Abstract
Purpose
Notch1 was previously shown to play a critical role in murine meibomian gland function and maintenance. In this study, we have examined the expression and activation of Notch pathway in human meibomian gland epithelial cells in vitro.
Methods
An immortalized human meibomian gland epithelial cell (HMGEC) line was cultured under proliferative and differentiative conditions. Expression of Notch receptors and ligands were evaluated by quantitative PCR and Western blot. The effect of Notch inhibition and induction on oil production was also assessed.
Results
Human meibomian gland epithelial cell expressed Notch1, Notch2, Notch3, Jagged1, Jagged2, Delta-like 1, and Delta-like 3. The level of cleaved (activated) Notch1 strongly increased with differentiation. The expression of Notch3 was inversely correlated with proliferation. Induction and inhibition of Notch1 led to an increase and decrease in the amount of oil production, respectively.
Conclusions
Notch signaling appears to play an important role in human meibomian gland epithelial differentiation and oil production. This may provide a potential therapeutic pathway for treating meibomian gland dysfunction.
Keywords: meibomian gland dysfunction, notch signaling, dry eye
Meibomian glands (MG) are a specialized form of sebaceous gland within the eyelids, which drain immediately behind the eyelashes.1 These holocrine glands contain epithelial cells that are responsible for the production and secretion of meibum, a lipid based substance that stabilizes the tear film and slows down its evaporation. An impairment in the meibomian glands or their lipid production can cause meibomian gland dysfunction (MGD) leading to a destabilized tear film and dry eye symptoms.1,2 Currently, the treatment of MGD is mostly aimed at enhancing the outflow of meibum and reducing bacterial growth and inflammation along the eyelid margin.3 There are very few specific treatments targeting the meibum/lipid production, in part because the pathways that regulate meibomian gland function remain largely unknown.
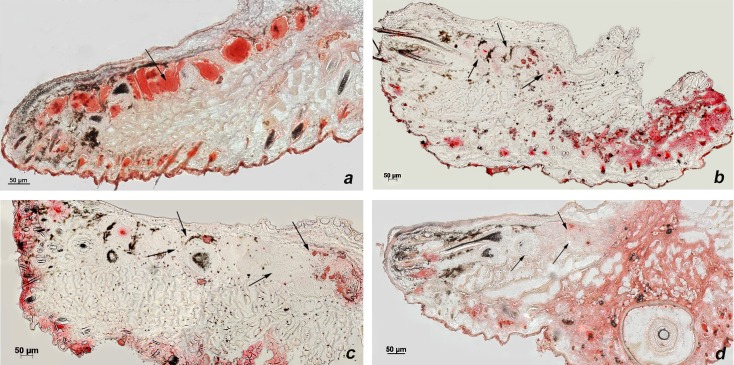
Previously, we and others have reported that mice with conditional deletion of Notch1 gene in the epithelium develop loss of MGs, which in turn contributes to their development of corneal inflammation and keratinization.4 In particular, deletion of Notch1 in adult mice first leads to the loss of lipid production and later complete degeneration of the meibomian glands5,6 (Fig. 1). Along the same lines, loss of Notch1 was shown to reduce oil production in sebaceous glands in the skin.7 Thus, Notch1 appears to play an important role in both lipid production and meibomian gland function.
Figure 1.
Oil red O staining in wild-type mice versus Notch1 knockout demonstrating intact meibomian oil production in wild-type (a) mice in contrast to the significantly reduced Oil red O staining in the meibomian glands of knockout mice 2 weeks after Notch1 deletion (b); at 3 and 4 weeks after Notch1 deletion, very little oil is seen in the meibomian glands (c, d). Black arrow heads indicate the meibomian glands: scale bar: 50 μm.
The Notch signaling pathway is a highly conserved cell to cell signaling mechanism that orchestrates cell fate decisions in many tissues and organisms. Notch proteins are membrane-bound receptors with corresponding membrane bound ligands, Delta and Jagged. Upon binding of the ligand, the Notch receptor is cleaved by the γ-secretase complex, which releases the Notch intracellular (NotchIC) fragment. NotchIC translocates into the nucleus and acts as a transcription factor regulating the expression of genes involved in many cell fate decisions such as proliferation, differentiation, and migration.8–10
In this study, we investigated the expression of Notch receptors and ligands in human meibomian gland epithelial cells (HMGEC) during proliferation and differentiation. We sought to determine whether Notch signaling pathway is involved in meibomian gland epithelial differentiation in vitro and whether induction of Notch1 expression or inhibition of Notch signaling in HMGEC can affect their lipid production.
