Abstract
Spatial normalization—applying standardized coordinates as anatomical addresses within a reference space—was introduced to human neuroimaging research nearly 30 years ago. Over these three decades, an impressive series of methodological advances have adopted, extended, and popularized this standard. Collectively, this work has generated a methodologically coherent literature of unprecedented rigor, size, and scope. Large-scale online databases have compiled these observations and their associated meta-data, stimulating the development of meta-analytic methods to exploit this expanding corpus. Coordinate-based meta-analytic methods have emerged and evolved in rigor and utility. Early methods computed cross-study consensus, in a manner roughly comparable to traditional (nonimaging) meta-analysis. Recent advances now compute coactivation-based connectivity, connectivity-based functional parcellation, and complex network models powered from data sets representing tens of thousands of subjects. Meta-analyses of human neuroimaging data in large-scale databases now stand at the forefront of computational neurobiology.
Keywords: human brain mapping, activation likelihood estimation, ALE, magnetic resonance imaging, MRI, fMRI
INTRODUCTION
Meta-analysis is most generally defined as the post hoc combination of numerical results from prior, independent studies. The original (and still most common) use of meta-analysis was to pool subsignificant effects from several small studies to determine which effects would achieve significance in larger samples (Pearson 1904). In particular, the method was developed to predict which adverse events were rare but real drug side effects and which were unrelated, random events. Neuroimaging meta-analysis, in contrast, pools statistically significant results to further improve predictive power, to build analytic tools and models, and to detect emergent properties of neural systems through large-scale data mining and computational modeling.
Coordinate-based meta-analysis (CBMA) methods collectively comprise a fairly recent, extremely powerful, rapidly evolving family of methods for mining and synthesizing the human neuroscience imaging literature. These methods rely on the widespread adoption by the neuroimaging research community of whole-brain analysis methods that reference a coordinate space, a unique and important accomplishment in its own right. In the earliest applications of CBMA, the primary objective was to report consensus locations and spatial probability distributions for specific functional areas and for widely used task-activation paradigms. The field has rapidly moved beyond this objective, now creating meta-analytic, synthetic images; performing experiments by contrasting these synthetic images in new ways; modeling the network properties of meta-analytic images; modeling interregional functional connectivity from very-large-scale meta-analyses; extracting highly plausible cortical parcellation schemes based on coactivation spatial probabilities; and, most recently, developing consensus-based functional attributions for regions and networks. In a very literal sense, meta-analyses are providing a steadily expanding, progressively enriched, potentially limitless consensus statement regarding the brain's structural and functional organization. That is, meta-analysis is creating the “collective mind.” A truly remarkable feature of these studies is the emergent properties they disclose. Studies that reported solely activation locations for a limited number of specific paradigms are being mined to model interregional connectivity and connectivity-based regional parcellation. The purpose of this review is to introduce the reader to this high-impact, rapidly evolving, highly exciting area of research.
COMMUNITY STANDARDS IN NEUROIMAGING
The neuroimaging community enjoys the enviable status of having developed analytic and reporting standards that not only provide excellent per-study sensitivity, but also enable a growing repertoire of spatial meta-analytic methods. The core analytic standards of the field are spatial normalization (coordinate-based anatomy) (see sidebar, Spatial Normalization), statistical parametric imaging, and local-maxima extraction.
Spatial Normalization
Spatial normalization is the most fundamental analytic standard underlying neuroimaging meta-analysis. The first human atlas that referenced a standardized, stereotaxic coordinate space was created by Jean Talairach, a French neurosurgeon and pioneer of quantitative human brain mapping. Talairach published a series of stereotactic atlases, the first of which (Talairach et al. 1967) reported functional locations (from electrical cortical stimulation) and structural boundaries (from pneumoencephalography) in standardized coordinates. Talairach was also the first person to describe the human brain in terms of functional and structural “probability distributions” (Talairach et al. 1967). The most widely used template is the Talairach & Tournoux (1988) atlas, produced specifically for image-based registration. The origin of Talairach's x-y-z coordinate space is the anterior commissure through which three orthogonal planes are oriented. The principal axis is the line connecting the anterior and posterior commissures (AC–PC line: the y axis). The remaining two axes are the x (right–left) and the z (superior–inferior).
The first algorithm for spatial normalization of tomographic brain images was a nine-parameter affine transformation published by Fox et al. (1985), with the goal of “facilitating direct comparison of experimental results from different laboratories” (p. 149), i.e., in anticipation of CBMA. Shortly thereafter, Fox and colleagues introduced functional-image averaging, which applied spatial standardization to combine images across subjects, decreasing image noise and improving the signal-to-noise ratio (Fox et al. 1988, Fox & Mintun 1989). Since that time, methods have steadily improved, expanding to nonaffine transformations including deformation field methods, which compute a unique warping vector for each image voxel (Toga 1998). Averaging functional images in standardized space is now the norm in human neuroscience imaging; tens of thousands of studies have been reported according to this convention (Fox 1995b).
Statistical Parametric Maps
Statistical parametric maps (SPMs) or statistical parametric images (SPIs) are 3-D arrays of group-wise statistical parameters computed from primary (“raw”), per-subject neuroimaging data. The original and still most widely reported type of SPM is computed from functional imaging data. SPMs can be generated from various types of functional imaging data, including H215O positron emission tomography (PET) and functional magnetic resonance imaging (fMRI). PET, the original SPM data source, is now rarely used, having been replaced by fMRI.
To generate functional SPMs, functional images are acquired under contrasting behavioral conditions that induce different brain activity patterns. Raw images are converted point-by-point (voxel-by-voxel) into images of statistical parameters that express the strength and consistency of the task-induced changes relative to an error term. Prior to the introduction of SPIs, analysis of functional images relied on regions of interest (ROIs), which sampled the data space in a predefined manner. Brain activations, however, can be quite discrete. Small errors in ROI placement can make large differences in the magnitude and statistical significance of observed effects. SPIs process the entire data matrix, allowing all task-induced changes to be detected. Thus, SPIs increase both the power and the objectivity of functional image analysis. Statistical parametric imaging was introduced to human brain mapping by Fox and colleagues (Fox et al. 1988, Fox & Mintun 1989). Friston and colleagues (1991, 1995) extended this image-analysis strategy to include multi-condition contrasts and computation of a wide range of statistical parameters, coining the term statistical parametric mapping. Correction for multiple comparisons (i.e., for the number of voxels tested) using Gaussian random field theory was a crucial additional advance. The SPM software packages, distributed by the Wellcome Trust Center for NeuroImaging (http://www.fil.ion.ucl.ac.uk), have been enormously influential and are among the most widely used worldwide.
Voxel-based morphometry (VBM) is the application of this same basic voxel-wise SPM strategy to high-resolution structural images to detect between-group anatomical differences (Ashburner & Friston 2000). Because the contrast computed (subtraction performed) is between-subjects, it does not have the advantage of within-subject task-control contrast (to eliminate background structure) permitted by functional SPM/SPI analyses. Without this within-subject control (used to eliminate background signal), VBM is intrinsically noisy and generally requires larger sample sizes to obtain significant effects and has a high false-positive rate. These limitations, however, make VBM even more likely to derive benefit from meta-analysis.
Local Maxima
As detected by SPM/SPI analysis, task-induced changes in brain activity or between-group differences in brain anatomy most often take the form of foci that are strongest (biggest effect size and most significant) at the center and fall off in an approximately Gaussian manner. A concise means of describing the location of an activated volume was necessary for analysis and publication. Fox and colleagues (1986, 1987) proposed that an area of activation could be viewed as a local maximum, the centroid of which could be estimated using a 3-D center-of-mass (COM) algorithm. Although it was new to brain imaging, this strategy had been known to astronomers for centuries as “vernier acuity” and to vision scientists for decades as “hyperacuity” (Fox et al. 1986). As applied to PET images with a spatial resolution (full-width at half maximum) of 1.8 cm, the spatial precision of hyperacute response localization was shown to be submillimeter (Mintun et al. 1989). This somewhat startling spatial precision argued strongly that response coordinates are a very valuable parameter to report and, subsequently, to compile in databases and to meta-analyze.
Current convention is to publish the locations of activation sites as the x-y-z coordinates of the COM or (less accurately) the peak voxel of each local maximum (or minimum). Additional data provided typically include the peak value of the statistical parameter forming the cluster (e.g., peak z value), the p-value of the peak voxel, the extent of the cluster (mm3) when thresholded to some p or z level, and various anatomical (e.g., hemisphere, lobe, gyrus) or functional (e.g., primary motor cortex) descriptors. The most widely used anatomical descriptors are hierarchical, volumetric labels that reference the 1988 Talairach atlas (Lancaster et al. 2000). An absolute requirement for inclusion of data in CBMA is that results are reported as COM (or peak voxel) addresses of local maxima detected in spatially standardized SPM/SPIs.
Data Volume
Papers following the above-described standards began appearing in the mid-1980s. Since then, the rate of publication has steadily risen. In 2005, we estimated the standards-compliant functional imaging literature to be no fewer than 4,000 articles (~16,000 experiments) with ~500 new articles (2,000 experiments) published per year (Fox et al. 2005a). As of 2007, Derrfuss & Mar (2009) estimated the conforming functional literature to be ~5,800 papers, with ~1,000 new conforming papers being published each year. The conforming literature now appears to be growing at a rate of more than 2,000 papers per year and likely exceeds 20,000 papers. With such a large volume of well-standardized data, the field of human neuroimaging provides uniquely fertile ground for meta-analysis.
Template Troubles
CBMA relies on the comparability of coordinates across studies. Satisfying this requirement does not mean that all studies must use the same brain template or the same normalization algorithm. Rather, it means that transforms capable of accurately converting between templates must be available, ideally validated prior to release of a new template. To correct some shortcomings of the Talairach & Tournoux (1988) atlas as a warping template, investigators at the Montreal Neurologic Institute (MNI) released a series of structural-MRI-derived templates created as averages of multiple subjects, seeking to create a template representative of a group rather than of an individual (reviewed in Evans et al. 2012). Unfortunately, the process used to create the template altered the origin and orientation (relative to the AC–PC standard) and expanded the brain size beyond the normal range, deviations which were not corrected prior to release. Compounding the problem, the discrepancies were not reported when the templates were released. Consequently, coordinates from studies using MNI templates (in MNI Space) did not correspond to coordinates or anatomical labels referenced to the Talairach space, although users (and reviewers and editors) were largely unaware of this discrepancy. When those in the field became aware of the problem, unvalidated (and ineffective) corrective transforms were made available online and variably applied, worsening the field's collective state of coordinate confusion. For a field at the forefront of computational biology, these were extremely unfortunate missteps. Eventually, Lancaster et al. (2007) quantified and corrected the discrepancies. The impact of these discrepancies on meta-analysis was quantified by Laird et al. (2010), specifically endorsing the use of Lancaster's transforms to move from MNI Space to Talairach Space or vice versa. Despite these efforts, the negative impact of coordinate confusion on database curation and meta-analysis is not fully resolved.
