions of the mouse genome with equivalent human genomic sequence. However, the technical challenges remain daunting and while current approaches to optimise different strategies are proving successful, for example generation of mice carrying a whole human chromosome, they are far from routine.
Here we look at the different approaches that have been developed for creating genomically humanised mice, why genomic humanisation remains a challenge, and which new technologies are the most promising. We also speculate on the direction of future advances in this field.
YAC and BAC transgenics
The first technologies for producing genomically humanised mice made use of yeast artificial chromosomes (YACs) and bacterial artificial chromosomes (BACs). Transgenic lines can be created by pronuclear injection of YAC and BAC DNAs, with these DNAs being integrated at random chromosomal positions (Figure 1Aa). The key advantage of BACs and YACs is their size, ranging up to 300 kilobases (kb) for BACs and up to Megabases (Mb) for YACs, thus enabling inclusion of all or some of the upstream and downstream cis-sequences regulating expression of a gene of interest. YAC and BAC transgenic insertions show position-independent and copy-number-dependent expression more frequently than smaller transgenes do, and they more faithfully recapitulate the anticipated expression profile4, 5. Compared to conventional cDNA transgenics, BAC and YAC transgenics also have the added advantage of containing low-copy-number integrations, which is important when studying the effect of gene dosage.
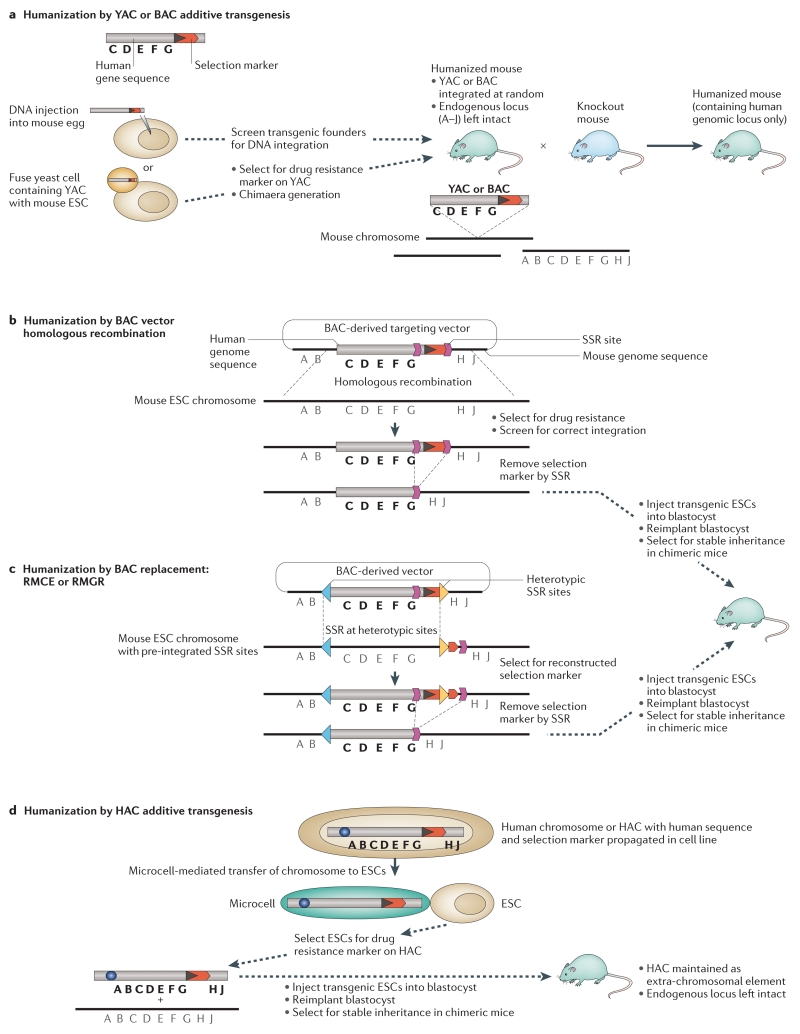
Figure 1. Methods of humanised mouse synthesis.
There are multiple ways of introducing the human genomic region of interest into the mouse germline. (a) Traditionally this has been via an additive process, where a YAC or BAC vector is introduced via pronuclear injection or cell fusion, resulting in random incorporation into mouse genome, while the endogenous mouse locus is unmodified. (b, c) An alternative is the specific targeting and replacement of genomic loci, either using (b) homologous recombination or (c) the SSR-based technologies RMCE and RMGR. Homologous recombination with a genomic fusion (mouse-human) BAC vector results in the endogenous mouse locus being replaced by equivalent human sequence, using the large regions of homology provided by the BAC vector for increased targeting efficiency (b). RMCE and RMGR require prior modification of the mouse genome, to introduce heterotypic SSR sites to flank the region of interest. A BAC vector containing equivalent human genomic region flanked by same SSR sites then acts as a donor for the swap of genetic material mediated by expression of recombinase (c). (d) A non-integrative approach to creating a humanised mouse is by the introduction of a HAC into ES cells via MMCT. The HAC vector is synthesised via a top-down or bottom-up approach, where the genomic region of interest is introduced by homologous recombination or SSR. The HAC is mitotically stable and maintained as an extra-chromosomal element, leaving the mouse genome unmodified.
