Abstract
Human phenotypes that are highly susceptible to radiation carcinogenesis have been identified. Sensitive phenotypes often display robust regulation of molecular features that modify biological response, which can facilitate identification of the pathways/networks that contribute to pathophysiological outcomes. Here we interrogate primary dermal fibroblasts isolated from Gorlin syndrome patients (GDFs), who display a pronounced inducible tumorigenic response to radiation, in comparison to normal human dermal fibroblasts (NHDFs). Our approach exploits newly developed thiol reactive probes to define changes in protein thiol profiles in live cell studies, which minimizes artifacts associated with cell lysis. Redox probes revealed deficient expression of an apparent 55 kDa protein thiol in GDFs from independent Gorlin syndrome patients, compared with NHDFs. Proteomics tentatively identified this protein as aldehyde dehydrogenase 1A1 (ALDH1A1), a key enzyme regulating retinoic acid synthesis, and ALDH1A1 protein deficiency in GDFs was confirmed by Western blot. A number of additional protein thiol differences in GDFs were identified, including radiation responsive annexin family members and lamin A/C. Collectively, candidates identified in our study have plausible implications for radiation health effects and cancer susceptibility.
Keywords: Gorlin syndrome, radiation, retinoic acid, carcinogenesis, protein thiol
INTRODUCTION
Gorlin syndrome is an autosomal dominant disease resulting in a dramatic predisposition to basal cell carcinoma (BCC) and developmental abnormalities [1,2]. Gorlin syndrome is characterized by a germline mutation in the Patched gene, which is hypothesized to render this phenotype haploinsufficient. The Patched gene encodes a putative tumor suppressing 12-span transmembrane receptor (PTCH) for hedgehog (Hh) ligands [3]. In the absence of HH ligands, PTCH inhibits the activity of a second transmembrane protein termed SMOOTHENED (SMO), and binding of Hh ligands to PTCH relieves this repression and results in activation of a signaling cascade whose output function is mediated by Gli transcription factors [3].
In addition to increased spontaneous cancer development in Gorlin syndrome, there are many clinical examples demonstrating a dramatic increase in radiation induced cancers in these patients. Radiotherapy for the treatment of medulloblastomas in children with Gorlin syndrome induces secondary intracranial tumors that are more aggressive than the initial tumor type [4]. Thousands of invasive BCCs after craniospinal irradiation are observed, which in some cases have led to patient death [5]. Radiotherapy is now contraindicated in Gorlin syndrome patients younger than 5 years of age [6]. In some occurrences, Gorlin syndrome patients exhibit multiple neoplasms (lung, liver, mesenteric, gastric and renal leiomyomas, lung typical carcinoid tumor, adenomatoid tumor of the pleura) with severe clinical presentation [7]. Latency for radiation induced tumors in Gorlin syndrome is generally 3–10 years after treatment [8].
Among susceptibility factors for carcinogenesis in Gorlin syndrome, Patched haploinsufficiency has received significant attention. However, it is important to note that loss of PTCH function occurs with high frequency in sporadic and Gorlin syndrome-associated BCCs [9]. PTCH haploinsufficiency could predispose Gorlin syndrome to genetic inactivation of the remaining allele by radiation induced DNA damage, but the fact that it is not unique to Gorlin syndrome patients suggests the existence of additional determinants of the radiation response. In this context, the Gorlin phenotype is defective in some types of DNA damage repair [10,11] and shows marked differences in DNA damage-induced p53 regulation [12]. Defective DNA damage repair may underlie human sensitivity to radiation carcinogenesis [13,14], and coupled with a haploinsufficient phenotype could contribute to genetic inactivation of Patched by mechanisms that remain incompletely understood. A third feature of Gorlin syndrome warranting consideration encompasses developmental abnormalities common to this phenotype. Paradigm shifts in toxicology and teratology have implicated epigenetic changes during developmental phases in later stage disease processes, including cancer [15]. A murine model of Gorlin syndrome (Ptch+/− mouse) has been developed that also displays sensitivity to radiation carcinogenesis [16]. The Ptch+/− mouse displays a defect in the radiation-induced activation of the ATR-Chk1 cell cycle checkpoint [17], suggesting aberrant cell cycle regulation might contribute to the tumorigenic response. Collectively, these observations indicate that the molecular basis for the dramatic increase in spontaneous and radiation-induced carcinogenesis in Gorlin syndrome is multifactorial.
In the present study, we investigate protein thiol status in primary dermal fibroblasts isolated from Gorlin syndrome patients (GDFs), compared to primary normal human dermal fibroblasts (NHDFs) used as control. Fibroblasts isolated from healthy photo-shielded skin of Gorlin syndrome patients exhibit a carcinoma-associated fibroblast phenotype [18], indicating a fundamental baseline change in the cell system. Fibroblasts play an active role in remodeling the tissue microenvironment to promote carcinogenesis [19] and display sensitive responses to both radiation [20] and H2O2 [21]. This phenotypic change in the stromal compartment of Gorlin syndrome patients is hypothesized to contribute to spontaneous BCC predisposition [18] and could also contribute to radiation-induced cancers. Protein thiols play an important role in mediating redox reactions and the same properties render them extremely sensitive to oxidation by free radicals. Altered cellular redox status and redox sensitive thiols have been implicated in clinical radiation resistance [22] and alterations in protein thiol status impacting cell survival after irradiation can be demonstrated in vitro under controlled conditions [23]. Oxidative protein thiol modifications can also play crucial roles in regulating the activity of molecular targets that promote tumorigenesis, such as the Nrf-2/ERK pathway [24,25]. Therefore, thiol reactive probes can identify protein thiols whose expression or thiol status is altered for prioritization of functional assessment. Thiol oxidation occurs rapidly upon cell lysis [26], and to circumvent this complication we are applying probes to live cells. We have examined iodoacetamide (IAM-RP; less reactive) and maleimide (Mal-RP; highly reactive) redox probe chemistries [27] that have been engineered to possess a flexible click chemistry alkyne functional group [28]. Copper-mediated click chemistry cycloadditions facilitate attachment of fluorescent tags to probe-labeled proteins for imaging applications or affinity isolation tags (e.g., biotin) for enrichment and protein identification by mass spectrometry [29]. We report a number of protein thiol differences from live cell studies comparing GDFs with NHDFs, including enzymes mediating retinoic acid (RA) synthesis, annexins, lamin A/C, and ribosomal proteins which have plausible multifaceted implications for spontaneous and radiation-induced carcinogenesis.
MATERIALS AND METHODS
Cell Culture
Adult NHDFs were purchased from Lonza (lot #00000293971; isolated from a 51-yr-old donor; Allendale, NJ) and were used as controls. GDFs were obtained with informed written consent of patients [18]. The two GDF stocks (designated AS573 and AS587; from 45 and 43-yr-old donors, respectively) used in the present study were isolated from healthy photo-shielded skin of different Gorlin syndrome patients and express distinct Patched mutations [12]. Fibroblasts were maintained in RPMI supplemented with 10% FBS, 1 ng/mL bFGF, 2 mM Glutamax, 100 U/mL penicillin, 100 mg/mL streptomycin, 25 mg/mL amphotericin B in 5% CO2/95% air at 37°C, and were subcultured by trypsinization.
X-Ray System
Irradiations were performed using an X-rad 320 machine (300 kV, Agfa NDT Pantak Seifert GmbH & Co. KG, Ahrensburg, Germany). Irradiated samples received doses of 10, 50, 100, 200, or 500 cGy at the following respective dose rates (6.3, 16.6, 33.3, 58.8, and 58.1 cGy/min). Sham controls were placed in the irradiator but not exposed.