DATABASE DEVELOPMENT
Neuroimaging databases can be classified by the level of processing the data sets have received and the number of subjects per data set. Primary data set repositories contain per-subject images that typically have been processed to remove artifacts but not to compute statistical effects. At the other extreme are coordinate-data repositories, which share reduced data (tables of coordinates) extracted from SPMs computed from groups of subjects. An intermediate option is to provide group-wise SPMs as image volumes, i.e., without reduction to COM coordinates. The advantages and disadvantages of managing and utilizing each data type fall beyond the scope of this review, which is focused on meta-analyses of reported coordinates.
Coordinate Data Databases
BrainMap® (Fox et al. 1994; Fox & Lancaster 1994, 2002) was the first online database of neuroimaging results. BrainMap was initially created as a personal compilation (a spreadsheet) of brain-activation results in standardized coordinates from published and pilot (unpublished) studies carried out by Fox and colleagues using H215O PET in the laboratory of Marcus Raichle and Michel Ter-Pogossian. Funded in 1988 by the J.S. McDonnell Foundation, BrainMap was unveiled in November 1992, at the first of seven BrainMap workshops (Gibbons 1992). From the outset, the BrainMap strategy aimed to provide coordinate-based results linked to experimental meta-data—emphasizing experimental designs and behavioral conditions—to enable coordinate-based meta-analysis (Fox & Lancaster 1994). One of the core conceptual developments was the BrainMap coding scheme, a taxonomy of experimental design that provides meta-data descriptors intended not simply for retrieval of like studies but also for data-driven inferences concerning the functional properties of specific brain regions and networks (Fox et al. 1994). The BrainMap taxonomy has continued to evolve as the field has matured, with various conceptual validations (Fox et al. 2005b). The BrainMap taxonomy has also helped inform comparable taxonomies for primary, raw data repositories (Turner & Laird 2011).
At BrainMap's inception, the developers’ intent was for it to be a community-curated database. It quickly became clear that community curation was antithetical to quality control. Thus, the model evolved to encourage submission of data sets coded in BrainMap terms using software available at http://www.BrainMap.org, but all papers would be reviewed and edited by the BrainMap development team. Community members most active in submitting papers for entry are those in the process of performing a meta-analysis, which is symbiotic because it results in a peer-reviewed publication citing the resource (Laird et al. 2005b). At the time of this writing, BrainMap contained 11,103 functional-imaging experiments (88,000 coordinates) from 2,336 peer-reviewed publications and contained 2,444 structural-imaging experiments (16,311 coordinates) from 796 publications.
Derrfuss & Mar (2009) argued that a comprehensive database of the neuroimaging literature was a highly desirable objective and opined that the best way to achieve this objective was in a commercial, subscription-based publication format. Although both points are likely correct, neither has been realized. As an alternative, Neurosynth was created as an online, open-source, uncurated resource that automatically extracts coordinates from neuroimaging articles for meta-analysis (Yarkoni et al. 2011). Rather than use manual coding (à lá BrainMap), Neurosynth uses text-mining and machine-learning techniques to provide frequency-based weightings for behavioral and cognitive terms appearing in the coordinate-containing articles. These weightings are used to drive meta-analyses that can be performed directly from the Neurosynth web-interface, an offline computation in the BrainMap model. At time of writing, Neurosynth contained 5,809 studies (see Related Resources).
Primary Data Databases
Progress toward open sharing of primary (per-subject) experimental neuroimaging data (a community objective dating back to the late 1980s) has been slow, limited by the variability among sites in instrumentation and data-acquisition parameters, by the sheer size of the data sets, by patient confidentiality issues, and by the desire of investigators to protect their invested effort (and grant money) by maintaining access control. Despite these barriers, large, primary data sets are becoming steadily more accessible. The International Consortium for Brain Mapping (ICBM) was a pioneer in this effort (Mazziotta et al. 1995); ICBM data are still being actively downloaded. The Alzheimer's Disease Neuroimaging Initiative (ADNI; Butcher 2007) is a similarly managed, disease-specific initiative. A common data-sharing model is to provide open access to online descriptions of available data, with comprehensive access being managed by an oversight committee.
COORDINATE-BASED META-ANALYSIS
As indicated above, the traditional use of meta-analysis was to achieve statistical significance across studies for effects that failed to achieve significance in individual studies. In neuroimaging, however, the primary use of meta-analysis has been to synthesize the published literature (of significant results) to compute consensus effects and, thereby, place constraints on the interpretation, design, and analysis of subsequent studies (Fox et al. 1998). The first neuroimaging meta-analyses were reported in the context of primary data. That is, coordinates from extant reports were tabulated and plotted to constrain interpretations of new, primary data (Frith et al. 1991). Shortly thereafter, stand-alone neuroimaging meta-analyses began to appear (Tulving et al. 1994, Fox 1995a, Picard & Strick 1996), serving as quantitative reviews and for hypothesis generation. Although the first neuroimaging meta-analyses were statistically informal, this soon changed.
Functional Volumes Modeling
Anticipating the long-term impact of quantitative meta-analysis, Fox, Lancaster, and colleagues launched the BrainMap database in advance of (but hoping to stimulate) the invention of suitable meta-analytic algorithms. The first step in the evolution of quantitative CBMA tools was taken by Paus (1996), who computed and interpreted means and standard deviations of the x-y-z addresses in a review of studies of the frontal eye fields. This initiative was extended by correcting raw estimates of spatial location and variance for sample size to create scalable models of location probabilities [functional volumes models (FVM)] and to suggest uses of such models for data analysis (Fox et al. 1997, 1999, 2001). A specific limitation of FVM was that users needed to select responses that should be grouped, based on expert knowledge of the tasks performed and the region being studied. Although the models produced could be used by nonexperts, creating the models required considerable expertise.
Activation Likelihood Estimation
Activation likelihood estimation (ALE) (Turkeltaub et al. 2002) (Figure 1) and related algorithms (Chein et al. 2002, Wager et al. 2003) moved CBMA a quantum leap forward. ALE input data are activation-location coordinates from conceptually related studies (e.g., all Stroop tasks). ALE models the uncertainty in localization of activation foci using Gaussian probability density distributions. The voxel-wise union of these distributions yields the ALE value, a voxel-wise estimate of the likelihood of activation, given the input data. As with FVM, a great advantage of ALE is that the tables of coordinates routinely reported by neuroimaging studies are its input data: Raw data are not required. Unlike FVM, however, ALE requires no user selection of comparable coordinates for modeling; rather, once a set of experiments (e.g., a group of experiments using a similar paradigm) is selected for meta-analysis, the entire set of reported coordinates is used, thereby greatly increasing the reproducibility and objectivity of the analysis.
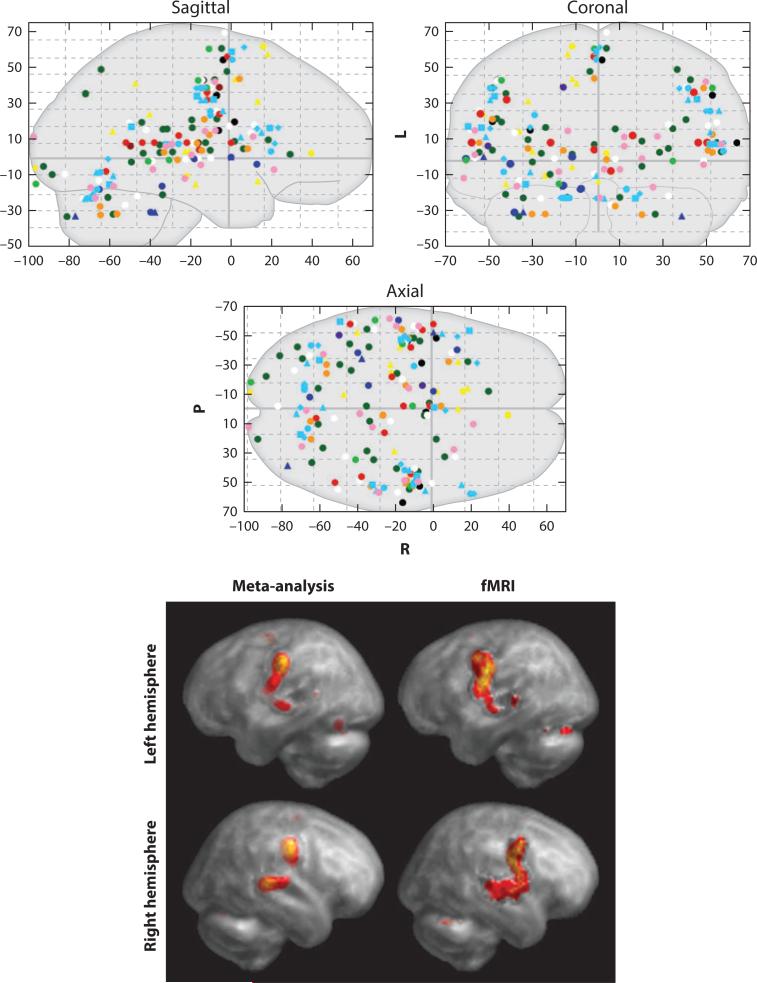
Figure 1.
ALE meta-analysis. Activation likelihood estimation (ALE) meta-analysis is illustrated. The top panels show spatial coordinates for single-word reading tasks reported by 11 PET studies (16 experiments; 117 subjects). The bottom panel shows a meta-analytic image computed using the ALE algorithm (left column) and an fMRI study performed using the same task (single-word reading). Images are reproduced with permission from Turkeltaub and colleagues (2002), the original ALE publication.
In the original implementation of ALE, the investigators acknowledged several limitations. For example, while applying the false discovery rate (FDR) method to compute voxel-wise significance, Turkeltaub used a fixed-effects analysis that did not correct for multiple comparisons; the size of the modeled Gaussian distribution was rationalized on the basis of the spatial resolution of the input images, rather than on a formal estimate of spatial uncertainty; a method for comparing two ALE maps was lacking; and there was no correction for the variable number of activations reported per experiment or the number of experiments per paper. Many of these limitations have subsequently been addressed by Turkeltaub and others. Laird et al. (2005a) provided an FDR correction for multiple comparisons and a method for ALE–ALE statistical contrasts. Eickhoff et al. (2009) introduced empirical estimates of between-subject and between-template spatial variability (a modification of the FVM spatial probability model) in place of user-selected Gaussian filtering. In addition, Eickhoff et al. (2009) modified the permutation test for above-chance clustering between experiments in an anatomically constrained space (gray-matter only), a transition from fixed-effects to random-effects inference. Turkeltaub et al. (2012) added corrections for the variable numbers of foci per experiment and experiments per paper to prevent undue weighting of ALE maps by individual experiments (e.g., with large numbers of foci) or individual papers (e.g., with multiple similar experiments). Eickhoff et al. (2012) provided an explicit solution for determining statistical significance rather than relying on FDR. Each of these improvements increased the statistical rigor and specificity of ALE.