The extent of humanisation achievable with YACs can be increased by exploiting homologous recombination in yeast to join two existing YACs via a region of shared homology into a single larger recombinant YAC. The recombinant YAC can then be transferred into ES cells by fusion with yeast spheroplasts carrying the YAC, followed by selection for a drug-resistance selection marker (Figure 1Aa). ES cells can then be used to generate chimeras to achieve germline transmission. Transgenic mice containing the entire functional human TCRαβ gene loci were created in this way6. BACs can also be manipulated by homologous recombination in E.coli, for example to insert reporter genes7.
Genomically humanised YAC and BAC transgenic strains may be bred onto a null background for the gene of interest such that the humanised locus is the only version of the gene that is expressed (Figure 1Aa)8. Production of transgenic mice expressing only human antibodies has been achieved by breeding transgenic lines with knock-out alleles of heavy and light chain constant regions (for example, see Ref.9).
BACs can also be designed to integrate at specific and ubiquitously expressed chromosomal loci, such as the hypoxanthine-guanine phosphoribosyltransferase (Hprt) locus, by homologous recombination10 or by site-specific recombination (SSR)11 in ES cells. Site-specific integration avoids the problems due to deletions and concatemerisation that can occur during random integration by non-homologous recombination and, as the site of integration is predetermined, also achieves a more reproducible expression profile.
Targeted Genomic Replacement
Targeted integration of a human sequence into the equivalent region of the mouse genome in ES cells is the most precise method for humanisation, enabling a single copy of human sequence to reside at a natural site for its expression, while simultaneously replacing the corresponding mouse sequence. In principle it is possible to generate a homozygous humanised mouse strain by inter-crossing heterozygotes carrying the human replacement.
Traditionally, this approach has involved small sized changes (up to ~10 kb) replacing some or all of the exons and introns of a mouse gene with corresponding human sequence; the human sequence therefore remains under the control of mouse transcriptional regulatory sequences. Mice in which the genomic region encompassing exons 4–9 of the p53 gene was replaced by the orthologous human genomic region have been constructed by this approach and have a correct splicing pattern, producing a chimeric protein in which the p53 core domain is human. These animals have proved valuable for determining the tumorigenic role of human p53 mutations12, 13. However, replacements are now possible in which an entire mouse locus, including non-coding upstream and downstream sequences, is substituted by equivalent human sequence using constructs derived from BACs. In principle, this approach could be extended to encompass a larger region of shared synteny. There are two ways to achieve this aim (described below and shown in Figure 1 Ab, c): either directly by using homologous recombination, or through a multi-step approach involving a combination of homologous recombination and SSR.
BAC-vector homologous recombination
Genomic replacements can be achieved by homologous recombination with a hybrid BAC vector (Figure 1Ab). This vector is assembled from mouse and human BAC clones by recombineering technology in E.coli, to create a construct with a large region of human sequence and a drug-resistance marker inserted between long regions of mouse genomic sequence (>100 kb in total). As a consequence of the length of the mouse sequences that form the homology arms this targeting vector results in efficient homologous recombination in ES cells14. Targeted integration is detected using quantitative PCR to assay for the reduction in copy number of mouse autosomal sequences from two to one. The advantage of this strategy is that it can be applied directly to unmodified ES cells and is essentially a single step procedure, although a second step to delete the selection marker using SSR is usually desirable.
A high throughput version of this technique has been developed, termed ‘VelociGene’, which allows rapid generation of large numbers of genetically modified mouse lines. So far, most mouse lines made using VelociGene contain null alleles, in that the entire mouse locus is replaced with a reporter cassette driven by the endogenous promoter. This high-throughput technology is being used by the NIH as part of their KnockOut Mouse Program (KOMP).
