Abstract
Successful disease management requires a rapid and sensitive diagnosis method that can recognize early infection even before the manifestation of its clinical signs. The only available field diagnostic tests for foot-and-mouth disease (FMD) are lateral flow devices, commonly known as chromatographic strips. Low sensitivity and inability to detect FMD virus (FMDV) at the serotype level are limitations of lateral flow devices. Therefore, a reverse transcriptase loop-mediated isothermal amplification (RT-LAMP) was standardized using universal and sero-type specific genes in a single tube. This test does not require sophisticated equipment and can detect FMDV at serotype level in about 60 min. In addition, the sensitivity and specificity of this test is comparable to conventional reverse transcriptase PCR and real time PCR (rRT-PCR).
Key Words: FMD, RT-LAMP, Sensitivity
Introduction
Foot-and-mouth disease (FMD) is a trans-boundary viral disease of cloven footed animals which spreads easily in susceptible populations (Grubman and Baxt, 2004 ▶). FMD viruses (FMDV) belong to the Aphthovirus genus of the Picornaviridae family with seven serotypes (Alexandersen et al., 2003 ▶) and no cross-protection among the serotypes (Cao et al., 2014 ▶). The disease causes severe economic loss besides its impact on national and international trade of livestock animals and their products (Thompson et al., 2002 ▶). Rapid, accurate and timely diagnosis is the key factor in controlling trans-boundary animal diseases including FMD.
Various diagnostic tests are available for FMD including ELISA and PCR (Reid et al., 2000 ▶; Reid et al., 2002 ▶). These tests are laborious and expensive and require sophisticated equipment and clinical specimen submissions. Therefore, a simple, rapid and cheap diagnostic assay which can be easily performed in the field is required to detect FMDV in animals and their products (Chen et al., 2011a ▶).
A novel nucleic acid amplification method, known as reverse transcription loop-mediated isothermal amplification (RT-LAMP), has been recently used to detection influenza A virus, Newcastle disease virus, classical swine fever virus and porcine reproductive and respiratory syndrome virus (Notomi et al., 2000 ▶; Pham et al., 2005 ▶; Chen et al., 2009 ▶). RT-LAMP has also been used in China to detect FMDV infection (Chen et al., 2011b ▶). In the present study, RT-LAMP was standardized in order to detect FMDV RNA and its sero-types for the first time in Pakistan.
Materials and Methods
FMDV isolates
Characterized FMDV field strains were used in this study. These FMDV strains were taken from the virus repository maintained at the Animal Health Program, Animal Sciences Institute, National Agricultural Research Center, Islamabad. The FMDV isolates were already confirmed by rRT-PCR (Reid et al., 2000 ▶, 2002) and typed by indirect sandwich ELISA (Roeder and Smith, 1987 ▶; Ferris and Dawson, 1988 ▶). They included FMDV serotypes O (n=21), A (n=16) and Asia-1 (n=8).
RNA extraction
FMDV specific RNA was extracted from the above mentioned viruses using QIAamp® Viral RNA Minikit (Qiagen, GmBh, Germany) following the manufacturer’s instructions. RNA was extracted from the controls (strong positive, weak positive, negative and no-template control).
RT-LAMP
The RT-LAMP was carried out in a volume of 25 µL reaction mixture containing 2.5 µL ThermoPol reaction buffer (New England Bio Labs Inc., Beverly, MA, USA), 1 µL MgSO4 (100 mM), 2 µL dNTP set (Fermentas, 10 mM), 5 µL betaine (Sigma-Aldrich, St. Louis, MO, USA, 5 M), 2 µL nuclease-free water, 1 µL hydroxy naphthol blue (Sigma Aldrich, 3 mM), 1 µL Bst DNA polymerase Lg fragment (New England Bio Labs Inc., Beverly, MA, USA), 0.2 µL AMV reverse transcriptase (New England Bio Labs Inc., Beverly, MA, USA), 0.1 µL forward and backward outer primers, each with a 5 pmol concentration, 1 µL forward and backward inner primers each with a 50 pmol concentration, 0.5 µL forward and backward loop primers each with a 25 pmol concentration and 2 µL of extracted RNA from the samples (with a 4 µg/µL concentration). The FMDV specific primers used in the RT-LAMP for amplification of FMDV RNA are given in Table 1. Serotype specific primers (A, O, Asia-1) used in this study were previously published (Madhanmohan et al., 2013 ▶). This mixture was incubated at 60°C for 15-60 min in a water bath.