Since its introduction, ALE has been applied to many aspects of normal brain function (Decety & Lamm 2007, Costafreda et al. 2008, Spreng et al. 2009). It has also been applied to functional and structural data in numerous disorders, including autism spectrum disorders (Duerden et al. 2012, Nickl-Jockschat et al. 2012), schizophrenia (Glahn et al. 2005, 2008; Ellison-Wright et al. 2008; Ellison-Wright & Bullmore 2009; Minzenberg et al. 2009; Ragland et al. 2009), epilepsy (Barron et al. 2012), Huntington's disease (Dogan et al. 2012, Lambrecq et al. 2013), obsessive-compulsive disorder (Menzies et al. 2008), depression (Fitzgerald et al. 2008), and developmental stuttering (Brown et al. 2005). The most interesting ALE applications do not merely merge previous results, but instead include highly previously ignored regions, resolve conflicting views, validate new paradigms, and generate new hypotheses for experimental testing. A more extensive list of ALE studies and algorithms is available at http://www.brainmap.org/pubs.
WITHIN-PARADIGM NETWORKS: CLIQUE ANALYSIS
Graph theory network modeling constructs—framing models as nodes connected by edges—have proven extremely well adapted for primary neuroimaging data sets of various types (Lohmann & Bohn 2002, Bullmore & Sporns 2009; see also sidebar, Graph Theory and Neuroimaging). For meta-analytic data sets, they have proven no less apt. In an early foray in this domain, Neumann applied replicator dynamics—a network discovery technique from theoretical biology based on principles of natural selection—to model the network properties of the Stroop task (Neumann et al. 2005). Noting that Neumann's method was limited to a single, dominant clique, Lancaster extended the algorithm to multiple subnets, adapting the Jaccard similarity measure for meta-analytic use, and quantitatively contrasted the two approaches (Lancaster et al. 2005). Note that for both these within-paradigm network-modeling approaches and the more advanced methods that followed (discussed below), graph “edges” are emergent properties, computed as coactivation patterns across studies with no comparable parameter being reported by the included studies.
BETWEEN-PARADIGM NETWORKS: META-ANALYTIC CONNECTIVITY MODELING
Meta-analytic approaches to assessing interregional connectivity are conceptually similar to functional non-meta-analytic approaches (e.g., resting state fMRI; Biswal et al. 1995) because they use temporal covariations in regional activation to detect connectivity. Whereas in a typical fMRI study the unit of time is the second, in the meta-analytic approach the unit of time is the study. Regions in which activations co-occur across studies (i.e., regions that are mutually predictive) are functionally connected; regions that do not co-occur are not connected. Higher probability of co-occurrence should reflect greater strength of functional connectivity.
Region to Region
The concept of a coactivation-based, meta-analytic connectivity mapping (MACM) was introduced by Koski & Paus (2000). To identify frontal lobe projections to the anterior cingulate gyrus, they manually collected and examined data from 107 studies, reporting differential connection patterns within different subregions of the anterior cingulate. Although they regarded their new approach as intrinsically plausible, Koski & Paus acknowledged that it lacked formal validation. In view of this shortcoming, they recommended replications using larger data sets, the development of statistically more sophisticated approaches, and validation of the approach against alternative connectivity measures, all of which eventually came to pass. Note that this application was region-to-region because it limited its scope to connections between frontal lobe and anterior cingulate gyrus.
Region to Whole Brain
The first region-to-whole-brain coactivation meta-analysis was reported by Postuma & Dagher (2006). Having identified 126 peer-reviewed, whole-brain studies with activations in caudate or putamen, the authors computed the first whole-brain, meta-analytic functional connectivity images. In these images, observed coactivation patterns were “consistent with the concept of spatially segregated cortico-striatal connections as predicted by previous anatomical labeling studies in nonhuman primates” (p. 1513). As with Koski & Paus (2000), no validation other than plausibility was offered.
The region-to-whole-brain analysis strategy was adopted and extended by Robinson et al. (2010). Using the Harvard/Oxford atlas to define the amygdala, the entire BrainMap database as the data source, and ALE to compute co-occurrence spatial probabilities, Robinson mapped the coactivation profile of the left and right amygdala. At that time, the BrainMap database contained 170 and 156 experiments for the left and right amygdala, respectively. To validate the approach she termed “meta-analytic connectivity modeling” (MACM), Robinson compared the amygdala MACM results with those obtained by various tract-tracing methods in rhesus monkeys, as reported in the CoCoMac database, finding startlingly good correspondence. Robinson et al. (2012) applied a similar strategy to the caudate nuclei, adding functional filtering using the BrainMap behavioral domain meta-data (Figure 2). Projection patterns were confirmed by diffusion tensor imaging (DTI) probabilistic tractography. Following Robinson's validation of MACM by comparing it with primate connectivity and DTI tractography, multiple other validations have been published, of which three are briefly presented here.
Figure 2.
Meta-analytic connectivity modeling (MACM) with behavioral domain filtering. The images illustrate the regional and behavioral specificity of connections of the caudate nucleus. Connectivity was computed as coactivation frequency with a seed region (caudate nucleus), sampling across the entire BrainMap database. Activation likelihood estimation (ALE) was used to compute statistical significance of co-occurrences. Behavioral filtering used the top tier of the BrainMap behavioral domain hierarchy. The projection patterns closely matched those established in the primate literature and were confirmed by diffusion tensor imaging (DTI) tractography. Images are reproduced from Robinson and colleagues (2012), the original report of behaviorally filtered MACM connectivity mapping, with permission.
MACM-derived connectivity patterns were compared with those obtained using DTI tractography by Eickhoff and colleagues (2010). Investigators compared connectivity patterns for two subdivisions of the human parietal operculum previously established using postmortem cytoarchitectonics (OP1 and OP4; Eickhoff et al. 2010). For MACM, the opercular regions of interest jointly extracted 245 experiments from the BrainMap database. For DTI, 18 healthy volunteers were studied. Comparison between techniques showed close (but not perfect) correspondence (Figure 3). Also, DTI tractography will provide connectivity limited to first-order (direct) connections, whereas MACM—showing all co-occurrences—would be expected to yield both direct and indirect connections. Furthermore, DTI will be intrinsically biased toward heavily myelinated connections, whereas MACM should preclude this bias.
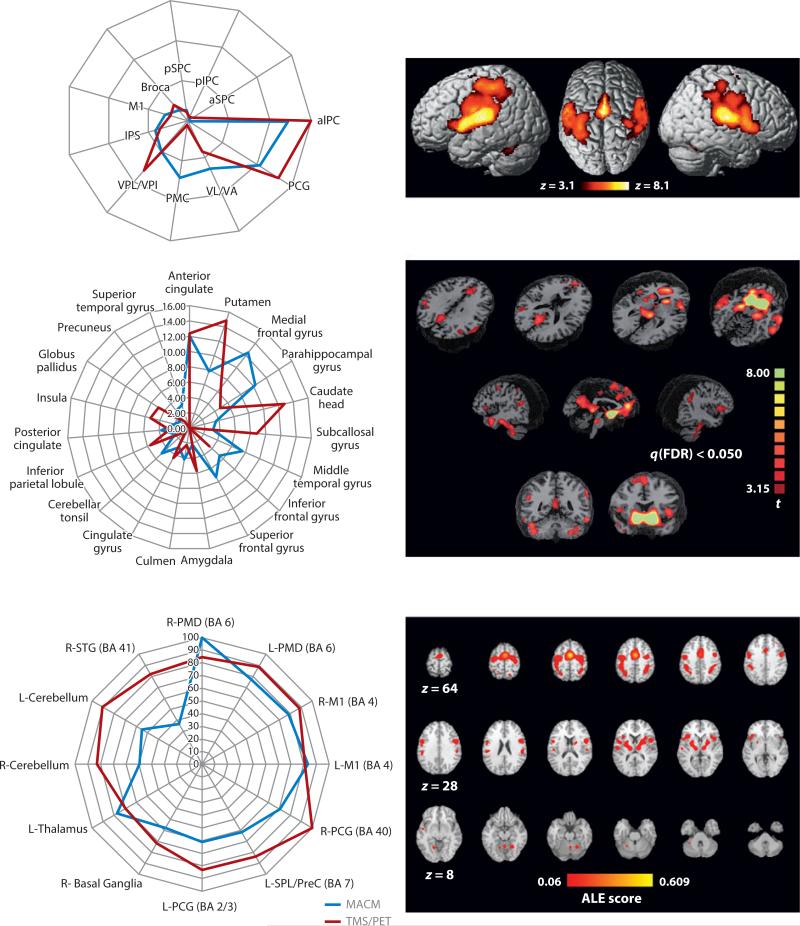
Figure 3.
Meta-analytic connectivity mapping (MACM) validations. The images illustrate three independent validations of MACM-derived connectivity patterns as forming one two-panel row. In each row, the left panel compares the strength of an MACM-derived projection with that of an alternative connectivity-mapping method. The right panels show the MACMs derived applying three different seed regions to the BrainMap database. The top row used DTI tractography as the validation methodology, seeding two regions within the parietal operculum (OP1, OP4; Eickhoff et al. 2010). The middle row used resting-state fMRI as the validation methodology, seeding nucleus accumbens (Cauda et al. 2011). The bottom row used concurrent transcranial magnetic stimulation (TMS)/PET as the validation methodology, seeding the supplementary motor area (SMA) (Narayana et al. 2012). Other abbreviations: aIPC, anterior inferior parietal cortex; aIPS, anterior intraparietal sulcus; aSPC, anterior superior parietal cortex; Broca, Broca's area; L-M1, left primary motor cortex; L-PCG/BA2/3, left postcentral gyrus, Brodmann areas 2 and 3; L-PMD, left dorsal premotor cortex; L-SPL/PreC, left superior parietal lobule/precuneus; M1, primary motor cortex; PCG, postcentral gyrus; pIPC, posterior inferior parietal cortex; PMC, premotor cortex; pSPC, posterior superior parietal cortex; R-M1, right primary motor cortex; R-PCG, right postcentral gyrus; R-PMD, right dorsal premotor cortex; R-STG (BA 4), right superior temporal gyrus, Brodmann area 41; VL/VA, ventrolateral nuclei/ventrolateral anterior nuclei; VPL/VPI, ventroposterior lateral and inferior nuclei. Images are reproduced with permission from each manuscript.
For the nucleus accumbens, Cauda et al. (2011) compared MACM-derived connectivity with the resting-state fMRI connectivity. For the nucleus accumbens region of interest, BrainMap provided 57 experiments, a relatively small input data set. For resting-state fMRI, 17 healthy subjects were studied. Despite the limited amount of BrainMap data utilized, the MACM proved robust (Figure 3), as did the resting-state connectivity map. Overall, the two techniques converged, with resting-state connectivity showing somewhat greater sensitivity than did MACM. In this context, it is important to note that the sensitivity of MACM is strongly influenced by the size of the seed region and the volume of data in the BrainMap database. As the database becomes more densely populated, MACM will become more sensitive and allow progressively finer anatomical connectivity parcellations.
Narayana et al. (2012) compared MACM-derived connectivity with cortical-stimulation-based connectivity, using concurrent transcranial magnetic stimulation (TMS) and H215O PET to map remote projections of the supplementary motor area (SMA). For the SMA seed region, BrainMap provided 266 experiments from 187 papers. As with prior validation, the two techniques converged nicely (Figure 3). It should be noted that Robinson, Eickhoff, Cauda, and Narayana all used BrainMap's behavioral domain and/or paradigm class meta-data (discussed further below) to interpret the functional roles of both the seed regions and their connections.