Redox Probe Labeling for SDS–PAGE Analysis
Monolayer cultures were treated with redox probes for 1 h in serum-free medium. Cells were then washed 3× in ice cold PBS and scraped into lysis buffer (PBS, 2% nonidet P40, complete EDTA-free protease inhibitor [Roche Diagnostics Corporation, Indianapolis, IN] and 2 mM sodium orthovanadate). Metal chelators (EDTA/EGTA) interfere with click chemistry and were omitted from lysis buffers. Lysates were sonicated for 10 s (Fisher Scientific sonic dismembrator 60, Pittsburgh, PA) followed by centrifugation at 18 000g for 5 min at 4°C and the supernatant used for SDS–PAGE. Protein concentration was determined by a BCA assay (Pierce, Rockford, IL) and equal amounts of protein were subjected to the click chemistry reaction to attach azido-tetramethylrhodamine fluorophore [29]. Labeled proteins were separated by SDS–PAGE and visualized on a FluorchemQ imager (Alpha Innotch, San Leandro, CA). To determine the impact of thiol oxidation on probe binding, we treated fibroblasts with 300 μM H2O2 for 1 h followed by 20 μM IAM-RP for 1 h and processed cell lysates for SDS–PAGE analysis. For quantification of probe binding individual gels were normalized to NHDF controls and results from three independent experiments were pooled (n = 3). Differences in probe labeling due to peroxide or radiation treatments were then presented as % control. Quantification included either whole lane analysis or selected individual bands.
Redox Probe Labeling for Cellular Imaging
Fibroblasts were seeded into Nunc eight-well chamber slides. Following treatment with thiol reactive probes for 1 h, fibroblasts were washed 3× in PBS followed by fixation (3.6% paraformaldehyde, 0.024% saponin in PBS) for 15 min at room temperature. Cells were then washed 3× in PBS and thiol reactive probes were labeled with an Alexa647 fluor using a commercial kit (Click-iT EdU Alexa Fluor 647 Imaging Kit; Life Technologies, Grand Island, NY). The click chemistry reaction was set up according to manufacturer's directions using a final volume of 250 μL/well for 30 min at room temperature. No probe controls represent cells that were not exposed to thiol reactive probes, but were processed under identical conditions to define non-specific backgrounds. Images were acquired by epifluorescence microscopy as described [30].
Redox Probe Labeling for Quantitative Mass Spectrometry-Based Identification
Fibroblasts were treated with 20 μM IAM-RP for 1 h in serum-free medium, washed 3× with PBS, scraped into PBS lysis buffer and solubilized by gentle sonication. For mass spectrometry analysis, lysates were not subjected to centrifugation to remove detergent insoluble constituents, which might contain probe-labeled protein thiols. Biotin-azide was attached by click chemistry [29], and protein values were normalized to 700 μg prior to enrichment. Probe-labeled proteins were enriched on streptavidin resin, reduced with TCEP, alkylated with IAM, and digested on-resin with trypsin [29,31]. Peptides were separated by high-resolution, reversed phase constant pressure capillary liquid chromatography as previously described [31]. MS analysis was performed using a Thermo Electron ion trap LTQ MS outfitted with a custom ion funnel and electrospray ionization (ESI) interface. Data was acquired for 100 min, beginning 65 min after sample injection (15 min into gradient). LTQ spectra were collected from 400 to 2000 m/z at a resolution of 100k, followed by data-dependent ion trap MS/MS spectra of the six most abundant ions using a 35% collision energy. A dynamic exclusion time of 30 s was used to discriminate against previously analyzed ions.
Generated MS/MS spectra were searched using the SEQUEST algorithm (V27, revision 12) [32] against the publicly available Homo sapiens translated genome sequence, and re-scored using the MS-GF approach. Identified peptides were required to be at least six amino acids in length having MS-GF scores ≤1E–10, which corresponds to an estimated FDR <1%. Using peptides that match these criteria, in order to classify a protein as specifically labeled by a redox probe we required the following criteria [33]: (i) ≥2 unique peptides per protein; (ii) ≥2 peptides measured per protein in at least two replicates; (iii) the protein exhibits ≥3-fold more abundance in the redox probe-labeled samples relative to the “no probe” controls. All filter passing peptide identifications were tallied to provide quantitative spectral count data for each protein. Relative protein abundance was estimated by averaging peptide spectral counts across three probe-labeled biological replicates.
Western blot Analysis
Western blot analysis was performed as described [34]. Final titers were: ALDH1A1 (1:2000), Annexin A2 (1:4000), Annexin A1 (1:3000), actin (1:10,000), Lamin A/C (1:3000), secondary antibodies (1:3000).
Cell Viability Assays
Equal numbers of cells were treated with H2O2 at the indicated concentrations for 24 h and cell viability was quantified using a neutral red assay [35]. For clonogenic survival assays, 1 × 104 cells were seeded into 10 cm dishes and allowed to attach for 24 h. Cells were then treated with peroxide or irradiated and colonies quantified on day 10 as described [36].
Statistical Analysis
Individual comparisons were made using the Student's t-test or ANOVA with a post hoc Student's Newman–Keul test, as appropriate. The P < 0.05 level was accepted as significant.
RESULTS
Redox Probes Identify Different Targets and Respond to Oxidative Perturbation
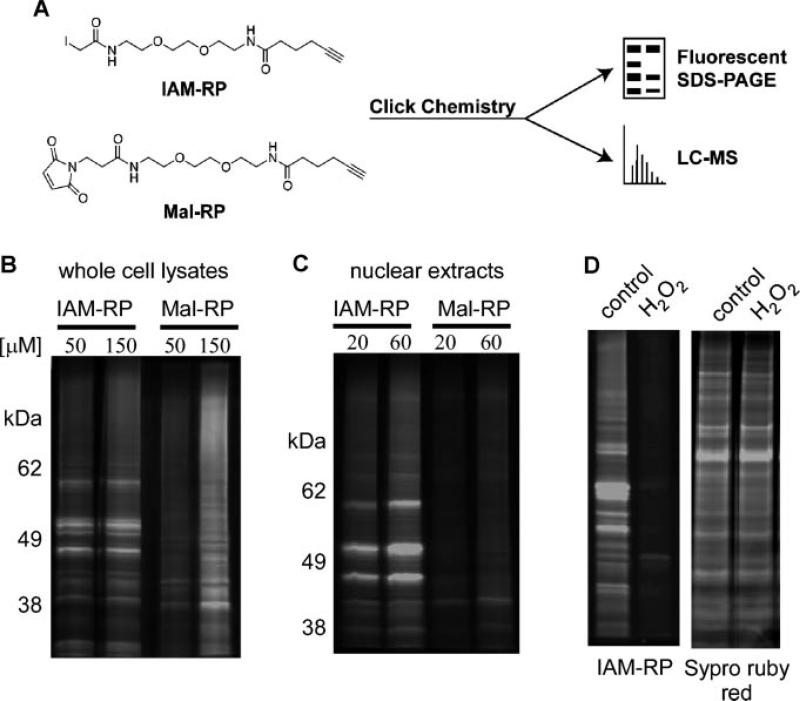
Prior to application of redox probes, validation studies were conducted. The click chemistry functional group enables addition of fluorescent tags which allows visualization of protein thiols by SDS–PAGE (Figure 1A). To determine the robustness of our approach, cell lysates prepared from fibroblasts exposed to IAM-RP or Mal-RP for 1 h were subjected to click chemistry to azido-tetramethylrhodamine followed by SDS–PAGE analysis [28]. We observed a qualitative difference between protein bands detected by IAM-RP and Mal-RP probes (Figures 1B). We also evaluated nuclear extracts as an initial indication of cell permeability. The IAM-RP produced robust labeling of proteins in nuclear extracts, relative to the Mal-RP (Figure 1C). In both cases, increasing probe concentration increased signal intensity. Oxidation of protein thiols should inhibit the covalent binding of probe to reduced thiol and we tested this hypothesis using the IAM-RP probe. Pretreatment with 300 μM H2O2 eliminated detection of the majority of protein bands with the IAM-RP (Figure 1D), suggesting that the observable bands are predominantly protein thiols. Analysis of protein loading with Sypro ruby red (non-specific fluorescent protein stain to measure abundance) confirmed equivalent protein loading levels on the SDS–PAGE gel (Figure 1D).
