Abstract
Enzyme Linked Immunosorbent Assay (ELISA) is the gold standard assay for detecting and identifying biomolecules using antibodies as the probe. Improving ELISA is crucial for detecting disease-causing agents and facilitating diagnosis at the early stages of disease. Biotinylated antibody and streptavidin-conjugated horse radish peroxide (streptavidin-HRP) often are used with ELISA to enhance the detection of various kinds of targets. In the present study, we used a competition-based strategy in which we pre-mixed free biotin with streptavidin-HRP to generate high-performance system, as free biotin occupies some of the biotin binding sites on streptavidin, thereby providing more chances for streptavidin-HRP to bind with biotinylated antibody. ESAT-6, which is a protein secreted early during tuberculosis infection, was used as the model target. We found that 8 fM of free biotin mixed with streptavidin-HRP anchored the higher detection level of ESAT-6 by four-fold compared with detection without free biotin (only streptavidin-HRP), and the limit of detection of the new method was 250 pM. These results suggest that biotin-streptavidin competition can be used to improve the diagnosis of analytes in other types of sensors.
Introduction
Characteristics of desired analyte and ligand molecules, which include enzymes, proteins, antibodies, nucleic acids, and glycans, are the primary criteria to be considered when designing sensing mechanisms. Measurement of the signal generated upon analyte and ligand interaction is the basis of such sensing devices [1–7]. This approach has been applied to disease diagnosis, environmental monitoring, drug discovery, drug screening, therapeutics, and extension of the human life span [8–15]. A good biosensing system must have both high sensitivity and selectivity. A system that can detect low levels of an analyte in crude samples such as serum or urine is crucial for identifying diseases at the early stage, which is important because treatment and control are easier when the disease is caught early. In addition, an effective system requires use of the right molecules or biomarkers to detect a given disease [4,16–20]. Various immunoassays with high sensitivity have been developed to diagnose the presence of analyte molecules using antibodies as the probe [21–25]. Among these, Enzyme Linked Immunosorbent Assay (ELISA) is one of the most efficient methods available to identify disease-causing agents [26–31]. ELISA is an easy-to-use, sensitive, high-throughput method that requires only a simple equipment [21,32,33].
The ELISA method can be improved to facilitate better level of detections and to be adaptable to a wide range of applications. For example, researchers have used different approaches, such as molecular complementation, to improve the limit of detection (LOD) of ELISA [33]. Sensitivity of ELISA depends on factors such as binding strength of biomolecules, surface functionalization, and molecular assembly. In particular, the detection limit of the system depends greatly on the number of capturing molecules bound to the ELISA surface. Molecular capturing and immobilization vary with different conditions, including pH, temperature, and charge on the sensor surface and protein [34]. Thus, the use of modified surface molecules with proper orientation of the analyte and capturing molecules can improve the sensitivity of the system. Biomolecules are immobilized on the ELISA plate mainly through chemical, physical, or electrostatic interaction. The ELISA plate is made of polystyrene (PS), so the antibody or protein generally is immobilized through the COOH-link on the PS. However, it is difficult to immobilize small molecules in this manner.
Vashist et al. (2014) [34] developed a method of one-step immobilization of antibody on the ELISA plate and showed that it enhanced the detection limit of the system. Nanoparticle-conjugated antibody or protein have also been shown to improve the LOD of ELISA; in particular, antibody-conjugated gold nanoparticles (GNPs) were found to improve the system’s sensitivity [35]. Similarly, the biotin-streptavidin conjugation strategy is commonly used in ELISA protocols to increase the LOD. Biotin-streptavidin is a powerful non-covalent interaction with high affinity and a dissociation constant of 2.3 x 1013 M–1 [36]. Each streptavidin molecule has four binding sites for biotin, and these binding opportunities are useful in different biological applications. Streptavidin also can be tagged with biomolecules such as enzymes, antibodies, or GNPs to improve detection of the system. The biotin-streptavidin interaction has been used in many biological applications, including sensor development, bio-imaging, drug delivery, and protein purification.
In this study, we utilized the biotin-streptavidin interaction with ELISA to include a competition-based strategy for enhancing the detection. Horseradish peroxide (HRP) conjugated streptavidin (streptavidin-HRP) was used to detect the analyte in the final step by reacting the substrate for HRP. Due to high-affinity between biotin and streptavidin, there will be a good sensitivity. Further, to enhance the sensitivity, we made competition between biotinylated antibody and streptravidin-HRP by externally adding free biotin. The improvement in sensitivity of this competition is due to four binding sites for biotin on streptavidin. More importantly, we pre-mixed free biotin with streptavidin-HRP prior to the experiments. Free biotin blocks some of the binding sites for biotin on the streptavidin-HRP molecules, thus providing more chances for streptavidin-HRP to bind with the biotinylated secondary antibody already immobilized on the ELISA plate. After immobilize the biotinylated antibody on the ELISA plate, we introduced this pre-mix and promoted the sensitivity. We used ESAT-6 (molecular weight 6 kDa [37]), a protein secreted early during tuberculosis infection, as the model protein. We postulated that this approach would be able to detect the presence of low levels of the target.