Coactivation-Based Parcellation
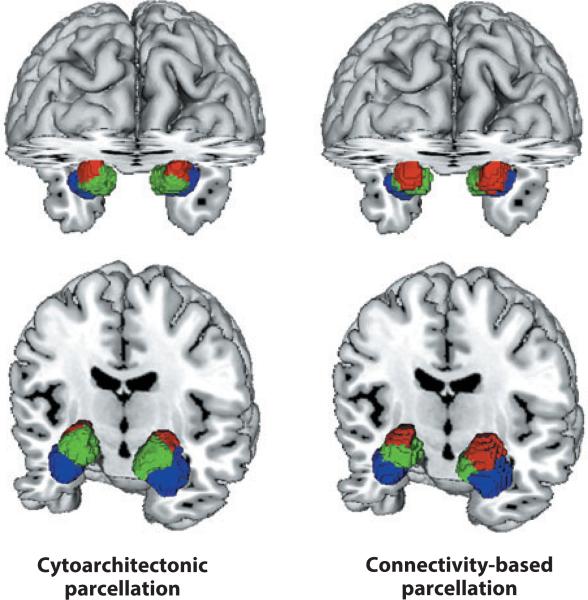
Coactivation-based parcellation (CBP) is an extremely exciting extension of the region-to-whole-brain meta-analytic connectivity-mapping approach. Connectivity-based parcellation of structural (DTI) data has provided close correspondence between structurally and functionally defined borders, using the boundary between SMA and pre-SMA as a demonstration case (Johannes-Berg et al. 2004). Eickhoff and colleagues (2011) applied connectivity-based parcellation to BrainMap data for the same brain regions (SMA and pre-SMA) and obtained virtually identical borders (Eickhoff et al. 2011). Providing further validation, Bzdok et al. (2012) applied connectivity-based parcellation to the amygdala, demonstrating close correspondence to previously defined cytoarchitectonic borders (Amunts et al. 2005) (Figure 4). This technique has subsequently been applied to differentiate “what” and “where” pathways in parietal cortex (Rottschy et al. 2012) and to classify subregions of dorsolateral prefrontal cortex (DLPFC; Cieslik et al. 2013), the medial prefrontal cortex (Bzdok et al. 2013), the cingulate cortex (Torta et al. 2013), and Broca's area (Brodmann area 44; Clos et al. 2013). In each of these applications, concurrent resting-state functional connectivity (using fMRI) corroborated the BrainMap-derived connectivity patterns.
Figure 4.
Co-activation-based parcellation. Connectivity-based parcellation of the amygdala as derived using the BrainMap database (right) proved highly similar to that observed (in a separate postmortem sample) using cytoarchitecture (left). Laterobasal nuclei group (blue); centromedial nuclei group (red ); superficial nuclei group (green). Images were reproduced with permission from Bzdok et al. (2012).
INDEPENDENT COMPONENTS ANALYSIS
In the two preceding sections, we have reviewed methods that drew connectivity inferences between two seed regions (less complex) and between a seed region and the whole brain (more complex). In the next two sections we review methods that draw connectivity inferences by comparing coactivations between every brain voxel with every other voxel (all-to-all). The two most general classes of network-discovery methods addressed are independent components analysis (ICA) and graph theoretical modeling. Both approaches have been applied to the entire BrainMap database (or large subsets thereof ), i.e., across behaviorally inhomogeneous paradigms.
Toro et al. (2008) used the BrainMap database to generate a comprehensive atlas of the brain's functional connectivity. At the time, BrainMap included 3,402 experiments (conditional contrasts) reporting a total of 27,909 activated locations. For each experiment, a binary, per-study activation volume was generated. From these, the co-occurrence pattern likelihood was computed between all voxels using likelihood ratios. This generated 45,000 unique coactivation maps (one for each 4 mm3 voxel in the brain). Reproducibility of the coactivation map was assessed by estimating the similarity between pairs of partial coactivation maps that used disjoint random subsamples of experiments for group sizes of 500, 1,000, 1,500, 2,000, 2,500, and 3,000 experiments. The correlation between maps was significant and increased asymptotically with the number of experiments and was strong with even the 500-experiment group. Thus, the coactivation maps did not depend on a particular choice of experiments, and a robust structure in the meta-analytic functional connectivity can be recovered even with a moderate number of studies.
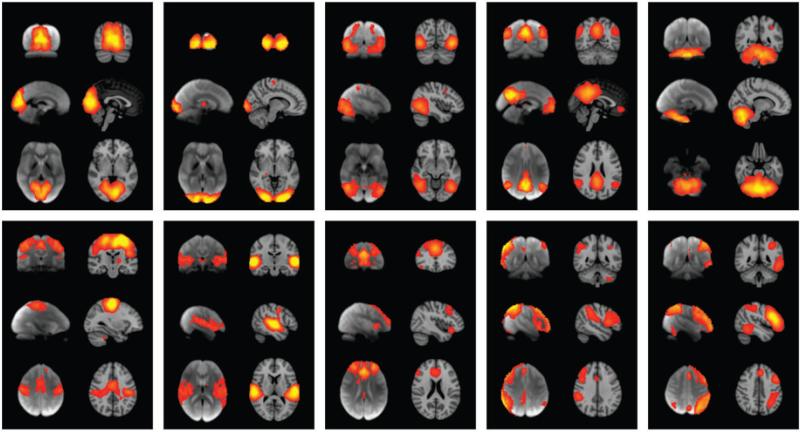
Intrigued by Toro's observations, Smith et al. (2009) took this strategy a step further and applied ICA to the entire BrainMap data volume. ICA has been widely used to demonstrate intrinsic connectivity networks in the resting brain using fMRI [i.e., resting-state networks (RSNs)]. Although observed at rest, Fox & Raichle (2007) proposed that RSNs represent basic organizational units of the brain and that they are “functional networks” used during task performance. Smith et al. (2009) tested this hypothesis by comparing ICA decompositions of resting-state fMRI to those derived from the BrainMap data. At the time of this data extraction (Fox & Raichle 2007), BrainMap contained 7,432 experiments, representing imaging studies from 29,671 human subjects. In parallel, ICA was performed using resting-state fMRI data from 36 healthy volunteers. Decompositions were performed into both 20 and 70 components.
Of the 20 components generated separately from the two data sets, ten maps from each set were unambiguously paired between data sets, with a minimum correlation r = 0.25 (p <10−5, corrected for multiple comparisons and for spatial smoothness). These ten well-matched pairs of networks are shown in Figure 5. With an ICA dimensionality of 70, the primary networks split into subnets in similar (but not identical) ways, continuing to show close correspondence between BrainMap and RSN components. This finding argues that the full repertoire of functional networks utilized by the brain in action (coded in BrainMap) is continuously and dynamically active even when at rest and, vice versa, that RSNs represent an intrinsic functional architecture of the brain that is drawn on to support task performance.
Figure 5.
The comparability of independent components derived from meta-analysis and resting-state networks is illustrated. Each of the ten panels shows one well-matched pair of networks from two, 20-component independent components analyses (ICA). In each panel, the left-side images are derived from a meta-analysis of the BrainMap database (~30,000 subjects); the right-side images are from a 36-subject resting-state fMRI database. Images are reproduced with permission from Smith and colleagues (2009).
GRAPH THEORY MODELING APPROACHES
As noted above, graph theory network modeling constructs have proven to be well suited for modeling both primary and reduced neuroimaging data. In recent work, these graph theoretical constructs have been applied meta-analytically for voxel-wise, all-to-all (whole-brain to whole-brain) network discovery (see sidebar, Graph Theory and Neuroimaging).
Bayesian Network Discovery
In 2005, Neumann et al. introduced the use of graph theory modeling techniques to within-paradigm meta-analysis (above) using the Stroop paradigm. In 2008, Neumann et al. introduced the use of hierarchical Gaussian analysis to ALE output data (again using the Stroop paradigm). In 2010, Neumann et al. introduced the use of Bayesian network graphs to represent statistical dependencies using all-to-all voxel-wise analyses as a starting point. Bayesian network graphs are probabilistic models that represent a set of random variables and their probabilistic interdependencies. More formally, a Bayesian network is a directed acyclic graph (DAG) that comprises a set of nodes (vertices) and directed links (edges) connecting these nodes. Bayesian networks were chosen for three reasons. First, they belong to the class of directed graphical models, which permits investigation of directed interdependencies between the activation(s) of multiple brain regions. Second, the structure of Bayesian networks can be inferred from observed data. Thus, statistical interdependencies between the brain regions can be inferred from observations across multiple imaging experiments. Third, the theory for learning Bayesian networks from data is well established.
In application to neuroimaging meta-analysis, Neumann's approach used coactivation patterns of brain regions across imaging studies to learn the structure of the underlying DAGs. This was done first by computing an ALE map of a large subset (2,505 experiments) of the BrainMap data. This map was then restricted to the 49 most commonly occurring regions and further restricted to the 13 most commonly co-occurring regions using three separate applications of the replicator dynamics process, each of which identified subsets of regions. The regions included part of the posterior medial frontal cortex primarily covering SMAs and pre-SMAs, the anterior cingulate cortex, posterior parts of the lateral prefrontal cortex bilaterally, the dorsal premotor cortex bilaterally, the left and right anterior insula, the left and right thalamus, the left and right anterior intraparietal sulcus, and the left cerebellum. For these regions, DAGs were computed for groupings provided by each run of replicator dynamics and for the collection of all regions. Although this approach began with the entire BrainMap corpus, the analytic strategy promoted serial, data-driven reductions in the scope of the analysis to specific brain regions. This is distinctly unlike ICA (above), which groups voxels into components without ever reducing the total volume under consideration from the whole brain.
