Abstract
Pathogens infecting mammalian cells have developed various strategies to suppress and evade their hosts’ defensive mechanisms. In this line, the intracellular bacteria that are able to survive and propagate within their host cells must have developed strategies to avert their host’s killing attitude. Studying the interface of host-pathogen confrontation can provide valuable information for defining therapeutic approaches. Brucellosis, caused by the Brucella strains, is a zoonotic bacterial disease that affects thousands of humans and animals around the world inflicting discomfort and huge economic losses. Similar to many other intracellular dwelling bacteria, infections caused by Brucella are difficult to treat, and hence any attempt at identifying new and common therapeutic targets would prove beneficial for the purpose of curing infections caused by the intracellular bacteria. In THP-1 macrophage infected with Brucella melitensis we studied the expression levels of four host’s genes, i.e. EMP2, ST8SIA4, HCP5 and FRMD5 known to be involved in pathogenesis of Mycobacterium tuberculosis. Our data showed that at this molecular level, except for FRMD5 that was downregulated, the other three genes were upregulated by B. melitensis. Brucella melitensis and M. tuberculosis go through similar intracellular processes and interestingly two of the investigated genes, i.e. EMP2 and ST4SIA8 were upregulated in THP-1 cell infected with B. melitensis similar to that reported for THP-1 cells infected with M. tuberculosis. At the host-pathogen interaction interface, this study depicts overlapping changes for different bacteria with common survival strategies; a fact that implies designing therapeutic approaches based on common targets may be possible.
Key Words: Brucella melitensis, EMP2, FRMD5, HCP5, ST8SIA4
Introduction
Multicellular organisms such as mammalians have evolved sophisticated and complex mechanisms to make themselves immune to invasion by a huge number of opportunists they encounter on a daily basis. On the other hand, for survival, many microbes are either completely dependent on other living organisms or at least part of their life cycle must involve taking advantage of the resources available from the other organisms. Therefore, to survive, these microorganisms have had to develop strategies to at least suppress their prospective host’s defensive mechanisms. In this context, the intracellular pathogenic bacteria not only effectively evade the host’s various components of immune system, but have evolved mechanisms to use the cells meant to kill them as hiding niches or as dwellings (Imbuluzqueta et al., 2010 ▶). Moreover, since many antibiotics either do not enter the cells or their concentrations inside the cells do not reach a therapeutic level, infections caused by the intracellular dwelling bacteria are either very difficult to treat or are refractory to many treatment procedures (Carryn et al., 2002 ▶).
Brucella genus includes ten gram-negative species of which B. abortus, B. melitensis, B. suis, and B. canis are known to affect humans (Grilló et al., 2012 ▶; He, 2012 ▶). Human infections with B. abortus, B. melitensis and B. suis, which are considered as category B priority pathogens by the US Center for Disease Control (CDC), are more commonly reported, affecting 500,000 people around the world per year (He, 2012 ▶). Moreover, brucellosis is a cause of huge economic losses worldwide; for example, it is estimated to lay a burden of 600 million dollars on Latin America’s economy (Barbier et al., 2011 ▶).
Brucella as an intracellular pathogen has developed sophisticated mechanisms to subvert its host defensive strategies rendering them ineffective. Inside the macrophage cells, Brucella is protected and trafficked within a membrane-bound compartment called Brucella-containing vacuole (BCV) (Roop II et al., 2009 ▶). It is known that factors from the bacteria either directly, by changing the BCV properties, or indirectly, by controlling and changing the expression of various host factors, help the intracellular pathogen survive within the host cells (Roop II et al., 2009 ▶). In this line, it has been shown that the transcriptome of macrophages infected with a pathogenic strain of B. melitensis is significantly different from that of the cells infected with the attenuated strain which is cleared more easily by the macrophages (Wang et al., 2011 ▶). It is expected that this difference should maintain the factors that help the pathogenic strain survive within the macrophage cells.