Materials and Methods
Cell Culture and Treatment
An immortalized human meibomian gland epithelial cell line, kindly provided by David Sullivan (Schepens Eye Research Institute, Boston, MA, USA) was used for all of the studies. The cell were maintained in keratinocyte serum-free medium supplemented with 5 ng/mL of epidermal growth factor (EGF) and 50 μg/mL of bovine pituitary extract (BPE; Life Technologies, Grand Island, NY, USA), which maintains the cells in a proliferative undifferentiated state. After reaching 80% to 90% confluence, the media was switched to Dulbecco modified Eagle medium/Ham F-12 nutrient 50:50 mixture (Sigma-Aldrich, St. Louis, MO, USA), supplemented with 10% fetal bovine serum (FBS), which promotes differentiation over the course of 5 to 7 days.11 The EGF-BPE– and 10% FBS–containing media are known to promote epithelial cell proliferation and differentiation, respectively, in this cell line.12
Inhibition of Notch Signaling
To inhibit Notch signaling, differentiated HMGECs at day 5 were treated with 20 μM of the γ-secretase inhibitor (N-[N-(3, 5-difluorophenacetyl)-l-alanyl]-S-phenylglycinet-butyl ester; DAPT; Farmingdale, NY, USA) or vehicle control (dimethyl sulfoxide; DMSO maximum concentration 0.02%). DAPT is a potent inhibitor of the Notch signaling pathway that abrogates proteolytic processing of the intracellular domain, both in cell culture and in vivo.13 Protein analyses were performed 48 hours after the addition of DAPT.
Induction of Notch1 Expression
To exogenously induce Notch1 expression in HMGEC, we screened several published natural compounds (data not shown) and determined that resveratrol can induce mRNA expression of Notch1 in HMGEC.14–16 Cells were switched from proliferative to differentiative media then starting immediately treated with resveratrol 100 μM (Santa Cruz Biotechnology, Dallas, TX, USA),17 or with vehicle control (DMSO maximum concentration 0.1%) for 3 days.
Real-Time PCR
The differential expression of selected genes was evaluated by quantitative PCR (qPCR) methods. Total RNA was extracted from the cells using extraction reagent (TRIzol; Life Technologies) according to the manufacturer's protocol. After spectrophotometric assessment for quality and concentration (Nanodrop ND-1000; Thermo Scientific, Wilmington, DE, USA) the cDNAs were generated using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) using the manufacturer's protocol. For each reaction, 2 μg of total RNA were used. The qPCR reactions were carried out in triplicate with FastStart Universal SYBR Green Master (Roche, Mannheim, Germany) in a total volume of 20 μL using thermal cycling condition of 10 minutes at 90°C, 10 seconds at 95°C followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. The primer sequences are available in the appendix. The average threshold cycle (Ct) value for 18S were used as an endogenous reference to correct for the differences in the amount of total RNA added to each reaction. The amount of target gene in each sample was normalized to the endogenous control by subtracting the Ct of 18S from the Ct of target gene.
Western Blot Analysis
The protein expression of Notch receptors and ligands were studied using immunoblotting. Following proliferation, differentiation, and treatment with resveratrol, DAPT or DMSO, the cells were directly lysed in SDS RIPA buffer (Sigma-Aldrich) supplemented with 1% protease inhibitor cocktail (Sigma-Aldrich), 5% beta-mercaptoethanol (Sigma-Aldrich), and ×2 Laemmli buffer (Bio-Rad Laboratories, Philadelphia, PA, USA). Lysates were heated at 95°C for 10 minutes, separated by SDS-PAGE on 4% to 12% Bis-Tris mini gels (Life technologies), and transferred to polyvinylidene difluoride membranes. Monoclonal antibodies specific for Notch1 Rabbit Ab (D1E11; Cell Signaling, Danvers, MA, USA), Cleaved-Notch1 Rabbit Ab (Val1744; Cell Signaling), Notch2 Rabbit Ab (D67C8; Cell Signaling), Notch3 Rabbit Ab (D11B8; Cell Signaling), and GAPDH (D16H11; Cell Signaling) were used. Membranes were blocked with 5% bovine serum albumin in Tris-buffered saline containing 0.1% Tween-20 (TBS/T); for all antibodies. All primary antibodies were diluted 1:1000 in blocking buffer except for GAPDH (1:5000). The secondary antibody was an HRP-linked goat anti-rabbit IgG (Cell Signaling). The proteins were visualized with commercial Western blotting substrate (Pierce ECL Western Blotting Substrate; Thermo Fisher Scientific).