Large-Scale Topological Modeling
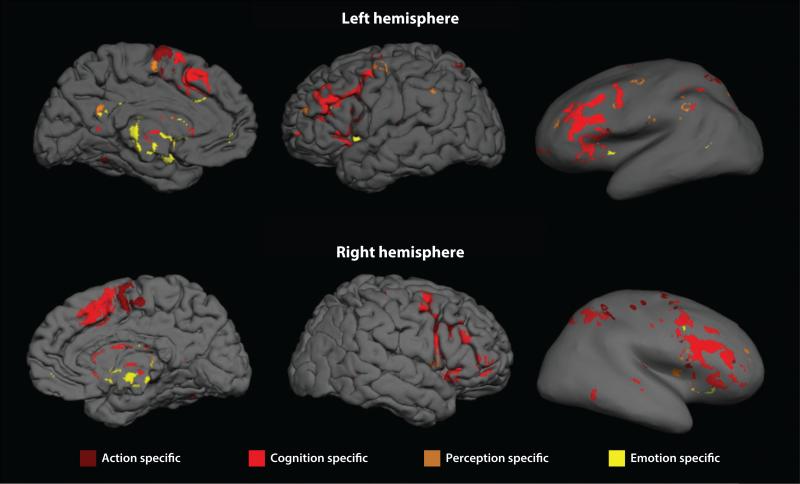
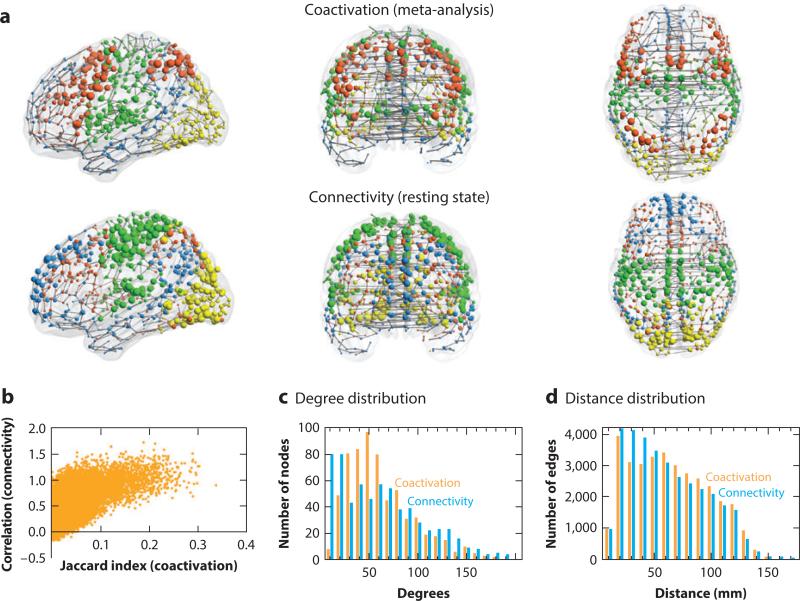
Expanding the all-to-all graph theory approach to neuroimaging meta-analysis, Crossley et al. (2013) estimated the relative frequency at which each pair of regions in standard space was coactivated by multiple tasks reported in the primary literature. Nodes that frequently coactivated were connected by an edge. The resulting functional coactivation graph (Figure 6) had complex topological properties such as modules, hubs, and a rich club. The community structure of the meta-analytic network could be linked to the tasks in the primary literature, demonstrating that the modules were functionally specialized (for perception, action, emotion, etc.), whereas the rich club was more diversely coactivated by tasks requiring both action and cognition. It was also notable that the modules defined by this graph theoretical analysis of the BrainMap database were almost exactly supersets of the independent components identified by ICA-based meta-analysis (Smith et al. 2009), which indicates that different methods of network meta-analysis can generate convergent rather than contradictory insights. This also demonstrates the important role that open-access databases can play in supporting comparative evaluation of alternative methodologies.
Figure 6.
Topological analysis. The comparability of topological networks derived from meta-analysis of the BrainMap database (top row) and resting-state fMRI in 27 subjects (middle row) is illustrated. In anatomical space (a, upper two rows) the size of the nodes is proportional to their weighted degree (strength), and their color corresponds to module membership. The relationship between the coactivation metric (Jaccard index; b) and the connectivity metric for every pair of regions is graphed (b). The degree and distance distributions for both networks are plotted (c and d, respectively). Images are reproduced with permission from Crossley et al. (2013).
FUNCTIONAL ONTOLOGIES
For systems modeling, meta-analysis has the substantial advantage of being able to filter its findings with the behavioral meta-data associated with each experiment in the BrainMap database. Behavioral filtering has been widely used when selecting papers for inclusion in a meta-analysis (quantified and discussed in Fox et al. 2005b). A more recently developed use of behavioral meta-data is to characterize the behavioral properties of specific networks (Robinson et al. 2010, Cauda et al. 2011). Statistical methods to test for between-region differences in behavioral domain profiles have been developed (Lancaster et al. 2012) and are currently being extended to paradigm class data. Using this approach, given sufficient numbers of experiments and well-developed behavioral meta-data, unique behavioral characterization of individual brain regions may be a viable possibility. Characterization will be done, however, using complex behavioral profiles rather than using a one- or two-word term (“put,” “get,” “move,” “selection for action”) assigning a mental operation to each brain region, as Posner et al. (1988) had suggested. Our approach is more concordant with the views of Price & Friston (2005), who argued that the mapping between mental operations and brain regions is a many-to-many mapping in which a single region can be involved in many cognitive processes and a single elementary process engages multiple regions. It is also concordant with Poldrack's (2006) argument that the cognitive “reverse inference” (i.e., that a specific mental operation is necessarily engaged if a particular brain region is activated) is intrinsically weak, owing in part to participation of individual regions in multiple cognitive operations. An extension of the behavioral domain profile approach is to extract profiles for multiple regions jointly, i.e., to characterize a functional network. This strategy was employed by Laird et al. (2009) in work that behaviorally categorized the default mode network (DMN), examining behavioral domain profiles of individual areas and of groups of areas (i.e., subnetworks).
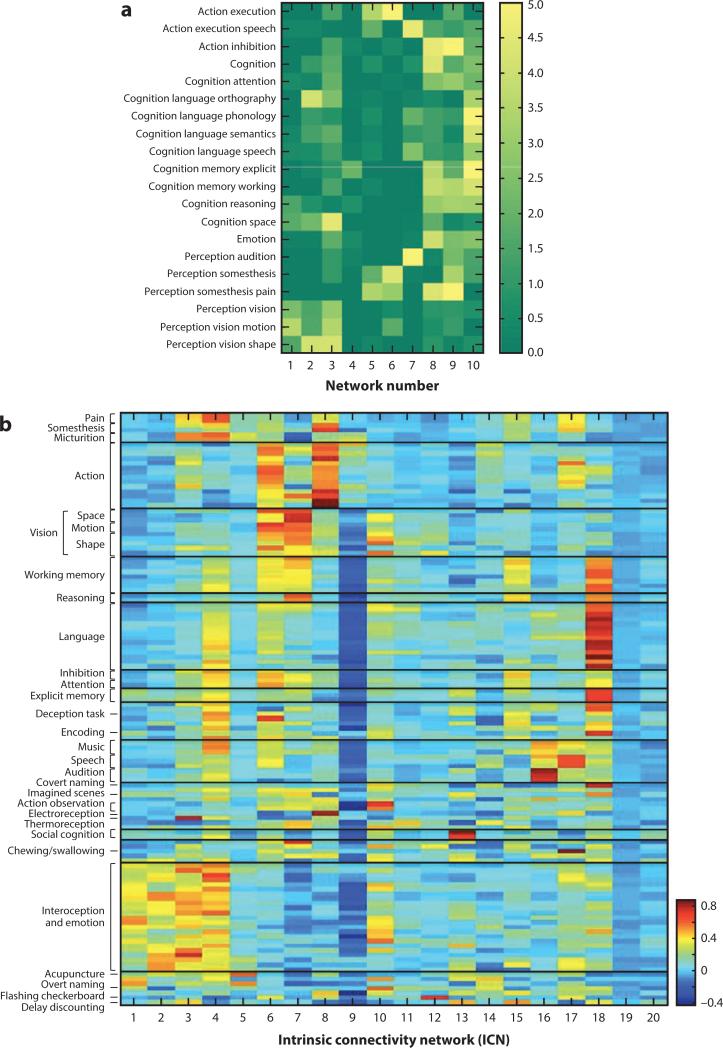
Another strategy for meta-analytic structure-function inference was pioneered by Smith and colleagues (2009), in the context of applying ICA to the BrainMap database. Figure 7 (left side) is a heat map showing the respective contributions of BrainMap behavioral domains to individual components in the ICA shown in Figure 5. Close inspection reveals that some components have very high behavioral specificity, whereas other components have contributions from a wide range of behavioral domains. The ICA-based strategy of Smith and colleagues has been extended by Laird et al. (2011b) both by enriching the meta-data included in the analysis and by applying hierarchical clustering analysis to sort components into functionally related groupings (Figure 7). Even though this approach provides a much more refined association of components with behaviors, some components still show limited behavioral specificity. The most likely explanations for this lack of behavioral specificity in some networks are twofold: First, the behavioral specificity of some regions and networks (“hubs” in the terminology of topological modeling) is almost certainly low. Hub regions are engaged in a wide variety of tasks and will defy precise behavioral characterization. Second, a more evolved functional ontology is needed, as other studies have argued (Price & Friston 2005, Poldrack 2006). Relative to the second cause, we suggest that the approaches illustrated here provide the tools for ontology development to proceed programmatically. Such development can be determined by targeting networks that show limited behavioral domain specificity and enriching the meta-data, e.g., by adding levels to the coding hierarchy. This work is ongoing (Fox et al. 2005b, Laird et al. 2011a). Ultimately, behavior categorizations that are reflected in the network properties of the brain will have superior intrinsic validity and utility as compared with those based solely on cognitive theory.
Figure 7.
BrainMap meta-data behavioral interpretations. Mapping of BrainMap meta-data onto ICA components is shown. Note that behavioral meta-data form discrete groupings, which functionally characterize the spatial groupings provided by ICA. (a) Twenty behavioral domain categorizations were correlated with the ten ICA-derived components shown in Figure 5. (b) The meta-data analysis has been extended to include 50 behavioral domain categories and 75 paradigm class categories. Hierarchical clustering was used to group the ICA into spatially and behaviorally related clusters for all 20 ICA components. Figures are reproduced with permission from Smith et al. (2009) (panel a) and Laird et al. (2011b) (panel b).
A closing point of some importance is that meta-analysis offers the most versatile, most powerful extant approach for discovering the behavioral significance of networks mapped using DTI tractography, cortical thickness covariances, or resting-state fMRI. DTI and cortical-thickness covariances, being anatomical techniques, contain no behavioral information. Resting-state fMRI, being performed at rest, is not under experimental control, leaving the behavior unspecified. Both DTI and resting-state fMRI provide very similar connectivity maps to MACM. Consequently, behavioral characterizations provided for MACM-defined pathways should be reasonably applied to pathways defined by the other techniques.
META-ANALYSES AS TOOLS
The family of CBMA methods described above may appear to be conceptually discrete methods. In practice, however, they tend to be applied serially, with simpler forms of meta-analysis providing input for more advanced forms. For example, clique analyses (described above) take an ALE volume as input and compute a paradigm-specific system model. Similarly, Neumann et al. (2010) used ALE to identify nodes as preparations for doing Bayesian network discovery. The MACM approach of Robinson et al. (2010) used ALE images as priors. Although these are examples of CBMAs providing priors for CBMAs, the strategy is more general. Karlsgodt et al. (2010), for example, used ALE to select ROIs for an analysis of brain-behavior pleiotropy (a one-to-many mapping) of visual working memory. Perhaps the most advanced and impactful use of CBMA to provide priors is in the domain of graphical modeling.
Many system-level modeling approaches commonly applied to functional neuroimaging data (e.g., structural equation modeling and dynamic causal modeling) are confirmatory methods that require strong a priori hypotheses about the regions involved (nodes) and their interdependencies (edges). Well-chosen priors improve model fit (Stephan et al. 2009). Given the ability of the several approaches described above to provide fairly complete, data-driven models, their use as priors for graphical modeling seems quite promising. Perhaps the first application of this strategy was reported by Laird and colleagues (2008), who used an ALE meta-analysis of TMS/PET studies of the primary motor cortex to inform a structural equations model (SEM) analysis of a TMS/PET data set. The goodness-of-fit of the ALE-based model to the data was quite striking, endorsing the value of this strategy. A subsequent application of the strategy used previously published ALE meta-analyses of stuttered and nonstuttered speech (Brown et al. 2005) as priors for fitting PET data during cued speech in persons with and without stuttering (Price et al. 2009). Again, the goodness-of-fit of the ALE-based models to data sets was striking. Furthermore, this strategy allowed excellent between-group (stuttering versus nonstuttering) discrimination with group sizes as small as 15 (power > 0.8). This finding strongly suggests a role for this analysis and modeling approach to treatment trials using graphical models to characterize the brain mechanisms of action of treatments in patient groups.