In a recent interesting work, using macrophages differentiated from THP-1 cells. Kumar et al. (2010) ▶ used RNAi to target all host genes, one by one, and studied the consequences with regard to the changes in replication capacity of the intracellular pathogen M. tuberculosis. These investigators identified 74 host genes whose downregulation resulted in reduced pathogen propagation within the cells, irrespective of the Mycobacterium strain they tested. Further, these investigators demonstrated that upon infection with M. tuberculosis the expression of certain genes, whose earlier downregulation was shown to result in lower M. tuberculosis replication capacity are indeed induced; a fact that may indicate this intracellular pathogen directs creation of a tolerating or permissive milieu within the host cell. In order to verify whether similar changes happen when the cells encounter different intracellular pathogens, a subset of the aforementioned genes were selected and their expression behaviors were studied using qPCR in differentiated THP-1 cells infected with B. melitensis. Previous research works show that Brucella and Mycobacterium survive within macro-phages by limiting the fusion of lysosomes with the phagosomes harboring the bacteria (Yuk et al., 2012 ▶; Mostowy, 2013 ▶; Huang and Brumell, 2014 ▶). Therefore, for therapeutic purposes it would be interesting to verify whether, irrespective of pathogen type, common pathways may be triggered by these intracellular pathogens to be used as common drug target(s).
Materials and Methods
Candidate gene selection
Using RNAi to knockdown genes in differentiated THP-1 cells infected with M. tuberculosis, Kumar et al. (2010) identified a list of 74 genes whose down-regulation restricted M. tuberculosis propagation irrespective of the infecting bacterial strain. These investigators further showed that a subset of these genes are upregulated upon THP-1 infection with M. tuberculosis, suggesting a mechanism by which M. tuberculosis induces expression of host factors that favor its survival and propagation within the host cells. Having in mind the goal of pinpointing possible therapeutic targets, from the list of 74 genes identified by Kumar et al. (2010) ▶ we chose four genes, i.e. EMP2 (epithelial membrane protein 2) and ST8SIA4 (2-8 sialyl transferase), HCP5 (HLA complex P5), FRMD5 (FERM domain 5) that, based on available knowledge did not seem essential for macrophage function and their downregulation had resulted in over 50% reduction in M. tuberculosis propagation within THP-1 cells.
Internal control gene selection for qPCR
Based on the data from Mae et al. (2010) ▶ that indicated ACTB and RPL37A as the most stable genes upon THP-1 differentiation among 21 other potential internal control genes and the data from Cao et al. (2012) ▶ that showed PPIB and PGK1 as the most stable genes during THP-1 stimulation with LPS, we chose RPL37A and PPIB as potential normalizers for this work. Subsequently, based on the expression values for these two genes in our THP-1 cells, PPIB was selected to normalize the qPCR data.
Cell culture, macrophage differentiation and infection
The THP-1 human monocytic cell line was obtained from the Persian Type Culture Collection (Pasteur Inst., Iran). Brucella melitensis was obtained from the Microbiology Lab at the School of Veterinary Medicine, Shiraz University, Shiraz, Iran. This bacterium had been isolated from an aborted sheep fetus and biochemically characterized in the aforementioned lab.
