Abstract
This study aims at molecular identification of Salmonella Infantis isolated from backyard chickens and the detection of their antibiotic resistance genes. A total of 46 Salmonella-suspected samples isolated from backyard chickens of northern Iran were collected. Serotyping was done by the traditional method and then confirmed by PCR. Antimicrobial susceptibility of the isolates against 13 antimicrobial agents was determined by the standard disk diffusion method. There were 44 samples identified as Salmonella. Serotyping results showed that all 44 isolates belonged to serogroup C1 and serovar Infantis. The most resistance observed was to tetracycline and doxycycline (100%), chloramphenicol (79%) and florfenicol (72%). The floR, catI, tetA and tetG genes were used for the detection of florfenicol chloramphenicol and tetracycline resistance. In order to identify the phenotypic resistance in strains which showed resistance genes by PCR, colony PCR and culture on plates each containing antibiotic was performed simultaneously. All the Salmonella Infantis resistant to florfenicol and chloramphenicol harbored floR and catI. None of the Salmonella resistant to tetracycline carried tetA or tetG. The result of colony PCR and culture in antibiotic medium confirmed the results of PCR and indicated phenotypic resistance in these samples.
Key Words: Backyard chickens, Resistance genes, Salmonella Infantis
Introduction
Salmonella which can be isolated from numerous animal species is a Gram-negative rod-shaped bacterium in the family of Entrobacteriacea. The intestinal tract is the primary reservoir of these zoonotic bacteria where colonisation is favoured by intensive animal production. Poultry products are frequent vehicles in the transmission of Salmonella dominating other foods of animal origin as a potential source of infection (Antunes et al., 2003 ▶; Shahada et al., 2006 ▶). In addition to its zoonotic importance, the economic loss in poultry industry is also significant (Zahraei Salehi et al., 2005a ▶; Nogrady et al., 2007 ▶). Infected animals are a threat to others and should be identified and separated so as to prevent the disease from spreading. Therefore, rapid identification of this pathogen has to be performed (Zahraei Salehi et al., 2005a ▶). The identification of Salmonella serovars by slide and tube agglutination tests using O and H antigen-specific anti-sera is both hard and time-consuming (Akiba et al., 2011 ▶). Molecular methods because of their specificity, rapidity and simplicity compared with traditional serotyping prove strong for this detection.
Salmonella Infantis is a host-unspecific serovar that can infect human and numerous animal species. Salmonellosis by this serovar in human mainly affects children but adults also suffer from it. The major symptom of the disease is septicaemia and the significant feature of this germ is its persistence in hospitals over a long period of time (Ranjbar et al., 2012 ▶).
In recent years, the incidence of resistance to antimicrobial agents among pathogens has been steadily rising in food as well as in clinical isolates (Van Hoeka et al., 2005 ▶). Food-producing animals are administered antibiotics for therapeutic, prophylactic, and production purposes to promote animal health, welfare, their growth rate and feed conversion (Schwarz and Chaslus-Dancla, 2001 ▶). This widespread use of antimicrobial agents in food animal production has contributed to the occurrence of resistant bacteria in animals, including principle zoonotic pathogens such as Salmonella (Shahada et al., 2006 ▶). Since these pathogens are difficult to treat, antibiotic resistance associated with food-borne diseases has become a major public health issue (Van Hoeka et al., 2005 ▶).
The occurrence of antibiotic resistance genes is increasing in Salmonella Infantis the same as other serovars. There are reports of drug resistance in Salmonella Infantis isolates of poultry origin in many countries such as Turkey, Hungary and Japan (Shahada et al., 2006 ▶; Nogrady et al., 2007 ▶; Abbasoglu and Akcelik, 2011 ▶) but there is little knowledge about this in poultry of Iran.
Considering the importance of Salmonella Infantis in backyard chickens and its potential to be transmitted to human via food chain, we tried to identify Salmonella Infantis from chicken samples by traditional serotyping and PCR. Besides, the antimicrobial susceptibility test and antibiotic resistance genes detection were carried out.
Materials and Methods
Sample collection and serotyping
46 Salmonella-suspected samples isolated from backyard chickens in the north of Iran, Mazandaran province, were collected. They were cultured in McConkey and Salmonella Shigella agar (Merck, Germany). The suspected colonies on these two media were cultured in Chrom agar (Merck, Germany) and then were identified by Urea and TSI medium (Merck, Germany).