SUMMARY POINTS.
Spatial normalization (use of standardized coordinates) has produced a large (tens of thousands of peer-reviewed papers), well-standardized neuroimaging literature reporting hundreds of thousands of functional and structural experimental observations. BrainMap® and other online databases make these data readily available.
Sophisticated meta-analytic methods have been developed specifically for this data type or adapted to it from other neuroimaging applications.
The most basic meta-analytic method, ALE, demonstrates cross-study reliability of regional observations, filtering out nonreplicating findings within a group of similar experiments. This approach works equally well on functional (task-activation) and structural (between-group anatomical differences) data.
Between-experiment coactivation patterns are a reliable index of functional and structural connectivity, with multiple cross-methodology validations in the literature.
Coactivation patterns differ between subfields of larger cortical and nuclear structures, allowing connectivity-based parcellation to be computed meta-analytically.
Network modeling approaches originally developed for primary-image data sets (e.g., ICA, graph-analytic modeling) are proving to be well suited for analysis of large-scale, reduced-image (standardized coordinates) data sets, with coactivation being the driving observation. Similarity of observations between primary-data and reduced-data (meta-analytic) analyses is the rule and provides additional validation of this strategy.
Behavioral meta-data linked per-experiment to coordinate data in online databases provide a rich interpretive framework not otherwise available for network modeling.
Meta-analytically vetted regional effects and meta-analytically derived network models can be used as tools (a priori constraints) to analyze primary data sets, decreasing (or eliminating) the need for corrections for multiple comparisons and increasing the likelihood of finding significant effects.
FUTURE ISSUES.
Advanced meta-analytic network modeling methods (e.g., high-dimensionality ICA) would benefit from substantially larger data sets than are currently available.
More high-quality data are available in the literature than can be effectively curated. Greater efficiency of data entry is needed but without sacrificing quality control of data and meta-data.
As the scope and sophistication of neuroimaging experimental designs progress, the meta-data taxonomies used by BrainMap and other online databases will need to expand accordingly.
Greater care needs to be taken by software providers when releasing new templates and methods to ensure comparability of coordinates (same coordinate = same brain location) with prior literature.
Data-driven network analyses conjointly using coactivation patterns and behavioral meta-data need to be developed, advancing beyond using coactivations for network extraction and meta-data for network interpretation.
Use of meta-analysis-derived products (ROIs, spatial templates, network models) as priors for data analysis and modeling should be expanded.
SPATIAL NORMALIZATION.
Spatial normalization is the process of transforming a brain image from its natural size and shape (native space) into a standardized form that references a 3-D template image and coordinate space (template space). This allows brain images to be averaged across individuals and compared across groups.
GRAPH THEORY AND NEUROIMAGING.
Graph theory models connected systems as nodes (locations) and edges (their connections). Graph theory modeling constructs are increasingly being used to infer brain networks from neuroimages (Lohmann & Bohn 2002, Bullmore & Sporns 2009, Bullmore & Bassett 2011). Brain network models have been constructed from diffusion tensor image (DTI) tractography (Hagmann et al. 2007), from cortical thickness measures (Alexander-Bloch et al. 2013), and from resting-state neurophysiological fluctuations (Bullmore & Sporns 2009). Brain graphs have nonrandom topological properties such as small-worldness (Achard et al. 2006), modularity (Meunier et al. 2009), and hubs (Sporns et al. 2007). Graph theory has been generalized recently to meta-analysis of neuroimaging data (Neumann et al. 2010, Crossley et al. 2013). The convergence of meta-analysis and graph theory is an advance important for understanding human connectomics in health and disease.
Meta-analysis: retrospective combination of previously reported results to better estimate the reliability of those results
Coordinate-based meta-analysis (CBMA): meta-analysis method(s) developed specifically for use with functional and structural neuroimaging data reported within a standardized coordinate space
Coordinate space: any of a number of reference spaces defined by an anatomical template and used to analyze and report neuroimaging data, the two most widely used being Talairach Space and MNI Space
Stereotactic coordinates: three-dimension anatomical addresses (x, y, z) defined relative to a reference space and used to analyze and report functional and structural brain-image-derived observations
Voxel: a VO-lume pi-XEL, or data point within a 3-D image array. Statistical analyses of 3-D image volumes can be performed at corresponding voxels across data sets, i.e., in a voxel-wise fashion
Statistical parametric map/image (SPM/SPI): transforming brain images from raw, individual (per-subject) data sets in native space to 3-D images of statistical parameters (Z score, T scores, F-values, R-values, p-values, etc.) in a standard coordinate space
Voxel-based morphometry (VBM): a voxel-wise analysis method for detecting between-group (e.g., patients versus controls) differences in brain anatomy within a standardized coordinate space
Meta-data: standardized descriptors of data sets stored in electronic databases to retrieve data and filter studies for inclusion in a meta-analysis
ALE: activation likelihood estimation (for functional meta-analyses) or anatomical likelihood estimation (for anatomical meta-analyses)
Activation likelihood estimation: a widely used family of algorithms for CBMA of functional and structural neuroimaging observations
MACM: meta-analytic connectivity mapping
ICA: independent components analysis
ACKNOWLEDGMENTS
This work was supported by awards from the National Institutes of Health (MH74457, RR024387, MH084812, NS062254, AA019691, EB015314) and the Congressionally Directed Medical Research Program (W81XWH0820112). The sidebar on graph theory was contributed by Ed Bullmore and Nicolas Crossley. Portions of this review were adapted from Fox & Friston (2012).
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- Achard S, Salvador R, Whitcher B, Suckling J, Bullmore E. A resilient, low-frequency, small-world human brain functional network with highly connected association cortical hubs. J. Neurosci. 2006;26:63–72. doi: 10.1523/JNEUROSCI.3874-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander-Bloch A, Giedd JN, Bullmore E. Imaging structural co-variance between human brain regions. Nat. Rev. Neurosci. 2013;14:322–36. doi: 10.1038/nrn3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amunts K, Kedo O, Kindler M, Pieperhoff P, Mohlberg H, et al. Cytoarchitectonic mapping of the human amygdala, hippocampal region and entorhinal cortex: intersubject variability and probability maps. Anat. Embryol. 2005;210:343–52. doi: 10.1007/s00429-005-0025-5. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Voxel-based morphometry—the methods. NeuroImage. 2000;11(6):805–21. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- Barron DS, Fox PM, Laird AR, Robinson JL, Fox PT. Thalamic medial dorsal nucleus atrophy in medial temporal lobe epilepsy: a VBM meta-analysis. NeuroImage Clin. 2012;2:25–32. doi: 10.1016/j.nicl.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn. Reson. Med. 1995;34(4):537–41. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- Brown S, Ingham RJ, Ingham JC, Laird AR, Fox PT. Stuttered and fluent speech production: an ALE meta-analysis of functional neuroimaging studies. Hum. Brain Mapp. 2005;25:105–17. doi: 10.1002/hbm.20140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullmore ET, Bassett DA. Brain graphs: graphical models of the human brain connectome. Annu. Rev. Clin. Psychol. 2011;7:113–40. doi: 10.1146/annurev-clinpsy-040510-143934. [DOI] [PubMed] [Google Scholar]
- Bullmore ET, Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat. Rev. Neurosci. 2009;10(3):186–98. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- Butcher J. Alzheimer's researchers open the doors to data sharing. Lancet Neurol. 2007;6:480–81. doi: 10.1016/S1474-4422(07)70118-7. [DOI] [PubMed] [Google Scholar]
- Bzdok D, Laird AR, Zilles K, Fox PT, Eickhoff SB. An investigation of the structural, connectional, and functional subspecialization in the human amygdala. Hum. Brain Mapp. 2013;34:3247–66. doi: 10.1002/hbm.22138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bzdok D, Langner R, Schilback L, Engemann DA, Laird AR, et al. Segregation of the human medial prefrontal cortex in social cognition. Front. Hum. Neurosci. 2013;7:232. doi: 10.3389/fnhum.2013.00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauda F, Cavanna AE, D'agata F, Sacco K, Duca S, Geminiani GC. Functional connectivity and coactivation of the nucleus accumbens: a combined functional connectivity and structure-based meta-analysis. J. Cogn. Neurosci. 2011;23:2864–77. doi: 10.1162/jocn.2011.21624. [DOI] [PubMed] [Google Scholar]
- Cieslik EC, Zilles K, Caspers S, Roski C, Kellermann TS, et al. Is there “one” DLPFC in cognitive action control? Evidence for heterogeneity from co-activation-based parcellation. Cereb. Cortex. 2013;23:2677–89. doi: 10.1093/cercor/bhs256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chein JM, Fissell K, Jacobs S, Fiez JA. Functional heterogeneity within Broca's area during verbal working memory. Physiol. Behav. 2002;77:635–39. doi: 10.1016/s0031-9384(02)00899-5. [DOI] [PubMed] [Google Scholar]
- Clos M, Amunts K, Laird AR, Fox PT, Eickhoff SB. Tackling the multifunctional nature of Broca's region meta-analytically: co-activation-based parcellation of area 44. NeuroImage. 2013;83:174–88. doi: 10.1016/j.neuroimage.2013.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costafreda SG, Brammer MJ, David AS, Fu CH. Predictors of amygdala activation during the processing of emotional stimuli: a meta-analysis of 385 PET and fMRI studies. Brain Res. Rev. 2008;58:57–70. doi: 10.1016/j.brainresrev.2007.10.012. [DOI] [PubMed] [Google Scholar]