Figure 1.
Protein thiol detection by SDS–PAGE analysis. Panel A: Schematic of redox probes with terminal alkyne group and click chemistry modification to fluorescent or affinity isolation tags for imaging or proteomic applications. Panel B: Protein thiol profiles following treatment of GDFs with 50–150 μM IAM-RP or Mal-RP. Panel C: Protein thiol profiles associated with nuclear extracts prepared from GDFs treated with 20–60 μM IAM-RP or Mal-RP. Panel D: Pretreatment of GDFs with 500 μM H2O2 eliminated the majority of probe binding without impacting total protein levels, suggesting that the detected bands represent protein thiols. Similar qualitative results were observed in two separate experiments.
Cellular Imaging of Thiol Reactive Probes In Situ
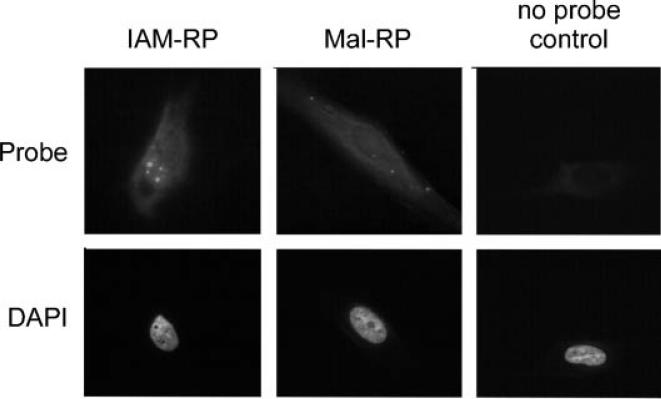
SDS–PAGE suggests that thiol reactive probes are reaching intracellular sites, including a strong nuclear localization for IAM-RP. To further investigate this possibility, fibroblasts were treated with 20 μM IAM-RP or Mal-RP for 1 h in serum-free medium followed by fixation and click chemistry to an Alexa647 fluor as described in methods. Both probes showed diffuse cellular staining. IAM-RP labeling of nuclear foci was apparent (Figure 2), consistent with IAM-RP nuclear localization indicated by SDS–PAGE (see Figure 1C). Cells processed under identical conditions without exposure to thiol reactive probes, termed no probe controls, showed marginal fluorescent signal at equal exposure times. These observations support the cell permeability of thiol reactive probes in live cell studies which helps circumvent oxidative artifacts induced by lysis. Additional studies characterizing imaging applications in 2D and 3D model systems can be found in Supplementary data (S1).
Figure 2.
Qualitative images of thiol reactive probe labeling in situ. Fibroblasts were treated with 20 μM IAM-RP or Mal-RP probe for 1 h followed by fixation and click chemistry to an Alexa647 fluorophore. Similar subcellular distribution patterns were observed in three separate experiments.
Fibroblast Response to Peroxide and Radiation
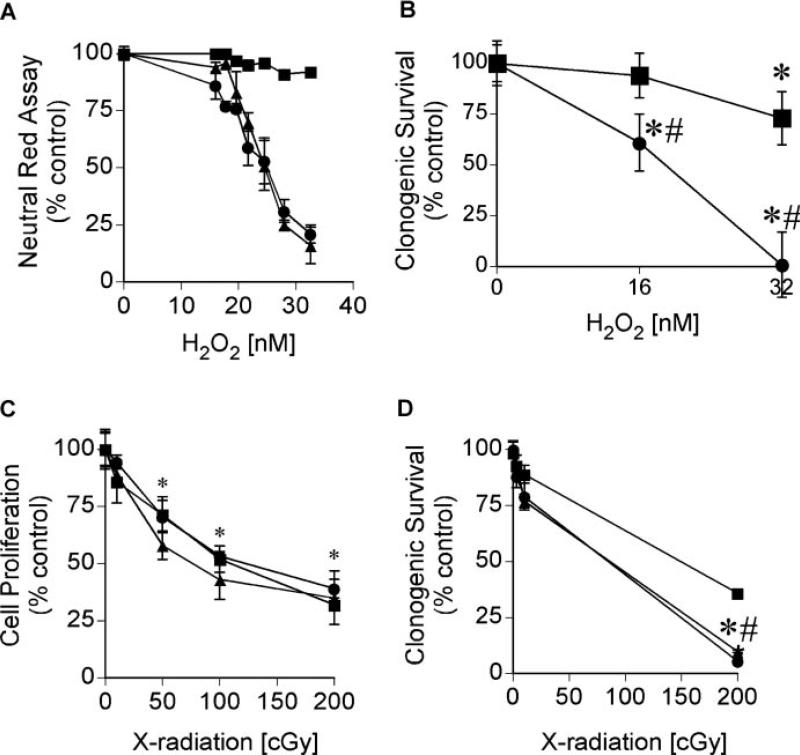
Dose–response curves for hydrogen peroxide (H2O2)- and X-radiation-induced toxicity were established to test sensitivity of SDS–PAGE analysis for detection of perturbation-induced changes in protein thiols. The neutral red assay measures lysosomal integrity as an index of cell viability and provides a sensitive index of oxidative damage, as compared with other cell viability assays [37]. We defined the transition between no observable effect and a toxic response measured 24 h after H2O2 treatment. GDFs displayed a dose-dependent decrease in cell viability over the concentration range of 16–32 nM H2O2, while NHDFs showed negligible toxicity over this range (Figure 3A). Increased sensitivity of GDFs to H2O2 toxicity, relative to NHDFs, was also determined using a clonogenic survival assay (Figure 3B). A similar shift in peroxide sensitivity was observed, however, H2O2 toxicity in NHDFs was apparent in the clonogenic survival assay, indicating that neutral red was underestimating toxicity. X-radiation (3–200 cGy) inhibited the proliferation of NHDFs and GDFs to a comparable extent (Figure 3C), consistent with reports demonstrating comparable initial DNA damage responses to ionizing radiation in cells from Gorlin syndrome patients versus controls [10]. GDFs showed slightly but significantly reduced clonogenic survival at the highest dose (200 cGy), compared with NHDFs (Figure 3D).
Figure 3.
Dose–response curves for H2O2 and X-radiation. Panel A: NHDFs (square) and GDFs (AS573—circle; AS587—triangle) were treated with 16–32 nM H2O2 for 24 h and cell viability was determined using a neutral red assay. Values represent the mean ± SE (n = 8). Similar results were observed in two separate experiments. Panel B: Clonogenic survival was determined 10 days postperoxide treatment for NHDFs (square) and AS587 GDFs (circle) as described in Methods section. Values represent the mean ± SE (n = 3). *Significantly different from respective control; #significantly different from NHDFs, P < 0.05. Panel C: NHDFs (square) and GDFs (AS573—circle; AS587—triangle) were exposed to 3–200 cGy X-radiation and cell number was quantified at 72 h using a Coulter Counter. Values represent the mean ± SE (n = 3). *Significantly different from respective control and is representative for all three experimental groups. There were no statistically significant differences between groups. Similar results were observed in two separate experiments. Panel D: Clonogenic survival was determined 10 days postradiation exposure as described in Methods. Values represent the mean ± SE (n = 3). *Significantly different from respective control; #significantly different from NHDFs for both GDF strains, P < 0.05. Similar results were observed in two separate experiments.