For serotyping, the Salmonella isolates were first cultured on to TSI slant medium and grown overnight at 37°C, and then were tested using antisera O (B, D, C1 to C4) and H (Difco, USA) based on slide and tube agglutination tests to determine O and H antigens, respectively (Waltman et al., 1998 ▶).
DNA extraction
A single colony of each isolate on LB agar (Merck, Germany) plate was picked up and suspended in 250 µL of distilled water. After vortexing, the suspension was boiled for 10 min and 100 µL of the supernatant was collected after centrifuging at 6000 × g for 7 min.
PCR amplification of fljB gene for the identifica-tion of Salmonella Infantis
Molecular identification of Salmonella Infantis was performed as described previously (Kardos et al., 2007 ▶). In this study, two primer pairs were used to detect the fljB gene in Salmonella Infantis (Table 1). The reaction was carried out in a volume of 25 µL which contained 8 mM MgCl2 (Sinaclon, Iran), 200 µM dNTPs (Sinaclon, Iran), 0.2 µM each primer (Sinaclon, Iran), 10 X PCR buffer (Sinaclon, Iran) and 1 U Taq DNA polymerase (Sinaclon, Iran). The amplification program was done by thermocycler (Techne TC-512, UK) as follows: 95°C initial denaturation for 6 min, 35 cycles of 95°C for 1 min, 58°C for 15 s, 72°C for 1 min and a final amplification at 72°C for 4 min. The amplified products were electrophoresed on a 1.2% agarose gel (Sinaclon, Iran) stained with ethidium bromide (Sinaclon, Iran) and visualized under ultraviolet light.
Table 1.
Primers used in this study
| Primers | Target gene | Primer sequence (5´ to 3´) | Amplification product (bp) |
|---|---|---|---|
| 558f 1275r |
fljB | AACAACGACAGCTTATGCCG CCACCTGCGCCAACGCT |
727 |
| 878f 1275r |
fljB | TTGCTTCAGCAGATGCTAAG CCACCTGCGCCAACGCT |
413 |
| tetA | tetA | GCTACATCCTGCTTGCCT CATAGATCGCCGTGAAGA |
210 |
| tetG | tetG | CCGGTCTTATGGGTGCTCTA CCAGAAGAACGAAGCCAGTC |
603 |
| floR | floR | AACCCGCCCTCTGGATCAAGTCAA CAAATCACGGGCCACGCTGTATC |
548 |
| cat1 | Cat1 | CCTATAACCAGACCGTTCAG TGAAACTCACCCAGGGATTG |
370 |
Antibiotic susceptibility testing
Antibiotic susceptibility of the isolates was determined by the disc diffusion method (Kirby-Bauer, 1996) on Mueller-Hinton agar using antibiotic discs (MAST, UK). The following antimicrobial agents were tested: 20 μg ampicillin (AMP), 30 µgcefazolin (CEF), 30 µgcefotaxime (CTX), 5 µgcefixime (CFM), 30 µgceftraxion (CRO), 5 µgenrofloxacin (ENF), 25 µg difloxacin (DIF), 30 μg chloramphenicol (CHL), 30 µgflorfenicol (FFC), 30 μg tetracycline (TE), 30 µgdoxycycline (DXT), 10 μg gentamicin (G), and 25 μgtrimethoperim-sulfamethoxazol (SXT). The inhibition zones were measured and scored as sensitive, intermediate susceptibility or resistant according to the CLSI recommendations. Escherichia coli ATCC 25922 was used as a reference strain for antibiotic disc control.
Detection of resistance genes
Antimicrobial resistance genes were characterized by polymerase chain reaction. PCR amplification of tetA, tetG and floR genes was performed as described previously (Randall et al., 2004 ▶; Abbasoglu and Akcelik, 2011 ▶).
For the detection of cat gene, primer sequences were based on the sequence in GeneBank (accession No. LK056646.1). Each 25 μL of reaction mixture contained 10 X PCR buffer, 2 mM MgCl2, 0.5 μL of 10 mMdNTPs, 0.4 μM of each of the primer, 1 U of Taq DNA polymerase and 1 μL of DNA template. The PCR amplification involved 30 cycles of denaturation at 95°C for 1 min, annealing at 58°C for 1 min, and elongation at 72°C for 1 min in a thermocycler (Techne TC 512, UK).