- Crossley NA, Mechelli A, Vértes PE, Winton-Brown TT, Patel AX, et al. Cognitive relevance of the community structure of the human brain functional coactivation network. Proc. Natl. Acad. Sci. USA. 2013;110:11583–88. doi: 10.1073/pnas.1220826110. [Presented the first large-scale graph-analytic meta-analysis; derived similar networks for BrainMap and resting-state fMRI data sets.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decety J, Lamm C. The role of the right temporoparietal junction in social interaction: how low-level computational processes contribute to meta-cognition. Neuroscientist. 2007;13:580–93. doi: 10.1177/1073858407304654. [DOI] [PubMed] [Google Scholar]
- Derrfuss J, Mar RA. Lost in localization: the need for a universal coordinate database. NeuroImage. 2009;48:1–7. doi: 10.1016/j.neuroimage.2009.01.053. [DOI] [PubMed] [Google Scholar]
- Dogan I, Eickhoff SB, Schulz JB, Shah JN, Laird AR, et al. Consistent neurodegeneration and its association with clinical progression in Huntington's disease: a coordinate-based meta-analysis. Neurodegener. Dis. 2013;12(1):23–35. doi: 10.1159/000339528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerden EG, Mak-Fan KM, Taylor MJ, Roberts SW. Regional differences in grey and white matter in children and adults with autism spectrum disorders: an activation likelihood estimate (ALE) meta-analysis. Autism Res. 2012;5:49–66. doi: 10.1002/aur.235. [DOI] [PubMed] [Google Scholar]
- Eickhoff SB, Bzdok D, Laird AR, Kurth F, Fox PT. Activation likelihood estimation meta-analysis revisited. NeuroImage. 2012;59:2349–61. doi: 10.1016/j.neuroimage.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickhoff SB, Bzdok D, Laird AR, Roski C, Caspers S, et al. Co-activation patterns distinguish cortical modules, their connectivity and functional differentiation. NeuroImage. 2011;57(3):938–49. doi: 10.1016/j.neuroimage.2011.05.021. [Introduced coactivation-based parcellation as a meta-analytic method.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickhoff SB, Jbabdi S, Caspers S, Laird AR, Fox PT, et al. Anatomical and functional connectivity of cytoarchitectonic areas within the human parietal operculum. J. Neurosci. 2010;30:6409–21. doi: 10.1523/JNEUROSCI.5664-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickhoff SB, Laird AR, Grefkes C, Wang LE, Zilles K, Fox PT. Coordinate-based activation likelihood estimation meta-analysis of neuroimaging data: a random-effects approach based on empirical estimates of spatial uncertainty. Hum. Brain Mapp. 2009;30:2907–26. doi: 10.1002/hbm.20718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison-Wright I, Bullmore E. Meta-analysis of diffusion tensor imaging studies in schizophrenia. Schizophr. Res. 2009;108:3–10. doi: 10.1016/j.schres.2008.11.021. [DOI] [PubMed] [Google Scholar]
- Ellison-Wright I, Glahn DC, Laird AR, Thelen SM, Bullmore E. The anatomy of first-episode and chronic schizophrenia: an anatomical likelihood estimation meta-analysis. Am. J. Psychiatry. 2008;165:1015–23. doi: 10.1176/appi.ajp.2008.07101562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans AC, Janke AL, Collins DL, Bailet S. Brain templates and atlases. NeuroImage. 2012;62(2):911–22. doi: 10.1016/j.neuroimage.2012.01.024. [DOI] [PubMed] [Google Scholar]
- Fitzgerald PB, Laird AR, Maller J, Daskalakis ZJ. A meta-analytic study of changes in brain activation in depression. Hum. Brain Mapp. 2008;29:683–95. doi: 10.1002/hbm.20426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Raichle ME. Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat. Rev. Neurosci. 2007;8(9):700–11. doi: 10.1038/nrn2201. [DOI] [PubMed] [Google Scholar]
- Fox PT. Broca's area: motor encoding in somatic space. Behav. Brain Sci. 1995a;18:344–45. [Google Scholar]
- Fox PT. Spatial normalization origins: objectives, applications, and alternatives. Hum. Brain Mapp. 1995b;3:161–64. [Google Scholar]
- Fox PT, Friston KJ. Distributed processing; distributed functions? NeuroImage. 2012;61:407–26. doi: 10.1016/j.neuroimage.2011.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox PT, Huang AY, Parsons LM, Xiong J-H, Rainey L, Lancaster JL. Functional volumes modeling: scaling for group size in averaged images. Hum. Brain Mapp. 1999;8:143–50. doi: 10.1002/(SICI)1097-0193(1999)8:2/3<143::AID-HBM12>3.0.CO;2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox PT, Huang A, Parsons LM, Xiong J-H, Zamarippa F, et al. Location-probability profiles for the mouth region of human primary motor–sensory cortex: model and validation. NeuroImage. 2001;13:196–209. doi: 10.1006/nimg.2000.0659. [DOI] [PubMed] [Google Scholar]
- Fox PT, Laird AR, Fox SP, Fox PM, Uecker AM, et al. BrainMap taxonomy of experimental design: description and evaluation. Hum. Brain Mapp. 2005a;25:185–98. doi: 10.1002/hbm.20141. [Described and validated the meta-data taxonomy of the BrainMap database.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox PT, Laird AR, Lancaster JL. Coordinate-based voxel-wise meta-analysis: dividends of spatial normalization. Report of a virtual workshop. Hum. Brain Mapp. 2005b;5:1–5. doi: 10.1002/hbm.20139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox PT, Lancaster JL. Neuroscience on the net. Science. 1994;266:994–96. doi: 10.1126/science.7973682. [DOI] [PubMed] [Google Scholar]
- Fox PT, Lancaster JL. Mapping context and content: the BrainMap model. Nat. Rev. Neurosci. 2002;3:319–21. doi: 10.1038/nrn789. [Announced the concept, purpose, structure, and online status of the BrainMap database.] [DOI] [PubMed] [Google Scholar]
- Fox PT, Lancaster JL, Parsons LM, Xiong JH, Zamarripa F. Functional volumes modeling: theory and preliminary assessment. Hum. Brain Mapp. 1997;5:306–11. doi: 10.1002/(SICI)1097-0193(1997)5:4<306::AID-HBM17>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Fox PT, Miezin FM, Allman JM, Van Essen DC, Raichle ME. Retinotopic organization of human visual cortex mapped with positron-emission tomography. J. Neurosci. 1987;7(3):913–22. doi: 10.1523/JNEUROSCI.07-03-00913.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox PT, Mikiten S, Davis G, Lancaster JL. BrainMap: a database of human functional brain mapping. In: Thatcher RW, Hallet M, Zeffiro T, John ER, Huerta M, editors. Functional Neuroimaging: Technical Foundations. Academic; San Diego: 1994. [Google Scholar]
- Fox PT, Mintun MA. Noninvasive functional brain mapping by change-distribution analysis of average PET images of H 152O tissue activity. J. Nucl. Med. 1989;30:141–49. [PubMed] [Google Scholar]
- Fox PT, Mintun MA, Raichle ME, Miezin FM, Allman JM, Van Essen DC. Mapping human visual cortex with position emission tomography. Nature. 1986;323:806–9. doi: 10.1038/323806a0. [DOI] [PubMed] [Google Scholar]
- Fox PT, Mintun MA, Reiman EM, Raichle ME. Enhanced detection of focal brain responses using intersubject averaging and change-distribution analysis of subtracted PET images. J. Cereb. Blood Flow Metab. 1988;8:642–53. doi: 10.1038/jcbfm.1988.111. [DOI] [PubMed] [Google Scholar]
- Fox PT, Parsons LM, Lancaster JL. Beyond the single study: function/location meta-analysis in cognitive neuroimaging. Curr. Opin. Neurobiol. 1998;8:178–87. doi: 10.1016/s0959-4388(98)80138-4. [DOI] [PubMed] [Google Scholar]
- Fox PT, Perlmutter JS, Raichle ME. A stereotactic method of anatomical localization for positron emission tomography. J. Comput. Assist. Tomogr. 1985;9:141–53. doi: 10.1097/00004728-198501000-00025. [Introduced the use of spatial normalization (standardized coordinates) to human brain mapping.] [DOI] [PubMed] [Google Scholar]
- Friston KJ, Ashburner J, Frith CD, Poline J-B, Heather JD, Frackowiak RSJ. Spatial registration and normalization of images. Hum. Brain Mapp. 1995;3:165–89. [Google Scholar]
- Friston KJ, Frith CD, Liddle PF, Frackowiak RSJ. Comparing functional (PET) images: the assessment of significant change. J. Cereb. Blood Flow Metab. 1991;11:690–99. doi: 10.1038/jcbfm.1991.122. [DOI] [PubMed] [Google Scholar]
- Frith CD, Friston KJ, Liddle PF, Frackowiak RS. Willed action and the prefrontal cortex in man: a study with PET. Proc. R. Soc. B. 1991;244:241–46. doi: 10.1098/rspb.1991.0077. [DOI] [PubMed] [Google Scholar]
- Gibbons A. Databasing the brain. Science. 1992;258:1872–73. doi: 10.1126/science.1470907. [DOI] [PubMed] [Google Scholar]
- Glahn DC, Laird AR, Ellison-Wright I, Thelen SM, Robinson JL, et al. Meta-analysis of gray matter anomalies in schizophrenia: application of anatomic likelihood estimation and network analysis. Biol. Psychiatry. 2008;64:774–81. doi: 10.1016/j.biopsych.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glahn DC, Ragland JD, Abramoff A, Barrett J, Laird AR, et al. Beyond hypofrontality: a quantitative meta-analysis of functional neuroimaging studies of working memory in schizophrenia. Hum. Brain Mapp. 2005;25:60–69. doi: 10.1002/hbm.20138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagmann P, Kurant M, Gigandet X, Thiran P, Wedeen VJ, et al. Mapping human whole-brain structural networks with diffusion MRI. PLoS ONE. 2007;2:e597. doi: 10.1371/journal.pone.0000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen-Berg H, Behrens TEJ, Robson MD, Drobnjak I, Rushworth MFS, et al. Changes in connectivity profiles define functionally distinct regions in human medial frontal cortex. Proc. Natl. Acad. Sci. USA. 2004;101(36):13335–40. doi: 10.1073/pnas.0403743101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsgodt KH, Kochunov P, Winkler AM, Laird AR, Almasy L, et al. A multimodal assessment of the genetic control over working memory. J. Neurosci. 2010;30:8197–202. doi: 10.1523/JNEUROSCI.0359-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koski L, Paus T. Functional connectivity of the anterior cingulate cortex within the human frontal lobe: a brain-mapping meta-analysis. Exp. Brain Res. 2000;133:55–65. doi: 10.1007/s002210000400. [Introduced the concept that meta-analysis of coactivations was an index of functional connectivity.] [DOI] [PubMed] [Google Scholar]
- Laird AR, Eickhoff SB, Fox PM, Uecker AM, Ray KL, et al. The BrainMap strategy for standardization, sharing and meta-analysis of neuroimaging data. BMC Res. Notes. 2011a;4:349. doi: 10.1186/1756-0500-4-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird AR, Eickhoff SB, Li K, Robin DA, Glahn DC, Fox PT. Investigating the functional heterogeneity of the default mode network using coordinate-based meta-analytic modeling. J. Neurosci. 2009;29:14496–505. doi: 10.1523/JNEUROSCI.4004-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird AR, Fox PM, Eickhoff SB, Turner JA, Ray KL, et al. Behavioral interpretations of intrinsic connectivity networks. J. Cogn. Neurosci. 2011b;23:4022–37. doi: 10.1162/jocn_a_00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird AR, Robinson JL, McMillan KM, Tordesillas-Gutiérrez D, Moran ST, et al. Comparison of the disparity between Talairach and MNI coordinates in functional neuroimaging data: validation of the Lancaster transform. NeuroImage. 2010;51:677–83. doi: 10.1016/j.neuroimage.2010.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird AR, Robbins JM, Li K, Price LR, Cykowski MD, et al. Modeling motor connectivity using TMS/PET and structural equation modeling. NeuroImage. 2008;41:424–36. doi: 10.1016/j.neuroimage.2008.01.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird AR, Fox PM, Price CJ, Glahn DC, Uecker AM, et al. ALE meta-analysis: controlling the false discovery rate and performing statistical contrasts. Hum. Brain Mapp. 2005a;25:155–64. doi: 10.1002/hbm.20136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird AR, Lancaster JL, Fox PT. BrainMap: the social evolution of a human brain mapping database. Neuroinformatics. 2005b;3:65–78. doi: 10.1385/ni:3:1:065. [DOI] [PubMed] [Google Scholar]