Materials and Methods
Reagents and biomolecules
ESAT-6 was purchased from Sino Biological Inc. (Beijing, China). Anti-ESAT-6 was obtained from Santa Cruz Biotechnology (USA), biotinylated anti-mouse-IgG was from Invitrogen (USA), streptavidin-HRP was procured from Thermo-Scientific (Japan), bovine serum albumin (BSA) was from Promega (USA), and. 16 kDa and Ag85B proteins were obtained from CalBioreagents (USA) and abcan (USA), respectively. ELISA coating buffer (5x) was from Biolegend (Japan). It composed of 0.1 M sodium carbonate (7.13 g NaHCO3, 1.59 g Na2CO3 in 1.0 L; pH 9.5). Substrate for HRP was purchased from Promega and biotin was obtained from Thermo Fisher Scientific, Malaysia. ELISA plates were purchased from Becton Dickinson (France), and the ELISA reader and Tween-20 were from R & M Chemicals (U.K.).
Optimization of the biotinylated anti-mouse-IgG and streptavidin-HRP interaction
To determine the optimal dilution of biotinylated anti-mouse-IgG (biotinylated antibody) and streptavidin-HRP, ELISA plates were directly coated with biotinylated antibody (1 mg/mL) at a dilution of 1:1000 or 1:500 in 1X coating buffer. Each plate was incubated for 3 h at room temperature (RT). The remaining surfaces were blocked with 2% BSA for 1 h at RT, and then three different concentrations of streptavidin-HRP [from the stock; 2 mg/mL, diluted at 1:1000 (1 μg/mL), 1:5000 (200 ng/mL), and 1:10,000 (100 ng/mL)] were added and allowed to incubate for 1 h at RT. The wells were washed five times with the washing buffer (PBS containing 0.05% of Tween-20) between each step. Finally, the substrate for HRP was added to detect the interaction between biotin and streptavidin. The optical density (OD) measurement was done using the ELISA reader at the wavelength of 405 nm. Photographs were taken 10 min after adding the HRP substrate.
Improvement of the biotin-streptavidin-based ELISA by the addition of free biotin
To evaluate whether the addition of free biotin improves the biotin-streptavidin assay, the optimized dilution (1:500) of biotinylated antibody (diluted in the 1X coating buffer) was coated onto the ELISA plate for 3 h at RT. The remaining binding places on the wells then were blocked with 2% BSA for 1 h. Different concentrations of free biotin (0 to 60 fM) were pre-mixed with the streptavidin-HRP (1:1000) and kept separately for 30 min. The molar ratios between free biotin and biotinylated antibody were calculated as 1:0.4x106 for 0.25 fM (lowest concentration) and 1:8x107 for 60 fM (highest concentration) of free biotin. The pre-mixtures of biotin and streptavidin-HRP then were added to the biotinylated antibody immobilized ELISA plate and allowed to incubate for 1 h. The wells were washed five times with washing buffer between each step. Finally, the substrate for HRP was added to detect the interaction between biotin and streptavidin. All experimental steps were performed at RT. The OD was read using the ELISA reader at 405 nm. Photographs were taken 10 min after adding the HRP substrate.
Optimization of the interaction between anti-ESAT- 6 and ESAT-6
To determine the optimal dilution of anti-ESAT-6, 200 nM of ESAT- 6 were diluted in 1X coating buffer, coated onto the ELISA plate, and allowed to incubate for 3 h at RT. Next, 2% BSA was added to each well to cover the remaining binding places on the wells for 1 h. Three different dilutions of anti-ESAT-6 [from the stock; 1 mg/mL, diluted at 1:5000 (200 ng/mL), 1:1000 (1 μg/mL), and 1:500 (2 μg/mL)] then were added to the wells. The optimized concentration of biotinylated antibody (1:500) was added to each well and allowed to incubate for 1 h, and then the 1:1000 dilution of streptavidin-HRP was added to each well. A control experiment was performed without ESAT-6. Wells were washed five times with the washing buffer between each step. Finally, the substrate for HRP was added to detect the interaction between ESAT-6 and anti-ESAT-6. All experimental steps were performed at RT. The OD was read using the ELISA reader at 405 nm. Photographs were taken 10 min after adding the HRP substrate.
Improvement in detection of ESAT-6 by free biotin with streptavidin-HRP
To confirm that the addition of free biotin to the system improves detection, ESAT-6 (100 nM) was diluted in the 1X coating buffer coated onto the ELISA plate and allowed to incubate for 3 h at RT. The ELISA wells were blocked with 2% BSA for 1 h. The optimized dilution of anti-ESAT-6 (1:500) then was added, followed by the optimized dilution of biotinylated antibody (1:500) and then finally the 1:1000 dilution of streptavidin-HRP pre-mixed with different concentrations of free biotin (streptavidin-HRP incubated with 0 to 60 fM of free biotin for 10 min). A control experiment was performed without ESAT-6. Wells were washed five times with washing buffer between each step. Finally, the substrate for HRP was added to detect the interaction between ESAT-6 and anti-ESAT-6. All experimental steps were performed at RT. The OD was read using the ELISA reader at 405 nm. Photographs were taken 10 min after adding the HRP substrate.
Detection limit: Comparison between ELISA with and without free biotin
To determine the LOD of ESAT-6, ESAT-6 was titrated from 0.1 to 120 nM. The steps were as follows: (a) ESAT-6 was coated onto the ELISA plate; (b) 2% BSA blocking was performed; (c) 1:500 dilution of anti-ESAT-6 was added; (d) 1:500 dilution of biotinylated antibody was added; (e) 1:1000 dilution of streptavidin-HRP with and without free biotin was added; and (f) substrate for HRP was added. Wells were washed five times with the washing buffer between each step. Finally, the substrate for HRP was added to detect the binding of ESAT-6 on the ELISA well. All experimental steps were performed at RT. The OD was read using the ELISA reader at 405 nm. Photographs were taken 10 min after adding the HRP substrate.