Table 1.
Details of oligonucleotide primers used in the current study for RT-LAMP amplification of the FMD viruses 3D polymerase gene. Genome position is arranged according to the FMD virus strain O UKG 35/2001 (GenBank Accession No. AJ539141)
| Virus serotype | Primers | Sequence (5´-3´) | Target (genome position) |
|---|---|---|---|
| O, A, Asia-1 | F | GGAACTGGGTTTTACAAACCTG | 1183-1382 (199 bp) Current study |
| B | CGCAGGTAAAGTGATCTGTAGC | ||
| FIP | CTGCCACGGAGATCAACTTCTCCTGGATCCGACCCTCGAGGCTATCCTCT | ||
| BIP | CTCGCCGTCCACTCTGGACCTGGATCCTGGAATCTCAAAGAGGCCCTG | ||
| FLP | GTATGGTCCCACGGCGTGC | ||
| BLP | GAGTACCGGCGTCTCTTTGAGC | ||
| O | F3 | CATCCTCACCACCCGTAAC | 66-297 (232 bp) Madhanmohan et al. (2013) |
| B3 | GACACCTTTGTGGTCGGTC | ||
| FIP | GGAAGTGTTCGGTCCGCTCACTTTTCCCAGTCAAGCGTTGGAG | ||
| BIP | CAGAGTTGTGCAGGCAGAACGGTTTTAACGTCCGAATGAGTCACTG | ||
| FLP | GGAGTCACATACGGGTACG | ||
| BLP | CACCTCTTCGACTGGGTC | ||
| A | F3 | CTACACTGCGCCTAACCG | 1728-1936 (209 bp) Madhanmohan et al. (2013) |
| B3 | TGGGGCAGTAGAGTTCGG | ||
| FIP | TGCGACTGCCCCTAGGTCACTTTTTAACAGTGTACAACGGGACG | ||
| BIP | GCCCAACTTCCTGCCTCTTTCATTTTCTTCATGCGCACAAGAAGC | ||
| FLP | CAAGTACTCCGCGGCCAGTG | ||
| BLP | GGTGCAATCAAGGCTGACG | ||
| Asia-1 | F3 | CCCCACTGAACACAAAGGC | 287-483 (187 bp) Madhanmohan et al. (2013) |
| B3 | GTGGGGAAAGAGAGTCAGC | ||
| FIP | AGTCACCTCTACGTCCCATCCATTTTGTGTACGGCAGTCTCATGG | ||
| BIP | TTGGAAACCAATTCAACGGCGGTTTTGTCAAGGCTCTTCAGCTCTG | ||
| FLP | CTCGTACGCCTACATGAGGAA | ||
| BLP | GCCTCCTTGTCGCACTTGTG |
F: Forward outer primer, B: Backward outer primer, FIP: Forward inner primer, BIP: Backward inner primer, FLP: Forward loop primer, and BLP: Backward loop primer
Analysis of the RT-LAMP products
Detection of gene amplification was accomplished by visual inspection and agarose gel electrophoresis. A positive reaction changed the color from violet to blue, while a negative reaction failed to turn blue and remained violet in color. Color change was either observed with the naked eye under natural light or with the aid of UV light at 365 nm. In addition, amplified RT-LAMP products were electrophoresed on a 2% molecular-grade agarose gel prepared in 0.5 × Tris-borate EDTA buffer stained with 0.5 µg/ml ethidium bromide.