Green Neutral Lipid Staining
Immortalized HMGEC were cultured on coated chamber slides (Lab-Tek II Chamber Slide System; Nalge Nunc International, Rochester, NY, USA) in FBS-containing medium in the presence or absence of resveratrol, DAPT, or DMSO. Media were replaced every 2 days for a total of 4 days, at which time the cells were washed and fixed in 4% paraformaldehyde for 30 minutes. After additional washes, cells were stained with green neutral lipid stain (1:800; LipidTOX; Life Technologies) in a humidified chamber for 30 minutes. Coverslips were mounted on slides with an antifade reagent containing 4′, 6-diamidino-2-phenylindole nuclear stain (ProLong Gold; Life Technologies) and permitted to dry overnight. Cells were imaged with LSM 710 Laser scanning confocal microscope (Carl Zeiss 710, Jena, Germany), ×20 magnification, under the same settings (including laser power, detector gain/offset, pin hole, and pixel time).
Fluorescence intensities of images were measured with Image-J software (version 1.47; http://imagej.nih.gov/ij/; provided in the public domain by the National Institutes of Health, Bethesda, MD, USA) and the averages for each group were compared in Excel 2013 (Microsoft, Redmond, WA, USA).
Lipid Extraction and Analysis
Differentiated HMGECs cultured for 3 days in serum-containing medium were harvested with trypsin/EDTA solution (Sigma-Aldrich), dispersed into single cell suspension and treated with Soybean Trypsin Inhibitor (Gibco, Grand Island, NY, USA). Cell numbers were counted after which the cells were pelleted by centrifugation in glass tubes, resuspended in 0.5mL Hanks' Balanced Salt Solution (Gibco) and immediately frozen on dry ice and stored at −80°C until lipid extraction. Lipid extraction was performed using a modified method of Bligh and Dyer18 and final lipid extracts were dissolved in methanol/chloroform (1:2, vol/vol) in such a way that each extract has the same lipid concentration based on cell numbers taken into the extraction procedure. Neutral lipids were separated by high-performance thin-layer chromatography (TLC) using the hexane/diethyl ether/acetic acid (85:15:1, vol/vol) system and visualized by spraying the plate with 10% sulfuric acid in methanol and plate charring at 200°C.
Statistical Analyses
For a descriptive statistical analysis we used one-way ANOVA and Student's t-test. Results are provided as the mean ± SD. Statistical significance was set at P less than or equal to 0.05.
Results
Notch Pathway Genes are Differentially Expressed During Proliferation and Differentiation
The mRNA expression of Notch receptors and ligands in HMGECs was examined during both proliferative and differentiative conditions at 7 days (Fig. 2).
Figure 2.