The THP-1 cells were maintained and cultured in RPMI 1640 (GIBCO) supplemented with 10% fetal bovine serum (FBS), 50 µM 2-mercaptoethanol, 5 µM L-glutamine and 100 IU/100 µg/ml penicillin/streptomycin at 37°C in a tissue culture incubator. The media were refreshed every 2-3 days and the cells were split before the cell number reached 1 × 106/ml. When the passage number reached 10, a new frozen vial was used to start a fresh culture. On each round of experiment, THP-1 cells were counted in the presence of trypan blue to evaluate cell viability and six tissue cultures dishes (35 mm) were seeded with 5 × 105 cells in 2 ml complete RPMI containing 5 ng/ml phorbol 12-myristate 13-acetate (PMA) (Cat # P 1585, SIGMA). The following day, when the differentiated cells had become adherent as confirmed by light microscopy, the cells were washed thrice gently with warm RPMI without antibiotic. The bacteria (B. melitensis) were grown to late logarithmic phase (OD600 = 1) in Tryptic Soy Broth (Merck, Germany), centrifuged at 12000 RPM for 10 min and resuspended in warm RPMI 1640 without antibiotic. For the test group, the media containing the bacteria were added to three cultures at the multiplicities of infection (MOI) of 10. The infected plates were centrifuged at 500 g for 10 min and were subsequently transferred to a tissue culture incubator. After 4 h of incubation at 37°C, the cells were gently washed thrice with complete RMPI containing 100 µg/ml gentamicin followed by 2 h of incubation in the same medium to kill extracellular bacteria. The cells were subsequently washed with complete RPMI containing 10 µg/ml gentamicin and were incubated in the same medium until used to extract RNA at ~20 h post infection. The remaining cultures were used as controls and hence were treated exactly as the infected cultures were, but without being exposed to the bacteria. Each round of experiment contained three replicates for each condition (i.e. infected and uninfected), and the whole experiment was also independently repeated two more times.
RNA isolation and cDNA synthesis
Cells were used to extract total RNA using Qiagen RNeasy plus Mini kit (Cat # 74134 Qiagen, Germany) that included a built-in on-column DNase treatment step. The extracted RNAs were qualitatively and quantitatively assessed by, respectively, running on agarose gel and spectrophotometry. The extracted RNAs were stored at -80°C until used.
For cDNA synthesis PrimeScript kit (perfect real time) (Cat # RR037A Takara, Japan) and the provided instruction was used. For each sample, 300 ng RNA with 25 picomole oligodT and 50 picomole random hexamer in 10 µL final volume was reverse transcribed. The cDNA product was diluted one in half and transferred to -20°C in aliquots until used.
Primer design and qPCR experiment
The primers were designed using Primer3 software (Untergasser et al., 2012 ▶) (Table 1). All primer pairs were designed to have similar melting temperatures and were checked for not having common SNPs using UCSC genome browser (Karolchik et al., 2003 ▶). The primers were synthesized by Bioneers Ltd. (South Korea). The primers were tested by running a melt curve analysis and also running the related PCR products on agarose gel to confirm the products size. In order to determine the best matching primer set concentrations, a primer optimization procedure was conducted for each set of the primer sets (Table 2) and the efficiencies of amplification were also determined by standard cure method.
Table 1.
Specifications of the primers used in this work
| Gene | Primers sequences | Tm (°C) | Product size (bps) |
|---|---|---|---|
| EMP2 | F: CTGTGTGTCATGATTGCGGC R: ATCATGCCGCTGATGAAGGT |
60.18 59.82 |
158 |
| ST8SIA4 | F: CTGGGGCAACCAGGACTTTC R: ATGAGTTGCGTCTCCTGGTG |
60.90 60.04 |
143 |
| HCP5 | F: AAGGAGAGTTGATCAAGGCCG R: AGGCCCTACTTCTCTCAGGC |
60.07 60.69 |
145 |
| FRMD5 | F: CAATGAGCGAGTAGCTGTG R: TCCTCCTCAATGCTGCAGGT |
59.35 61.20 |
142 |
| PPIB | F: AGATGTAGGCCGGGTGATCT R: CTCCGCCCTGGATCATGAAG |
60.11 60.25 |
152 |
Table 2.
Primer optimization data. The best performing concentrations for each primer sets as determined in our work are presented
| Primer set | Optimum concentration (nM) |
|---|---|
| EMP2 | F:400 / R:600 |
| FRMD5 | F:400 / R:600 |
| HCP5 | F:400 / R:600 |
| PPIB | F:600 / R:300 |
| ST8SIA4 | F:600 / R:400 |
Quantitative PCR was conducted on a Miniopticon instrument (BioRad, USA) using the SYBR Premix Ex Taq II (Cat # RR820Q, Takara, Japan) according to the instructions provided by the manufacturer. qPCR cycling conditions were as follows: 30 s pre-incubation at 95°C, 40 cycles of 95°C/5 s and 58°C/30 s. After completion of the amplification step a melting curve analysis was also conducted.
