Abstract
Despite the recent breakthrough advances in GPCR crystallography, structure determination of protein-protein complexes involving chemokine receptors and their endogenous chemokine ligands remains challenging. Here we describe disulfide trapping, a methodology for generating irreversible covalent binary protein complexes from unbound protein partners by introducing two cysteine residues, one per interaction partner, at selected positions within their interaction interface. Disulfide trapping can serve at least two distinct purposes: (i) stabilization of the complex to assist structural studies, and/or (ii) determination of pairwise residue proximities to guide molecular modeling. Methods for characterization of disulfide-trapped complexes are described and evaluated in terms of throughput, sensitivity, and specificity towards the most energetically favorable cross-links. Due to abundance of native disulfide bonds at receptor:chemokine interfaces, disulfide trapping of their complexes can be associated with intramolecular disulfide shuffling and result in misfolding of the component proteins; because of this, evidence from several experiments is typically needed to firmly establish a positive disulfide crosslink. An optimal pipeline that maximizes throughput and minimizes time and costs by early triage of unsuccessful candidate constructs is proposed.
1. Introduction
Chemokines promote cell migration in the context of development, immunity, inflammation and many other pathological and physiological processes (Baggiolini, 1998; Charo & Ransohoff, 2006; Gerard & Rollins, 2001; Griffith, Sokol, & Luster, 2014; Murdoch & Finn, 2000; Ransohoff, 2009). They do so by the virtue of binding to and activating seven transmembrane (7TM) receptors on the surface of migrating cells. In humans, there are approximately 45 chemokines that, based on the pattern of the conserved cysteine motif in their N-terminus, are divided into CC, CXC, CX3C, or XC families (Bachelerie et al.). The 22 chemokine receptors that are expressed in human tissues exhibit remarkable specificity in their recognition of the chemokines of different families, e.g. some receptors exclusively bind and are activated by CC chemokines while others strictly prefer CXC chemokines; based on this observation, the receptors are also classified into the same four subfamilies. Some receptors interact with multiple chemokines within their subfamily, while others have but a single endogenous chemokine ligand. Finally, several members of the Herpesviridae (herpesvirus) family encode chemokines and/or chemokine receptors in their genomes (Montaner, Kufareva, Abagyan, & Gutkind, 2013); these viral proteins interact with human receptors or chemokines, respectively, frequently demonstrate broad specificity spanning both CC and CXC families, and hijack chemokine receptor signaling cascades in host cells for the replicative advantage of the virus.
Knowledge of the structural basis of the high affinity, specificity, and pharmacology of receptor:chemokine interactions is extremely important, both from the standpoint of understanding the biology and for the development of therapeutics. Yet crystallography of chemokine receptors and especially their complexes with chemokines has proved to be quite challenging. As most members of the seven transmembrane (7TM) receptor family, chemokine receptors are unstable outside their native membrane environment and conformationally heterogeneous; they also lack hydrophilic surfaces for crystal formation. Due to advances in protein engineering, screening and crystallization (Bill et al., 2011; Ghosh, Kumari, Jaiman, & Shukla, 2015; Liu, Wacker, Wang, Abola, & Cherezov, 2014; Moraes, Evans, Sanchez-Weatherby, Newstead, & Stewart, 2014), the last few years were marked by dramatic progress in structure determination of 7TM receptors. However, even with engineered receptor constructs and with novel crystallization techniques, structure determination of protein-protein complexes involving chemokine receptors and their endogenous chemokine ligands remains difficult. The binding affinity of chemokines to detergent-solubilized receptors may be reduced in comparison to that observed in cell membranes, contributing to lower stability of the complexes. Further, some chemokines bind with high affinity only to select conformational (e.g. G protein-coupled, active) states of their receptors (Nijmeijer, Leurs, Smit, & Vischer, 2010) and these states are challenging to reproduce in detergent-solubilized conditions and in the absence of intracellular effectors and scaffolding proteins. Finally, crystallization of a 7TM receptor with any ligand frequently relies on slow complex dissociation kinetics (Zhang, Stevens, & Xu, 2015); such kinetics may be an inherent property of some receptor:chemokine pairs (e.g. the virally encoded receptor US28 and human CX3CL1/fractalkine (Burg et al., 2015)), but not others.
Here we describe disulfide trapping (also called cysteine trapping or disulfide crosslinking) – an experimental methodology for generating irreversible covalent binary protein complexes from unbound protein partners by introducing two cysteine residues (one per interaction partner) at strategically selected positions within their interaction interface. Disulfide trapping can serve at least two distinct purposes: (i) stabilization of the complex to prevent spontaneous dissociation and to trap a specific conformation, both of which can facilitate structural studies, and/or (ii) evaluation of residue proximities that, in conjunction with molecular modeling, can provide insight into the structural basis of the interaction even if the complex does not yield to crystallization efforts.
With respect to the second application (determining residue proximities), the disulfide trapping approach has been successfully applied to complexes of several receptors with small molecules and peptides (Buck & Wells, 2005; Dong et al., 2012; Hagemann, Miller, Klco, Nikiforovich, & Baranski, 2008; Kufareva et al., 2014; Monaghan et al., 2008). Residue proximities are established on the basis of crosslinks of varying strength and specificity and used to guide molecular modeling and interface mapping. This is in contrast to disulfide trapping for structure determination where only the strongest, energetically favorable crosslinks help stabilization and crystallization of the complexes. The first application of disulfide trapping for crystallization of a GPCR was achieved in 2011 by Rasmussen et al. (Rasmussen et al., 2011) when they crystallized an irreversible complex between a small molecule agonist and a nano-body stabilized β2 adrenergic receptor. Last year was marked by a successful application of the methodology to a complex between a receptor and a chemokine, yielding the first high resolution structural insight into chemokine recognition by an intact receptor (Qin et al., 2015).
Disulfide trapping has a number of advantages compared to other types of intermolecular crosslinking. First, unlike unnatural amino acids (Grunbeck et al., 2012; Grunbeck & Sakmar, 2013; Huber, Naganathan, Tian, Ye, & Sakmar, 2013; Kim, Axup, & Schultz, 2013), cysteine can be incorporated into recombinant proteins using straightforward cloning techniques and a wide range of expression systems. Next, when implemented this way, the incorporation is 100% efficient. Finally, unlike bulky moieties used in photoaffinity labeling (Chen, Pinon, Miller, & Dong, 2009, 2010; Coin et al., 2013; Dong et al., 2007; Dong et al., 2011; Miller et al., 2011; Pham & Sexton, 2004; Wittelsberger et al., 2006), a cysteine residue side chain is small and does not tend to induce artificial distortions of complex geometry.
Although a powerful approach, disulfide trapping is prone to artefacts and false positives, especially when cysteines are introduced at interfaces that are rich in native intramolecular disulfide bonds as in chemokine receptor complexes. Because of that, combined evidence from several assays is typically required to establish a strong positive intramolecular disulfide crosslink. This chapter provides an account of methods for generating disulfide crosslinked complexes and for experimentally characterizing the efficiency and quality of the crosslinks. A strategy for streamlining the approaches into a cost-effective pipeline for screening multiple candidates and quickly triaging unsuccessful pairs is also discussed.
2. Architecture of receptor:chemokine interfaces
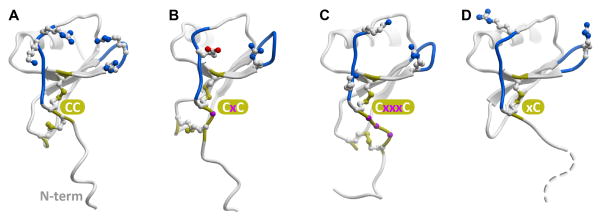
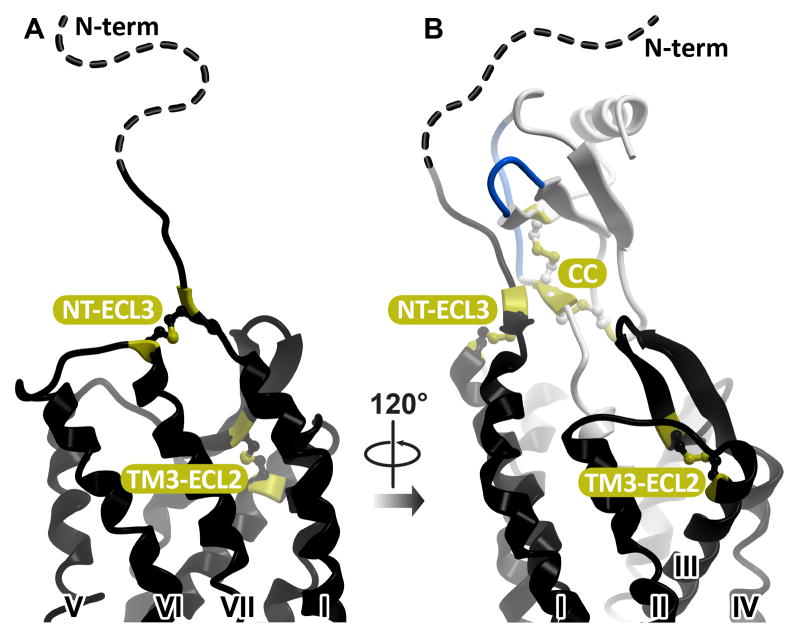
All chemokines share a conserved topology that consists of a flexible N-terminus, a double-cysteine motif, a three-strand β-sheet, and a C-terminal helix (Figure 1). The cysteines in the double-cysteine motif may be adjacent to one another (in the CC chemokine family), separated by one residue (in the CXC family), or separated by three residues (in the CX3C family that only has one member, CX3CL1/fractalkine). In two human chemokines, XCL1/lymphotactin and XCL2/SCM-1, only one cysteine is present in this region. The cysteines are involved in intra-molecular disulfide bonds connecting the N-terminus to the loop between the first and the second β-strands (in CC, CXC, and CX3C families) and to the third β-strand (in all chemokines) (Figure 1). Chemokine receptors, like all members of the 7TM receptor superfamily, share a conserved topology with seven membrane-spanning helices and a disulfide bond between TM3 and the extracellular loop (ECL) 2. All chemokine receptors except CXCR6 also have a disulfide bond connecting their flexible N-terminus to ECL3 (Figure 2).