RT-PCR
The samples analyzed using RT-LAMP were re-analyzed using RT-PCR and real time RT-PCR to compare the results. RT-PCR was performed following Reid et al. (2000) ▶. The RT-PCR was carried out in a 25 µL total reaction volume using a one step RT-PCR kit (Invitrogen, Life Technologies) with 50 pmol of the forward and reverse primers and 2 µL of the sample extracted RNA (with a 4 µg/µL concentration). The cycling reaction comprised of 50°C for 30 min and 95°C for 2 min, followed by 40 cycles of 94°C for 20 s, 55°C for 30 s and 72°C for 30 s, and a final extension cycle of 72°C for 10 min. After amplification, 5 µL of the PCR products (217 bp) were analyzed by electrophoresis on an ethidium bromide-stained 2% agarose gel.
Real time TaqMan RT-PCR
The FMDV real time TaqMan RT-PCR assay performed on an ABI 7500 Sequence Detection System (Applied Bio-Systems, Branchburg, NJ, USA) was used as reference. The 25 µL reaction mixture contained 2.5 µL extracted RNA, 5 µL of 5 × One Step reaction buffer, 2.5 µL 25 mM concentration, 3 µL dNTPs with a 10 mM concentration, 0.1 µL forward and reverse primers each with a concentration of 50 µM and 10 µM of TaqMan probe. The following thermal profile was used: an initial reverse transcription step at 60°C for 10 min, followed by 95°C for 2 s and 60°C for 60 s 45 cycles of amplification in plate on ABI Prism 7500 real time PCR machine.
The extracted RNA from the sample was subjected to real time PCR (rRT-PCR) using primer set (1 F/1 R) located in the 5´ un-translated region (Reid et al., 2000 ▶; Callahan et al., 2002 ▶). Core reagents kits (Taq Man®, EZ-RT-PCR core reagent and N808-0236) were used in the rRT-PCR. The final volume of the reaction mix was adjusted to 25 µL by adding 2.5 µL template RNA. The reaction plate was loaded and run on an ABI 7500 real time PCR system (Applied Bio-System) using ABI Prism SDS 7500 software and Ct values were recorded.
Detection limit of RT-LAMP
The detection limit of RT-LAMP was tested using the same RNA templates with identical concentrations performed in triplicate at each template concentration. Viral RNA was quantitated using UV spectrophotometry (UNICAM 3000, US). Serial dilutions of 1, 10, 102, 103 and 104 copies per reaction from FMDV strains were used in the assay. The specificity of RT-LAMP, cross reactions with the RNA of the strains’ PPRV and other important prevalent viruses was assessed.
Results
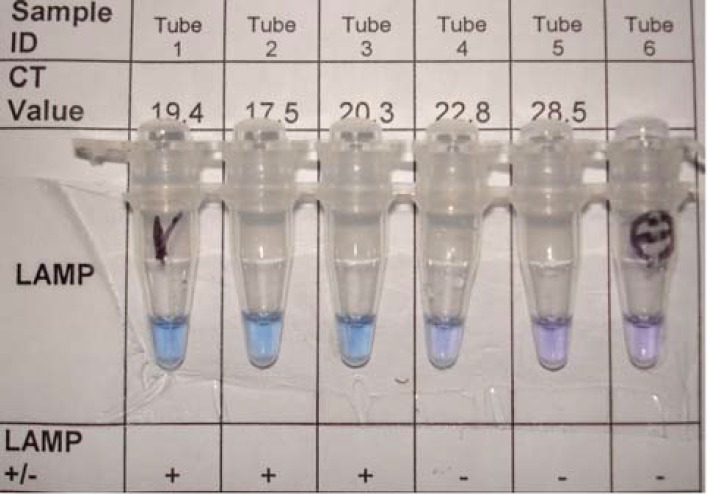
RT-LAMP was optimized initially using the 3D gene specific primers of the FMDV serotype O (GenBank ID: AJ539141). The RT-LAMP successfully amplified the 199 bp target sequence of the 3D polymerase gene of serotypes A, O and Asia-1 at 60°C during 15 to 60 min while the FMDV type-specific RT-LAMP amplified 232 bp, 209 bp and 187 bp of sero-type O, A and Aisa-1, respectively. Amplification products were visible to the naked eye. Upon the visual inspection of the LAMP reaction tubes color changes were noted: positive samples changed to sky blue, while negative samples remained violet as shown in Fig. 1 (tube 1-3 are positive while tube 4-6 are negative).