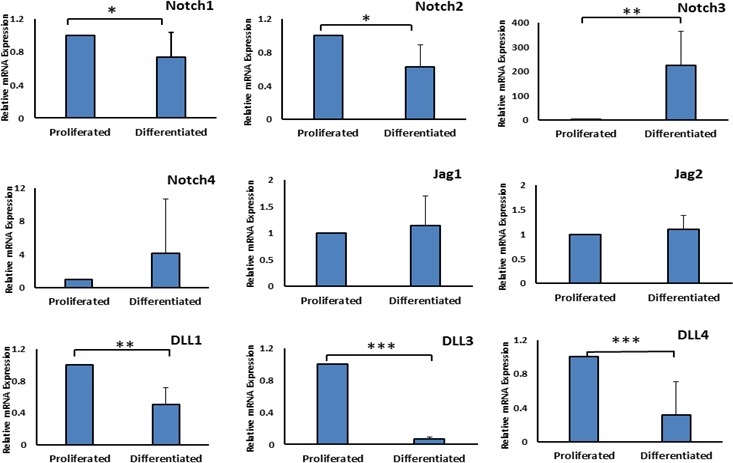
Relative mRNA expression of Notch receptors and ligands during proliferating and differentiating culture conditions. mRNA expression of Notch1, Notch2, Delta like1, and Delta like3 decreased after 7 days of differentiation. mRNA expression of Notch4 and Delta like4 were slightly lower with differentiation. Notch3 mRNA expression increased significantly as cells were switched from proliferating to differentiating conditions. Jag1 and Jag2 ligands did not show any significant changes with differentiation. *P value < 0.05, **P value < 0.0025, and ***P value < 00015 all by t-test. Error bars represent mean ± SD.
The results showed changes in the expression of Notch family genes with serum/calcium induced differentiation. In particular, the mRNA expression of Notch1, Notch2, DLL1, and DLL3 was slightly lower in differentiated cells while the expression of Notch3 was significantly higher in differentiated cells (200-fold induction). The mRNA expression of Notch4 and DLL4 was found to be extremely low (cycle number 31–35 by qPCR) nonetheless there was a slight change with differentiation (Fig. 2).
To analyze the expression and activation state of Notch receptors (Notch1, Notch2, and Notch3) at the protein level, we performed Western blot analyses (Fig. 3). In undifferentiated proliferating cells, Notch1 was predominantly in a full-length (inactive) form while in differentiated cells it was mostly in an active (cleaved) form (Figs. 3a, 3b). The expression of full length Notch2 protein was lower with differentiation while the Notch transmembrane (NTM) fragment was only weakly expressed in both conditions (Fig. 3c). Similar to the mRNA expression, the protein expression of Notch3 was generally absent in the proliferative condition and only expressed as the cells switched from proliferative to differentiating condition. The NTM fragment, which in part includes cleaved Notch3, was absent during proliferation indicating that in HMGECs Notch3 is inactive during proliferative conditions (Fig. 3d).
Figure 3.
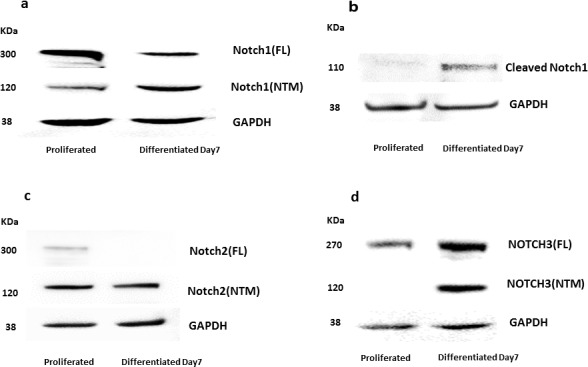
Protein expression of Notch1, cleaved Notch1, Notch2, and Notch3 under proliferative and differentiative culture conditions. By Western blot analysis (a) the Notch1 full length (FL) domain decreased with differentiation while the NTM domain increased. (b) Cleaved Notch1 was found to increase with differentiation. (c) The FL domain of Notch2 was weakly expressed under proliferative conditions while its NTM domain increased slightly with differentiation, (d) The FL domain of Notch3 was higher under differentiative condition while its NTM domain was not expressed under proliferative conditions.
Inhibition of Notch Signaling Blocks Lipid Production in HMGECs
To inhibit Notch signaling broadly, we used the γ-secretase inhibitor DAPT. Inhibition of Notch1 activation was confirmed by a reduction in cleaved Notch1 protein after exposure of differentiated HMGEC to DAPT for 2 days (Figs. 4a, 4b). Lipitox staining further demonstrated that DAPT significantly decreased the production of neutral lipids in HMGECs (Figs. 4c, 4d).
Figure 4.