Figure 1.
Representative chemokines of CC (A), CXC(B), CX3C (C) and XC (D) families. The conserved cysteine pattern is shown in yellow sticks; residues separating the two N-terminal cysteines are shown as magenta balls (B, C). Loops believed to be involved in coordinating the N-terminus of the receptor (N-loop and 40s-loop) are colored blue and charged residues in these regions are shown as sticks.
Figure 2.
(A) Conserved intramolecular disulfide bonds in the extracellular part of CXCR4. (B) Both disulfides are a part of the receptor:chemokine interface, with one of the disulfides found in direct proximity to the conserved N-terminal disulfide pattern of the chemokine.
From biochemical, biophysical, mutagenic, modeling and structural studies of the chemokine receptor system to-date (Brelot, Heveker, Montes, & Alizon, 2000; Burg et al., 2015; Gupta, Pillarisetti, Thomas, & Aiyar, 2001; Kofuku et al., 2009; Kufareva et al., 2014; Qin et al., 2015; Saini et al., 2011; Zhou & Tai, 2000) it is clear that the interaction between the receptor and the chemokine involves two distinct epitopes (Kufareva, Salanga, & Handel, 2015). In the so-called chemokine recognition site 1 (CRS1, (Scholten et al., 2011)), the extended N-terminus of the receptor binds to the globular core of the chemokine. In CRS2, the flexible N-terminus of the chemokine reaches into the TM domain binding pocket of the receptor. For most chemokines, their distal N-termini are recognized as critical signaling domains that directly induce the activation-related conformational changes in the TM domains of the receptors via CRS2 interaction (Chevigne, Fievez, Schmit, & Deroo, 2011; Proost et al., 2008; Van Damme et al., 1989). The first two structures of the receptor:chemokine complexes (Burg et al., 2015; Qin et al., 2015) demonstrated that the two epitopes are joined by an intermediate region that we refer to as CRS1.5 and that brings into close proximity the conserved N-terminal cysteine of the receptor with the conserved cysteine motif of the chemokine (Figure 2). This understanding is especially important in the context of disulfide trapping, because introducing additional artificial cysteines at disulfide-rich interfaces has high potential of shuffling the native disulfide connectivity and misfolding the proteins while still exhibiting, at least in some assays, the behavior of a positive cross-link.
While retaining the conserved overall architecture, the multiple receptor:chemokine pairs appear to utilize distinct structural determinants and conformational mechanisms to achieve specificity of binding and signaling (Burg et al., 2015; Choi et al., 2005; Duchesnes, Murphy, Williams, & Pease, 2006; Pakianathan, Kuta, Artis, Skelton, & Hebert, 1997; Qin et al., 2015; Saini et al., 2011). This diversity hinders transfer of structural knowledge between different complexes by homology and emphasizes the importance of detailed studies of individual receptor:chemokine pairs; and while this task may be cost-prohibitive for X-ray crystallography, it is more tractable by other approaches such as molecular modeling in conjunction with disulfide trapping.
3. Cysteine as a natural crosslinking agent
Cysteine is the most chemically reactive of the 20 natural amino acids: its thiol side chain is easily oxidized due to its nucleophilic nature. Oxidation of two spatially proximal cysteine residues leads to formation of disulfide bonds that play a major role in protein folding and stability. The dissociation energy of a typical disulfide bond is about 50–60 kcal/mol, which makes it 10–20 times stronger than the strongest hydrogen bond, but still much weaker than a typical covalent bond (e.g. carbon-carbon). Disulfide bonds are susceptible to breakage in the presence of other nucleophiles, but thiol-disulfide exchange is inhibited at physiological and acidic pH.
An ideal disulfide bond has a Sγ-Sγ distance of 2.04±0.07Å and a specific relative orientation of remaining atoms within the two cysteines (Cβ-Sγ-Sγ-Cβ dihedral angle of 90±12°) (Pellequer & Chen, 2006). Bonds with dihedral angles of 0° to 180° occur in protein structures, but are significantly weaker than those with ideal geometry.
Most cellular compartments are rich in glutathione and thus represent a reducing environment in which disulfide bonds are not stable. Consequently, cysteine residues are usually present in their free form in soluble cytosolic and nuclear proteins. However, the oxidizing environment in the extracellular space, in the lumen of the rough endoplasmic reticulum, and in the mitochondrial intermembrane space favors formation of disulfide bonds. As a result, intramolecular disulfide bonds are common in secreted proteins, with chemokines being a perfect example (Figure 1). Intramolecular disulfide bonds are also frequently found in the extracellular domains of transmembrane proteins, which, as illustrated in Figure 2, includes the chemokine receptors. Contrary to the extracellular fragments, cysteine residues deeper in the TM domain of the receptor or in intracellular loops appear to be reduced and not involved in disulfide formation. In live cells, the interaction between chemokines and their receptors occurs in the extracellular oxidizing environment and thus additional disulfide bonds introduced at their interface are expected to form and remain stable when the cysteines are in the correct proximity and orientation. However, when receptors are extracted from their natural cellular context, there is a danger of transmembrane and especially intracellular cysteine residues, which are normally reduced, to become oxidized and lead to the formation of non-specific covalent adducts with chemokines or each other.
4. Disulfide trapping
4.1 Overall strategy
Disulfide trapping serves the goal of generating an irreversible covalently bound receptor:chemokine complex from otherwise dissociable protein partners. To accomplish this goal, a single non-native cysteine residue is introduced into the receptor and into the chemokine. Depending on the method used to generate the binding partners (e.g. bacterial expression, eukaryotic cell expression, or chemical synthesis), the mutated proteins can be either expressed separately and mixed, or co-expressed in the same cells. In both approaches, the main assumption is that if formation of the native complex positions the two engineered cysteine residues in favorable proximity and geometry, a disulfide bond will spontaneously form thus locking the complex and preventing its dissociation. The resulting irreversible complexes can be detected using non-reducing SDS page and/or Western blotting and further characterized in terms of overall yield, crosslinking efficiency, stability, and monodispersity. In our own applications for generating receptor:chemokine complexes, we had the best success with co-expressing the mutated receptor and the mutated chemokine in Sf9 insect cells. Among other advantages, this strategy avoids the need to make purified chemokine, which, due to an extra free cysteine residue, may be complicated by intramolecular disulfide shuffling and formation of covalent chemokine oligomers.
4.2 Selection of candidate residue pairs
Finding optimal position pairs for introducing receptor:chemokine disulfide crosslinks might at first seem like an impossible endeavor; however, in our implementation, the disulfide trapping method involves 3D model-guided “cherry-picking” of residue pairs for cysteine mutagenesis. Starting with a model of the complex, we assign a numeric score to each pair of receptor:chemokine residues that reflects the likelihood of them spontaneously forming a disulfide bond when mutated to cysteine. In a hypothetical situation when a high-resolution structure or model is already available, the residues are mutated in silico to cysteines, a disulfide bond is imposed, the side-chains of the two bonded residues and the neighboring residues are optimized by conformational sampling, and the disulfide bond energy is evaluated by
where dS:S, dSi:Cβj, and dCβ:Cβ stand for pairwise distances between the indicated atoms in the disulfide-bonded residues. This energy term is combined with the overall energy involving steric compatibility of the in silico introduced disulfide bond with its immediate environment, and used for scoring and ranking of the residue pairs. In typical real-life situations where only an approximate model is available, relaxed criteria should be used, mostly based on appropriate Cα-Cα and Cβ-Cβ distances between the residues in question. In general, due to the size and geometry of cysteine residues, disulfide crosslinking works best for residue pairs that are involved in backbone-backbone contacts; residues with only side chain-side chain contacts are usually poor crosslinking candidates.
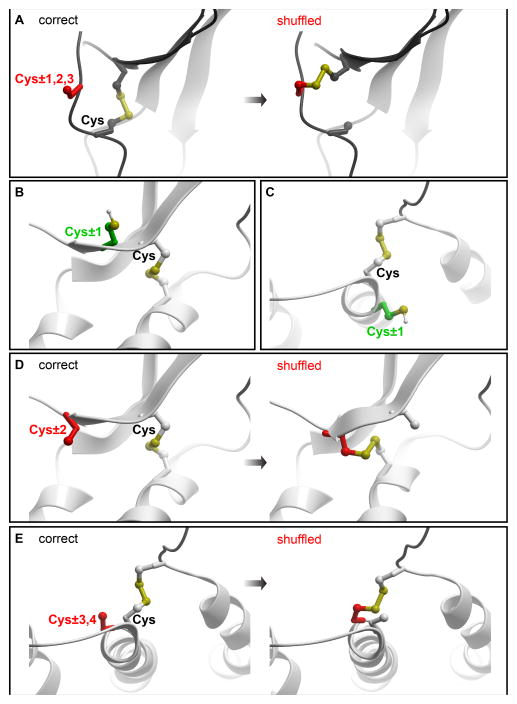
Receptor:chemokine residue pairs are rank-ordered by decreasing score. After that, penalties are introduced to account for possible liabilities of introducing a reactive cysteine at each candidate location. For example, a penalty is imposed if one of the designed cysteine residues is located in a fragment without a defined secondary structure and is adjacent or proximal (within two to three residue positions) to a native cystine (disulfide-bonded cysteine) in the sequence. Introducing an artificial cysteine in such positions increases the probability of protein misfolding by formation of an unwanted disulfide bond between the counterpart of the native and the newly introduced cysteine (shuffling, Figure 3A). An immediately adjacent native cystine within a helix or a β-strand may be free of such liabilities because (i) these regions are expected to have less flexibility and (ii) adjacent residues in these secondary structures point in different directions (Figure 3C, in the case of an α-helix) or strictly opposite directions (in the case of a β-strand, Figure 3B). On the other hand, taking into account residue geometry within α-helices and β-strand (Figure 3D and E), we usually penalize an artificial cysteine introduced two residues up or down from a native cysteine in a β-strand, and three to four residues up and down from a native cysteine in an α-helix.
Figure 3.
Representative geometries of polypeptide chains where introduction of a cysteine poses high risk of intramolecular disulfide shuffling. (A) within 1–3 residues from a native cysteine in a loop without defined secondary structure. (B–C) within an α-helix and a β-strand, positions proximal to a native cysteine are generally safe. Instead, positions two residues up or down from a native cysteine within a β-strand (D) or 3–4 residues up or down from a native cysteine within an α-helix (E) are subject to disulfide shuffling liability.