For the identification of phenotypic resistance in strains which showed resistance genes by PCR, colony PCR and culture on plates, each containing antibiotic, was performed simultaneously. PCR from colony and plating bacteria from the same colony on LB agar plates containing chloramphenicol (20 µg/ml) (Sigma, Aldrich, USA) and florfenicol (30 µg/ml) (SERVA, Heidelberg) was performed. The concentration of each antibiotic in culture medium was considered according to CLSI standards (2011).
Results
Forty-four samples were confirmed as Salmonella based on the cultural and biochemical methods. The results of serotyping with O and H antisera demonstrated that all the samples belonged to serogroup C1 and serovar Infantis with the antigenic formula of 6,7: r: 1,5.
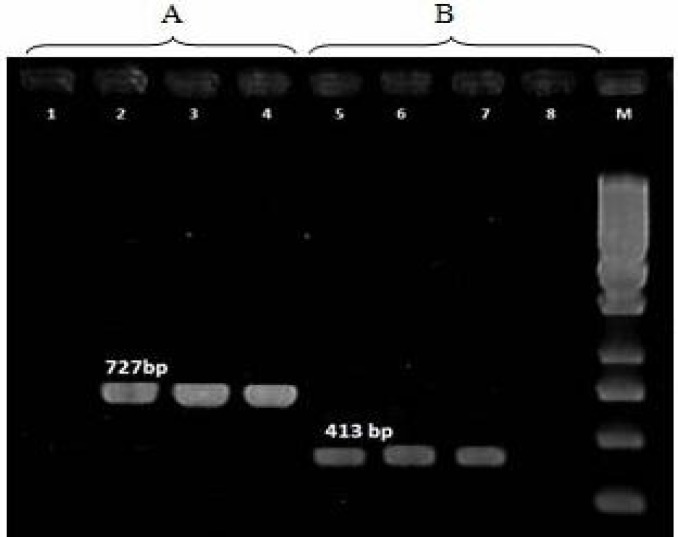
Both assays which targeted fljB gene of Salmonella Infantis illustrated products of the expected size in all 44 S. Infantis isolates (Fig. 1).
Fig. 1.
Results of PCR assays used to identify S. Infantis with the primer pairs 558f-1275r (A) and 878f-1275r (B). M: 250 bp ladder. Lane 1: S. Paratyphi C as negative control, Lane 2: Positive control, Lanes 3-4: Positive detection, Lane 5: Positive control, Lanes 6-7: Positive detection, and Lane 8: S. Paratyphi C as negative control
The most observed resistance resistance was to tetracycline (100%), doxycycline (100%), chloramphenicol (79%) and florfenicol (72%) respectively. All the isolates were sensitive to other antibiotics with two exceptions. Of those two exceptions, one was resistant to gentamicin and both of them were resistant to enrofloxacin.
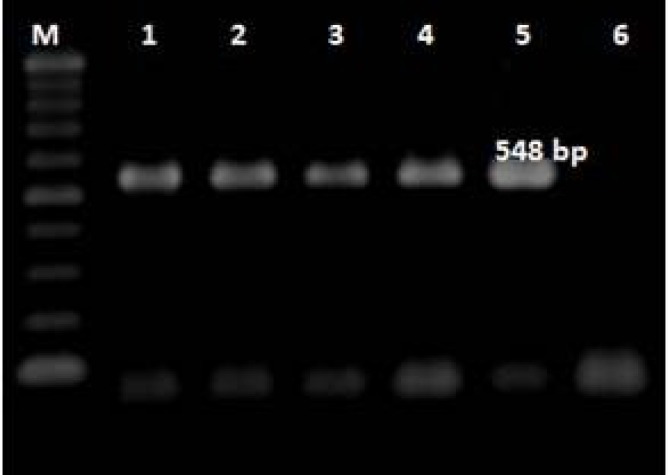
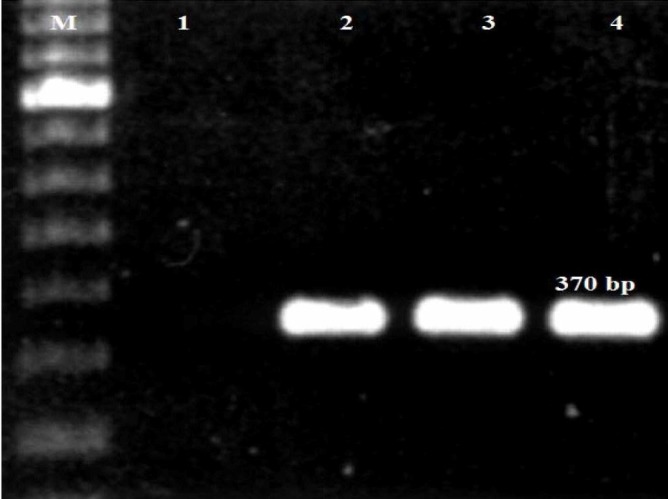
All the S. Infantis isolates resistant to florfenicol contained floR gene (Fig. 2) and the chloramphenicol-resistant isolates showed resistance gene cat1 with the expected bands (Fig. 3).