Fig. 1.
Visual detection of RT-LAMP amplification product of FMDV
RT-LAMP specificity was evaluated by cross-reactivity studies using the RNA templates of O with A and Asia-1 primer sets and vice versa. All samples were tested against the three primer sets. Individual primer sets were found to be strain-specific and did not show any cross-reactivity with other serotypes of the viruses included in this study. Furthermore, the specificity of the FMDV reaction was determined by checking the cross-reactivity of the assay with other viruses such as NDV, IBV and PPRV; the primer sets were found to be specific to FMDV.
These observations were consistent with the results of gel electrophoresis, where no bands were seen. Positive LAMP products were observed as ladder-like patterns on the agarose gel due to the formation of a mixture of stem-loop DNAs with various stem lengths and cauliflower-like structures with multiple loops formed by annealing between alternately inverted repeats of the target.
The results of field and experimental samples evaluation using RT-LAMP, virus isolation, con-ventional and real time PCR are presented in Table 2.
Table 2.
Comparison of results of conventional PCR, RT-LAMP and real time PCR for the detection of FMDV
| FMDV sero-type | Number | Number of positive samples |
||
|---|---|---|---|---|
| Conventional PCR | RT-LAMP | Real time PCR | ||
| O | 21 | 12 (57.1) | 17 (80.9) | 21 (100) |
| A | 16 | 10 (62.5) | 13 (81.2) | 16 (100) |
| Asia-1 | 8 | 5 (62.5) | 7 (87.5) | 8 (100) |
| Negative | 16 | 16 (100) | 16 (100) | 16 (100) |
| Total | 61 | 43 (70.5) | 53 (86.9) | 61 (100) |
The values in parenthesis are percentages
The colorimetric detection limit of RT-LAMP was found to be 10 copies of DNA.
Discussion
Accurate and timely diagnosis is the key to the effective control of trans-boundary animal pathogens. Clinical diagnosis is not completely accurate; therefore, diagnostic tests with high sensitivity and specificity are required. Various molecular tests are used to diagnose FMDV. These tests are difficult to perform and require expansive laboratory equipment and chemicals. On the other hand, RT-LAMP is capable of detecting FMDV and its various serotypes within 60 min. The present study was, therefore, conducted to standardize RT-LAMP which can be performed in the field.
During the present study, non-structural protein 3D pol RNA polymerase was targeted for 1 h at 60°C which is highly conserved among various FMDV subtypes and serotypes (Reid et al., 2009 ▶). In this study, six sets of primers identified 8 distinct regions of FMDV RNA making RT-LAMP more specific and sensitive. An isothermal auto-cycling strand-displacement DNA synthesis assay can recognize six regions of the target genome, making this assay more specific and sensitive (Notomi et al., 2000 ▶). The creation of loop arrangements allows for volatile polymerase-based enzymatic amplification, generating double-stranded, multi-sized amplicons. This test does not require electrophoresis since color changes in reaction tubes indicate the presence or absence of FMDV in the sample (Shaw et al., 2007 ▶; Reid et al., 2009 ▶). The dye, hydroxynaphthol blue, was used for colorimetric analysis of the LAMP reaction due to its remarkable advantage over other color-based assays. That is to say it can be mixed prior to amplification, and therefore reduce the risk of cross-contamination (Wang et al., 2012 ▶).