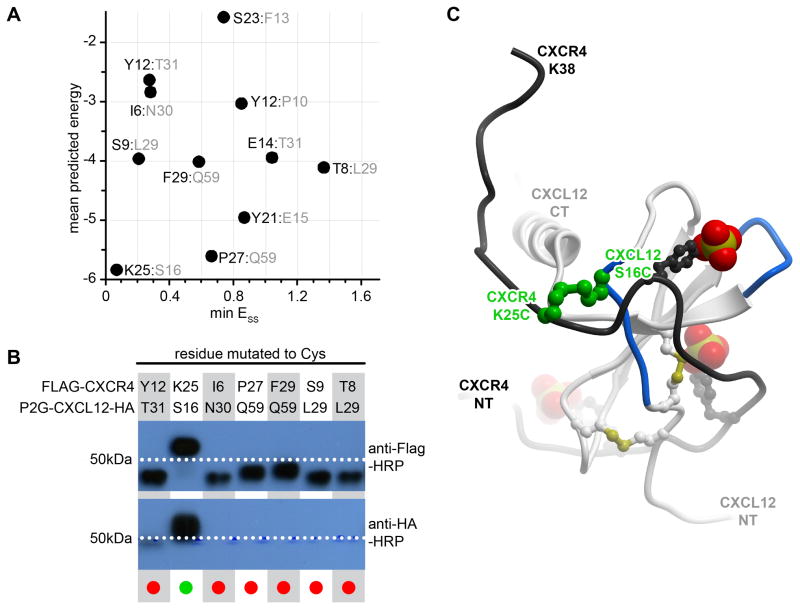
Our early attempts in designing a disulfide crosslink between CXCR4 and CXCL12 were guided by the NMR structure (Veldkamp et al., 2008) of a CXCL12 dimer in complex with a 38 residue peptide isolated from the CXCR4 N-terminus (CRS1). The in silico disulfide bonds introduced between all possible proximal CXCR4:CXCL12 residue pairs were ranked by both the disulfide bond energy, ESS, according to the equation above, and by the overall energy of the disulfide in the context of the complex; the latter included van der Waals, electrostatics, hydrogen bonding, hydrophobicity and polar surface area terms (Kufareva et al., 2014). Figure 4A illustrates the scatter plot of ESS and overall energy values for the 11 residue pairs selected for experimental validation. Mutant pair characterization performed as described in the next section confirmed that the pair predicted to form the best crosslink, CXCR4(K25C):CXCL12(S16C), did indeed crosslink very efficiently (Figure 4B), while the remaining pairs showed no or ambiguous crosslinking. This single CRS1 crosslink (Figure 4C) was later used in molecular docking to build and refine the model of the complex between the full-length receptor and the chemokine (Kufareva, Handel, & Abagyan, 2015; Kufareva et al., 2014).
Figure 4.
Identification of the first CRS1 crosslink between CXCR4 and CXCL12 from the CXCR4:CXCL12 NMR structure (PDB 2k05). (A) 11 crosslinks were designed in silico and ranked according to the overall predicted energy (vertical) and disulfide bond energy (horizontal). (B) Following co-expression of the mutant pairs in insect cells, the best ranking pair, CXCR4(K25C):CXCL12(S16C) demonstrated efficient crosslinking in Western blotting as evidenced by the double (anti-receptor and anti-chemokine) staining and the molecular weight shift of the material in the respective lane. (C) The identified crosslink (green) is shown in the context of the NMR structure from which it was generated: CXCL12 is in white ribbon and the N-terminal peptide of CXCR4 is in black ribbon. The sulfate groups on CXCR4 sulfotyrosines sTyr7, sTyr12, and sTyr21 are shown as CPK. Panel (B) is adapted from (Kufareva et al., 2014).
4.3 Generation of disulfide-trapped receptor:chemokine complexes
The methodology for cloning, expressing and purifying disulfide-crosslinked receptor:chemokine complexes is no different from that for non-covalent complexes, which is described in detail by Gustavsson et al in Chapter 11 of this Methods of Enzymology volume. Below we provide a brief summary of the major steps while stressing the nuances important in the context of disulfide crosslinking.
4.3.1 Cloning
Baculovirus infected Spodoptera frugiperda (Sf9) insect cells represent an appropriate expression system for production of membrane proteins and complexes in many applications including biophysical studies and structure determination. The Bac-to-Bac Baculovirus Expression System (Invitrogen) is used to generate bacmids and baculovirus.
First, the receptors are cloned into a pFastBac™ vector (Invitrogen) under either a gp64 or a polH promoter, and are N-terminally or C-terminally tagged with a FLAG tag (DYKDDDD, for detection). The placement of the tag has to be optimized for each individual receptor as it may differentially affect yield and trafficking even for homologous receptors (e.g. CXCR4 tolerates the tag on the N-terminus while ACKR3 constructs express substantially better when the tag is placed at the C-terminus). Additionally, a His10 tag is placed at the C-terminus for metal affinity purification. The chemokine constructs include the native signal sequences, are C-terminally tagged with an HA-tag (YPYDVPDYA, for detection) and are cloned into a pFastBac™ vector under a polH promoter. Codon-optimization reflecting the inherent bias of the expression system may provide an additional way to optimize the expression constructs. Cysteine mutations are introduced at the desired positions in the receptor and chemokine constructs using standard QuikChange site-directed mutagenesis protocols.
Recombinant bacmids separately carrying the genes for the receptor and the chemokine are generated by transforming the pFastBac™ constructs into DH10Bac™ competent E. coli cells (Invitrogen) according to the manufacturer's instructions. Propagation of a single selected white colony with subsequent bacmid purification is described in detail by Gustavsson et al in Chapter 11.
4.3.2 Generation of baculovirus stocks
To generate high-titer (>109 viral particles per mL) recombinant baculovirus, recombinant bacmids (5 μL of the purified material at the concentration obtained through purification) containing the target genes are transfected separately into Sf9 cells (2.5 mL at a density of 1.2×106 cells/mL) using 3 μL of Xtreme Gene Transfection Reagent (Roche) and 100 μL of Transfection Medium (Expression Systems). Cell suspensions are incubated for 96 h with shaking at 27 °C. P0 viral stocks are isolated and used to infect larger Sf9 cultures for generation of P1 viral stock. Typically, 400 uL of P0 virus is used to infect 40 mL of culture at this step. Virus titer can be quantified by flow cytometry following cell staining with PE-conjugated anti-gp64 antibody (Expression Systems).
4.3.3 Sf9 co-expression
Co-expression of disulfide-trapped complexes between receptors and chemokines in insect Sf9 cells is similar to co-expression of non-covalent complexes and is described in detail by Gustavsson et al in Chapter 11. Briefly, the P1 stocks of the two types of particles (those carrying receptor and those carrying chemokine) are used to co-infect Sf9 cells at a density of 2–2.6×106 cells/mL. Even if the multiplicity of infection (MOI) has been previously separately optimized for each vector, it frequently needs to be re-optimized in the context of the co-expression experiment, likely because the cell expression machinery gets taxed to a different degree when producing individual components vs the complex (Fig. 5).
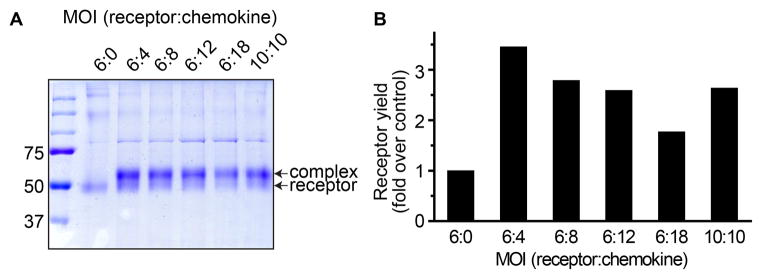
Figure 5.
Effect of varying the multiplicity of infection (MOI) on the overall levels of expression and the efficiency of crosslinking between a pair of single cysteine mutants of ACKR3 and CXCL12. (A) Non-reducing 10% SDS-PAGE of the samples following metal affinity purification by the tag on the receptor. The lower and the upper bands indicate non-crosslinked receptor and crosslinked complex, respectively. (B) Band densitometry for quantification of receptor expression levels. Receptor yield is expressed as fold increase over the control sample (receptor only, no chemokine). Although the calculated fraction of crosslinked complex is close to 80% across all samples, the receptor yield is maximal at the MOI of 6:4 (receptor:chemokine).
The cells are allowed to grow while expressing the two proteins for a period that may also need optimization, but typically for 44–48 hours. Biomass is harvested by centrifugation and stored at −80 °C until further use.
It is unknown how and when the receptor:chemokine complexes form when the two proteins are co-expressed in Sf9 cells. One possibility is that the chemokine molecules are secreted in the cell culture medium and from there, bind to the receptors that are expressed and trafficked to the plasma membrane. Alternatively, the proteins may bind one another at the time of folding in the endoplasmic reticulum and be trafficked to the cell surface together as a complex. In either case, the interaction interface is in an oxidizing environment which promotes disulfide bond formation for those cysteine residue pairs that are in spatial proximity and favorable orientation. As a result, covalently trapped complexes may be harvested from the cell membranes along with non-covalent complexes and uncomplexed receptors.
4.3.4 Membrane preparation
Harvesting and metal affinity purification of disulfide-trapped receptor:chemokine complexes is performed in the same way as purification of isolated receptors or non-covalent complexes, as described in detail by Gustavsson et al in Chapter 11. Briefly, cell biomass is thawed, lysed in hypotonic buffer (10 mM HEPES pH 7.5, 10 mM MgCl2, 20 mM KCl, and EDTA-free protease inhibitor cocktail (Roche)), and membranes are purified by repeated (3×) Dounce homogenization (40 strokes per round) followed by centrifugation at 50,000×g at 4 °C for 30 min. Membrane pellets are resuspended in the above hypotonic buffer during round 1 and in a high salt buffer (10 mM HEPES pH 7.5, 10 mM MgCl2, 20 mM KCl, 1 M NaCl and EDTA-free protease inhibitor cocktail) during rounds 2–3. Following the last centrifugation, membranes are resuspended and homogenized in hypotonic buffer supplemented with 30% glycerol (v/v) and flash-frozen for storage at −80 °C until further use.