VelociGene has been successfully used to create a series of humanised knock-ins, to improve xenogeneic transplantation mouse models for studying in vivo human haematopoiesis and immune function. Four different cytokines have been humanised, in three separate targeting events. Both thrombopoietin (TPO), an essential cytokine for haematopoietic stem cell maintenance, and colony stimulating factor–1 (CSF1), a cytokine important for differentiation and function of human macrophages, were humanised in single targeting events15, 16. For both loci, the mouse promoter was left intact but the coding region and sequence extending to 3 kb downstream of the polyA signal was replaced with the human equivalent (changes of 5 kb and 18 kb, respectively). Granulocyte-macrophage colony-stimulating factor (CSF2) and Interleukin-3 (IL3) were humanised in the same targeting event due to their close proximity (<10 kb) in both the mouse and human genome17. The dual targeting construct replaced the mouse loci with the human equivalents (a change of 20 kb): While IL3 retained the mouse promoter, CSF2 is controlled by its human regulatory elements. The largest humanisation project carried out using VelociGene technology was the VelocImmune mouse, in which 6 Mb of the variable portion of the mouse immunoglobulin (Ig) loci were humanised, to allow production of human monoclonal antibodies for antibody therapeutics18.
Recombinase-Mediated Cassette Exchange (RMCE) and Recombinase-Mediated Genomic Replacement (RMGR)
The second approach to ‘humanisation by replacement’ is to use SSR to achieve efficient exchange of chromosomal sequence with sequence on an incoming plasmid or BAC. This involves a strategy termed recombinase-mediated cassette exchange (RMCE). RMCE is a two stage process: firstly, a selection marker cassette that is flanked by heterospecific SSR sites is integrated into the genome by homologous recombination, deleting the region which is to be replaced. Secondly, another donor vector with the human genomic sequence flanked by the same SSR sites is introduced, in the presence of a recombinase (usually expressed from a co-transfected plasmid) resulting in the human sequence recombining into the genome (Figure 1Ac). RMCE has been applied in various cell lines, including ES cells19, 20. The advantage of RMCE is that once the initial selection marker cassette is integrated into the desired chromosomal position, the resulting cell line can be used recursively for unlimited rounds of cassette exchange via the inserted heterospecific sites, and the desired events are readily recovered by genetic selection.
In principle, RMCE could be applied to any gene in mouse ES cells. For example, RMCE was used to place a human cDNA for a cardiac sodium channel in exon 2 of the mouse orthologue21. For-large scale replacement of mouse genomic sequence with human sequence from a BAC clone, an elaboration of the RMCE strategy can be used called Recombinase-Mediated Genomic Replacement (RMGR)22. In RMGR, heterospecific SSR sites and linked positive and negative selection markers are integrated into ES cells by sequential rounds of homologous recombination to delineate the region for replacement. The human BAC clone is modified by recombineering to insert heterospecific SSR sites at the corresponding human sequences and following co-transfection with a Cre recombinase expressing plasmid, SSR events at both ends are selected (Figure 1Ac). This method was used to replace the 87 kb genomic region encompassing the α-globin regulatory domain of mouse with the equivalent human sequence of 117 kb. An additional round of recombineering of the BAC was used to delete the major regulatory element before introduction into ES cells and consequently create a mouse model of human α-thalassaemia22. Theoretically, RMGR could encompass replacements of any size up to the size limit of the BAC donor. Recently, Hasegawa and co-workers published a modified version of RMGR in which they flanked the mouse cytochrome P450 Cyp3a gene cluster (a region of 820 kb) with a pair of homospecific loxP sites that allowed deletion of this cluster in an intermediate step; these sites were present in addition to the heterospecific sites required to insert human CYP3A4 and CYP3A7, contained within a 100 kb region of the donor BAC. The resulting humanised mouse has been created to study drug interactions23.
Transchromosomic and chromosome-engineered mice
Copy number variation (CNV) is highly polymorphic in the human population and it is likely that many CNVs give rise to phenotypic effects because of differences in sequence dose between individuals. Genomic CNV can be modelled through chromosome engineering of the mouse genome24, 25, but to model the effects of human CNVs that span extensive regions of non-coding sequence, genomically humanised mice carrying different copy numbers of the human sequence will need to be generated.
Humanised mouse models carrying a freely-segregating partial or whole human chromosome have been created by using microcell-mediated chromosome transfer (MMCT) into mouse ES cells26-28. These transchromosomic ES cells are then used in conventional approaches to establish strains of transchromosomic mice that transmit the human chromosome through the germline (Figure 1Ad), for example, the Tc1 mouse carries a freely segregating human chromosome 21 and models trisomy 21 in humans34.