Fig. 2.
Results of PCR assays for identification of floR gene. M: 100 bp ladder. Lane 1: Positive control, Lanes 2-5: Positive detection, and Lane 6: Negative control
Fig. 3.
Results of PCR assays for identification of cat1 gene. M: 100 bp ladder. Lane 1: Negative control, Lane 2: Positive control, and Lanes 3-4: Positive detection
None of the tetracycline and doxycycline-resistant strains carried tetA or tetG gene.
For the detection of phenotypic resistance, PCR and culture in antibiotic medium was performed at the same time. Their outcome confirmed the PCR results and demonstrated phenotypic resistance in these samples.
Discussion
Salmonella enterica is one of the most important food-borne pathogens throughout the world (Nogrady et al., 2008 ▶). The prevalence of each serovar of Salmonella in human and animal is changing and one may replace another at any time (Zahraei Salehi, 1999 ▶). In this research, all the Salmonella isolates belonged to serovar Infantis. Previous studies showed that the most pre-dominant serogroups isolated from poultry in Iran were of D and B and the main serovars were Enteritidis and Typhimurium (Zahraei Salehi et al., 2005a ▶; Jaffari et al., 2007 ▶; Emaddi Chashni et al., 2009 ▶; Mirzaei et al., 2010 ▶). However, the present investigation indicates a higher prevalence of serogroup C1 and serovar Infantis. In European countries, the prevalence of S. Enteritidis and S. Typhimurium has decreased due to their vaccination programs but the serovar Infantis is still increasing and is a major problem in some countries such as Hungary (Miller et al., 2010 ▶).
The survey of Salmonella Infantis in hospitals of Iran indicates an increase in the prevalence of this serovar. Naghoni et al. (2010) ▶ and Ranjbar et al. (2011) ▶ found the serovar Infantis as the second most prevalent in the human cases of salmonellosis. In the survey conducted in the years 2007-2010, 19% of the isolates were S. Infantis (Ranjbar et al., 2012 ▶). Since the late 1970s, the incidence of the human cases of serovar Infantis has been increasingly recorded worldwide in countries like Argentina, Australia, Brazil, The Netherlands, Finland, Canada, Hungary, Japan, New Zealand and Russia (Miller et al., 2010 ▶). In recent years, the rate of Salmonella infections with this serovar among poultry has been increasing in some countries such as Hungary and Japan and is the main cause of salmonellosis in their poultry (Kardos et al., 2010 ▶; Miller et al., 2010 ▶). The reason for such a significant increase in humans is that this pathogen is food-borne and can be transmitted through poultry products.
Little is known about the presence of this serovar in the poultry of Iran and we have screened the backyard chickens for its the occurrence. Also, molecular methods have been used for rapid identification of Salmonella Infantis for the reasons mentioned above. The results of PCR confirmed the serotyping and all the Salmonella isolates were recognized as Salmonella Infantis.
Our study depicts a high percentage of antibiotic resistance in Salmonella Infantis isolated from poultry showing extensive usage of antibiotic in them while backyard flocks are not treated with antibiotics like the industrial poultry. Such a high percentage of antibiotic resistance observed in this research could be due to the transmission of resistant bacteria from industrial poultry by different vehicles such as human, free flying birds and poultry products (Emaddi Chashni et al., 2009 ▶).
One the common resistance observed in S. Infantis was to tetracycline and doxycycline. Tetracycline has been one of the most commonly used antibiotics for the production of animals. As a result, the very frequent occurrence of resistance among almost all bacterial species is probably a consequence of this (Gebreyes and Altier, 2002 ▶). The incidence of tetracycline resistance has been described recently by other authors in Salmonella isolates of poultry origin in Iran (Zahraei Salehi et al., 2005b ▶; Jafari et al., 2007 ▶; Mirzaei et al., 2010 ▶; Morshed et al., 2010 ▶).