Specificity: Spiking human serum with ESAT-6
Specific detection of biomolecules in the crude samples such as serum is important to identify the particular target. To demonstrate the specificity, ESAT-6 was spiked into the human serum and detected the ESAT-6. This experiment will reveal the genuine detection with serum containing high concentration of BSA, Globulin and other proteins. The specificity of different concentrations of ESAT-6 (0 to 120 nM) spiked into a 1:1000 dilution of human serum was measured. The mixed sample was coated onto the ELISA plate and the other procedures were the same as those described above. To confirm the specific detection of ESAT-6, 250 pM of ESAT-6 was also spiked into human saliva, urine and with the mixture of other TB proteins (16 kDa, Ag85B), and detected as described above.
Results and Discussion
ELISA is the gold standard method for detecting a given analyte molecule using a suitable antibody. Biotinylated secondary antibody and streptavidin-HRP are commonly used in the ELISA to improve the limit of detection (LOD). Streptavidin is a tetrameric protein with a molecular weight of 60 kDa; it has a high binding affinity to biotin (in the low femtomolar range) and four biotin binding sites [36]. Because of these properties, the biotin-streptavidin assay is commonly used in many biotechnological applications, including imaging [38], purification [39], drug delivery [40], and bioanalytical immunoassays [23,41].
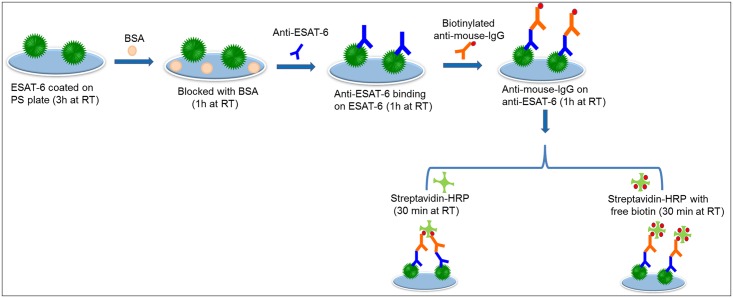
The interaction between biotin and streptavidin can be further utilized to improve biomolecule detection by ELISA. Researchers have experimented with modifying the structure of streptavidin, especially the biotin binding sites, to increase the binding of biotin and streptavidin [42–44]. The four binding sites can be occupied completely by biotin immobilized on the sensing surface, leading to comparatively less binding than without free biotin [9]. In this study we blocked some of the biotin binding sites on streptavidin-HRP using free biotin in order to provide more chances for streptavidin-HRP to bind with biotinylated antibody. We found that free biotin pre-mixed with streptavidin-HRP improved the detection of ESAT-6. Fig 1 shows a schematic representation of the improved detection of ESAT-6 with the ELISA system modified by the addition of free biotin. After the interaction of anti-ESAT-6 and biotinylated anti-mouse-IgG, we introduced the mixture of streptavidin-HRP and free biotin, it gives more chance of streptavidin-HRP binding on the biotinylated anti-mouse-IgG, leads enhanced detection.
Fig 1. Schematic representation of ESAT-6 detection by the biotin-streptavidin interaction.
Complete schematics are displayed. ESAT-6 protein was coated onto the plate followed by anti-ESAT-6 and biotinylated anti-mouse-IgG. Streptavidin-HRP with and without free biotin then was added.
Interaction between biotinylated anti-mouse-IgG and streptavidin-HRP
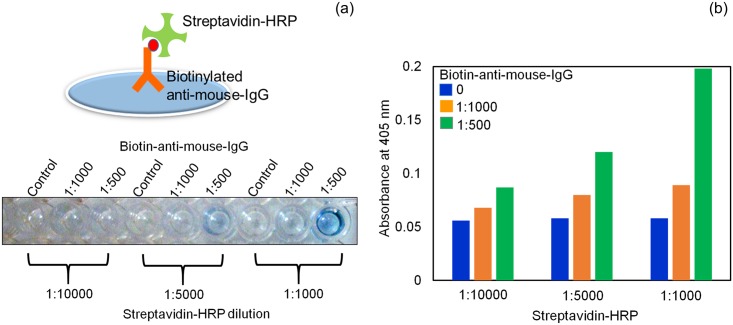
Due to the importance of concentration of streptavidin-HRP and to attain the maximum sensitivity of the ELISA, we first determined the optimal dilution of biotinylated antibody and streptavidin-HRP. The combination of the 1:500 dilution of biotinylated antibody and the 1:1000 dilution of streptavidin-HRP resulted in the maximum OD (0.2), and no non-specific signal was detected in the control experiment (without biotinylated antibody) (Fig 2). Therefore, we used these two dilutions in the subsequent experiments.
Fig 2. Optimization of biotinylated antibody and streptavidin-HRP.
Two different dilutions of biotinylated antibody (1:500 and 1:1000) were coated onto ELISA plates. Different dilutions of streptavidin-HRP (1:1000, 1:5000, and 1:10000) were added and the interaction between biotinylated antibody and streptavidin-HRP was detected. (a) Color change in the solution after the addition of HRP substrate. (b) Graphical representation of the optimization.