Human artificial chromosomes (HACs) offer alternatives as vectors for gene delivery and to create animal models29, 30. HACs are non-integrating vectors that can be engineered to contain desired sequences and then moved by MMCT into mouse ES cells to make transchromosomic mice31. HACS are synthesised by two methods, either bottom-up (de novo synthesis) or top-down (engineered). The bottom-up approach involves introduction of two vectors, BAC or YAC, into cells permissive for recombination. One vector contains centromeric alphoid DNA, the other the genomic region of interest. Multiple copies of each are randomly assembled with no control on size or composition. The resultant HACs are usually circular and 1-10 Mb in size. The top-down approach involves shortening of human chromosomes by introduction of telomeric sequences via homologous recombination, to form mini-chromosomes. Efficiency of recombination is increased if this step is carried out in chicken DT40 cells which are unusually permissive for homologous recombination. More recently SSR sites have been introduced into engineered HACs, allowing introduction of any genomic region flanked by SSR sites in a donor vector (BAC or YAC). The latest generation of engineered HACs, designed with gene therapy and synthesis of animal models in mind contain multiple different SSR sites32, allowing introduction of multiple genes of interest on the same vector, while the tet-O HAC33 is conditional and once introduced can be selectively removed from cells (the current generation of HAC vectors are reviewed in ref 34). The merits of creating transchromosomic mice are that they carry low or single copy number human chromosomes or HACs, they are generally freely segregating and thus do not disrupt endogenous sequences, and, presumably, the maximum size is that of a chromosome34. A possible disadvantage of this approach may be instability of the transchromosomes, though this has not been reported as a problem so far.
Mouse models for developing therapies
In addition to understanding pathogenic processes, genomically humanised mouse models are likely to be important for developing therapies, particularly gene therapies. In a recent example, a genomically humanised mouse strain was created by knocking into the ubiquitously expressed Rosa26 locus a minigene for human factor 9 (F9), which included intron 1 and the Y155stop mutation, known to cause haemophilia B35. This humanised F9 mouse was then used to develop in vivo gene therapy, using zinc finger nucleases plus a promoterless therapeutic gene fragment consisting of wildtype F9 cDNA exons 2–8 preceded by a splice acceptor site. The site-specific nucleases corrected the mutant F9 gene by inserting the gene fragment into the first intron of F9. It is difficult to imagine how this therapeutic approach for haemophilia B could have been validated without the use of such mice.
Existing and new resources
Comparison between existing strains of mutant mice and humanised genomic models can be highly instructive for understanding species-specific biology36, 37. Additive multi-copy transgenics will also continue to be necessary for modelling disorders, such as some late-onset neurodegenerative diseases, in which expression of human sequences needs to attain a critical level to manifest a phenotype38. Furthermore, existing mouse genetic resources can be used to understand the effects of genetic modifiers, by breeding humanised loci onto different inbred mouse lines 39 or into genetically sensitised mouse strains. Similarly, we may be able to dissect the effects of the environment on human genetic disease by altering the conditions in which genetically identical humanised mouse models are maintained.
Our ability to humanise mice is likely to increase greatly in the near future. New applications might include iterative application of the VelociGene technology along the length of a chromosome, to create contiguous regions of humanisation. Humanisation of multiple loci, linked or unlinked, could also be achieved in an ES cell line by extending RMGR technology. For example, targeting vectors already generated by the International Knockout Mouse Consortium could be adapted to insert heterotypic SSR sites and/or selection markers into the mouse genome, and corresponding human BACs overlapping these loci could be used as donors. Alternatively, BACs could joined by recombineering into a single mega-BAC to span the desired region. New SSR systems, such as Dre/rox recombination will undoubtedly be useful to implement this strategy40.
If large-scale changes are required, it should also be possible to use chromosome engineering to apply RMGR on a megabase scale. In this scenario, human artificial chromosomes (HACs) maintained in DT40 cells could be first modified by homologous recombination to insert SSR sites and selection markers at the desired end points of the prospective replacement interval. Subsequently the HACs could be transferred by MMCT into mouse ES cells previously modified by targeted insertion of SSR sites and selection markers at the equivalent chromosomal positions. Expression of recombinase could then be induced to mediate the replacement event between the HAC and the mouse chromosome. This is an exciting prospect for modelling large CNVs and aneuploidy.
Current experience with transchromosomic mice is limited. Recognised issues include difficulty in obtaining germline transmission of the freely segregating human chromosome and its subsequent mosaicism in the animal27, 41. One way around these problems may be to engineer the human chromosome such that it is translocated onto a mouse chromosome, although inevitably this means deletion of some human and mouse sequences at the translocation breakpoints.
Conclusion
As the technologies necessary for producing genomically humanised models develop further, it will become possible to introduce greater amounts of human genetic material into mice and other species. This prospect has promoted discussion about the ethical implications of such humanisations, for example in a report from the UK Academy of Medical Sciences, published in July 2011, which is freely available on-line42.
A key question about genomically humanised mice concerns the extent to which the human DNA sequence is read correctly and efficiently by the mouse transcriptional machinery. Little information is available, partly because there are relatively few genomically humanised models. It is reassuring that in the α-globin humanised mouse model, the human genes are expressed in an appropriate developmental stage- and cell type-specific manner22. However, in humanised heterozygotes human α-globin RNA is expressed at 40% of the level of endogenous mouse α-globin, possibly because the relevant mouse transcription factors bind cognate human sequences with sub-optimal efficiency or stability. Nevertheless, the humanised α-globin locus accurately recapitulates many of the important features of the human α-globin gene chromatin state, most notably chromosomal looping, transcription factor binding and polycomb (PcG) recruitment at α-globin CpG islands in non-erythroid cells, and subsequent eviction of PcG from the α-globin CpG island in mature erythroid cells39. Consistent with this observation, a recent study showed that when a human chromosome is placed in a mouse environment, the human DNA sequence directs a human rather than mouse pattern of chromatin modifications43.