4.3.5 Purification of receptor complexes
Purified membranes are thawed on ice, homogenized with an equal volume of hypotonic buffer, mixed with an equal volume of 2× solubilization buffer (100 mM HEPES pH 7.5, 800 mM NaCl, 1.5% n-dodecyl-β-D-maltopyranoside (DDM, Anatrace), 0.3% cholesteryl hemisuccinate (CHS, Sigma)), incubated for 3 h at 4 °C, and then centrifuged at 25,000×g for 30 min. The supernatant is incubated overnight at 4 °C with TALON IMAC resin (Clontech) and 20 mM imidazole. After binding, the resin is washed with twenty column volumes of wash buffer (25 mM HEPES pH7.5, 400 mM NaCl, 0.025% DDM, 0.005% CHS, 10% glycerol). Resin is then resuspended in 6 column volumes of wash buffer and incubated with 10 μL of PreScission Protease at 15 mg/mL (purified in house in our case) for 3 h at 4 °C. Purified protein is collected as flow through.
The above composition of solubilization, wash, and elution buffers is such that target proteins remain folded, and tightly bound slow dissociating or covalent complexes remain intact. In case modifications to the buffer compositions are necessary, it is important that nucleophiles and reducing agents are avoided to ensure stability of any intra- or intermolecular disulfide bonds through the purification process.
4.4 Characterization of disulfide-trapped receptor:chemokine complexes
Identification of an efficient and energetically favorable disulfide crosslink can be performed using multiple experimental approaches. Furthermore, because of the complexities of the disulfide-rich receptor:chemokine interfaces, and because of general and implementation-specific variations in assay throughput, material requirements, sensitivity, and specificity, no single assay is ideal and crosslink characterization typically requires combined evidence from several assays. We were successful in crosslink characterization using the following methods:
flow cytometry, to detect complexes directly on cells
denaturing, but non-reducing SDS polyacrylamide get electrophoresis of the purified protein material, to evaluate the presence and relative abundance of the covalently linked complexes
Western blotting, to confirm the nature of the observed bands
differential scanning fluorimetry using cysteine-reactive CPM dye (CPM-DSF) to characterize thermal stability of the purified complexes
size-exclusion chromatography (SEC), to study monodispersity of the complexes
Table 1 summarizes time and material requirements for the five assays as implemented in our hands, and provides an estimate of their throughput as well as specificity and sensitivity in separating strong energetically favorable disulfide crosslinks from weak non-specific ones. According to these parameters, the assays can be prioritized in the workflow in order to quickly triage unsuccessful pairs. Notably, SDS-PAGE and Western blotting appear to be the central and indispensable experiments in crosslink characterization despite their moderate throughput, sensitivity, or specificity. This is because they can provide proper context for interpreting results of other experiments, especially when these results are ambiguous or inconclusive.
Table 1.
Comparison of experimental approaches for characterization of disulfide-trapped receptor:chemokine complexes
| Amount of sample needed (μg) | Optimal protein concentration (μg/ul) | Preparation time(a) | Throughput(b) | Specificity(c) | Sensitivity(c) | |
|---|---|---|---|---|---|---|
| Flow cytometry | n/a (performed on cells) | ~1h | +++ | − | +++ | |
| SDS-PAGE | 5–10 | 0.3–0.5 | ~1h | ++ | ++ | ++ |
| Western blotting | 0.5–1 | 0.1–0.5 | ~4–6 h | ++ | ++ | +++ |
| CPM-DSF | 0.2–0.4 | 0.2–0.4 | ~1h | +++ | +++ | − |
| SEC | 5–10 | 0.3–0.5 | ~0.5–1h | + | +++ | − |
The estimated preparation time does not include protein purification. Protein purification is required for all experiments except flow cytometry and, in some implementations, Western blotting, and typically takes at least 3 days.
Number of samples that can be reasonably tested in parallel in a single experiment, including controls: up to 5–6 (+), up to 12–15 (++), up to 48–96 (+++)
A qualitative estimate of specificity and sensitivity of each assay towards strong crosslinks favorably compatible with the native complex geometry.
4.4.1 Detection of covalent complexes on cells using flow cytometry
Flow cytometry allows the initial detection of disulfide-trapped complexes directly in the cells, prior to purification of receptor material. It requires staining of complex-expressing cells with fluorescent antibodies against a tag on the chemokine. At least with some chemokines (including CXCL11, CXCL12 and CXCL14), C-terminal placement of the tag on the chemokine prevents interference between the receptor:chemokine and chemokine:antibody binding and ensures that the receptor-bound chemokine is detectable on the cell surface with anti-tag antibodies. This may or may not be the case for other chemokines; for example, so far, we have been unsuccessful in detection of cell surface-bound vMIP-II using this method.
4.4.1.1. Experiment
For flow cytometry experiments, Sf9 cells co-expressing the disulfide-trapped complexes are washed twice in TBS+0.5% BSA and stained in 1:100 dilution of FITC-conjugated anti-tag antibody. We use mouse monoclonal anti-FLAG® M2-FITC antibody and mouse monoclonal anti-HA-FITC antibodies, both from Sigma-Aldrich (item numbers F4049 and H7411, respectively). Following 20 min incubation on ice in the dark, the cells are diluted in TBS and analyzed using a Guava bench top mini-flow cytometer (Millipore). Controls must include untransfected cells in order to quantify the levels of non-specific antibody binding; in cases where non-specific binding is non-negligible, repeated washing of the stained cells with TBS+0.5 BSA may help. Controls should also include cells co-expressing WT receptor and WT chemokine that are presumed to form efficient but dissociable complexes (Figure 6).
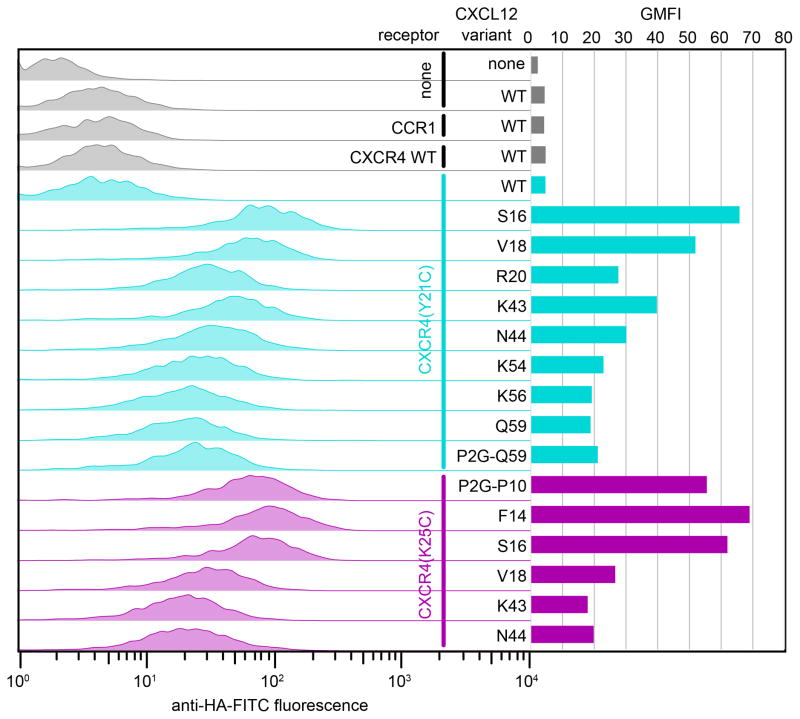
Figure 6.
Detection of crosslinked CXCR4:CXCL12 and CXCR4:P2G-CXCL12 complexes by flow cytometry. Each of two receptor variants (CXCR4(Y21C), cyan or CXCR4(K25C), magenta) was co-expressed with multiple chemokine variants. Left: chemokine on the cell surface is detected by fluorescently labeled antibody against a tag on its C-terminus. Right: geometric mean fluorescence intensity is plotted for each receptor:chemokine pair. Notably, for CXCR4:CXCL12 complexes, staining of non-crosslinked complexes (CXCR4 WT:CXCL12 WT, gray and CXCR4 (Y21C):CXCL12 WT, top cyan) is low and comparable to staining of the control sample that does not have the receptor (sample 2); this indicates lower complex affinity and/or fast dissociation and makes the experiment possible without unlabeled competitor. This is in contrast to Figure 7 showing a similar experiment for a slower dissociating complex of ACKR3:CXCL12.
For some receptor:chemokine complexes (e.g. ACKR3:CXCL12), the affinity and dissociation kinetics is such that large amounts of cell surface bound chemokine are present even if the covalent disulfide crosslink between the receptor and the chemokine is not formed. To separate disulfide-trapped complexes from long-lived non-covalent complexes, prolonged incubation of samples with excess unlabeled competitor (a chemokine or a small molecule) is helpful. Such pre-incubation dissociates the non-covalent complexes, but has no effect on the covalent disulfide-trapped complexes (Figure 7).
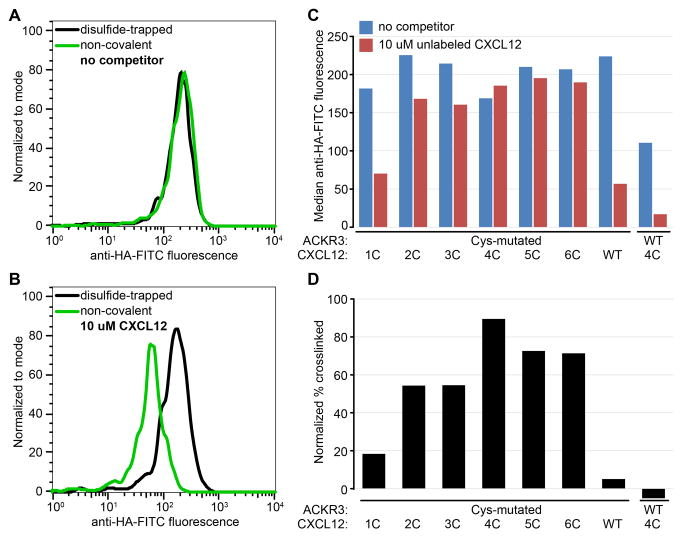
Figure 7.