Statistical analysis
The differences in expression values between test and control conditions for each gene were statistically analyzed using REST software (http://rest.gene-quantification.info) (Pfaffl et al., 2002 ▶). A cut-off of P<0.05 was considered as significant.
Results
Reference gene selection
As explained in the materials and methods section, based on published data (Maeß et al., 2010 ▶; Cao et al., 2012 ▶) PPIB and RPL37A genes were initially selected as potential reference genes for the current work. A qPCR experiment was conducted using the primer sets amplifying PPIB or RPL37A and after analyzing the expression values for these two genes, PPIB was chosen over RPL37A for this work. This is because RPL37A was highly expressed in THP-1 cells and PPIB expression level was within the same range as the other four experimental genes.
Differential regulation of EMP2 , ST8SIA4 , HCP5 and FRMD5 in THP-1 macrophages infected with Brucella melitensis
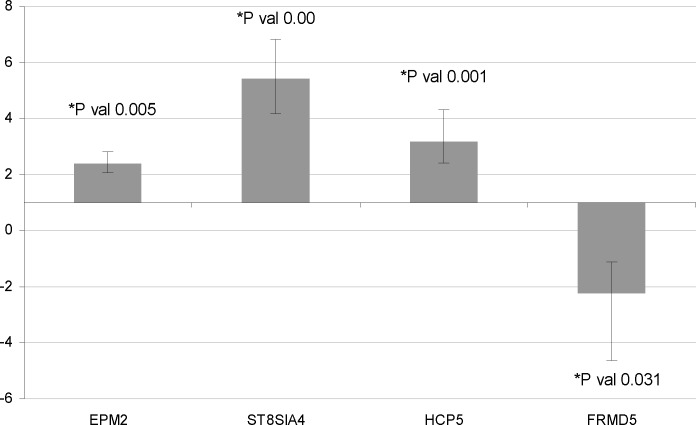
qPCR data for EMP2, ST8SIA4, HCP5 and FRMD5 showed that these genes are significantly dysregulated upon THP-1 infection with B. melitensis (Fig. 1). EMP2 (Fold change (FC) = 2.402, P=0.005), ST8SIA4 (FC = 5.421, P=0.00) and HCP5 (FC = 3.18, P=0.001) are upregulated while FRMD5 (FC = -2.222, P=0.031) is downregulated when the THP-1 macrophages are infected with B. melitensis.
Fig. 1.
qPCR data on genes changed by THP-1 macrophages infected with B. melitensis. The qPCR analysis of four genes in THP-1 macrophages infected with B. melitensis showed that compared to the uninfected cells, three of them were significantly upregulated while one gene was significantly downregulated. Bars show standard error of mean
Discussion
The characteristics of eukaryotic cells and their intracellular conditions are governed by their functional genomics. Therefore, it is rational to expect that intracellular dwelling parasites must have developed strategies to change their host properties to their advantage by inducing changes in the functional genomic aspects of their host. In this line, Kumar et al. (2010) ▶ took an interesting approach to interrogate the aspects of the transcriptome of THP-1 macrophages that influence the persistence of the infection caused by M. tuberculosis. These investigators showed that many hundreds of the host’s genes were involved in the process of development and persistence of the infection, and therefore manipulating these factors may be helpful in maintaining the infection under control. Interestingly, they verified that several of the genes that had earlier shown their downregulation by RNAi suppressed the progress of the infection were indeed upregulated in the infected cells, implying M. tuberculosis induces changes in the host’s transcriptome in favour of its survival and replication. Here, we chose B. melitensis with similar life cycle to that of Mycobacterium regarding its persistence in phagosomes and changes to the levels of expression of four genes, i.e. EMP2, ST8SIA4, HCP5 and FRMD5 taken from Kumar’s study were determined in the infected THP-1 macrophages. The goal here was to verify whether similar alterations in gene transcription may be induced by these two different intracellular pathogens. Our data confirmed a similar pattern in expression changes in two of the selected genes, i.e. EMP2 and ST8SIA4 between THP-1 cells infected with B. melitensis and those infected with M. tuberculosis as reported by Kumar et al. (2010) ▶. Based on the previous studies, B. melitensis and M. tuberculosis use similar strategies to survive within the host by limiting the fusion of phagosomes harbouring them with lysosomes (Arenas et al., 2000 ▶; Knodler et al., 2001 ▶; Yuk et al., 2012 ▶). Therefore, the genes commonly changed in THP-1 cells infected with either B. melitensis or M. tuberculosis may be involved in some stages of normal phagosome maturation such as fusion with lysosomes.