We have learned a significant amount from genetically humanised mice already. The tools now available, and those to come, will allow more accurate manipulation of the mouse genome with engineered human genomic material. This holds great promise for the better understanding of disease, development of effective therapies, more accurate models of drug metabolism as well as enhancing our understanding of mammalian and human genome function.
Acknowledgements
AD, RB-S, VJLT, EMCF were funded by the UK Medical Research Council, and VLJT and EMCF also received funding from the Wellcome Trust and the AnEUploidy consortium, an EU FP6 Integrated Project. We thank Mark Pepys, Graham Taylor, Steve Wood for information about SAP, and Jean-Francois Bureau, Peter D’Eustachio, David Hughes, Ravinesh Kumar, Melissa Landrum, Jeff Milner, Harry Noyes for information about human-specific genes. We apologise if significant papers have been omitted due to space constraints.
Glossary
- Knock-out mouse strain
A mouse strain in which the functional protein-coding capacity of a particular gene has been disrupted. This could be via a targeted deletion, gene trap, or targeted conditional knock-out.
- Knock-in mouse strain
A mouse strain carrying a targeted replacement or insertion; for example, an exchange of nucleotide sequence to change the encoded protein sequence or an insertion to create a tagged protein.
- Conditional mutation
A mutation whose expression is under experimental control. In a conditional knock-out, deletion of a critical exon flanked by recombination sites can be induced by expressing a recombinase, for example under a tissue-specific promoter.
- Inducible mutation
A class of conditional mutation that is expressed by using a ligand-inducible recombinase. Inducible mutations allow recombinase-driven change to be induced at a particular timepoint by delivering the ligand into the animal by injection, diet or using viral vectors.
- Transchromosomic mouse strain
A mouse strain into which either an entire chromosome or a chromosome fragment from another species has been transferred. This chromosome or chromosome fragment may be freely segregating or inserted into an existing mouse chromosome.
- Pronuclear injection
A process by which DNA is added to the genome, generally to create transgenic mice. Linearised DNA is injected into one of the two pronuclei of the fertilized egg, is stably incorporated into the genome and the eggs develop to term in a pseudopregnant female mouse.
- Gene targeting
A method of exchanging genetic information from a donor vector into a recipient genome, exploiting a DNA-repair mechanism that recognises homology between the donor DNA and the recipient locus. It results in the replacement of the original genomic sequence with the donor sequence.
- Genome-wide association study (GWAS)
A whole-genome scan testing the statistical association between finely spaced polymorphic genetic markers and a trait under study. Success depends on large numbers of phenotypically well-classified DNA samples in order to have the statistical power to detect such associations.
- Recombineering
Recombineering (derived ‘recombination-mediated genetic engineering’) uses recombination functions encoded by λ phage to mediate in-vivo recombination using short stretches of homologous sequence (30-50 nucleotides long) that can be readily appended to the termini of any desired DNA.
- KOMP
An NIH-funded program contributing to the international mouse knockout consortium (IKMC) to generate a null mutation in every mouse gene. Other contributors are the Texas Institute for Genomic Medicine (TIGM), the North American Conditional Mouse Mutagenesis Program (NorCOMM) and the European Conditional Mouse Mutagenesis Programme (EUCOMM).
- Xenogeneic transplantation
The transplantation of tissues or cells derived from one species into another. For example, the introduction of human stem cells into an immune-deficient mouse strain.
- Microcell mediated chromosome transfer (MMCT)
A method for transferring entire chromosomes or chromosome fragments from a donor cell line into a recipient cell line.
- Minigene
A gene construct that contains fewer exons and introns than its original full gene counterpart. Minigenes have been used to investigate regulatory sequences but may also be used because of their more convenient size.
Biographies
Author biographies
Anny Devoy received her PhD in Biomedical Science from The University of Nottingham, UK. She is presently a postdoctoral scientist at the Institute of Neurology, University College London, UK, in the laboratory of Elizabeth Fisher. Her current research focuses on the creation and analysis of mouse models of amyotrophic lateral sclerosis.
Rosie K.A. Bunton-Stasyshyn is a PhD student at the Institute of Neurology, University College London, UK. She received her undergraduate degree from Goldsmith’s College, London. She is working on creating new mouse models for amyotrophic lateral sclerosis in Elizabeth Fisher’s laboratory.