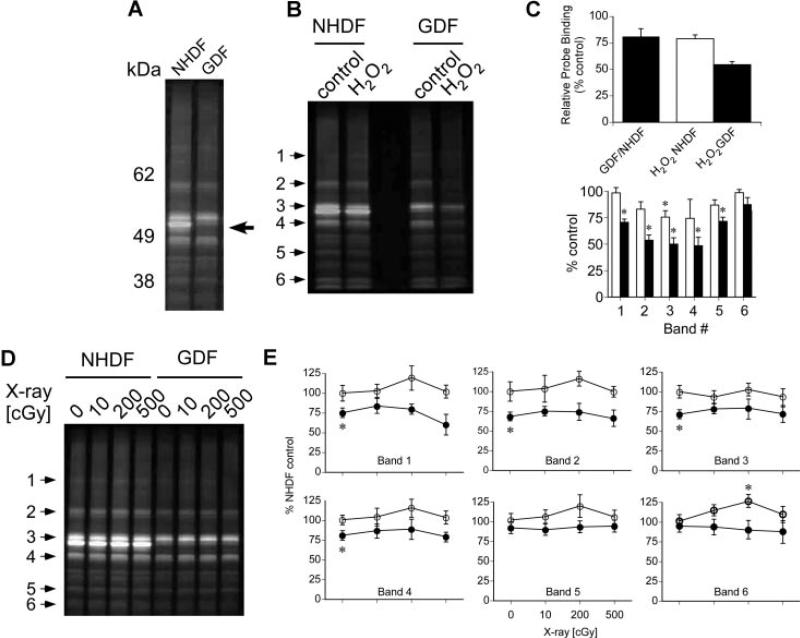
We then asked whether SDS–PAGE could detect (1) baseline differences in protein thiol profiles measurable by redox probes, and (2) perturbation of protein thiol profiles by H2O2 or X-radiation. These experiments employed the IAM-RP due to its enhanced cell and nuclear permeability. NHDFs and GDFs were incubated with 20 μM IAM-RP followed by preparation of cell lysates and fluorescent tagging via click chemistry. Figure 4A highlights a qualitative difference in the detection of an apparent 55 kDa protein that is reduced in GDFs (see arrow), compared to NHDFs. NHDFs and GDFs were then treated with 32 nM H2O2 for 1 h, which induces toxicity to a different extent in NHDFs and GDFs (see Figure 3A and B) and probe binding was quantified. Probe-labeled proteins visualized by SDS–PAGE are shown in Figure 4B. Probe binding to proteins was decreased by approximately 21% in control GDF preparations, relative to control NHDF preparations (Figure 4C, GDF/NHDF ratio of whole lane). H2O2 reduced probe binding by approximately 24% in NHDFs and 48% in GDFs, relative to respective controls (H2O2/NHDF and H2O2 GDF ratios). Six individual bands spanning low to high molecular weight proteins (see arrows in Figure 4B) were analyzed and were generally consistent with the differential response of GDFs to H2O2, compared with NHDFs (Figure 4C). In a similar study, cells were exposed to X-radiation (10–500 cGy) followed by a 1 h pulse with 20 μM IAM-RP and fluorescence detection. In contrast to H2O2, high radiation doses producing marked reductions in clonogenic survival (see Figure 3C) showed little impact on protein thiol profiles detected by SDS–PAGE (Figure 4D and E), with the exception of a slight but statistically significant increase in detection of band #6 (estimated MW ~30 kDa) in NHDFs at 200 cGy.
Figure 4.
Protein thiol profiles for NHDFs versus GDFs labeled with IAM-RP. Panel A: Baseline comparison of IAM-RP-sensitive protein thiols for NHDFs and GDFs. Arrow indicates a protein thiol that is abundant in NHDFs and apparently absent or markedly reduced in GDFs. Similar profiles were observed in three separate experiments. Panel B: Treatment of cells with 32 nM H2O2 for 1 h reduces protein thiol detection to a greater extent in GDFs, as compared with NHDFs. Similar results were observed in two separate experiments. Panel C: Quantification of protein thiol from peroxide-treated cells. Top graph represents whole lane analysis and illustrates a decrease in control GDF protein thiol levels, compared with control NHDFs (GDF/NHDF ratio). The decrease in protein thiol detection induced by peroxide is presented as the % respective control. In addition, six individual protein thiol bands were selected (see arrows in Figure 4B) and results from three independent experiments were pooled for statistical analysis (bottom graph). Values represent the mean ± SE (n = 3). *Significantly different from respective control; #significantly different from NHDF counterpart. Panel D: Qualitative protein thiol profiles visualized by SDS–PAGE for irradiated NHDFs and GDFs. Cells were incubated with redox probe immediately after irradiation for 1 h prior to preparation of cell lysates for SDS–PAGE analysis. Panel E: six individual protein thiol bands from NHDF (open circle) or AS587 GDFs (solid circle) (see arrows in Figure 4D) were quantified and results from three independent experiments were pooled for statistical analysis. Values represent the mean ± SE (n = 3). *Significantly different from NHDF control.
Proteomic Characterization of Redox Probe Targets
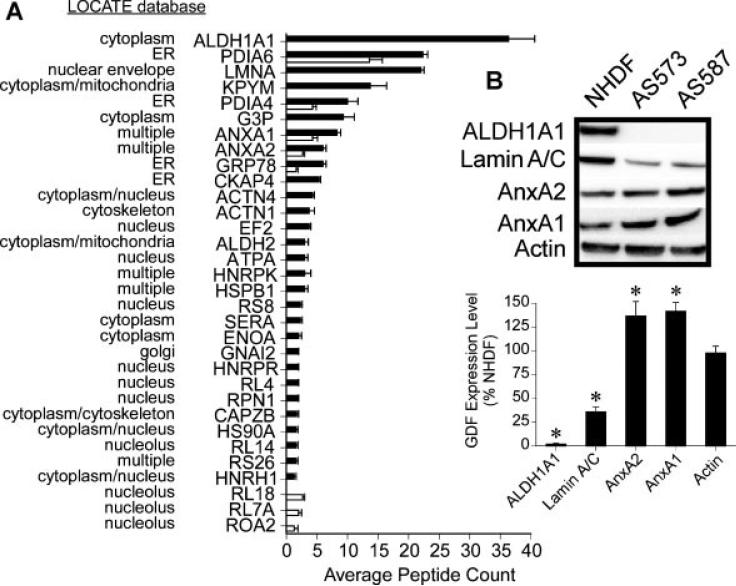
SDS–PAGE only permits the detection of a small fraction of the proteome, and does not provide identification of probe-labeled proteins. Following probe labeling in situ, we modified the click chemistry to a biotin label for streptavidin-resin based affinity isolation of probe-labeled proteins and identification by mass spectrometry [28]. A no probe control group was processed under identical conditions in parallel to define non-specific backgrounds. To assure an accurate quantification of protein thiols, GDF and NHDF proteomes were normalized by protein concentration prior to streptavidin-mediated enrichment of probe-labeled proteins. Upon subtraction of non-specific backgrounds in the proteomic data, a total of 48 probe-labeled proteins were detected, of which 45 were present in NHDFs and 16 were present in GDFs. Figure 5A illustrates the subset of probe-labeled proteins that displayed statistically significant differences between NHDF and GDF experimental groups. Protein subcellular distribution was predicted using the LOCATE database (http://locate.imb.uq.edu.au/), and predictions suggested that the IAM-RP probe reached multiple subcellular compartments, consistent with SDS–PAGE and imaging analysis of probe subcellular distribution (Figures 1 and 2). A number of proteins were specifically enriched by the IAM-RP probe that did not show significant differences between NHDFs and GDFs, including PDIA3, PDIA1, CLIC1, RRBP1, IMDH2, ACTBL, RL7, CLIC4, LDHB, PPIB, ANXA6, PPIA (proteins arranged in order from highest to lowest average peptide counts/protein; ranging from 30 → 2). Protein thiol candidates will be expanded in Discussion section.