Detection of crosslinked ACKR3:CXCL12 complexes by flow cytometry following incubation with an unlabeled competitor. (A) Due to high affinity and/or slow dissociation of the chemokine, anti-chemokine staining is indistinguishable between the non-covalent (green) and disulfide-trapped (black) complexes, (B) Prolonged incubation with excess unlabeled CXCL12 dissociates non-covalent (green) but not disulfide-trapped (black) complexes, effectively reducing surface staining. (C) Flow-cytometry screening of multiple disulfide-trapped candidate complexes side-by-side with controls (ACKR3(Cys):CXCL12 WT and ACKR3 WT:CXCL12(4C)). (D) Percent crosslinked for each complex was estimated by dividing median anti-HA-FITC fluorescence in the presence of competitor by that in the absence of competitor and subtracting non-specific signal from control samples.
4.4.1.2. Interpretation
The result of a flow cytometry experiment is a histogram of cell distribution by the total amount of surface-bound chemokine, with or without an unlabeled competitor ligand. This assay provides an easy way to quickly screen numerous candidate disulfide-trapped construct combinations without the complexities of sample purification; however, the utility of this method is limited to chemokines that are detectable on the cell surface by a C-terminal tag. Furthermore, even for chemokines that are efficiently detected, the average levels of specific anti-tag antibody fluorescence on cells may vary significantly as a result of varying surface expression of the receptor; therefore, flow cytometry at best allows one to qualitatively estimate the presence of disulfide-trapped complexes or to rank on a relative scale several chemokine mutants that are co-expressed with a single receptor mutant as a part of the same experiment. In this sense, the assay has relatively low specificity. On the other hand, as long as antibody detection of the surface-bound chemokine is established, the assay has high sensitivity and thus rarely produces false negatives: the absence of anti-chemokine staining in these cases signals an unsuccessful crosslink that can be eliminated from further consideration.
4.4.2 Quantification and characterization of disulfide-trapped complexes by SDS-PAGE and Western blotting
Following metal affinity purification, promising candidate complexes are analyzed for yield and crosslinking efficiency by non-reducing SDS-PAGE and Western blotting. These assays appear central in crosslink characterization as they provide the proper context for interpretation of all other experiments.
4.4.2.1. Experiment
For non-reducing SDS-PAGE analysis, 5–10 ug of purified protein is mixed with Laemmli buffer containing no reducing agents and loaded onto a 10% polyacrylamide gel. With some membrane receptors, it is important that the SDS-denatured samples are not heated prior to loading. Room temperature is a reasonable tradeoff between avoiding protein aggregation (that occurs when samples are heated) and maintaining the solubility of SDS (that precipitates if samples are kept on ice). Gels are stained with Coomassie stain, and destained 3x with a 50%/40%/10% (v/v/v) mixture of H2O, methanol, and acetic acid. Molecular weight shift and the relative band intensity are used as indicators of the presence and relative abundance of the disulfide trapped complex, respectively. In a separate control experiment, the same samples are run using standard reducing Laemmli buffer.
Western blot detection is used to specifically identify Flag-tagged receptor and HA-tagged chemokine bands. In this case, non-reducing SDS-PAGE is performed as above and, without staining, transferred to a nitrocellulose membrane at 100 V for 1 h. The membrane is incubated in 10 mL TBS-T (1x TBS pH 7.4, 0.1% Tween (v/v)) supplemented with 5% milk for 1 h at room temperature. 1 μL of primary antibodies (mouse anti-Flag M2 primary antibody (Sigma, for receptor) and rat anti-HA 3F10 primary antibody (Roche, for chemokine)) is added to 10 mL of fresh TBS-T with 5% milk and again incubated at room temperature for 1 h. 0.5 μL of secondary antibodies (IRDye 680 donkey anti-mouse IgG and IRDye 800 goat anti-rat IgG (LI-COR Biosciences)) is added to 10 mL of TBS-T with 5% BSA and incubated for 1 h at room temperature. Following incubation, the membrane is washed 3× with 10 mL of fresh TBS-T for 10 min. The membrane is finally rinsed with 1× sterile PBS and imaged using the Odyssey IR imaging system (LI-COR Bioscience). Membranes can also be dried overnight between layers of filter paper, and then kept and imaged dry for up to 2 weeks with minimal loss in signal provided they are protected from light.
We usually conduct Western blot analysis on purified samples and interpret it in conjunction with other assays described herein: SDS-PAGE, SEC, and DSF. However, unlike these other assays, Western blotting does not require the samples to be purified and thus can be conducted on whole cell lysates as an alternative first line of analysis to quickly screen for promising disulfide-trapped constructs.
4.4.2.2. Interpretation
Following a non-reducing SDS-PAGE, most frequently observed bands are located at the following molecular weights (from smallest to largest):
Uncomplexed chemokine monomer
Uncomplexed chemokine dimer
Uncomplexed receptor monomer (Figure 8A)
Receptor:chemokine complex (Figure 8A)
Receptor:chemokine dimer complex
Uncomplexed receptor dimer
Receptor dimer:chemokine complex
Aggregated protein material
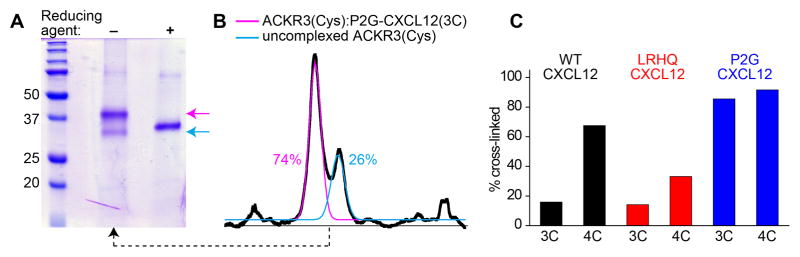
Figure 8.
SDS-PAGE analysis of a crosslinked complex between ACKR3(Cys) and N-terminal mutants of CXCL12. (A) relative intensity of the bands on a non-reducing and a reducing SDS-PAGE for P2G-CXCL12 (3C), (B) Band intensity from (A) quantified by densitometry. (C) The crosslinking efficiency with ACKR3(Cys) is quantified for cysteine residues introduced at two proximal positions of CXCL12, each in the context of three N-terminally modified chemokine variants; the observed difference in crosslinking efficiency of homologous mutants reflects conformational variations between the chemokine variants.
Additional proximal bands may appear as a result of proteolytic degradation of receptor (effectively lowering its MW) or heterogeneous post-translation modifications (increasing MW). In all cases, it is important to include the following controls on the same gel:
A homologous well-behaving insect cell expressed receptor construct, if available
A non-crosslinked insect cell expressed receptor:chemokine pair (e.g. including a non-mutated receptor and/or non-mutated chemokine)
A well-behaving disulfide-trapped pair, if available.
By ensuring that an equal amount of protein is loaded into each lane of the gel, and by providing adequate controls, relative ranking of the cross-link quality becomes possible.
An ideal crosslinked complex shows as a bright band on a non-reducing SDS-PAGE and is possibly accompanied by a much dimmer (if present at all) band corresponding to uncomplexed monomeric receptor, in the faint or absent background of other irrelevant bands (Figure 8A). Relative brightness of the two bands may serve as a measure of cross-linking efficiency (the ratio of cross-linked to non-cross-linked receptor, Figure 8B,C), although in many cases, it may overestimate the fraction of cross-linked receptor due to poor membrane extraction of non-crosslinked species. Western blot analysis of an ideal crosslinked pair will show clear and weak (if present at all) receptor-only staining of the lower MW band with clear and strong co-staining of the disulfide-trapped higher MW complex band (Figure 9). Further confirmation of the successful crosslink may be obtained by repeating the SDS-PAGE and Western blotting analysis under reducing conditions, which is expected to completely dissociate the higher MW complex band, to increase the density of the receptor-only band (Figure 8A), and to bring about the lower molecular weight chemokine-only band (assuming the gel percentage is sufficient to detect low MW proteins in the sample).
Figure 9.
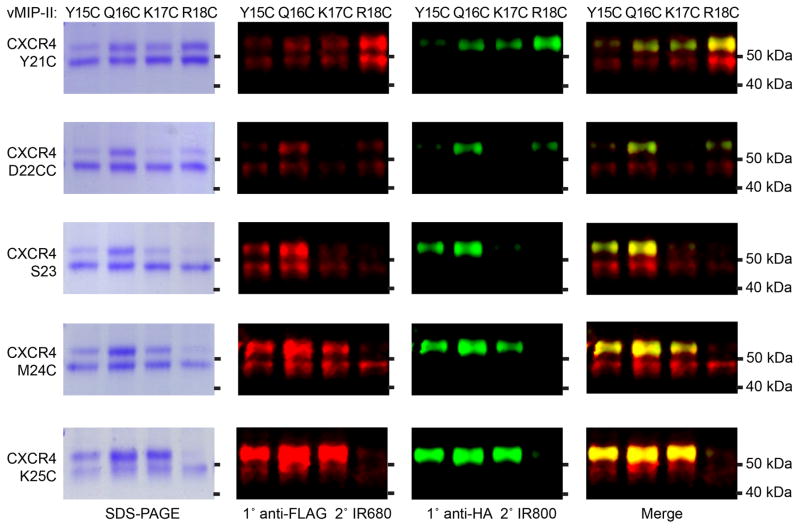
SDS-PAGE and Western blotting analysis of CRS1 disulfide crosslinked complexes of CXCR4 and vMIP-II. Left column: Coomassie stained SDS-PAGE gels of 5 different CXCR4 Cys mutants with 4 different vMIP-II Cys mutants. 2nd column: detection of the same SDS-PAGE gel by Western with an anti-FLAG antibody against a tag on the receptor. 3rd column: detection with an anti-HA antibody against a tag on the chemokine. 4th column: merge of the anti-Flag and anti-HA Westerns. Figure adapted from (Qin et al., 2015).
We consider successful clear-cut intermolecular crosslinks as evidence of spatial proximity and favorable orientation of the mutated residues in the context of the native complex geometry, even if these crosslinks do not withstand the scrutiny of the more stringent SEC and DSF assays described below. As such, they can be used for validation and refinement of 3D models of the complex (Kufareva, Handel, et al., 2015), at least in the form of inter-residue distance restraints. Stronger evidence may be required in order to integrate the crosslink in the modeling procedure in the form of an explicit inter-molecular disulfide bond.