Addition of free biotin improved the interaction between biotinylated antibody and streptavidin-HRP
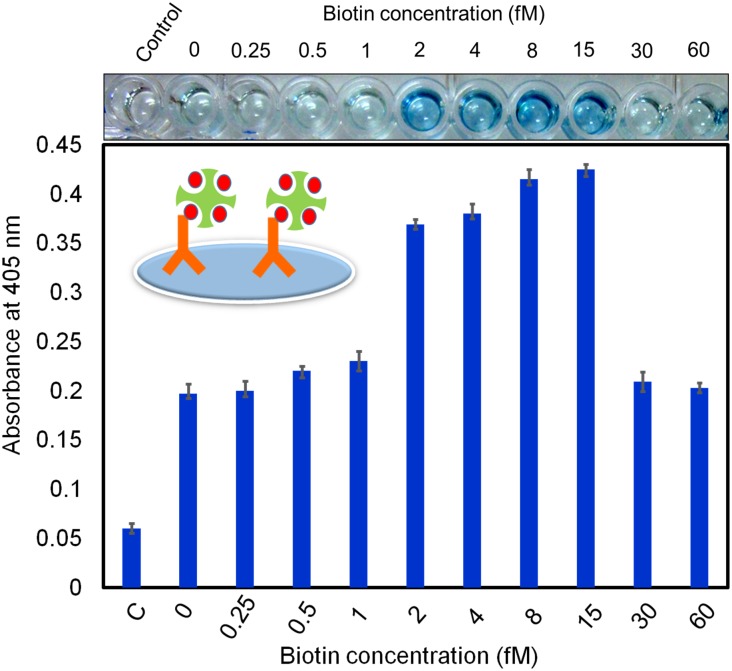
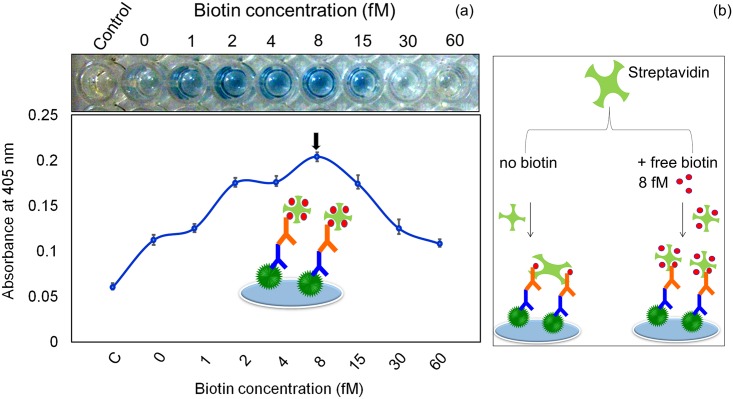
Because streptavidin has four binding sites for biotin, our goal was to block one or two of these binding sites with free biotin to provide more chances for streptavidin-HRP to bind with biotinylated antibody. We pre-mixed different concentrations of free biotin with streptavidin-HRP to determine the optimal concentration of free biotin. Biotin and streptavidin exhibit high affinity and a low dissociation constant [36], thus we expected saturation to occur quickly when we added the optimal concentration of biotin. If we added too much free biotin, it would occupy all four binding sites and not allow binding with the biotinylated antibody. To determine the optimal level, we carefully titrated the free biotin from 0 to 60 fM concentration. As shown in Fig 3, without biotin the OD was ~0.2 for the 1:500 dilution of anti-ESAT-6. As the free biotin concentration was increased, the OD also gradually increased. At 8 and 15 fM, the OD was ~0.425 for the 1:500 dilution of anti-ESAT-6 (i.e., almost 2.5 times higher than the OD for no biotin). This result illustrates that the addition of free biotin to streptavidin-HRP enhanced the binding of biotin and streptavidin. At the higher concentrations of biotin (30 and 60 fM), the OD was gradually decreased because more biotin binding sites on streptavidin-HRP were occupied by free biotin. The ELISA photographs clearly showed that with free biotin concentrations ranging from 2 to 15 fM the color of the solution changed to blue, which is indicative of higher OD (Fig 3). At 30 and 60 fM, however, the blue color is faint, indicating lower OD. These results show that 8 fM of free biotin is optimal for improving the interaction of biotinylated antibody and streptavidin-HRP.
Fig 3. Addition of free biotin improved the interaction between biotinylated antibody and streptavidin-HRP.
The 1:500 dilution of biotinylated antibody was detected by streptavidin-HRP mixed with different concentrations of free biotin (0 to 60 fM). Concentrations ranging from 2 to 15 nM of free biotin improved the detection, as indicated by the visible color change of the solution.
Interaction of anti-ESAT-6 and ESAT-6 in the absence of free biotin
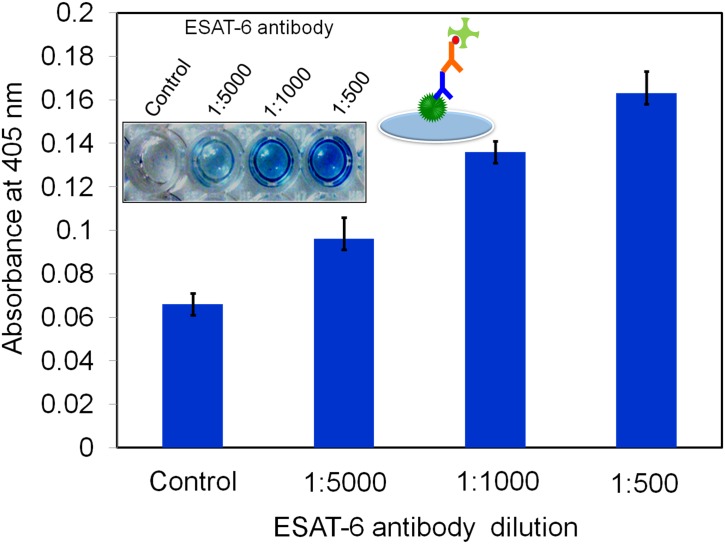
The next step was to determine the optimal dilution of anti-ESAT-6 for detecting ESAT-6. Different dilutions of anti-ESAT-6 were tested. The OD of the 1:500 dilution was higher (0.18) than that of the 1:5000 and 1:1000 dilutions for 200 nM of ESAT-6 (Fig 4). Photographs showed the dark blue color of the 1:500 dilution of anti-ESAT- 6. Thus, the 1:500 dilution of anti-ESAT- 6 was used in the subsequent experiments.