Figure 5.
Protein thiol identification by mass spectrometry. Panel A: NHDFs (black) and GDFs (white) were incubated with 20 μM IAM-RP for 1 h followed by click chemistry to a biotin tag for affinity isolation as described in Methods section. Isolated proteins were identified following tryptic digestion and peptide sequencing by mass spectrometry. No probe controls were used to subtract non-specific backgrounds and results represent the subset of proteins showing statistically significant differences between NHDFs and GDFs. Values represent the mean ± SE (n = 3, biological replicates). Panel B: Cell lysates were prepared from NHDFs and GDFs (AS573, AS587) and equal amounts of protein were subjected to Western blot analysis of ALDH1A1, Lamin A/C, AnxA2, and AnxA1 as validation. Results from two independent experiments were pooled for quantitative analysis shown in graph (n = 4). Values represent the mean ± SE. *Significantly different from NHDF control.
Differences in probe-dependent enrichment of protein thiols in situ could arise from multiple mechanisms, including oxidation of the protein thiol, changes in protein expression or changes in thiol accessibility. Several candidate protein thiols (ALDH1A1, Lamin A/C, AnxA2, AnxA1) were selected for validation by Western blot. ALDH1A1 and Lamin A/C showed reduced protein expression (Figure 5B), consistent with their decreased detection by mass spectrometry (Figure 5A). In contrast, AnxA2 and AnxA1 protein levels were significantly increased in GDFs, relative to NHDFs (Figure 5B), suggesting loss of their detection by thiol reactive probe could not be accounted for by a reduction in protein levels.
DISCUSSION
We have investigated the application of cell permeable thiol reactive probes to reveal differences in protein thiol profiles between NHDFs and GDFs, the latter representing a genetically susceptible human phenotype for radiation carcinogenesis. Our discussion is divided into two major topics, the first focused on data validating application of clickable thiol reactive probes and the second on the probe-identified protein thiols and their implication in Gorlin syndrome and radiation health effects.
Thiol Reactive Probe Validation
Pretreatment of cells with H2O2 eliminates the majority of probe-dependent protein bands detected by SDS–PAGE (Figure 1D), suggesting low non-specific backgrounds for protein thiol detection. However, non-specific background is detectable in all applications and it is prudent to include no probe controls. We observed loss of an apparent 55 kDa protein thiol in GDFs by SDS–PAGE (Figure 4A, arrow), which was tentatively identified as ALDH1A1 (Figure 5) by mass spectrometry based on predicted molecular weight correlation. ALDH1A1 protein deficiency in GDFs was then confirmed by Western blot (Figure 5B), validating mass spectrometry results. Similar validation was observed for Lamin A/C (Figure 5). In contrast, redox probes identified reduced detection of AnxA2 and AnxA1 that was not due to decreased protein levels (Figure 5B). Therefore, the change in detection of annexins must be due to either a change in thiol oxidation status or thiol accessibility.
We examined acute impact of H2O2 on protein thiol profiles measured 1 h after peroxide treatment with longer term indices of cell viability. The dose–response curve for H2O2 was right-shifted in NHDFs compared with GDFs (Figure 3), indicating that GDFs are more sensitive to free radical damage. A similar trend was observed for protein thiol profiles detected by SDS–PAGE using redox probe where peroxide treatment reduced protein thiol detection to a greater extent in GDFs, compared with NHDFs (Figure 4B).
Oxidative damage induced by radiation versus endogenous free radicals is a continuing area of interest [38]. In contrast to peroxide effects on protein thiol profiles, a high radiation dose (200–500 cGy) producing massive cell death (Figure 3D), exhibited negligible effects on protein thiol profiles (Figure 4D and E). Radiation cell killing occurs through the generation of oxygen free radicals and we were somewhat surprised by this observation. It is important to recognize the limits of detection by SDS–PAGE, which inherently under samples the proteome. Thus, it is possible that future studies may identify radiation-sensitive protein thiols that were not detected by the SDS–PAGE analysis. The fact that a subset of protein thiols in GDFs were clearly impacted by H2O2, but not radiation, suggests that radiation-derived free radicals are targeting unique substrates. We did identify one protein thiol band (#6 on Figure 4D and E; ~30 kDa) that significantly changed in response to radiation treatment in NHDFs, but not GDFs. The identity of this protein is unknown but we can provide a tentative prediction based on MW correlation with MS results for proteins near the 30 kDa size (G3P, AnxA2, AnxA1, HSPB1, RS8, CAPZB, RL14, RL18, RL7A). Once identified, studies are warranted to determine why this protein shows a differential response to radiation in NHDFs, relative to GDFs, which may provide further insight into the Gorlin phenotype.
Radiation Responsive Annexins
The differential detection of AnxA1 and A2 by redox probes in a radiation susceptible human phenotype is intriguing. AnxA2 is a multifunctional protein that regulates molecular processes in many subcellular compartments, some of which contribute to tumorigenesis and metastasis [39,40]. We have previously identified AnxA2 as a radiation-responsive protein that influences the cell transformation response to radiation in vitro [41], as well as the radiation transcriptome and cell fate [34]. AnxA2 possesses redox sensitive cysteine(s) [39] and oxidation at these sites could reduce probe binding, however, the mechanism for reduced probe binding to AnxA2 cannot be determined from these studies. For example, conformational changes in AnxA2 tertiary structure or subcellular distribution could influence probe–protein interactions by mechanisms unrelated to thiol oxidation and cannot be ruled out. AnxA1 is also implicated in human carcinogenesis and metastasis [42,43], therefore, increased expression of AnxA1 in a cancer prone phenotype warrants further consideration. Future studies will begin dissecting compartmental regulation of annexins to identify which variants functionally contribute to radiation carcinogenesis/cell killing, and define the site-specific modifications that influence protein function.
Gorlin Syndrome and Radiation Carcinogenesis
Susceptibility factors for radiation carcinogenesis in Gorlin syndrome have been identified including, (1) haploinsufficiency at the Patched allele, (2) defective DNA damage repair, (3) defective cell cycle regulation, and (4) developmental defects. Our data suggest that retinoic acid (RA)-deficiency may be an additional feature of Gorlin syndrome that is relevant to, and likely intertwined with, these susceptibility features. RA deficiency in GDFs is indicated by deficient expression of ALDH1A1 (Figures 5). ALDH1A1 is one of three rate-limiting enzymes in the conversion of retinaldehyde to RA. RA synthesis is reduced by 77% in the Aldh1a1 knockout mouse [44], indicating ALDH1A1 accounts for the major fraction of RA physiologically. RA is also a key regulator of developmental biology [45] and developmental abnormalities in Gorlin syndrome patients provide clear phenotypic support for the significance of this observation.