Kumar et al. (2010) ▶ showed that EMP2 is not only upregulated when THP-1 cells are infected with M. tuberculosis, but its downregulation by RNAi restricts M. tuberculosis replication within these cells. Our data also showed that this gene is upregulated in THP-1 cells infected with B. melitensis (Fig. 1). EMP2 is a transmembrane protein and is a member of the GAS3/PMP22 subfamily that is involved in the delivery of certain proteins such as integrin isoform to the cell surface (Wadehra et al., 2005 ▶; Shimazaki et al., 2009 ▶). EMP2 is reported to be highly expressed at membrane sites of Chlamydia infection and blockage of this protein using anti-EMP2 antibody reduces the infectability of the genital tract of mouse model by Chlamydia (Shimazaki et al., 2009 ▶). Similarly, it has been shown that the HEC-1A cell line with reduced expression of EMP2 is refractory to infection by Chlamydia, but overexpression of this protein in this cell line makes it amenable to infection (Shimazaki et al., 2007 ▶). In the RNAi experiment conducted by Kumar et al. (2010) ▶, it was demonstrated that EMP2 downregulation limits the propagation capacity of M. tuberculosis within THP-1 cells. These investigators designed their experiment so that they targeted the gene after M. tuberculosis had entered the cells. Therefore, this design ruled out any possible influence from reduced THP-1 cells infectability by lower surface expression of EMP2 and suggested that this gene may be active somewhere in post-infection stages of Mycobacterium life cycle. Moreover, they showed that the expression level of EMP2 gene was, indeed, upregulated after THP-1 cells had been infected with Mycobacterium, indicating positive influence on Mycobacterium survival and propagation from EMP2 after infection. Interestingly, it has been shown that macrophages infected with Mycobacterium and activated with INFγ show enhanced killing activity (Gutierrez, 2004 ▶; Herbst, 2011 ▶) while INFγ stimulation of cells results in downregulation of EMP2 expression (Kumar et al., 2010 ▶). Therefore, it seems reasonable to assume that INFγ accomplishes its antibacterial effect at least partly through modulating the expression of a variety of genes, such as EMP2, to overtake the suppression state already induced by some bacteria.
ST8SIA4 encodes for a protein that is involved in the synthesis of polysialic acid (Nakayama et al., 1995 ▶). This gene that is overexpressed in whole blood compared to its average expression in all tissues shows substantial expression in immune related cells such as monocytes, neutrophils and B and T cells (Safran et al., 2010 ▶; Montague et al., 2014 ▶; Uhlén et al., 2015). In our experiment ST8SIA4 is upregulated in THP-1 macrophages infected with B. melitensis as it was also reported to be overexpressed in THP-1 macrophages infected with M. tuberculosis (Kumar et al., 2010 ▶). It has also been shown that upon infection with some viruses such as the SARS-associated coronavirus, the infected macrophages endure overexpression of ST8SIA4 (Peiris and Cheung, 2009 ▶). Downregulation of ST8SIA4 by RNAi lowers the capacity of THP-1 macrophages in supporting propagation of intracellular M. tuberculosis (Kumar et al., 2010 ▶). As mentioned earlier INFγ stimulation of the macrophages infected with M. tuberculosis increases the capacity of the infected cells in overtaking the suppression state induced by the bacteria (Gutierrez et al., 2004 ▶; Kumar et al., 2010 ▶; Herbst et al., 2011 ▶). Similar to the case for EMP2, the cells stimulated with INFγ undergo a downregulation in ST8SIA4 expression levels as well (Kumar et al., 2010 ▶). Moreover, the notions that for survival within macrophages, Brucella intercepts membrane trafficking from endoplasmic reticulum to Golgi apparatus (Atluri et al., 2011 ▶), and that ST8SIA4 is localized to Golgi apparatus (Angata and Fukuda, 2014 ▶) may suggest some other molecular mechanisms for the involvement of this gene in pathogenesis of Brucella.