Victor L. J. Tybulewicz is head of the Division of Immune Cell Biology at the Medical Research Council (MRC) National Institute for Medical Research, London, UK. He obtained his PhD at the MRC Laboratory of Molecular Biology, Cambridge, UK, studying ATP synthase, and then trained as a postdoctoral fellow at the Whitehead Institute, Cambridge, Massachusetts, USA, developing mouse gene targeting techniques. His current interests are in lymphocyte signal transduction and the genetics of Down Syndrome.
Andrew J. H. Smith received his PhD from the University of Cambridge, in the UK, and was a post-doctoral fellow at Stanford University in California, USA. He is presently a Research Group Leader and Lecturer at the Institute for Stem Cell Research, University of Edinburgh, UK, and also a staff scientist at the MRC Molecular Haematology Unit in the Weatherall Institute of Molecular Medicine, University of Oxford, UK. His research is focussed on the development of strategies for genome engineering in embryonic stem cells for functional genetics studies, and on creating mouse models of human disease.
Elizabeth M.C. Fisher received her PhD from Imperial College (St Mary’s Campus) in London, UK, and was a postdoctoral fellow in the laboratory of David Page at the Whitehead Institute, Cambridge, Massachusetts, USA. She is Professor of Neurogenetics at the Institute of Neurology, University College London, UK. Her current research focuses on the creation and analysis of mouse models of Down syndrome and of amyotrophic lateral sclerosis.
Footnotes
FURTHER INFORMATION
ENCylopedia Of DNA Elements, genome.ucsc.edu/ENCODE
International Knockout Mouse Consortium, www.knockoutmouse.org
KnockOut Mouse Project KOMP, www.komp.org
Contributor Information
Anny Devoy, Department of Neurodegenerative Disease, UCL Institute of Neurology, Queen Square, London WC1N 3BG, UK a.devoy@prion.ucl.ac.uk, +44 203 456 7890.
Rosie KA Bunton-Stasyshyn, Department of Neurodegenerative Disease, UCL Institute of Neurology, Queen Square, London WC1N 3BG, UK, r.bunton@prion.ucl.ac.uk, +44 203 456 7890.
Victor L.J. Tybulewicz, MRC National Institute for Medical Research, The Ridgeway, London NW7 1AA, UK, vtybule@nimr.mrc.ac.uk; +44 20 8816 2184
Andrew J.H. Smith, Institute for Stem Cell Research, University of Edinburgh, Edinburgh EH9 3JQ, UK; and the MRC Molecular Haematology Unit, Weatherall Institute of Molecular Medicine, University of Oxford, Oxford OX3 9DS, Andrew.smith@ed.ac.uk; +44 131 651 7244
Elizabeth M.C. Fisher, Department of Neurodegenerative Disease, UCL Institute of Neurology, Queen Square, London WC1N 3BG, UK, e.fisher@prion.ucl.ac.uk; +44 203 456 7890
Reference List
- 1.Nguyen D, Xu T. The expanding role of mouse genetics for understanding human biology and disease. Dis Model. Mech. 2008;1:56–66. doi: 10.1242/dmm.000232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fisher EMC, Lana-Elola E, Watson SD, Vassiliou G, Tybulewicz VLJ. New approaches for modelling sporadic genetic disease in the mouse. Disease Models & Mechanisms. 2009;2:446–453. doi: 10.1242/dmm.001644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giraldo P, Montoliu L. Size matters: Use of YACs, BACs and PACs in transgenic animals. Transgenic Research. 2001;10:83–103. doi: 10.1023/a:1008918913249. [DOI] [PubMed] [Google Scholar]
- 5.Heaney JD, Bronson SK. Artificial chromosome-based transgenes in the study of genome function. Mammalian Genome. 2006;17:791–807. doi: 10.1007/s00335-006-0023-9. [DOI] [PubMed] [Google Scholar]
- 6.Li LP, et al. Transgenic mice with a diverse human T cell antigen receptor repertoire. Nature Medicine. 2010;16:1029–U127. doi: 10.1038/nm.2197. [DOI] [PubMed] [Google Scholar]
- 7.Gong SC, Kus L, Heintz N. Rapid bacterial artificial chromosome modification for large-scale mouse transgenesis. Nature Protocols. 2010;5:1678–1696. doi: 10.1038/nprot.2010.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson SJ, Wade-Martins R. A BACwards glance at neurodegeneration: molecular insights into disease from LRRK2, SNCA and MAPT BAC-transgenic mice. Biochem Soc Trans. 2011;39:862–867. doi: 10.1042/BST0390862. [DOI] [PubMed] [Google Scholar]
- 9.Lonberg N, et al. Antigen-Specific Human-Antibodies from Mice Comprising 4 Distinct Genetic Modifications. Nature. 1994;368:856–859. doi: 10.1038/368856a0. [DOI] [PubMed] [Google Scholar]
- 10.Heaney JD, Rettew AN, Bronson SK. Tissue-specific expression of a BAC transgene targeted to the Hprt locus in mouse embryonic stem cells. Genomics. 2004;83:1072–1082. doi: 10.1016/j.ygeno.2003.12.015. [DOI] [PubMed] [Google Scholar]