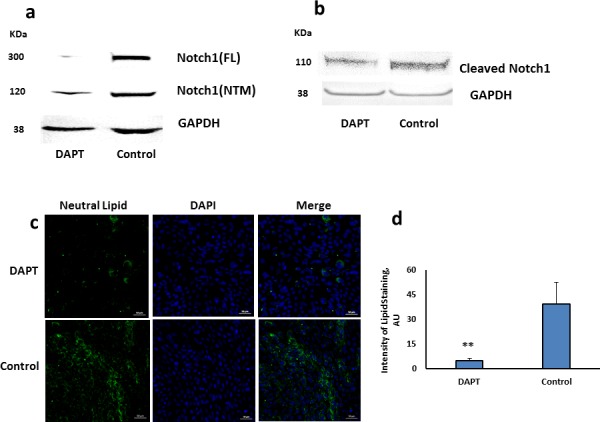
Effect of DAPT on Notch1 protein expression and lipid accumulation in HMGECs. Cells were differentiated for 5 days and then treated with DAPT or vehicle control (DMSO) for 2 days in serum-containing media. (a) Immunoblot images showing decreased Notch1 FL protein and NTM fragment in response to DAPT treatment. (b) Cleaved Notch1 likewise decreased with DAPT treatment. (c) Neutral lipids were stained with Lipitox (green) and nuclei with DAPI (blue), Magnification ×20. (d) Decrease in fluorescence intensity of Lipitox staining with DAPT was significant [(**) = (P value < 0.0005 by t-test)]. Error bars represent mean ± SD.
Resveratrol Induces Notch1 and Enhances Lipid Production in HMGECs
Based on previous studies, we screened several natural compounds that have been reported to induce Notch expression (data not shown). We identified resveratrol as a strong inducer of Notch1 and proceeded to examine Notch1 expression and lipid production in HMGECs following exposure to resveratrol 100 μM for 3 days. As shown in Figures 5a and 5b, lipid production was significantly higher in resveratrol treated cells compared to control (P value = 0.023). Resveratrol induced the mRNA expression of Notch1 by nearly 3-fold (P value = 0.000494; Fig. 5c). The induced expression and activation of Notch1 was also confirmed at the protein level (P value = 0.042).
Figure 5.
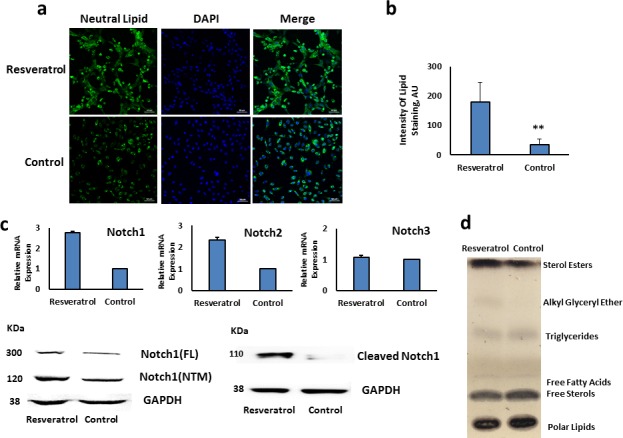
Resveratrol induces Notch1 expression and lipid accumulation in meibomian gland epithelial cells. Human meibomian gland epithelial cells treated with resveratrol (100 μM) or vehicle control (DMSO) (a) demonstrate increased neutral lipid accumulation in HMGEC, which treatment with resveratrol, Lipitox (green) and DAPI (blue), (b) Increase in fluorescence intensity of Lipitox staining with resveratrol was significant [(**) = (P value = 0.023)]. Error bars represent mean ± SD, (c) Increased mRNA expression of Notch1 and Notch2 as well as increase in FL, NTM, and cleaved Notch1 in resveratrol treated HMGEC compared with control (DMSO). The mRNA expression of Notch3 did not change with resveratrol. (d) Thin layer chromatography (TLC) demonstrated that resveratrol treatment increases sterol esters (neutral lipids) in HMGEC.
Further analysis of the lipids by TLC demonstrated that resveratrol augmented cellular levels of Sterol Esters and also induced a significant rise in the accumulation of neutral lipids (Fig. 5d).
Discussion
Notch signaling, which was originally identified as an important developmental pathway, has since been implicated in numerous cellular functions such as proliferation, differentiation, and migration.19,20 The results of this study further highlight the role of Notch signaling in meibomian gland homoeostasis and lipid production. In particular, we have shown activation of Notch signaling with in vitro differentiation of HMGECs. These findings are further corroborated by our results that inhibiting Notch signaling blocks lipid production while inducing Notch1 expression leads to enhanced lipid production.