Our data show that HCP5 gene is upregulated in THP-1 cells infected with B. melitensis. HCP5 codes for a non-coding RNA that is structurally related to human endogenous retroviruses (HERVs) group of transposable elements (Kulski and Dawkins, 1999 ▶). HCP5 locus is positioned within the MHC class I region and the sequence shows some homology to retroviral pol gene. Some genomic variations in this locus have been associated with viral load in HIV infection, and hence with delay in development of AIDS (Fellay et al., 2007 ▶), as well as with susceptibility to development of herpes zoaster (Crosslin et al., 2015 ▶).
Based on our data, in THP-1 macrophages infected with B. melitensis the expression level of FRMD5 gene is downregulated by over two-fold (Fig. 1). Except for some data on the association of FRMD5 and tumor progression (Wang et al., 2012 ▶) little information is available on the possible function of FRMD5 in development and progression of infection. It has been shown that FRMD5 shares 51% identity with another family member, i.e. FRMD3 (Susana et al., 2013 ▶) and that FRMD3 overexpression induces apoptosis (Haase et al., 2007 ▶). Therefore, if a similar pro-apoptotic function could be attributed to FRMD5, the notion that infection of human monocytes with B. suis inhibits apoptosis (Gross et al., 2000 ▶) can best be used to envision the interrelationship between bacterial infection, FRMD5 downregulation and apoptosis. Otherwise, in infected cells apoptosis would limit the bacteria propagation and persistence within the host. On the other hand, Kumar et al. (2010) ▶ demonstrate positive influence from FRMD5 downregulation by RNAi on bacterial replication within THP-1 macrophages. Though it may be argued that the level of suppression may have an influence, the scientific reason for this apparent contradiction remains to be explored.
Taken together, based on the previous studies and the data from this work it is obvious that intracellular pathogenic bacteria influence their host’s transcriptome for their own benefit. Therefore, taking approaches to revert the changes induced by these pathogens can prove beneficial for prophylactic and/or therapeutic purposes. Moreover, though the changes induced by the bacteria can be type specific, our data shows that the bacteria with similar life style such as Mycobacterium spp. and Brucella spp. direct overlapping molecular events that can be considered as common therapeutic targets.
Acknowledgment
This work was financially supported by School of Veterinary Medicine, Shiraz University, Shiraz, Iran.
Conflict of interest
The authors declare no conflict of interest.