Unfortunately, ideal crosslinks are rare, and in most cases, the relative density of the two bands is non-optimal and additional irrelevant bands are present. Such bands may indicate, for example, formation of covalent homodimers or higher order homo-oligomers from the mutated and possibly misfolded chemokines or receptors, or formation of covalent receptor:chemokine complexes of non-canonical stoichiometry, all of which can be deduced from Western blotting staining against the receptor and the chemokine. The prevalence of such bands indicates that the artificially introduced cysteine residue pair interferes with proper folding of the individual proteins or with their interactions, and warrants caution in application of the crosslink in modeling procedures.
Given the prevalence and the role of native intramolecular disulfide-bonds at the receptor:chemokine interface, any artificially introduced cysteine can potentially result in disulfide shuffling. This scenario may exhibit the behavior of an efficient crosslink on SDS-PAGE or a Western blot, although in reality, the artificially introduced cysteines are not proximal. Consequently, interpretation of the crosslinks in the context of the amino-acid sequence and the nascent 3D model of the complex may be instrumental in separating true hits from results of disulfide shuffling.
4.4.3 Characterization of monodispersity and thermal stability of disulfide-trapped complexes with SEC and CPM-DSF
SEC and CPM-DSF represent the most stringent and specific assays for characterization of disulfide trapped complexes. This is because even the most clear-cut crosslinks may introduce minor distortions to the native favorable positions and orientations of the receptor and the chemokine in the complex; such distortions are detectable by lowered thermal stability or increased aggregation of the purified complexes as compared to an ideal geometry crosslink.
4.4.3.1. SEC and CPM-DSF experimental protocols
Complex monodispersity is analyzed by analytical size exclusion chromatography (SEC) using a Sepax SRT-C 300 column. Details of the assay are described in Chapter 11 by Gustavsson et al. Absorbance at 280 nm (A280) is recorded and plotted as a function of elution time to obtain the SEC trace. As implemented in our hands, the samples are analyzed one-by-one with each run taking 15–20 minutes (assuming the column is properly equilibrated), i.e. it is lower throughput than other experiments described herein (Table 1).
Thermostabilities of purified complexes are analyzed by a differential scanning fluorimetry (DSF) assay adapted from previous publications (Alexandrov, Mileni, Chien, Hanson, & Stevens, 2008). Receptor:chemokine samples are mixed with a thiol-reactive 7-Diethylamino-3-(4'-Maleimidylphenyl)-4-Methylcoumarin (CPM) dye that increases in fluorescence upon interaction with internal (non-disulfide-bonded) cysteines of the receptor that gradually become exposed and accessible for labeling as a result of thermal denaturation. The melting curve is obtained by plotting CPM fluorescence as a function of temperature (Figures 10A, 11A); in the canonical case, this curve has a clear sigmoidal shape and its first derivative has a clear sharp peak whose x-coordinate is interpreted as the melting temperature (Tm) of the sample (selected curves in Figures 10B, 11B). We perform this assay using a RotorGene Q 6-plex RT-PCR machine (Qiagen), which allows miniaturization and relatively high throughput (up to 72 samples can be analyzed at a time and sample volume can be as low as 20 μL). Briefly, 0.2–0.5 μg of protein is mixed with 5 μM CPM dye in 25 mM HEPES pH 7.5, 400 mM NaCl, 0.025% DDM, 0.005% CHS, 10% glycerol to a final volume of 20 μL; samples are incubated for 5 min at room temperature and then heated gradually from 28 °C to 90 °C at a rate of 1.5 °C/min, with CPM fluorescence (excitation 365 nm, emission 460 nm) recorded at every 1 °C increase. Rotor-Gene Q – Pure Detection software is used to extract the Tm from the first derivative plot of the denaturation curve.
Figure 10.
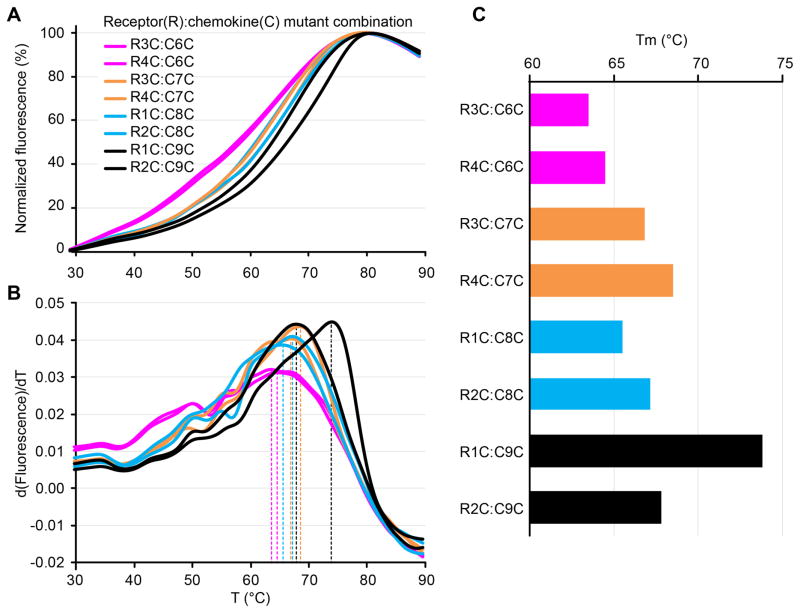
CPM-DSF characterization of several cysteine mutant pairs for a CC chemokine and its receptor. (A) Melting curves plotting the increasing fluorescence of the CPM dye as a function of temperature. (B) First derivative curves of (A) demonstrate the positions of the peaks used to calculate Tm. (C) Melting temperatures are calculated from the derivative curves. One mutant combination stands out as having a significantly higher Tm.
Figure 11.
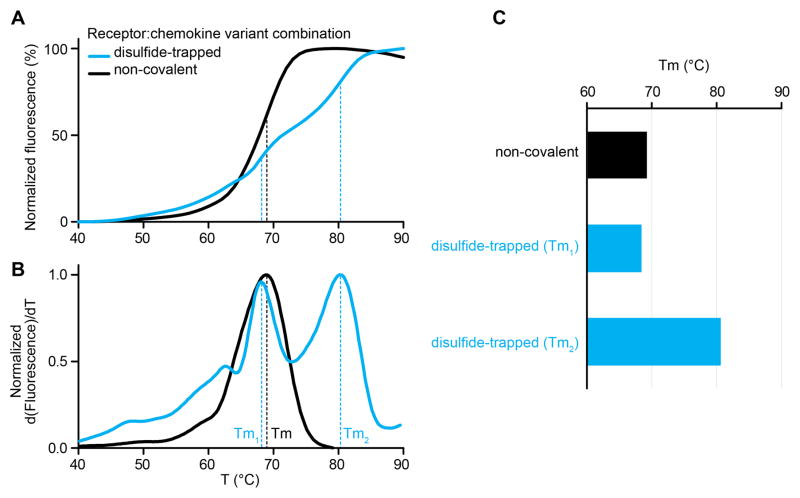
CPM-DSF characterization of non-covalent and disulfide-trapped complexes of a CXC chemokine and its receptor. (A) Melting curves plotting the increasing fluorescence of the CPM dye as a function of temperature. A biphasic transition is observed for the disulfide-trapped sample, likely due to the presence of both non-covalent and disulfide-trapped sub-populations. (B) First derivative curves of (A) demonstrate the positions of the peaks used to calculate Tm. A double-peak is clearly present for the disulfide-trapped sample. (C) Melting temperatures are calculated from the derivative curves. The disulfide-trapped sample shows two Tm’s one of which is indistinguishable from the non-covalent sample Tm.
4.4.3.2. Interpretation
An ideal SEC curve has a narrow sharp peak at the elution time that represents the hydrodynamic volume of the protein or complex in question. Protein:protein complexes typically elute earlier than the isolated components due to higher hydrodynamic volume. An ideal peak is tall (compared to controls), narrow, and sharp with no shoulders on either side, indicating high concentration and high degree of monodispersity (homogeneity) of the sample (e.g. Figure 12C). A widened peak may result from conformational heterogeneity or from formation of dimers and higher order oligomers. A shoulder on the left of the main peak is typically due to protein aggregates and is undesirable. The heights and shapes of SEC peaks obtained in parallel for several disulfide-trapped complex candidates (with proper controls) can be used to rank the candidates in terms of both protein concentration (or, with uniform treatment, yield) and monodispersity.
Figure 12.
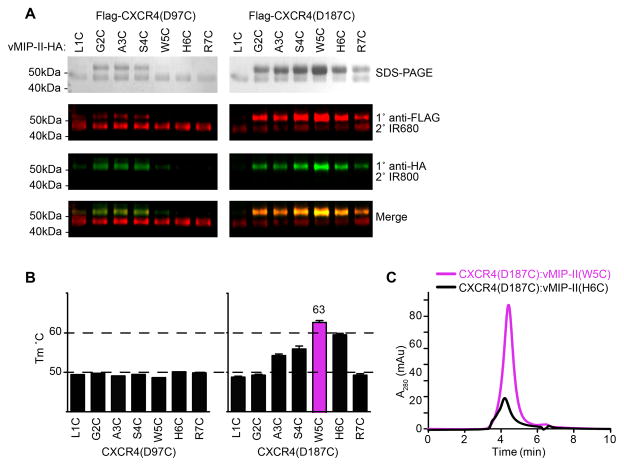
Candidate CRS2 crosslinks between CXCR4 and vMIP-II identified by SDS-PAGE and Western blotting (A) were further subjected to CPM-DSF (B) and SEC (C). Although spatially close and similar in both yield and crosslinking efficiency to the neighboring residues (A), pairs of CXCR4(D187C) with vMIP-II(W5C) and vMIP-II(H6C) stand out as having higher thermal stability (B). Furthermore, the complex with vMIP-II(W5C) has a much better peak shape and height than the complex with vMIP-II(H6C), indicating a higher yield and higher degree of monodispersity of vMIP-II(W5C) than other candidate pairs. Figure adapted from (Qin et al., 2015).