RA regulates cancer susceptibility, suggesting that ALDH1A1 deficiency in Gorlin syndrome is likely important to spontaneous and radiation-induced carcinogenesis. In support of this statement, epidemiological studies have correlated increased serum retinoid levels with reduced cancer risk [46], or conversely demonstrated that patients who have died of cancer have depressed retinoid levels [47]. These relationships have been confirmed in animal models where treatment with RA inhibits tumorigenesis, including basal cell carcinoma in a murine model for Gorlin syndrome [48] and skin carcinogenesis induced by ionizing radiation [49]. In contrast, retinoid deficient diets promote tumorigenesis at multiple sites [50]. Since RA levels are frequently depressed in cancer patients [51], it is plausible that this work may have broader implications such as retinoid deficiency contributing to the promotion of secondary cancers at the time of radiotherapy [52].
RA regulates many molecular and cellular processes that could impact radiation-induced DNA damage repair or unrelated processes functioning at different stages of carcinogenesis. Molecular examples are related to Ataxia telangiectasia, which is another human phenotype that is sensitized to radiation carcinogenesis [53]. Enhanced sensitivity is attributed to mutations in the Ataxia telangiectasia mutated (ATM) gene that functions in repair of radiation-induced DNA damage. RA can activate the ATM kinase [54], therefore, RA deficiency in Gorlin syndrome might contribute to documented deficiencies in DNA damage repair in this disease [10,11] through deficient ATM activation. Defective DNA damage repair could lead to a prolonged presence of lesions in DNA, which in turn, stabilize p53 levels, consistent with a marked increase in p53 stabilization in irradiated cells from Gorlin syndrome patients (8). Alternatively, the Ptch+/− mouse displays a defect in the radiation-induced activation of the ATR-Chk1 cell cycle checkpoint [17] and the presence or absence of RA can significantly influence Chk1 phosphorylation status and activity [55]. Therefore, retinoid deficiency can impact multiple molecular processes of importance to radiation carcinogenesis that could plausibly function in concert or independently.
At the cellular level retinoids have been shown to stimulate stem cell differentiation [56] or induce apoptosis in cancer cells [57]. ALDH1A1 is a reported marker of the stem cell niche and its inhibition prevents the differentiation of stem cells [58]. Therefore, ALDH1A1-deficiency might prevent differentiation of the cancer stem cell niche, consistent with the unusually high tumorigenic response to radiation in Gorlin syndrome patients, and reduced apoptotic responses due to depressed retinoid levels could exert a similar function.
The developmental basis of carcinogenesis is an emerging focal point [15]. Primary fibroblasts used in the present study have been isolated from healthy photo-shielded skin of Gorlin syndrome patients, yet phenotypically resemble cancer-associated fibroblasts [18]. Lamin A/C expression is frequently decreased in carcinogenesis [59] and proteomics analysis revealed decreased Lamin A/C expression in GDFs (Figure 5), which appears consistent with their phenotypic characterization as cancer-associated fibroblasts. This indicates that GDFs exhibit a fundamental change in phenotype, which could plausibly be influenced by RA deficiency impacting developmental programming and epigenetics. Lamin A/C-deficiency does not alter the DNA damage response to ionizing radiation in murine embryonic fibroblasts [60]. However, Lamin A/C functions in maintaining nuclear and cellular organization, as well as signal transduction [61]. In this context, there are several reports linking Lamin A/C to the regulation of pathways implicated in carcinogenesis [62]. Therefore, loss of Lamin A/C expression is expected to fundamentally alter cell signaling responses in the Gorlin phenotype.
Cancer-associated fibroblasts are involved in remodeling the extracellular matrix, suppressing immune responses, and secreting growth factors and cytokines that regulate differentiation, angiogenesis, and chronic inflammation [19]. It is known that low doses of radiation [20] and H2O2 [21] can induce a fibroblast senescent phenotype, which is also a secretory phenotype that promotes tumorigenesis [63]. GDFs were sensitized to H2O2 and radiation toxicity (Figure 3), raising the possibility that free radical perturbation of stromal–epithelial interactions may also be sensitized in Gorlin syndrome.
Finally, we observed enrichment of several ribosomal proteins in our proteomics analysis, including three candidates uniquely enriched in GDFs (Figure 5). Ribosomal biogenesis is hypothesized to play an important regulatory role in the p53 pathway [64]. We have previously documented abnormal p53 regulation in primary cells from Gorlin syndrome patients [12]. Thus, redox probes may provide new methods to interrogate ribosomal biogenesis and the extent to which ribosomal function in carcinogenesis is modulated by changes in thiol status.
In summary, we have validated thiol reactive probes for interrogating genetically susceptible human phenotypes and identify candidate protein thiols that are consistent with susceptibility to spontaneous and radiation-induced carcinogenesis in Gorlin syndrome. Protein thiols identified are known to be radioresponsive proteins (annexins) or enzymes (ALDH1A1; Lamin A/C) regulating pathways that significantly impact carcinogenesis. Collectively, the pathways identified have plausible implications for modifying radiation health effects. A multifactorial basis for unusual sensitivity to radiation carcinogenesis in humans appears likely based on our collective knowledge of the Gorlin phenotype. How these aberrant features interact to modify radiation carcinogenesis in such a dramatic fashion remains unclear.
Supplementary Material
ACKNOWLEDGEMENTS
We are indebted to Drs. Florence Brellier, Alexandre Valin, Stephanie Barnay, Mathilde Frechet, Emilie Warrick, Sabine Scarzello, Odile Chevallier-Lagente, and Pr. Marie-Françoise Avril for their expert and enthusiastic contribution to this work. This work was supported by The Biological and Environmental Research Program (BER)—U.S. Department of Energy [DE-AC06-76RLO], and the Laboratory Directed Research and Development program at PNNL. This work used instrumentation and capabilities developed under support from the NIGMS (8P41GM103493-10), and the U.S. DOE. Proteomic measurements were made in the Environmental Molecular Sciences Laboratory, a DOE-BER national scientific user facility at PNNL. T.M. lab was supported by the Fondation de l'Avenir (ET9-551), the Association pour la Recherche sur le Cancer (SFI201212055859), the Societe Fran-çaise de Dermatologie.