Our results have specifically implicated the Notch1 and Notch3 subtypes in meibomian gland differentiation. Notch1 has previously been shown to play a critical function in murine meibomian gland function and mice with conditional loss of Notch1 first stop producing meibum/oil and later undergo complete degeneration.21 On the other hand, conditional knockouts of Notch2 and Notch3 were not reported to have any observed phenotype involving the meibomian glands. Notch3 mutations have also been implicated in the human disease cerebral autosomal-dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL); however meibomian gland disease has not been reported as a feature of CADASIL. Overall, we hypothesize that Notch1 likely plays a more critical role for meibomian gland differentiation and significant change in the expression of Notch3 is likely related to its downregulation with proliferation (personal communication with David Sullivan in June, 2015). Overall, these results suggest that Notch signaling is a novel pathway that may have potential therapeutic implications for MGD.
The pathophysiology of MGD is complex and is in part influenced by hormonal, immunologic, and microbial factors. Current treatments of MGD are aimed mostly at improving the outflow of meibum through warm compresses and lid massages as well as controlling bacterial growth and inflammation with the use of antibiotics and anti-inflammatory therapies. However, treatments that specifically enhance meibomian gland function and lipid production are generally lacking. Most recently, topical azithromycin has been used as a treatment of MGD. In addition to its anti-inflammatory and antibacterial effects, azithromycin was also found to stimulate the growth and differentiation of immortalized human meibomian gland epithelial cells in vitro, which may, in part, explain its beneficial effect for the treatment of MGD.22
Based on the results of this study, we propose that agents that induce Notch expression and activation may be potentially useful therapeutically in MGD. Previously investigators using a carcinoid cell line identified resveratrol as one such agent, which was able to induce expression and activation of Notch. Resveratrol is a plant-derived polyphenol found in the skin of red grapes, peanuts, and berries and is currently available over the counter as a supplement. Its health benefits are thought to be mediated in part via sirtuins.23 The effects of resveratrol on Notch are most likely indirect, nonetheless the results support its potential use as a treatment of MGD. Future clinical trials are necessary to determine if local and/or systemic treatment with resveratrol can be beneficial in MGD.
Acknowledgments
Supported by R01 EY024349-01A1 (ARD) and a core Grant EY01792 both from National Eye Institute, NIH (Bethesda, MD, USA) and an unrestricted grant from Research to Prevent Blindness (New York, NY, USA). ARD is the recipient of a Career Development Award from Research to Prevent Blindness.
Disclosure: S. Gidfar, None; N. Afsharkhamseh, None; S. Sanjari, None; A.R. Djalilian, None
References
- 1. Thody AJ,, Shuster S. Control and function of sebaceous glands. Physiol Rev. 1989; 69: 383–416. [DOI] [PubMed] [Google Scholar]
- 2. Knop E,, Knop N,, Millar T,, Obata H,, Sullivan DA. The international workshop on meibomian gland dysfunction: report of the subcommittee on anatomy physiology, and pathophysiology of the meibomian gland. Invest Ophthalmol Vis Sci. 2011; 52: 1938–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Green-Church KB,, Butovich I,, Willcox M,, et al. The international workshop on meibomian gland dysfunction: report of the subcommittee on tear film lipids and lipid-protein interactions in health and disease. Invest Ophthalmol Vis Sci. 2011; 52: 1979–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lemp MA,, Nichols KK. Blepharitis in the United States 2009: a survey-based perspective on prevalence and treatment. Ocul Surf. 2009; 7 (suppl 2): S1–S14. [DOI] [PubMed] [Google Scholar]