A canonical CPM-DSF curve has a clear sigmoidal shape and its first derivative has a single peak, indicating a monophasic melting transition of the sample. The height of the derivative peak is not necessarily representative of the sample yield; instead, it is a characteristic of the sharpness of the melting transition. Poor (flat) transitions, with short and wide derivative peaks (Figure 10A, pink curves), may result not only from insufficient sample concentrations but also from high degree of heterogeneity or the overall low quality and stability of the sample; they may also be an inherent property of the protein in question (e.g. due to a very small number of buried cysteine residues that can potentially become exposed and accessible for CPM labeling). In high quality disulfide trapped samples, two distinct transitions are sometimes discernible: one corresponding to melting of the non-crosslinked population and another (at a higher Tm) corresponding to the crosslinked population (Figure 11A,B). As for individual samples above, the relative height of the derivative peaks should not be interpreted as relative abundance of one population over another, or as fraction crosslinked, but rather simply as the sharpness of the melting transition.
Compared to SDS-PAGE and Western blotting, SEC and DSF typically provide much better resolution in distinguishing optimal from neighboring suboptimal crosslinks. Depending on implementation, DSF and SEC may also be more or less time- and resource-consuming than SDS-PAGE and Western blotting. In our hands, the RotorGene Q 6-plex RT-PCR instrument allows one to run up to 72 samples in parallel, with each of them taking only about 5% of the protein that is typically used for SDS-PAGE, thus far exceeding both SDS-PAGE and Western blotting in throughput and miniaturization. In view of this, when complex stabilization (and not simply determination of residue proximities) is the primary goal, high throughput DSF can be used as a first line of screening of the candidate pairs, with only the complexes showing clear melting transitions and potentially increased melting temperatures being subject to SDS-PAGE.
Figure 12 illustrates an example where multiple promising crosslinks (all spatially close) were identified in a receptor:chemokine complex by gel electrophoresis and Western blotting (Figure 12A), but only two of them resulted in a significant increase in the complex melting temperature (Figure 12B), and only one of them stood out in terms of complex yield and monodispersity as demonstrated by the height and the quality of the SEC trace (Figure 12C). Following these stringent validation steps, the most promising crosslinks may serve as tools for structure determination of the complex, as was the case for a CXCR4:vMIP-II complex by Qin et al (Qin et al., 2015).
5. Conclusion and perspectives
Although a useful and relatively straightforward methodology for both modeling and structure determination of protein:protein complexes, disulfide trapping has numerous caveats, especially when applied to interfaces rich in native intramolecular disulfide bonds such as those of chemokines with their receptors. Combined evidence from several biophysical experiments as well as critical assessment of results in the context of 3D models enable one to separate false positives from energetically favorable crosslinks that stabilize the native geometry of the complex. Prioritizing high-throughput high-sensitivity assays helps optimize the pipeline by early triage of unsuccessful candidates. Iterative application of the methodology in conjunction with molecular modeling is a viable strategy for elucidating structural determinants of receptor:chemokine interactions.
Acknowledgments
Authors thank Dr. Ruben Abagyan (UCSD) for useful discussions. This work was partially supported by NIH grants U01 GM094612, U54 GM094618, R01 GM071872, R01 AI118985, and R01 AI37113.
References
- Alexandrov AI, Mileni M, Chien EYT, Hanson MA, Stevens RC. Microscale Fluorescent Thermal Stability Assay for Membrane Proteins. Structure. 2008;16(3):351–359. doi: 10.1016/j.str.2008.02.004. Retrieved from http://www.sciencedirect.com/science/article/B6VSR-4S1K22J-8/2/541b65b93c931f875a192deee2cf6575. [DOI] [PubMed] [Google Scholar]
- Bachelerie F, Ben-Baruch A, Combadiere C, Farber JM, Graham GJ, Horuk R, … Zlotnik A. Chemokine receptors, introduction. IUPHAR/BPS Guide to PHARMACOLOGY. 2015 Oct 08; Retrieved from http://www.guidetopharmacology.org/GRAC/FamilyIntroductionForward?familyId=14.
- Baggiolini M. Chemokines and leukocyte traffic. Nature. 1998;392(6676):565–568. doi: 10.1038/33340. Retrieved from http://dx.doi.org/10.1038/33340. [DOI] [PubMed] [Google Scholar]
- Bill RM, Henderson PJF, Iwata S, Kunji ERS, Michel H, Neutze R, … Vogel H. Overcoming barriers to membrane protein structure determination. Nat Biotech. 2011;29(4):335–340. doi: 10.1038/nbt.1833. Retrieved from http://dx.doi.org/10.1038/nbt.1833. [DOI] [PubMed] [Google Scholar]
- Brelot A, Heveker N, Montes M, Alizon M. Identification of residues of CXCR4 critical for human immunodeficiency virus coreceptor and chemokine receptor activities. J Biol Chem. 2000;275(31):23736–23744. doi: 10.1074/jbc.M000776200. Retrieved from http://www.hubmed.org/display.cgi?uids=10825158. [DOI] [PubMed] [Google Scholar]
- Buck E, Wells JA. Disulfide trapping to localize small-molecule agonists and antagonists for a G protein-coupled receptor. Proc Natl Acad Sci USA. 2005;102(8):2719–2724. doi: 10.1073/pnas.0500016102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burg JS, Ingram JR, Venkatakrishnan AJ, Jude KM, Dukkipati A, Feinberg EN, … Garcia KC. Structural basis for chemokine recognition and activation of a viral G protein–coupled receptor. Science. 2015;347(6226):1113–1117. doi: 10.1126/science.aaa5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charo IF, Ransohoff RM. The Many Roles of Chemokines and Chemokine Receptors in Inflammation. New England Journal of Medicine. 2006;354(6):610–621. doi: 10.1056/NEJMra052723. [DOI] [PubMed] [Google Scholar]
- Chen Q, Pinon DI, Miller LJ, Dong M. Molecular Basis of Glucagon-like Peptide 1 Docking to Its Intact Receptor Studied with Carboxyl-terminal Photolabile Probes. Journal of Biological Chemistry. 2009;284(49):34135–34144. doi: 10.1074/jbc.M109.038109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Pinon DI, Miller LJ, Dong M. Spatial Approximations between Residues 6 and 12 in the Amino-terminal Region of Glucagon-like Peptide 1 and Its Receptor: A REGION CRITICAL FOR BIOLOGICAL ACTIVITY. Journal of Biological Chemistry. 2010;285(32):24508–24518. doi: 10.1074/jbc.M110.135749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevigne A, Fievez V, Schmit JC, Deroo S. Engineering and screening the N-terminus of chemokines for drug discovery. Biochemical Pharmacology. 2011;82(10):1438–1456. doi: 10.1016/j.bcp.2011.07.091. http://dx.doi.org/10.1016/j.bcp.2011.07.091. [DOI] [PubMed] [Google Scholar]
- Choi WT, Tian S, Dong CZ, Kumar S, Liu D, Madani N, … Huang Z. Unique ligand binding sites on CXCR4 probed by a chemical biology approach: implications for the design of selective human immunodeficiency virus type 1 inhibitors. J Virol. 2005;79(24):15398–15404. doi: 10.1128/JVI.79.24.15398-15404.2005. Retrieved from http://www.hubmed.org/display.cgi?uids=16306611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coin I, Katritch V, Sun T, Xiang Z, Siu FY, Beyermann M, … Wang L. Genetically Encoded Chemical Probes In Cells Reveal the Binding Path of Urocortin-I to CRF Class B GPCR. Cell. 2013;155(6):1258–1269. doi: 10.1016/j.cell.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong M, Lam PCH, Gao F, Hosohata K, Pinon DI, Sexton PM, … Miller LJ. Molecular approximations between residues 21 and 23 of secretin and its receptor: development of a model for peptide docking with the amino terminus of the secretin receptor. Mol Pharmacol. 2007;72(2):280–290. doi: 10.1124/mol.107.035402. Retrieved from http://www.hubmed.org/display.cgi?uids=17475809. [DOI] [PubMed] [Google Scholar]
- Dong M, Lam PCH, Pinon DI, Hosohata K, Orry A, Sexton PM, … Miller LJ. Molecular basis of secretin docking to its intact receptor using multiple photolabile probes distributed throughout the pharmacophore. J Biol Chem. 2011;286(27):23888–23899. doi: 10.1074/jbc.M111.245969. Retrieved from http://www.hubmed.org/display.cgi?uids=21566140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong M, Xu X, Ball AM, Makhoul JA, Lam PCH, Pinon DI, … Miller LJ. Mapping spatial approximations between the amino terminus of secretin and each of the extracellular loops of its receptor using cysteine trapping. FASEB J. 2012 doi: 10.1096/fj.12-212399. Retrieved from http://www.hubmed.org/display.cgi?uids=22964305. [DOI] [PMC free article] [PubMed]
- Duchesnes CE, Murphy PM, Williams TJ, Pease JE. Alanine scanning mutagenesis of the chemokine receptor CCR3 reveals distinct extracellular residues involved in recognition of the eotaxin family of chemokines. Molecular Immunology. 2006;43(8):1221–1231. doi: 10.1016/j.molimm.2005.07.015. Retrieved from http://www.sciencedirect.com/science/article/B6T9R-4GWC0KX-2/2/4257c4d48a2d0ae36c4d6d45012b06d4. [DOI] [PubMed] [Google Scholar]
- Gerard C, Rollins BJ. Chemokines and disease. Nat Immunol. 2001;2(2):108–115. doi: 10.1038/84209. Retrieved from http://dx.doi.org/10.1038/84209. [DOI] [PubMed] [Google Scholar]