Fig 4. Optimization of ESAT-6 antibody.
100 nM of constant ESAT-6 coated on the ELISA well followed by adding the different dilutions (1:500, 1:1000 and 1:5000) of ESAT-6 antibody. And then detected by biotinylated antibody and streptavidin-HRP.1: 500 dilutions of ESAT-6 antibody showed the optimum dilution.
Improved detection of ESAT-6 when free biotin was present
To determine the effect of free biotin on detection of ESAT-6, a constant concentration of ESAT-6 (100 nM) was detected using different concentrations of free biotin mixed with streptavidin-HRP. When no free biotin was present, the OD was ~0.12, and the OD was increased with increasing free biotin concentration (Fig 5a). The mixture of 8 fM of free biotin with streptavidin had the highest OD (~0.2), so we used this concentration to determine the LOD of ESAT-6. At concentrations higher than 8 fM, the OD was decreased. The ELISA photographs showed clear color changes in the presence of 1 to 15 fM of free biotin. It was proved that blocking some binding sites on streptavidin increased the binding of more streptavidin-HRP on the biotinylated antibody, ultimately there is an enhancement in the signal. To check the suitable concentration of free biotin, we titrated again with different biotin concentrations and found that 8 fM is more suitable to occupy some binding site(s) on streptavidin. When we increased further, it occupies all the biotin binding sites on streptavidin. With 8 fM, biotin binds only two or three binding sites on streptavidin, so that the remaining biotin binding sites on streptavidin-HRP are free to capture biotinylated antibody. A model is created that explains the enhanced sensitivity of streptavidin-HRP pretreated with a fixed amount of free biotin (Fig 5b).
Fig 5. Addition of free biotin improved detection of ESAT-6.
100 nM of ESAT-6 were coated onto the ELISA plate, followed by ESAT-6 antibody and biotinylated anti-mouse-IgG. (a) Detection by streptavidin mixed with different concentrations of free biotin (0 to 60 fM) showed that 8 fM of free biotin resulted in the highest OD. (b) A model explains the enhanced sensitivity of streptavidin-HRP pretreated with a fixed amount (8 fM) of free biotin.
Limit of detection for ESAT-6
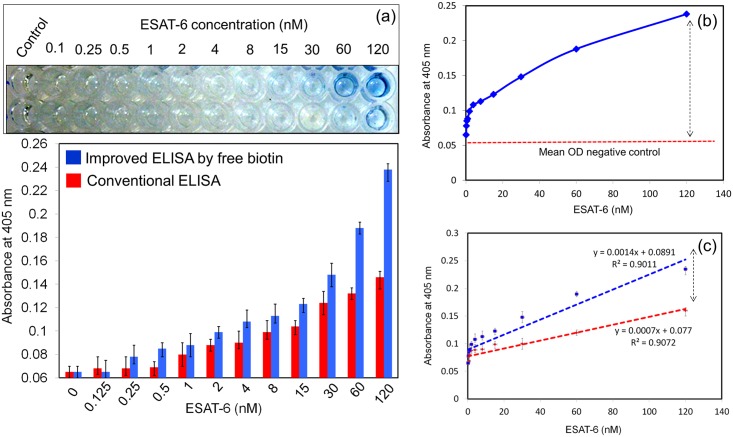
To determine the LOD of the ELISA system with the pre-mixture of 8 fM free biotin and streptavidin-HRP, we titrated ESAT-6 from 0 to 120 nM. The LOD was found to be 250 pM (Fig 6a), which was a four-fold improvement over the system without free biotin (1 nM). Photographs were also clearly indicated the color changes which depends on the ESAT-6 concentrations. The LOD was calculated using statistical approaches by the method of blank ± 3 standard deviation (3σ). As shown in Fig 6b, the mean OD values were also calculated. R2 values of both conventional and improved ELISA with biotin showed the significant (Fig 6c). In fact, the OD was higher for all concentrations of ESAT- 6 when free biotin was present. In the control experiment, it didn’t show any non-specificity in both cases (with and without free biotin). This method is attested for many biological applications that involve the biotin and streptavidin interaction.
Fig 6. Limit of detection of ESAT-6.
(a) ESAT-6 was titrated from 0 to 120 nM. LOD was compared between experiments with and without free biotin added to streptavidin-HRP. (b) Calculation of mean OD. (c) Statistical calculation on improved and conventional ELISA. R2 values were calculated and they indicate the significance.