During this study, the diagnostic sensitivity of RT-LAMP (86.9%) was lower than real time PCR (61%) but higher than conventional PCR (70.5%). However, the specificity of all these assays was 100%. Other studies have also reported higher sensitivity of RT-LAMP compared to conventional PCR (Dahlenborg et al., 2001 ▶; Ding et al., 2013 ▶). RT-LAMP, as standardized in this study, is simple, rapid, cost effective and highly sensitive and can act as a specific assay for FMDV diagnosis. This assay may be used for FMDV clinical diagnosis and surveillance in developing countries such as Pakistan, as it does not require sophisticated equipment or skilled technicians.
References
- Alexandersen S, Quan M, Murphy C, Knight J, Zhang Z. Studies of quantitative parameters of virus excretion and transmission in pigs and cattle experimentally infected with foot-and-mouth disease virus. J. Comp. Pathol. 2003;129:268–282. doi: 10.1016/s0021-9975(03)00045-8. [DOI] [PubMed] [Google Scholar]
- Callahan JD, Brown F, Osorio FA, Sur JH, Kramer E, Long GW, Lubroth J, Ellis SJ, Shoulars KS, Gaffney KL, Rock DL, Nelson WM. Use of a portable real-time reverse transcriptase-polymerase chain reaction assay for rapid detection of foot-and-mouth disease virus. J. Am. Vet. Med. Assoc. 2002;220:1636–1642. doi: 10.2460/javma.2002.220.1636. [DOI] [PubMed] [Google Scholar]
- Cao Y, Lu Z, Li D, Fan P, Sun P, Bao H, Fu Y, Li P, Bai X, Chen Y, Xie B, Liu Z. Evaluation of cross-protection against three topotypes of serotype O foot-and-mouth disease virus in pigs vaccinated with multi-epitope protein vaccine incorporated with poly(I:C) Vet. Microbiol. 2014;168:294–301. doi: 10.1016/j.vetmic.2013.11.023. [DOI] [PubMed] [Google Scholar]
- Chen H, Zhang J, Liu Y, Liu X. Detection of foot-and-mouth disease virus RNA by reverse transcription loop-mediated isothermal amplification. Virol. J. 2011a;8:510–514. doi: 10.1186/1743-422X-8-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zhang J, Liu Y, Liu X. Rapid typing of foot-and-mouth disease serotype Asia-1 by reverse transcription loop-mediated isothermal amplification. Virol. J. 2011b;8:489–492. doi: 10.1186/1743-422X-8-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zhang J, Ma L, Ma Y, Ding Y, Liu X, Chen L, Ma L, Zhang Y, Liu Y. Rapid pre-clinical detection of classical swine fever by reverse transcription loop-mediated isothermal amplification. Mol. Cell. Probes. 2009;23:71–74. doi: 10.1016/j.mcp.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlenborg M, Borch E, Rådström P. Development of a combined selection and enrichment PCR procedure for clostridium botulinum types B, E, and F and its use to determine prevalence in fecal samples from slaughtered pigs. Appl. Environ. Microbiol. 2001;67:4781–4788. doi: 10.1128/AEM.67.10.4781-4788.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Chen H, Zhang J, Zhou J, Ma L, Zhang L, Gu Y, Liu Y. An overview of control strategy and diagnostic technology for foot-and-mouth disease in China. Virol. J. 2013;10:78–83. doi: 10.1186/1743-422X-10-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris NP, Dawson M. Routine application of enzyme-linked immunosorbent assay in comparison with complement fixation for the diagnosis of foot-and-mouth and swine vesicular diseases. Vet. Microbiol. 1988;16:201–209. doi: 10.1016/0378-1135(88)90024-7. [DOI] [PubMed] [Google Scholar]