The resistance to tetracycline is associated with tet genes. Six classes of genes including tetA, B, C, D, E, and G were identified responsible for resistance to tetracyclines. We have selected tetA and tetG genes for the identification of resistance to tetracycline. The isolates carried neither tetA nor tetG. These isolates may carry other tet genes that have not been studied here. TetA was the most common gene detected in poultry responsible for resistance to this antibiotic (Shahada et al., 2006 ▶; Nogrady et al., 2007 ▶; Abbasoglu and Akcelik, 2011 ▶). Salmonella Infantis isolated from human cases in Iran carried both tetA and tetB genes (Tajbakhsh et al., 2012 ▶). The study of antibiotic resistance genes in Brazil showed the presence of tetD gene in Salmonella Infantis isolated from human cases (Fonseca et al., 2006 ▶). The tetG gene was mostly detected in Salmonella Typhimurium in previous studies (Walker et al., 2001 ▶; Randall et al., 2004 ▶).
Resistance to chloramphenicol and florfenicol as antibiotics of the same family was seen in more than 70% of the isolates. Chloramphenicol used to be applied for the treatment of salmonellosis in animals but due to becoming resistant to this antibiotic, florfenicol was introduced. However, the resistance to this antibiotic soon appeared (Nogrady et al., 2005 ▶). In the previous studies, there was antibiotic resistance to these two antibiotics in poultry Salmonella isolates (Zahraei Salehi et al., 2005b ▶; Emaddi Chashni et al., 2009 ▶; Morshed and Peighambari, 2010 ▶) but it seems these days this resistance is on the increase. The florfenicol resistant strains can show cross resistance to chloramphenicol (Nogrady et al., 2005 ▶).
Cat1 gene encoding chloramphenicol acetyl transferase was detected in all the chloramphenicol resistant isolates and the florfenicol resistant strains harbored floR gene. In other studies on Salmonella Infantis, cat1 gene was predominately observed in chloramphenicol resistant isolates (Fonseca et al., 2006 ▶; Dionisi et al., 2011 ▶). The floR gene was also identified in numerous Salmonella serovars such as S. Infantis, S. Typhimurium (Bolton et al., 1999 ▶), S. Agona (Cloeckaert et al., 2000 ▶), S. Paratyphi B (Meunier et al., 2002 ▶), S. Albany (Doublet et al., 2003 ▶) and many other serovars (Randall et al., 2004 ▶).
In conclusion, this study shows the high incidence of antibiotic resistance in Salmonella Infantis isolated from backyard chickens. In order to prevent antimicrobial resistance, antibiotic administration in animals has to be closely supervised. However, this is a short report about the occurrence of drug resistant Salmonella Infantis and there should be more studies concerning the prevalence of this pathogen in both poultry and humans.
Acknowledgment
This study was supported by the Faculty of Veterinary Medicine, University of Tehran.
Conflict of interest
We declare no conflict of interest.
References
- Abbasoglu D, Akcelik M. Phenotypic and genetic characterization of multidrug-resistant Salmonella Infantis strains isolated from broiler chicken meats in Turkey. Biologia. 2011;66:406–410. [Google Scholar]
- Akiba M, Kusumoto M, Iwata T. Rapid identification of Salmonella enterica serovars, Typhimurium, Choleraesuis, Infantis, Hadar, Enteritidis, Dublin and Gallinarum, by multiplex PCR. J. Microbiol. Methods. 2011;85:9–15. doi: 10.1016/j.mimet.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Antunes P, Reu C, Sousa JC, Peixe L, Pestana N. Incidence of Salmonella from poultry products and their susceptibility to antimicrobial agents. Int. J. Food Microbiol. 2003;82:97–103. doi: 10.1016/s0168-1605(02)00251-9. [DOI] [PubMed] [Google Scholar]
- Bauer A, Kirby WM, Sherris JC, Truck M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Path. 1996;45:493–496. [PubMed] [Google Scholar]