Specific detection of ESAT-6: Spiking experiments
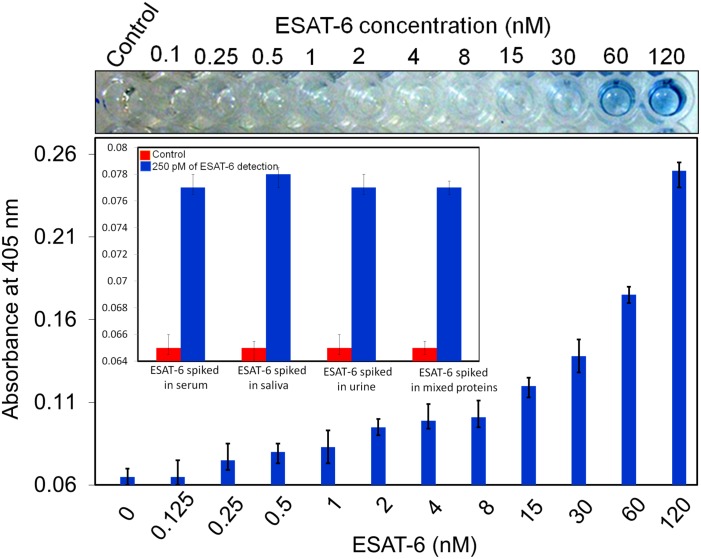
Specificity is very important in a diagnosis system, particularly for mixed samples or crude samples such as serum or urine. The ability of our system to specifically detect ESAT-6 was tested by spiking a 1:1000 dilution of human serum with different concentrations of ESAT-6. Serum contains protein albumin at a concentration of 0.045 mg/mL [9]. Fig 7 shows that 120 nM of ESAT-6 was detected at a high OD value (0.24) and with a LOD of 250 pM (i.e., the same as that recorded in the absence of serum). We also evaluated spiking experiments with other crude media and to confirm the specificity at 250 pM of ESAT-6, we spiked in human saliva, urine and the mixture of TB proteins (ESAT-6, 16 kDa, Ag85B). As shown in Fig 7 (Fig inset), the result obtained in serum sample with 250 pM of ESAT-6 was also displayed in other media. These findings illustrated that diagnosis of ESAT-6 was not affected by the presence of human serum, saliva and urine and it was proved that ESAT-6 at low levels can be specifically detected by our developed ELISA strategy.
Fig 7. Specific detection of ESAT-6 in human serum.
A 1:1000 dilution of human serum was spiked with different concentrations of ESAT-6. Specific detections were also confirmed by the spiking of 250 pM of ESAT-6 with human saliva, urine and mixture of other TB proteins (Fig inset).
Conclusion
ELISA is an immunoassay commonly used to detect different diseases using suitable antibodies. In this study, the ELISA method was improved to enhance the LOD for the given target using a competition-based strategy with biotin-streptavidin interaction. By blocking some of the biotin binding sites on streptavidin-HRP with free biotin, we enhanced the sensitivity of the system for detecting ESAT-6, an important protein secreted early during tuberculosis infection. The addition of pre-mixed free biotin (8 fM) with streptavidin-HRP improved the LOD four times relative to the system without free biotin (from 1 nM to 250 pM). Moreover, the LOD of ESAT-6 was also retained in the spiking experiments with human serum, saliva, urine and mixture of TB proteins. We could also reproduce the current strategy with other important targets and they are under progress. This competition-based strategy may also useful for other biotechnological applications in the future.
Acknowledgments
T. Lakshmipriya was supported by a post-doctoral research fellowship from Universiti Sains Malaysia.
Data Availability
All relevant data are within the paper.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Kumarevel T, Fujimoto Z, Karthe P, Oda M, Mizuno H, Kumar PKR. Crystal structure of activated HutP; an RNA binding protein that regulates transcription of the hut operon in Bacillus subtilis. Structure. 2004;12: 1269–80. 10.1016/j.str.2004.05.005 [DOI] [PubMed] [Google Scholar]
- 2.Kumarevel T, Tanaka T, Nishio M, Gopinath SCB, Takio K, Shinkai A, et al. Crystal structure of the MarR family regulatory protein, ST1710, from Sulfolobus tokodaii strain 7. J Struct Biol. 2008;161: 9–17. 10.1016/j.jsb.2007.08.017 [DOI] [PubMed] [Google Scholar]
- 3.Gopinath SCB, Balasundaresan D, Akitomi J, Mizuno H. An RNA aptamer that discriminates bovine factor IX from human factor IX. J Biochem. 2006;140: 667–676. 10.1093/jb/mvj203 [DOI] [PubMed] [Google Scholar]
- 4.Gopinath SCB, Shikamoto Y, Mizuno H, Kumar PKR. Snake-venom-derived Factor IX-binding protein specifically blocks the gamma-carboxyglutamic acid-rich-domain-mediated membrane binding of human Factors IX and X. Biochem J. 2007;405: 351–357. 10.1042/BJ20061737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gopinath SCB, Hayashi K, Kumar PKR. Aptamer That Binds to the gD Protein of Herpes Simplex Virus 1 and Efficiently Inhibits Viral Entry. J Virol. 2012;86: 6732–6744. 10.1128/JVI.00377-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gopinath SCB, Awazu K, Fujimaki M. Waveguide-mode sensors as aptasensors. Sensors. 2012;12: 2136–2151. 10.3390/s120202136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujimaki M, Nomura K, Sato K, Kato T, Gopinath SCB, Wang X, et al. Detection of colored nanomaterials using evanescent field-based waveguide sensors. Opt Express. 2010;18: 15732–15740. 10.1364/OE.18.015732 [DOI] [PubMed] [Google Scholar]
- 8.Gopinath SCB, Awazu K, Fujimaki M, Sugimoto K, Ohki Y, Komatsubara T, et al. Influence of nanometric holes on the sensitivity of a waveguide-mode sensor: Label-free nanosensor for the analysis of RNA aptamer-ligand interactions. Anal Chem. 2008;80: 6602–6609. 10.1021/ac800767s [DOI] [PubMed] [Google Scholar]