- Grubman MJ, Baxt B. Foot-and-mouth disease. Clin. Microbiol. Rev. 2004;17:465–493. doi: 10.1128/CMR.17.2.465-493.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhanmohan M, Nagendrakumar SB, Manikumar K, Yuvaraj S, Parida S, Srinivasan VA. Development and evaluation of a real-time reverse transcription-loop-mediated isothermal amplification assay for rapid serotyping of foot-and-mouth disease virus. J. Virol. Methods. 2013;187:195–202. doi: 10.1016/j.jviromet.2012.08.015. [DOI] [PubMed] [Google Scholar]
- Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham HM, Nakajima C, Ohashi K, Onuma M. Loop-mediated isothermal amplification for rapid detection of Newcastle disease virus. J. Clin. Microbiol. 2005;43:1646–1650. doi: 10.1128/JCM.43.4.1646-1650.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid SM, Ebert K, Bachanek-Bankowska K, Batten C, Sanders A, Wright C, Shaw AE, Ryan ED, Hutchings GF, Ferris NP, Paton DJ, King DP. Performance of real-time reverse transcription polymerase chain reaction for the detection of foot-and-mouth disease virus during field outbreaks in the United Kingdom in 2007. J. Vet. Diagn. Invest. 2009;21:321–330. doi: 10.1177/104063870902100303. [DOI] [PubMed] [Google Scholar]
- Reid SM, Ferris NP, Hutchings GH, Samuel AR, Knowles NJ. Primary diagnosis of foot-and-mouth disease by reverse transcription polymerase chain reaction. J. Virol. Methods. 2000;89:167–176. doi: 10.1016/s0166-0934(00)00213-5. [DOI] [PubMed] [Google Scholar]
- Reid SM, Ferris NP, Hutchings GH, Zhang Z, Belsham GJ, Alexandersen S. Detection of all seven serotypes of foot-and-mouth disease virus by real-time, fluorogenic reverse transcription polymerase chain reaction assay. J. Virol. Methods. 2002;105:67–80. doi: 10.1016/s0166-0934(02)00081-2. [DOI] [PubMed] [Google Scholar]
- Roeder PL, Smith PMLB. Detection and typing of foot-and-mouth disease virus by enzyme-linked immunosorbent assay: a sensitive, rapid and reliable technique for primary diagnosis. Res. Vet. Sci. 1987;43:225–232. [PubMed] [Google Scholar]
- Scott M, Reid SM, Ebert K, Bachanek-Bankowska K, Batten C, Sanders A, Wright C, Shaw AE, Ryan ED, Hutchings GH, Ferris NP, Paton DJ, King DP. Performance of real-time reverse transcription polymerase chain reaction for the detection of foot-and-mouth disease virus during field outbreaks in the United Kingdom in 2007. J. Vet. Diagn. Invest. 2009;21:321–330. doi: 10.1177/104063870902100303. [DOI] [PubMed] [Google Scholar]
- Shaw AE, Reid SM, Ebert K, Hutchings GH, Ferris NP, King DP. Implementation of a one-step real-time RT-PCR protocol for diagnosis of foot-and-mouth disease. J. Virol. Methods. 2007;143:81–85. doi: 10.1016/j.jviromet.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Thompson D, Muriel P, Russell D, Osborne P, Bromley A, Rowland M, Creigh-Tyte S, Brown C. Economic costs of the foot and mouth disease outbreak in the United Kingdom in 2001. Rev. Sci. Tech. Off. Int. Epiz. 2002;21:675–687. doi: 10.20506/rst.21.3.1353. [DOI] [PubMed] [Google Scholar]
- Wang X, Zhang Q, Zhang F, Ma F, Zheng W, Zhao Z, Bai Y, Zheng L. Visual detection of the human metapneumovirus using reverse transcription loop-mediated isothermal amplification with hydroxynaphthol blue dye. Virol. J. 2012;9:1–6. doi: 10.1186/1743-422X-9-138. [DOI] [PMC free article] [PubMed] [Google Scholar]