- 11.Prosser HM, Rzadzinska AK, Steel KP, Bradley A. Mosaic complementation demonstrates a regulatory role for myosin VIIa in actin dynamics of stereocilia. Molecular and Cellular Biology. 2008;28:1702–1712. doi: 10.1128/MCB.01282-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo JL, et al. Knock-in mice with a chimeric human/murine p53 gene develop normally and show wild-type p53 responses to DNA damaging agents: a new biomedical research tool. Oncogene. 2001;20:320–328. doi: 10.1038/sj.onc.1204080. [DOI] [PubMed] [Google Scholar]
- 13.Song H, Hollstein M, Xu Y. P53 gain-of-function cancer mutants induce genetic instability by inactivating ATM. Nature Cell Biology. 2007;9:573–U166. doi: 10.1038/ncb1571. [DOI] [PubMed] [Google Scholar]
- 14.Valenzuela DM, et al. High-throughput engineering of the mouse genome coupled with high-resolution expression analysis. Nature Biotechnology. 2003;21:652–659. doi: 10.1038/nbt822. [DOI] [PubMed] [Google Scholar]
- 15.Rathinam C, et al. Efficient differentiation and function of human macrophages in humanized CSF-1 mice. Blood. 2011;118:3119–3128. doi: 10.1182/blood-2010-12-326926. [DOI] [PubMed] [Google Scholar]
- 16.Rongvaux A, et al. Human thrombopoietin knockin mice efficiently support human hematopoiesis in vivo. Proc Natl Acad Sci USA. 2011;108:2378–2383. doi: 10.1073/pnas.1019524108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Willinger T, et al. Human IL-3/GM-CSF knock-in mice support human alveolar macrophage development and human immune responses in the lung. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:2390–2395. doi: 10.1073/pnas.1019682108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murphy A. VelocImmune: Immunoglobulin variable region humanized mice. In: Little M, editor. Recombinant antibodies for immunotherapy. Cambridge University Press; Cambridge: 2011. pp. 100–108. [Google Scholar]
- 19.Bouhassira EE, Westerman K, Leboulch P. Transcriptional behavior of LCR enhancer elements integrated at the same chromosomal locus by recombinase-mediated cassette exchange. Blood. 1997;90:3332–3344. [PubMed] [Google Scholar]
- 20.Khandelia P, Yap K, Makeyev EV. Streamlined platform for short hairpin RNA interference and transgenesis in cultured mammalian cells. Proc Natl Acad Sci U S A. 2011;108:12799–12804. doi: 10.1073/pnas.1103532108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu K, et al. Recombinase-mediated cassette exchange to rapidly and efficiently generate mice with human cardiac sodium channels. Genesis. 2006;44:556–564. doi: 10.1002/dvg.20247. [DOI] [PubMed] [Google Scholar]
- 22.Wallace HA, et al. Manipulating the mouse genome to engineer precise functional syntenic replacements with human sequence. Cell. 2007;128:197–209. doi: 10.1016/j.cell.2006.11.044. [DOI] [PubMed] [Google Scholar]
- 23.Hasegawa M, et al. Quantitative prediction of human pregnane X receptor and cytochrome P450 3A4 mediated drug-drug interaction in a novel multiple humanized mouse line. Mol Pharmacol. 2011;80:518–528. doi: 10.1124/mol.111.071845. [DOI] [PubMed] [Google Scholar]
- 24.Tybulewicz VL, Fisher EM. New techniques to understand chromosome dosage: mouse models of aneuploidy. Hum. Mol Genet. 2006;15(Spec No 2):R103–R109. doi: 10.1093/hmg/ddl179. [DOI] [PubMed] [Google Scholar]
- 25.van der WL, Shaw-Smith C, Bradley A. Chromosome engineering in ES cells. Methods Mol Biol. 2009;530:49–77. doi: 10.1007/978-1-59745-471-1_4. [DOI] [PubMed] [Google Scholar]
- 26.Nakatani J, et al. Abnormal behavior in a chromosome-engineered mouse model for human 15q11-13 duplication seen in autism. Cell. 2009;137:1235–1246. doi: 10.1016/j.cell.2009.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Doherty A, et al. An aneuploid mouse strain carrying human chromosome 21 with Down syndrome phenotypes. Science. 2005;309:2033–2037. doi: 10.1126/science.1114535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomizuka K, et al. Functional expression and germline transmission of a human chromosome fragment in chimaeric mice. Nature Genetics. 1997;16:133–143. doi: 10.1038/ng0697-133. [DOI] [PubMed] [Google Scholar]
- 29.Kazuki Y, et al. Refined human artificial chromosome vectors for gene therapy and animal transgenesis. Gene Ther. 2011;18:384–393. doi: 10.1038/gt.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki N, Itou T, Hasegawa Y, Okazaki T, Ikeno M. Cell to cell transfer of the chromatin-packaged human beta-globin gene cluster. Nucleic Acids Research. 2010;38 doi: 10.1093/nar/gkp1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuroiwa Y, et al. Manipulation of human minichromosomes to carry greater than megabase-sized chromosome inserts. Nat Biotechnol. 2000;18:1086–1090. doi: 10.1038/80287. [DOI] [PubMed] [Google Scholar]