- 9.Lakshmipriya T, Fujimaki M, Gopinath SCB, Awazu K, Horiguchi Y, Nagasaki Y. A high-performance waveguide-mode biosensor for detection of factor IX using PEG-based blocking agents to suppress non-specific binding and improve sensitivity. Analyst. 2013;138: 2863–2870. 10.1039/c3an00298e [DOI] [PubMed] [Google Scholar]
- 10.Lakshmipriya T, Fujimaki M, Gopinath SCB, Awazu K. Generation of anti-influenza aptamers using the systematic evolution of ligands by exponential enrichment for sensing applications. Langmuir. 2013;29: 15107–15115. 10.1021/la4027283 [DOI] [PubMed] [Google Scholar]
- 11.Lakshmipriya T, Horiguchi Y, Nagasaki Y. Co-immobilized poly(ethylene glycol)-block-polyamines promote sensitivity and restrict biofouling on gold sensor surface for detecting factor IX in human plasma. Analyst. 2014;139: 3977–85. 10.1039/c4an00168k [DOI] [PubMed] [Google Scholar]
- 12.Kim S, Lee J, Lee SJ, Lee HJ. Ultra-sensitive detection of IgE using biofunctionalized nanoparticle-enhanced SPR. Talanta. Elsevier B.V.; 2010;81: 1755–1759. 10.1016/j.talanta.2010.03.036 [DOI] [PubMed] [Google Scholar]
- 13.Perumal V, Hashim U, Gopinath SCB, Haarindraprasad R, Foo KL, Balakrishnan SR, et al. “Spotted Nanoflowers”: Gold-seeded Zinc Oxide Nanohybrid for Selective Bio-capture. Sci Rep. Nature Publishing Group; 2015;5: 12231 10.1038/srep12231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perumal V, Hashim U, Gopinath SCB, Haarindraprasad R, Poopalan P, Liu W-W, et al. A New Nano-worm Structure from Gold-nanoparticle Mediated Random Curving of Zinc Oxide Nanorods. Biosens Bioelectron. 2015; 10.1016/j.bios.2015.10.083 [DOI] [PubMed] [Google Scholar]
- 15.Haarindraprasad R, Hashim U, Gopinath SCB, Kashif M, Veeradasan P, Balakrishnan SR, et al. Low Temperature Annealed Zinc Oxide Nanostructured Thin Film-Based Transducers: Characterization for Sensing Applications. PLoS One. 2015;10: e0132755 10.1371/journal.pone.0132755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gopinath SCB, Hilda a., Lakshmi Priya T, Annadurai G, Anbu P. Purification of lipase from Geotrichum candidum: Conditions optimized for enzyme production using Box-Behnken design. World J Microbiol Biotechnol. 2003;19: 681–689. 10.1023/A:1025119222925 [DOI] [Google Scholar]
- 17.Anbu P, Gopinath SCB, Hilda a, Mathivanan N, Annadurai G. Secretion of keratinolytic enzymes and keratinolysis by Scopulariopsis brevicaulis and Trichophyton mentagrophytes: regression analysis. Can J Microbiol. 2006;52: 1060–1069. 10.1139/w06-067 [DOI] [PubMed] [Google Scholar]
- 18.Gopinath SCB. Anti-coagulant aptamers. Thromb Res. 2008;122: 838–847. 10.1016/j.thromres.2007.10.022 [DOI] [PubMed] [Google Scholar]
- 19.Gopinath SCB. Antiviral aptamers. Arch Virol. 2007;152: 2137–2157. 10.1007/s00705-007-1014-1 [DOI] [PubMed] [Google Scholar]
- 20.Chen Y, Chan CK, Kerishnan JP, Lau YL, Wong Y-L, Gopinath S. Identification of circulating biomarkers in sera of Plasmodium knowlesi-infected malaria patients—comparison against Plasmodium vivax infection. BMC Infect Dis. 2015;15: 49 10.1186/s12879-015-0786-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poongodi GL, Suresh N, Gopinath SCB, Chang T, Inoue S, Inoue Y. Dynamic change of neural cell adhesion molecule polysialylation on human neuroblastoma (IMR-32) and rat pheochromocytoma (PC-12) cells during growth and differentiation. J Biol Chem. 2002;277: 28200–28211. 10.1074/jbc.M202731200 [DOI] [PubMed] [Google Scholar]
- 22.Gopinath SCB, Awazu K, Fons P, Tominaga J, Kumar PKR. A sensitive multilayered structure suitable for biosensing on the BioDVD platform. Anal Chem. 2009;81: 4963–4970. 10.1021/ac802757z [DOI] [PubMed] [Google Scholar]
- 23.Nomura K, Gopinath SCB, Lakshmipriya T, Fukuda N, Wang X, Fujimaki M. An angular fluidic channel for prism-free surface-plasmon-assisted fluorescence capturing. Nat Commun. 2013;4: 2855 10.1038/ncomms3855 [DOI] [PubMed] [Google Scholar]
- 24.Balakrishnan SR, Hashim U, Gopinath SCB, Poopalan P, Ramayya HR, Iqbal Omar M, et al. A Point-of-Care Immunosensor for Human Chorionic Gonadotropin in Clinical Urine Samples Using a Cuneated Polysilicon Nanogap Lab-on-Chip. PLoS One. 2015;10: e0137891 10.1371/journal.pone.0137891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balakrishnan SR, Hashim U, Gopinath SCB, Poopalan P, Ramayya HR, Veeradasan P, et al. Polysilicon Nanogap Lab-on-Chip Facilitates Multiplex Analyses with Single Analyte. Biosens Bioelectron. Elsevier; 2015; 10.1016/j.bios.2015.10.075 [DOI] [PubMed] [Google Scholar]
- 26.Guimaraes AJ, Pizzini C V, De Matos Guedes HL, Albuquerque PC, Peralta JM, Hamilton AJ, et al. ELISA for early diagnosis of histoplasmosis. J Med Microbiol. 2004;53: 509–514. 10.1099/jmm.0.05469-0 [DOI] [PubMed] [Google Scholar]