References
- Angata K, Fukuda M. ST8 alpha-N-acetyl-neuraminide alpha-2, 8-Sialyltransferase 4 (ST8SIA4) In: Taniguchi, N, Honke, K, Fukuda, M, Narimatsu, H, Yamaguchi, Y, Angata, T, editors. Handbook of glycosyltransferases and related genes. 2nd Edn. Japan: Springer; 2014. pp. 805–812. [Google Scholar]
- Arenas GN, Staskevich AS, Aballay A, Mayorga LS. Intracellular trafficking of Brucella abortus in J774 macrophages. Infect. Immun. 2000;68:4255–4263. doi: 10.1128/iai.68.7.4255-4263.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atluri VL, Xavier MN, de Jong MF, den Hartigh AB, Tsolis RM. Interactions of the human pathogenic Brucella species with their hosts. Annu. Rev. Microbiol. 2011;65:523–541. doi: 10.1146/annurev-micro-090110-102905. [DOI] [PubMed] [Google Scholar]
- Barbier T, Nicolas C, Letesson J. Brucella adaptation and survival at the crossroad of metabolism and virulence. FEBS Lett. 2011;585:2929–2934. doi: 10.1016/j.febslet.2011.08.011. [DOI] [PubMed] [Google Scholar]
- Cao XM, Luo XG, Liang JH, Zhang C, Meng XP, Guo DW. Critical selection of internal control genes for quantitative real-time RT-PCR studies in lipopolysaccharide-stimulated human THP-1 and K562 cells. Biochem. Bioph. Res. Co. 2012;427:366–372. doi: 10.1016/j.bbrc.2012.09.066. [DOI] [PubMed] [Google Scholar]
- Carryn S, Van Bambeke F, Mingeot-Leclercq MP, Tulkens PM. Comparative intracellular (THP-1 macrophage) and extracellular activities of β-lactams, azithromycin, gentamicin, and fluoroquinolones against Listeria monocytogenes at clinically relevant con-centrations. Antimicrob. Agents Chemo. 2002;46:2095–2103. doi: 10.1128/AAC.46.7.2095-2103.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosslin D, Carrell D, Burt A, Kim D, Underwood J, Hanna D, Comstock B, Baldwin E, de Andrade M, Kullo I. Genetic variation in the HLA region is associated with susceptibility to herpes zoster. Genes Immun. 2015;16:1–7. doi: 10.1038/gene.2014.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellay J, Shianna KV, Ge D, Colombo S, Ledergerber B, Weale M, Zhang K, Gumbs C, Castagna A, Cossarizza A. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–947. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grilló MJ, Blasco JM, Gorvel JP, Moriyón I, Moreno E. What have we learned from brucellosis in the mouse model. Vet. Res. 2012;43:1–35. doi: 10.1186/1297-9716-43-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross A, Terraza A, Ouahrani-Bettache S, Liautard JP, Dornand J. In vitro Brucella suis infection prevents the programmed cell death of human monocytic cells. Infect. Immun. 2000;68:342–351. doi: 10.1128/iai.68.1.342-351.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- Haase D, Meister M, Muley T, Hess J, Teurich S, Schnabel P, Hartenstein B, Angel P. FRMD3, a novel putative tumour suppressor in NSCLC. Oncogene. 2007;26:4464–4468. doi: 10.1038/sj.onc.1210225. [DOI] [PubMed] [Google Scholar]
- He Y. Analyses of Brucella pathogenesis, host immunity, and vaccine targets using systems biology and bioinformatics. Front. Cell. Infect. Microb. 2012;2:1–6. doi: 10.3389/fcimb.2012.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbst S, Schaible UE, Schneider BE. Interferon gamma activated macrophages kill Mycobacteria by nitric oxide induced apoptosis. PloS one. 2011;6:e19105. doi: 10.1371/journal.pone.0019105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Brumell JH. Bacteria-autophagy interplay: a battle for survival. Nat. Rev. Microb. 2014;12:101–114. doi: 10.1038/nrmicro3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbuluzqueta E, Gamazo C, Ariza J, Blanco-Prieto MJ. Drug delivery systems for potential treatment of intracellular bacterial infections. Front. Biosci. 2010;15:397–417. doi: 10.2741/3627. [DOI] [PubMed] [Google Scholar]
- Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu Y, Roskin KM, Schwartz M, Sugnet CW, Thomas DJ. The UCSC genome browser database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knodler LA, Celli J, Finlay BB. Pathogenic trickery: deception of host cell processes. Nat. Rev. Mol. Cell Biol. 2001;2:578–588. doi: 10.1038/35085062. [DOI] [PubMed] [Google Scholar]