- Bolton LF, Kelly L, Lee MD, Cray PF, Maurer JJ. Detection of multidrug-resistant Salmonella enterica serotype Typhimurium DT104 based on a gene which confers cross-resistance to florfenicol and chloramphenicol. J. Clin. Microbiol. 1999;37:1348–1351. doi: 10.1128/jcm.37.5.1348-1351.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI) Performance standards for antimicrobial susceptibility testing; twenty first informational supplement. M100-S21. Wayne, PA: CLSI; 2011. [Google Scholar]
- Cloeckaert A, Boumedine KS, Flaujac G, Imberechts H, D’Hooghe I, Chaslus-Dancla E. Occurrence of a Salmonella enterica serovar Typhimurium DT104-like antibiotic resistance gene cluster including the floR gene in S enterica serovar Agona. Antimicrob. Agents Chemother. 2000;44:1359–1361. doi: 10.1128/aac.44.5.1359-1361.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionisi AM, Lucarelli C, Benedetti I, Owczarek S, Luzzi I. Molecular characterisation of multidrug-resistant Salmonella enterica serotype Infantis from humans, animals and the environment in Italy. Int. J. Antimicrob. Agents. 2011;38:384–389. doi: 10.1016/j.ijantimicag.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Doublet B, Lailler R, Meuneir D, Brisabois A, Boyd D, Mulvey MR. Variant Salmonella genomic island 1 antibiotic resistance gene cluster in Salmonella enterica serovar Albany. Emerg. Infect. Dis. 2003;9:585–591. doi: 10.3201/eid0905.020609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emaddi Chashni SH, Hassanzadeh M, Bozorgmehri Fard MH, Mirzaie S. Characterization of the Salmonella isolates from backyard chickens in north of Iran, by serotyping, multiplex PCR and antibiotic resistance analysis. Arch. Razi Inst. 2009;64:77–83. [Google Scholar]
- Fonseca EL, Mykytczuk OL, Asensi MD, Reis EMF, Ferraz LR, Paula FL, Ng LK, Rodrigues DP. Clonality and antimicrobial resistance gene profiles of multidrug-resistant Salmonella enterica serovar Infantis isolates from four public hospitals in Rio de Janeiro, Brazil. J. Clin. Microb. 2006;44:2767–2772. doi: 10.1128/JCM.01916-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebreyes WA, Altier C. Molecular characteriza-tion of multidrug-resistant Salmonella enterica subsp enterica serovar Typhimurium isolates from swine. J. Clin. Microbiol. 2002;40:2813–2822. doi: 10.1128/JCM.40.8.2813-2822.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafari RA, Ghorbanpour M, Jaideri M. An investigation in Salmonella status in backyard chicken in Iran. Int. J. Poult. Sci. 2007;6:227–229. [Google Scholar]
- Kardos G, Farkas T, Antal M, Nogrady N, Kiss I. Novel PCR assay for identification of Salmonella enterica serovar Infantis. Let. App. Microbiol. 2007;45:421–425. doi: 10.1111/j.1472-765X.2007.02220.x. [DOI] [PubMed] [Google Scholar]
- Meunier D, Boyd D, Mulvey MR, Baucheron S, Mammina C, Nastasi A. Salmonella enterica serovar Typhimurium DT104 antibiotic resistance genomic island 1 in serotype Paratyphi B. Emerg. Infect. Dis. 2002;8:430–433. doi: 10.3201/eid0804.010375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller T, Prager R, Rabsch W, Fehlhaber K, Voss M. Epidemiological relationship between Salmonella Infantis isolates of human and broiler origin. Loh. Inf. 2010;45:27–32. [Google Scholar]
- Mirzaie S, Hassanzadeh M, Ashrafi I. Identification and characterization of Salmonella isolates from captured house sparrows. Turk. J. Vet. Med. 2010;34:181–186. [Google Scholar]
- Morshed R, Peighambari SM. Drug resistance, plasmid profile and random amplified polymorphic DNA analysis of Iranian isolates of Salmonella Enteritidis. New Microbiologica. 2010;33:47–56. [PubMed] [Google Scholar]
- Naghoni A, Ranjbar R, Tabaraie B, Farshad S, Owlia P, Safiri Z. High prevalence of integron-mediated resistance in clinical isolates of Salmonella enterica. Japan. J. Infect. Dis. 2010;63:417–423. [PubMed] [Google Scholar]