- 32.Yamaguchi S, et al. A Method for Producing Transgenic Cells Using a Multi-Integrase System on a Human Artificial Chromosome Vector. PLoS One. 2011;6 doi: 10.1371/journal.pone.0017267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakano M, et al. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Developmental Cell. 2008;14:507–522. doi: 10.1016/j.devcel.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kazuki Y, Oshimura M. Human artificial chromosomes for gene delivery and the development of animal models. Mol Therapy. 2011;19:1591–1601. doi: 10.1038/mt.2011.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, et al. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature. 2011;475:217–221. doi: 10.1038/nature10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gross B, et al. Generation and characterization of a humanised PPARdelta mouse model. Br. J. Pharmacol. 2011;164:192–208. doi: 10.1111/j.1476-5381.2011.01359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uno S, et al. CYP1A1 and CYP1A2 expression: Comparing ‘humanized’ mouse lines and wild-type mice; comparing human and mouse hepatoma-derived cell lines. Toxicol Appl Pharmacol. 2009;237:119–126. doi: 10.1016/j.taap.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wong PC, Cai HB, Borchelt DR, Price DL. Genetically engineered mouse models of neurodegenerative diseases. Nature Neuroscience. 2002;5:633–639. doi: 10.1038/nn0702-633. [DOI] [PubMed] [Google Scholar]
- 39.Lehman EJ, et al. Genetic background regulates beta-amyloid precursor protein processing and beta-amyloid deposition in the mouse. Hum. Mol Genet. 2003;12:2949–2956. doi: 10.1093/hmg/ddg322. [DOI] [PubMed] [Google Scholar]
- 40.Anastassiadis K, et al. Dre recombinase, like Cre, is a highly efficient site-specific recombinase in E-coli, mammalian cells and mice. Disease Models & Mechanisms. 2009;2:508–515. doi: 10.1242/dmm.003087. [DOI] [PubMed] [Google Scholar]
- 41.Tomizuka K, et al. Double trans-chromosomic mice: maintenance of two individual human chromosome fragments containing Ig heavy and kappa loci and expression of fully human antibodies. Proc Natl Acad Sci U S A. 2000;97:722–727. doi: 10.1073/pnas.97.2.722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Academy of Medical Sciences . Animals containing human material. London: 2011. [Google Scholar]
- 43.Wilson MD, et al. Species-specific transcription in mice carrying human chromosome 21. Science. 2008;322:434–438. doi: 10.1126/science.1160930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waterston RH, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 45.Hawkins PN, Myers MJ, Epenetos AA, Caspi D, Pepys MB. Specific localization and imaging of amyloid deposits in vivo using 123I-labeled serum amyloid P component. J Exp Med. 1988;167:903–913. doi: 10.1084/jem.167.3.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grad LI, et al. Intermolecular transmission of superoxide dismutase 1 misfolding in living cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:16398–16403. doi: 10.1073/pnas.1102645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dorner M, et al. A genetically humanized mouse model for hepatitis C virus infection. Nature. 2011;474:208–211. doi: 10.1038/nature10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Enard W, et al. A humanized version of Foxp2 affects cortico-basal ganglia circuits in mice. Cell. 2009;137:961–971. doi: 10.1016/j.cell.2009.03.041. [DOI] [PubMed] [Google Scholar]
- 49.Stahl PD, Wainszelbaum MJ. Human-specific genes may offer a unique window into human cell signaling. Sci Signal. 2009;2:e59. doi: 10.1126/scisignal.289pe59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schorderet P, Duboule D. Structural and functional differences in the long non-coding RNA hotair in mouse and human. PLoS Genet. 2011;7:e1002071. doi: 10.1371/journal.pgen.1002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by Noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yeretssian G, et al. Gender differences in expression of the human caspase-12 long variant determines susceptibility to Listeria monocytogenes infection. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:9016–9020. doi: 10.1073/pnas.0813362106. [DOI] [PMC free article] [PubMed] [Google Scholar]