REFERENCES
- 1.Kimonis VE, Goldstein AM, Pastakia B, et al. Clinical manifestations in 105 persons with nevoid basal cell carcinoma syndrome. Am J Med Genet. 1997;69:299–308. [PubMed] [Google Scholar]
- 2.Samela PC, Tosi V, Cervini AB, Bocian M, Bujan MM, Pierini AM. Nevoid basal cell carcinoma syndrome: our experience in a pediatric hospital. Actas Dermosifiliogr. 2013;104:426–433. doi: 10.1016/j.adengl.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 3.Jiang J, Hui CC. Hedgehog signaling in development and cancer. Dev Cell. 2008;15:801–812. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choudry Q, Patel HC, Gurusinghe NT, Evans DG. Radiation-induced brain tumours in nevoid basal cell carcinoma syndrome: implications for treatment and surveillance. Childs Nerv Syst. 2007;23:133–136. doi: 10.1007/s00381-006-0178-4. [DOI] [PubMed] [Google Scholar]
- 5.Evans DG, Birch JM, Orton CI. Brain tumours and the occurrence of severe invasive basal cell carcinoma in first degree relatives with Gorlin syndrome. Br J Neurosurg. 1991;5:643–646. doi: 10.3109/02688699109002890. [DOI] [PubMed] [Google Scholar]
- 6.Amlashi SF, Riffaud L, Brassier G, Morandi X. Nevoid basal cell carcinoma syndrome: relation with desmoplastic medulloblastoma in infancy. A population-based study and review of the literature. Cancer. 2003;98:618–624. doi: 10.1002/cncr.11537. [DOI] [PubMed] [Google Scholar]
- 7.Garavelli L, Piemontese MR, Cavazza A, et al. Multiple tumor types including leiomyoma and Wilms tumor in a patient with Gorlin syndrome due to 9q22.3 microdeletion encompassing the PTCH1 and FANC-C loci. Am J Med Genet A. 2013;161:2894–2901. doi: 10.1002/ajmg.a.36259. [DOI] [PubMed] [Google Scholar]
- 8.Evans DG, Birch JM, Ramsden RT, Sharif S, Baser ME. Malignant transformation and new primary tumours after therapeutic radiation for benign disease: substantial risks in certain tumour prone syndromes. J Med Genet. 2006;43:289–294. doi: 10.1136/jmg.2005.036319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reifenberger J, Wolter M, Knobbe CB, et al. Somatic mutations in the PTCH, SMOH, SUFUH and TP53 genes in sporadic basal cell carcinomas. Br J Dermatol. 2005;152:43–51. doi: 10.1111/j.1365-2133.2005.06353.x. [DOI] [PubMed] [Google Scholar]
- 10.Newton JA, Black AK, Arlett CF, Cole J. Radiobiological studies in the naevoid basal cell carcinoma syndrome. Br J Dermatol. 1990;123:573–580. doi: 10.1111/j.1365-2133.1990.tb01473.x. [DOI] [PubMed] [Google Scholar]
- 11.Nagasawa H, Burke MJ, Little FF, McCone EF, Chan GL, Little JB. Multiple abnormalities in the ultraviolet light response of cultured fibroblasts derived from patients with the basal cell nevus syndrome. Teratog Carcinog Mutagen. 1988;8:25–33. doi: 10.1002/tcm.1770080104. [DOI] [PubMed] [Google Scholar]
- 12.Brellier F, Valin A, Chevallier-Lagente O, Gorry P, Avril MF, Magnaldo T. Ultraviolet responses of Gorlin syndrome primary skin cells. Br J Dermatol. 2008;159:445–452. doi: 10.1111/j.1365-2133.2008.08650.x. [DOI] [PubMed] [Google Scholar]
- 13.Lehmann AR, McGibbon D, Stefanini M. Xeroderma pigmentosum. Orphanet J Rare Dis. 2011;6:70. doi: 10.1186/1750-1172-6-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ambrose M, Gatti RA. Pathogenesis of ataxia-telangiectasia: the next generation of ATM functions. Blood. 2013;121:4036–4045. doi: 10.1182/blood-2012-09-456897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reamon-Buettner SM, Borlak J. A new paradigm in toxicology and teratology: altering gene activity in the absence of DNA sequence variation. Reprod Toxicol. 2007;24:20–30. doi: 10.1016/j.reprotox.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 16.Mancuso M, Pasquali E, Leonardi S, et al. Oncogenic bystander radiation effects in Patched heterozygous mouse cerebellum. Proc Natl Acad Sci USA. 2008;105:12445–12450. doi: 10.1073/pnas.0804186105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leonard JM, Ye H, Wetmore C, Karnitz LM. Sonic Hedgehog signaling impairs ionizing radiation-induced checkpoint activation and induces genomic instability. J Cell Biol. 2008;183:385–391. doi: 10.1083/jcb.200804042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valin A, Barnay-Verdier S, Robert T, et al. PTCH1 / dermal fibroblasts isolated from healthy skin of Gorlinþsyndrome patients exhibit features of carcinoma associated fibroblasts. PLoS ONE. 2009;4:e4818. doi: 10.1371/journal.pone.0004818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J, Liu J. Tumor stroma as targets for cancer therapy. Pharmacol Ther. 2012;137:200–215. doi: 10.1016/j.pharmthera.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsai KK, Stuart J, Chuang YY, Little JB, Yuan ZM. Low-dose radiation-induced senescent stromal fibroblasts render nearby breast cancer cells radioresistant. Radiat Res. 2009;172:306–313. doi: 10.1667/RR1764.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiyoshima T, Enoki N, Kobayashi I, et al. Oxidative stress caused by a low concentration of hydrogen peroxide induces senescence-like changes in mouse gingival fibroblasts. Int J Mol Med. 2012;30:1007–1012. doi: 10.3892/ijmm.2012.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Selenius M, Hedman M, Brodin D, et al. Effects of redox modulation by inhibition of thioredoxin reductase on radio-sensitivity and gene expression. J Cell Mol Med. 2012;16:1593–1605. doi: 10.1111/j.1582-4934.2011.01469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biaglow JE, Ayene IS, Koch CJ, et al. Radiation response of cells during altered protein thiol redox. Radiat Res. 2003;159:484–494. doi: 10.1667/0033-7587(2003)159[0484:rrocda]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 24.Patwardhan RS, Checker R, Sharma D, Sandur SK, Sainis KB. Involvement of ERK-Nrf-2 signaling in ionizing radiation induced cell death in normal and tumor cells. PLoS ONE. 2013;8:e65929. doi: 10.1371/journal.pone.0065929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He X, Ma Q. NRF2 cysteine residues are critical for oxidant/electrophile-sensing, Kelch-like ECH-associated protein-1-dependent ubiquitination-proteasomal degradation, and transcription activation. Mol Pharmacol. 2009;76:1265–1278. doi: 10.1124/mol.109.058453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindahl M, Mata-Cabana A, Kieselbach T. The disulfide proteome and other reactive cysteine proteomes: analysis and functional significance. Antioxid Redox Signal. 2011;14:2581–2642. doi: 10.1089/ars.2010.3551. [DOI] [PubMed] [Google Scholar]
- 27.Rogers LK, Leinweber BL, Smith CV. Detection of reversible protein thiol modifications in tissues. Anal Biochem. 2006;358:171–184. doi: 10.1016/j.ab.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 28.Sadler NC, Melnicki MR, Serres MH, et al. Live cell chemical profiling of temporal redox dynamics in a photoautotrophic cyanobacterium. ACS Chem Biol. doi: 10.1021/cb400769v. epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 29.Ansong C, Ortega C, Payne SH, et al. Identification of widespread adenosine nucleotide binding in Mycobacterium tuberculosis. Chem Biol. 2013;20:123–133. doi: 10.1016/j.chembiol.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weber TJ, Siegel RW, Markillie LM, Chrisler WB, Lei XC, Colburn NH. A paracrine signal mediates the cell transformation response to low dose gamma radiation in JB6 cells. Mol Carcinog. 2005;43:31–37. doi: 10.1002/mc.20092. [DOI] [PubMed] [Google Scholar]
- 31.Chauvigne-Hines LM, Anderson LN, Weaver HM, et al. Suite of activity-based probes for cellulose-degrading enzymes. J Am Chem Soc. 2012;134:20521–20532. doi: 10.1021/ja309790w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yates JR, III, Eng JK, McCormack AL, Schieltz D. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal Chem. 1995;67:1426–1436. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