- Nogrady N, Gado I, Zsoltfeke P, Pastzti J. Chloramphenicol resistance genes in Salmonella enterica subsp enterica serovar Typhimurium isolated from human and animal sources in Hungary. Vet. Med-Czech. 2005;50:164–170. [Google Scholar]
- Nogrady N, Kardos G, Bistyak A. Prevalence and characterization of Salmonella Infantis isolates originating from different points of the broiler chicken-human food chain in Hungary. Int. J. Food Microbiol. 2008;127:162–167. doi: 10.1016/j.ijfoodmicro.2008.07.005. [DOI] [PubMed] [Google Scholar]
- Nogrady N, Toth A, Kostyak A, Paszti J, Nagy B. Emergence of multidrug-resistant clones of Salmonella Infantis in broiler chickens and humans in Hungary. J. Antimicrob. Chemother. 2007;60:645–648. doi: 10.1093/jac/dkm249. [DOI] [PubMed] [Google Scholar]
- Randall LP, Cooles SW, Osborn MK, Piddock LJV, Woodward MJ. Antibiotic resistance genes, integrons and multiple antibiotic resistance in thirty-five serotypes of Salmonella enterica isolated from humans and animals in the UK. J. Antimicrob. Chemother. 2004;53:208–216. doi: 10.1093/jac/dkh070. [DOI] [PubMed] [Google Scholar]
- Ranjbar R, Giammanc GM, Farshad S, Owlia P, Aleo A, Mammina C. Serotypes, antibiotic resistance, and class 1 integrons in Salmonella isolates from pediatric cases of enteritis in Tehran, Iran. Foodborne Pathog. Dis. 2011;8:547–553. doi: 10.1089/fpd.2010.0736. [DOI] [PubMed] [Google Scholar]
- Ranjbar R, Sarshar M, Sadeghifard N. Characterization of genetic diversity among clinical strains of Salmonella enterica serovar Infantis by ribotyping method. Sci. J. Zanjan. 2012;20:75–84. [Google Scholar]
- Schwarz S, Chaslus-Dancla E. Use of anti-microbials in veterinary medicine and mechanisms of resistance. Vet. Res. 2001;32:201–225. doi: 10.1051/vetres:2001120. [DOI] [PubMed] [Google Scholar]
- Shahada F, Chuma T, Tobata T, Okamoto K, Sueyoshi M, Takase K. Molecular epidemiology of antimicrobial resistance among Salmonella enterica serovar Infantis from poultry in Kagoshima, Japan. Int. J. Antimicrob. Agents. 2006;28:302–307. doi: 10.1016/j.ijantimicag.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Tajbakhsh M, Hendriksen RS, Nochi Z, Zali M, Aarestrup FM, Garcia-Migura L. Anti-microbial resistance in Salmonella spp. recovered from patients admitted to six different hospitals in Tehran, Iran from 2007 to 2008. Folia Microbiol. 2012;57:91–97. doi: 10.1007/s12223-012-0099-4. [DOI] [PubMed] [Google Scholar]
- Van Hoeka AH, Scholtensa IM, Cloeckaert A, Aartsa HJ. Detection of antibiotic resistance genes in different Salmonella serovars by oligonucleotide micro-array analysis. J. Microbiol. Meth. 2005;62:13–23. doi: 10.1016/j.mimet.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Walker RA, Lindsay E, Woodward MJ, Ward LR, Threlfall EJ. Variation in clonality and antibiotic-resistance genes among multiresistant Salmonella enterica serotype Typhimurium phage-type U302 (MR U302) from humans, animals, and foods. Microb. Drug Resis. 2001;7:13–21. doi: 10.1089/107662901750152701. [DOI] [PubMed] [Google Scholar]
- Waltman WD, Gast RK, Mallinson ET. Salmonellosis. In: Glisson, JR, Swayne, DE, Jackwood, MW, Pearson, JE, Reed, WM, editors. A laboratory manual for the isolation and identification of avian pathogens. (4th Edn.) Kennett Square, PA: American Association of Avian Pathologists; 1998. [Google Scholar]
- Zahraei Salehi, T. Epidemiology. In: Zahrei Salehi, T., editor. Salmonella. 1st Edn. University of Tehran Press; 1999. pp. 101–102. [Google Scholar]
- Zahraei Salehi T, Mahzounieh M, Saeedzadeh A. Detection of invA gene in isolated Salmonella from broilers by PCR method. Int. J. Poult. Sci. 2005a;4:557–559. [Google Scholar]
- Zahraei Salehi T, Mahzounieh M, Saeedzadeh A. The isolation of antibiotic-resistant Salmonella from intestine and liver of poultry in Shiraz Province of Iran. Int. J. Poult. Sci. 2005b;4:320–323. [Google Scholar]