- 27.Li X, Li G, Teng Q, Yu L, Wu X, Li Z. Development of a blocking ELISA for detection of serum neutralizing antibodies against newly emerged duck Tembusu virus. PLoS One. 2012;7: e53026 10.1371/journal.pone.0053026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu QL, Yan XH, Yin XM, Situ B, Zhou HK, Lin L, et al. Electrochemical enzyme-linked immunosorbent assay (ELISA) for α-fetoprotein based on glucose detection with multienzyme-nanoparticle amplification. Molecules. 2013;18: 12675–12686. 10.3390/molecules181012675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dixit CK, Vashist SK, O’neill FT, O’reilly B, MacCraith BD, O’kennedy R. Development of a high sensitivity rapid sandwich ELISA procedure and its comparison with the conventional approach. Anal Chem. 2010;82: 7049–7052. 10.1021/ac101339q [DOI] [PubMed] [Google Scholar]
- 30.Dixit CK, Vashist SK, MacCraith BD, O’Kennedy R. Multisubstrate-compatible ELISA procedures for rapid and high-sensitivity immunoassays. Nat Protoc. 2011;6: 439–45. 10.1038/nprot.2011.304 [DOI] [PubMed] [Google Scholar]
- 31.Vashist SK. A sub-picogram sensitive rapid chemiluminescent immunoassay for the detection of human fetuin A. Biosens Bioelectron. 2013;40: 297–302. 10.1016/j.bios.2012.07.067 [DOI] [PubMed] [Google Scholar]
- 32.Gopinath SCB, Awazu K, Fujimaki M, Shimizu K. Evaluation of anti-A/Udorn/307/1972 antibody specificity to influenza A/H3N2 viruses using an evanescent-field coupled waveguide-mode sensor. PLoS One. 2013;8: e81396 10.1371/journal.pone.0081396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toh SY, Citartan M, Gopinath SCB, Tang T-H. Aptamers as a replacement for antibodies in enzyme-linked immunosorbent assay. Biosens Bioelectron. 2015;64: 392–403. 10.1016/j.bios.2014.09.026 [DOI] [PubMed] [Google Scholar]
- 34.Vashist SK, Marion Schneider E, Lam E, Hrapovic S, Luong JHT. One-step antibody immobilization-based rapid and highly-sensitive sandwich ELISA procedure for potential in vitro diagnostics. Sci Rep. 2014;4: 4407 10.1038/srep04407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saha K, Agasti SS, Kim C, Li X, Rotello VM. Gold nanoparticles in chemical and biological sensing. Chemical Reviews. 2012. pp. 2739–2779. 10.1021/cr2001178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Green NM. Advances in Protein Chemistry Volume 29 [Internet]. Advances in Protein Chemistry. 1975. 10.1016/S0065-3233(08)60411-8 [DOI] [Google Scholar]
- 37.Geluk A, van Meijgaarden KE, Franken KLMC, Subronto YW, Wieles B, Arend SM, et al. Identification and characterization of the ESAT-6 homologue of Mycobacterium leprae and T-cell cross-reactivity with Mycobacterium tuberculosis. Infect Immun. 2002;70: 2544–2548. 10.1128/IAI.70.5.2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cauchon N, Langlois R, Rousseau J a., Tessier G, Cadorette J, Lecomte R, et al. PET imaging of apoptosis with 64Cu-labeled streptavidin following pretargeting of phosphatidylserine with biotinylated annexin-V. Eur J Nucl Med Mol Imaging. 2007;34: 247–258. 10.1007/s00259-006-0199-y [DOI] [PubMed] [Google Scholar]
- 39.Tse J, Wang Y, Zengeya T, Rozners E, Tan-Wilson A. Peptide nucleic acid probe for protein affinity purification based on biotin–streptavidin interaction and peptide nucleic acid strand hybridization. Anal Biochem. 2015;470: 34–40. 10.1016/j.ab.2014.10.005 [DOI] [PubMed] [Google Scholar]
- 40.Sakahara H, Saga T. Avidin-biotin system for delivery of diagnostic agents. Adv Drug Deliv Rev. 1999;37: 89–101. 10.1016/S0169-409X(98)00101-X [DOI] [PubMed] [Google Scholar]
- 41.Wilchek M, Bayer E a., Livnah O. Essentials of biorecognition: The (strept)avidin-biotin system as a model for protein-protein and protein-ligand interaction. Immunol Lett. 2006;103: 27–32. 10.1016/j.imlet.2005.10.022 [DOI] [PubMed] [Google Scholar]
- 42.Duan X, Li Y, Rajan NK, Routenberg D a., Modis Y, Reed M a. Quantification of the affinities and kinetics of protein interactions using silicon nanowire biosensors. Nat Nanotechnol. 2012;7: 401–407. 10.1038/nnano.2012.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sano T, Vajda S, Smith CL, Cantor CR. Engineering subunit association of multisubunit proteins: a dimeric streptavidin. Proc Natl Acad Sci U S A. 1997;94: 6153–6158. 10.1073/pnas.94.12.6153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lim KH, Huang H, Pralle A, Park S. Engineered streptavidin monomer and dimer with improved stability and function. Biochemistry. 2011;50: 8682–8691. 10.1021/bi2010366 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.