- Lambrecq V, Langbour N, Guehl D, Biolac B, Burbaud P, Rotge JY. Evolution of gray matter loss in Huntington's disease: a meta-analysis. Eur. J. Neurol. 2013;20:315–21. doi: 10.1111/j.1468-1331.2012.03854.x. [DOI] [PubMed] [Google Scholar]
- Lancaster JL, Laird AR, Eickhoff SB, Martinez MJ, Fox MP, Fox PT. Automated regional behavioral analysis for human brain images. Front. Neuroinform. 2012;6:23. doi: 10.3389/fninf.2012.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster JL, Laird AR, Fox M, Glahn DE, Fox PT. Automated analysis of meta-analysis networks. Hum. Brain Mapp. 2005;25:174–84. doi: 10.1002/hbm.20135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster JL, Tordesillas-Gutiérrez D, Martinez M, Salinas F, Evans A, et al. Bias between MNI and Talairach coordinates analyzed using the ICBM-152 brain template. Hum. Brain Mapp. 2007;28:1194–205. doi: 10.1002/hbm.20345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster JL, Woldorff MG, Parsons LM, Liotti M, Freitas CS, et al. Automated Talairach atlas labels for functional brain mapping. Hum. Brain Mapp. 2000;10(3):120–31. doi: 10.1002/1097-0193(200007)10:3<120::AID-HBM30>3.0.CO;2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann G, Bohn S. Using replicator dynamics for analyzing fMRI data of the human brain. IEEE Trans. Med. Imaging. 2002;21(5):485–92. doi: 10.1109/TMI.2002.1009384. [DOI] [PubMed] [Google Scholar]
- Mazziotta JC, Toga TW, Evans A, Fox P, Lancaster J. A probabilistic atlas of the human brain: theory and rationale for its development. The International Consortium for Brain Mapping (ICBM). NeuroImage. 1995;2(2A):89–101. doi: 10.1006/nimg.1995.1012. [DOI] [PubMed] [Google Scholar]
- Menzies L, Chamberlain SR, Laird AR, Thelen SM, Sahakian BJ, Bullmore ET. Integrating evidence from neuroimaging and neuropsychological studies of obsessive-compulsive disorder: the orbitofrontostriatal model revisited. Neurosci. Biobehav. Rev. 2008;32:525–49. doi: 10.1016/j.neubiorev.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier D, Achard S, Morcom A, Bullmore E. Age-related changes in modular organization of human brain functional networks. NeuroImage. 2009;44:715–23. doi: 10.1016/j.neuroimage.2008.09.062. [DOI] [PubMed] [Google Scholar]
- Mintun MA, Fox PT, Raichle ME. A highly accurate method of localizing regions of neuronal activation in the human brain with positron emission tomography. J. Cereb. Blood Flow Metab. 1989;9(1):96–103. doi: 10.1038/jcbfm.1989.13. [DOI] [PubMed] [Google Scholar]
- Minzenberg MJ, Laird AR, Thelen SM, Carter CS, Glahn DC. Meta-analysis of 41 functional neuroimaging studies of executive function in schizophrenia. Arch. Gen. Psychiatry. 2009;66:811–22. doi: 10.1001/archgenpsychiatry.2009.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayana S, Laird AR, Tandon N, Franklin C, Lancaster JL, Fox PT. Electrophysiological and functional connectivity of the human supplementary motor area. NeuroImage. 2012;62:250–65. doi: 10.1016/j.neuroimage.2012.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann J, Fox PT, Turner R, Lohmann G. Learning partially directed functional networks from meta-analysis imaging data. NeuroImage. 2010;49:1372–84. doi: 10.1016/j.neuroimage.2009.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann J, Lohmann G, Derrfuss J, Yves von Cramon D. The meta-analysis of functional imaging data using replicator dynamics. Hum. Brain Mapp. 2005;25:165–73. doi: 10.1002/hbm.20133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann J, Yves von Cramon D, Lohmann G. Model-based clustering of meta-analytic functional imaging data. Hum. Brain Mapp. 2008;29:177–92. doi: 10.1002/hbm.20380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickl-Jockschat T, Habel U, Michel TM, Manning J, Laird AR, et al. Brain structure anomalies in autism spectrum disorder (ASD)—a meta-analysis of VBM studies using anatomic likelihood estimation (ALE). Hum. Brain Mapp. 2012;33:1470–89. doi: 10.1002/hbm.21299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paus T. Location and function of the human frontal eye-field: a selective review. Neuropsychologia. 1996;34:475–83. doi: 10.1016/0028-3932(95)00134-4. [DOI] [PubMed] [Google Scholar]
- Pearson K. Report on certain enteric fever inoculation statistics. Br. Med. J. 1904;3:1243–46. [PMC free article] [PubMed] [Google Scholar]
- Picard N, Strick PL. Motor areas of the medial wall: a review of their location and functional activation. Cereb. Cortex. 1996;6:342–53. doi: 10.1093/cercor/6.3.342. [DOI] [PubMed] [Google Scholar]
- Poldrack RA. Can cognitive processes be inferred from neuroimaging data? Trends Cogn. Sci. 2006;10:59–63. doi: 10.1016/j.tics.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Posner M, Petersen S, Fox PT, Raichle M. Localization of cognitive operations in the human brain. Science. 1988;240:1627–31. doi: 10.1126/science.3289116. [DOI] [PubMed] [Google Scholar]
- Postuma RB, Dagher A. Basal ganglia functional connectivity based on a meta-analysis of 126 positron emission tomography and functional magnetic resonance imaging publications. Cereb. Cortex. 2006;16:1508–21. doi: 10.1093/cercor/bhj088. [DOI] [PubMed] [Google Scholar]
- Price CJ, Friston KJ. Functional ontologies for cognition: the systematic definition of structure and function. Cogn. Neuropsychol. 2005;22:262–75. doi: 10.1080/02643290442000095. [DOI] [PubMed] [Google Scholar]
- Price LR, Laird AR, Fox PT. Modeling dynamic functional neuroimaging data using structural equation modeling. Struct. Equ. Modeling. 2009;16:147–62. doi: 10.1080/10705510802561402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragland JD, Laird AR, Ranganath C, Blumenfeld RS, Gonzales SM, Glahn DC. Prefrontal activation deficits during episodic memory in schizophrenia. Am. J. Psychiatry. 2009;166:863–74. doi: 10.1176/appi.ajp.2009.08091307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JL, Laird AR, Glahn DC, Blangero J, Sanghera MK, et al. The functional connectivity of the human caudate: an application of meta-analytic connectivity modeling with behavioral filtering. NeuroImage. 2012;60:117–29. doi: 10.1016/j.neuroimage.2011.12.010. [Introduced meta-analytic connectivity modeling with behavioral filtering, i.e., sorting connected networks by their functional meta-data.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JL, Laird AR, Glahn DC, Lovallo WR, Fox PT. Metaanalytic connectivity modeling: delineating the functional connectivity of the human amygdala. Hum. Brain Mapp. 2010;31:173–84. doi: 10.1002/hbm.20854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottschy C, Caspers S, Roski C, Reetz K, Dogan I, et al. Differentiated parietal connectivity of frontal regions for “what” and “where” memory. Brain Struct. Funct. 2012;218:1551–67. doi: 10.1007/s00429-012-0476-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Fox PT, Miller KL, Glahn DC, Fox PM, et al. Correspondence of the brain's functional architecture during activation and rest. Proc. Natl. Acad. Sci. USA. 2009;106:13040–45. doi: 10.1073/pnas.0905267106. [Presented the first ICA meta-analysis and demonstrated similarity of ICA networks for BrainMap and resting-state fMRI data sets.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O, Honey CJ, Kötter R. Identification and classification of hubs in brain networks. PLoS ONE. 2007;2:e1049. doi: 10.1371/journal.pone.0001049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spreng RN, Mar RA, Kim AS. The common neural basis of autobiographical memory, prospection, navigation, theory of mind and the default mode: a quantitative meta-analysis. J. Cogn. Neurosci. 2009;21:489–510. doi: 10.1162/jocn.2008.21029. [DOI] [PubMed] [Google Scholar]
- Stephan KI, Tittgemeyer M, Knosche TR, Moran RJ, Friston KJ. Tractography based priors for dynamic causal models. NeuroImage. 2009;47:1628–38. doi: 10.1016/j.neuroimage.2009.05.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talairach J, Szikla G, Tournoux P, Prosalentis A, Bordas-Ferrier M. Atlas d'Anatomie Stéréotaxique du Télencépahle. Masson; Paris: 1967. [Google Scholar]
- Talairach J, Tournoux P. Co-Planar Stereotaxic Atlas of the Human Brain. Thieme; New York: 1988. [Google Scholar]
- Toga AW. Brain Warping. Academic; San Diego: 1998. [Google Scholar]
- Toro R, Fox PT, Paus T. Functional coactivation map of the human brain. Cereb. Cortex. 2008;18:2553–59. doi: 10.1093/cercor/bhn014. [Presented the first large-scale (whole-database) application of coactivation-based meta-analysis.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torta DM, Costa T, Duca S, Fox PT, Cauda F. Parcellation of the cingulate cortex at rest and during tasks: a meta-analytic clustering and experimental study. Front. Hum. Neurosci. 2013;7:275. doi: 10.3389/fnhum.2013.00275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulving E, Kapur S, Craik FI, Moscovitch M, Huole S. Hemispheric encoding/retrieval asymmetry in episodic memory: positron emission tomography findings. Proc. Natl. Acad. Sci. USA. 1994;91:2016–20. doi: 10.1073/pnas.91.6.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkeltaub PE, Eden GF, Jones KM, Zeffiro TA. Meta-analysis of the functional neuroanatomy of single-word reading: method and validation. NeuroImage. 2002;16:765–80. doi: 10.1006/nimg.2002.1131. [Introduced the activation likelihood estimation method of coordinate-based meta-analysis.] [DOI] [PubMed] [Google Scholar]
- Turkeltaub PE, Eickhoff SB, Laird AR, Fox M, Wiener M, Fox PT. Minimizing within-experiment and within-group effects in activation likelihood estimation meta-analyses. Hum. Brain Mapp. 2012;33:1–13. doi: 10.1002/hbm.21186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JA, Laird AR. The cognitive paradigm ontology: design and application. Neuroinformatics. 2011;10:57–66. doi: 10.1007/s12021-011-9126-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wager TD, Phan KL, Liberzon I, Taylor SF. Valence, gender, and lateralization of functional brain anatomy in emotion: a meta-analysis of findings in neuroimaging. NeuroImage. 2003;19:513–31. doi: 10.1016/s1053-8119(03)00078-8. [DOI] [PubMed] [Google Scholar]
- Yarkoni T, Poldrack RA, Nichols TE, Van Essen DC, Wager TD. Large-scale automated synthesis of human functional neuroimaging data. Nat. Methods. 2011;8(8):665–70. doi: 10.1038/nmeth.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]