- Kulski JK, Dawkins RL. The P5 multicopy gene family in the MHC is related in sequence to human endogenous retroviruses HERV-L and HERV-16. Immunogenetics. 1999;49:404–412. doi: 10.1007/s002510050513. [DOI] [PubMed] [Google Scholar]
- Kumar D, Nath L, Kamal MA, Varshney A, Jain A, Singh S, Rao KV. Genome-wide analysis of the host intracellular network that regulates survival of Mycobacterium tuberculosis. Cell. 2010;140:731–743. doi: 10.1016/j.cell.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Maeß MB, Sendelbach S, Lorkowski S. Selection of reliable reference genes during THP-1 monocyte differentiation into macrophages. BMC Mol. Biol. 2010;11:90. doi: 10.1186/1471-2199-11-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montague E, Stanberry L, Higdon R, Janko I, Lee E, Anderson N, Choiniere J, Stewart E, Yandl G, Broomall W. MOPED 2. 5—an integrated multi-omics resource: multi-omics profiling expression database now includes transcriptomics data. Omics: J. Integ. Biol. 2014;18:335–343. doi: 10.1089/omi.2014.0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostowy S. Autophagy and bacterial clearance: a not so clear picture. Cell. Microbiol. 2013;15:395–402. doi: 10.1111/cmi.12063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Fukuda MN, Fredette B, Ranscht B, Fukuda M. Expression cloning of a human polysialyltransferase that forms the polysialylated neural cell adhesion molecule present in embryonic brain. Proc. Natl. Acad. Sci. U. S. A. 1995;92:7031–7035. doi: 10.1073/pnas.92.15.7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J, Cheung C. The macrophage in the pathogenesis of severe acute respiratory syndrome coronavirus infection. Med. J. Hong Kong. 2009;15:21–25. [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roop II RM, Gaines JM, Anderson ES, Caswell CC, Martin DW. Survival of the fittest: how Brucella strains adapt to their intracellular niche in the host. Med. Microbiol. Immunol. 2009;198:221–238. doi: 10.1007/s00430-009-0123-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safran M, Dalah I, Alexander J, Rosen N, Stein TI, Shmoish M, Nativ N, Bahir I, Doniger T, Krug H. GeneCards version 3: the human gene integrator. Database (Oxford) 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimazaki K, Chan AM, Moniz RJ, Wadehra M, Nagy A, Coulam CP, Mareninov S, Lepin EM, Wu AM, Kelly KA. Blockade of epithelial membrane protein 2 (EMP2) abrogates infection of Chlamydia muridarum murine genital infection model. FEMS Immunol. Med. Microbiol. 2009;55:240–249. doi: 10.1111/j.1574-695X.2008.00525.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimazaki K, Wadehra M, Forbes A, Chan AM, Goodglick L, Kelly KA, Braun J, Gordon LK. Epithelial membrane protein 2 modulates infectivity of Chlamydia muridarum (MoPn) Microb. Infect. 2007;9:1003–1010. doi: 10.1016/j.micinf.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Susana M, Andrew TL, Liselotte A, Frank GM, Paul AR. The expanding family of FERM proteins. Biochem. J. 2013;452:183–193. doi: 10.1042/BJ20121642. [DOI] [PubMed] [Google Scholar]
- Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A. Tissue-based map of the human proteome. Science. 2015;347:1260419–1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadehra M, Forbes A, Pushkarna N, Goodglick L, Gordon LK, Williams CJ, Braun J. Epithelial membrane protein-2 regulates surface expression of αvβ3 integrin in the endometrium. Dev. Biol. 2005;287:336–345. doi: 10.1016/j.ydbio.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Wang F, Hu S, Liu W, Qiao Z, Gao Y, Bu Z. Deep-sequencing analysis of the mouse transcriptome response to infection with Brucella melitensis strains of differing virulence. PloS one. 2011;6:e28485. doi: 10.1371/journal.pone.0028485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Pei X, Zhan J, Hu J, Yu Y, Zhang H. FERM-containing protein FRMD5 is a p120-catenin interacting protein that regulates tumor progression. FEBS Lett. 2012;586:3044–3050. doi: 10.1016/j.febslet.2012.07.019. [DOI] [PubMed] [Google Scholar]
- Yuk JM, Yoshimori T, Jo EK. Autophagy and bacterial infectious diseases. Exp. Mol. Med. 2012;44:99–108. doi: 10.3858/emm.2012.44.2.032. [DOI] [PMC free article] [PubMed] [Google Scholar]