- 33.Kim S, Gupta N, Pevzner PA. Spectral probabilities and generating functions of tandem mass spectra: a strike against decoy databases. J Proteome Res. 2008;7:3354–3363. doi: 10.1021/pr8001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waters KM, Stenoien DL, Sowa MB, et al. Annexin A2 modulates radiation-sensitive transcriptional programming and cell fate. Radiation Res. 2013;179:53–61. doi: 10.1667/RR3056.1. [DOI] [PubMed] [Google Scholar]
- 35.Weber TJ, Monks TJ, Lau SS. DDM-PGE(2)-mediated cytoprotection in renal epithelial cells by a thromboxane A(2) receptor coupled to NF-kappaB. Am J Physiol Renal Physiol. 2000;278:F270–278. doi: 10.1152/ajprenal.2000.278.2.F270. [DOI] [PubMed] [Google Scholar]
- 36.Sowa MB, Goetz W, Baulch JE, et al. Lack of evidence for low-LET radiation induced bystander response in normal human fibro-blasts and colon carcinoma cells. Int J Radiat Biol. 2010;86:102–113. doi: 10.3109/09553000903419957. [DOI] [PubMed] [Google Scholar]
- 37.Mertens JJ, Gibson NW, Lau SS, Monks TJ. Reactive oxygen species and DNA damage in 2-bromo-(glutathion-S-yl) hydroquinone-mediated cytotoxicity. Arch Biochem Biophys. 1995;320:51–58. doi: 10.1006/abbi.1995.1341. [DOI] [PubMed] [Google Scholar]
- 38.Roti Roti JL. Radiation-induced versus endogenous DNA damage and assays that measure parameters reflecting DNA damage on cell by cell basis: comments on the article by Pollycove and Feinendegen. Hum Exp Toxicol. 2003;22:309–313. doi: 10.1191/0960327103ht367oa. discussion 321–303. [DOI] [PubMed] [Google Scholar]
- 39.Madureira PA, Hill R, Miller VA, Giacomantonio C, Lee PW, Waisman DM. Annexin A2 is a novel cellular redox regulatory protein involved in tumorigenesis. Oncotarget. 2011;2:1075–1093. doi: 10.18632/oncotarget.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kwon M, MacLeod TJ, Zhang Y, Waisman DM. S100A10, annexin A2, and annexin a2 heterotetramer as candidate plasminogen receptors. Front Biosci. 2005;10:300–325. doi: 10.2741/1529. [DOI] [PubMed] [Google Scholar]
- 41.Weber TJ, Opresko LK, Waisman DM, et al. Regulation of the low-dose radiation paracrine-specific anchorage-independent growth response by annexin A2. Radiat Res. 2009;172:96–105. doi: 10.1667/RR1220.1. [DOI] [PubMed] [Google Scholar]
- 42.Cheng TY, Wu MS, Lin JT, et al. Annexin A1 is associated with gastric cancer survival and promotes gastric cancer cell invasiveness through the formyl peptide receptor/extracellular signal-regulated kinase/integrin beta-1-binding protein 1 pathway. Cancer. 2012;118:5757–5767. doi: 10.1002/cncr.27565. [DOI] [PubMed] [Google Scholar]
- 43.Sato Y, Kumamoto K, Saito K, et al. Up-regulated Annexin A1 expression in gastrointestinal cancer is associated with cancer invasion and lymph node metastasis. Exp Ther Med. 2011;2:239–243. doi: 10.3892/etm.2011.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Molotkov A, Duester G. Genetic evidence that retinaldehyde dehydrogenase Raldh1 (Aldh1a1) functions downstream of alcohol dehydrogenase Adh1 in metabolism of retinol to retinoic acid. J Biol Chem. 2003;278:36085–36090. doi: 10.1074/jbc.M303709200. [DOI] [PubMed] [Google Scholar]
- 45.Schilling TF, Nie Q, Lander AD. Dynamics and precision in retinoic acid morphogen gradients. Curr Opin Genet Dev. 2012;22:562–569. doi: 10.1016/j.gde.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Asgari MM, Brasky TM, White E. Association of vitamin A and carotenoid intake with melanoma risk in a large prospective cohort. J Invest Dermatol. 2012;132:1573–1582. doi: 10.1038/jid.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ostrowski J, Jarosz D, Butruk E. Liver vitamin A concentration in patients who died of cancer. Neoplasma. 1989;36:353–355. [PubMed] [Google Scholar]
- 48.So PL, Fujimoto MA, Epstein EH., Jr. Pharmacologic retinoid signaling and physiologic retinoic acid receptor signaling inhibit basal cell carcinoma tumorigenesis. Mol Cancer Ther. 2008;7:1275–1284. doi: 10.1158/1535-7163.MCT-07-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burns FJ, Tang MS, Frenkel K, et al. Induction and prevention of carcinogenesis in rat skin exposed to space radiation. Radiat Environ Biophys. 2007;46:195–199. doi: 10.1007/s00411-007-0106-3. [DOI] [PubMed] [Google Scholar]
- 50.Suphakarn VS, Newberne PM, Goldman M. Vitamin A and aflatoxin: effect on liver and colon cancer. Nutr Cancer. 1983;5:41–50. doi: 10.1080/01635588309513777. [DOI] [PubMed] [Google Scholar]
- 51.Moulas AN, Gerogianni IC, Papadopoulos D, Gourgoulianis KI. Serum retinoic acid, retinol and retinyl palmitate levels in patients with lung cancer. Respirology. 2006;11:169–174. doi: 10.1111/j.1440-1843.2006.00833.x. [DOI] [PubMed] [Google Scholar]
- 52.Simone CB II, Kramer K, O'Meara WP, et al. Predicted rates of secondary malignancies from proton versus photon radiation therapy for stage I seminoma. Int J Radiat Oncol Biol Phys. 2011;82:242–249. doi: 10.1016/j.ijrobp.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 54.Fernandes ND, Sun Y, Price BD. Activation of the kinase activity of ATM by retinoic acid is required for CREB-dependent differentiation of neuroblastoma cells. J Biol Chem. 2007;282:16577–16584. doi: 10.1074/jbc.M609628200. [DOI] [PubMed] [Google Scholar]
- 55.Wang X, Lui VC, Poon RT, Lu P, Poon RY. DNA damage mediated s and g(2) checkpoints in human embryonal carcinoma cells. Stem Cells. 2009;27:568–576. doi: 10.1634/stemcells.2008-0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tang XH, Gudas LJ. Retinoids, retinoic acid receptors, and cancer. Annu Rev Pathol. 2011;6:345–364. doi: 10.1146/annurev-pathol-011110-130303. [DOI] [PubMed] [Google Scholar]
- 57.Hormi-Carver K, Feagins LA, Spechler SJ, Souza RF. All transretinoic acid induces apoptosis via p38 and caspase pathways in metaplastic Barrett's cells. Am J Physiol Gastrointest Liver Physiol. 2007;292:G18–G27. doi: 10.1152/ajpgi.00237.2006. [DOI] [PubMed] [Google Scholar]
- 58.Muramoto GG, Russell JL, Safi R, et al. Inhibition of aldehyde dehydrogenase expands hematopoietic stem cells with radioprotective capacity. Stem Cells. 2010;28:523–534. doi: 10.1002/stem.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Belt EJ, Fijneman RJ, van den Berg EG, et al. Loss of lamin A/C expression in stage II and III colon cancer is associated with disease recurrence. Eur J Cancer. 2011;47:1837–1845. doi: 10.1016/j.ejca.2011.04.025. [DOI] [PubMed] [Google Scholar]
- 60.Singh M, Hunt CR, Pandita RK, et al. Lamin A/C depletion enhances DNA damage-induced stalled replication fork arrest. Mol Cell Biol. 2013;33:1210–1222. doi: 10.1128/MCB.01676-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang L, Munck M, Swaminathan K, Kapinos LE, Noegel AA, Neumann S. Mutations in LMNA modulate the lamin A—nesprin-2 interaction and cause LINC complex alterations. PLoS ONE. 2013;8:e71850. doi: 10.1371/journal.pone.0071850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Broers JL, Ramaekers FC, Bonne G, Yaou RB, Hutchison CJ. Nuclear lamins: laminopathies and their role in premature ageing. Physiol Rev. 2006;86:967–1008. doi: 10.1152/physrev.00047.2005. [DOI] [PubMed] [Google Scholar]
- 63.Krtolica A, Parrinello S, Lockett S, Desprez PY, Campisi J. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proc Natl Acad Sci USA. 2001;98:12072–12077. doi: 10.1073/pnas.211053698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Y, Lu H. Signaling to p53: ribosomal proteins find their way. Cancer Cell. 2009;16:369–377. doi: 10.1016/j.ccr.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.