- 5. Vauclair S,, Majo F,, Durham AD,, Ghyselinck NB,, Barrandon Y,, Radtke F. Corneal epithelial cell fate is maintained during repair by Notch1 signaling via the regulation of vitamin A metabolism. Dev Cell. 2007; 13: 242–253. [DOI] [PubMed] [Google Scholar]
- 6. Zhang Y,, Lam O,, Nguyen MT,, et al. Mastermind-like transcriptional co-activator-mediated Notch signaling is indispensable for maintaining conjunctival epithelial identity. Development. 2013; 140: 594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Movahedan A,, Afsharkhamseh N,, Sagha HM,, et al. Loss of Notch1 disrupts the barrier repair in the corneal epithelium. PLoS One. 2013; 8: e69113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pan Y,, Lin MH,, Tian X,, et al. gamma-secretase functions through Notch signaling to maintain skin appendages but is not required for their patterning or initial morphogenesis. Dev Cell. 2004; 7: 731–743. [DOI] [PubMed] [Google Scholar]
- 9. Fre S,, Huyghe M,, Mourikis P,, Robine S,, Louvard D,, Artavanis-Tsakonas S. Notch signals control the fate of immature progenitor cells in the intestine. Nature. 2005; 435: 964–968. [DOI] [PubMed] [Google Scholar]
- 10. Conboy IM,, Conboy MJ,, Smythe GM,, Rando TA. Notch-mediated restoration of regenerative potential to aged muscle. Science. 2003; 302: 1575–1577. [DOI] [PubMed] [Google Scholar]
- 11. Djalilian AR,, Namavari A,, Ito A,, et al. Down-regulation of Notch signaling during corneal epithelial proliferation. Mol Vis. 2008; 14: 1041–1049. [PMC free article] [PubMed] [Google Scholar]
- 12. Sullivan DA,, Liu Y,, Kam WR,, et al. Serum-induced differentiation of human meibomian gland epithelial cells. Invest Ophthalmol Vis Sci. 2014; 55: 3866–3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu S,, Hatton MP,, Khandelwal P,, Sullivan DA. Culture, immortalization, and characterization of human meibomian gland epithelial cells. Invest Ophthalmol Vis Sci. 2010; 51: 3993–4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Movahedan A,, Majdi M,, Afsharkhamseh N,, et al. Notch inhibition during corneal epithelial wound healing promotes migration. Invest Ophthalmol Vis Sci. 2012; 5: 7476–7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yu XM,, Jaskula-Sztul R,, Ahmed K,, Harrison AD,, Kunnimalaiyaan M,, Chen H. Resveratrol induces differentiation markers expression in anaplastic thyroid carcinoma via activation of notch1 signaling and suppresses cell growth. Mol Cancer Ther. 2013; 12: 1276–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lin H,, Xiong W,, Zhang X,, et al. Notch-1 activation-dependent p53 restoration contributes to resveratrol-induced apoptosis in glioblastoma cells. Oncol Rep. 2011; 26: 925–930. [DOI] [PubMed] [Google Scholar]
- 17. Wang Q,, Li H,, Liu N,, et al. Correlative analyses of notch signaling with resveratrol-induced differentiation and apoptosis of human medulloblastoma cells. Neurosci Lett. 2008; 438: 168–173. [DOI] [PubMed] [Google Scholar]
- 18. Zhang P,, Li H,, Yang B,, et al. Biological significance and therapeutic implication of resveratrol-inhibited Wnt, Notch and STAT3 signaling in cervical cancer cells. Genes Cancer. 2014; 5 (5-6): 154–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Morris, K. Techniques of Lipidology: Isolation, Analysis and Identification of Lipids. Amsterdam: North-Holland Publishing Company; 1972. [Google Scholar]
- 20. Williams SE,, Beronja S,, Pasolli HA,, Fuchs E. Asymmetric cell divisions promote Notch-dependent epidermal differentiation. Nature. 2011; 470: 353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ezratty EJ,, Stokes N,, Chai S,, Shah AS,, Williams SE,, Fuchs E. A role for the primary cilium in Notch signaling and epidermal differentiation during skin development. Cell. 2011; 145: 1129–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu Y,, Kam WR,, Ding J,, Sullivan DA. Effect of azithromycin on lipid accumulation in immortalized human meibomian gland epithelial cells. JAMA Ophthalmol. 2014; 132: 226–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gertz M,, Nguyen GT,, Fischer F,, et al. A molecular mechanism for direct sirtuin activation by resveratrol. PLoS One. 2012; 7: e49761. [DOI] [PMC free article] [PubMed] [Google Scholar]