- Ghosh E, Kumari P, Jaiman D, Shukla AK. Methodological advances: the unsung heroes of the GPCR structural revolution. Nat Rev Mol Cell Biol. 2015;16(2):69–81. doi: 10.1038/nrm3933. http://www.nature.com/nrm/journal/v16/n2/abs/nrm3933.html#supplementary-information. [DOI] [PubMed] [Google Scholar]
- Griffith JW, Sokol CL, Luster AD. Chemokines and Chemokine Receptors: Positioning Cells for Host Defense and Immunity. Annual Review of Immunology. 2014;32(1):659–702. doi: 10.1146/annurev-immunol-032713-120145. [DOI] [PubMed] [Google Scholar]
- Grunbeck A, Huber T, Abrol R, Trzaskowski B, Goddard WA, Sakmar TP. Genetically Encoded Photo-cross-linkers Map the Binding Site of an Allosteric Drug on a G Protein-Coupled Receptor. ACS Chemical Biology. 2012;7(6):967–972. doi: 10.1021/cb300059z. [DOI] [PubMed] [Google Scholar]
- Grunbeck A, Sakmar TP. Probing G Protein-Coupled Receptor—Ligand Interactions with Targeted Photoactivatable Cross-Linkers. Biochemistry. 2013;52(48):8625–8632. doi: 10.1021/bi401300y. [DOI] [PubMed] [Google Scholar]
- Gupta SK, Pillarisetti K, Thomas RA, Aiyar N. Pharmacological evidence for complex and multiple site interaction of CXCR4 with SDF-1alpha: implications for development of selective CXCR4 antagonists. Immunology Letters. 2001;78(1):29–34. doi: 10.1016/s0165-2478(01)00228-0. http://dx.doi.org/10.1016/S0165-2478(01)00228-0. [DOI] [PubMed] [Google Scholar]
- Hagemann IS, Miller DL, Klco JM, Nikiforovich GV, Baranski TJ. Structure of the Complement Factor 5a Receptor-Ligand Complex Studied by Disulfide Trapping and Molecular Modeling. Journal of Biological Chemistry. 2008;283(12):7763–7775. doi: 10.1074/jbc.M709467200. [DOI] [PubMed] [Google Scholar]
- Huber T, Naganathan S, Tian H, Ye S, Sakmar TP. Unnatural Amino Acid Mutagenesis of GPCRs Using Amber Codon Suppression and Bioorthogonal Labeling. In: Conn PM, editor. Methods in Enzymology. Vol. 520. Academic Press; 2013. pp. 281–305. [DOI] [PubMed] [Google Scholar]
- Kim CH, Axup JY, Schultz PG. Protein conjugation with genetically encoded unnatural amino acids. Current Opinion in Chemical Biology. 2013;17(3):412–419. doi: 10.1016/j.cbpa.2013.04.017. http://dx.doi.org/10.1016/j.cbpa.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofuku Y, Yoshiura C, Ueda T, Terasawa H, Hirai T, Tominaga S, … Shimada I. Structural Basis of the Interaction between Chemokine Stromal Cell-derived Factor-1/CXCL12 and Its G-protein-coupled Receptor CXCR4. J Biol Chem. 2009;284(50):35240–35250. doi: 10.1074/jbc.M109.024851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufareva I, Handel TM, Abagyan R. Experiment-Guided Molecular Modeling of Protein–Protein Complexes Involving GPCRs. In: Filizola M, editor. G Protein-Coupled Receptors in Drug Discovery. Vol. 1335. Springer; New York: 2015. pp. 295–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufareva I, Salanga CL, Handel TM. Chemokine and chemokine receptor structure and interactions: implications for therapeutic strategies. Immunol Cell Biol. 2015;93(4):372–383. doi: 10.1038/icb.2015.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufareva I, Stephens BS, Holden LG, Qin L, Zhao C, Kawamura T, … Handel TM. Stoichiometry and geometry of the CXC chemokine receptor 4 complex with CXC ligand 12: Molecular modeling and experimental validation. Proc Natl Acad Sci USA. 2014;111(50):E5363–E5372. doi: 10.1073/pnas.1417037111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Wacker D, Wang C, Abola E, Cherezov V. Femtosecond crystallography of membrane proteins in the lipidic cubic phase. Philos Trans R Soc Lond B Biol Sci. 2014;369(1647):20130314. doi: 10.1098/rstb.2013.0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller LJ, Chen Q, Lam PCH, Pinon DI, Sexton PM, Abagyan R, Dong M. Refinement of Glucagon-like Peptide 1 Docking to Its Intact Receptor Using Mid-region Photolabile Probes and Molecular Modeling. Journal of Biological Chemistry. 2011;286(18):15895–15907. doi: 10.1074/jbc.M110.217901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan P, Thomas BE, Woznica I, Wittelsberger A, Mierke DF, Rosenblatt M. Mapping Peptide Hormone Receptor Interactions Using a Disulfide-Trapping Approach†. Biochemistry. 2008;47(22):5889–5895. doi: 10.1021/bi800122f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montaner S, Kufareva I, Abagyan R, Gutkind JS. Molecular Mechanisms Deployed by Virally Encoded G Protein-Coupled Receptors in Human Diseases. Annu Rev Pharmacol Toxicol. 2013;53(1):331–354. doi: 10.1146/annurev-pharmtox-010510-100608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraes I, Evans G, Sanchez-Weatherby J, Newstead S, Stewart PDS. Membrane protein structure determination — The next generation. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2014;1838(1 Part A):78–87. doi: 10.1016/j.bbamem.2013.07.010. http://dx.doi.org/10.1016/j.bbamem.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdoch C, Finn A. Chemokine receptors and their role in inflammation and infectious diseases. Blood. 2000;95(10):3032–3043. Retrieved from http://www.bloodjournal.org/bloodjournal/95/10/3032.full.pdf. [PubMed] [Google Scholar]
- Nijmeijer S, Leurs R, Smit MJ, Vischer HF. The Epstein-Barr virus-encoded G protein-coupled receptor BILF1 hetero-oligomerizes with human CXCR4, scavenges G{alpha}i proteins, and constitutively impairs CXCR4 functioning. J Biol Chem. 2010;285(38):29632–29641. doi: 10.1074/jbc.M110.115618. Retrieved from http://www.hubmed.org/display.cgi?uids=20622011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakianathan DR, Kuta EG, Artis DR, Skelton NJ, Hebert CA. Distinct but overlapping epitopes for the interaction of a CC-chemokine with CCR1, CCR3 and CCR5. Biochemistry. 1997;36(32):9642–9648. doi: 10.1021/bi970593z. Retrieved from http://www.hubmed.org/display.cgi?uids=9289016. [DOI] [PubMed] [Google Scholar]
- Pellequer JL, Chen S-wW. Multi-template approach to modeling engineered disulfide bonds. Proteins: Structure, Function, and Bioinformatics. 2006;65(1):192–202. doi: 10.1002/prot.21059. [DOI] [PubMed] [Google Scholar]
- Pham V, Sexton PM. Photoaffinity scanning in the mapping of the peptide receptor interface of class II G protein—coupled receptors. Journal of Peptide Science. 2004;10(4):179–203. doi: 10.1002/psc.541. [DOI] [PubMed] [Google Scholar]
- Proost P, Loos T, Mortier A, Schutyser E, Gouwy M, Noppen S, … Van Damme J. Citrullination of CXCL8 by peptidylarginine deiminase alters receptor usage, prevents proteolysis, and dampens tissue inflammation. The Journal of Experimental Medicine. 2008;205(9):2085–2097. doi: 10.1084/jem.20080305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L, Kufareva I, Holden LG, Wang C, Zheng Y, Zhao C, … Handel TM. Crystal structure of the chemokine receptor CXCR4 in complex with a viral chemokine. Science. 2015;347(6226):1117–1122. doi: 10.1126/science.1261064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransohoff RM. Chemokines and Chemokine Receptors: Standing at the Crossroads of Immunobiology and Neurobiology. Immunity. 2009;31(5):711–721. doi: 10.1016/j.immuni.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SGF, Choi HJ, Fung JJ, Pardon E, Casarosa P, Chae PS, … Kobilka BK. Structure of a nanobody-stabilized active state of the b2 adrenoceptor. Nature. 2011;469(7329):175–180. doi: 10.1038/nature09648. Retrieved from http://dx.doi.org/10.1038/nature09648, http://www.nature.com/nature/journal/v469/n7329/abs/nature09648.html#supplementary-information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini V, Staren DM, Ziarek JJ, Nashaat ZN, Campbell EM, Volkman BF, … Majetschak M. The CXC Chemokine Receptor 4 Ligands Ubiquitin and Stromal Cell-derived Factor-1alpha Function through Distinct Receptor Interactions. J Biol Chem. 2011;286(38):33466–33477. doi: 10.1074/jbc.M111.233742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholten DJ, Canals M, Maussang D, Roumen L, Smit MJ, Wijtmans M, … Leurs R. Pharmacological modulation of chemokine receptor function. Br J Pharmacol. 2011;165(6):1617–1643. doi: 10.1111/j.1476-5381.2011.01551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme J, Decock B, Lenaerts JP, Conings R, Bertini R, Mantovani A, Billiau A. Identification by sequence analysis of chemotactic factors for monocytes produced by normal and transformed cells stimulated with virus, double-stranded RNA or cytokine. European Journal of Immunology. 1989;19(12):2367–2373. doi: 10.1002/eji.1830191228. [DOI] [PubMed] [Google Scholar]
- Veldkamp CT, Seibert C, Peterson FC, De la Cruz NB, Haugner JC, III, Basnet H, … Volkman BF. Structural Basis of CXCR4 Sulfotyrosine Recognition by the Chemokine SDF-1/CXCL12. Sci Signal. 2008;1(37):ra4. doi: 10.1126/scisignal.1160755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittelsberger A, Corich M, Thomas BE, Lee BK, Barazza A, Czodrowski P, … Rosenblatt M. The Mid-Region of Parathyroid Hormone (1 34) Serves as a Functional Docking Domain in Receptor Activation†. Biochemistry. 2006;45(7):2027–2034. doi: 10.1021/bi051833a. [DOI] [PubMed] [Google Scholar]
- Zhang X, Stevens RC, Xu F. The importance of ligands for G protein-coupled receptor stability. Trends in Biochemical Sciences. 2015;40(2):79–87. doi: 10.1016/j.tibs.2014.12.005. http://dx.doi.org/10.1016/j.tibs.2014.12.005. [DOI] [PubMed] [Google Scholar]
- Zhou H, Tai HH. Expression and functional characterization of mutant human CXCR4 in insect cells: role of cysteinyl and negatively charged residues in ligand binding. Arch Biochem Biophys. 2000;373(1):211–217. doi: 10.1006/abbi.1999.1555. Retrieved from http://www.hubmed.org/display.cgi?uids=10620340. [DOI] [PubMed] [Google Scholar]